Abstract
Background
Drug resistance caused by G1202R/G1202del mutation in anaplastic lymphoma kinase (ALK) represents a great challenge in the clinic. The effect of other mutation(s) at G1202 on the available tyrosine kinase inhibitors (TKIs) in the clinic remains unknown.
Case Presentation
A 50-year-old Chinese male non-smoker with lung adenocarcinoma progressed with spinal metastasis after receiving chest radiation together with Pemetrexed and Cisplatin as adjuvant chemotherapy. Targeted next generation sequencing (NGS) identified EML4-ALK gene fusion in the resected left lung tissue. Local radiation followed by Crizotinib were used in the following treatment and the spinal metastasis was found to shrink, but the progression free survival (PFS) only lasted for 2 months with the appearance of brain metastasis. Afterwards, the patient benefited from the therapy of Alectinib with a PFS of 8 months. Then he progressed with metastases in right lung and pleural, and did not show response to the chemotherapy with Docetaxel plus Bevacizumab. The targeted sequencing consistently identified EML4-ALK gene fusion in both plasma and pleural effusion (PE), as well as a novel ALK G1202K mutation (c.3604_3605delGGinsAA). Given the lack of established or known drug treatment for this novel mutation, we implemented molecular dynamics (MD) simulation-guided drug sensitivity prediction, which results suggested Lorlatinib remains potent against G1202K mutant ALK. Therefore, Lorlatinib was used as the fourth-line therapy, which lead to the considerable efficacy with improved performance status (PS) score and reduced lung metastases. The structural mechanism underlying G1202K-induced drug resistance to different ALK-TKIs was also discussed.
Conclusion
Our case suggested the ALK-G1202K mutation may serve as a novel mechanism underlying the resistance to Alectinib, and provide direct evidence to support its sensitization to Lorlatinib. Our work represented an example of integrating in silico predictions into clinical practice.
Keywords: lung cancer, drug resistance, in silico prediction, binding energy calculation, Alectinib, Lorlatinib
Background
Since the discovery of anaplastic lymphoma kinase (ALK) fusion oncogenes (including EML4, KIF5B, KLC1, and TPR) in non-small-cell lung cancer (NSCLC),1,2 the targeted therapy with tyrosine kinase inhibitors (TKIs) has significantly improved the clinical outcomes for these patients. Despite their efficiency, almost all patients have eventually experienced disease progression owing to the seemingly inevitable resistance acquired upon drug treatment in the clinic.3–6
The secondary mutations of ALK kinase domain (ALK-KD) serve as the most common mechanism underlying drug resistance7,8 and were likely to increase to 56% after treatments with the second-generation ALK-TKIs, among which G1202R is the most prevalent one.9 Besides, recent study also suggested ALK-G1202del is able to confer moderate resistance to Ceritinib, Alectinib, and Brigatinib,10 suggesting the important role of G1202. However, the effect of other unknown mutation(s) at G1202 on the available ALK-TKIs remains inconclusive. Here we described a case with advanced lung adenocarcinoma bearing a novel ALK secondary mutation G1202K (c.3604_3605delGGinsAA) who presented resistance to Alectinib but remained sensitive to the third -generation ALK-TKI Lorlatinib.
