Highlights
-
•
Early aberrant expression of actin subunits can serve as a biomarker for neoplastic transformation.
-
•
Abnormal actin isoform expression can lead to increased migration, cell proliferation and drug resistance.
-
•
Actin can serve as a potential therapeutic target for chemotherapy and also as an indicator for the efficacy of currently used chemotherapeutic agents.
Keywords: Actin, Cancer, Neoplasia, Transformation, Migration, Cytoskeleton
Abstract
Actin is a key structural protein that makes up the cytoskeleton of cells, and plays a role in functions such as division, migration, and vesicle trafficking. It comprises six different cell-type specific isoforms: ACTA1, ACTA2, ACTB, ACTC1, ACTG1, and ACTG2. Abnormal actin isoform expression has been reported in many cancers, which led us to hypothesize that it may serve as an early biomarker of cancer. We show an overview of the different actin isoforms and highlight mechanisms by which they may contribute to tumorigenicity. Furthermore, we suggest how the aberrant expression of actin subunits can confer cells with greater proliferation ability, increased migratory capability, and chemoresistance through incorporation into the normal cellular F-actin network and altered actin binding protein interaction. Studying this fundamental change that takes place within cancer cells can further our understanding of neoplastic transformation in multiple tissue types, which can ultimately aid in the early-detection, diagnosis and treatment of cancer.
Graphical abstract
Introduction
Actin is an important and prominent structural protein that is found in all cells of the body and has a role in multiple cellular functions such as division, migration, chromatin remodelling, and vesicle trafficking. This cytoskeletal protein is composed of several isoforms, all which contribute to its diverse range of functions. Notably, these isoforms mainly differ from one another at in their N-terminal amino acid sequence, but it is currently unclear whether these regions are responsible for the unique function of each isoform (Fig. 1) [1], [2]. In birds and mammals, four of these isoforms are primarily expressed in skeletal, smooth, and cardiac muscle: αskeletal-actin 1 (ACTA1), αsmooth-actin 2 (ACTA2), αcardiac-actin 1 (ACTC1), and γsmooth-actin 2 (ACTG2). The other two isoforms, which are ubiquitously expressed, are βcyto-actin (ACTB) and γcyto-actin 1 (ACTG1) [2]. The varying α, β, and γ subunits play important roles in cell motility, structure, integrity, and intracellular signalling [1], [2]. Interestingly, dysregulation of their functions is often found during malignant transformation of normal cells [3]. This led us to hypothesize that altered actin subunit expression is a hallmark of neoplasia with implications in migration and cell survival. In this review, we look at how an abnormal actin cytoskeleton interacts with tropomyosin, tropomodulin and actin binding proteins (ABPs) to confer neoplastic properties to normal cells [4], [5]. Additionally, we elucidate how early changes to the actin cytoskeleton composition alters rates of actin turnover, F-actin stability, and interactions with actin binding proteins. Lastly, we suggest how this enables oncogenic processes such as increased migration, cell proliferation, and survival in multiple tissue types.
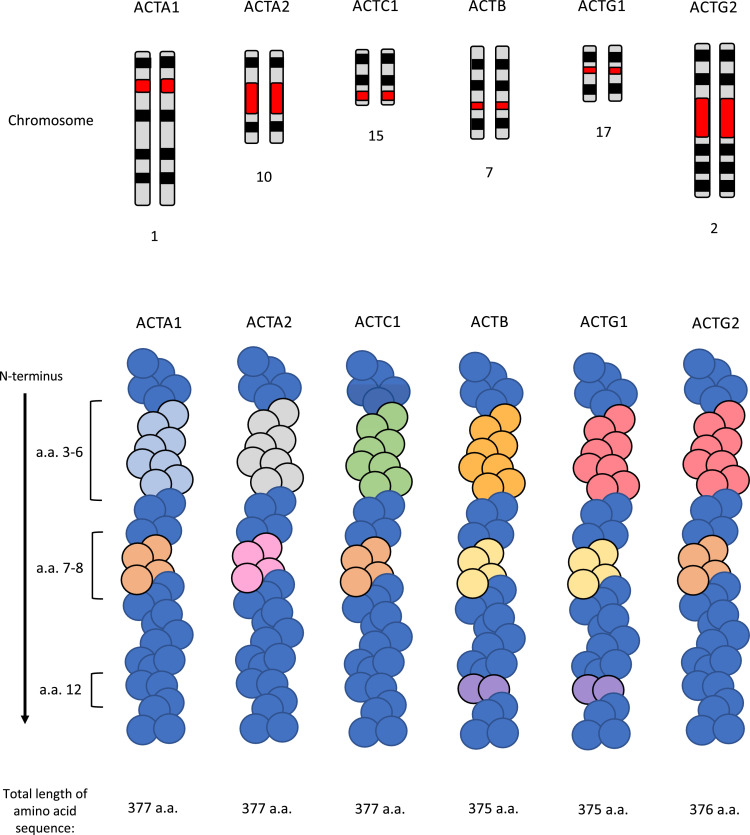
Fig. 1.
A schematic representation of the different actin isoforms. Top panel - karyotype marking of the particular chromosome with the locus of each actin gene marked in red. Bottom panel – actin fibre schematic with regions of variability at the N-terminus of each actin isoform. Regions of the same colour represent an identical amino acid sequence .
