Abstract
There has recently been a rapid progress in computational methods for determining protein targets of small molecule drugs, which will be termed as compound protein interaction (CPI). In this review, we comprehensively review topics related to computational prediction of CPI. Data for CPI has been accumulated and curated significantly both in quantity and quality. Computational methods have become powerful ever to analyze such complex the data. Thus, recent successes in the improved quality of CPI prediction are due to use of both sophisticated computational techniques and higher quality information in the databases. The goal of this article is to provide reviews of topics related to CPI, such as data, format, representation, to computational models, so that researchers can take full advantages of these resources to develop novel prediction methods. Chemical compounds and protein data from various resources were discussed in terms of data formats and encoding schemes. For the CPI methods, we grouped prediction methods into five categories from traditional machine learning techniques to state-of-the-art deep learning techniques. In closing, we discussed emerging machine learning topics to help both experimental and computational scientists leverage the current knowledge and strategies to develop more powerful and accurate CPI prediction methods.
Keywords: Compound-protein interaction, Data representation, Interpretable learning, Chemical descriptors, Protein descriptors, Machine learning, Deep learning, Pharmacophore discovery
1. Introduction
Successful drug discovery requires technological advances in various research fields to meet the standards in pharmaceutical industries [1], [2]. High-throughput screening (HTS), a widely used technology, is a large-scale experimental platform to enrich chemical compound candidates. The HTS technology has been widely used to identify lead compounds of the desired properties [3], [4], [5], [6]. Such high throughput experimental techniques generate a huge amount of Compound-Protein Interaction (CPI) data which are curated and accumulated in several databases. For example, 1.9 million binding affinity information is currently compiled in BindingDB [7]. In recent years, the drug discovery pipeline has undergone a paradigm shift by leveraging computational techniques to accelerate discovery of active hits before entering into clinical trials [8], [9].
Machine learning (ML) techniques have been extensively used in the field of computer-aided drug discovery (CADD) and they have increased the success rate of candidate drugs significantly [10], [11], [12]. Recent ML-based approaches focus on predicting affinity or interactions between small molecule drugs and protein targets, CPI, using techniques such as kernel-based, tree-based classifications, and neural network variations [13], [14], [15]. Among them, neural networks are the mainstream in cheminformatics such as Quantitative Structure–Activity Relationship (QSAR), ADMET or etc [16], [17], [18], [19].
The availability of chemical knowledge and databases accelerated the advances in computational CPI prediction models [20], [21]. Both pharmacophores in chemical compounds and ligand binding sites in amino acid (AA) sequences were efficiently modeled to explore the CPI data space [22], [23]. Recently developed deep learning technologies have significant impact on drug discovery. Zhavoronkov et al. [24] used variational autoencoder (VAE) with strong prior computed by tensor train decomposition to design drug candidates for DDR1 kinase only in 46 days. Another study by Stokes et al. [25] demonstrated that halicin can be used as a new antibiotic through a message-passing neural network (MPNN) and extensive experimental validations.
To fully utilize the power of computational methods and data, we need to understand how certain CPI prediction methods or analysis pipelines have evolved along with databases used for the prediction. An important goal of this survey paper is to outline current CPI prediction methods in the context of data preparation and model construction. To achieve generalization of our current knowledge on CPI prediction using AI methods, we grouped the computational methods into five categories: tree-based ML, network- and kernel-based ML, and three deep learning (DL) based architectures. Tree-based models (Section 4.1) learn data hierarchically in a rule-based manner, thus interpretation of decision process is natural in terms of the feature space. Network- and kernel-based methods (Section 4.2) transform input data into feature map and make prediction. Deep learning (DL) technologies such as RNN and CNN (Section 4.3) have demonstrated the prediction power in capturing local sequence/structure patterns that can be used for CPI prediction. Graph-based neural networks (Section 4.4) represent input compounds as structured graphs and assign features on atoms (nodes). These techniques can effectively capture essential CPI information in terms of graphs by embedding features of neighboring atoms into the central one when learning chemical representations. There are relatively new technologies that can be very useful for predicting CPIs (Section 4.5). Generative models such as Variational autoencoder (VAE) or generative adversarial network (GAN) are extensively used, together with reinforcement learning. These techniques are particularly useful for predicting novel CPI.
In summary, we organized major topics of CPI as data, formats, representations, databases, and machine learning models. Thus, the review starts with data and format for small-molecule ligands and target proteins in terms of data formats and encoding schemes. Then, we conducted an extensive survey on databases for CPI prediction in perspectives of both chemical compounds and proteins. CPI prediction methods are categorized in five groups based on the ML methods used for CPI prediction. In closing, we discussed issues including new computational strategies for expanding our knowledge on CPI prediction and interpretability of prediction results, in particular, attention mechanism that is widely used to complement the black-box nature of deep learning-based methods.
2. Data formats and encoding schemes
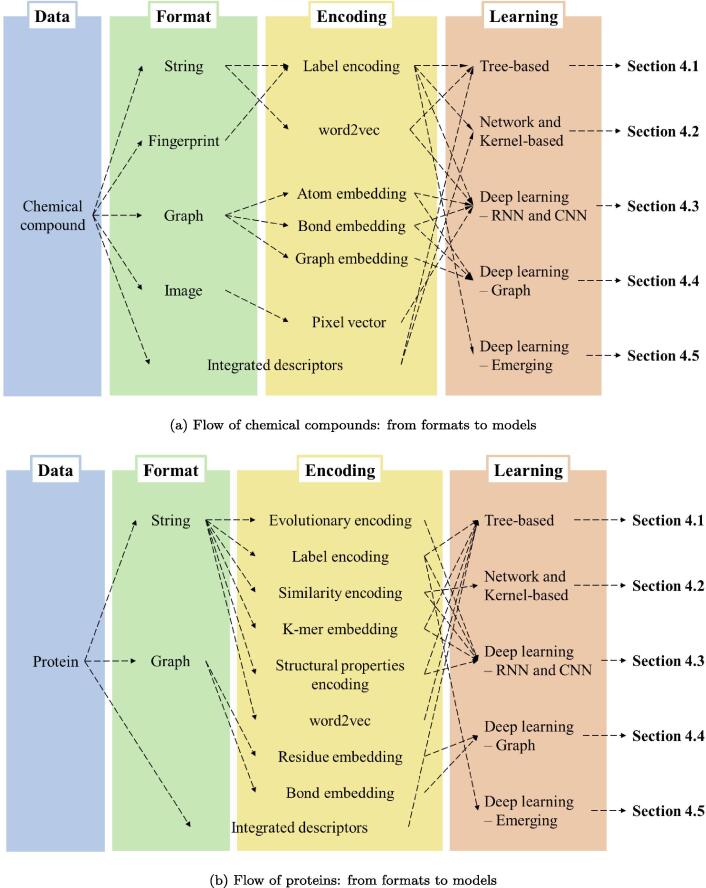
Computational methods for training CPI prediction models require data on compounds, targets, and their interaction profiles. This section summarizes data formats and encoding schemes of data as input to ML models. We then discuss databases for CPI prediction in Section 3. see Fig. 1.
Fig. 1.
Overview of how chemical compound and protein data that are processed to perform CPI prediction tasks. Data encoding depends on data type and how the data can be prepared as input to ML models. Data processed by network-based methods are not allocated here and will be discussed in detail in 4.2).
2.1. Chemical compounds (small molecules)
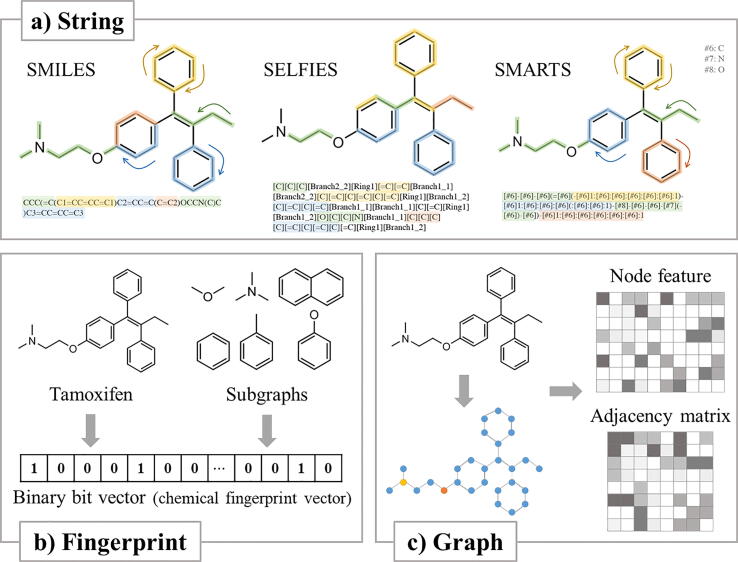
Chemical compounds can be described naturally in a human-readable format such as strings, graphs, or images. The most widely used string format is Simplified Molecular-Input Line-Entry System (SMILES) [26]. SMILES describes a chemical compound as a linear string. Starting from one atom, it visits all atoms by trimming bonds of the closed ring system. Once the order of atoms is determined, it extends the line-entry with specific rules for atoms, bonds, cycles, branches, and stereochemistry. There are several canonicalization schemes, depending on algorithms that uniquely match the SMILES representation to a compound. For example, RDKit, the most commonly used Python library for cheminformatics, implements an algorithm that considers both stereochemistry and symmetry of molecules for generating SMILES [27]. Some ML tools utilize augmentation of SMILES to compensate its non-bijective nature and to generate less biased information [28]. There are other types of string-formatted representations, such as SMARTS and SELFIES, to either highlight substructures or mirror semantic constraints better (Fig. 2) [29], [30].
Fig. 2.
Formats and encoding schemes of chemical compounds. a) String-based methods translate compounds into context-aware strings - SMILES, SMARTS, or SELFIES. SMARTS, based on SMILES, focuses on localized substructures. SELFIES produces a string that guarantees valid molecular structure. b) Chemical fingerprint encodes a compound into a binary bit vector by comparing the compound with pre-defined set of substructures. c) Graphical representation of chemical compounds transforms the input molecule into a set of matrices - adjacency and node features.
SMILES can be encoded as a mixture of one-hot and multi-hot vectors. As an example, Hirohara et al. [31] normalized the number of valence electrons (VEs), and encoded chemical structures such as chirality, aromaticity with one-hot encoding to assign the atom value. This scheme effectively encoded a SMILES into a computable format. In most cases, encoded SMILES vectors are fed into deep learning models that construct latent vectors for representing the chemical space [32]. Word2vec is another way to encode SMILES that constructs word embeddings by mapping characters to vectors of real numbers [33]. Coupled with sequential models such as RNN, word2vec can generates powerful embedding of the entire chemical sentences by treating the fixed length of characters as ‘a word’ [34].
