Abstract
Proliferative vitreoretinopathy (PVR) is a disease leading to the formation of contractile preretinal membranes (PRMs) and is one of the leading causes of blindness. Connective tissue growth factor (CTGF) has been identified as a possible key determinant of progressive tissue fibrosis and excessive scarring. Therefore, the present study investigated the role and mechanism of action of CTGF in PVR. Immunohistochemical staining was performed to detect the expression of CTGF, fibronectin and collagen type III in PRMs from patients with PVR. The effects and mechanisms of recombinant human CTGF and its upstream regulator, TGF-β1, on epithelial-mesenchymal transition (EMT) and the synthesis of extracellular matrix (ECM) by retinal pigment epithelium (RPE) cells were investigated using reverse transcription-quantitative PCR, western blotting and a [3H]proline incorporation assay. The data indicated that CTGF, fibronectin and collagen type III were highly expressed in PRMs. In vitro, CTGF significantly decreased the expression of the epithelial markers ZO-1 and E-cadherin and increased that of the mesenchymal markers fibronectin, N-cadherin and α-smooth muscle actin in a concentration-dependent manner. Furthermore, the expression of the ECM protein collagen type III was upregulated by CTGF. However, the trends in expression for the above-mentioned markers were reversed after knocking down CTGF. The incorporation of [3H]proline into RPE cells was also increased by CTGF. In addition, 8-Bromoadenosine cAMP inhibited CTGF-stimulated collagen synthesis and transient transfection of RPE cells with a CTGF antisense oligonucleotide inhibited TGF-β1-induced collagen synthesis. The phosphorylation of PI3K and AKT in RPE cells was promoted by CTGF and TGF-β1 and the latter promoted the expression of CTGF. The results of the present study indicated that CTGF may promote EMT and ECM synthesis in PVR via the PI3K/AKT signaling pathway and suggested that targeting CTGF signaling may have a therapeutic or preventative effect on PVR.
Keywords: connective tissue growth factor, epithelial-mesenchymal transition, extracellular matrix, retinal pigment epithelium cells, proliferative vitreoretinopathy
Introduction
Proliferative vitreoretinopathy (PVR), which is characterized by the formation of contractile preretinal membranes (PRMs) in the vitreous and on the inner and outer surfaces of the detached retina, is one of the leading causes of blindness (1–3). PVR is associated with recurrent retinal detachment and is the main cause of failure of retinal reattachment surgery (1). Despite the development of advanced surgical techniques and devices, there remain unresolved issues regarding the pathogenesis and treatment of PVR (1). Therefore, the identification of key molecules involved in PVR is required for improving the clinical outcome of this disease.
PRMs result from cell dedifferentiation, migration, adherence and proliferation and the secretion of extracellular matrix (ECM) proteins, including collagen, following rhegmatogenous retinal detachment (3,4). PVR represents an excessive wound-healing response during tissue repair following eye injury. Previous studies have demonstrated that retinal pigment epithelium (RPE) cells, Müller glial cells and blood-derived lymphocytes are present in the PRMs of patients with PVR (5–7). Among these cell types, RPE cells appear to be the primary proliferative cells that can change their phenotype and undergo the process of epithelial-mesenchymal transition (EMT) (2,8). It has been reported that EMT of RPE cells is triggered by a host of growth factors, including TGF-β1, platelet-derived growth factor and epidermal growth factor (2,9–11). Activated RPE cells can proliferate and migrate into the vitreous or the inner layer of the retina, producing ECM components and being transformed from epithelial cells to fibroblast-like cells, a biological phenomenon termed EMT, and ultimately forming a PRM (2,12). When the PRM contracts it results in traction to the retina (1). Apert from stripping the PRM during surgery, no other target-oriented interventions are available to decrease the EMT of RPE cells and the production of ECM, thereby increasing the success rate of a PVR operation (1,3).
Previous studies have found that growth factors are necessary for the repair of damaged tissue; however, prolonged production or dysregulated expression of growth factors can lead to persistent EMT and excessive wound healing, as found in PVR (4,13,14). Among these growth factors, TGF-β is a key factor in EMT and matrix remodeling (2). The role of TGF-β is controversial in eye diseases and is associated with the specific condition/system. Studies have demonstrated that topical application of a TGF-β1-loaded liposomal suspension could protect retinal tissue in rat models of age-related macular degeneration (15,16). In PVR, TGF-β is a pivotal contributor to tissue fibrosis (2,17). Previous studies have focused on a downstream mediator of TGF-β signaling, connective tissue growth factor (CTGF) (2,18–20). CTGF, which is rich in cysteine residues, is a secreted growth factor and was originally identified in a conditioned medium from human umbilical vein endothelial cells using affinity chromatography (21). CTGF is identified as a possible key determinant of progressive tissue fibrosis and excessive scarring, which also serves an important role in wound repair, neoangiogenesis, tumor growth and embryonic development (18,22,23). A number of studies have demonstrated that CTGF can stimulate the growth and adhesion of fibroblasts and vascular endothelial cells, as well as promoting the secretion of ECM proteins (2,18,22,23).
Our previous study confirmed that CTGF increases the migratory potential of RPE cells by stimulating the calcium signaling system and serves an important role in the pathogenesis of PVR (19). Previous studies have also demonstrated that CTGF is a major mediator of retinal fibrosis and potentially an effective therapeutic target for PVR (17,18). However, the role of CTGF in the progress of PVR and the downstream signaling mechanisms remains to be elucidated.
The present study aimed to investigate the expression of CTGF, fibronectin and collagen type III in PVR membranes, as well as the effects and mechanisms of CTGF and its upstream regulator TGF-β1 in the process of EMT and ECM synthesis by RPE cells in vitro. The findings may provide insights into the mechanistic details of intraocular proliferative diseases including PVR.
Materials and methods
PRM collection and preparation
A total of 26 PRM specimens were obtained from patients who underwent a vitreoretinal surgery after having been diagnosed with rhegmatogenous retinal detachment, accompanied by PVR, at the Department of Ophthalmology, Xijing Hospital (Xian, China) between March 2017 and March 2019. In accordance with the tenets of the Declaration of Helsinki, informed consent of the patients and the approval by the institutional review board were obtained before sample collection. The patients ranged in age between 19 and 67 years (12 female and 14 male) and their course of disease ranged between 4 months and 2 years. Among the specimens collected, six were classified as grade B membranes [moderate PVR: Wrinkling of the inner retinal surface, a rolled edge(s) of a retinal break(s), retinal stiffness and vascular tortuosity]; eleven were classified as grades C1-C3 (marked PVR: Full-thickness retinal folds in 1–3 quadrants, respectively); and nine were classified as grades D1-D3 (massive PVR: Fixed retinal folds in four quadrants) according to the classification of PVR grades established by the International Retinal Society in 1983 (24). All 26 specimens were fixed in 4% (w/v) neutral formalin for 24 h at room temperature, and dehydrated sequentially as follows: 1X into 70% ethanol (1 h at 4°C), 1X into 85% ethanol (1 h at 4°C), 2X into 95% ethanol (1 h each at 4°C), and 2X into 100% ethanol (1 h each at 4°C). The tissues were immersed 2X into the paraffin bath (2 h each at 60°C), then transported to the mold with paraffin, and incubated at room temperature until set. The tissues were prepared as 6-µm-thick sections.
Immunohistochemical staining of PRMs
Sections were deparaffinized in multiple changes of xylene (Sangon Biotech Co., Ltd.) and rehydrated in a decreasing graded ethanol series (Sangon Biotech Co., Ltd.). Endogenous peroxidase was blocked by incubation of the sections in 3% H2O2 (Beyotime Institute of Biotechnology) for 10 min at room temperature. For heat-induced epitope retrieval, the sections were immersed in 0.01 M citrate buffer, pH 6.0 (Sangon Biotech Co., Ltd.) and heated for 10 min in a microwave, followed by cooling for 10 min and reheating for 5 min. The sections were then cooled for 30 min at room temperature and washed with water and phosphate-buffered saline (PBS; Beyotime Institute of Biotechnology). Following incubation with blocking serum (Beyotime Institute of Biotechnology) for 30 min, the sections were incubated with the following primary antibodies at 4°C overnight: CTGF (1:100; cat. no. ab5097; Abcam), collagen type III (1:100; cat. no. 22734-1-AP; ProteinTech Group, Inc.) and fibronectin (1:100; cat. no. 26836; Cell Signaling Technology, Inc.). A streptavidin/biotin complex immunostaining kit was purchased from Wuhan Boster Biological Technology, Ltd. Incubation with a biotinylated secondary antibody (1:500; cat. no. ab205718; Abcam) was followed by incubation with peroxidase-labeled streptavidin at room temperature for 1 h (Sangon Biotech Co., Ltd.). Staining was visualized via the reaction of samples with a diaminobenzidine substrate-chromogen solution (Sangon Biotech Co., Ltd.). The sections were additionally counterstained with hematoxylin at room temperature for 2 min (Sangon Biotech Co., Ltd.), dehydrated and mounted in Paramount (Agilent Technologies, Inc.). The primary antibodies were omitted in negative controls and the sections were incubated only in the diluent (PBS). All subsequent steps of the control and experimental procedures were identical.