Clinical Description
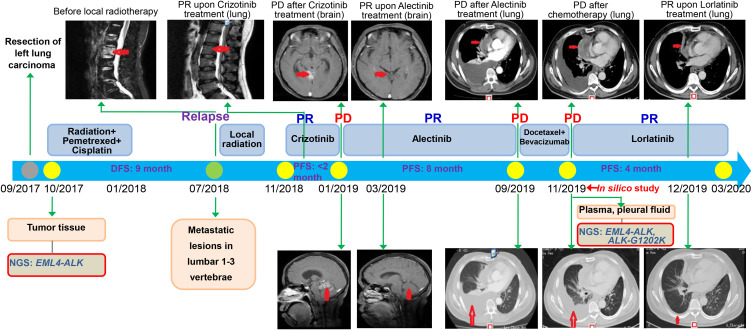
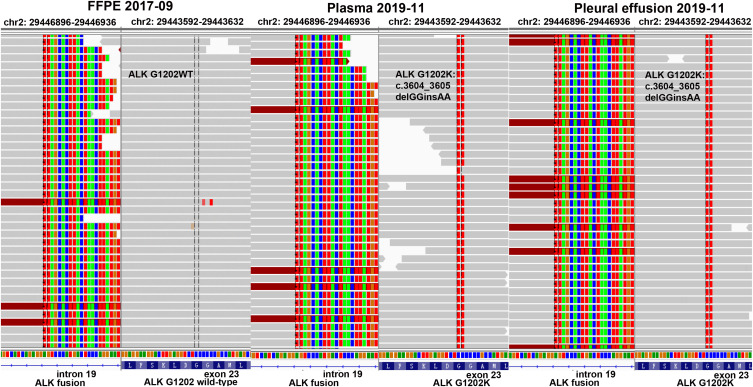
The patient in the present study was a 50-year-old Chinese male non-smoker, who was diagnosed with stage IIIA (pT1N2M0) lung adenocarcinoma after the thoracoscopic radical resection of left lung cancer in September 2017. The EML4-ALK gene fusion (VAF: 24.2%), together with CDKN2A p.V28E (VAF: 8%) and TP53 splicing (VAF: 6%), were detected by next generation sequencing (NGS) using the surgical tissue specimen in October 2017 (Figure 1, Table 1). During the period between October 2017 and January 2018, he received chest radiation (56 Gy/28 fractions) together with 4 cycles of Pemetrexed plus Cisplatin.11 In July 2018, the patient attended the hospital with Lumbar pain, and then the metastatic lesions of lumbar 1–3 vertebrae were found by magnetic resonance imaging (MRI). Local radiation (40 Gy/20 fractions to the lumbar 1–3 vertebrae) was administered and from November 2018 the patient started oral administration of Crizotinib, after that his lumbar pain was released and tumor was reduced (Figure 1). Two months later, brain metastatic lesions were found by MRI without other new metastatic lesions. Afterwards, Alectinib (600 mg twice a day) was administered as the second-line therapy from January 2019. Despite no guidance from NGS before starting Alectinib treatment, almost complete response of brain metastatic lesions was encouragingly observed 2 months later (Figure 1). The progression free survival (PFS) lasted for 8 months. In September 2019, the patient presented with chest pain and a cough. The metastases in right lung, pleural were confirmed with computed tomography (CT),12 including massive right pleural effusion (PE) (Figure 1). The performance status (PS) score for the patient was 3, and 2 cycles of Docetaxel plus Bevacizumab as well as intermittent PE drainage were administered from September 2019. After 2 months (November 2019), he showed a progression of disease (PD) in the lung lesions (Figure 1). In November 2019, we detected the acquired ALK-G1202K mutation (c.3604_3605delGGinsAA) in both plasma (VAF: 2.1%) and PE (VAF: 23.4%) of this patient (Figure 2), as well as the previously identified EML4-ALK gene fusion (VAF: 5.8% and 22% for plasma and PE), CDKN2A p.V28E (VAF: 3.9% and 34% for plasma and PE), TP53 splicing and stop-gained (VAF: 5.5% and 48.7% for plasma and PE) (Table 1). To the best of our knowledge, G1202K represents the novel secondary ALK mutation (Figure 2). Although its identification was not made at the time-point of Alectinib PD immediately, we supposed that G1202K could play an essential role in Alectinib resistance based on the previous reports on the effect of G1202 mutations.9,10 Since there is no established or known drug that is able to target this novel mutation and reverse G1202K-induced drug resistance, we designed in silico experiments to predict the effects of ALK-G1202K on the first-, second-, and third-generation ALK-TKIs (Crizotinib, Alectinib, and Lorlatinib), which results suggested that Lorlatinib is able to show activity against the Alectinib-resistant mutant ALK-G1202K (see below). On this basis, the off-label treatment with the fourth-line therapy of Lorlatinib for the same patient was restarted (100mg once daily) from November 2019. Encouragingly, this patient was observed with a rapid and dramatic clinical response, exemplified by the fact that he soon felt symptomatic improvement in chest pain and cough, and his PS score also improved to 1. After 4 weeks of the treatment with Lorlatinib (December 2019), scanning results demonstrated a dramatic response in his progressive lung disease (Figure 1). Afterwards, this patient continued to benefit from the Lorlatinib treatment with a PFS of 4 months.
Figure 1.
