Abstract
The high attrition rate in drug development processes calls for additional human-based model systems. However, in the context of brain disorders, sampling live neuronal cells for compound testing is not applicable. The use of human induced pluripotent stem cells (iPSCs) has revolutionized the field of neuronal disease modeling and drug discovery. Thanks to the development of iPSC-based neuronal differentiation protocols, including tridimensional cerebral organoids, it is now possible to molecularly dissect human neuronal development and human brain disease pathogenesis in a dish. These approaches may allow dissecting patient-specific treatment efficacy in a disease-relevant cellular context. For drug discovery approaches, however, a highly reproducible and cost-effective cell model is desirable. Here, we describe a step-by-step process for generating robust and expandable neural progenitor cells (NPCs) from human iPSCs. NPCs generated with this protocol are homogeneous and highly proliferative. These features make NPCs suitable for the development of high-throughput compound screenings for drug discovery. Human iPSC-derived NPCs show a metabolism dependent on mitochondrial activity and can therefore be used also to investigate neurological disorders in which mitochondrial function is affected. The protocol covers all steps necessary for the preparation, culture, and characterization of human iPSC-derived NPCs.
Graphic abstract:
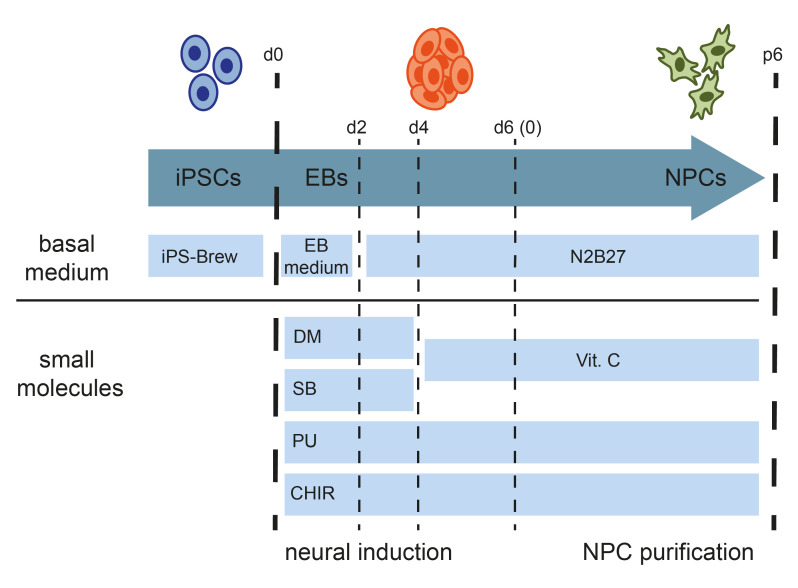
Schematic of the protocol for the generation of human iPSC-derived NPCs
Keywords: Human iPSCs, Neural progenitor cells, Drug discovery, Stem cell differentiation, Neuronal disease modeling, Mitochondrial disorders
Background
In recent years, the downsides of target-centered drug discovery have become evident, in particular for programs addressing neurological diseases ( Paul et al., 2010 ). The challenges of target-based drug discovery led to the revival of phenotypic drug discovery. Phenotypic screenings require the identification of a disease-specific trait (phenotype) that can be modulated within the physiological environment of a cell or organism ( Khurana et al., 2015 ). This approach has the advantage of identifying compounds that show an effect within complex biological environments. At the same time, phenotypic drug discovery requires the presence of a robust disease-relevant phenotype that can be efficiently modulated using a reliable high-throughput detection method. Current drug discovery pipelines strongly rely on cancer-derived immortalized cell lines that lack the functional properties of brain cells. Moreover, immortalized cell lines are mostly glycolytic and less sensitive to mitochondrial impairment than neuronal cells ( Bénit et al., 2016 ). Therefore, immortalized cells are not an effective drug discovery model for neurological diseases and diseases in which mitochondrial oxidative phosphorylation (OXPHOS) is dysfunctional, such as mitochondrial disorders and neurodegeneration ( Cunnane et al., 2020 ).
An important breakthrough in the field of neuronal disease modeling and drug discovery is represented by cellular reprogramming technologies. Cellular reprogramming allows the conversion of the identity of a given cell, thereby enabling the generation of brain cells from patient-specific somatic cells. Cellular conversion can be accomplished through the derivation of induced pluripotent stem cells (iPSCs) ( Takahashi et al., 2007 ) or through the process of direct conversion, which can reprogram skin cells directly into post-mitotic induced neurons (iNs) ( Vierbuchen et al., 2010 ). iPSCs can also be used to derive three-dimensional (3D) cerebral organoids that may recapitulate the organization of the developing human brain ( Lancaster et al., 2013 ).
In order to perform meaningful phenotypic screens, it is necessary to use disease-relevant cellular models. Among reprogramming-derived cell types, patient-specific neural progenitor cells (NPCs) appear particularly well suited for phenotypic drug discovery of neurological diseases, given their homogeneous features, cost-effective culture conditions, and mild proliferative state. Indeed, iPSC-derived NPCs have been successfully employed for drug discovery in the context of neurological and neuropsychiatric disorders (Lorenz et al., 2017; Readhead et al., 2018 ; Walter et al., 2019 ).
During the process of neurogenesis, which is characterized by a metabolic shift from glycolysis to OXPHOS ( Zheng et al., 2016 ), NPCs represent the first cell type to depend on mitochondrial respiration (Beckervordersandforth, 2017; Lorenz and Prigione, 2017). Since NPCs show sensitivity to conditions that impair OXPHOS, they can be used as an effective cell model to carry out high-throughput screenings for neurological and mitochondrial diseases. Accordingly, iPSC-derived NPCs from patients affected by mitochondrial diseases exhibited meaningful phenotypes that could be harnessed to perform phenotypic compound screenings ( Lorenz et al., 2017 ). Altogether, iPSC-derived NPCs might be instrumental for identifying novel treatments in the context of neurological and mitochondrial diseases that are hard to model using conventional drug discovery systems ( Inak et al., 2017 ).
