Abstract
Cinnamomum zeylanicum Blume is an endemic Sri Lankan species commonly known as Ceylon cinnamon or true cinnamon. It is considered the king of spices in addition to its medicinal benefits. Despite recent scientific evidence on its medicinal properties and the industrial demand, cinnamon breeding and crop improvement are not been improved to the expectation. It is mainly due to the limited availability of the genomic information of cinnamon, linked with technical challenges caused by abundant secondary metabolites in all plant parts. Therefore, obtaining high-quality RNA is the fundamental step of transcriptomic analysis and the gene discovery process of cinnamon. We have optimized a CTAB based protocol for high-quality RNA extraction from different cinnamon tissues at various maturity stages collected from the field. Regular pH around 8 and the presence of Polyvinylpyrrolidone (PVP) in CTAB buffer increased the viscosity of the cinnamon lysate. Adjusting the pH of the lysis buffer to 6–6.5 reduced the viscosity of lysate while chloroform precipitates protein efficiently at the adjusted pH with no phenol. Therefore, this protocol excludes PVP and phenol extraction steps. Nanodrop spectrophotometer, gel electrophoresis, and bioanalyzer readings confirmed the quality of extracted RNA. RNA-seq libraries prepared were sequenced with Illumina Sequencing by synthesis technology and obtained good quality data to be used for transcriptomic analysis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02756-1.
Keywords: Ceylon cinnamon, Cinnamaldehyde, Gene expression, Phenol, Polyvinylpyrrolidone (PVP)
Introduction
Gene expression analyses have become the standard laboratory procedure to understand how plants grow and complete the life cycle, and to learn their response to biotic and abiotic stresses and environmental conditions. While the procedures are optimized for model plant species such as Arabidopsis (Kaul et al. 2000), rice (Matsumoto et al. 2005) and tomato (Mueller et al. 2009), no standard procedures for many non-model species. Cinnamomum zeylanicum Blume also known as Ceylon cinnamon or true cinnamon is one such species that gained recent attention due to its health benefits and culinary value. Both leaf and bark of cinnamon are commercially used for culinary purposes, pharmaceutical, nutraceutical, perfume, and incense industries (Pathirana and Senaratne 2020).
While the C. zeylanicum bark consists of cinnamaldehyde, cinnamyl acetate, cinnamic acid, cinnamyl alcohol, and β-Phellandrene (Atish Gursale et al. 2010; Zachariah and Leela 2018; Damasceno et al. 2019) its leaf consists of eugenol, cinnamaldehyde, cinnamyl acetate, cinnamyl alcohol and linalool (Paranagama et al. 2001; Shimna et al. 2017; Liyanage et al. 2020a). C. zeylanicum possess a diverse chemical profile even in the seedling stage grown in greenhouse conditions (Liyanage et al. 2020b). Further, there is a considerable diversity of the chemical composition among different tissues of C. zeylanicum, with abundant cinnamaldehyde in bark, and eugenol is in leaves. Similarly, cinnamyl acetate is dominated in fruits, while camphor is in its root (Kaul et al. 1974; Rao and Gan 2014). The chemical properties of C. zeylanicum have challenged the typical RNA extraction procedures such as Trizol, CTAB and commercial kits.
Nevertheless, extracting high-quality RNA is essential for identifying specific biosynthetic genes involved with unique chemical composition, plant behavior in response to environmental conditions, and identification of gene-specific markers for breeding programs and for studying evolutionary mechanisms of uniqueness. Therefore, we optimized a cetyltrimethylammonium bromide (CTAB) based protocol for extracting good quality RNA from cinnamon bark and leaf at different maturity stages. This protocol was further extended to other C. zeylanicum tissues such as flower, stem, and roots.
As a cationic surfactant, CTAB solubilizes the plant cell walls and lipid membranes of internal organelles simultaneously (Sánchez et al. 2016; Barbier et al. 2019). Since CTAB interacts with anionic nucleic acids to reduce the yield of RNA, higher concentrations of Sodium chloride (NaCl) prevent the CTAB-nucleic acid complexes formations and generate an environment to precipitate nucleic acids but polysaccharides to persist soluble (Barbier et al. 2019). Lithium chloride (LiCl) is added for the precipitation as it efficiently precipitates RNA (Sánchez et al. 2016). As the first step, we modified the CTAB lysis buffer composition, content, and its pH to facilitate quality RNA isolation. Although the RNA extraction protocols optimized for the majority of recalcitrant woody species consist of Polyvinylpyrrolidone (PVP) and phenol (Gasic et al. 2004; Farrell 2017; Rahmani and Amraee 2020), we have excluded both in our new protocol. The optimized protocol resulted in good quality and quantity of RNA for RNA-seq experiments. More importantly, this protocol was optimized with field-collected materials.