Constitutive substructures/scaffolds or common functional groups occur frequently in chemical compounds [35] and they are used to construct chemical fingerprints that describe chemical compounds as boolean representations of their substructures. There are several ways to generate different fingerprint schemes such as ECFP, Morgan, PubChem, and MACCS. We can group these chemical fingerprint generation schemes into topology-based schemes (Morgan, ECFP, 2D pharmacophore and etc.), and SMARTS-based schemes (MACCS, PubChem and etc.). Topology-based fingerprints characterize atoms and bonds by calculating the topological distance in molecules while SMARTS-based fingerprints consider the presence of SMARTS patterns that describe bond orders and bond aromaticity. In either of the two schemes, the presence and absence of a substructure can be used to generate a boolean array for a chemical compound, which can be utilized as a search strategy for similar compounds. Since fingerprints are intuitive and informative, fingerprint-based schemes have been successfully used in cheminformatics [36]. Similarity of two compounds can be easily calculated by comparing the corresponding fingerprint vectors using the Tanimoto algorithm [37], [38], [39], [40].
Graph-based representations, such as weave or graph neural fingerprints, have recently been successful in reflecting the chemical properties [11], [41]. To use graph-based learning strategies, compounds need to be converted to graphs, typically adjacency matrices representation of graphs with atom/bond information. These matrices are then provided as input to graph convolution networks (GCNs). The main role of GCN is to generate the context of a node by considering neighboring nodes. There are two major GCN types for updating neighboring information. The spectral GCN is to consider a graph as a whole while the spatial GCN uses only neighboring nodes, thus considering local subgraphs. The spatial GCN is more popular and scalable due to its power that learns graphs by handling atoms in batches. More details on the graph encoding of chemical compounds can be found in a recent review paper [11].
2.2. Target proteins
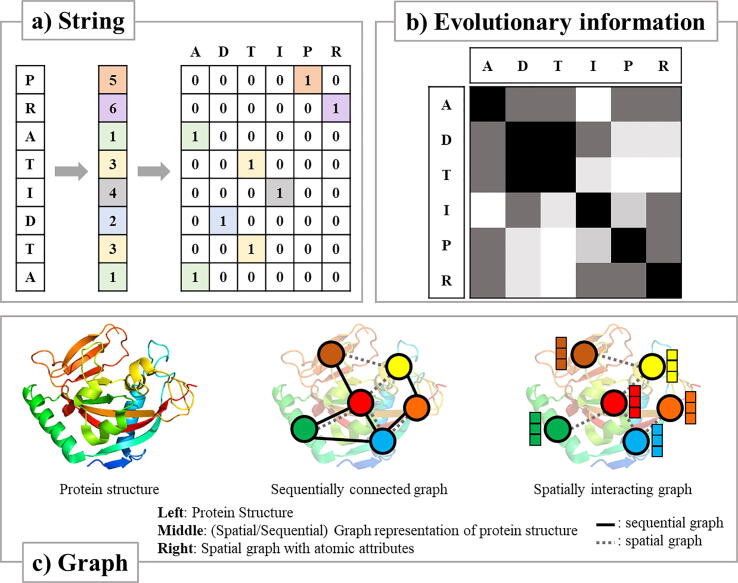
Various formats and encoding schemes are available for protein sequences and structures. Protein is basically a sequence of amino acid residues that are highly-conserved evolutionary information. Thus, a protein can be encoded in a sequential way considering its evolutionary, structural property, or inter-similarity with one-hot, word2vec, or k-mer-based methods (Fig. 3). Among them, one-hot encoding is to transform a character into a binary-bit vector [42], [43]. One-hot encoding scheme is popular since deep learning models require grid-like input with numbers. For protein structures, it is common to convert a protein structure to a chemically attributed spatial graph with nodes for residues and edges between two residues within a preset distance. Structural information of a protein can be coordinates, electrostatic properties, or surface-area at individual amino-acid level [44], [45], [46]. UniProt and Protein Data Bank (PDB) are major resources for protein sequence and structural information, respectively [47], [48]. PDB contains compound-protein interaction information including ligand-specific spatial conformation. One issue with PDB is that the number of structurally characterized proteins are much smaller than proteins with amino acid sequence determined [49]. Thus, use of protein structure information for computer-aided drug design (CADD) is limited. However, AlphaFold [50] and AlphaFold2 demonstrated that use of protein evolutionary information with protein structural information can be very powerful for predicting protein structure from protein sequence. This recent remarkable advance will have a deep impact on CADD.
Fig. 3.
Formats and corresponding encoding schemes of proteins. a) String: Represent protein as amino acid sequence. b) Evolutionary information: Encode protein considering its evolutionary information. c) Graph: Encode protein structure whether by considering sequentially connected relations (Middle), or by calculating the spatial distance between the residues (Right).
3. Databases for CPI prediction
Commonly used databases for drug discovery tasks are summarized in the following subsections as either chemistry-centric, protein-centric, or integrated databases (Table 1). Chemistry-centric databases include FDA-approved drugs in TTD, BindingDB, PharmKGB and DrugBank, and commercially available drugs in ZINC. Protein Databases contain mostly sequence-based information. CPI information is present in either of databases or as standalone databases including chemical/protein entity-based attributes.
Table 1.
List of databases used in CPI prediction. The databases are organized in three separate categories: chemistry-centric, protein-centric and integrated databases. (a) Chemistry-centric databases mostly focus on integrating the information from chemical experiments. They comprise SMILES, InChI key, or other accession data and their interacting/targeting proteins with corresponding affinities. (b) Protein databases provide sequence information in general. They rarely contain information linked with chemical compounds. (c) Other databases include integrated information in addition to compounds or proteins, such as association with genes, diseases, or phenotypes.
| Database | Coverage (Number of entities) |
ML methods to use DB |
Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compounds | Proteins | Interactions | T | F | G | S | P | D | ||||
| PubChem | 111 m | 99 k | 273 m | – | [51], [63], [64] | [44], [65], [66] | [44], [66] | [44] | [67], [66] | [68], [69] | ||
| ChEMBL | 1,961,462 | 13,382 | 16,066,124 | [53] | [70], [71] | [70] | [52] | [53], [54] | [72] | |||
| DUD-E | 22,886 | 102 | 22.8 k* | [45], [62], [71], [73] | [62] | [52], [45], [46], [73] | [46] | [55] | ||||
| DrugBank | 13,791 | 5,696 | 27,954 | [53], [74], [75], [76] | [51], [63], [64], [74], [76], [77], [78] | [44], [65], [70], [73], [79] | [34], [44], [70], [80] | [44], [46], [73] | [46], [53], [54], [77], [79] | [81], [82] | ||
| STITCH | 0.5 m | 9.6 m | 1.6b | [75], [76] | [76] | [66] | [66] | – | [66], [67] | [83], [84] | ||
| TTD | 2,251 | 3,473*** | 43,875 | [53] | – | – | – | – | [53], [54] | [85] | ||
| PharmGKB | 708 | – | – | [53] | [53], [54] | [86] | ||||||
| Matador | 801 | 2,901 | 15,843 | [74] | [74] | [73], [79] | [73] | [79] | [87] | |||
| DrugCentral | 2,529 | 2,003 | 17,390 | [53] | – | – | – | – | [53] | [88] | ||
| SuperTarget | 195,770 | 6,219 | 332,828 | [76], [89] | [76], [78] | – | [80] | – | [87], [90] | |||
| Metz | 3,858 | 172 | 258,094 | – | – | – | – | – | [91] | [92] | ||
| MUV | 93 k | 17 | – | – | – | [71] | – | [46] | [46] | [93] | ||
| ZINC | 750 m** | 2,864 (for eukaryotes) | 638,174 | – | – | [71] | – | – | – | [94] | ||
| Protein-centric databases | ||||||||||||
| Compounds | Proteins | Interactions | T | F | G | S | P | D | ||||
| UniProt | – | 20,385 | – | – | [51], [63] | [70] | [70] | – | – | [47] | ||
| Protein Data Bank | – | 170,597 | – | – | [64] | [45] | – | [45], [46] | [46] | [48], [95] | ||
| PDBbind | 11,762 | 3,566 | 17,679* | – | – | [45] | – | [45], [52], [96] | – | [97], [98] | ||
| Pfam | – | 18,259 | – | – | [51], [63], [64] | – | – | – | [67] | [99], [100], [101] | ||
| BRENDA | 46 | 8083** | 500 k | [74], [89] | [74], [78] | [65] | – | – | – | [102] | ||
| Integrated databases | ||||||||||||
| Compounds | Proteins | Interactions | T | F | G | S | P | D | ||||
| KEGG | 18,749*** | 31,224,482**** | – | [74], [75], [76], [89] | [74], [76], [77], [78] | [79] | [80] | – | [77], [79] | [103] | ||
| BindingDB | 910,479 | 8,161 | 2.1 m | – | – | [45], [61], [62], [65], [66] | [34], [61], [62], [66], [104] | [45], [61], [104] | [54], [66] | |||
| Davis | 72 | 442 | 30 k | – | [44], [105] | [44] | [44] | [91], [106] | [107] | |||
| K KIBA | 229 | 211 | 118 k | [44], [105] | [44] | [44] | [91], [106] | [108] | ||||
| IUPHAR/BPS | 10,053 | 2,943 | 48,902 | [53] | – | [79] | – | – | [53], [54], [79] | [109] | ||
positives.
213 m 3D information available.
raw file downloaded on Nov 11th, 2020.
protein-ligand complexes.
EC numbers (online).
drugs:11.3 k
human proteins:19.7 k
3.1. Chemistry-centric databases
As a chemistry-centric database, PubChem contains 2D and 3D structural information of compounds and interacting protein information from various bio/chemical experiments and the literature. It also compiles benchmark fingerprints of 881 chemical substructures that are widely used to transform chemicals into learnable forms [51]. ChEMBL, one of the most comprehensive databases in cheminformatics, contains a large amount of CPI information including potentially druggable compounds. Using the chemical compound and activity assay data from ChEMBL, Lim et al. [52] used IC value to compile a dataset for model evaluation. DrugBank provides more detailed information on drugs including annotations such as approved, experimental, and nutraceutical. Using this annotated information, Zeng et al. [53] constructed a dataset for target-driven drug repurposing on GPCR proteins. Drug-drug interaction, drug indications, and FDA-approved from DrugBank were also used by Zeng et al. [54] to construct a drug-gene-disease network which was used for identifying novel antagonists of a nuclear receptor (ROR-t). DUD-E [55] provides active-interact molecules and sets of decoy molecules that are similar in physical properties but dissimilar in topology with the active molecules. These active compounds and decoys can be used as positive and negative samples for CPI prediction [46]. In DUD-E, interaction labels and binding affinities are provided.