Here, we describe in detail the generation and maintenance of human iPSC-derived NPCs. Different protocols can be used to obtain these cells. Some protocols give rise to NPC populations with limited proliferative features ( Elkabetz et al., 2008 ), while others may depend on the addition of human LIF, which makes culturing the cells very cost-intensive ( Li et al., 2011 ). However, most useful approaches rely solely on small molecule to produce highly proliferative NPC populations ( Reinhardt et al., 2013 ). A key characteristic of these NPCs is their capability of robust and homogenous expansion. At the same time, NPCs should be able to efficiently differentiate into neuronal and glial cell types ( Reinhardt et al., 2013 ; Lorenz et al., 2017).
Our approach is based on the protocol of Reinhardt et al. ( 2013) and was published in Lorenz et al. (2017) for drug discovery applications in the context of neurological mitochondrial diseases. The protocol allows the generation of NPCs that maintain their characteristics under chemically defined conditions over several passages and can be readily differentiated into the desired brain cell types. The protocol is highly reproducible and leads to homogenous cell populations. It is similarly effective starting from human embryonic stem cells, healthy control iPSCs, and patient-specific iPSCs ( Lorenz et al., 2017 ).
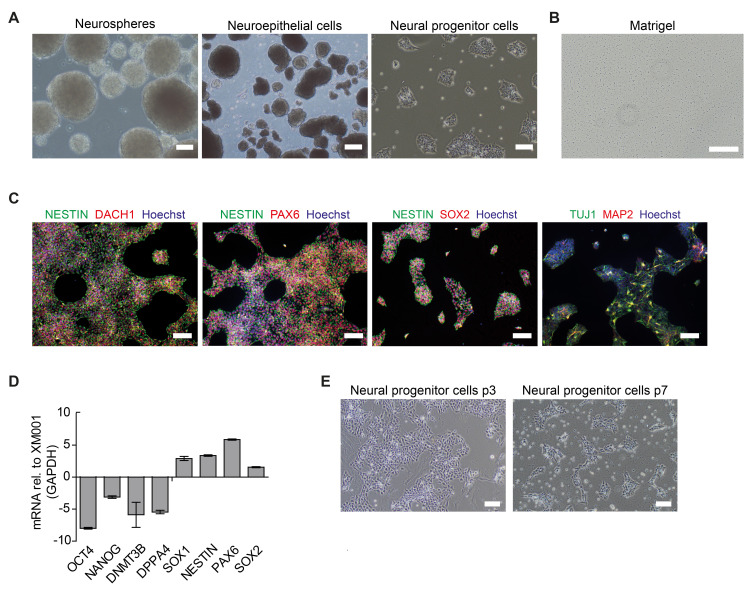
To initiate neural induction, bone morphogenic protein (BMP) inhibitor dorsomorphin (DM), and transforming growth factorβ (TGFβ)-signaling inhibitor SB43152 (SB) are supplemented to the basic NPC media (BM1). Purmorphamine (PU) and CHIR99021 (CHIR) are added to stimulate the sonic hedgehog (SHH) signaling pathway, inhibit glycogen synthase kinase-3 (GSK3), and activate canonical WNT signaling ( Chambers et al., 2009 ; Kim et al., 2010 ). After the first days in the induction medium, embryoid bodies (EBs) form neuroepithelial structures (Figure 1A) . After 6 days, the EBs are gently dissociated and seeded on Matrigel-coated plates for purification. Neural progenitor morphology becomes homogenous after 4-6 passages post neural induction (Figure 1A) . NPCs should express typical NPC markers such as NESTIN, PAX6, SOX1, SOX2, and DACH1. Spontaneous differentiation of cells expressing the pan-neuronal marker TUJ1 is always present (Figure 1C-1D) . Conversely, pluripotency markers like OCT4, NANOG, DNMT3B, and DPPA4 should be downregulated compared to the parental iPSC lines. NPCs retain their multipotent identity over multiple passages (> 30) as well as after freezing and thawing without changes in cell morphology.
Figure 1. Generation and characterization of human iPSC-derived NPCs.
A. Representative images of neurospheres, neuroepithelial cells, and neural progenitor cells (NPCs). B. Image of a matrigel coated well. Scale bar: 200 µm. C. Immunocytochemistry of NPCs obtained from the control iPSC line XM001 ( Wang et al., 2018 ) stained for the neural progenitor markers NESTIN, DACH1, PAX6, and SOX2. Spontaneous differentiation of cells expressing TUJ1 or MAP2 can be present. Scale bar: 100µm. D. Quantitative real-time reverse transcription PCR (qRT-PCR) of neural progenitor markers SOX1, NESTIN, PAX6, and SOX2, and of pluripotency-associated markers OCT4, NANOG, DNMT3B, and DPPA4. Transcription levels in NPCs were normalized using GAPDH housekeeper gene (mean=SD +/-; n=3 technical replicates) and reported in relation to the transcription level of the respective control iPSC line XM001. E. Representative images of neural progenitor cells (NPCs) from passage 3 (p3) and passage 7 (p7) demonstrating the change in morphology towards homogenous NPC cultures. Scale bar: 100 µm.
The following step-by-step protocol allows the experimenter to obtain human iPSC-derived NPCs with a high degree of homogeneity within 4-8 weeks, and to culture and characterize the cells for subsequent use in disease modeling or drug discovery applications.