After the colorimetric reaction was completed, the sections were observed under a light microscope (Eclipse E100, Nikon Corporation). The area and intensity of staining for each antibody was independently estimated by two researchers. A clear background, yellow or pale brown in the cytoplasm, was a positive signal. The number of positive cells per 100 cells was counted in each field (magnification, ×400) and the average was calculated for three fields in each specimen. i) The samples were scored according to the presence of color and its darkness in the proliferative membrane tissue as follows: 0 points for no coloration; 1 point for light yellow; 2 points for yellow or pale brown; and 3 points for brown. ii) The samples were scored according to the proportion of positive cells: 0 points, <5%; 1 point, 5–30%; 2 points, 30–60%; and 3 points, >60%. The final score for each specimen was (i + ii)/2 and the samples were classified according to the scores as follows: score 0, no labeling (−); score 0.5–1, weak, focal staining (+); score 1.5–2, moderate to strong staining (++); and score 2.5–3, intense staining (+++). No differences were found in the data obtained by the two observers.
Immunofluorescence staining
PRMs were fixed with 4% paraformaldehyde (PFA; Sangon Biotech Co., Ltd.) at 4°C for 1 h, incubated in 30% sucrose at 4°C overnight, embedded in Tissue-Tek optimal cutting temperature compound (Sakura Finetek USA, Inc.) and frozen to prepare 8-µm-thick sections (cat. no. CM1800; Leica Microsystems GmbH). The sections were dried at room temperature for 2 h and blocked with PBS containing 1% bovine serum albumin (BSA, Zeta-life Company) and 0.5% Triton X-100 for 1 h. The sections were incubated with primary antibodies at 4°C overnight, followed by incubation with secondary antibodies at room temperature for 1 h. Each step was followed by washing three times with PBS for 5 min each. The primary antibodies included CTGF (1:100; cat. no. ab5097; Abcam) and retinal pigment epithelium-specific 65 kDa protein (RPE65; 1:100; cat. no. MA1-16578; Invitrogen; Thermo Fisher Scientific, Inc.). The secondary antibodies included Cy3-conjugated goat anti-rabbit IgG (H+L; 1:100; cat. no A10522; Invitrogen; Thermo Fisher Scientific, Inc.) and Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L; 1:100; cat. no. ab150113; Abcam).
Human RPE cells were fixed with 4% PFA at 4°C for 30 min and then blocked with PBS containing 1% BSA and 0.5% Triton X-100 at room temperature for 1 h. The primary antibody was RPE65 (1:100; cat. no. MA1-16578, Invitrogen; Thermo Fisher Scientific, Inc.) and the secondary antibody was Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L; 1:100; cat. no. ab150113; Abcam). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 5 min (Bioworld Technology, Inc.). The cells were incubated with antibodies as described above.
All samples were observed and images captured under a confocal scanning laser microscope (FV1000; Olympus Corporation, magnification, ×400); three randomly selected fields from each sample were examined. Red and green staining indicated the positive reaction. ImageJ software (version 1.49p; National Institutes of Health) was used for analysis.
Human RPE cell culture, small interfering RNA (siRNA) transfection and treatment
The human RPE cell line ARPE19 was obtained from the Chinese Academy of Sciences cell bank. Cells were routinely cultured in low-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Zeta-Life Inc.) and 1% penicillin/streptomycin (Sigma-Aldrich; Merck KGaA) in a humidified atmosphere of 5% CO2 at 37°C. ARPE19 cells were used between passages 4 and 6 and a specific marker (RPE65) was detected using immunofluorescence (25) (Fig. S1). Cells were fixed with 4% PFA at 4°C for 30 min and then blocked with PBS containing 1% BSA and 0.5% Triton X-100 at room temperature for 1 h. Cells were incubated the primary and secondary antibodies as above. Cell nuclei were stained with DAPI at room temperature for 5 min (Bioworld Technology, Inc.).
RPE cells were cultured to 60–70% confluence and then transfected with a negative control (NC) siRNA or siCTGF (SIGS0002293-1; RiboBio, Guangzhou, China) at a concentration of 50 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Target sequences for siCTGF were as follows: si-1, GCACCAGCATGAAGACATA; si-2, GTGCATCCGTACTCCCAAA; si-3, CTCCAAGCCTATCAAGTTT. The sequences of negative control siRNA are as follows: Sense 5′-3′UUCUCCGAACGUGUCACGUTT, antisense 5′-3′ACGUGACACGUUCGGAGAATT.
RPE cells were treated with 15, 30 and 60 ng/ml recombinant human CTGF (cat. no. CM22; Shanghai Novoprotein Technology Co., Ltd.) or 30 ng/ml TGF-β1 (cat. no. CA59; Shanghai Novoprotein Technology Co., Ltd.) for 24 h to perform reverse transcription- quantitative (RT-q) PCR and western blotting. Based on the results of a preliminary experiment and the reference concentration, the most suitable concentration of TGF-β1 (30 ng/ml) was chosen for this experiment (20). Cells were used for subsequent experimentation 24 h following transfection.
RT-qPCR
RPE cells were cultured to 95% confluence. Total RNA was purified using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using a reverse transcription kit (Takara Bio, Inc.) according to the manufacturer's protocols. RT-qPCR was performed in a 20-µl reaction, containing the cDNA and specific primers, using a PCR kit (SYBR Premix Ex Taq; Takara Bio, Inc.) according to the manufacturer's instructions. The thermocycling conditions were as follows: 98°C for 3 min, followed by 40 cycles at 98°C for 15 sec, 55°C for 15 sec and 72°C for 15 sec. Gene expression levels were determined using the 2−ΔΔCq method (26) with β-actin used as an internal reference. The primer sequences are shown in Table I. Each experiment was performed three times.
Table I.
Primer sequences used in the study.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| β-actin | TGTTACCAACTGGGACGACA | CTTTTCACGGTTGGCCTTAG |
| CTGF | CAGCATGGACGTTCGTCTG | AACCACGGTTTGGTCCTTGG |
| Fibronectin | CGGTGGCTGTCAGTCAAAG | AAACCTCGGCTTCCTCCATAA |
| ZO-1 | CAACATACAGTGACGCTTCACA | CACTATTGACGTTTCCCCACTC |
| N-cadherin | CAGAATCGTGTCTCAGGCTCCAAG | CTGCGTTCCAGGCTGGTGTATG |
| E-cadherin | TACAATGCCGCCATCGCTTACAC | TGACGGTGGCTGTGGAGGTG |
| Collagen type III | GGAGCTGGCTACTTCTCGC | GGGAACATCCTCCTTCAACAG |
| α-SMA | CTATGAGGGCTATGCCTTGCC | GCTCAGCAGTAGTAACGAAGGA |
CTGF, connective tissue growth factor; α-SMA, α-smooth muscle actin.
Western blotting
RPE cells were lysed with RIPA buffer (Beyotime Institute of Biotechnology) containing a complete protease inhibitor cocktail (Roche Molecular Biochemicals). A bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) was used to determine protein concentrations. Proteins (30 µg) were separated by 8–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad Laboratories, Inc.) and transferred to polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% skimmed milk [diluted with Tris-buffered saline containing 0.05% Tween-20 (TBST)] at room temperature for 1 h, followed by incubation with primary antibodies in TBST at 4°C overnight. After three washes with TBST for 5 min each, the membranes were incubated with a secondary antibody for 1 h at room temperature, followed by three washes. The membranes were scanned using an enhanced chemiluminescence assay (Beijing 4A Biotech Co., Ltd.). ImageJ software (version 1.49p; National Institutes of Health) was used to calculate the gray value and analysis of the protein bands.