Basic overview of actin dynamics
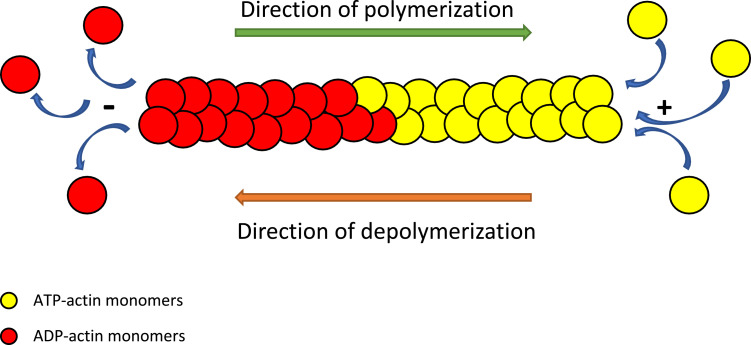
The inherent ability of actin to exert the aforementioned functions within cells depends on the polymerization and depolymerization capabilities of actin fibers. Briefly, the genes encoding actin are first transcribed and translated into globular monomers (G-actin). These actin monomers join the rapidly growing barbed (+) end of the filament in the more stable ATP-state to form fibrous strands (F-actin) which function to maintain structural integrity, support migration, and accomplish cell division [1], [2]. Then, ATP-hydrolysis takes place within the F-actin, and this causes the ADP-monomers to begin to dissociate from the pointed (-) end faster than the ATP-bound ones [1]. This process of steady-state polymerization and depolymerization is known as treadmilling (Fig. 2) [1].
Fig. 2.
Illustration of actin polymerization and steady-state treadmilling. ATP-actin subunits becomes added to the “+” or barbed end whereas the ADP-actin monomers dissociate from the “-” end of the F-actin filament.
However, the actual rates of filament assembly and disassembly are modulated by actin binding proteins (ABPs). One of these ABPs is Arp2/3 whose main function is to create branch points by nucleating the assembly of filaments and also acts to form cross-links [6]. Another well-known actin nucleator is a class of proteins known as formins which have an instrumental role in enhancing the nucleation of actin filament assembly, the bundling of filaments, and binding to the barbed ends to influence the rate of elongation of actin fibers [7]. The actin depolymerizing factor (ADF)/cofilin family of proteins serve many functions within the cell. It is responsible for producing the high rate of turnover which actin monomers are known to experience. Additionally, it functions to depolymerize, nucleate, promote the disassembly of ADP-actin monomers, and also sever actin filaments [6]. While ADF/cofilin dissociates ADP-actin from the pointed end, an ABP called profilin adds ATP-actin to the barbed end, thereby growing the actin filament [6]. Conversely, Gelsolin is an ABP that functions to sever the actin filament as well as to cap the severed ends, thereby preventing re-annealing of the monomers [6]. As the monomers within the polymerized filament “age”, their ATP becomes hydrolysed to ADP, thereby causing them to dissociate [6]. Thus, capping proteins such as profilin, gelsolin and CapZ bind to the barbed-end of the actin filament to regulate actin dynamics by blocking the addition and loss of actin subunits [8]. On the other hand, tropomodulin (Tmod) is a capping protein that binds to the pointed-end of the actin filament (strongly in the presence of tropomyosin) to regulate filament length and assembly [6].
The last actin binding protein that this review will highlight is tropomyosin, because its altered expression levels are closely associated with the rearrangement of microfilament bundles, altered cell morphology, as well as increased cell motility [9]. Its primary role is to stabilize actin filaments to promote filament elongation by reducing the probability of filament severing [10]. It also plays a very important role in determining the steady-state polymerized actin levels within the cell [10]. Notably, the location of tropomyosin on the actin fibers suggests the possibility of competitive binding between tropomyosin and other actin binding proteins [11]. Overall, there are many ABPs present within the cell and they all have an important role in modulating actin dynamics under normal physiological conditions.
Non-redundant roles of the different actin isoforms
Several knockout experiments have been performed in order to elucidate the role of each actin isoform. ACTA1, ACTB and ACTC1 knockouts were embryonic, perinatal or postnatal lethal, whereas knocking out ACTG1, ACTG2, and ACTA2 led to a viable organism with many cardiac, muscular, and vascular developmental defects [12], [13], [14], [15], [16], [17], [18]. In ACTC1 knockout mice, a partial rescue was possible by heart-specific ACTG2 expression, but the hearts were hypodynamic, considerably enlarged, and had lower contractility [16], [19]. Similarly, ACTG1 can be expressed in ACTA1-knockout mice for a partial rescue that yields some developmental defects, still without completely restoring ACTA1 function [15], [19]. Knockout phenotypes of ACTB and ACTG1 are vastly different despite these non-muscle actins showing similar sequence homology, biochemical properties, and coexisting within the same cells [12], [13], [14], [18], [19]. Interestingly, exogenously expressed smooth muscle actin incorporates into stress fibers in smooth muscle cells, but not in endothelial cells [20]. Furthermore, non-muscle actin subunit expression in both muscle and non-muscle cell types results in altered cell shape and poorly formed stress fiber network [20]. These experiments support the hypothesis that actin isoforms have distinct functions depending on subunit type and the cell type in which they are expressed.