Materials and Reagents
The following list provides examples of the materials and equipment that we routinely use in our laboratory. Nevertheless, reagents and equipment with similar specifications will work as well.
Cellstar cell culture multiwell plates (6-24 well) (Greiner Bio-One, catalog numbers: 657 160 [6-well; 665 180 [12-well]; 662 160 [24-well])
Corning 60 mm Ultra-low attachment culture dish (Corning, catalog number: 3261)
Cell spatula (TPP, catalog number: 99010)
Serological pipettes (Greiner Bio-One, catalog numbers: 760180 [2 ml]; 606180 [5 ml]; 607180 [10 ml]; 760107 [25 ml]; 768160 [50 ml])
BemisTM Curwood ParafilmTM M Laboratory Film (Fisher Scientific, product code HS234526C, catalog number: 10018130)
Greiner centrifuge tubes (50 ml and 15 ml) (Sigma-Aldrich, manufacturer: Greiner; catalog number for 15 ml: T1818, for 50 ml: T2318)
Liquid nitrogen
Corning ® Matrigel ® Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenolred-free, LDEV-free (Corning, CAS number: 356231, format: 10 ml, store at -20 °C and follow instructions from supplier for preparation)
CHIR 99021 (Caymen Chemical, CAS number: 252917-06-9, catalog number: 13122; product format: 2.5 mg/ml in DMSO; storage at -20 °C as 6 mM stock; stability: ≥ 2 years)
SB431542 in solution (MACS Miltenyi, CAS number: 130-106-543, catalog number: 130-106-543; product format: 10 mM in DMSO; storage at -20 °C as 10 mM stock, protected from light; stability: 6 months)
Dorsomorphin (StemCell; CAS number: 866405-64-3, catalog number: 72102; product format 10 mg solid; storage at -20 °C as 5 mM stock in DMSO (heat applied) or ethanol; stability: 12 months)
Purmorphamin (MACS Miltenyi, CAS number: 483367-10-8; catalog number: 130-104-465 product format: 5 mg solid; storage at -20 °C as 10mM stock in DMSO, protected from light)
(+)-Sodium L-ascorbate (Vitamin C) (Merck, Sigma-Aldrich, catalog number: A4034; product format solid powder; store as 200 µM stock in water at -20 °C)
N-2 Supplement (100×) (Thermo Fisher Scientific, Gibco, catalog number: 17502-048; product format: 5 ml liquid; store at -20 °C, protected from light; stability: 18 months)
B27 without Vitamin A (50×) (Thermo Fisher Scientific, Gibco, catalog number: 12587010; product format: 10 ml liquid; store at -20 °C protected from light)
Neurobasal® Medium (Thermo Fisher Scientific, Gibco, catalog number: 21103049; product format: 500 ml liquid; store at 2-8 °C, protected from light; shelf life: 12 months)
DMEM/F12, HEPES (Thermo Fisher Scientific, Gibco, catalog number: 31330038; product format: 500 ml liquid; store at 2-8 °C, protected from light; shelf life: 12 months)
KnockOut DMEM (Thermo Fisher Scientific, Gibco, catalog number: 10829018; product format: 500 ml liquid; store at 2-8 °C protected from light
KnockOut Serum replacement (Thermo Fisher Scientific, Gibco, catalog number: 10828010; product format: 500 ml liquid; store at -20 °C protected from light
StemMACSTM iPS-Brew XF, human (MACS Miltenyi Biotec, catalog number: 130-104-368)
mTeSRTM1 (STEMCELL Technologies, catalog number for 500 ml kit: 85850)
Penicilin-Streptomycin (P/S; 10,000 U/ml Penicilin, 10,000 µg/ml Streptomycin) (Thermo Fisher Scientific, Gibco, catalog number 15140122; product format: 100 ml liquid; store at -20 °C; shelf life: 12 months)
L-Glutamine (200 mM) (Thermo Fisher Scientific, Gibco, Catalog number: 25030081; product format: 100 ml liquid; store at -20 °C, protected from light; shelf life: 24 months)
Non-essential amino acids (NEAA, 100×) (Thermo Fisher Scientific, Gibco, catalog number: 11140035, product format: 100 ml, store at 2-8 °C; shelf life: 12 months)
Sodium pyruvate (100 mM) (Thermo Fisher Scientific, Gibco, catalog number: 11360070; product format: 100 ml; store at 2-8 °C protected from light; shelf life: 12 months)
MycoZap Plus-CL (Lonza, catalog number VZA-2012; product format: 20 ml liquid; store at -20 °C, protected from light)
StemProTM AccutaseTM Cell Dissociation Reagent (Thermo Fisher Scientific, Gibco/StemPro, catalog number: A1110501; product format: 100 ml liquid, store at -20 °C, protected from light)
DPBS (without calcium and magnesium) (Thermo Fisher Scientific, Gibco, catalog number: 14190250; product format: 500 ml liquid; store at 15-30 °C; shelf life: 36 months)
ROCK inhibitor (RI), Y-27632, dihydrochloride (Enzo, CAS number: 129830-38-2; catalog number: ALX-270-333-M005; product format 5 mg powder; store at -20 °C; soluble in DMSO as 25 mg/ml; stability: stock solution stable for 1 month at -20 °C)