Materials and methods
Vegetative propagated C. zeylanicum variety Sri gamunu bark and leaf samples were collected from three fields, representing different agro-ecological zones of Sri Lanka; National Cinnamon Research and Training Centre, Department of Export Agriculture, Palolpitiya (GPS coordinates: 6.0241, 80.5654), Mid country research station, Department of Export Agriculture, Dalpitiya, Atabage (GPS coordinates: 7.133162, 80.590005) and Intercropping and Betel Research Station, Department of Export Agriculture, Dampelassa, Narammala (GPS coordinates: 7.404936, 80.205481). Cinnamon peel or the bark comprises parts of the cortex and secondary phloem (Ravindran and Babu 2004). We harvested cinnamon stems at the correct commercial harvestable stage and peeled the bark using a wood chisel and scraper. A harvested stem was divided into three parts as immature, intermediate, and mature along the stem longitudinally. Two maturity stages of cinnamon leaf were assessed based on the leaf color and collected using secateurs and scissor. Moreover, we also collected few cinnamon flowers, stem (without bark) and root samples (Fig. 1). All the collected tissues were flash-frozen in the field using 50 mL RNA-free tubes containing liquid nitrogen, transported in the same, and stored at − 80 °C until RNA extraction.
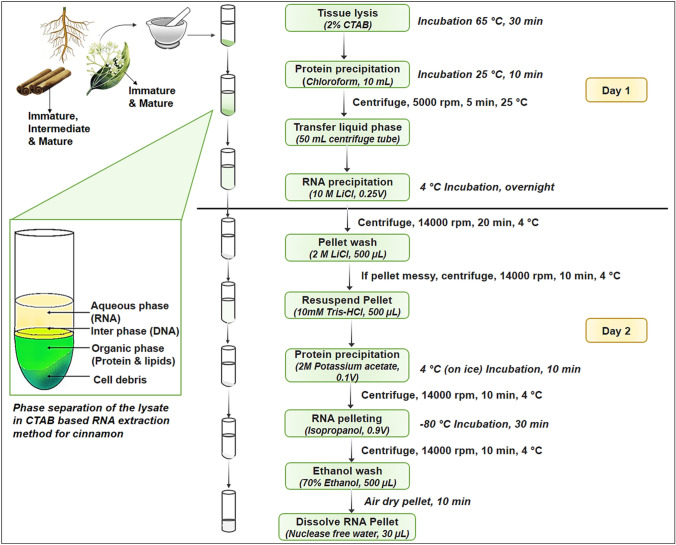
Fig. 1.
Workflow of a modified RNA extraction method for cinnamon. †Cinnamon tissue types used—Bark; immature, intermediate and mature: leaf; immature and mature: inner stem: Flower: Root
All the solutions, buffers, materials, and equipment used for the RNA extraction are described in detail in supplementary file 1. Cinnamon samples were ground to powder using liquid nitrogen and stored at − 80 °C and kept frozen until the addition of lysis buffer. Either two grams of cinnamon bark tissue or 1.5 g of cinnamon leaf tissue were used for the RNA extraction.
The original CTAB based method (Chang et al. 1993) was modified to a considerable extent (Fig. 1). The protocol has two parts; nucleic acid precipitation and RNA isolation. It was performed in two days as the first part in the evening of the first day and the second part in the morning of the next day with an overnight incubation period. CTAB extraction buffer was prepared as given in the supplementary file 01, and pH was adjusted to 6.5 using concentrated HCl. PVP is considered as an essential component in the CTAB buffer optimized protocols for plants with high polyphenol content. However, we have excluded PVP for cinnamon. Then, 2% β-mercaptoethanol (200 µL/ per sample) and 0.5% W/V spermidine (50 µL/ per sample) was freshly added to the CTAB buffer and it was preheated to 65 °C to aid pipetting. Samples were weighed and transferred to 10 mL of preheated CTAB buffer and then incubated at 65 °C for 30 min with shaking and vortexed at 10 min intervals. An equal volume of chloroform (10 mL) was added, shaken vigorously for 10 s and incubated for 10 min at room temperature with 3–5 times vortexing. The samples were then centrifuged at 5000 rpm for 10 min at room temperature and 4–5 mL of liquid phase was pipetted out to a new 50 mL centrifuge tube. Extra care is needed to pipette out only the upper layer of the lysate to avoid possible DNA and impurity contaminations (Fig. 1). If the supernatant was not a clear solution and disturbed by the debris, tubes were centrifuged for additional 5–8 min. Opaque supernatants were common for cinnamon leaf and immature bark samples, probably due to its high abundance of secondary metabolites. As the last step of the first part, 0.25 volume of 10 M LiCl (1 mL to make 2.5 M final concentration) was added to each sample followed by overnight incubation on ice.