3.2. Protein-centric databases
UniProt is the representative protein sequence database that compiles 563,552 reviewed proteins in Swiss-Prot (version: UniProtKB 2020_05). PDB assembles a large number of 3D structural data that are obtained by X-ray crystallography or other methods. PDBbind provides a comprehensive experimentally-measured binding affinity data between protein and ligand complexes. Ballester and Mitchell [56] suggested a new machine learning-based scoring function and PDBbind benchmark was used for validation of CPI predictions [57], [58]. However, compared with the number of AA sequences, protein 3D structure data is much smaller partly because of technical difficulties for establishing crystallization. Moreover, compound-protein interactions usually take place at preferred sites on the protein surface named ‘pockets’. The utilization of protein pocket information can generate more precise CPI prediction with structural insights [59]. In a work by Torng and Altman [46] that considers protein pockets as graphs of key residues, FEATURE [60] software was used to model local protein pocket using 480 physicochemical properties into protein-encoding vectors.
3.3. Integrated databases
There are databases that provide integrated annotations with extra curation efforts. BindingDB collects detailed binding data from experiments, such as enzyme inhibition or calorimetry, and curated the literature information from PubChem and ChEMBL. Gao et al. [61] compiled the CPI information of 39,747 positive and 31,218 negative records by IC50 value from BindingDB. This customized dataset was utilized by Zheng et al. [62].
4. AI methods for CPI prediction
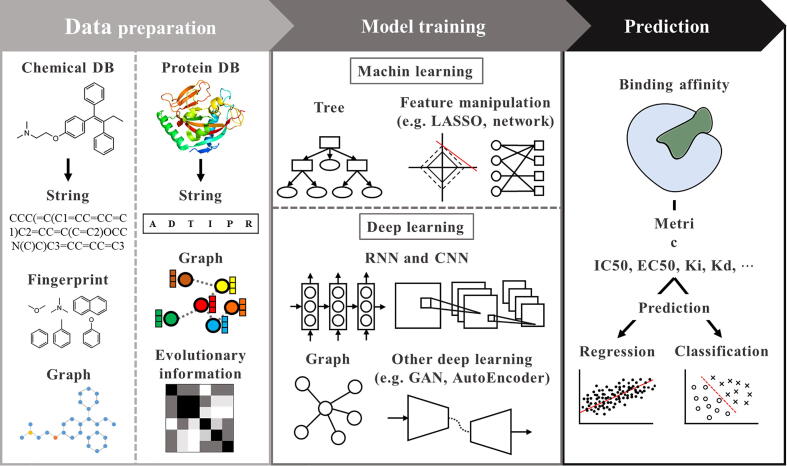
Computational models for CPI prediction have been extensively developed over the decades. As shown in Fig. 4, CPI prediction models can be grouped into five categories according to the computational techniques used. In the subsequent subsections, we discuss models in each category in detail.
Fig. 4.
Overview of the process of CPI prediction. After obtaining the original chemical or protein data from databases, various encoding techniques are used to prepare the data with model-readable vector formats (left). These encoded data are then submitted to a model or a combination of several models to learn the pattern of data. We categorize these models into two main groups (machine learning and deep learning) that contain five subgroups together (middle). After training models with the data, different metrics are chosen to evaluate the model. Note that CPI is predicted whether in a regression style by predicting the affinity value or in a classification style by predicting the interaction label (right).
4.1. Tree-based methods
Technical background. The decision tree (DT) is a flowchart-like tree structure, where each node denotes a test on an attribute value, each branch represents an outcome of the test, and tree leaves represent classes or class distributions [110]. Because a decision tree shows how decision is made clearly as ‘if-then’ rules, DT is one of the most widely used classification techniques. When the number of dimensions is higher than the number of data samples, DT is limited in predictive power. To increase the predictive power of the decision tree, a group of decision trees can be used. Popular techniques are random forest (RF) and tree-boosting algorithms. In fact, model performance can be improved by aggregating a group of prediction ‘trees’ as a ‘forest’ which is a widely used ensemble learning strategy [111]. Another important issue of using the decision tree for CPI prediction is generation of features since the use of nodes or edges in the compound graph or protein structure as features results in too many features and can also lose contextual information.
CPI data is naturally of very high dimensions since the search space is a cartesian product of two large dimensions, dimension for compounds and dimension for protein targets. Unfortunately, the number of samples, i.e., CPI examples, is relatively small. Thus, the generalization power of a decision tree is quite limited for CPI prediction. For this reason, random forests are used for CPI to avoid over-fitting to the training set [15], [53], [76]. Beyond the simple use of RF as a model predictor, Zeng et al. [53] proposed a network-based computational framework called AOPEDF to infer CPI prediction. Inspired by the work of Zhou and Feng 00[112], they constructed a heterogeneous network by uniquely integrating 15 networks covering chemical, genomic, phenotypic, network profiles among drugs, proteins, and disease. The network features are used as input to the cascade deep forest classifier to infer new drug-target interactions. The entire system was termed as an arbitrary-order proximity embedded deep forest approach (AOPEDF). In addition to RF, other boosting methods are also widely used for protein–ligand prediction [74], [75], [89]. Using the Bayesian approach as a prior, Li et al. [75] built a Bayesian Additive Regression Trees (BART) [113] based model that provides a reliable posterior mean of the results instead of simply producing a binary answer for prediction. XGBoost is another tree boosting system that follows a similar procedure as the Gradient Boosting Tree (GBT) algorithm. XGBoost uses the regularized learning objective to improve the model efficiency [114]. Mahmud et al. [74] used XGBoost on the reduced features to train the computational model for CPI prediction, whose result shows that the XGBoost classifier outperforms other three learning methods.
4.2. Network-based and Kernel-based methods
Technical background. Compounds are naturally a graph with nodes of chemical elements and edges that connect chemical elements. Protein structure is also a graph of nodes of amino acids. This natural representation of graphs requires computational methods to generate features for compounds and proteins. A widely used feature generation method is to use random walks on a graph, which results in generation of many sub-graphs as features. Once features are determined, the decision boundary between true CPIs and non-CPIs needs to be constructed by machine learning methods such as support vector machine (SVM), Lasso regression-based classifiers, and canonical correlation analysis. Since the decision boundary for CPI can be complicated, kernel trick or kernel-based methods are frequently used for handling non-linear decision boundaries.
Network-based and kernel-based ML methods have long been used for CPI prediction. A number of computational methods utilized the CPI network of known/identified edges between compounds and proteins to identify novel targets [116], [117], [118]. For CPI prediction at the network level, Lo et al. [119] developed a scoring function and used a concept of ‘chemotype’ to reduce the size of CPI space by measuring the fingerprint-based pairwise similarity of chemical compounds. The search space for this problem is called bow-pharmacological space to integrate both chemical and target spaces [75], [120]. By building the CPI space with known interactions, new interactions are predicted. A seminal work on CPI space exploration was done by Chen et al. [121] that they proposed network-based random walk with restart on two heterogeneous network (NRWRH) of drugs and target proteins. They built a network by integrating similarity information among homogeneous compound/protein entities and compound-protein interactions from four different databases (EGG BRITE, BRENDA, SuperTarget, and DrugBank databases. See Table 1) where four separate protein subcategories (enzyme, ion channel, GPCR and nuclear receptor) were dealt individually. This work provided a novel perspective on CPI that considers topological importance with nearby entities. Another interesting study measured chemical similarity using fingerprints for predicting drug responses [122].
Rather than using fingerprints as is, kernel-based methods are frequently used to determine complex non-linear decision boundaries for CPI. A representative kernel-based method is support vector machine (SVM) that it maps data points in the high-dimensional space into the feature space and then constructs decision boundaries in the feature space. SVM-based methods have been used for CPI prediction extensively [63], [74], [76]. Although SVM itself is a powerful classification method, selection of features (herein chemical/protein features) is very important for constructing decision boundaries and also for interpretability. Tabei et al. [63] used chemical fingerprints and protein domains as features. Yu et al. [76] chose description methods to extract protein features from amino acid sequences for the representation of structural and physicochemical information. ML methods like LASSO (Least Absolute Shrinkage and Selection Operator) [123] are also widely used for feature extraction. Shi et al. [15] proposed a LASSO-DNN model, where multiple LASSO models are used to integrate different combinations of feature sets of protein and compound features, reducing the effect of less significant features. Other types of feature manipulation methods such as sparse CCA [51] were used to match chemical substructures with protein domains.
4.3. Deep learning – RNN and CNN
Technical background. Recently, deep learning (DL) technologies have advanced rapidly. A class of DL methods are designed for handling sequential information and had remarkable success in language translation and speech recognition. Recurrent neural network (RNN) is a classical feed-forward neural network that uses a sequence of building blocks or states to process a sequence of input. Recent progress of RNN is due to the change in the architecture of building blocks for modeling sequential dependency and also due to the attention mechanism to model arbitrary inter-dependence among building blocks. For CPI, a chemical compound can be represented in a sequential format, e.g, SMILES, and target protein as a sequence of amino acids. Thus, sequential deep learning technologies are aggressively tried for CPI prediction. Convolutional Neural Network (CNN) is a class of feed-forward neural networks to extract relevant features from input data using a series of convolution operations and, optionally, pooling operations. CNN is originally developed for processing and analyzing image data, thus CNN usually takes data in the 2D format. When linear representations of compounds and proteins are used, it is necessary to transform the linear representation into a 2D format. This is usually done by representing sequences in one-hot or multi-hot encoding, which becomes a 2D format. CNN has the power of highlighting sub-images of an image that correspond to objects, e.g., tree, human or dog in a photo image of a public park. For CPI, CNN can identify subsequences of compounds and proteins that can interact each other for CPI.
Recurrent Neural Networks. In [61], RNN was used to project sequential input of amino acid sequences to dense vector representations by building embedding lookups in terms of both GO annotations and amino acids sequence. Considering dependencies between residues or atoms that may be close in 3D structure, Karimi et al. [66] used RNN-based seq2seq autoencoder to learn embedding vectors and subsequently used the attention mechanism to learn binding site information between a compound and a protein while training the CPI prediction model with convolution neural network (CNN). LSTM (Long Short-Term Memory), a variant of RNN, uses memory blocks instead of summation units, which results in good performance in [80]. In addition, by replacing two gates of LSTM (input and forget gates) with the updated gate, GRU (gated recurrent unit) was proposed in [129] and used to capture local and global context information in molecular or protein strings [45], [128]. Shin et al. [44] used a BERT [130] model that model the word-like embeddings and the position embeddings of molecule sequences, for CPI prediction. Transformer, another sequence-based method, is widely used in CPI prediction tasks [44], [70]. Transformer has both an encoder and a decoder, unlike BERT only with an encoder, so that training can be possible to improve prediction accuracies.