UltraPureTM 0.5 M EDTA (Invitrogen, catalog number: 15575020)
Bambanker (NIPPON Genetics, catalog number: BB03)
ShandonTM Immu-MountTM (Thermo ScientificTM, catalog number: 77-86-1; reference number 9990402; product formal: 20 ml)
Anti-Nestin Antibody, clone 10C2 (Millipore, catalog number: MAB5326; Brand family: Chemicon®)
Anti-Pax-6 Antibody (Covance, catalog number: 901301)
SOX2 antibody (R&D Systems, catalog number: AF2018)
DACH1 antibody (ProteinTech, catalog number: 10914-1-AP)
Anti-β-Tubulin III (TUJ-1) (Sigma-Aldrich, catalog number: T8578)
MAP2 (Synaptic system, catalog number: 188004)
Hoechst 33342 (Hoechst) (InvitrogenTM, catalog number: H3570)
16% Paraformaldehyde Aqueous Solution (PFA) (Electron Microscopy Sciences; catalog number: 50-980-487)
Tween-20 (Sigma-Aldrich, catalog number: P9416)
TritonTM-X100 (Sigma-Aldrich, catalog number: T8532)
Alexa FluorTM 488 donkey anti-rabbit IgG (H+L) secondary antibody (Thermo Fisher Scientific, catalog number: A-21206)
Alexa FluorTM 647 donkey anti-mouse IgG (H+L) secondary antibody (Thermo Fisher Scientific, catalog number: A-31571)
Donkey anti-goat Cy 3 (Sigma-Aldrich, catalog number: AP180C)
Donkey anti-guinea pig Cy 3 (Sigma-Aldrich, catalog number: AP193C)
SYBRTM Green PCR Master Mix (Thermo Fisher Scientific, catalog number: 4309155)
M-MLV Reverse Transkriptase (Invitrogen, catalog number: 28025013)
RNeasy Mini Kit (50) (Qiagen, catalog number: 74104)
Rnase-Free Dnase Set (50) (Qiagen, catalog number: 79254)
Oligo(dT) 12-18 Primer (Invitrogen, catalog number: 18418012)
dNTP-Mix (10 mM) (Invitrogen, catalog number: 18427088)
MicroAmpTM Optical 348-well-reaction plate with barcode (Thermo Fisher Scientific, catalog number: 4343814)
Basic medium 1 (BM1) (see Recipes, Table 1)
BM2 (sm-) medium (see Recipes, Table 1)
M1 medium (see Recipes, Table 2)
M2 medium (see Recipes, Table 2)
M3 (sm+) medium (see Recipes, Table 2)
Table 1. Basic media formulation.
| Medium | Components | Volume | Stock conc. | Final conc. |
|---|---|---|---|---|
|
Basic Medium 1 (BM1) |
KnockOut-DMEM | 40 ml | 1× | 1× |
| KnockOut-SR | 10 ml | 5× | 1× | |
| Pen/Strep | 500 µl | 10 mg/ml; 10.000 U/ml |
0.1 mg/ml; 100 U/ml |
|
| Glutamine | 500 µl | 200 mM | 2 mM | |
| NEAA | 500 µl | 100× | 1× | |
| Pyruvate | 500 µl | 100 mM | 1 mM | |
| MycoZap | 100 µl | 500× | 1× | |
|
Basic Medium sm- (base for sm+) |
DMEM/F12 | 240 ml | 1× | 0.5× |
| Neurobasal | 240 ml | 1× | 0.5× | |
| N-2 Supplement | 2.5 ml | 100× | 0.5× | |
| B-27 Supplement without vitamin A | 5 ml | 50× | 0.5× | |
| Pen/Strep | 5 ml | 10 mg/ml; 10.000 U/ml |
0.1 mg/ml; 100 U/ml |
|
| Glutamine | 5 ml | 200 mM | 2 mM | |
| MycoZap | 1 ml | 500× | 1× |
Table 2. Culture media formulation.
| Medium | Components | Volume | Stock conc. | Final conc. |
|---|---|---|---|---|
|
Medium 1 (M1); day 0-day 2 |
BM1 | 10 ml | 1× | 1× |
| CHIR | 5 µl | 6 mM | 3 µM | |
| SB | 10 µl | 10 mM | 10 µM | |
| Dorsomorphin | 2 µl | 5 mM | 1 µM | |
| Purmorphamine | 7.69 µl | 0.65 mM | 500 nM | |
|
Medium 2 (M2); day 2-day 4 |
sm- | 10 ml | 1× | 1× |
| CHIR | 5 µl | 6 mM | 3 µM | |
| SB | 10 µl | 10 mM | 10 µM | |
| Dorsomorphin | 2 µl | 5 mM | 1 µM | |
| Purmorphamine | 7.69 µl | 0.65mM | 500 nM | |
|
M3/sm+ (maintenance of NPCs) |
sm- | 10 ml | 1× | 1× |
| CHIR | 5 µl | 6 mM | 3 µM | |
| Purmorphamine | 7.69 µl | 0.65 mM | 500 nM | |
| Vitamin C | 7.5 µl | 200 mM | 150 µM |
Equipment
Centrifuge (Eppendorf, model: 5810R)
Water bath (Thermo Fisher Scientific, FisherbrandTM IsotempTM Digital-Control Water Baths: Model 220)
CO2 incubator for cell culture (Thermo Fisher Scientific, model: Heraeus BBD 6220)
Hypoxia incubator for iPSC culture (Binder GmbH, model: Binder BCB 160)
qPCR machine (Thermo Fisher Scientific, model: ViiA 7)
Confocal Microscope system (Zeiss, model: Axio Imager Z1)
Freezing container (Nalgene, model: Mr. Frosty; Corning CoolCellTM)
-80°C Freezer (New Brunswick, model: Innova 4725)
Laminar flow hood (BDK, Luft und Reinraumtechnik GMBH)
Sterile scissor (for example: Surgical scissors, sharp, Sigma-Aldrich, catalog number: Z265977)
Shaker (Rocky, LTF Labortechnik)
Procedure
In general:
Work under sterile conditions, e.g., under a laminar flow hood.
Disinfect reagents and consumables before transferring under the hood.
All media should be prepared fresh and used within one week.