Notably, the directions for the functional complexity of each isoform may lie in their mRNA sequence. Work by Vedula et al. showed that although the amino acid sequence at the N-termini of both ACTB and ACTG1 are very similar, editing the nucleotide sequence of ACTG1 to encode an ACTB-like protein can compensate for the loss of ACTB [21]. This suggests that although the major differences between the isoforms are found at the level of their amino acids, the mRNA sequence may be more important in determining the ultimate protein function since it directs the rate of synthesis and type of secondary protein modifications [21], [22]. Ultimately, as this review further elaborates, the differences present within the genes encoding each isoform are associated with a particular set of neoplastic functions.
ACTA1
ACTA1 is a gene that encodes the skeletal muscle α-actin which is the main isoform found in skeletal muscle, and is essential for muscle contraction [23]. Bioinformatics analyses have revealed that ACTA1 gene expression is altered in many cancers. For example, it is downregulated in head and neck squamous cell carcinomas and this is associated with tumorigenesis [24], [25]. This decrease in ACTA1 expression can serve as a prognostic marker for the poor clinical outcome of head and neck squamous cell carcinoma patients (Table 1) [24], [25]. Similarly, it is downregulated in colorectal cancer, prostate cancer, and pancreatic adenocarcinoma through DNA hypermethylation and this is associated with aggressive carcinogenesis (Table 1) [26], [27], [28], [29]. In prostate cancers with the PTEN mutation, ACTA1 expression is significantly correlated with shorter patient survival [30], [31]. Interestingly, stromal fibroblasts in prostate cancer tissue samples show increased expression of the cancer-associated fibroblast marker ACTA1 (Table 1) [31]. The presence of ACTA1 expressing fibroblasts in the prostate cancer stroma is associated with increased tumour metastatic potential [31]. In the case of oral squamous cell carcinoma, high expression of ACTA1 is associated with shorter survival (Table 1) [32]. Additionally, in basal-like breast cancer, ACTA1 is a biomarker that is associated with chemoresistance (Table 1) [33].
Table 1.
Summary of aberrant actin isoform expression in various cancers.
| Actin Isoform | Cancer(s) in which it is aberrantly expressed | Upregulated or Downregulated | mRNA or Protein | Tumour cells or tumour tissue analysed | Clinical Outcome | Reference |
|---|---|---|---|---|---|---|
| ACTA1 | Head and neck squamous cell carcinomas | Downregulated | mRNA and protein | Cells and tissue | Correlated with greater aggressiveness and shorter survival | [24], [25] |
| Colorectal cancer | Downregulated | mRNA | Tissue | – | [26] | |
| Prostate | Downregulated in the cancer, Upregulated in associated cancer-associated fibroblasts | mRNA | Cells and tissue | – | [29], [31] | |
| Pancreatic adenocarcinoma | Downregulated | mRNA | Cells and tissue | – | [28] | |
| Oral squamous cell carcinoma | Upregulated | mRNA | Tissue | Shorter survival | [32] | |
| Basal-like breast cancer | Upregulated | mRNA | Tissue | Associated with drug resistance | [33] | |
| ACTA2 | Lung adenocarcinoma | Upregulated | mRNA and protein | Cells and tissue | Shorter survival and early distant metastasis | [44] |
| HER2+ breast cancer | Upregulated | mRNA and protein | Cells and tissue | Shorter survival | [43] | |
| Colorectal cancer | Upregulated in the cancer, Upregulated in associated cancer-associated fibroblasts | mRNA and protein | Cells and tissue | Shorter disease-free survival | [45], [49] | |
| Bladder cancer | Upregulated | mRNA | Tissue | Shorter survival | [42] | |
| Non-small cell lung cancer | Upregulated | Protein | Cells | – | [46] | |
| Head and neck cancers | Upregulated in associated cancer-associated fibroblasts | mRNA and protein | Cells and tissue | Shorter survival | [47] | |
| Pancreatic cancer | Upregulated in associated cancer-associated fibroblasts | mRNA and protein | Cells and tissue | – | [48] | |
| ACTB | Sarcoma | Upregulated | Protein | Cells | – | [56] |
| Colon adenocarcinoma | Upregulated | Protein | Cells | – | [53] | |
| Hepatoma | Upregulated | Protein | Cells | Greater metastatic potential | [54] | |
| Melanoma | Upregulated | mRNA | Cells | – | [58] | |