The following primary antibodies were used: Fibronectin (1:1,000; cat. no. 26836; Cell Signaling Technology, Inc.); ZO-1 (1:1,000; cat. no. 13663S; Cell Signaling Technology, Inc.); N-cadherin (1:1,000; cat. no. 13116T; Cell Signaling Technology, Inc.); E-cadherin (1:1,000; cat. no. 3195T; Cell Signaling Technology, Inc.); collagen type III (1:1,000; cat. no. 22734-1-AP, ProteinTech Group, Inc.); α-smooth muscle actin (α-SMA; 1:1,000; cat. no. ab5694, Abcam); phosphorylated (p)-PI3K (1:1,000; cat. no. 4228; Cell Signaling Technology, Inc.); PI3K (1:1,000; cat. no. 4257; Cell Signaling Technology, Inc.); p-AKT (1:1,000; cat. no. 4060; Cell Signaling Technology, Inc.); AKT (1:1,000; cat. no. 4691; Cell Signaling Technology, Inc.); CTGF (1:1,000; cat. no. ab5097, Abcam); GAPDH (1:1,000; cat. no. 10494-1-AP, ProteinTech Group, Inc.). Horseradish peroxidase-conjugated goat anti-rabbit IgG (1:3,000; cat. no. SA00001-2; ProteinTech Group, Inc.) was used as the secondary antibody.
[3H]proline incorporation by RPE cells
The [3H]proline incorporation assay was employed as an indicator of the rate of collagen synthesis (27). RPE cells (5×104/ml) were seeded into 96-well plates and grown to 85% confluence. The medium was replaced with a serum-free medium and incubation continued for 24 h before the cells were treated with CTGF (5, 15, 30 and 60 ng/ml), 8-Bromoadenosine (8-Br) cAMP (0.1 mM; Sigma-Aldrich; Merck KGaA) and 8-Br-cAMP (0.1 mM) + CTGF (60 ng/ml). 8-Br-cAMP can inhibit the expression of CTGF (28). The CTGF-oligonucleotide-transfected cells were treated with 15 ng/ml TGF-β1 for 24 h, followed by incubation with 2 µCi of [3H]proline (China Institute of Atomic Energy) for an additional 24 h. Subsequently, the cells were washed three times with PBS for 5 min each and lysed with trypsin (Sigma-Aldrich; Merck KGaA) at room temperature for 2 min. The incorporation of [3H]proline was determined by scintillation counting. Each experimental condition was performed in triplicate wells and the experiments were repeated three times. The results are expressed as cpm/103 cells ± standard error of the mean (SEM) after counting trypsinized cell monolayers; the tested reagents caused only minor changes in the cell numbers of these growth-arrested cultures over a 2-day assay period.
CTGF-oligonucleotide ODN (CTGF-ODN) transfection of RPE cells
Oligonucleotide (ODN) is a polymer consisting of a small number of nucleotides (29). The ODNs used to bind to CTGF included CTGF sense oligonucleotide (SODN; 5′-ATGACCGCCGCCAGTA-3′) and antisense oligonucleotide (ASODN, to inhibit the expression of CTGF; 5′-TACTGGCGGCGGTCAT-3′), as previously designed (30). The ODNs included a phosphorothioate modification synthesized by Shanghai Shenggong Biotech Co., Ltd. RT-qPCR was used to detect the knockdown efficiency of CTGF-ASODN (Fig. S2). RPE cells (5×104/ml) were seeded in 96-well plates and incubated in a 5% CO2 incubator at 37°C for 2 days until they almost reached 85% confluence. Prior to transfection, the medium was replaced with a serum-free medium without antibiotics. For transfection, each ODN was diluted with serum-free DMEM to a final concentration of 400 nM and Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was diluted 1:25 with serum-free DMEM. The solution of each ODN was mixed with diluted Lipofectamine® and incubated at room temperature for 20 min, followed by aliquoting 50 µl of the resulting complexes into each well. Cells mixed with the ODN/Lipofectamine® 2000 were incubated in a 5% CO2 incubator at 37°C for 12 h. Cells with >90% viability, as determined using trypan blue exclusion, were maintained in culture. The cells that took up the trypan blue were considered non-viable. Cells (1 ml) were stained with 0.1 ml of trypan stock solution (Sigma-Aldrich; Merck KGaA). Cells (10 µl) were added to a hemocytometer (Paul Marienfeld GmbH & Co. KG) and examined immediately under a light microscope (Eclipse E100, Nikon Corporation; magnification, ×10). The number of blue staining cells and the number of total cells were counted. The transfection medium was replaced with a serum-free medium containing 100 µg/ml vitamin C (Tianjin Aoran Fine Chemical Research Institute) and 15 ng/ml TGF-β1 and the cells were cultured for another 24 h.
Statistical analysis
Data analysis was performed using GraphPad Prism 8.0 (GraphPad Software, Inc.). All quantitative results are presented as the mean ± SEM. An unpaired Student's t-test was used to determine the statistical significance of differences between the two groups. To determine the statistical significance of the differences among multiple groups, one-way analysis of variance (ANOVA) with Dunnett's or Tukey's post hoc test was employed. P<0.05 was considered to indicate a statistically significant difference.
Results
CTGF induces EMT and the ECM synthesis
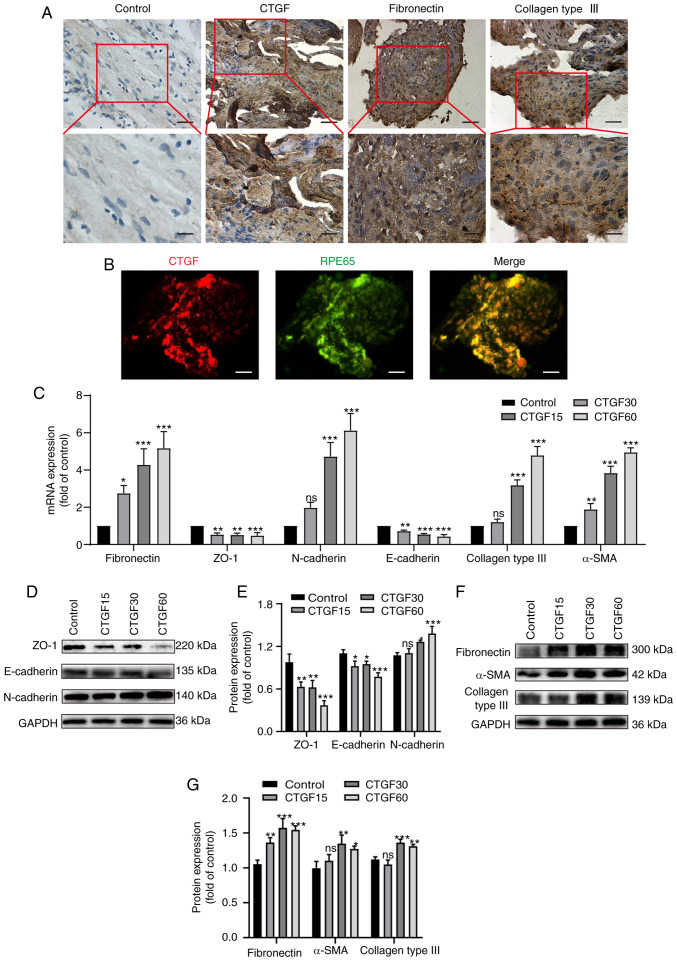
Immunohistochemical staining was employed to detect the expression of CTGF, fibronectin and collagen type III in PRM sections. These proteins were detected in all PRMs (Fig. 1A). CTGF-positive cells were scattered or clustered in PRMs and were found in fibrotic regions of the membranes (Fig. 1A). CTGF-positive cells were mainly epithelioid cells with a long elliptic or polygonal phenotype and abundant cytoplasm. Immunofluorescence co-staining with a specific marker of RPE cells demonstrated that CTGF-positive cells were also RPE65 positive, which confirmed that these cells were RPE cells (Fig. 1B). However, a smaller population of CTGF-positive cells included fibroblast-like cells, which were spindle shaped and large bodied.
Figure 1.