Do not thaw N-2 Supplement and B-27 Supplement in the water bath. Instead thaw them at room temperature (RT, 20-22 °C) for 2-4 h or in the fridge (4 °C) over night.
All media should be pre-warmed before applying to the cells. Do not keep the media at 37 °C for long periods but rather pre-warm at RT.
-
Culture the NPCs and iPSCs in a controlled environment with 37 °C and 5% CO2.
Optional: If possible, culture the iPSCs under hypoxic conditions (37 °C, 5% CO2, 5% O2), which more closely mimics in vivo conditions.
We culture iPSCs under feeder-free conditions in either StemMACSTM iPS-Brew XF, or mTeSRTM1 on Matrigel-coated 6-well plates.
We passage iPSCs using 0.5 mM EDTA/PBS in ratios ranging from 1:4-1:12. To increase cell survival, we recommend adding ROCK inhibitor (RI) after each splitting in a concentration of 10 µM.
-
Step-by-step protocol for NPC generation
Day 0 (Monday): Harvesting of the iPSCs and transfer on Corning Ultra-Low Attachment 60mm/15mm dishes.
Prepare the basic medium 1 (BM1) (Table 1).
Prepare M1 medium (Table 2).
Choose one to two wells of iPSCs (~80% confluent).
Remove the media and rinse the wells twice with DPBS to remove dead cells and debris.
Detach cells from the plate using Accutase (1 mg/ml). Add 0.5-1 ml/well and incubate for 2-5 min at 37 °C, check visually in between.
Continue when colonies are mainly detaching from the plate.
Add 4-10 ml PBS or medium to reduce Accutase activity.
Optional: Mechanically detach cells using a cell spatula if cells are not completely detached by Accutase activity.
Transfer the cell suspension to a 15 ml Falcon tube (pool both wells), let the cells settle to the bottom (10-15 min) in water bath.
Optional: Centrifuge at 120 × g for 5 min (potentially more damaging but faster and results in a more solid pellet).
Remove as much medium as possible from the sedimented cells by pipetting (for example with a 5 ml serological pipette). Make sure not to disperse or suck up the cells.
Add 5 ml of M1 to the cells, transfer the cells to a non-treated Corning Ultra-Low Attachment 60 mm/15 mm dish (1:1; or X number of wells in case of high confluence).
Incubate at 37 °C, 5% CO2 for 2 days.
Incubation with M1 will form embryoid bodies (EBs) floating in the media. If they attach at day 1 to the bottom of the Corning Ultra-Low attachment 60 mm/15 mm dish, detach them by gently pipetting using a 1 ml pipette (cut off the tip with sterile scissors!). Make sure not to shred the floating EBs. Alternatively, use a cell spatula. Keep the media in the well and gently scrape throughout the well using a cell spatula to detach EBs.
Day 2 (Wednesday): 50% media exchange to M2- Prepare BM2 (sm-) medium (Table 1).
- Prepare M2 medium (Table 2).
- Cut off the tip of a 1 ml pipet tip using a sterile scissor.
- Transfer EBs to a 15 ml Falcon tube using a 1 ml pipette tip (cut off the tip!).
- Optional: If some EBs attached, detach EBs using a cell spatula.
- Let EBs settle for 15 min in the water bath at 37 °C.
-
Remove 50% of M1 and add 50% of M2 instead.Optional: Remove all M1 and add 100% M2 (but potentially more damaging for the cells).
- Transfer the EBs back to the same Corning Ultra-Low Attachment 60 mm/15 mm dish using a 1 ml pipette tip (cut off the tip using a sterile scissor!).
Day 3 (Thursday): 100% media exchange to M2- Prepare M2 medium (Table 2).
- Cut off the tip of a 1 ml pipet tip using a sterile scissor.
- Transfer EBs to a 15 ml Falcon tube using a 1 ml pipette tip (cut off the tip!).
- Optional: If necessary, detach EBs using a cell spatula as described before.
- Let EBs settle for 15 min in the water bath at 37 °C.
- Exchange the medium completely to 100% of M2 medium (5-6 ml final volume).
- Transfer EBs back to the same Corning Ultra-Low Attachment 60 mm/15 mm dish using a 1 ml pipette tip (cut off the tip using a sterile scissor!).
Day 4 (Friday): 100% media exchange to M3 (sm+) and preparation of Matrigel-coated plates
- Prepare M3 (sm+) medium (Table 2).
- Cut off the tip of a 1 ml pipet tip using a sterile scissor.
- Transfer EBs to a 15 ml Falcon tube using a 1 ml pipette tip (cut off the tip!).
- Optional: If necessary, detach EBs using a cell spatula as described before.
- Let EBs settle for 15 min in the water bath at 37 °C.
- Exchange the media completely to 100% of M3 (sm+) medium (5-6 ml final volume).
- Transfer EBs back to the same Corning Ultra-Low Attachment 60 mm/15 mm dish using a 1 ml pipette tip (cut off the tip using a sterile scissor!).
- Coat a 6-well plate with Matrigel (procedure described separately) as preparation for day 7.
Day 7 (Monday): Transfer the cells to Matrigel-coated plates
- Prepare M3 (sm+) medium (Table 2).
-
Transfer EBs from the Corning Ultra-Low Attachment 60 mm/15 mm dish to a 15 ml Falcon tube. Use a 1 ml tip (cut off the tip!).If necessary, detach neural tube-like formations using a cell spatula as described for EBs.
- Let EBs settle down in the water bath at 37 °C (10-15 min).
- Remove old media.
- Add new sm+ medium.
- Shred the cell formations by pipetting 5-10 times using a normal 1 ml tip.
- Transfer the whole suspension to the Matrigel-coated 6-well plate, distribute equally to 2-X wells depending on the amount of EBs.