| Gastric cancer | Upregulated | mRNA | Tissue | – | [62] | |
| oesophageal carcinoma | Upregulated | Protein | Tissue | – | [59] | |
| Non-small cell lung cancer | Upregulated | mRNA | Cells and tissue | – | [61], [63] | |
| Breast cancer | Upregulated | mRNA | Cells | – | [60] | |
| Lymphoma | Upregulated | Protein | Cells | – | [66] | |
| Cervical cancer | Downregulated | Protein | Cells | [65] | ||
| ACTC1 | Head and neck cancer | Upregulated | Protein | Cells | Greater distant metastasis | [76] |
| Bladder cancer | Upregulated | mRNA | Tissue | – | [79] | |
| Urothelial cancer | Upregulated | mRNA | Tissue | – | [79] | |
| Prostate cancer | Upregulated | mRNA | Tissue | Predictor of the diseased/cancer state | [78] | |
| Non-small cell lung cancer | Upregulated | mRNA | Cells | – | [80] | |
| Breast cancer | Upregulated | mRNA | Cells | Multi-drug resistance | [81], [82] | |
| Glioblastoma | Upregulated | mRNA | Cells | Greater distant metastasis | [77], [83] | |
| ACTG1 | Skin cancer | Upregulated | mRNA | Tissue | – | [85] |
| Hepatocellular carcinoma | Upregulated | Protein | Cells and tissue | Shorter survival and high expression is correlated to advanced tumour stage | [87] | |
| Non-clear cell renal cell carcinoma | Upregulated | mRNA | Tissue | – | [88] | |
| Colorectal cancer | Upregulated | Protein | Cells | Greater distant metastasis | [90] | |
| Lung cancer | Upregulated | mRNA | Cells | – | [89] | |
| Cervical cancer | Upregulated | mRNA | Tissue | Can serve as a biomarker for malignancy. | [91] | |
| Acute lymphoblastic leukaemia | Downregulated | Protein | Cells | Resistance to microtubule-targeting agents | [93] | |
| Neuroblastoma | Downregulated | Protein | Cells | Greater distant metastasis | [86] | |
| Breast cancer | Downregulated | Protein | Cells | Drug resistance to anti-mitotic agents | [94] | |
| Lung cancer | Upregulated | Protein | Tissue | – | [92] | |
| Colon cancer | Upregulated | Protein | Tissue | – | [92] | |
| Prostate cancer | Upregulated | Protein | Cells | – | [92] | |
| Pancreatic cancer | Upregulated | Protein | Cells | – | [92] | |
| Testicular cancer | Upregulated | Protein | Cells | – | [92] | |
| Colorectal cancer | Upregulated | Protein | Cells | – | [92] | |
| Oral squamous cell carcinoma | Upregulated | Protein | Cells | – | [92] | |
| ACTG2 | Hepatocellular carcinoma | Upregulated | mRNA and protein | Cells and tissue | Correlated with EMT and aggressiveness | [96], [97] |
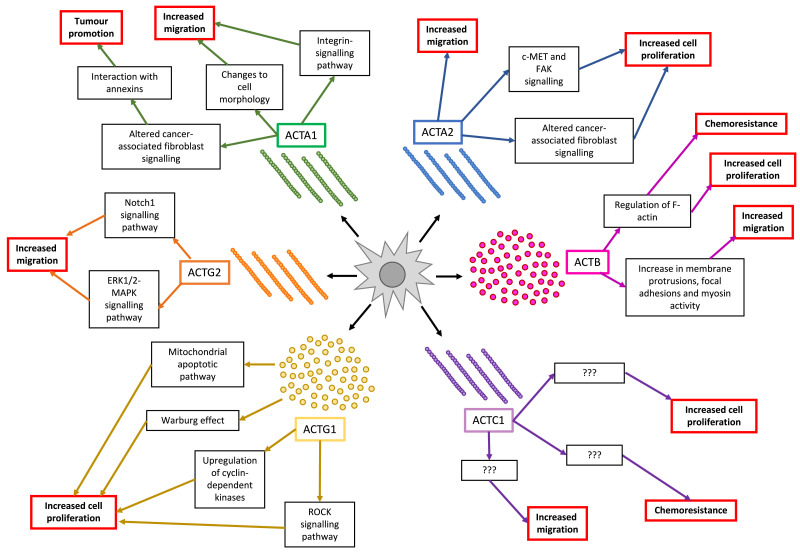
There are many possible mechanisms by which the ACTC1 protein promotes oncogenesis. One possibility could be through annexins which are Ca2+-dependant, phospholipid-binding proteins that function in vesicle trafficking, cell proliferation and apoptosis. Annexin expression is correlated with shorter survival, tumorigenesis, and the progression of malignant ovarian cancer. Notably, annexins have extensive physical interactions with ACTA1 [34]. ACTA1 could also regulate cell migration through its involvement in the integrin-signalling pathway (Fig. 2) [35]. More specifically, during conditions of mechanical strain on normal skeletal muscle, integrin β1-mediated signalling results in the activation of RhoA which induces the activity of several promoters, one being that of ACTA1, thereby increasing the cytoplasmic levels of the ACTA1 subunit [36]. Another downstream protein that is activated by RhoA is RhoA kinase (ROCK), which will result in the inhibition of the F-actin-severing ability of the protein cofilin [37]. This in turn will promote stress fiber formation [37]. When ACTA1 subunits are aberrantly expressed in the cytoplasm, more of them are readily available to form F-actin and stress fibers, which are implicated in cytoskeletal stabilization, cell survival, migration and proliferation, all of which are downstream of the integrin-signalling pathway [37].
In the case of pancreatic ductal adenocarcinoma, expression of ACTA1 is a characteristic feature of cancer-associated fibroblasts (Fig. 2) [38]. Stromal progenitor cells and fibroblast-like cells show increased ACTA1 expression when undergoing a morphological change from a spherical to flattened phenotype (Fig. 2) [39]. Coincidentally, this change in cell shape also occurs during the epithelial-mesenchymal transition of cells undergoing malignant transformation, and confers a more migratory phenotype to cells [40].