On the following morning, tubes were centrifuged at 14,000 rpm for 20 min, to pellet nucleic acid. Pellet was washed with 1 mL of 2 M LiCl and centrifuged again for 10 min at 14,000 rpm. Next, the pellet was dissolved in 500 µL of 10 mM Tris–HCl (pH 7.5) and it was transferred to a new 1.5 mL microcentrifuge tube. Then, 0.1 of 2 M potassium acetate (pH 5.5), was added, to the dissolved pellet, mixed inversely, and incubated on ice for 10 min. After centrifugation at 14,000 rpm for 10 min, the supernatant (450–500 µL) was collected into a new microcentrifuge tube. Then, 0.9 volume of cold isopropanol (about 450 mL) was added to the supernatant, mixed inversely, incubated at -80 °C for half an hour and centrifugated at 14,000 rpm for 10 min to pellet RNA. The pellet was washed with 500 µL of 70% ethanol and air-dried for 10 min. Finally, the RNA pellet was resuspended with 30 µL of nuclease-free water and stored at − 80 °C in aliquots after assessing quality and quantity. RNA samples were treated with DNA free DNA removal kit (Cat. No. AM1907; Invitrogen Inc., Carlsbad, California, USA) as 0.5 µL per 1 µg of total RNA to get rid of any DNA contamination presents in samples.
The quality of the extracted RNA was determined using 1.2% agarose gel with 250 µL of sodium hypochlorite (Aranda et al. 2012) to eliminate the RNase activity during the gel electrophoresis. Safe-Green™ dye (Cat. No. G108-G, Applied Biological Materials Inc., Richmond, British Columbia, Canada) was added to 5 µL of RNA before loading to the gel. The gels were visualized under a UV gel documentation system (Chemi Doc TM XRS + Molecular imager, Bio-Rad Laboratories Inc., Hercules, California, USA) and photographed.
The quantity and quality of the RNA samples were also analyzed using a Nanodrop spectrophotometer (Nano2000, Thermo scientific, Wilmington, Delaware, USA). The quality of the RNA was also assessed with Agilent’s bioanalyzer with the Plant RNA Pico (6000) chip assay following the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA).
RNA sequencing was done at Macrogen, South Korea (Macrogen Inc., Seoul, South Korea). Libraries were constructed using TrueSeq Stranded mRNA LT Sample Prep Kit (Cat. No. RS-122-2101, RS-122-2102, and RS-122-2103; Illumina Inc., San Diego, California, USA). Illumina sequencing by synthesis (SBS) technology was used for RNA-seq to generate paired end reads using the NovaSeq6000 platform. The raw reads were subjected to standard trimming and quality control using Trimomatic (Lohse et al. 2012). Then the quality of the sequenced reads was assessed by FastQC (Andrews 2014). The MultiQC program was used to summarize the analyzed quality results (Ewels et al. 2016).
Results and discussion
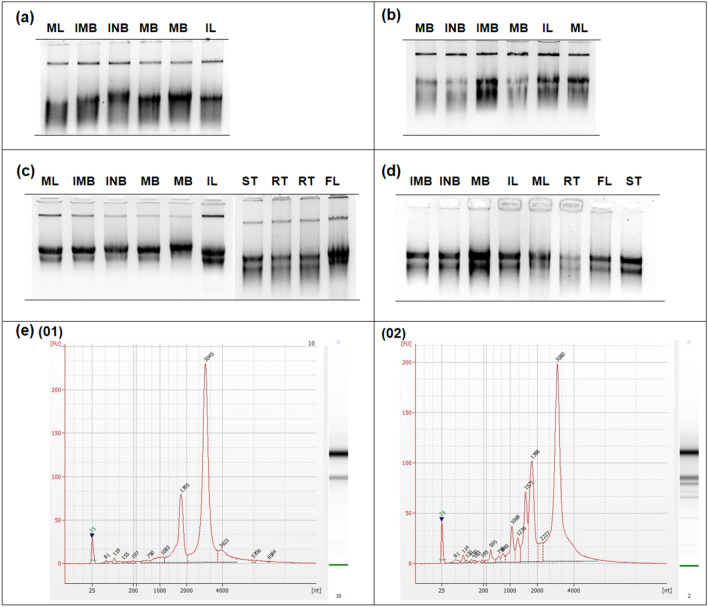
Generally, RNA extraction is more challenging than DNA extraction due to the widely present steady RNase enzyme. Nevertheless, high-quality plant RNA can be extracted if the RNase activities are inhibited (Daohong et al. 2004). There is no standard RNA extraction protocol, which suites all plants. Because, plants possess multifarious tissue types, each has specific challenges in RNA extraction compared to other organisms. Even though several protocols and commercial kits were available, high-quality RNA extraction protocols are keep updating and modifying to meet the specific challenges in plant tissues due to its diverse chemical profiles (Jaakola et al. 2001; Daohong et al. 2004; Wang et al. 2009; Kalinowska et al. 2012; de Lima et al. 2016; Maceda-López et al. 2021). The optimized CTAB based RNA extraction method resulted in good quality RNA with higher yields from all the cinnamon tissues collected from the field (Fig. 2c, Table 1).