- Fill up to 2 ml sm+ each 6-well.
-
Add ROCK inhibitor (RI) (10 µM final concentration).Note: To increase cell survival, we recommend adding RI to the cells for the first 1-2 splittings. Afterwards, RI is not needed!
- Try to keep the cells in the first well for 1 week.
- If necessary, split the cells as described in the maintenance protocol.
Note: NPCs should appear homogenous after 4-5 passages, expressing NESTIN, PAX6, and SOX2. NPCs usually also express the immature pan-neuronal marker TUJ1. Spontaneous differentiation can occur, and some cells can express the neuronal marker MAP2. Cells do not necessarily need to be cultured in 6-well plates.
-
Matrigel preparation
Thaw frozen Matrigel (stored at -20 °C) OVERNIGHT (ON) on ice at +4 °C.
Check protein concentration (stock concentration) of Matrigel as provided in Lot datasheet.
Calculation: Add a specific volume (=x) of cold KnockOut-DMEM to the 10 ml thawed Matrigel in the vial to dilute it to 5 mg/ml final concentration. The stock concentration of your vial (= y) should be written on the lot datasheet. Calculate the needed volume using the formula: x = y * 2-10 ml.
Perform next steps under the hood and on ice!
Prepare a pre-chilled 50 ml Falcon tube with specific volume of cold KnockOut-DMEM.
Prepare pre-chilled 15 ml Falcon tubes on ice labeled with 1 M.
Transfer Matrigel quickly to KnockOut-DMEM and mix carefully.
Aliquot 1 ml of the diluted Matrigel in each pre-chilled 15 ml Falcon tube.
Store Aliquots immediately at -20 °C.
Final concentration in 6/12-well-plates: 200 µg/ml.
-
Coating plates with Matrigel
Transfer 30 ml cold KnockOut-DMEM into a 50 ml Falcon tube.
Dilute 1 ml aliquot 1:30 in cold KnockOut-DMEM.
Resuspend the 1 ml Matrigel aliquot (15 ml Falcon tube, -20 °C) with 5 ml of the 30 ml cold KnockOut-DMEM in the 15 ml Falcon tube until pellet is resolved! (It may take a while).
-
1 ml Matrigel aliquot resuspended in 30 ml cold KnockOut-DMEM as described in C2 is enough for 4 plates.
6-well: 1.25 ml.
12-well: 500 µl.
24-well: 300 µl.
Add Matrigel-medium to the wells (see point 4).
-
Polymerization:
Store at 4 °C in the fridge, ON (preferentially!).
2 h at 37 °C is possible.
Check polymerization in phase contrast before use! The evenly distributed formation of an extracellular matrix should be visible (Figure 1B). Coated plates can be stored for 1-2 weeks (4 °C). For storage seal the plates with Parafilm.
Before use, pre-warm the plate either at RT or 37 °C.
-
Maintenance of NPC cultures
Media needs to be changed every 2nd day.
Media should be pre-warmed before applying to the cells. Do not keep media at 37 °C for long periods but rather pre-warm at RT.
Typically, cells were split using Accutase in ratios of 1:3 up to 1:10 every 4-5 days depending on the cell density.
-
Splitting NPCs
1.Aspirate growth media.
2.Add 500 μl Accutase and incubate for 2-5 min at RT or 37 °C if cells do not detach easily.
3.Dilute Accutase activity by addition of 2 ml sm- (without any supplementation) or PBS.
4.Gently detach the cells by pipetting.
5.Transfer the cells to a 15 ml Falcon tube and centrifuge at 200 × g for 3 min at RT.
6.In the meantime, prepare Matrigel-coated plates for plating the cells.
Aspirate Matrigel-mix.
Add 1 ml sm+ and store in the incubator until further use.
-
Fill blank wells with 2 ml DPBS.
Note: Matrigel coating should not dry out.
7.Aspirate the supernatant.
8.Resuspend the pellet in sm+ medium.
9.Distribute cells to Matrigel-coated 6-well plates.
10.Fill up with media to 2 ml sm+.
Note: To increase cell survival, we recommend adding RI (10 µM) to the cells for the first 1-2 splittings. Afterwards, RI is not needed!
-
Freezing NPCs
1.Detach NPCs according to the splitting procedure.
2.Resuspend NPCs in 0.5 ml of sm+. Add freezing medium (2× sm+ freeze:sm+, 1:1).
Note: 10 ml of 2× sm+ freeze is prepared by mixing 2 ml of sm+, 6 ml of KnockOut-SR, and 2 ml of DMSO.
3.Alternatively, use commercial freezing media according to manufacturer’s protocol.
Note: For example, Bambanker (Nippon Genetics, catalog number: BB03).
4.Transfer the resuspended NPCs into a cryovial and place the vials at -80 °C in a cryo-freezing container.
Note: Use a freezing container such as Mr. FrostyTM (Nalgene) to freeze the cells, which allows slow freezing at a rate of -1 °C per minute.
5.Store NPCs at -80 °C or in a liquid nitrogen tank for long term storage (1-3 years).
-
Thawing NPCs
1.Quickly but gently thaw the cryovial containing NPCs in a 37 °C pre-warmed water bath.
2.Transfer NPCs into a 15 ml Falcon tube set with 1 ml of sm+ and up to 10 ml of pre-warmed PBS.
3.Centrifuged the cells at 200 × g for 3 min.
4.Remove the supernatant and resuspend NPCs in sm+ medium.
5.Seed NPCs on previously prepared Matrigel-coated plates 1:1 (see Coating plates with Matrigel). Note: Add 10 µM RI after thawing to support cell survival.