ACTA2
ACTA2 is a gene that encodes for smooth-muscle actin found in the vasculature. It is primarily located in the microfilaments bundles of smooth muscle cells and functions in its contractility (i.e. during body temperature homoeostasis) [41]. Interestingly, increased ACTA2 expression is also associated with more distant metastasis and unfavourable prognosis of lung adenocarcinoma, HER2+ breast cancer, early-onset colorectal cancer, bladder cancer, and non-small cell lung cancer (Table 1) [42], [43], [44], [45], [46]. Furthermore, the acquisition of ACTA2 is found in “activated” myofibroblastic cancer-associated fibroblasts of head and neck cancers, colorectal cancer, and pancreatic cancer (Table 1) [47], [48], [49].
Mechanistically, ACTA2 plays an important role in the contractility of myofibroblasts, and its elevated levels serve as a marker for oncogenic transformation of these cancer-associated fibroblasts (Fig. 2). [50]. Lee et al [44]. previously reported that ACTA2 regulates c-MET and FAK expression, as well as positively influences the metastatic potential of lung adenocarcinoma, and affects the prognosis (Fig. 2) [44], [51]. Work by Milewicz et al [52]. showed that ACTA2 proliferative and secretory activities as well as transition from a contractile to a synthetic phenotype was the underlying mechanism of vasculogenesis [41], [52]. Furthermore, explanted smooth muscle cells and myofibroblasts from patients with mutations in ACTA2 revealed that the increased proliferation of smooth muscle cells contributed to occlusive diseases. Mutations to the ACTA2 gene can also lead to smooth muscle cell proliferation and subsequent remodelling of the vascular wall [41]. Taken together, this suggests that deregulated α-subunit expression can lead to uncontrolled proliferation and the activation of metastatic genes, which can thereby result in neoplastic transformation (Fig. 2).
ACTB
ACTB is a ubiquitously expressed cytoskeletal protein that plays a role in a wide variety of cellular functions such as cell growth, cell division, cell motility, immune response, gene expression, maintenance of cell stability and cytoskeletal formation [13], [53], [54], [55]. These functions suggest that the deregulation of ACTB can play a role in cancer pathogenesis. Interestingly, increased ACTB levels have been reported in a number of highly metastatic tumour cell lines - sarcomas, colon adenocarcinomas, and hepatomas (Table 1) [53], [54], [56]. Its expression is found to be much higher than normal in invasive melanomas, gastric cancer, oesophageal carcinomas, non-small cell lung cancers, renal, aggressive breast cancers, and cervical cancers (Table 1) [57], [58], [59], [60], [61], [62], [63], [64], [65]. Notably, ACTB has also been found to be upregulated in drug-resistant lymphomas (Table 1) [66]. This isoform is essential for the migratory capability of normal cells as knockdown results in decreased membrane protrusions at the leading edge of migrating cells, increased focal adhesion formation and increased expression of genes that regulate myosin activity [13].
The dynamic polymerization of actin is shown to contribute to tumour malignancy in some cancers [67], [68], [69], [70]. In three adenocarcinoma cell lines with high ACTB expression, G-actin levels were found to have decreased while F-actin levels increased (Fig. 3). These cells demonstrate high metastatic potential and invasion, which is in line with the idea that the high level of actin polymerization is necessary for the formation of pseudopods and cancer cell invasion to the surrounding tissues (Fig. 3) [70], [71]. ACTB may also play an essential role in maintaining cell growth potential. Work by Kwiatkowski et al [72]. showed that loss of ACTB methylation by a protein called SET Domain Containing 3 (an actin histidine methyltransferase) leads to the depletion of F-actin and subsequent loss of cytoskeleton integrity (Fig. 3). This destabilization of the actin cytoskeleton through increased degradation of F-actin is a massive energy drain in the cell (causing up to 50% of total ATP consumption), and can in turn cause the cell to resort to anaerobic metabolism, which increases lactate production [72]. The accelerated degradation of the hypomethylated F-actin fibers is the reason for the increased demand of ATP and shift in cellular metabolism towards glycolysis [72]. The interplay between F-actin stability and ATP consumption is an important phenomenon to considergiven the increased metabolic demands in cancer cells [73].
Fig. 3.
A flow diagram of the different actin isoforms and how they contribute to tumorigenicity. ACTA1 = αskeletal-actin 1, ACTA2 = αsmooth-actin 2, ACTB = βcyto-actin, ACTC1 = αcardiac-actin 1, ACTG1 = γcyto-actin 1 and ACTG2 = γsmooth-actin 2.