Effects of CTGF on EMT and the ECM synthesis. (A) Immunohistochemistry of surgically excised human PVR specimens stained for CTGF, fibronectin and collagen III. Original magnification, ×400 and ×800 (enlargement of boxed area). (B) Immunofluorescence co-staining of PVR membranes for CTGF (red) and RPE65 (green). Original magnification, ×400 (C). Relative mRNA expression levels of biomarkers for EMT and the ECM synthesis in ARPE19 cells treated with CTGF (15, 30 and 60 ng/ml), including specific markers of epithelial cells (ZO-1 and E-cadherin), mesenchymal cells (fibronectin, N-cadherin and α-SMA) and the ECM (collagen type III). (D) Representative western blot analysis of the expression levels of EMT markers in ARPE19 cells treated with CTGF (15, 30 and 60 ng/ml). (E) Quantification of relative protein expression of ZO-1, E-cadherin and N-cadherin. (F) Representative western blot analysis of the expression levels of EMT and ECM synthesis markers in ARPE19 cells treated with CTGF (15, 30 and 60 ng/ml). (G) Relative protein expression levels of fibronectin, α-SMA and collagen type III; n=3. *P<0.05, **P<0.01, ***P<0.001 vs. control group; ns, not significant; CTGF, connective tissue growth factor; EMT, epithelial-mesenchymal transition; ECM, extracellular matrix; PVR, proliferative vitreoretinopathy; RPE65, retinal pigment epithelium-specific 65 kDa protein; α-SMA, α-smooth muscle actin.
Tables II and III demonstrate that the intense positive rate of CTGF expression in the grade B membranes (16.7%) was lower compared with the grade C and D membranes (54.5 and 77.8%, respectively). ECM proteins, fibronectin and collagen type III, were expressed in all PRMs and evidently expressed in CTGF intense-positive PRMs. The positive rates of fibronectin expression in the grade B and C membranes (66.7 and 36.4%, respectively) were higher compared with the grade D membranes (22.2%). By contrast, the positive rates of collagen type III expression in the grade B and C membranes (33.3 and 54.5%, respectively) were significantly lower compared with the grade D membranes (66.7%). These results support the hypothesis that fibronectin is involved in scar formation during the early phase of wound healing, while collagens become the primary constituents of the ECM during the advanced stages of PVR.
Table II.
CTGF, fibronectin and collagen type III expression in the PRMs of PVR.
| No. | Sex (M/F) | Age (year) | Duration of symptoms (month) | PVR (C/D) | CTGF | FN | Collagen III |
|---|---|---|---|---|---|---|---|
| 1 | M | 42 | 12 | D2 | +++ | ++ | +++ |
| 2 | F | 29 | 6 | B | +++ | +++ | +++ |
| 3 | M | 67 | 14 | C2 | ++ | ++ | + |
| 4 | M | 39 | 18 | D2 | +++ | ++ | +++ |
| 5 | F | 52 | 10 | C1 | +++ | ++ | +++ |
| 6 | M | 27 | 20 | C3 | +++ | +++ | +++ |
| 7 | F | 24 | 15 | D2 | ++ | + | ++ |
| 8 | M | 38 | 12 | D1 | +++ | +++ | ++ |
| 9 | F | 49 | 5 | B | ++ | +++ | ++ |
| 10 | F | 33 | 4 | B | + | ++ | + |
| 11 | M | 19 | 9 | D1 | ++ | ++ | ++ |
| 12 | F | 55 | 15 | D1 | +++ | +++ | +++ |
| 13 | M | 48 | 12 | C1 | + | + | + |
| 14 | M | 34 | 8 | B | ++ | +++ | ++ |
| 15 | F | 25 | 9 | C1 | ++ | ++ | ++ |
| 16 | M | 36 | 24 | D3 | +++ | + | +++ |
| 17 | F | 58 | 18 | D1 | +++ | ++ | +++ |
| 18 | M | 28 | 12 | C3 | +++ | +++ | +++ |
| 19 | M | 26 | 10 | C2 | +++ | ++ | +++ |
| 20 | F | 30 | 6 | C2 | ++ | ++ | ++ |
| 21 | M | 22 | 14 | C3 | +++ | +++ | +++ |
| 22 | M | 21 | 4 | B | + | + | + |
| 23 | F | 36 | 9 | C2 | ++ | ++ | ++ |
| 24 | F | 39 | 15 | C3 | +++ | +++ | +++ |
| 25 | M | 43 | 18 | D2 | +++ | + | +++ |
| 26 | F | 49 | 7 | B | ++ | +++ | +++ |
M, male; F, female; PVR, proliferative vitreoretinopathy; CTGF, connective tissue growth factor; FN, fibronectin; PRMs, preretinal membranes; 1+, weak, focal staining (<10% of cells); 2++, moderate to strong staining (up to 50% of cells); 3+++, intense staining (>50% of cells).
Table III.
Expression of CTGF, fibronectin, collagen type III protein in the PRMs of PVR.
| PVR B | PVR C1-C3 | PVR D1-D3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n (%) | + (%) | ++ (%) | +++ (%) | + (%) | ++ (%) | +++ (%) | + (%) | ++ (%) | +++ (%) |
| CTGF | 2 (33.3) | 3 (50) | 1 (16.7) | 1 (9.1) | 4 (36.4) | 6 (54.5) | 0 | 2 (22.2) | 7 (77.8) |
| Fibronectin | 1 (16.7) | 1 (16.7) | 4 (66.7) | 1 (9.1) | 6 (54.5) | 4 (36.4) | 3 (33.3) | 4 (44.4) | 2 (22.2) |
| Collagen III | 2 (33.3) | 2 (33.3) | 2 (33.3) | 2 (18.2) | 3 (27.3) | 6 (54.5) | 0 | 3 (33.3) | 6 (66.7) |
PVR, proliferative vitreoretinopathy; CTGF, connective tissue growth factor; PRMs, preretinal membranes.
The present study further investigated whether CTGF promotes EMT, as well as ECM synthesis, by treating RPE cells with different concentrations of CTGF (15, 30 and 60 ng/ml) using RT-qPCR and western blotting (Fig. 1C-G). The results demonstrated that the mRNA (Fig. 1C) and protein (Fig. 1D and E) expression levels of the epithelial markers ZO-1 and E-cadherin decreased in a CTGF concentration-dependent manner. In addition, the mRNA (Fig. 1C) and protein (Fig. 1D-G) expression levels of the mesenchymal markers fibronectin, N-cadherin and α-SMA were elevated. Furthermore, the expression of the ECM protein collagen type III was upregulated with the increased concentrations of CTGF (Fig. 1C, F and G). These results indicated that CTGF may promote EMT and the ECM synthesis by RPE cells.
Knocking down CTGF reverses EMT and the ECM synthesis by RPE cells
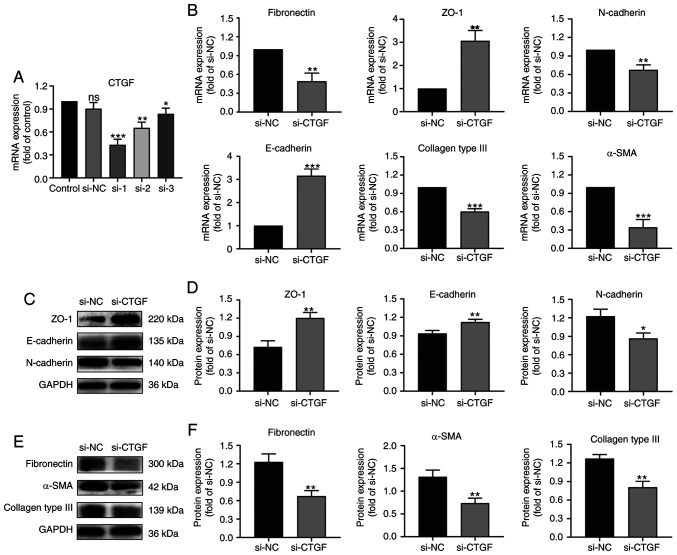
To verify the effect of CTGF on EMT and the ECM synthesis by RPE cells, the present study screened for a siCTGF with the highest knockdown efficiency using RT-qPCR (Fig. 2A). Thereafter, RPE cells were treated with an NC siRNA and siCTGF and the expression of EMT and ECM biomarkers was determined using RT-qPCR (Fig. 2B) and western blotting (Fig. 2C-F).
Figure 2.