Fig. 2.
RNA extracted from different cinnamon tissue types. Representative RNA gel images of Cinnamon from different maturity stages (Bark and leaf): a obtained by traditional CTAB method with partially degraded RNA; b without PVP in CTAB buffer; c and different tissue types after pH optimization of CTAB buffer; d after DNase treatment: e Representative bioanalyzer electropherograms of (01) Immature bark (02) Mature leaf. RNA Gels were resolved on 1.2% Agarose gel. †IMB immature bark, INB intermediate bark, MB mature bark, IL immature leaf, ML mature leaf, RT root, FL flower, ST inner stem. ‡Electropherograms were obtained from Agilent Bioanalyzer with RNA Pico chip assay (Agilent Technologies, Santa Clara, CA, USA)
Table 1.
Quality and quantity measure of Cinnamon RNA
| Cinnamon Tissue types | Nanodrop | †Bioanalyzer | |||||
|---|---|---|---|---|---|---|---|
| No. of samples | Concentration (ng/µL) | 260/230 | 260/280 | No. of samples | RIN | rRNA ratio | |
| Immature Leaf | 27 | 2872 ± 1408 | 2 ± 0 | 1.9 ± 0.1 | 07 | 7.9 ± 0.2 | 5.4 ± 2.3 |
| Mature Leaf | 27 | 2601 ± 1331 | 2 ± 0 | 2.0 ± 0.1 | 19 | 8.0 ± 0.2 | 3.7 ± 0.6 |
| Immature Bark | 27 | 1800 ± 481 | 2 ± 0 | 1.9 ± 0.1 | 07 | 9.0 ± 0.8 | 2.7 ± 1.5 |
| Intermediate Bark | 27 | 1763 ± 503 | 2 ± 0 | 1.8 ± 0.2 | 06 | 8.4 ± 1.4 | 1.5 ± 1.5 |
| Mature Bark | 27 | 1885 ± 635 | 2 ± 0 | 1.7 ± 0.2 | 13 | 6.9 ± 2.1 | 2.5 ± 2.2 |
| Inner stem | 01 | 724 | 2 | 1.9 | 01 | 8.8 | 2.0 |
| Flower | 01 | 3523 | 2 | 2.0 | 01 | 7.9 | 2.0 |
| Root | 01 | 747 | 2 | 2.0 | 01 | 5.6 | 2.0 |
†Bioanalyzer readings were taken after the sample shipment, before RNA-seq by the sequencing service provider
Since cinnamon leaf and bark are the main tissue types used for commercial and medicinal purposes, we have considered cinnamon leaf and bark tissues at different development stages for the method optimization. Furthermore, inner stem, flower, and root samples were also included for completeness and expected future needs.
Removal of PVP from the CTAB buffer and adjusting its pH were the main modifications of the buffers originally described by Chang et al. (1993). In addition, the quantity of the samples, the incubation times, centrifugation time, periods, and speeds were changed in some steps. PVP is a common reducing agent in the CTAB method (Jaakola et al. 2001; Tan and Yiap 2009; Farrell 2017). While many CTAB based extraction methods consist of 1–2% of PVP (Wan and Wilkins 1994; Jaakola et al. 2001; Gasic et al. 2004; Moser et al. 2004; Ono et al. 2012; Inglis et al. 2018; Barbier et al. 2019; Rahmani and Amraee 2020) in the lysis buffer, the PVP content has increased up to 4% for fruits like maqui berry (Sánchez et al. 2016). On the other hand, cinnamon extracts are slightly sticky and slimy due to various viscous polysaccharides, and cinnamon leaf and immature bark samples are slimier than mature bark samples. Hence, it was difficult to separate the cinnamon lysate in the first attempts with traditional Trizol, CTAB, and some commercial kits. The root, stem, and flowers were also less slimy compared to others. In slimy samples, the separation of the liquid phase at the lysis step was difficult compared to the others. The addition of PVP in the CTAB buffer intensified the slimy nature of cinnamon lysate and it was even more difficult to transfer the liquid phase after cell lysis. Moreover, PVP is responsible for take-out lots of RNA, before RNA precipitating by LiCl (Daohong et al. 2004). Consequently, DNA and partially degraded RNA was resulted from the traditional CTAB method, for all cinnamon tissue types considered (Fig. 2a). Excluding PVP from the buffer resulted in RNA bands, but with considerable DNA contaminations (Fig. 2b). A previous study on succulent plant species with high phenolic compounds and polysaccharides had shown that PVP is less effective on quality RNA extraction when high mucilage contents are present (Gehrig et al. 2000). Our results also suggest that PVP is not effective as a contaminant absorbent for plants such as cinnamon.