Convolutional Neural Networks. Inspired by the success in the computer vision domain, CNN was used to make structure-based binding affinity prediction in [124]. Ragoza et al. [125] used CNN to score CPI with the structural information of protein–ligand complexes. In addition, CNN is also used for feature extraction: 1D protein-sequence-encoded vector [44], [66], [73], [105], [126], [127], or molecular SMILES encoded vector [66], [105], [127], or combined vector of protein and small molecule [65]. In Lee et al. [79], local residue patterns of generalized protein classes are captured from AA sub-sequences of various lengths. To utilize evolutionary information of protein data, protein sequences are encoded with BLOSUM62 matrix [131] and further processed with CNN module in the work of Li et al. [45]. Attention mechanism [66], [73] or RNN [66] are also coupled with CNN to provide interpretation or unsupervised pre-training to achieve better performance. However, considering only 1D information is limited in reflecting 3D structures of a protein. In the work of Zheng et al. [62], 2D distance map of a protein was used to provide structural information of a protein. Given a 2D distance map as input, a CNN-based Visual Question Answering (VQA) system can be used to generate the answer to ’whether a pair of compound and protein interact with each other’ when taking molecular linear notations as a query. Recently, to reduce information loss during the process of data transformation, 2D images of compounds also used as input by Rifaioglu et al. [71] to predict interactions between compounds and proteins.
4.4. Deep learning – graph based methods
Technical background. The compound and the protein can be naturally represented as a graph with nodes of chemical elements or amino acids and edges between nodes. Handling graphs is a complicated task. Fortunately, DL methods for graph learning, specifically Graph Neural Network (GNN), have recently advanced dramatically. The basic strategy is to learn embedding vectors of a compound graph and a protein graph separately and combine two embedding vectors for CPI prediction, which is called the late integration strategy. Alternatively, embedding vectors can be learned simultaneously for compounds and proteins, which is called the early integration strategy. Among various GNN methods, Graph Convolutional Network (GCN) uses convolution operations on adjacent nodes to update the central node. Message Passing Neural Network (MPNN) learns the structure of a graph topology by propagating information of each node to neighboring nodes via the edges, which results in considering edge and node features simultaneously.
GCN was used to learn embedding vectors of molecular graphs in [61], [66]. Torng and Altman [46] used two graph autoencoder, one for molecular graph structure and the other for protein pockets, to construct embedding vectors that are combined to determine the interaction patterns. Protein–ligand complexes are considered as input to embed 3D graph representation similarly in the work of Lim et al. [52]. In addition, attention mechanisms are often coupled with GCN to provide better interpretability while achieving better CPI prediction performance [44], [52], [73]. One limitation of GCN is that GCN considers local neighboring nodes only and has difficulty in reflecting the global 3D structure and edge information. To overcome the limitation, Karlov et al. [96] used MPNN to embed drug compounds by considering both nodes and edges. In a recent study, ensembles of DL methods were used for CPI prediction Li et al. [45]. Both MPNN and GWU (Graph Wrap Unit) were used to generate chemical graph features.
4.5. Deep learning – emerging methods
In addition to DL models that learn latent representation (e.g. autoencoder), generative models such as variational autoencoder (VAE) or generative adversarial network (GAN) are extensively used. Autoencoder (AE) is an artificial neural network model that compresses input data effectively and reconstructs data as compressed reduced representation in an unsupervised manner. VAE is for learning parameters that estimate the distribution of input data. On the other hand, GAN is based on game theory that one network (generator) generates fake data in order to deceive the other (descriptor).
The features of the input data can be extended using the aforementioned models. AE uses the output of the encoder network as a required latent representation. GAN uses the discriminator network as a feature extraction network while the last classification layer of the discriminator is useless and usually be removed. In a recent study of Mao et al. [132], researchers have shown that GAN can be used to extract features of the input sequence. In the GAN model, a discriminator network can be used as a feature extractor which can be decomposed into feature extractor layers and a classification layer. Between these two compositions, the feature extraction layer can effectively learn the latent representation of the input sequence.
5. Discussions
5.1. Major issues
For the successful CPI interaction, there are two major issues. The one is data representation and the other is decision boundaries with negative samples.
Data representation. Widely used representations of compounds and proteins are human-readable formats such as SMILES and AA sequences. However, these human-readable formats often fail to carry critical information such as neighborhood in the 3D space. Thus, various data formats have been designed and tried for CPI prediction. The selection of methods for representing compounds and proteins depends on the technologies used for CPI prediction. For example, DL technologies use latent vector representation of compounds and proteins. This is because DL methods are not designed to handle symbolic information such as chemical elements and AA. Instead, DL methods generate latent vectors and combine these latent vectors to predict CPI. It is a merit of DL strategies that, since the amount of data for CPI is small compared to the joint interaction space of compounds and proteins, embedding vectors can have more generalization power for to predict CPIs beyond the training data. For compounds, Sanchez-Lengeling and Aspuru-Guzik [133] classified molecular representations into three categories: discrete, continuous, and weighted graphs. For compounds, SMILES string is a typical 1D representation of molecular graphs, fingerprints of compounds are useful for quantifying the molecular environment [69], [134], [135], and other representations such as Coulomb matrix [136] or electron density [137] can mimic electrostatic environment among nuclei. For representing proteins, AA sequences are mostly widely used. Instead of using AA sequence as is, many of current methods also consider evolutionary information of proteins by encoding AA sequence with PSSM or BLOSUM62 [45], [74]. In addition, sequence-based features with PseAAC (Pseudo Amino Acid Composition Chou [138]), or structure-based features with 3D protein information [74] can be used together with AA sequences.
Decision boundary with negative examples. Constructing decision boundary for true CPIs requires sophisticated computational methods such as DL-based latent vector representations of compounds and proteins. In addition to the computational methods, it is important to filter true negative interactions when predicting compound-protein interactions [38]. Based on the converse negative proposition with the assumption that similar compounds are likely to interact with similar target proteins and vice versa, Liu et al. [139] presented a systematic method of screening reliable negative samples. They computed chemical structural similarity and protein structural similarity from various chemogenomic resources (e.g. Chemical fingerprints, side effects, sequence similarity, GO annotations and protein domain). These similarities are integrated to calculate feature divergence for further screening negative samples from validated/predicted interactions. With different experiment settings on classical classifiers and existing predictive models, they demonstrated that screened negative samples by their framework are highly credible and helpful for identifying CPIs. Recently the human and C.elegans datasets that were screened by the work are successfully used in the prediction of CPIs achieving a significant performance improvement [79], [73].
5.2. Interpretable learning
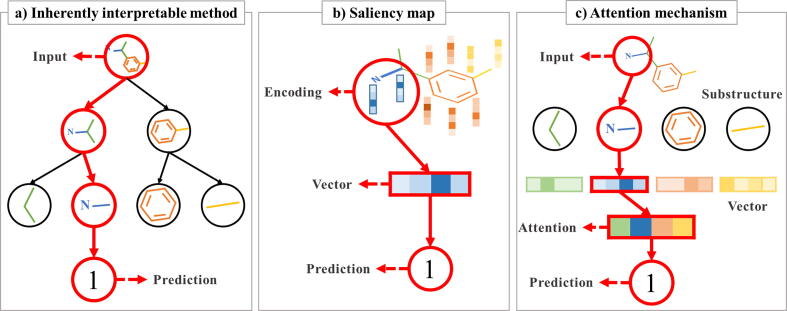
ML model perspective. Two main approaches should be considered for interpretable learning from the ML point of view: 1) design an algorithm that is inherently interpretable; 2) build an effective encoding scheme that helps human-level interpretation of data, and subsequently uses a separate set of re-representation techniques to assist the user in understanding the prediction results from the algorithm [140]. (Fig. 5) One way for interpretable learning is to use structural information. PDB database is the representative database that provide co-crystallized information of protein–ligand combinations. Leveraging PDB database was well demonstrated in a recent work by Torng and Altman [46]. Motivated by the fact that CPI is a docking process between a ligand and a small part of a target protein, the authors treated compounds as graphs and target proteins as a pocket graph, with node features retrieved from their own program using PDB database. Generation of salient features at the trained graph convolution layers provides atom/residue-level contributions on molecular docking for interpretation [141]. Existing works also demonstrated the improved interpretability of GCN convolutional filters by progressively narrowing down features extracted from GCN [142]. see Table 2, Table 3, Table 4, Table 5, Table 6.
Fig. 5.
Methods for interpretation of deep learning models divided into 3 types: inherently interpretable method, saliency map, and attention mechanism. (a) Inherently interpretable method refers to the method whose components can be further used to comprehend how the machine makes decisions. Hierarchical division of molecular structures can classify compounds in terms of the existing substructures determine given class labels (b) Saliency map is widely used to reveal the most contributed part of an input that activates the specific layer of the network. (c) Attention mechanism is mainly applied to neural networks, revealing where the model focuses on the input representation when making predictions.
Table 2.
Tree-based methods for CPI prediction.
| Tool |
Year |
Format |
Encoding |
|---|---|---|---|
| Description | |||
| Yu et al. [76] | 2012 | C: - | DRAGON |
| P: - | PROFEAT WEBSEVER | ||
| A method that integrates the chemical, genomic, and pharmacological information to predict CPI. | |||
| Zhang et al. [89] | 2017 | C: - | *- |
| P: AA properties | AAindex1 | ||
| An ensemble of REPTree classifiers by random projection to identify drug-target interactions. | |||
| Li et al. [75] | 2019 | C: - | MACCS |
| P: AA seq | AAC and * | ||
| A method that applies Bayesian Additive Regression Trees on uniform proteochemical space to predict protein–ligand interactions. | |||
| Shi et al. [15] | 2019 | C: FP2 | Pubchem (binary vector) |
| P: AA seq | PSSM matrix | ||
| A method that uses LASSO to remove redundant information from protein PsePSSM and molecular FP2 description and makes prediction with Random Forest. | |||
| Mahmud et al. [74] | 2020 | C: SMILES | MSF |
| P: AA seq | PseAAC and ** | ||
| A computational model that uses balancing techniques and applies feature eliminator to extract features for CPI prediction. | |||
| Zeng et al. [53] | 2020 | C: - | interaction and *** |
| P: - | interaction and *** | ||
| A network-based computational framework that learns low-dimensional vector representation of features and predicts CPI with cascade deep forest. | |||
C: Compound, P: Protein, : Physicochemical features, Property groups.
MSF: Molecular Substructure Fingerprint, : PSSM-Bigram and SPIDER2.
: association and similarity matrices.
Table 3.