Effects of CTGF knockdown on EMT and the ECM synthesis by ARPE19 cells. (A) Screening for a siCTGF with the highest knockdown efficiency by reverse transcription-quantitative PCR. (B) Relative mRNA expression levels of biomarkers for EMT and the ECM synthesis in ARPE19 cells transfected with si-NC and siCTGF, including specific markers of epithelial cells (ZO-1 and E-cadherin), mesenchymal cells (fibronectin, N-cadherin and α-SMA) and the ECM (collagen type III). (C) Representative western blot analysis of the expression levels of EMT markers in ARPE19 cells transfected with si-NC and siCTGF. (D) Quantification of relative expression levels of ZO-1, E-cadherin and N-cadherin. (E) Representative western blot analysis of the expression levels of EMT and ECM synthesis markers in ARPE19 cells transfected with si-NC and siCTGF. (F) Quantification of relative expression levels of fibronectin, α-SMA and collagen type III; n=3. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC group; ns, not significant; CTGF, connective tissue growth factor; EMT, epithelial-mesenchymal transition; ECM, extracellular matrix; siCTGF, CTGF-specific small interfering RNA (siRNA); si-NC, negative control siRNA; α-SMA, α-smooth muscle actin.
The results demonstrated that the expression of ZO-1 and E-cadherin (epithelial markers) increased after knocking down CTGF compared with the NC group (Fig. 2B-D). By contrast, the expression of fibronectin, N-cadherin and α-SMA (mesenchymal markers) decreased after knocking down CTGF compared with the NC group (Fig. 2B-F). Knockdown of CTGF significantly downregulated the expression of collagen type III compared with the NC group (Fig. 2B, E and F). These results indicate that knocking down CTGF can reverse the EMT process and ECM synthesis by RPE cells.
TGF-β1-stimulated synthesis of collagen is partially mediated by CTGF
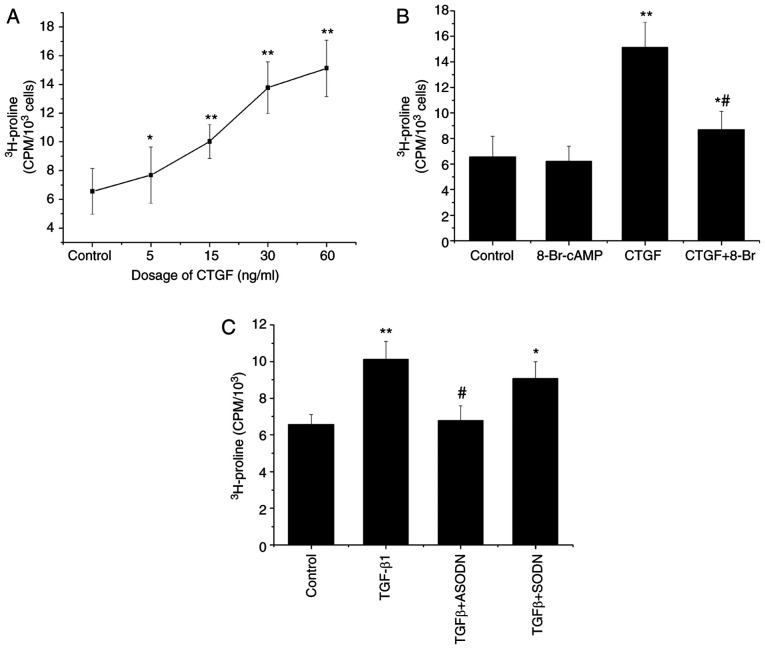
[3H]proline incorporation assay was used to determine the effects of CTGF on collagen synthesis. The results demonstrated that the incorporation of [3H]proline by RPE cells increased in a CTGF concentration-dependent manner (Fig. 3A). CTGF also significantly enhanced the synthesis of collagen by RPE cells compared with that by control cells. In combination with CTGF, 8-Br-cAMP inhibited the CTGF-stimulated incorporation of [3H]proline by RPE cells (Fig. 3B). Compared with CTGF treatment (60 ng/ml), 8-Br-cAMP reduced the [3H]proline incorporation, but no significant difference in collagen production was observed between cells exposed to 8-Br-cAMP and control cells.
Figure 3.
Mediation of TGF-β1-stimulated collagen synthesis by CTGF. (A) Incorporation of [3H]proline by ARPE19 cells treated with CTGF (5, 15, 30 and 60 ng/ml). (B) Incorporation of [3H]proline by untreated ARPE19 cells and cells treated with 8-Br-cAMP (0.1 mM), CTGF (60 ng/ml) and CTGF (60 ng/ml) + 8-Br-cAMP (0.1 mM). *P<0.05, **P<0.01 vs. control; #P<0.05 vs. the CTGF group. (C) Incorporation of [3H]proline by CTGF-ODN-transfected ARPE19 cells treated with 15 ng/ml TGF-β1. The results are expressed as cpm/103 cells ± standard error of the mean; n=3. *P<0.05, **P<0.01 vs. control; #P<0.05 vs. the TGF-β1 group. CTGF, connective tissue growth factor; 8-Br, 8-Bromoadenosine; ODN, oligonucleotide; S, sense; A, antisense; cpm, counts per min.
The incorporation of [3H]proline significantly increased following treatment of RPE cells with 15 ng/ml TGF-β1 compared with control cells. By contrast, RPE cells transfected with the CTGF antisense oligonucleotide and then treated with TGF-β1 (30 ng/ml) exhibited a decrease in the rate of [3H]proline incorporation, which indicated that the CTGF antisense oligonucleotide could partially block the TGF-β1-induced synthesis of collagen (Fig. 3C). By contrast, the CTGF sense oligonucleotide demonstrated no such effect under the same experimental conditions, resulting in no significant difference in [3H]proline incorporation compared with the TGF-β1 group (Fig. 3C). Taken together, these results indicate that the TGF-β1-stimulated production of ECM proteins is partially mediated by CTGF.
The PI3K/AKT pathway is regulated by CTGF
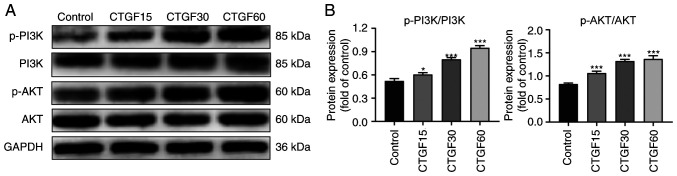
To explore the potential pathway involved in the CTGF-mediated EMT process and ECM synthesis by RPE cells, the phosphorylation levels of PI3K and AKT in CTGF-treated RPE cells were determined using western blotting (Fig. 4). The results revealed marked phosphorylation of PI3K and AKT in a CTGF concentration-dependent manner, whereas the total PI3K and AKT levels did not significantly change (Fig. 4). These data suggested that the observed regulation of EMT and the ECM synthesis by CTGF was associated with PI3K/AKT signaling in RPE cells.
Figure 4.
Regulation of the PI3K/AKT pathway in ARPE19 cells by CTGF. (A) Representative western blot analysis of p-PI3K/PI3K and p-Akt/Akt in ARPE19 cells treated with CTGF (15, 30 and 60 ng/ml). (B) Quantification of p-PI3K/PI3K levels; n=3. *P<0.05, ***P<0.001 vs. control group. CTGF, connective tissue growth factor; p-, phosphorylated.
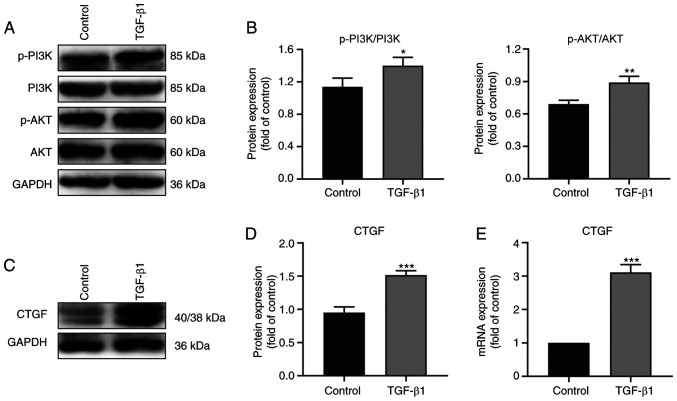
TGF-β1 regulates CTGF expression and the activation of the PI3K/AKT pathway
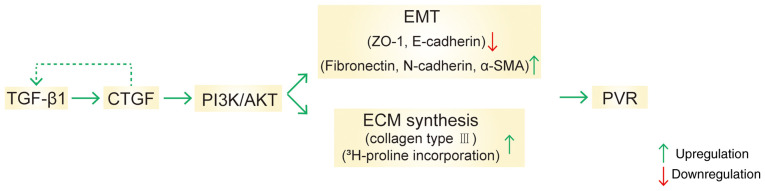
To study upstream factors involved in the regulation by CTGF of EMT and ECM synthesis, RPE cells were treated with TGF-β1 and the expression of CTGF and phosphorylation levels on the signaling pathway proteins determined by western blotting (Fig. 5). Notably, the expression of CTGF and the phosphorylation of PI3K and AKT were higher in the TGF-β1 group compared with the control group (Fig. 5A-D). RT-qPCR analysis also demonstrated that the mRNA expression of CTGF was higher in the TGF-β1 group (Fig. 5E). Taken together, the data indicated that TGF-β1 regulated CTGF expression and the activation of the PI3K/AKT pathway, thereby further promoting EMT and the ECM synthesis in PVR (Fig. 6).