Timeline summarizing the therapeutic history for a non-small-cell lung cancer patient. MRI scans show lesions of the lumbar 1–3 vertebrae or brain (red arrows). CT imaging shows multiple right lung, pleural metastasis and massive right pleural effusion (red arrows).
Abbreviations: PR, partial response; PD, progressive disease; DFS, disease-free survival; PFS, progression-free survival; NGS, next-generation sequencing.
Table 1.
Cancer-Related Genetic Variants Detected Across Different Samples by NGS
| Date | Sample Type | Gene | Variant Type | cDNA | Amino Acid | VAF |
|---|---|---|---|---|---|---|
| Oct/2017 | Tissue | ALK | Fusion | EML4-ALK | EML4: exon6~ALK: exon20 | 24.20% |
| Oct/2017 | Tissue | CDKN2A | Mutation | c.83T>A | p.V28E | 8.00% |
| Oct/2017 | Tissue | TP53 | Splicing | c.376–2A>G | 6.00% | |
| Nov/2019 | Plasma | ALK | Fusion | EML4-ALK | EML4: exon6~ALK: exon20 | 5.80% |
| Nov/2019 | Plasma | ALK | Mutation | c.3604_3605delGGinsAA | p.G1202K | 2.10% |
| Nov/2019 | Plasma | CDKN2A | Mutation | c.83T>A | p.V28E | 3.90% |
| Nov/2019 | Plasma | TP53 | Splicing | c.376–2A>G | 2.20% | |
| Nov/2019 | Plasma | TP53 | Stop_gained | c.825T>A | p.C275 | 3.30% |
| Nov/2019 | Pleural effusion | ALK | Fusion | EML4-ALK | EML4: exon6~ALK: exon20 | 22.00% |
| Nov/2019 | Pleural effusion | ALK | Mutation | c.3604_3605delGGinsAA | p.G1202K | 23.40% |
| Nov/2019 | Pleural effusion | CDKN2A | Mutation | c.83T>A | p.V28E | 34.00% |
| Nov/2019 | Pleural effusion | TP53 | Stop_gained | c.825T>A | p.C275 | 48.70% |
Abbreviation: VAF, variant allele frequency.
Figure 2.
The identification of ALK-G1202K mutation. EML4: exon6~ALK: Exon20 fusion was detected in the tissue sample (NO: FB19AN0069), and no mutation of ALK-G1202K was found. Both EML4: exon6~ALK: Exon20 fusion and mutation of ALK-G1202K was detected in pleural effusion (NO: CA19AB0015) and plasma (NO: PA19AB0014) samples.
Abbreviation: WT, wild type.
In silico Experiment of G1202k Effects on ALK-TKI
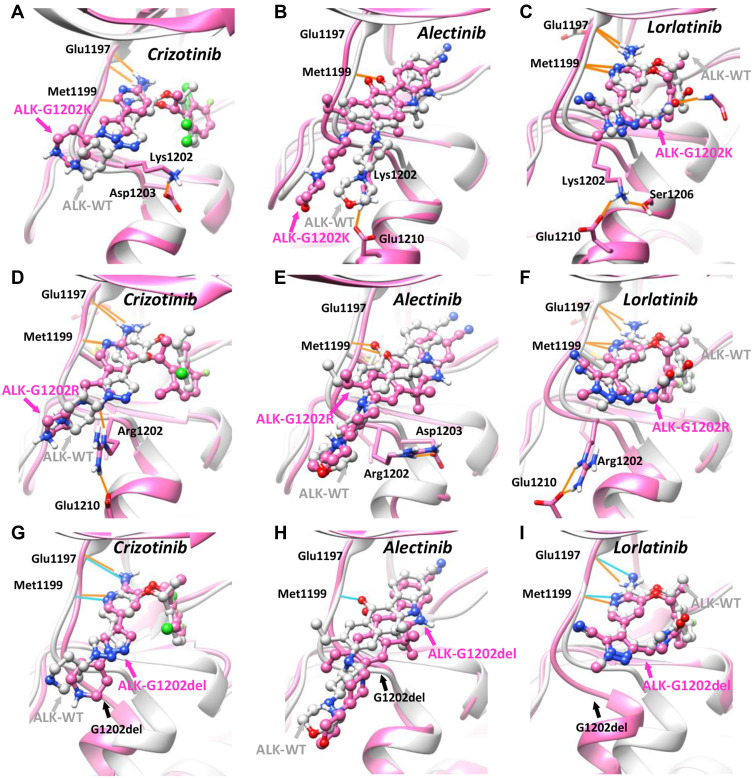
We performed in silico modeling and simulation experiments on the complexes of selected drug and ALK proteins. To compare the mutant effects, we also studied other different variants at the same site, including G1202R and G1202del. Structurally, G1202 residue is located in the hinge region (gatekeeper+6 residue) at the opening of ATP-binding pocket (Figure 3A–I). From our results, it can be seen the solvent-front mutation G1202K, similar to G1202R, orients outside of the pocket and forms favorable charge–charge interaction with surrounding residue Asp1203 or Glu1210 (Figure 3A–F). Importantly, by comparing the binding mode of 3 different ALK-TKIs in context of ALK-G1202K with that of