Network- and Kernel-based machine learning methods for CPI prediction.
| Tool |
Year |
Format |
Encoding |
|---|---|---|---|
| Description | |||
| Yamanishi et al. [51] | 2011 | C: FP | Pubchem * |
| P: domains | binary coding scheme | ||
| A method that utilizes sparse CCA to extract chemical substructures and protein domains. | |||
| Cheng et al. [78] | 2012 | C: - | - |
| P: - | – | ||
| A network-based inference method to create compound-protein network and predict new CPI. | |||
| Tabei et al. [63] | 2012 | C: FP | Pubchem * |
| P: domains | binary coding scheme | ||
| A classifier-based approach to identify chemogenomic features that are involved in compound-protein interaction networks. | |||
| Yu et al. [76] | 2012 | C: - | DRAGON |
| P: - | PROFEAT WEBSEVER | ||
| A method that integrates the chemical, genomic, and pharmacological information to predict CPI. | |||
| Zu et al. [64] | 2015 | C: FP | Pubchem * |
| P: domains | binary coding scheme | ||
| A statistical model to evaluate substructure-domain interactions globally and infer interactions. | |||
| Hu et al. [77] | 2016 | C: FP | Pubchem * |
| P: AA seq | binary coding scheme | ||
| A hybrid model based on stacked sparse autoencoder and SVM. | |||
| You et al. [115] | 2019 | C: structural info | OCHEM |
| P: AA seq | AAC ** | ||
| A LASSO-DNN model for compound and protein feature extraction and CPI prediction. | |||
| Mahmud et al. [74] | 2020 | C: SMILES | MSF |
| P: AA seq | PseAAC *** | ||
| A computational model that uses balancing techniques and applies feature eliminator to extract features for CPI prediction. | |||
C: Compound, P: Protein, MSF: Molecular Substructure Fingerprint, *: binary vector.
**: DC, TC, ajacency matrix, ***: PSSM-Bigram and SPIDER2.
Table 4.
RNN and CNN methods for DTI prediction.
| Tool |
Year |
Format |
Encoding |
|---|---|---|---|
| Description | |||
| Wallach et al. [124] | 2015 | co-complex structure | * with 1Åspacing |
| The first structure-based CNN model to predict CPI. | |||
| Ragoza et al.[125] | 2017 | co-complex structure | * with 0.5Åresolution |
| A CNN based model to predict protein–ligand interaction with 3D depiction of co-complex structure. | |||
| Gao et al. [61] | 2018 | C: SMILES | chemical structure graph |
| P: AA seq, GO term | lookup embedding | ||
| An end-to-end deep neural network that embedded with two-way attention mechanism for identifying compound-protein interactions. | |||
| Öztürk et al. [105] | 2018 | C: SMILES | label encoding ** |
| P: AA seq | label encoding ** | ||
| An end-to-end CNN-based CPI prediction model that eliminates the need for feature engineering. | |||
| Feng et al. [65] | 2018 | C: SMILES | MGC, ECFP |
| P: AA seq | PSC descriptor | ||
| A feature-engineering free deep learning-based model for CPI prediction. | |||
| Karimi et al. [66] | 2019 | C: SMILES | seq2seq ** |
| P: AA seq | seq2seq (SPS) | ||
| A semi-supervised unified RNN-CNN model for jointly learning protein/compound representations and predicting affinity. | |||
| Karimi et al. [104] | 2019 | C: SMILES | chemical structure graph |
| P: AA seq | k-mers (SSPro/ACCPro) | ||
| An intrinsically explainable neural network architecture for predicting compound-protein interactions. | |||
| Lee et al. [79] | 2019 | C: SMILES | Morgan/Circular Fingerprint |
| P: AA seq | lookup embedding | ||
| A CNN-based model for detecting local residue patterns and predicting CPI. | |||
| Nguyen et al. [126] | 2019 | C: SMILES | chemical structure graph |
| P: AA seq | label encoding *** | ||
| A deep learning based network for capturing compound structural information and predict binding affinity. | |||
| Öztürk et al. [127] | 2019 | C: SMILES | label encoding * |
| P: AA seq, motifs, domains | label encoding | ||
| A deep-learning based prediction model that employs chemical and biological textual sequence information to predict binding affinity. | |||
| Shin et al. [44] | 2019 | C: SMILES | word embedding |
| P: AA seq | label encoding ** | ||
| A self-attention-based molecular transformer for CPI prediction. | |||
| Tsubaki et al. [73] | 2019 | C: SMILES | chemical structure graph |
| P: AA seq | overlapping 3-gram AA vector | ||
| A deep learning based CPI prediction model that captures interaction sites between compound and protein with neural attention mechanism. | |||
| Huang et al. [70] | 2020 | C: SMILES | one-hot encoding **** |
| P: AA seq | one-hot encoding **** | ||
| An end-to-end biologically inspired transformer based framework for CPI modeling. | |||
| Li et al. [45] | 2020 | C: SMILES | chemical structure graph |
| P: AA seq | BLOSUM62 matrix | ||
| A multi-objective neural network to predict non-covalent interaction and binding affinity. | |||
| Peng et al. [128] | 2020 | C: SMILES | word embedding |
| P: - | – | ||
| An end-to-end deep learning-based framework to learn molecular representation and predict toxicity. | |||
| Rifaioglu et al. [71] | 2020 | C: SMILES | label encoding |
| P: AA seq | Physicochemical features | ||
| An end-to-end parallel convolution neural networks to obtain 1D representations from protein sequences and compounds SMILES. | |||
| Rifaioglu et al. [71] | 2020 | C: SMILES | 2D compound image |
| P: - | P: - | ||
| A CPI prediction system that takes compound 2D image as input. | |||
| Wang et al. [80] | 2020 | C: SMILES | MSF |
| P: AA seq | PSSM matrix | ||
| A DeepLSTM-based method for representing and compressing features for compound-protein pairs interaction prediction. | |||
| Zhang et al. [34] | 2020 | C: SMILES | SMILES2Vec **** |
| P: AA seq | encoded with ProtVec | ||
| A word2vec-inspired featrue representation method for CPI prediction | |||
| Zheng et al. [62] | 2020 | C: SMILES | token embedding |
| P: AA seq | pairwise distance matrix | ||
| A Visual Question Answering (VQA)-inspired interpretable deep learning model for compound-protein interaction prediction. | |||
C: Compound, P: Protein, MGC: Molecular Graph Convolutio, MSF: Molecular Substructure Fingerprint.
*: fixed-size grid, **: fixed-size vector, SPS: SSPro/ACCPro.
***: SMILES, Max Common Substructure, ****: substructure representation.
Table 5.
Graph based methods for CPI prediction.
| Tool |
Year |
Format |
Encoding |
|---|---|---|---|
| Description | |||
| Gao et al. [61] | 2018 | C: SMILES | CSG |
| P: AA seq, GO term | lookup embedding | ||
| An end-to-end deep neural network that embedded with two-way attention mechanism for identifying compound-protein interactions. | |||
| Karimi et al. [104] | 2019 | C: SMILES | CSG |
| P: AA seq | k-mers (k-mers) | ||
| An intrinsically explainable neural network architecture for predicting compound-protein interactions. | |||
| Lim et al. [52] | 2019 | co-complex structure | ajacency matrix (graph embedding) |
| A GNN-based model that predict CPI with 3D structure-embedded graph representation of protein–ligand complex. | |||
| Shin et al. [44] | 2019 | C: SMILES | word embedding |
| P: AA seq | label encoding | ||
| A self-attention-based molecular transformer for CPI prediction. | |||
| Torng and Altman [46] | 2019 | C: SMILES | CSG |
| P: PDB file | FEATURE * | ||
| A GNN-based method to learn fixed-size representations of protein pockets and chemical structural graph synchronously and predict CPI. | |||
| Tsubaki et al. [73] | 2019 | C: SMILES | CSG |
| P: AA seq | overlapping 3-gram AA vector | ||
| A deep learning based CPI prediction model that captures interaction sites between compound and protein with neural attention mechanism. | |||
| Li et al. [45] | 2020 | C: SMILES | CSG |
| P: AA seq | BLOSUM62 matrix | ||
| A multi-objective neural network to predict non-covalent interactions and binding affinites. | |||
| Karlov et al. [96] | 2020 | co-complex structure | 3D grid representation map |
| An MPNN framework for learning protein–ligand complex features and predicting binding affinity. | |||
C: Compound, P: Protein, CSG: Chemical Structure Graph, SPS: SSPro/ACCPro.
: graph of key residues.
Table 6.
Emerging methods for CPI prediction.
| Tool |
Year |
Format |
Encoding |
|---|---|---|---|
| Description | |||
| Hu et al. [77] | 2016 | C: FP | PubChem FP |
| P: AA seq |
binary coding scheme |
||
| A hybrid model based on stacked sparse autoencoder and SVM. | |||
| Tian et al. [67] | 2016 | C: FP | PubChem FP |
| P: domains | binary coding scheme | ||
| A DNN model to extract features from chemical subsrtucture and protein domain and predict CPI. | |||
| Karimi et al. [66] | 2019 | C: SMILES | seq2seq * |
| P: AA seq | seq2seq (SPS) | ||
| A semi-supervised unified RNN-CNN model for jointly learning protein/compound representations and predicting affinities. | |||
| Lee et al. [79] | 2019 | C: SMILES | Morgan/Circular Fingerprint |
| P: AA seq | lookup embedding | ||
| A CNN-based model for detecting local residue patterns and predicting CPI. | |||
| Zhao et al. [106] | 2019 | C: SMILES | text embedding |
| P: AA seq | text embedding | ||
| A semi-supervised GAN-based GANs to learn representations from the raw sequence data of proteins and compounds and predict affinity. | |||
| Agyemang et al. [91] | 2020 | C: SMILES | various descriptor schemes |
| P: AA seq | various descriptor schemes | ||
| A multi-view self-attention-based architecture for learning the representation of compounds and targets from different unimodal descriptor schemes. | |||
| Zeng et al. [53] | 2020 | C: - | interaction and ** |
| P: - | interaction and ** | ||
| A network-based computational framework that learns low-dimensional vector representation of features and predict CPI with cascade deep forest. | |||
| Zeng et al. [54] | 2020 | C: - | probabilistic co-occurrence matrix |
| P: - | probabilistic co-occurrence matrix | ||
| A network-based deep learning methodology for CPI prediction that embeds various types of chemical, genomic, phenotypic, and cellular networks. | |||
C: Compound, P: Protein, : fixed-size vector, SPS: SSPro/ACCPro.
: association and similarity matrics.
Attention mechanism. DL methods are often criticized for making black-box decisions in that interpretation on how the final decision was made is difficult. Attention mechanism [143] is suggested as the most promising way to address this issue. Attention mechanism basically tries to capture instance-level importance to the final classification result by highlighting weights of features that are most relevant to prediction decisions. It has been widely used for image processing or speech recognition [144], [145]. Attention is extensively used for CPI prediction as summarized in Table 7. ML models with attention mechanism can capture atom-level contributions for CPI prediction. For example, Gao et al. [61] used attention on protein and compound latent vectors in LSTM and GCN layers, respectively. Their method enabled visual investigation on the contribution of atoms related to target proteins which can characterize pharmacophores. Karimi et al. [66] used attention mechanism for training model, identifying ligand binding sites, and also predicting corresponding protein segments. Shin et al. [44] proposed a molecular transformer that models SMILES strings into better representation vectors with a self-attention mechanism. To capture interaction sites between a subgraph of compound and a subsequence of protein, Tsubaki et al. [73] used the neural attention mechanism on GNN and CNN output, to measure the molecule-protein pair interaction strength represented by attention weight for CPI prediction. In addition, Agyemang et al. [91] used the multi-head self-attention mechanism to generate an information-rich representation of compounds and targets by combining various unimodal representations.