Further optimizations were needed to avoid slimy nature and severe DNA contaminations. The pH is an important parameter in the Trizol method (Chomczynski and Sacchi 2006). However, adjusting the pH of the CTAB buffer has not been considered in previous optimizations. Generally, DNA collects in 8–9 pH, and RNA collects in 6–7 pH. Because, in acidic conditions, RNA remains in the upper liquid phase of the lysate, while DNA and proteins remain in the interphase or lower organic phase (Chomczynski and Sacchi 2006; Sánchez et al. 2016; Farrell 2017) (Fig. 1). The unavoidable slimy nature of cinnamon led us to test the pH of the CTAB buffer and recorded around 8. Therefore, we have adjusted the buffer pH 6–6.5 by adding concentrated HCl with the objective to retain RNA in the upper aqueous phase. Lowering pH value reduced both the viscosity of the supernatant and severe DNA contaminations. The optimization step reduced the DNA contaminations in cinnamon mature bark, intermediate bark, stem, root, and flower samples, while cinnamon leaf and immature bark still had a considerable amount of DNA in extracted RNA (Fig. 2c). However, the DNase treatment resulted in no detectable DNA in RNA samples (Fig. 2d). Although many RNA extraction protocols of recalcitrant species depend on toxic and expensive phenol for the protein precipitation step, the optimized method excludes it. Instead, chloroform was efficiently precipitated proteins within samples as the modified buffer (PVP excluded) pH reduced the viscosity for a clear phase separation.
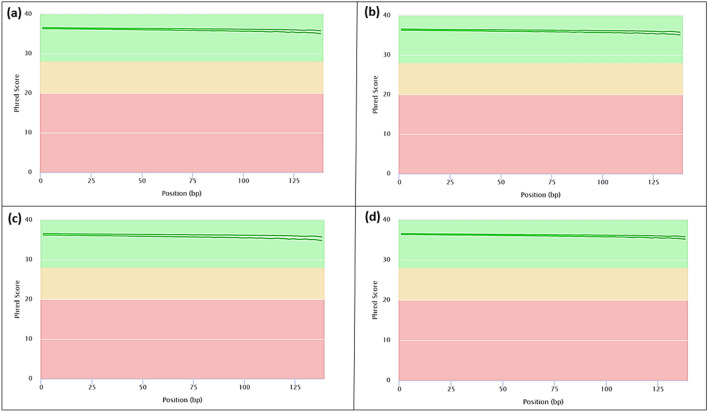
The optimized protocol was used for the extraction of 138 samples collected from the field and all of them were successful in the first attempt. It confirmed the reliability and repeatability of the protocol. The quality and quantity measured with Nanodrop spectrophotometer and gel electrophoresis confirmed sufficiency for continuation (Fig. 2d, Table 1). Nevertheless, leaf and flower samples had higher concentration values while it was low in bark, inner stem, and root samples (Table 1). Variability in cellular composition and secondary metabolites in stem and roots compared to other tissue types explains the observed differences. The bioanalyzer readings qualified the suitability of selected 57 samples for RNA-seq work (Fig. 2e, Table 1). Good quality RNA-seq data validated the suitability of extracted RNA for such sensitive work (Fig. 3). The FastQC analysis indicated good phred quality with uniform high-quality base calls to the end of the forward and reverse sequence reads (Fig. 3). The phred quality is a measure of the quality of the identification of the nucleobases generated by automated sequencing (Zhang et al. 2017). This further confirms the appropriateness of the modified RNA extraction method for sensitive downstream applications. The time taken for extraction is about three hours except for the overnight incubation. We suggest starting on the evening of the first day and continuing the following morning. Nevertheless, eliminating abundant polyphenol compounds prior to the extraction as Maceda-López et al. (2021) described for polyphenolic roots may further increase the quality of Isolated RNA.
Fig. 3.