-
Characterization of NPCs by immunofluorescence
After 4-6 passages, seed NPCs on Matrigel-coated 24-well plates containing 12 mm coverslips. When reaching approximately 80% of confluence, rinse NPCs with PBS, and fix the cells with a 4% paraformaldehyde (PFA)/PBS solution for 15 min at RT.
Remove the PFA and rinse NPCs with PBS calcium/magnesium free (PBS -/-) 3 times for 5 min.
-
Add blocking solution to the cells for 1 h at RT.
Note: Blocking solution is prepared with 10% donkey serum and 1% Triton X-100 in PBS -/- with 0.1% Tween-20.
-
Incubate the cells with specific primary antibodies overnight at 4 °C on an orbital shaker (60-80 rpm).
Note: Dilute primary antibody in blocking solution. Following antibodies are commonly used in our laboratory: NESTIN (Millipore, 1:200), PAX6 (Covance, 1:200), SOX2 (Santa Cruz, 1:100), DACH1 (ProteinTech, 1:100), TUJ-1 (Sigma-Aldrich, 1:1,000), MAP2 (Synaptic system, 1:1,000).
Rinse the cells with PBS -/-, 3 times for 5 min.
-
Incubate the cells with a specific secondary antibody and Hoechst staining (1:2500) for nucleus visualization for at least 1 h at RT.
Note: Dilute secondary antibodies 1:300 using blocking solution. Following secondary antibodies are commonly used in our laboratory: Alexa FluorTM 488 donkey anti-rabbit IgG (H+L) (Thermo Fisher Scientific), Alexa FluorTM 647 donkey anti-mouse IgG (H+L) (Thermo Fisher Scientific), donkey anti-goat Cy 3 (Sigma-Aldrich) and donkey anti-guinea pig Cy 3 (Sigma-Aldrich).
Rinse the cells with PBS -/-, 3 times for 5 min.
Invert coverslips and mount them to a glass slide using mounting medium (IMMU-MOUNTTM or similar). After the mounting medium is dry the samples can be analyzed by confocal imaging.
-
Characterization of NPCs by quantitative real-time reverse transcription PCR (qRT-PCR)
Prepare SYBR Green PCR Master Mix according to the manufacturer’s description.
-
Pipette the PCR master mix and the cDNA samples into the 384-Well Optical Reaction Plates (Applied Biosystems).
Note: RNA was extracted from cell pellets using the RNeasy Mini Kit (50) from Qiagen according to the manufacturers protocol. cDNA was generated using the Moloney Murine Leukemia Virus (M-MLV) Reverse Transcriptase kit from Thermo Fisher Scientific according to the manufacturers protocol. For each target gene, cDNA samples and negative controls should be measured in biological triplicates.
Set the PCR cycle conditions as follows: 1 cycle 50 °C 2 min, 1 cycle 95 °C 10 min, 40 cycles 95 °C 15 s → 60 °C 30 s → 72 °C 30 s, 1 cycle 72 °C 10 min.
-
Primers used were:
For OCT4
F: GTGGAGGAAGCTGACAACAA and R: ATTCTCCAGGTTGCCTCTCA
For NANOG
F: CCTGTGATTTGTGGGCCTG and R: GACAGTCTCCGTGTGAGGCAT
For SOX2
F: GTATCAGGAGTTGTCAAGGCAGAG and R: TCCTAGTCTTAAAGAGGCAGCAAAC
For DNMT3B
F: GCTCACAGGGCCCGATACTT and R: GCAGTCCTGCAGCTCGAGTTTA
For DPP4
F: TGGTGTCAGGTGGTGTGTGG and R: CCAGGCTTGACCAGCATGAA
For VIM
F: GGAGCTGCAGGAGCTGAATG and R: GACTTGCCTTGGCCCTTGAG
For NESTIN
F: TTCCCTCAGCTTTCAGGAC and R: GAGCAAAGATCCAAGACGC
For PAX6
F: GAATTCTGCAGACCCATGC and R TCTCGTAATACCTGCCCAG
For SOX1
F: TTGGCATCTAGGTCTTGGCTCA and R: CGGGCGCACTAACTCAGCTT
For SOX2
F: GTATCAGGAGTTGTCAAGGCAGAG and R: TCCTAGTCTTAAAGAGGCAGCAAAC
For GAPDH
F: CTGGTAAAGTGGATATTGTTGCCAT and R: TGGAATCATATTGGAACATGTAAACC
Data analysis
Fluorescent images were visualized using the Zeiss Axio Imager Z1 confocal microscope in combination with AxioVision V4.6.3.0 software. Images were merged either directly during image acquisition using AxioVision V4.6.3.0 software, or afterwards using Adobe Photoshop CS6 (Adobe, California, USA).
Gene expression analysis was performed by qRT-PCR using SYBR Green PCR Master Mix and the ViiATM 7 Real-Time PCR System (Applied Biosystems, California, USA). Relative transcript levels of each gene were calculated based on the 2−ΔΔCT method. Data were normalized to the housekeeping gene GAPDH and presented as mean LOG2 ratios in relation to control iPSCs.
Recipes
Media formulation (Table 1 and Table 2):
1. All media should be prepared fresh and used within one week.
2. Do not thaw N-2 Supplement and B-27 Supplement in the water bath. Instead thaw them at RT for 2-4 h or in the fridge (4 °C) over night.
3. All media should be pre-warmed before applying to the cells. Do not keep media at 37 °C for long periods but rather pre-warm them at RT.
Acknowledgments
We acknowledge support from the Deutsche Forschungsgemeinschaft (DFG) (PR1527/5-1 to A.P.), Spark-Berlin Institute of Health (BIH Validation Funds to A.P.), National Science Center, Poland (No. 2016/22/M/NZ2/00548; No. 2017/27/B/NZ1/02401 to P.L), the United Mitochondrial Disease Foundation (UMDF to A.P.), and the German Federal Ministry of Education and Research (BMBF) (AZ.031A318 & 031L0211 to A.P.). The original research paper was: Lorenz et al.(2017).