Table 7.
Applications of the attention mechanism in CPI methods.
| a) molecular string, protein string | |
|---|---|
| Study | Description |
| Karimi et al. [66] | Included different attention mechanisms in the unified RNN-CNN models to quantify the contribution of compound and protein. |
| Shin et al. [44] | Proposed a Molecule Transformer that models molecular SMILES strings into better representation vectors with self-attention mechanism. |
| Tsubaki et al. [73] | Used a neural attention mechanism to weight for hidden vectors of subsequences in protein considering molecular vector. |
| b) molecular graph, protein string | |
| Study | Description |
| Gao et al. [61] | Used two-way attention mechanism to estimate how CPI pair interacts. |
| Agyemang et al. [91] | Used multi-head self-attention mechanism to learn most significant segments (segment refers to an atom in molecule or a residue in target) that may be vital to protein–ligand recognition. |
| c) molecular graph, protein graph | |
| Study | Description |
| Lim et al. [52] | Devised distance-aware graph attention mechanism to find the significant nodes and differentiate the contribution of each interaction to binding affinity. |
5.3. Emerging technologies
Data description. Most CPI methods provide interpretation on either chemical or protein space. Interaction fingerprint (IFP) is a method to represent and analyze 3D protein–ligand complex that it encodes the presence or absence of specific interactions of binding sites with one-dimensional vector. Deng et al. [146] pioneered the use of IFPs to identify and cluster docking poses with similar binding modes, revealing distinct binding interactions and demonstrating that IFP is useful for visualizing and analyzing CPI. Inspired by this work, Chupakhin et al. [147] devised a novel type of fixed size fingerprint called SILIRID (Simple Ligand-Receptor Interaction Descriptor). SILIRID is calculated from IFPs by summing up bits corresponding to identical AAs. It consists of 168 integer values that describe the complex of ligand-receptor (compound-protein) by considering the set of eight types of interaction for a pair of AA and an atom. In addition, Nguyen et al. [148] reviewed in detail how biomolecular data of high complexity and dimensionality are converted to features using mathematical methods.
Generative model with reinforcement learning. As discussed in Section 4.5, we can use the hidden representation of data to generate new compounds for specific targets. Zhavoronkov et al. [24] developed an innovative software framework to generate compounds for DDR1 kinase inhibitor. They used several strategies for exploring CPI space. First, they used VAE to model compound space for DDR1 kinase inhibitor. Generation of compounds is guided with a strong prior for VAE that was learned from the Zinc Clean Leads collection [94] by tensor train decomposition. In this work, the exploration of compound space by VAE is confined by limiting the target gene space to DDR1 kinase. To guide search for commercially valid compounds, they used a strong prior from the Zinc Clean Leads collection. Second, they used reinforcement learning (RL) to explore the target gene space of kinase inhibitors by evaluating compounds generated by VAE using three self-organizing maps (SOM) as a reward function. They developed and used a search framework, called GENTRY, to discover potent inhibitors of discoidin domain receptor 1 (DDR1), a kinase target implicated in fibrosis and other diseases. This entire discovery process was done only in 21 days. This work is an outstanding example of exploring the compound space and the target gene space in terms of CPI interaction. In other recent studies, VAE and RL were used independently or in combination to explore the data space to design a compound with desired properties [149], [150], [151], [152].
Challenges and issues. There is a challenge named ‘D3R Grand Challenge’, a worldwide competition to test state-of-the-art methods for compound design, which has been organized by the Drug Design Data Resource since 2015 [153]. In each year, a number of co-crystal structures of protein–ligand and affinity data were provided to estimate pose, affinity, and free energy of ligands. Nguyen et al. [154] developed the winning model to predict the free energy of Cathepsin S (set 1) in D3R Grand Challenges 3. The latest D3R challenge was held in December 2018 where Nguyen et al. [155] shows the top-placed performances in estimating the pose of BACE ligands by GAN- and CNN-based deep learning model. For protein structure prediction, CASP13 is one of the well-known competition series in the protein structure prediction technology field. Last year, AlphaFold2, an advanced version of AlphaFold [50] by Google’s Deepmind demonstrated that AI techniques can infer structures of proteins from AA sequences with high accuracy.
Furthermore, the selection of suitable evaluating metrics is an important issue. Performance of the protein–ligand scoring metrics was extensively compared in terms of scoring power, ranking power, docking power, and screening power in Comparative Assessment of Scoring Functions (CASF) [58], [98], [156].
CRediT authorship contribution statement
Sangsoo Lim: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Yijingxiu Lu: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Chang Yun Cho: Data curation. Inyoung Sung: Visualization, Investigation. Jungwoo Kim: Investigation. Youngkuk Kim: Investigation. Sungjoon Park: Investigation. Sun Kim: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Collaborative Genome Program for Fostering New Post-Genome Industry of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2014M3C9A3063541); Bio & Medical Technology Development Program of the NRF (NRF-2019M3E5D4065965); and by the Ministry of Food and Drug Safety (DY0002258224), Republic of Korea.
References
- 1.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discovery. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Mayorga K., Madariaga-Mazon A., Medina-Franco J.L., Maggiora G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin Drug Discov. 2020;15(3):293–306. doi: 10.1080/17460441.2020.1696307. [DOI] [PubMed] [Google Scholar]
- 3.Bleicher K.H., Böhm H.-J., Müller K., Alanine A.I. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2(5):369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 4.Brideau C., Gunter B., Pikounis B., Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screening. 2003;8(6):634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 5.Mayr L.M., Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9(5):580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Schmidtke P., Barril X. Understanding and predicting druggability. A high-throughput method for detection of drug binding sites. J Medicinal Chem. 2010;53(15):5858–5867. doi: 10.1021/jm100574m. [DOI] [PubMed] [Google Scholar]
- 7.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucl Acids Res. 2007;35(supp_1):D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B., Butte A. Leveraging big data to transform target selection and drug discovery. Clin Pharmacol Therapeut. 2016;99(3):285–297. doi: 10.1002/cpt.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews H., Hanison J., Nirmalan N. ”Omics”-informed drug and biomarker discovery: opportunities, challenges and future perspectives. Proteomes. 2016;4(3):28. doi: 10.3390/proteomes4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo Y.-C., Rensi S.E., Torng W., Altman R.B. Machine learning in chemoinformatics and drug discovery. Drug Discovery Today. 2018;23(8):1538–1546. doi: 10.1016/j.drudis.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., Li B., Madabhushi A., Shah P., Spitzer M. Applications of machine learning in drug discovery and development. Nat Rev Drug Discovery. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Tan J., Han D., Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discovery Today. 2017;22(11):1680–1685. doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Costello J.C., Heiser L.M., Georgii E., Gönen M., Menden M.P., Wang N.J., Bansal M., Hintsanen P., Khan S.A., Mpindi J.-P. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32(12):1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapins M., Arvidsson S., Lampa S., Berg A., Schaal W., Alvarsson J., Spjuth O. A confidence predictor for logD using conformal regression and a support-vector machine. J Cheminformatics. 2018;10(1):17. doi: 10.1186/s13321-018-0271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H., Liu S., Chen J., Li X., Ma Q., Yu B. Predicting drug-target interactions using Lasso with random forest based on evolutionary information and chemical structure. Genomics. 2019;111(6):1839–1852. doi: 10.1016/j.ygeno.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Ghasemi F., Mehridehnavi A., Perez-Garrido A., Perez-Sanchez H. Neural network and deep-learning algorithms used in QSAR studies: merits and drawbacks. Drug Discov Today. 2018;23(10):1784–1790. doi: 10.1016/j.drudis.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg EN, Joshi E, Pande VS, Cheng AC. Improvement in ADMET Prediction with Multitask Deep Featurization, J Medicinal Chem. [DOI] [PubMed]
- 18.Zakharov A.V., Zhao T., Nguyen D.-T., Peryea T., Sheils T., Yasgar A., Huang R., Southall N., Simeonov A. Novel consensus architecture to improve performance of large-scale multitask deep learning QSAR models. J Chem Inf Model. 2019;59(11):4613–4624. doi: 10.1021/acs.jcim.9b00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira L.L., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discovery Today. 2019;24(5):1157–1165. doi: 10.1016/j.drudis.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Yan C.C., Zhang X., Zhang X., Dai F., Yin J., Zhang Y. Drug–target interaction prediction: databases, web servers and computational models. Briefings Bioinform. 2016;17(4):696–712. doi: 10.1093/bib/bbv066. [DOI] [PubMed] [Google Scholar]
- 21.Bagherian M, Sabeti E, Wang K, Sartor MA, Nikolovska-Coleska Z, Najarian K. Machine learning approaches and databases for prediction of drug–target interaction: a survey paper, Briefings in bioinformatics. [DOI] [PMC free article] [PubMed]
- 22.Wang X., Pan C., Gong J., Liu X., Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chem Inform Modeling. 2016;56(6):1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Liu J, Luo F, Deng Z, Hu Q-N. Predicting target-ligand interactions using protein ligand-binding site and ligand substructures. In: BMC systems biology, vol. 9, Springer, S2; 2015. [DOI] [PMC free article] [PubMed]
- 24.Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., Terentiev V.A., Polykovskiy D.A., Kuznetsov M.D., Asadulaev A. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol. 2019;37(9):1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 25.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., French S., Carfrae L.A., Bloom-Ackerman Z. A deep learning approach to antibiotic discovery. Cell. 2020;180(4):688–702. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inform Comput Sci. 1988;28(1):31–36. [Google Scholar]
- 27.Schneider N., Sayle R.A., Landrum G.A. Get Your Atoms in Order – An Open-Source Implementation of a Novel and Robust Molecular Canonicalization Algorithm. J Chem Inform Modeling. 2015;55(10):2111–2120. doi: 10.1021/acs.jcim.5b00543. [DOI] [PubMed] [Google Scholar]
- 28.Bjerrum EJ. SMILES enumeration as data augmentation for neural network modeling of molecules, arXiv preprint arXiv:1703.07076.
- 29.SMARTS – A Language for Describing Molecular Patterns, URL: https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, [Accessed: 2020-11-26]; 2007
- 30.Krenn M, Häse F, Nigam A, Friederich P, Aspuru-Guzik A. SELFIES: a robust representation of semantically constrained graphs with an example application in chemistry, arXiv preprint arXiv:1905.13741.
- 31.Hirohara M., Saito Y., Koda Y., Sato K., Sakakibara Y. Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinform. 2018;19(19):526. doi: 10.1186/s12859-018-2523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Méndez-Lucio O., Baillif B., Clevert D.-A., Rouquié D., Wichard J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-019-13807-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space, arXiv preprint arXiv:1301.3781.