Sequence quality of Cinnamon RNA extracted using the modified CTAB RNA extraction protocol. RNA was assessed by producing high-throughput RNA sequencing (RNA-seq) libraries of the different cinnamon tissue samples. The RNA-Seq libraries were pair-end sequenced (150 bp) on the Illumina NovaSeq6000 Platform. Sequencing quality assessment using FastQC version 0.10.1 (Andrews 2014) is represented in graphs describing quality across all bases from every sequence read at cinnamon a Bark; b Leaf; c Flower; d Root tissues. †Phred quality score numerically expresses the accuracy of each nucleotide. Higher Q number signifies higher accuracy. For example, if Phred assigns a quality score of 30 to a base, the chances of having base call error are 1 in 1000. The background of the graph divides the y-axis into very good quality calls (green), reasonable quality (orange), and poor quality (red). The graphs are representative of the forward and reverse reads
Together with its culinary value and medicinal importance (Rao and Gan 2014; Ribeiro-Santos et al. 2017; Hajimonfarednejad et al. 2019) C. zeylanicum is a candidate for nutraceuticals and therapeutics and many industries. However, no transcriptomic and gene expression data are available for cinnamon species except Cinnamomum camphora (Chen et al. 2018; Hou et al. 2020). Cinnamomum camphora is botanically and chemically distinct from other Cinnamomum species traded; Cinnamomum cassia Presl (Chinese cassia), Cinnamomum tamala Nees (Indian cassia), Cinnamomum aromaticum Nees (Chinese cinnamon), Cinnamomum urmanii Nees (Indonesian cassia) and Cinnamomum loureirii Nees (Saigon cassia) Cinnamomum (Islam et al. 2009; Li et al. 2010; Pragadheesh et al. 2013; Sharma and Rao 2014; Fajar et al. 2019). In addition, there are wild Cinnamomum species including several endemic in Sri Lanka are having potential future applications (Chandrasekara et al. 2021). As we identified, the sticky and slimy nature of the cinnamon is the major barrier for quality nucleic acid isolation. Since Cinnamomum species, in general, possess similar chemical composition and aroma to C. zeylanicum (Lee et al. 2015; Damasceno et al. 2019; Pandey et al. 2020) except coumarin content (Liyanage et al. 2020a), our modifications would result in quality RNA from other Cinnamomum species.
Conclusion
In summary, a CTAB based RNA isolation procedure was modified to isolate high-quality RNA in sufficient quantity from different cinnamon tissues at different maturity stages. The modified protocol can reliably use for isolating high-quality RNA from different maturity stages of cinnamon bark and leaf and different tissue types such as; roots, inner stem and flowers. The extracted RNA is suitable for sensitive downstream applications such as next-generation sequencing. Overall, our method has improved purity, integrity and yield of RNA. This could be used for extracting quality RNA from field-collected materials of other Cinnamomum species and plants with similar chemical compositions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors acknowledge the financial assistance provided by the Ministry of Primary Industries and Social Empowerment through the National Science Foundation of Sri Lanka under the special Cinnamon project—Grant No: NSF SP/CIN/2016/01. The authors thank the National Cinnamon Research and Training Center, Department of Export Agriculture, Thihagoda, Palolpitiya (NCRTC), Mid country research station, Department of Export Agriculture, Dalpitiya, Atabage, specially, Mr. R.A.A.K. Ranawaka, Assistant Director Research, and Intercropping & Betel Research Station, Department of Export Agriculture, Dampelassa, Narammala for providing cinnamon samples. The authors would like to thank specially Ms. R. M. M. Wijerathne for her continuous technical assistance and the staff of the Agricultural Biotechnology Centre, Faculty of Agriculture, University of Peradeniya for the support extended throughout.
Author contributions
RMMW technical assistance to RNA extraction. HMBMH Technical assistance to sample processing. PGAJ Technical assistance to sample collection, sample processing and preparing. PSAKA Technical assistance to RNA sample preparation. RAJR, YMADKY and BSB Technical assistance to sample collection. SL Assisting and troubleshooting in RNA sequencing.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Nishadi M. N. Liyanage, Email: nishadi3652.liy@gmail.com, https://www.researchgate.net/profile/Nishadi_Liyanage
Bhagya C. H. W. M. Chandrasekara, Email: chmbhagya@gmail.com, https://www.researchgate.net/profile/C_Chandrasekara
Pradeepa C. G. Bandaranayake, Email: pradeepag@agri.pdn.ac.lk, Email: pgunathilake@pdn.ac.lk, https://www.researchgate.net/profile/Pradeepa_Gunathilake_Bandaranayake