Competing interests
The authors declare no competing financial and non-financial competing interests.
Ethics
The study was approved by the ethic committee of the Medical Faculty of Heinrich Heine University (study number 2019/681).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Beckervordersandforth R.(2017). Mitochondrial Metabolism-Mediated Regulation of Adult Neurogenesis. Brain Plast 3(1): 73-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bénit P., Schiff M., Cwerman-Thibault H., Corral-Debrinski M. and Rustin P.(2016). Drug development for mitochondrial disease: recent progress, current challenges, and future prospects. Expert Opin Orphan Drugs 4: 83-92. [Google Scholar]
- 3. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M. and Studer L.(2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27(3): 275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cunnane S. C., Trushina E., Morland C., Prigione A., Casadesus G., Andrews Z. B., Beal M. F., Bergersen L. H., Brinton R. D., de la Monte S., Eckert A., Harvey J., Jeggo R., Jhamandas J. H., Kann O., la Cour C. M., Martin W. F., Mithieux G., Moreira P. I., Murphy M. P., Nave K. A., Nuriel T., Oliet S. H. R., Saudou F., Mattson M. P., Swerdlow R. H. and Millan M. J.(2020). Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov 19(9): 609-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N. D., Tabar V. and Studer L.(2008). Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev 22(2): 152-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Inak G., Lorenz C., Lisowski P., Zink A., Mlody B. and Prigione A.(2017). Concise Review: Induced Pluripotent Stem Cell-Based Drug Discovery for Mitochondrial Disease. Stem Cells 35(7): 1655-1662. [DOI] [PubMed] [Google Scholar]
- 7. Khurana V., Tardiff D. F., Chung C. Y. and Lindquist S.(2015). Toward stem cell-based phenotypic screens for neurodegenerative diseases. Nat Rev Neurol 11(6): 339-350. [DOI] [PubMed] [Google Scholar]
- 8. Kim D. S., Lee J. S., Leem J. W., Huh Y. J., Kim J. Y., Kim H. S., Park I. H., Daley G. Q., Hwang D. Y. and Kim D. W.(2010). Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev Rep 6(2): 270-281. [DOI] [PubMed] [Google Scholar]
- 9. Lancaster M. A., Renner M., Martin C. A., Wenzel D., Bicknell L. S., Hurles M. E., Homfray T., Penninger J. M., Jackson A. P. and Knoblich J. A.(2013). Cerebral organoids model human brain development and microcephaly. Nature 501(7467): 373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W., Sun W., Zhang Y., Wei W., Ambasudhan R., Xia P., Talantova M., Lin T., Kim J., Wang X., Kim W. R., Lipton S. A., Zhang K. and Ding S.(2011). Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A 108(20): 8299-8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Aure K., Leong M., Zabiegalov O., Lyras E. M., Pfiffer V., Fauler B., Eichhorst J., Wiesner B., Huebner N., Priller J., Mielke T., Meierhofer D., Izsvak Z., Meier J. C., Bouillaud F., Adjaye J., Schuelke M., Wanker E. E., Lombes A. and Prigione A.(2017). Human iPSC-Derived Neural Progenitors Are an Effective Drug Discovery Model for Neurological mtDNA Disorders. Cell Stem Cell 20(5): 659-674 e659. [DOI] [PubMed] [Google Scholar]
- 12. Lorenz C. and Prigione A.(2017). Mitochondrial metabolism in early neural fate and its relevance for neuronal disease modeling. Curr Opin Cell Biol 49: 71-76. [DOI] [PubMed] [Google Scholar]
- 13. Paul S. M., Mytelka D. S., Dunwiddie C. T., Persinger C. C., Munos B. H., Lindborg S. R. and Schacht A. L.(2010). How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 9: 203-214. [DOI] [PubMed] [Google Scholar]
- 14. Readhead B., Hartley B. J., Eastwood B. J., Collier D. A., Evans D., Farias R., He C., Hoffman G., Sklar P., Dudley J. T., Schadt E. E., Savic R. and Brennand K. J.(2018). Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nat Commun 9(1): 4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C. S., Hoing S., Moritz S., Parga J. A., Wagner L., Bruder J. M., Wu G., Schmid B., Ropke A., Klingauf J., Schwamborn J. C., Gasser T., Scholer H. R. and Sterneckert J.(2013). Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One 8(3): e59252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S.(2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5): 861-872. [DOI] [PubMed] [Google Scholar]
- 17. Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Sudhof T. C. and Wernig M.(2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463(7284): 1035-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walter J., Bolognin S., Antony P. M. A., Nickels S. L., Poovathingal S. K., Salamanca L., Magni S., Perfeito R., Hoel F., Qing X., Jarazo J., Arias-Fuenzalida J., Ignac T., Monzel A. S., Gonzalez-Cano L., de Almeida Pereira, Skupin L., Tronstad A., Schwamborn K. J., J. C. (2019). Neural Stem Cells of Parkinson’s Disease Patients Exhibit Aberrant Mitochondrial Morphology and Functionality. Stem Cell Reports 12(5): 878-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Sterr M., Burtscher I., Chen S., Hieronimus A., Machicao F., Staiger H., Haring H. U., Lederer G., Meitinger T., Cernilogar F. M., Schotta G., Irmler M., Beckers J., Hrabe de Angelis M., Ray M., Wright C. V. E., Bakhti M. and Lickert H.(2018). Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Mol Metab 9: 57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F. H. and Hunter T.(2016). Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife 5: e13374. [DOI] [PMC free article] [PubMed] [Google Scholar]