- 34.Zhang Y.-F., Wang X., Kaushik A.C., Chu Y., Shan X., Zhao M.-Z., Xu Q., Wei D.-Q. SPVec: a Word2vec-inspired feature representation method for drug-target interaction prediction. Front Chem. 2020;7:895. doi: 10.3389/fchem.2019.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reymond J.-L., Van Deursen R., Blum L.C., Ruddigkeit L. Chemical space as a source for new drugs. MedChemComm. 2010;1(1):30–38. [Google Scholar]
- 36.Faulon J.-L., Misra M., Martin S., Sale K., Sapra R. Genome scale enzyme–metabolite and drug–target interaction predictions using the signature molecular descriptor. Bioinformatics. 2008;24(2):225–233. doi: 10.1093/bioinformatics/btm580. [DOI] [PubMed] [Google Scholar]
- 37.Steffen A., Kogej T., Tyrchan C., Engkvist O. Comparison of molecular fingerprint methods on the basis of biological profile data. J Chem Inform Modeling. 2009;49(2):338–347. doi: 10.1021/ci800326z. [DOI] [PubMed] [Google Scholar]
- 38.Ding H., Takigawa I., Mamitsuka H., Zhu S. Similarity-based machine learning methods for predicting drug–target interactions: a brief review. Briefings Bioinform. 2014;15(5):734–747. doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 39.Bajusz D., Rácz A., Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminformatics. 2015;7(1):20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Boyle N.M., Sayle R.A. Comparing structural fingerprints using a literature-based similarity benchmark. J Cheminformatics. 2016;8(1):1–14. doi: 10.1186/s13321-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z., Ramsundar B., Feinberg E.N., Gomes J., Geniesse C., Pappu A.S., Leswing K., Pande V. MoleculeNet: a benchmark for molecular machine learning. Chem Sci. 2018;9(2):513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin K., May A.C., Taylor W.R. Amino acid encoding schemes from protein structure alignments: Multi-dimensional vectors to describe residue types. J Theoret Biol. 2002;216(3):361–365. doi: 10.1006/jtbi.2001.2512. [DOI] [PubMed] [Google Scholar]
- 43.ElAbd H., Bromberg Y., Hoarfrost A., Lenz T., Franke A., Wendorff M. Amino acid encoding for deep learning applications. BMC Bioinform. 2020;21(1):1–14. doi: 10.1186/s12859-020-03546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin B, Park S, Kang K, Ho JC. Self-attention based molecule representation for predicting drug-target interaction, arXiv preprint arXiv:1908.06760.
- 45.Li S., Wan F., Shu H., Jiang T., Zhao D., Zeng J. MONN: a multi-objective neural network for predicting compound-protein interactions and affinities. Cell Systems. 2020;10(4):308–322. [Google Scholar]
- 46.Torng W., Altman R.B. Graph convolutional neural networks for predicting drug-target interactions. J Chem Inf Model. 2019;59(10):4131–4149. doi: 10.1021/acs.jcim.9b00628. [DOI] [PubMed] [Google Scholar]
- 47.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucl Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucl Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin X., Yang J., Xiao F., Yang Y., Shen H.-B. MemBrain: an easy-to-use online webserver for transmembrane protein structure prediction. Nano-micro Lett. 2018;10(1):2. doi: 10.1007/s40820-017-0156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.AlQuraishi M. AlphaFold at CASP13. Bioinformatics. 2019;35(22):4862–4865. doi: 10.1093/bioinformatics/btz422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamanishi Y., Pauwels E., Saigo H., Stoven V. Extracting sets of chemical substructures and protein domains governing drug-target interactions. J Chem Inform Modeling. 2011;51(5):1183–1194. doi: 10.1021/ci100476q. [DOI] [PubMed] [Google Scholar]
- 52.Lim J., Ryu S., Park K., Choe Y.J., Ham J., Kim W.Y. Predicting drug–target interaction using a novel graph neural network with 3D structure-embedded graph representation. J Chem Inform Modeling. 2019;59(9):3981–3988. doi: 10.1021/acs.jcim.9b00387. [DOI] [PubMed] [Google Scholar]
- 53.Zeng X., Zhu S., Hou Y., Zhang P., Li L., Li J., Huang L.F., Lewis S.J., Nussinov R., Cheng F. Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics. 2020;36(9):2805–2812. doi: 10.1093/bioinformatics/btaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y., Fang J., Huang Y., Guo H., Li L. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci. 2020;11(7):1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Medicinal Chem. 2012;55(14):6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballester P.J., Mitchell J.B. A machine learning approach to predicting protein–ligand binding affinity with applications to molecular docking. Bioinformatics. 2010;26(9):1169–1175. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ain Q.U., Aleksandrova A., Roessler F.D., Ballester P.J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2015;5(6):405–424. doi: 10.1002/wcms.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng T., Li X., Li Y., Liu Z., Wang R. Comparative assessment of scoring functions on a diverse test set. J Chem Inform Modeling. 2009;49(4):1079–1093. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 59.Gao M., Skolnick J. A comprehensive survey of small-molecule binding pockets in proteins. PLoS Comput Biol. 2013;9(10) doi: 10.1371/journal.pcbi.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagley S.C., Altman R.B. Characterizing the microenvironment surrounding protein sites. Protein Sci. 1995;4(4):622–635. doi: 10.1002/pro.5560040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao K.Y., Fokoue A., Luo H., Iyengar A., Dey S., Zhang P. Interpretable Drug Target Prediction Using Deep Neural Representation. IJCAI. 2018;2018:3371–3377. [Google Scholar]
- 62.Zheng S., Li Y., Chen S., Xu J., Yang Y. Predicting drug–protein interaction using quasi-visual question answering system. Nat Mach Intell. 2020;2(2):134–140. [Google Scholar]
- 63.Tabei Y., Pauwels E., Stoven V., Takemoto K., Yamanishi Y. Identification of chemogenomic features from drug–target interaction networks using interpretable classifiers. Bioinformatics. 2012;28(18):i487–i494. doi: 10.1093/bioinformatics/bts412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zu S., Chen T., Li S. Global optimization-based inference of chemogenomic features from drug–target interactions. Bioinformatics. 2015;31(15):2523–2529. doi: 10.1093/bioinformatics/btv181. [DOI] [PubMed] [Google Scholar]
- 65.Feng Q, Dueva E, Cherkasov A, Ester M. Padme: A deep learning-based framework for drug-target interaction prediction, arXiv preprint arXiv:1807.09741.
- 66.Karimi M., Wu D., Wang Z., Shen Y. DeepAffinity: interpretable deep learning of compound–protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35(18):3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian K., Shao M., Wang Y., Guan J., Zhou S. Boosting compound-protein interaction prediction by deep learning. Methods. 2016;110:64–72. doi: 10.1016/j.ymeth.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 68.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. PubChem 2019 update: improved access to chemical data. Nucl Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. PubChem: integrated platform of small molecules and biological activities. Annual reports in computational chemistry Elsevier. 2008;vol. 4:217–241. [Google Scholar]
- 70.Huang K, Xiao C, Glass L, Sun J. MolTrans: Molecular Interaction Transformer for Drug Target Interaction Prediction, arXiv preprint arXiv:2004.11424. [DOI] [PMC free article] [PubMed]
- 71.Rifaioglu A.S., Nalbat E., Atalay V., Martin M.J., Cetin-Atalay R., Doğan T. DEEPScreen: high performance drug–target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem Sci. 2020;11(9):2531–2557. doi: 10.1039/c9sc03414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrián-Uhalte E. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsubaki M., Tomii K., Sese J. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35(2):309–318. doi: 10.1093/bioinformatics/bty535. [DOI] [PubMed] [Google Scholar]
- 74.Mahmud S.H., Chen W., Meng H., Jahan H., Liu Y., Hasan S.M. Prediction of drug-target interaction based on protein features using undersampling and feature selection techniques with boosting. Anal Biochem. 2020;589 doi: 10.1016/j.ab.2019.113507. [DOI] [PubMed] [Google Scholar]
- 75.Li L., Koh C.C., Reker D., Brown J., Wang H., Lee N.K., Liow H.-H., Dai H., Fan H.-M., Chen L. Predicting protein-ligand interactions based on bow-pharmacological space and Bayesian additive regression trees. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-43125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu H., Chen J., Xu X., Li Y., Zhao H., Fang Y., Li X., Zhou W., Wang W., Wang Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PloS One. 2012;7(5) doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu P.-W., Chan K.C., You Z.-H. 2016 International Joint Conference on Neural Networks (IJCNN) IEEE. 2016. Large-scale prediction of drug-target interactions from deep representations; pp. 1236–1243. [Google Scholar]
- 78.Cheng F., Liu C., Jiang J., Lu W., Li W., Liu G., Zhou W., Huang J., Tang Y. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8(5) doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee I., Keum J., Nam H. DeepConv-DTI: Prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput Biol. 2019;15(6) doi: 10.1371/journal.pcbi.1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y.-B., You Z.-H., Yang S., Yi H.-C., Chen Z.-H., Zheng K. A deep learning-based method for drug-target interaction prediction based on long short-term memory neural network. BMC Med Inform Decis Mak. 2020;20(2):1–9. doi: 10.1186/s12911-020-1052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets, Nucl Acids Res 36(suppl_1) (2008) D901–D906. [DOI] [PMC free article] [PubMed]
- 82.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhn M., Szklarczyk D., Franceschini A., Von Mering C., Jensen L.J., Bork P. STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res. 2012;40(D1):D876–D880. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whirl-Carrillo M., McDonagh E.M., Hebert J., Gong L., Sangkuhl K., Thorn C., Altman R.B., Klein T.E. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Therapeut. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Günther S., Kuhn M., Dunkel M., Campillos M., Senger C., Petsalaki E., Ahmed J., Urdiales E.G., Gewiess A., Jensen L.J. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2007;36(suppl_1):D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ursu O., Holmes J., Bologa C.G., Yang J.J., Mathias S.L., Stathias V., Nguyen D.-T., Schürer S., Oprea T. DrugCentral 2018: an update. Nucleic Acids Res. 2019;47(D1):D963–D970. doi: 10.1093/nar/gky963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang J., Zhu M., Chen P., Wang B. Drugrpe: Random projection ensemble approach to drug-target interaction prediction. Neurocomputing. 2017;228:256–262. [Google Scholar]
- 90.Hecker N., Ahmed J., von Eichborn J., Dunkel M., Macha K., Eckert A., Gilson M.K., Bourne P.E., Preissner R. SuperTarget goes quantitative: update on drug–target interactions. Nucleic Acids Res. 2012;40(D1):D1113–D1117. doi: 10.1093/nar/gkr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agyemang B, Wu W-P, Kpiebaareh MY, Lei Z, Nanor E, Chen L. Multi-View Self-Attention for Interpretable Drug-Target Interaction Prediction, arXiv preprint arXiv:2005.00397. [DOI] [PubMed]
- 92.Metz J.T., Johnson E.F., Soni N.B., Merta P.J., Kifle L., Hajduk P.J. Navigating the kinome. Nat Chem Biol. 2011;7(4):200–202. doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 93.Rohrer S.G., Baumann K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J Chem Inform Modeling. 2009;49(2):169–184. doi: 10.1021/ci8002649. [DOI] [PubMed] [Google Scholar]
- 94.Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J Chem Inform Modeling. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karlov D.S., Sosnin S., Fedorov M.V., Popov P. graphDelta: MPNN Scoring Function for the Affinity Prediction of Protein-Ligand Complexes. ACS Omega. 2020;5(10):5150–5159. doi: 10.1021/acsomega.9b04162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang R., Fang X., Lu Y., Yang C.-Y., Wang S. The PDBbind database: methodologies and updates. J Medicinal Chem. 2005;48(12):4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 98.Su M., Yang Q., Du Y., Feng G., Liu Z., Li Y., Wang R. Comparative assessment of scoring functions: the CASF-2016 update. J Chemical Inform Modeling. 2018;59(2):895–913. doi: 10.1021/acs.jcim.8b00545. [DOI] [PubMed] [Google Scholar]
- 99.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2004;32(suppl_1):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C., Huhn G. BRENDA the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32(suppl_1):D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(suppl 1):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karimi M, Wu D, Wang Z, Shen Y. Explainable Deep Relational Networks for Predicting Compound-Protein Affinities and Contacts, arXiv preprint arXiv:1912.12553. [DOI] [PMC free article] [PubMed]
- 105.Öztürk H., Özgür A., Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34(17):i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao L, Wang J, Pang L, Liu Y, Zhang J. GANsDTA: Predicting Drug-Target Binding Affinity Using GANs, Frontiers in Genetics 10. [DOI] [PMC free article] [PubMed]
- 107.Davis M.I., Hunt J.P., Herrgard S., Ciceri P., Wodicka L.M., Pallares G., Hocker M., Treiber D.K., Zarrinkar P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 108.Tang J., Szwajda A., Shakyawar S., Xu T., Hintsanen P., Wennerberg K., Aittokallio T. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J Chem Inf Model. 2014;54(3):735–743. doi: 10.1021/ci400709d. [DOI] [PubMed] [Google Scholar]
- 109.Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Southan C., Sharman J.L., Campo B., Cavanagh D.R., Alexander S.P., Davenport A.P. The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 2020;48(D1):D1006–D1021. doi: 10.1093/nar/gkz951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han J, Kamber M, Pei J. Data mining concepts and techniques third edition, The Morgan Kaufmann Series in Data Management Systems 5(4) (2011) 83–124.