ALK-WT, we observed significant structural deviation for the second-generation TKIs Alectinib, with a root-mean-square deviation (RMSD) of 2.791 angstrom (Figure 3B, Table 2), followed by the first-generation Crizotinib (RMSD: 1.802 angstrom) (Figure 3A, Table 2). In contrast, the third-generation TKI Lorlatinib, was found occupy the ATP-binding site of ALK-G1202K most stably (RMSD: 0.601 angstrom) among all these drugs (Figure 3C, Table 2). Similar results were also observed in the simulations of G1202R and G1202del (Figure 3D–I). It should also be noted that the established favorable hydrogen bonds (HBs) between ALK-WT hinge region and Alectinib, as well as Crizotinib, were either disrupted or weakened by the G1202K mutation (Figure 3A and B), which however, was well maintained in the results of Lorlatinib (Figure 3C), suggesting G1202K mutation is able to destabilize the binding of Alectinib and Crizotinib, but not Lorlatinib. Moreover, the results of binding energy calculation were in agreement with the above structural observations for all 3 mutant types (G1202R/del/K), suggesting their consistent roles in mediating drug resistance against both the 1st- and 2nd-generations of ALK-TKIs. Taking G1202K as an example, the most significant difference in calculated binding energy was observed in Alectinib, increased from −32.22±4.71 kcal/mol (ALK-WT) to −28.22±3.66 kcal/mol (ALK-G1202K) (Table 2), supporting the role of G1202K-induced resistance to Alectinib. In contrast, comparable binding energy was found for Lorlatinib (ALK-WT: −33.41±3.83 kcal/mol, ALK-G1202K: −34.68±4.87 kcal/mol), and small increase was found for Crizotinib (ALK-WT: −32.74±4.61 kcal/mol, ALK-G1202K: −29.01±4.57 kcal/mol) (Table 2).
Figure 3.
In silico study of wild-type and G1202-mutant ALK proteins in complex with the first-, second-, third-generation of ALK-TKIs. Comparison of the binding mode of (A) Crizotinib, (B) Alectinib, (C) Lorlatinib in the context of ALK-G1202K model (pink color) and ALK-WT crystal structure (grey color, PDB ID: 2XP2/3AOX/4CLI). Comparison of the binding mode of (D) Crizotinib, (E) Alectinib, (F) Lorlatinib in the context of ALK-G1202R model (pink color) and ALK-WT crystal structure (grey color, PDB ID: 2XP2/3AOX/4CLI). Comparison of the binding mode of (G) Crizotinib, (H) Alectinib, (I) Lorlatinib in the context of ALK-G1202del model (pink color) and ALK-WT crystal structure (grey color, PDB ID: 2XP2/3AOX/4CLI). The small-molecule drugs are depicted in ball-and-stick model, the protein structure and selected residues are represented in surface/ribbon and stick model, and the hydrogen bonding interactions are shown in orange and cyan lines.
Abbreviation: WT, wild type.
Table 2.