Figure 5.
Regulation of CTGF expression and the PI3K/AKT pathway by TGF-β1 in ARPE19 cells. (A) Representative western blot analysis of p-PI3K/PI3K and p-Akt/Akt in the control and TGF-β1 groups. (B) Quantification of p-PI3K/PI3K and p-Akt/Akt in the control and TGF-β1 groups. (C) Representative western blot analysis of CTGF in the control and TGF-β1 groups. (D) Quantification of the relative expression of CTGF in the control and TGF-β1 groups. (E) Relative mRNA expression of CTGF in the control and TGF-β1 groups; n=3. *P<0.05, **P<0.01, ***P<0.001 compared with the control group. CTGF, connective tissue growth factor; p-, phosphorylated.
Figure 6.
Schematic representation of the study findings. TGF-β1 regulates CTGF expression and the PI3K/AKT pathway, thus promoting EMT and the ECM synthesis in PVR. CTGF, connective tissue growth factor; EMT, epithelial-mesenchymal transition; ECM, extracellular matrix; PVR, proliferative vitreoretinopathy; α-SMA, α-smooth muscle actin.
Discussion
PVR is a common disease, which occurs as a result of a dysregulated healing process of an intraocular wound and is one of the leading causes of blindness (1,14). Prevention of the formation of contractile PRMs in the vitreous and on the inner and outer surfaces of a detached retina contributes to the clinical treatment of PVR (2). CTGF is a member of a new immediate early gene family and encodes a protein that has been classified into a group of structurally related molecules, termed CCN (CTGF/Cyr61/Nov) (31). CTGF is a growth factor that has been demonstrated to stimulate the cell adhesion, chemotactic activity and proliferation of multiple cell types; it also induces angiogenesis and promotes EMT as well as the synthesis of ECM components (2). CTGF is highly expressed in fibrotic lesions of the lung and kidney, in the cardiac muscle during atherosclerosis and in tumors (18,31,32).
In the present study, CTGF was found to be highly expressed in PRMs. In addition, it was found that the expression of fibronectin and collagen type III was upregulated compared with the control group. The results supported the hypothesis that fibronectin is involved in scar formation during the early phase of wound healing, whereas collagens become the primary constituents of the ECM in the advanced stages of PVR. Analysis of the cell morphology and immunofluorescence staining demonstrated that CTGF-expressing cells were RPE and fibroblast-like cells. In previous studies, double staining of specimens demonstrate that CTGF-positive cells are also positive for keratin expression, indicating that the cells are derived from RPE cells (13,18). Therefore, the present study hypothesized that CTGF-positive cells are mainly RPE cells. Previous studies have demonstrated that RPE cells can undergo changes in differentiation signaling and the phenotype, acquiring resemblance to mesenchymal or fibroblast-like cells (EMT), which are involved in the generation of a PVR lesion (2,8,14). Furthermore, the spindle cells positive for CTGF expression were determined to be transformed RPE cells (33). CTGF immunoreactivity of RPE cells has been demonstrated to increase from early- to late-stage PVR and CTGF expression is the highest during late-stage PVR (13,18,34). The results of the present study are consistent with these previous results. It is hypothesized that at the later stage, the inflammatory period of PVR development and upregulation of CTGF can further aggravate RPE cell proliferation, adherence, EMT and collagen secretion to form a PRM.
Previous studies have demonstrated that the EMT process of RPE cells is a major pathological characteristic of PVR (2,35). EMT refers to morphological changes that involve the transformation of epithelial cells into fibroblasts or mesenchymal cells and is associated with tissue regeneration, damage repair and organ fibrosis (36,37). The ability of fibrotic RPE cells to proliferate, migrate and synthesize the ECM is significantly increased (1,2). The present study found that CTGF was highly expressed in the proliferative membranes of patients with PVR.
It has been reported that CTGF is closely associated with the process of EMT and the synthesis of an ECM (2). However, whether CTGF induces EMT of RPE cells has not been thoroughly investigated. There are two types of molecular markers associated with the process of EMT; one includes the markers of epithelial-like cells, including E-cadherin, ZO-1 and β-catenin, and the other type includes mesenchymal-like cell markers, including fibronectin, N-cadherin, α-SMA and vimentin (36,38). The present study further demonstrated that CTGF significantly decreased the expression of epithelial markers, including ZO-1 and E-cadherin, and increased that of mesenchymal markers, including fibronectin, N-cadherin and α-SMA, in a concentration-dependent manner. Meanwhile, the expression trends of the above markers were reversed after knocking down CTGF.
In the microenvironment wherein cells survive, a number of factors, including the ECM, hypoxia and macrophages, serve a significant role in inducing EMT (36,39). The ECM is not only a cytoskeleton for maintaining the structure and function, but it also mediates processes including cell growth, differentiation, migration and wound healing (40,41). Studies of PRMs have demonstrated that the primary ECM constituents are fibronectin and collagen subtypes I, III and IV, which are most consistently associated with RPE and fibroblast-like cells (42–44).
The present study also found that the expression of the ECM protein collagen type III was upregulated in RPE cells. In addition, the incorporation of [3H]proline by RPE cells increased in a CTGF concentration-dependent manner, while 8-Br-cAMP inhibited the CTGF-stimulated collagen synthesis. Proline residues account for almost 21% of the total amino acid content of collagens, whereas they are rarely found in other proteins (27). Consequently, [3H]proline incorporation by cells can be used as an indirect measure of the rate of collagen synthesis (27). An increase in the synthesis of collagen can reflect an increase in the cell proliferation (45,46). Accordingly, the present study suggested that CTGF may promote EMT and the ECM synthesis by RPE cells, thus contributing to the formation and contraction of proliferative membranes in PVR.
A number of studies have focused on the possible signaling pathways of EMT (2,38,47). The precise regulatory mechanism of the EMT process involves extracellular signal transduction to the cell via specific binding to a receptor on the membrane, leading to the initiation of a variety of signaling pathways to activate transcription factors in the nucleus and finally regulate the expression of EMT-related genes (36,37). EMT involves a number of regulatory signaling pathways, including PI3K/AKT, TGF-β, Wnt and Notch pathways, and there is also a complex crosstalk between pathways during the EMT process (37,38). However, the signaling pathway by which CTGF regulates EMT of RPE cells remains to be elucidated. The present study demonstrated that the phosphorylation of PI3K and AKT significantly increased with the increase of the CTGF concentration. Therefore, the data suggested that the regulation of EMT and the ECM synthesis by CTGF is associated with the PI3K/AKT signaling pathway in RPE cells.
Several studies have indicated that TGF-β is a strong inducer of CTGF expression in tubular epithelial cells and in a variety of other cells, including fibroblasts, mesangial cells, endothelial cells and chondrocytes, and that CTGF serves an important role in TGF-β signaling (2,17,18,20). Previous studies have demonstrated that TGF-β and CTGF are highly expressed in the vitreous humor and subretinal fluid of patients with PVR and exert stimulatory effects on the contraction of gels seeded with RPE cells (18,34,48,49). TGF-β is also considered to be an important factor in the phenotypic transformation of RPE cells and in the fibrosis of PRMs (2,50). CTGF has been demonstrated to stimulate the synthesis of collagen by mediating the TGF-β or autocrine signaling pathways (17). Transient transfection has been employed to express a CTGF antisense oligonucleotide and block CTGF gene expression in RPE cells (51). The CTGF antisense oligonucleotide, but not the CTGF sense oligonucleotide, was found to block an increase in the TGF-β1-induced [3H]proline incorporation in the present study. The results suggested that the TGF-β1-stimulated production of ECM proteins is partially mediated by CTGF. In addition, the present study found that the expression of CTGF and the phosphorylation of PI3K and AKT were higher in the TGF-β1 group compared with the control group. It has been proposed that TGF-β induces EMT of RPE cells via classical, Smad-dependent and non-classical, Smad-independent, signaling (2,52). Gressner et al (53) demonstrated that CTGF acts as a Smad2-dependent sensitizer of TGF-B actions in hepatocytes, not a direct activator of Smad2. Our unpublished data indicated that CTGF can induce the phosphorylation of Smad2/3, which needs further exploration.