- 111.Svetnik V., Liaw A., Tong C., Culberson J.C., Sheridan R.P., Feuston B.P. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inform Comput Sci. 2003;43(6):1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 112.Zhou Z-H, Feng J. Deep forest, arXiv preprint arXiv:1702.08835.
- 113.Chipman H.A., George E.I., McCulloch R.E. BART: Bayesian additive regression trees. Ann Appl Stat. 2010;4(1):266–298. [Google Scholar]
- 114.Chen T., Guestrin C. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016. Xgboost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 115.You J., McLeod R.D., Hu P. Predicting drug-target interaction network using deep learning model. Comput Biol Chem. 2019;80:90–101. doi: 10.1016/j.compbiolchem.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 116.Bleakley K., Yamanishi Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics. 2009;25(18):2397–2403. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Buza K., Peška L., Koller J. Modified linear regression predicts drug-target interactions accurately. Plos One. 2020;15(4) doi: 10.1371/journal.pone.0230726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wan F., Hong L., Xiao A., Jiang T., Zeng J. NeoDTI: neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics. 2019;35(1):104–111. doi: 10.1093/bioinformatics/bty543. [DOI] [PubMed] [Google Scholar]
- 119.Lo Y.-C., Senese S., Li C.-M., Hu Q., Huang Y., Damoiseaux R., Torres J.Z. Large-scale chemical similarity networks for target profiling of compounds identified in cell-based chemical screens. PLoS Comput Biol. 2015;11(3) doi: 10.1371/journal.pcbi.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamanishi Y., Araki M., Gutteridge A., Honda W., Kanehisa M. Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24(13):i232–i240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen X., Liu M.-X., Yan G.-Y. Drug–target interaction prediction by random walk on the heterogeneous network. Mol BioSyst. 2012;8(7):1970–1978. doi: 10.1039/c2mb00002d. [DOI] [PubMed] [Google Scholar]
- 122.Jeon M., Park D., Lee J., Jeon H., Ko M., Kim S., Choi Y., Tan A.-C., Kang J. ReSimNet: drug response similarity prediction using Siamese neural networks. Bioinformatics. 2019;35(24):5249–5256. doi: 10.1093/bioinformatics/btz411. [DOI] [PubMed] [Google Scholar]
- 123.Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Stat Soc: Ser B (Methodol) 1996;58(1):267–288. [Google Scholar]
- 124.Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery, arXiv preprint arXiv:1510.02855.
- 125.Ragoza M., Hochuli J., Idrobo E., Sunseri J., Koes D.R. Protein–ligand scoring with convolutional neural networks. J Chem Inform Modeling. 2017;57(4):942–957. doi: 10.1021/acs.jcim.6b00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nguyen T., Le H., Venkatesh S. GraphDTA: prediction of drug–target binding affinity using graph convolutional networks. BioRxiv. 2019 [Google Scholar]
- 127.Öztürk H, Ozkirimli E, Özgür A. WideDTA: prediction of drug-target binding affinity, arXiv preprint arXiv:1902.04166.
- 128.Peng Y, Zhang Z, Jiang Q, Guan J, Zhou S. TOP: A Deep Mixture Representation Learning Method for Boosting Molecular Toxicity Prediction, Methods. [DOI] [PubMed]
- 129.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling, arXiv preprint arXiv:1412.3555.
- 130.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding, arXiv preprint arXiv:1810.04805.
- 131.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc Nat Acad Sci. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mao X, Su Z, Tan PS, Chow JK, Wang Y-H. Is Discriminator a Good Feature Extractor?, arXiv preprint arXiv:1912.00789.
- 133.Sanchez-Lengeling B., Aspuru-Guzik A. Inverse molecular design using machine learning: Generative models for matter engineering. Science. 2018;361(6400):360–365. doi: 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- 134.Duvenaud DK, Maclaurin D, Iparraguirre J, Bombarell R, Hirzel T, Aspuru-Guzik A, Adams RP. Convolutional networks on graphs for learning molecular fingerprints. In: Advances in neural information processing systems, 2224–2232; 2015.
- 135.Rogers D., Hahn M. Extended-connectivity fingerprints. J Chem Inform Modeling. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 136.Rupp M., Tkatchenko A., Müller K.-R., Von Lilienfeld O.A. Fast and accurate modeling of molecular atomization energies with machine learning. Phys Rev Lett. 2012;108(5) doi: 10.1103/PhysRevLett.108.058301. [DOI] [PubMed] [Google Scholar]
- 137.Hirn M., Mallat S., Poilvert N. Wavelet scattering regression of quantum chemical energies. Multiscale Modeling Simul. 2017;15(2):827–863. [Google Scholar]
- 138.Chou K.-C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins: Struct Funct Bioinform. 2001;43(3):246–255. doi: 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- 139.Liu H., Sun J., Guan J., Zheng J., Zhou S. Improving compound–protein interaction prediction by building up highly credible negative samples. Bioinformatics. 2015;31(12):i221–i229. doi: 10.1093/bioinformatics/btv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu X., Wang X., Matwin S. 2018 International Joint Conference on Neural Networks (IJCNN) IEEE. 2018. Interpretable deep convolutional neural networks via meta-learning; pp. 1–9. [Google Scholar]
- 141.Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps, arXiv preprint arXiv:1312.6034.
- 142.Preuer K, Klambauer G, Rippmann F, Hochreiter S, Unterthiner T. Interpretable deep learning in drug discovery. In: Explainable AI: Interpreting, Explaining and Visualizing Deep Learning, Springer, 331–345; 2019.
- 143.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need, in: Advances in neural information processing systems, 5998–6008; 2017.
- 144.Wang F., Jiang M., Qian C., Yang S., Li C., Zhang H., Wang X., Tang X. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Residual attention network for image classification, in; pp. 3156–3164. [Google Scholar]
- 145.Dong L., Xu S., Xu B. 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE. 2018. Speech-transformer: a no-recurrence sequence-to-sequence model for speech recognition; pp. 5884–5888. [Google Scholar]
- 146.Deng Z., Chuaqui C., Singh J. Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein- ligand binding interactions. J Medicinal Chem. 2004;47(2):337–344. doi: 10.1021/jm030331x. [DOI] [PubMed] [Google Scholar]
- 147.Chupakhin V., Marcou G., Gaspar H., Varnek A. Simple Ligand-Receptor Interaction Descriptor (SILIRID) for alignment-free binding site comparison. Computat Struct Biotechnol J. 2014;10(16):33–37. doi: 10.1016/j.csbj.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nguyen D.D., Cang Z., Wei G.-W. A review of mathematical representations of biomolecular data. PCCP. 2020;22(8):4343–4367. doi: 10.1039/c9cp06554g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwon Y., Yoo J., Choi Y.-S., Son W.-J., Lee D., Kang S. Efficient learning of non-autoregressive graph variational autoencoders for molecular graph generation. J Cheminformatics. 2019;11(1):70. doi: 10.1186/s13321-019-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jin W, Barzilay R, Jaakkola T. Junction tree variational autoencoder for molecular graph generation, arXiv preprint arXiv:1802.04364.
- 151.Griffiths R.-R., Hernández-Lobato J.M. Constrained Bayesian optimization for automatic chemical design using variational autoencoders. Chem Sci. 2020;11(2):577–586. doi: 10.1039/c9sc04026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhou Z., Kearnes S., Li L., Zare R.N., Riley P. Optimization of molecules via deep reinforcement learning. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-47148-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gathiaka S., Liu S., Chiu M., Yang H., Stuckey J.A., Kang Y.N., Delproposto J., Kubish G., Dunbar J.B., Carlson H.A. D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J Comput-Aided Molecular Des. 2016;30(9):651–668. doi: 10.1007/s10822-016-9946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nguyen D.D., Cang Z., Wu K., Wang M., Cao Y., Wei G.-W. Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J Comput-Aided Molecular Des. 2019;33(1):71–82. doi: 10.1007/s10822-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nguyen D.D., Gao K., Wang M., Wei G.-W. MathDL: mathematical deep learning for D3R Grand Challenge 4. J Comput-Aided Molecular Des. 2020;34(2):131–147. doi: 10.1007/s10822-019-00237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li Y., Su M., Liu Z., Li J., Liu J., Han L., Wang R. Assessing protein–ligand interaction scoring functions with the CASF-2013 benchmark. Nat Protocols. 2018;13(4):666–680. doi: 10.1038/nprot.2017.114. [DOI] [PubMed] [Google Scholar]