References
- Andrews S (2014) FastQC a quality-control tool for high-throughput sequence data http://www.Bioinformaticsbabrahamacuk/projects/fastqc
- Aranda PS, Lajoie DM, Jorcyk CL. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier FF, Chabikwa TG, Ahsan MU, et al. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods. 2019;15:9–12. doi: 10.1186/s13007-019-0447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekara CHWMRB, Naranpanawa DNU, Bandusekara BS, et al. Universal barcoding regions, rbcL, matK and trnH-psbA do not discriminate Cinnamomum species in Sri Lanka. PLoS One. 2021;16:e0245592. doi: 10.1371/journal.pone.0245592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Chen C, Zheng Y, Zhong Y, et al. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genomics. 2018;19:550. doi: 10.1186/s12864-018-4941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- Damasceno CSB, Fabri Higaki NT, Dias JDFG, et al. Chemical composition and biological activities of essential oils in the family lauraceae: a systematic review of the literature. Planta Med. 2019;85:1054–1072. doi: 10.1055/a-0943-1908. [DOI] [PubMed] [Google Scholar]
- Daohong W, Bochu W, Biao L, et al. Extraction of total RNA from Chrysanthemum containing high levels of phenolic and carbohydrates. Colloids Surfaces B Biointerfaces. 2004;36:111–114. doi: 10.1016/j.colsurfb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- de Lima JC, Füller TN, de Costa F et al (2016) A modified protocol for high-quality RNA extraction from oleoresin-producing adult pines. In: Methods in Molecular Biology. Humana Press Inc., pp 27–33 [DOI] [PubMed]
- Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajar A, Abdillah Ammar G, Hamzah M, et al. Effect of tree age on the yield, productivity, and chemical composition of essential oil from Cinnamomum burmannii. Curr Res Biosci Biotechnol. 2019;1:17–22. [Google Scholar]
- Farrell RE (2017) Going green: RNA and the molecular biology of plants. In: Robert E. Farrell J (ed) RNA Methodologies. Elsevier, pp 145–165
- Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report. 2004 doi: 10.1007/BF02772687. [DOI] [Google Scholar]
- Gehrig HH, Winter K, Cushman J, et al. An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Mol Biol Report. 2000;18:369–376. doi: 10.1007/BF02825065. [DOI] [Google Scholar]
- Gursale A, Dighe V, Parekh G. Simultaneous quantitative determination of cinnamaldehyde and methyl eugenol from stem bark of Cinnamomum zeylanicum Blume Using RP-HPLC. J Chromatogr Sci. 2010;48:60–62. doi: 10.1093/chromsci/48.1.59. [DOI] [PubMed] [Google Scholar]
- Hajimonfarednejad M, Ostovar M, Raee MJ, et al. Cinnamon: a systematic review of adverse events. Clin Nutr. 2019;38:594–602. doi: 10.1016/j.clnu.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Hou J, Zhang J, Zhang B, et al. Transcriptional analysis of metabolic pathways and regulatory mechanisms of essential oil biosynthesis in the leaves of Cinnamomum camphora (L.) Presl. Front Genet. 2020 doi: 10.3389/fgene.2020.598714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PW, Marilia de Castro RP, Resende LV, Grattapaglia D. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS ONE. 2018;13:e0206085. doi: 10.1371/journal.pone.0206085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R, Khan RI, Al-Reza SM, et al. Chemical composition and insecticidal properties of Cinnamomum aromaticum (Nees) essential oil against the stored product beetle Callosobruchus maculatus (F.) J Sci Food Agric. 2009;89:1241–1246. doi: 10.1002/jsfa.3582. [DOI] [Google Scholar]
- Jaakola L, Pirttilä AM, Halonen M, Hohtola A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol. 2001;19:201–203. doi: 10.1385/MB:19:2:201. [DOI] [PubMed] [Google Scholar]
- Kalinowska E, Chodorska M, Paduch-cichal E, Mroczkowska K. An improved method for RNA isolation from plants using commercial extraction kits. Acta Biochim Polonica. 2012;59:391–394. doi: 10.18388/abp.2012_2127. [DOI] [PubMed] [Google Scholar]
- Kaul PN, Bhattacharya AK, Rajeswara Rao BR, et al. Volatile constituents of leaf, stem and root oils of cinnamon (Cinnamomum zeylanicum) J Sci Food Agric. 1974;25:1211–1220. doi: 10.1002/jsfa.2740251004. [DOI] [Google Scholar]
- Kaul S, Koo HL, Jenkins J, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee DG, Park JY, et al. Analysis of the trans-Cinnamic acid content in Cinnamomum spp. and commercial cinnamon powder using HPLC. J Agric Chem Environ. 2015;4:102–108. doi: 10.4236/jacen.2015.44011. [DOI] [Google Scholar]
- Li R, Wang Y, Jiang ZT, Jiang S. chemical composition of the essential oils of Cinnamomum loureirii nees. From China obtained by hydrodistillation and microwave-assisted hydrodistillation. J Essent Oil Res. 2010;22:129–131. doi: 10.1080/10412905.2010.9700281. [DOI] [Google Scholar]