There are some limitations to the present study. The potential role of microRNAs in preventing PVR by regulating EMT in RPE cells has been extensively studied and this has been helpful in developing new biological markers and therapeutic targets (6,54,55). The present study only found some indirect mechanisms; thus, it is necessary to further explore novel direct mechanisms of CTGF regulation of PVR, including the regulation of CTGF by microRNAs.
In summary, the present study demonstrated that CTGF promoted EMT and the ECM synthesis in PVR via the PI3K/AKT signaling pathway. Its findings suggested that the inhibition of CTGF expression may represent a possible target for the prevention of an excessive wound-healing response by RPE cells, thereby preventing PVR. Further detailed research on the role of CTGF in PVR, along with the latest mechanism including microRNAs and more in vivo studies, will enable the use of CTGF in future clinical applications to alleviate the sufferings of patients with PVR.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Chaowei Tian (Department of Ophthalmology, Xijing Hospital, Fourth Military Medical University, Xi'an, Shaanxi, China) for his technical assistance in collecting PVR membranes.
Glossary
Abbreviations
- α-SMA
α-smooth muscle actin
- BSA
bovine serum albumin
- cDNA
complementary DNA
- CTGF
connective tissue growth factor
- DMEM
Dulbecco's modified Eagle's medium
- ECM
extracellular matrix
- EMT
epithelial-mesenchymal transition
- NC
negative control
- PBS
phosphate-buffered saline
- PFA
polyformaldehyde
- PRM
preretinal membrane
- PVR
proliferative vitreoretinopathy
- RPE
retinal pigment epithelium
- RT-qPCR
reverse transcription-quantitative PCR
- SEM
standard error of the mean
- siRNA
small interfering RNA
- TBST
Tris-buffered saline with Tween-20
Funding Statement
This project was supported by the National Natural Science Foundation of China (grant nos. 81470655, 81570856, 81670863, 81770936 and 81970814).
Funding
This project was supported by the National Natural Science Foundation of China (grant nos. 81470655, 81570856, 81670863, 81770936 and 81970814).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
CG, YH and YW conceived and designed the study. YW, TC, TW and HD performed experiments. YW, TC, TW, WY and GD analyzed the data and wrote the manuscript. CG and YW reviewed and edited the manuscript. GD, YW, and YH supervised the project, provided thoughtful comments and suggestions during this study. YW and TC confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All donors provided written informed consent prior to tissue donation. The study was performed in accordance with the tenets of the Declaration of Helsinki and the protocol was approved by the institutional research ethics committee of the Fourth Military Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tosi GM, Marigliani D, Romeo N, Toti P. Disease pathways in proliferative vitreoretinopathy: An ongoing challenge. J Cell Physiol. 2014;229:1577–1583. doi: 10.1002/jcp.24606. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Li H, Li M, Wang F. Mechanisms of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Discov Med. 2015;20:207–217. [PubMed] [Google Scholar]
- 3.Campochiaro PA. Pathogenic mechanisms in proliferative vitreoretinopathy. Arch Ophthalmol. 1997;115:237–241. doi: 10.1001/archopht.1997.01100150239014. [DOI] [PubMed] [Google Scholar]
- 4.Abu El-Asrar AM, Van den Steen PE, Al-Amro SA, Missotten L, Opdenakker G, Geboes K. Expression of angiogenic and fibrogenic factors in proliferative vitreoretinal disorders. Int Ophthalmol. 2007;27:11–22. doi: 10.1007/s10792-007-9053-x. [DOI] [PubMed] [Google Scholar]
- 5.Sethi CS, Lewis GP, Fisher SK, Leitner WP, Mann DL, Luthert PJ, Charteris DG. Glial remodeling and neural plasticity in human retinal detachment with proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2005;46:329–342. doi: 10.1167/iovs.03-0518. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko H, Terasaki H. Biological involvement of MicroRNAs in proliferative vitreoretinopathy. Transl Vis Sci Technol. 2017;6:5. doi: 10.1167/tvst.6.4.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastor JC. Proliferative vitreoretinopathy: An overview. Surv Ophthalmol. 1998;43:3–18. doi: 10.1016/S0039-6257(98)00023-X. [DOI] [PubMed] [Google Scholar]
- 8.Lee SC, Kwon OW, Seong GJ, Kim SH, Ahn JE, Kay ED. Epitheliomesenchymal transdifferentiation of cultured RPE cells. Ophthalmic Res. 2001;33:80–86. doi: 10.1159/000055648. [DOI] [PubMed] [Google Scholar]
- 9.Dvashi Z, Goldberg M, Adir O, Shapira M, Pollack A. TGF-β1 induced transdifferentiation of rpe cells is mediated by TAK1. PLoS One. 2015;10:e0122229. doi: 10.1371/journal.pone.0122229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Li H, Liu X, Xu D, Wang F. MicroRNA-29b regulates TGF-β1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells by targeting AKT2. Exp Cell Res. 2016;345:115–124. doi: 10.1016/j.yexcr.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Pennock S, Kazlauskas A. Vascular endothelial growth factor A competitively inhibits platelet-derived growth factor (PDGF)-dependent activation of PDGF receptor and subsequent signaling events and cellular responses. Mol Cell Biol. 2012;32:1955–1966. doi: 10.1128/MCB.06668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umazume K, Barak Y, McDonald K, Liu L, Kaplan HJ, Tamiya S. Proliferative vitreoretinopathy in the Swine-a new model. Invest Ophthalmol Vis Sci. 2012;53:4910–4916. doi: 10.1167/iovs.12-9768. [DOI] [PubMed] [Google Scholar]
- 13.Cui JZ, Chiu A, Maberley D, Ma P, Samad A, Matsubara JA. Stage specificity of novel growth factor expression during development of proliferative vitreoretinopathy. Eye (Lond) 2007;21:200–208. doi: 10.1038/sj.eye.6702169. [DOI] [PubMed] [Google Scholar]
- 14.Ciprian D. The pathogeny of proliferative vitreoretinopathy. Rom J Ophthalmol. 2015;59:88–92. [PMC free article] [PubMed] [Google Scholar]
- 15.Platania CBM, Fisichella V, Fidilio A, Geraci F, Lazzara F, Leggio GM, Salomone S, Drago F, Pignatello R, Caraci F, Bucolo C. Topical ocular delivery of TGF-β1 to the back of the eye: Implications in age-related neurodegenerative diseases. Int J Mol Sci. 2017;18:2076. doi: 10.3390/ijms18102076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisichella V, Giurdanella G, Platania CB, Romano GL, Leggio GM, Salomone S, Drago F, Caraci F, Bucolo C. TGF-β1 prevents rat retinal insult induced by amyloid-β (1–42) oligomers. Eur J Pharmacol. 2016;787:72–77. doi: 10.1016/j.ejphar.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, Nguyen D, Ouyang H, Zhang XH, Chen XM, Zhang K. Inhibition of RhoA/Rho-kinase pathway suppresses the expression of extracellular matrix induced by CTGF or TGF-beta in ARPE-19. Int J Ophthalmol. 2013;6:8–14. doi: 10.3980/j.issn.2222-3959.2013.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, Chen Y, Khankan R, Barron E, Burton R, Zhu D, Ryan SJ, Oliver N, Hinton DR. Connective tissue growth factor as a mediator of intraocular fibrosis. Invest Ophthalmol Vis Sci. 2008;49:4078–4088. doi: 10.1167/iovs.07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo CM, Wang YS, Hu D, Han QH, Wang JB, Hou X, Hui YN. Modulation of migration and Ca2+ signaling in retinal pigment epithelium cells by recombinant human CTGF. Curr Eye Res. 2009;34:852–862. doi: 10.3109/02713680903128935. [DOI] [PubMed] [Google Scholar]