ACTC1
ACTC1 is a gene that encodes the major protein of the cardiac sarcomere thin filaments, which are responsible for cardiac muscle contraction [74]. Although it is expressed mainly in the heart and at lower levels in skeletal muscle, there is evidence for cardiac actin isoform expression during early mammalian neurodevelopment [75]. Interestingly, ACTC1 expression is shown to recur in many cancer types such as brain, head and neck, bladder, urothelial, prostate, lung, and breast cancers (Table 1) [76], [77], [78], [79], [80], [81], [82]. Furthermore, in patients with the malignant brain tumour glioblastoma (GBM), high ACTC1 mRNA expression may serve as a novel prognostic and invasion marker since it has been correlated with shorter survival and more frequent distant recurrence (Table 1) [77]. Wanibuchi et al. showed that ACTC1 mRNA knockdown in the U87 glioblastoma cells leads to impaired migration, which indicates its potential role in cell motility [83]. High ACTC1 expression has been associated with a lack of sensitivity to the mitotic inhibitor Paclitaxel in a lung cancer cell line and is also highly correlated with altered cell surface receptor linked signal transduction through sonic hedgehog [80]. Microtubule-binding inhibitors of mitosis such as taxanes result in altered ACTC1 gene expression in breast and lung cancers [81], [82]. It has also been previously reported that ACTC1 is a hub gene that confers chemoresistance in various tumours and its expression is upregulated in multi-drug resistant breast cancer cells (Fig. 3) [82]. Thus, the presence of elevated levels of ACTC1 may actually play a role in cancer cell survival, but the mechanism by which it does so remains to be elucidated.
ACTG1
ACTG1 encodes for the cytoskeleton protein γ-actin which functions in non-muscle cells and is abundant in the auditory hair cells of the cochlea. This actin isoform is essential for the shape and function of stereocilia of hair cells, and plays a role in the internal cell motility [84], [85]. Suppression of this isoform results in impaired migration of SH-EP neuroblastoma cells [86]. Interestingly, work by Dong et al [85]. showed that ACTG1 is expressed at significantly higher levels in skin cancer tissue (Table 1). It was also found that overexpressing ACTG1 in a squamous cell carcinoma cell line promotes growth and migration via inhibition of the ROCK pathway, leading to the reduction of myosin light chain phosphorylation (Fig. 3) [85]. In hepatocellular carcinomas, ACTG1 is overexpressed compared to adjacent normal tissues, and this promotes proliferation by upregulating cyclin-dependant kinases, inhibiting the mitochondrial apoptotic pathway, and promoting the Warburg effect by increasing cellular aerobic glycolysis, all which promote tumorigenicity (Fig. 3) [87]. A potential mechanism through which this works was found in non-clear cell renal cell carcinomas in which ACTG1 fuses with micropthalmia-associated transcription factor (MITF), and this dimer drives the transcription of downstream genes to promote anchorage-independent growth [88]. Corroborating with this finding, high ACTG1 expression is associated with greater metastatic potential and worse prognosis in hepatocellular carcinomas, colorectal cancer, lung cancer, cervical cancer (Table 1) [87], [89], [90], [91]. The overexpression of ACTG1 has been associated with many malignant and invasive cancer types such as breast, lung, colon, prostate, pancreatic, testicular, colorectal, and oral squamous cancers (Table 1) [92].
Work by Dugina et al [92]. shows that, the upregulation of γ-actins is induced by the downregulation of β-actins, while keeping the total cytoplasmic actin levels relatively the same, which supports the presence of endogenous compensatory mechanisms to maintain the homoeostasis of actin isoforms. Interestingly, the downregulation of β-actin leads to a more motile cell phenotype while the downregulation of γ-actin induced a more contractile one. Overexpressing β-actin lead to a more “normal” phenotype and increasing γ-actin led to a more transformed one with a change in morphology to become more fan-like [92]. This suggests that the endogenous balance of actin isoforms can play a role in altering cell morphology and producing certain cancer phenotypes.
Interestingly, at the microtubule level, a decrease in ACTG1 levels in acute lymphoblastic leukaemia is associated with resistance to anti-microtubule drugs and protects microtubules from the action of both stabilizing and destabilizing agents (Table 1) [93]. This shows that the expression level of ACTG1 plays a role in regulating microtubule integrity. Microtubule disruption can activate RhoA and induce actomyosin contraction via myosin phosphorylation, which can in turn trigger tumorigenesis (Fig. 3) [93]. An experiment was performed in which ACTG1 was depleted in neuroblastoma and breast cancer cells, and this resulted in centrosome amplification, subsequent formation of multipolar spindles, as well as improper chromosome segregation, all which resulted in mitotic abnormalities [94]. Thus, γ-actin plays a role in maintaining centrosome integrity and regulating mitotic progression.
ACTG2
The final actin isoform that will be discussed is ACTG2, which is specifically expressed in smooth muscle cells of the intestinal and urogenital tracts, and functions in the contraction of smooth muscle in these organs [95]. In the context of hepatocellular carcinoma, ACTG2 is associated with epithelial to mesenchymal transition and the stem like properties of those cells. It also serves as a marker for aggressiveness of this cancer (Table 1) [96], [97]. A reason for the increased oncogenic potential/aggressiveness and invasive ability of these cancer cells would be the activation of Notch1 signalling as well as ERK1/2 activation by ACTG2 (Fig. 3) [27], [92], [97]. Activation of the ERK-MAPK pathway leads to lamellipodial protrusions, which then leads to the subsequent activation of the WAVE2/Arp2/3 polymerization complex, resulting in actin reorganization [98], [99], [100], [101]. Although this particular isoform has an important physiological role, literature on its dysregulation in the context of cancer is limited.