In silico Prediction Results of Wild-Type and G1202K/R/Del Mutant ALK Proteins in Complex with Different Generations of ALK-TKIs
| ALK-TKIs | Binding Energy (kcal/mol) | RMSD (Angstrom) | Binding Energy (kcal/mol) | RMSD (Angstrom) | Binding Energy (kcal/mol) | RMSD (Angstrom) | |
|---|---|---|---|---|---|---|---|
| ALK-WT | ALK-G1202K | G1202K vs WT | ALK-G1202R | G1202R vs WT | ALK-G1202del | G1202del vs WT | |
| Crizotinib (1st) | −32.74±4.61 | −29.01±4.57 | 1.802 | −27.32±5.5 | 2.600 | −25.28±5.56 | 1.438 |
| Alectinib (2nd) | −32.22±4.71 | −28.22±3.66 | 2.791 | −28.96±4.12 | 1.635 | −28.43±4.40 | 1.411 |
| Lorlatinib (3rd) | −33.41±3.83 | −34.68±4.87 | 0.601 | −34.04±4.26 | 1.172 | −34.27±4.35 | 0.680 |
Abbreviations: RMSD, root-mean-square deviation; WT, wild type; ALK, anaplastic lymphoma kinase.
Methods
Targeted NGS
Tissue genomic DNA (gDNA) was extracted using QIAamp DNA FFPE Tissue kit (Qiagen, USA) and cfDNA was purified from plasma and PE with QIAamp Circulating Nucleic Acid kit (Qiagen, USA). Libraries for sequencing were prepared following instructions with KAPA Hyper Prep Kit (KAPA Biosystems) and quantified using KAPA Library Quantification kit (KAPA Biosystems). Target-enriched library was sequenced using HiSeq4000 platform (Illumina) with the mean coverage depth of 1000X and 3000X for the FFPE and cfDNA samples, respectively.
Data Processing and Analysis
Reads passed quality control processing were mapped to the human genome (GRCh37) using BWA (v0.7.12).13 Local realignment and recalibration of base quality scores was performed with GATK (v3.4–0).14 SNVs and INDELs were identified using VarScan2 (v2.3.9)15 with the minimum VAF threshold of 1% and annotated with ANNOVAR,16 followed with manually checking.
In silico Prediction
We computationally constructed the structural model of G1202 mutant ALK-KD in complex with 3 different small-molecule ALK-TKIs based on the crystal structure of ALK-WT in complex with Crizotinib (PDB ID: 2XP2), Alectinib (PDB ID: 3AOX), Lorlatinib (PDB ID: 4CLI). WT and mutant (G1202K/R/del) ALK-KD structural models were built and sampled with a focus on the missing loops in the crystal structure using Modeller17 The modeled structure of ALK-KD in complex with small-molecule ALK-TKI was fully optimized with the package of AMBER18 and then was used for production run in the molecular dynamic (MD) simulation. The binding affinities was estimated with modified end-point binding free energy calculation methods previously reported.19,20
Discussion
Since November 2017, Alectinib has been approved by the Food and Drug Administration (FDA) for initial treatment of ALK-positive lung cancer. Although Alectinib has shown more potent and highly selective, the acquired drug resistance is still inevitable, which seems worsen as the expanded use would result in more unrecognized drug-resistant mutations. Among the known ones, ALK-G1202R is the most frequent resistance mutation among patients with ALK-rearranged lung cancer and progression on the second-generation ALK-TKIs treatment, against which only Lorlatinib maintains activity.10,21 Recent studies also suggested that ALK-G1202del is able to confer moderate resistance to Ceritinib, Alectinib, and Brigatinib.10 All these previous results indicate the important role of the specific residue G1202, which is located at the opening of ATP site of ALK and thus governs the process of TKIs binding. In this study, we identified a novel secondary mutation of ALK-G1202K in a patient progressed upon Alectinib treatment, which could serve as a novel mechanism underlying the drug resistance against Alectinib.
Despite the similar effect across 3 different mutants at G1202 (G1202R, G1202del, G1202K), varying structural basis of mechanism can be found in our simulations. G1202K, much like G1202R, induced the drug resistance against the 1st- and 2nd-generations of ALK-TKIs by introducing bulky and charged substitution in the residue. The favorable charge–charge interaction between G1202K/R and surrounding residues (e.g., D1203 and E1210) is likely to stabilize the bulky group of sidechain at the opening of ATP pocket, which consequently impairs the binding of Crizotinib and Alectinib, both of which extends to this solvent front space (Figure 3A–F). In contrast, G1202del largely changes the conformation of target protein, e.g., displaces the first helix right next to the hinge region, as suggested by the atomic distance of backbone Ca (residue RMSD: 3.028Å) (Figure 3G–I). This movement disturbed the drug binding of Crizotinib and Alectinib, both of which are in contact with this specific motif.