- Liyanage NMN, Bandusekara BS, Kanchanamala RWMK, et al. Identification of superior Cinnamomum zeylanicum Blume germplasm for future true cinnamon breeding in the world. J Food Compos Anal. 2020;96:103747. doi: 10.1016/j.jfca.2020.103747. [DOI] [Google Scholar]
- Liyanage NMN, Ranawake AL, Bandaranayake PCG. Cross-pollination effects on morphological, molecular, and biochemical diversity of a selected cinnamon (Cinnamomum zeylanicum Blume) seedling population. J Crop Improv. 2020;00:1–17. doi: 10.1080/15427528.2020.1795769. [DOI] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceda-López LF, Villalpando-Aguilar JL, García-Hernández E, et al. Improved method for isolation of high-quality total RNA from Agave tequilana Weber roots. 3 Biotech. 2021;11:1–10. doi: 10.1007/s13205-020-02620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Wu J, Kanamori H, et al. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Moser C, Gatto P, Moser M, et al. Isolation of functional RNA from small amounts of different grape and apple tissues. Mol Biotechnol. 2004;26:95–99. doi: 10.1385/MB:26:2:95. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Lankhorst RK, Tanksley SD, et al. A snapshot of the emerging tomato genome sequence. Plant Genome. 2009 doi: 10.3835/plantgenome2008.08.0005. [DOI] [Google Scholar]
- Ono NN, Bandaranayake PCG, Tian L. Establishment of pomegranate (Punica granatum) hairy root cultures for genetic interrogation of the hydrolyzable tannin biosynthetic pathway. Planta. 2012;236:931–941. doi: 10.1007/s00425-012-1706-y. [DOI] [PubMed] [Google Scholar]
- Pandey DK, Chaudhary R, Dey A, et al (2020) Current knowledge of Cinnamomum species: a review on the bioactive components, pharmacological properties, analytical and biotechnological studies. In: Bioactive Natural Products in Drug Discovery. Springer Singapore, pp 127–164
- Paranagama PA, Wimalasena S, Jayatilake GS, et al. A comparison of essential oil constituents of bark, leaf, root and fruit of cinnamon (Cinnamomum zeylanicum Blume) grown in Sri Lanka. J Natl Sci Found Sri Lanka. 2001;29:147–153. doi: 10.4038/jnsfsr.v29i3-4.2613. [DOI] [Google Scholar]
- Pathirana R, Senaratne R. An introduction to Sri Lanka and its Cinnamon industry. In: Pathirana R, Senaratne R, editors. Cinnamon. Cham: Springer International Publishing; 2020. pp. 1–38. [Google Scholar]
- Pragadheesh VS, Saroj A, Yadav A, et al. Chemical characterization and antifungal activity of Cinnamomum camphora essential oil. Ind Crops Prod. 2013;49:628–633. doi: 10.1016/j.indcrop.2013.06.023. [DOI] [Google Scholar]
- Rahmani F, Amraee L. Modified CTAB protocol for RNA extraction from Lemon balm (Melissa officinalis L.) Acta Agric Slov. 2020;115:53. doi: 10.14720/aas.2020.115.1.692. [DOI] [Google Scholar]
- Rao PV, Gan SH (2014) Cinnamon: a multifaceted medicinal plant. Evidence-based Complement. Altern Med 2014 [DOI] [PMC free article] [PubMed]
- Ravindran PN, Babu KN. Introduction. In: Ravindran PN, Babu NK, Shylaja M, editors. Cinnamon and Cassia. Florida: CRC PRESS; 2004. pp. 1–13. [Google Scholar]
- Ribeiro-Santos R, Andrade M, Madella D, et al. Revisiting an ancient spice with medicinal purposes: Cinnamon. Trends Food Sci Technol. 2017;62:154–169. doi: 10.1016/j.tifs.2017.02.011. [DOI] [Google Scholar]
- Sánchez C, Villacreses J, Blanc N, et al. High quality RNA extraction from Maqui berry for its application in next-generation sequencing. Springerplus. 2016;5:1–7. doi: 10.1186/s40064-016-2906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Rao LJM. An overview on chemical composition, bioactivity and processing of leaves of Cinnamomum tamala. Crit Rev Food Sci Nutr. 2014;54:433–448. doi: 10.1080/10408398.2011.587615. [DOI] [PubMed] [Google Scholar]
- Shimna K, Krishnamurthy KS, Shamina A. Coumarin, essential oil and total phenol levels in bark and leaves of Cinnamomum species. J Plant Crop. 2017;45:200–205. doi: 10.19071/JPC.2017.V45.I3.3345. [DOI] [Google Scholar]
- Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol. 2009;2009:1–10. doi: 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Wang H, Yin W, Wang C, To K. Isolation of functional RNA from different tissues of tomato suitable for developmental profiling by microarray analysis. Bot Stud. 2009;50:115–125. [Google Scholar]
- Zachariah TJ, Leela NK. Spices: secondary metabolites and medicinal properties. In: Sharangi A, editor. Indian Spices. Cham: Springer International Publishing; 2018. pp. 277–316. [Google Scholar]
- Zhang S, Wang B, Wan L, Li LM. Estimating Phred scores of Illumina base calls by logistic regression and sparse modeling. BMC Bioinform. 2017;18:335. doi: 10.1186/s12859-017-1743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.