The role of actin in mitochondrial apoptotic signalling
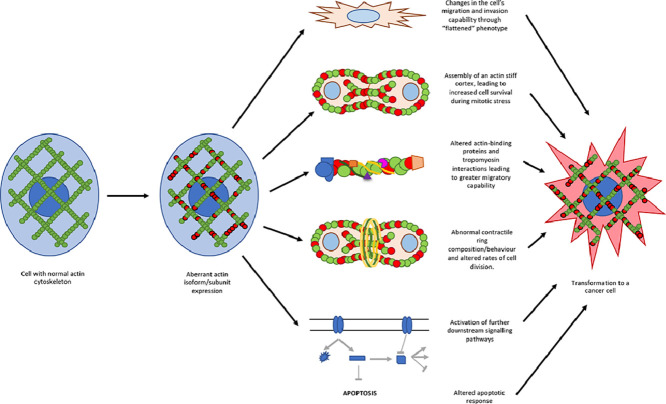
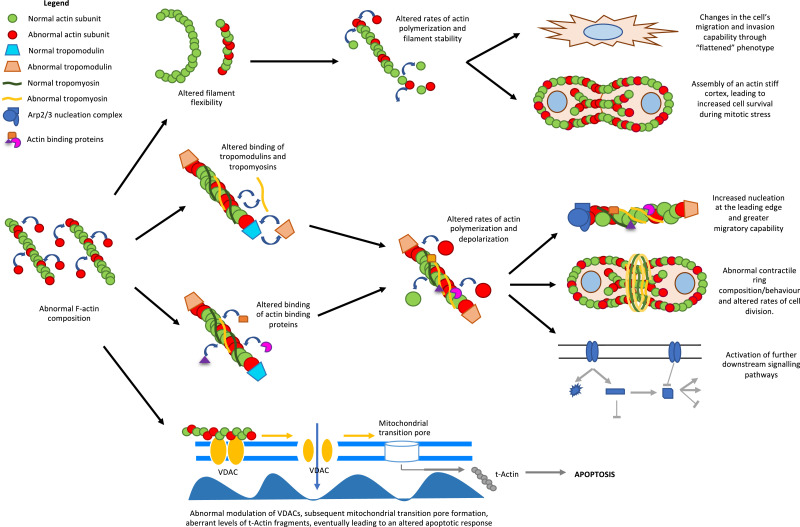
Actin filaments can facilitate apoptosis signalling in cells. In fact, the polymerization of actin can regulate apoptosis via the intrinsic pathway. The disruption of actin filament dynamics by the drug Cytochalasin D, which binds the barbed end of F-actin preventing polymerization, can induce caspase 3-mediated apoptosis in T-lymphocytes [102]. Additionally, the release of pro-apoptotic Bmf from myosin V-actin complex upon Cytochalasin D treatment has also been found to induce mitochondria mediated apoptosis [103]. In mouse fibrosarcoma cells, Cytochalasin D induces mitochondrial cytochrome c release, which is a marker of mitochondrial permeability transition pore formation [104]. In yeast cells, actin point mutations that result in reduced actin dynamics or F-actin stabilizing drug Jasplakinolide increase the cell susceptibility to apoptosis due to mitochondrial membrane depolarization and accumulation of reactive oxygen species (ROS) [105]. α G-actin from muscle is more effective than β or γ G-actin at reducing the conductance of mitochondrial voltage-dependant anion channels (VDACs) in the yeast Neurospora crassa [106]. This modulation of VDAC is lost in the presence of F-actin stabilization (Fig. 4) [106]. These observations indicate that alterations in actin dynamics and actin subunit expression may regulate the mitochondrial signal for intrinsic apoptosis.
Fig. 4.
Flow diagram of the potential mechanisms by which altered F-actin composition can promote tumorigenicity. .
Another possible mechanism whereby actin isoforms regulate cellular apoptosis is through modulating the opening of VDACs through direct or indirect protein-protein interactions [106], [107]. Apart from regulating mitochondrial membrane potential, actin could also act downstream of mitochondrial transition pore formation. To elaborate, actin has been shown to be cleaved by caspases to 32-kDa (Fractin) and 15-kDa (tActin) fragments [108]. The tActin fragment then undergoes N-myristoylation to target it to the mitochondria where it leads to mitochondrial transition pore formation, the caspase signalling cascade, and cellular morphological changes that resemble those observed in apoptotic cells [108], [109]. Here, the change in isoform levels would alter formation of the tActin fragment in response to mitotic stress. Therefore, aberrant expression of actin isoforms may alter the propensity of cancer cells to undergo apoptosis (Fig. 4).
The effect of subunit composition on actin polymerization
The incorporation of new subunits into the already established F-actin network may disrupt its normal function (Fig. 4). It is believed that muscle and non-muscle actins do not incorporate into the same filaments in-vivo due to their higher amino acid divergence [110]. However, β- and γ-isoforms, which only differ by 4 amino acids in the N-terminal region distant from polymerization interfaces, can co-polymerize in mixtures of purified recombinant protein (Fig. 1) [111]. Under normal conditions, it has been found that ACTC1 is able to incorporate into ACTA2 polymers in-vitro whereas ACTB and ACTG1 cannot [20]. The different localization of the ACTA1, ACTA2, ACTC1 and ACTG2 isoforms suggest that they may not co-polymerize in cells. For example, ACTC1 is localized at the sarcomere of cardiac (and muscle) cells, and co-expression of ACTA1 with ACTC1 and ACTA2 with ACTG2 did not affect localization of sarcomeric proteins [20], [112], [113], [114].