In silico simulation and binding free energy calculation have been widely used to gain insights into the mechanism of molecular interactions.22 Recent studies have proved the power of these state-of-the-art calculations in profiling drug resistant mutations for selected protein kinases as well as predicting the strategy of drug repurposing for the specific mutation(s).23–26 Taking advantage of modern computer, it is possible to perform in silico studies and make rational predictions for the specific mutation(s) within 1 or few days.19,20 In our own case, a MD simulation of 5 nanoseconds for a drug-target (e.g., Lorlatinib-ALKG1202K) complex model, which simultaneously samples the drug molecule along with the binding-site residues in the explicit water environment,27 takes approximately a day on an eight-core CPU. Therefore, it provides a powerful tool in precision medicine in the clinic setting, e.g., in the presence of rare or new mutation(s) against which no drug or treatment knowledge is available.
Limitations
There are several limitations in the present study. First, there is lack of sequential and comprehensive molecular analyses using NGS at every relevant time-point of disease history such like at disease progression indicating resistance to the specific systemic therapy (e.g., Crizotinib, Alectinib). Meanwhile, given the limited mutation detecting scope of currently used sequencing panel, it was not possible to exclude other potential mechanism underlying the drug resistance observed in the clinic. The progression of disease shortly after Crizotinib treatment observed in our case is not in agreement with previous studies.28,29 For instances, Solomon et al reported a median PFS of 10.9 months for Crizotinib among 343 advanced ALK-positive nonsquamous NSCLC patients in a international and multicenter Phase 3 study (PROFILE 1014).29 Similarly, Liu et al reported the estimated PFS of 13.0 months (95% CI 9.0–17.0 months) among 104 patients with ALK rearrangement NSCLC in a single Chinese cancer center.28 All these results suggested there could be other resistant mutation/variation(s) against Crizotinib, which however, is beyond our panel scope. Additionally, given the rare case of G1202K and lack of established or known drug treatment, our current report could provide an example but not guidance for the clinical intervention, which clearly demands more extensive investigations. Given the facts that Lorlatinib is already standard of care for Alectinib-resistant population and it can be used without examining resistant mechanism, as well as that herein we only performed in silico prediction for the drugs already used in the clinic, more extensive and elaborate work is definitely demanded to support the potential use of computational methods in the clinical setting, e.g., changing the clinical scenario.
Conclusion
Collectively, our results suggested ALK-G1202K could function as a novel acquired Alectinib-resistant mechanism, which however, can be targeted by the third-generation ALK-TKI Lorlatinib, suggesting a possible solution against drug resistance in the clinic. Our work also provides structural insights into the G1202K induced drug resistance.
Funding Statement
This work was supported by grants from WUJIEPING Medical Foundation (No. 320675017171).
Abbreviations
ALK, anaplastic lymphoma kinase; EML4, echinoderm microtubule associated protein-like 4; TKI, tyrosine kinase inhibitor; KD, kinase domain; NGS, next generation sequencing; VAF, variant allele frequency; PFS, progression free survival; MD, molecular dynamics; PS, performance status; MRI, magnetic resonance imaging; CT, computed tomography; PD, progression of disease; RMSD, root-mean-square deviation; HB, hydrogen bond; FDA, food and drug administration.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author, Dr. Ping Yang, upon reasonable request.
Ethics statement
This study was approved by Ethics Committee of Tungwah Hospital of Sun Yat-Sen University.
Consent for publication
The written informed consent for the publication of the case report was obtained from the patient under the present study.
Disclosure
Ran Cao, Hua Bao, Xue Wu, Lingling Yang, Dongqin Zhu, and Yang Shao are employees of Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, China. The authors report no other conflicts of interest in this work.