- 20.Khankan R, Oliver N, He S, Ryan SJ, Hinton DR. Regulation of fibronectin-EDA through CTGF domain-specific interactions with TGFbeta2 and its receptor TGFβRII. Invest Ophthalmol Vis Sci. 2011;52:5068–5078. doi: 10.1167/iovs.11-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradham DM, Igarashi A, Potter RL, Grotendorst GR. Connective tissue growth factor: A cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blom IE, Goldschmeding R, Leask A. Gene regulation of connective tissue growth factor: New targets for antifibrotic therapy? Matrix Biol. 2002;21:473–482. doi: 10.1016/S0945-053X(02)00055-0. [DOI] [PubMed] [Google Scholar]
- 23.Moussad EE, Brigstock DR. Connective tissue growth factor: What's in a name? Mol Genet Metab. 2000;71:276–292. doi: 10.1006/mgme.2000.3059. [DOI] [PubMed] [Google Scholar]
- 24.Hilton G, Machemer R, Michels R, Okun E, Schepens C, Schwartz A. The classification of retinal detachment with proliferative vitreoretinopathy. Ophthalmology. 1983;90:121–125. doi: 10.1016/S0161-6420(83)34588-7. [DOI] [PubMed] [Google Scholar]
- 25.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst GR. Connective tissue growth factor mediates transforming growth factor beta-induced collagen synthesis: Down-regulation by cAMP. FASEB J. 1999;13:1774–1786. doi: 10.1096/fasebj.13.13.1774. [DOI] [PubMed] [Google Scholar]
- 28.Kothapalli D, Hayashi N, Grotendorst GR. Inhibition of TGF-beta-stimulated CTGF gene expression and anchorage-independent growth by cAMP identifies a CTGF-dependent restriction point in the cell cycle. FASEB J. 1998;12:1151–1161. doi: 10.1096/fasebj.12.12.1151. [DOI] [PubMed] [Google Scholar]
- 29.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 30.Hishikawa K, Oemar BS, Tanner FC, Nakaki T, Lüscher TF, Fujii T. Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J Biol Chem. 1999;274:37461–37466. doi: 10.1074/jbc.274.52.37461. [DOI] [PubMed] [Google Scholar]
- 31.Daniels JT, Schultz GS, Blalock TD, Garrett Q, Grotendorst GR, Dean NM, Khaw PT. Mediation of transforming growth factor-beta(1)-stimulated matrix contraction by fibroblasts: A role for connective tissue growth factor in contractile scarring. Am J Pathol. 2003;163:2043–2052. doi: 10.1016/S0002-9440(10)63562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JT, Foster WJ. Substrate stiffness influences the time dependence of CTGF protein expression in muller cells. Int Physiol J. 2018;1:1. doi: 10.14302/issn.2578-8590.ipj-17-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer P, Wunderlich K, Kain HL, Prunte C, Flammer J. Human connective tissue growth factor mRNA expression of epiretinal and subretinal fibrovascular membranes: A report of three cases. Ophthalmologica. 2002;216:284–291. doi: 10.1159/000063843. [DOI] [PubMed] [Google Scholar]
- 34.Hinton DR, He S, Jin ML, Barron E, Ryan SJ. Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy. Eye (Lond) 2002;16:422–428. doi: 10.1038/sj.eye.6700190. [DOI] [PubMed] [Google Scholar]
- 35.Takayama K, Kaneko H, Hwang SJ, Ye F, Higuchi A, Tsunekawa T, Matsuura T, Iwase T, Asami T, Ito Y, et al. Increased ocular levels of microRNA-148a in cases of retinal detachment promote epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci. 2016;57:2699–2705. doi: 10.1167/iovs.15-18660. [DOI] [PubMed] [Google Scholar]
- 36.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Theocharis AD, Manou D, Karamanos NK. The extracellular matrix as a multitasking player in disease. FEBS J. 2019;286:2830–2869. doi: 10.1111/febs.14818. [DOI] [PubMed] [Google Scholar]
- 41.Polanco TO, Xylas J, Lantis JC., II Extracellular matrices (ECM) for tissue repair. Surg Technol Int. 2016;28:43–57. [PubMed] [Google Scholar]
- 42.Miller CG, Budoff G, Prenner JL, Schwarzbauer JE. Minireview: Fibronectin in retinal disease. Exp Biol Med (Maywood) 2017;242:1–7. doi: 10.1177/1535370216675245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morino I, Hiscott P, McKechnie N, Grierson I. Variation in epiretinal membrane components with clinical duration of the proliferative tissue. Br J Ophthalmol. 1990;74:393–399. doi: 10.1136/bjo.74.7.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Priglinger SG, Alge CS, Kreutzer TC, Neubauer AS, Haritoglou C, Kampik A, Welge-Luessen U. Keratinocyte transglutaminase in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2006;47:4990–4997. doi: 10.1167/iovs.06-0273. [DOI] [PubMed] [Google Scholar]
- 45.Kita T, Hata Y, Miura M, Kawahara S, Nakao S, Ishibashi T. Functional characteristics of connective tissue growth factor on vitreoretinal cells. Diabetes. 2007;56:1421–1428. doi: 10.2337/db06-1644. [DOI] [PubMed] [Google Scholar]
- 46.Zhao X, Zhang LK, Zhang CY, Zeng XJ, Yan H, Jin HF, Tang CS, Du JB. Regulatory effect of hydrogen sulfide on vascular collagen content in spontaneously hypertensive rats. Hypertens Res. 2008;31:1619–1630. doi: 10.1291/hypres.31.1619. [DOI] [PubMed] [Google Scholar]
- 47.Jia P, Hu Y, Li G, Sun Y, Zhao J, Fu J, Lu C, Liu B. Roles of the ERK1/2 and PI3K/PKB signaling pathways in regulating the expression of extracellular matrix genes in rat pulmonary artery smooth muscle cells. Acta Cir Bras. 2017;32:350–358. doi: 10.1590/s0102-865020170050000004. [DOI] [PubMed] [Google Scholar]
- 48.Chen XF, Du M, Wang XH, Yan H. Effect of etanercept on post-traumatic proliferative vitreoretinopathy. Int J Ophthalmol. 2019;12:731–738. doi: 10.18240/ijo.2019.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abu El-Asrar AM, Imtiaz Nawaz M, Kangave D, Siddiquei MM, Geboes K. Osteopontin and other regulators of angiogenesis and fibrogenesis in the vitreous from patients with proliferative vitreoretinal disorders. Mediators Inflamm. 2012;2012:493043. doi: 10.1155/2012/493043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamulescu MA, Chen Y, He S, Spee C, Jin M, Ryan SJ, Hinton DR. Transforming growth factor beta2-induced myofibroblastic differentiation of human retinal pigment epithelial cells: Regulation by extracellular matrix proteins and hepatocyte growth factor. Exp Eye Res. 2006;83:212–222. doi: 10.1016/j.exer.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Yamanaka O, Saika S, Ikeda K, Miyazaki K, Kitano A, Ohnishi Y. Connective tissue growth factor modulates extracellular matrix production in human subconjunctival fibroblasts and their proliferation and migration in vitro. Jpn J Ophthalmol. 2008;52:8–15. doi: 10.1007/s10384-007-0497-3. [DOI] [PubMed] [Google Scholar]
- 52.Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal. 2019;12:eaav5183. doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gressner OA, Lahme B, Siluschek M, Rehbein K, Weiskirchen R, Gressner AM. Connective tissue growth factor is a Smad2 regulated amplifier of transforming growth factor beta actions in hepatocytes-but without modulating bone morphogenetic protein 7 signaling. Hepatology. 2009;49:2021–2030. doi: 10.1002/hep.22850. [DOI] [PubMed] [Google Scholar]
- 54.Usui-Ouchi A, Ouchi Y, Kiyokawa M, Sakuma T, Ito R, Ebihara N. Upregulation of Mir-21 levels in the vitreous humor is associated with development of proliferative vitreoretinal disease. PLoS One. 2016;11:e0158043. doi: 10.1371/journal.pone.0158043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Ye S, Xiao W, Luo L, Liu Y. Differentially expressed microRNAs in TGFβ2-induced epithelial-mesenchymal transition in retinal pigment epithelium cells. Int J Mol Med. 2014;33:1195–1200. doi: 10.3892/ijmm.2014.1688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.