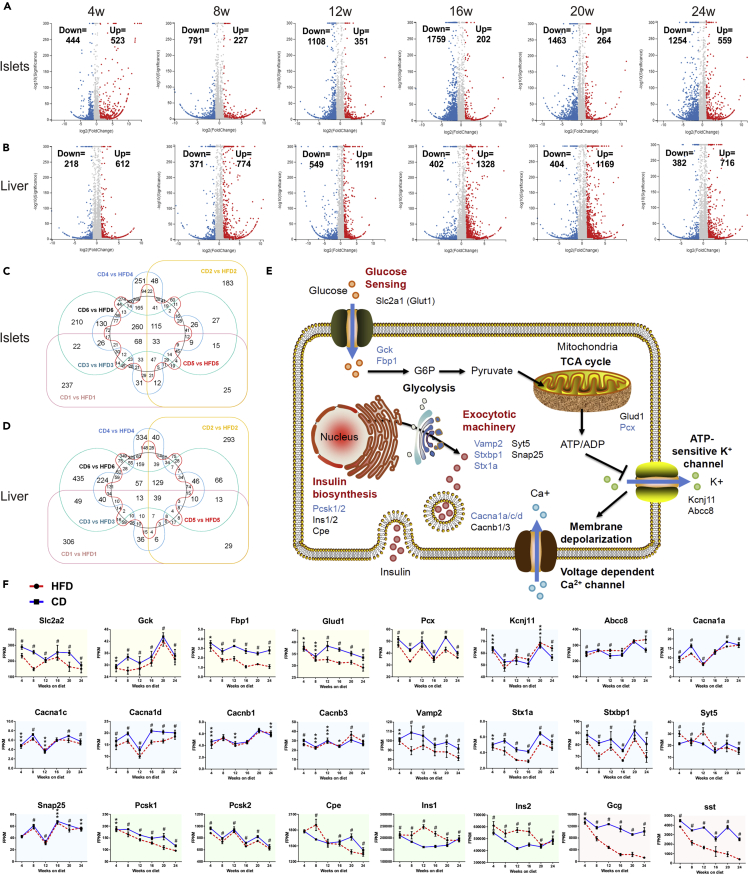
Figure 4.
Transcriptomic profiles and pathway dynamics of islets and liver during diabetes progression
(A and B) Islet (A) and liver (B) volcano plots. The numbers of upregulated (red dots) and downregulated (blue dots) genes are marked in each graph.
(C and D) Islet (C) and liver (D) Venn diagrams of identifiable and quantifiable DEGs.
(E) Schematic representation illustrating the physiology and key genes of GSIS in pancreatic β-cells. Genes that were significantly downregulated are colored in blue.
(F) The time course expression data for genes involved in GSIS. Yellow, blue, green, and red shaded areas, respectively refer to genes participating in glucose sensing, exocytotic machinery, insulin biosynthesis, and the secretion of other islet cells.
All data are expressed as mean ± SEM and analyzed using unpaired two-tailed t test. ∗adjusted p < 0.05, ∗∗adjusted p < 0.01, ∗∗∗adjusted p < 0.001, #adjusted p < 0.0001. See also Figures S4–S6.