References
- 1.Du X, Shao Y, Qin H-F, Tai Y-H, Gao H-J. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac Cancer. 2018;9(4):423–430. doi: 10.1111/1759-7714.12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 3.Choi YL, Soda M, Yamashita Y, et al.; A.L.K.L.C.S. Group. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363(18):1734–1739. doi: 10.1056/NEJMoa1007478 [DOI] [PubMed] [Google Scholar]
- 4.Yu Y, Ou Q, Wu X, et al. Concomitant resistance mechanisms to multiple tyrosine kinase inhibitors in ALK-positive non-small cell lung cancer. Lung Cancer. 2019;127:19–24. doi: 10.1016/j.lungcan.2018.11.024 [DOI] [PubMed] [Google Scholar]
- 5.Shi R, Filho SNM, Li M, et al. 600E mutation and MET amplification as resistance pathways of the second-generation anaplastic lymphoma kinase (ALK) inhibitor alectinib in lung cancer. Lung Cancer. 2020;146:78–85. doi: 10.1016/j.lungcan.2020.05.018 [DOI] [PubMed] [Google Scholar]
- 6.Fukuda K, Takeuchi S, Arai S, et al. Epithelial-to-mesenchymal transition is a mechanism of ALK inhibitor resistance in lung cancer independent of ALK mutation status. Cancer Res. 2019;79(7):1658–1670. doi: 10.1158/0008-5472.CAN-18-2052 [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Savooji J, Liu D. Second- and third-generation ALK inhibitors for non-small cell lung cancer. J Hematol Oncol. 2016;9(1):19. doi: 10.1186/s13045-016-0251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davare MA, Vellore NA, Wagner JP, et al. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2015;112(39):E5381–90. doi: 10.1073/pnas.1515281112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7(2):137–155. doi: 10.1158/2159-8290.CD-16-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK -rearranged lung cancer. Cancer Discov. 2016;6(10):1118. doi: 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu VW, Cui JJ, Fernandez-Rocha M, Schrock AB, Ali SM, Ou SI. Identification of a novel T1151K ALK mutation in a patient with ALK-rearranged NSCLC with prior exposure to crizotinib and ceritinib. Lung Cancer. 2017;110:32–34. doi: 10.1016/j.lungcan.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 12.Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374(1):54–61. doi: 10.1056/NEJMoa1508887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Šali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 18.Case DA, Cheatham TE 3rd, Darden T, et al. The amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao R, Huang N, Wang Y. Evaluation and application of MD-PB/SA in structure-based hierarchical virtual screening. J Chem Inf Model. 2014;54(7):1987–1996. doi: 10.1021/ci5003203 [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Liu M, Yin M, Liu Q, Wang Y, Huang N. Discovery of novel tubulin inhibitors via structure-based hierarchical virtual screening. J Chem Inf Model. 2012;52(10):2730–2740. doi: 10.1021/ci300302c [DOI] [PubMed] [Google Scholar]
- 21.Lin YT, Yu CJ, Yang JC, Shih JY. Anaplastic lymphoma kinase (ALK) kinase domain mutation following ALK inhibitor(s) failure in advanced ALK positive non-small-cell lung cancer: analysis and literature review. Clin Lung Cancer. 2016;17(5):e77–e94. doi: 10.1016/j.cllc.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 22.Kollman PA, Massova I, Reyes C, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- 23.Hauser K, Negron C, Albanese SK, et al. Predicting resistance of clinical Abl mutations to targeted kinase inhibitors using alchemical free-energy calculations. Commun Biol. 2018;1(1):70. doi: 10.1038/s42003-018-0075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldeghi M, Gapsys V, de Groot BL. Accurate estimation of ligand binding affinity changes upon protein mutation. ACS Cent Sci. 2018;4(12):1708–1718. doi: 10.1021/acscentsci.8b00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aldeghi M, Gapsys V, de Groot BL. Predicting kinase inhibitor resistance: physics-based and data-driven approaches. ACS Cent Sci. 2019;5(8):1468–1474. doi: 10.1021/acscentsci.9b00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikemura S, Yasuda H, Matsumoto S, et al. Molecular dynamics simulation-guided drug sensitivity prediction for lung cancer with rare EGFR mutations. Proc Natl Acad Sci U S A. 2019;116(20):10025–10030. doi: 10.1073/pnas.1819430116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao R, Wang Y, Huang N. Discovery of 2-acylaminothiophene-3-carboxamides as multitarget inhibitors for BCR-ABL kinase and microtubules. J Chem Inf Model. 2015;55(11):2435–2442. doi: 10.1021/acs.jcim.5b00540 [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Yu H, Long Q, et al. Real world experience of crizotinib in 104 patients with ALK rearrangement non-small-cell lung cancer in a single chinese cancer center. Front Oncol. 2019;9:1116. doi: 10.3389/fonc.2019.01116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon BJ, Mok T, Kim D-W, et al. First-line crizotinib versus chemotherapy in ALK -positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]