To test the different stabilities of polymers formed by actin isoforms, rheological experiments were performed which revealed that the ACTA1 gel was most elastic, smooth muscle ACTB and ACTG2 gels were less elastic, and the cytoplasmic ACTB gel did not form an elastic gel at all [113], [115]. Furthermore, the polymers formed by β- and γ-actin have a lower stability compared to sarcomeric α-actin, and in yeast, this lower stability of F-actin results in greater filament fragmentation [113]. The rate of filament turnover has a role in the mitotic stress response as well as in migration and invasion of cancer cells (Fig. 4) [116], [117]. To support this further, it has been demonstrated that polyploidal giant cancer cells as well as breast cancer cells have more stress fibers of greater thickness and length [118], [119]. These cells also show an upregulation of actin cytoskeletal elements, which results in stiffer gross-tumour rheological properties as well as increased migratory capability [119].
It has been found that α and β-actin as well as α and γ-actin copolymers show a linear relationship between the mixing ratio of actin subunits with respect to each other, as well as their resulting ATPase activity [120]. This hybrid F-actin behaves correspondingly to its isoform composition suggesting the independent function of each actin compared to one another [120]. This is interesting because β- and γ- actin polymers differ in their polymerization kinetics, inorganic phosphate release and treadmilling [111]. These biochemical differences between the two isoforms were exacerbated when Ca2+ was used with the actin rather than Mg2+ [19], [111]. The preference for certain cations may allow cells to rapidly respond to signals by using a specific actin isoform [19], [111]. A limitation that should be noted is that the co-polymerization of actin isoforms has only been studied in select cell types as it is generally assumed that copolymers do not commonly exist in vivo [120].
The rates of polymerization of F-actin appear to also depend on the isoform of the constituent subunits. α-, β- and γ-actins all have different rates of polymerization, resulting in different filament stabilities, elongation and turnover rates [111], [121], [122], [123]. This in turn could affect cell survival during mitosis since cells undergoing mitosis experience increased plasma membrane tension and actively assemble a stiff actin cortex (Fig. 4) [124], [125], [126]. Taken together, the subunit composition of the F-actin network plays an important role in maintaining cell integrity during mitosis and abnormal constituents within the polymerized filament may result in a disease phenotype.
The role of actin isoforms in altering interactions with tropomyosin
Changes in tropomyosin (TM) isoform expression have been shown to accompany neoplastic cell transformation [127]. Furthermore, different TM isoforms show binding specificity to different F-actin isoforms [127]. When a human, rodent or chick fibroblasts are transfected with a non-cell-type specific isoform, the expression of its associated TMs is altered, suggesting that actin microfilament composition regulates tropomyosin expression [128], [129], [130]. It has been found that reversing this actin-initiated altered tropomyosin expression can also revert the cancer phenotype (i.e. cell spreading, microfilament organization and contact-inhibited cell growth) to a large extent in many cell types [127], [131], [132], [133], [134], [135]. However, the synergistic restoration of the proper expression levels of multiple TM isoforms reverted the actin organization and ultimately the cancer phenotype of neuroblastomas as well as Ras-transformed NIH3T3 cells to a much greater extent than just one isoform alone [127], [135], [136].
Mechanistically, aberrant actin isoform expression in cancer can result in abnormal actin-tropomyosin binding interactions within the cells, thereby causing greater actin polymerization at the leading edge and subsequent cellular contraction (Fig. 4) [137]. Dysregulated actin dynamics can be seen when an alteration to the filament occurs since this disrupts the electrostatic contacts between the F-actin and TM [138]. Altering TM-binding would affect the binding of ABPs such as cofilin as well as the loss of inhibition of actin filament branching and nucleation at the leading edge by the Arp2/3 complex. This will lead to altered polymerization/depolymerization kinetics and promote cell migration (Fig. 4) [11], [139], [140], [141]. (Fig. 4) [141]. Furthermore, the resulting altered TM expression can work in conjunction with actin to affect rates of mammalian cell division and regulate cytokinesis through TM localization in the contractile ring of dividing cells to maintain cell stiffness (Fig. 4) [142], [143]. Lastly, the abnormal expression of TM can impact organelle trafficking and the contacts of different myosin motors with actin, all which all which affects downstream cellular pathways (Fig. 4) [144], [145], [146].
By the same token, different tropomodulin (Tmod) isoforms are also found in regions where their preferred actin isoform binding partner is located. For example, Tmod1 is abundant in post-mitotic, terminally differentiated cells [147]. Tmod3 is mainly found in dynamic cellular contexts such as those of the dendritic actin network in the leading lamellipodia of migrating endothelial cells as well as in proliferating erythroblasts [147]. Tmods1 and 3 cap the pointed ends of ACTA1, ACTB and ACTG1 [147]. However, in the absence of tropomyosin, only Tmod3 could sequester ACTB and ACTG1, but not ACTA2 [147]. Tmod1 can induce conformation changes in α/βTM and ACTA1, thereby potentially activating downstream molecular pathways (Fig. 3) [147].
Altered ABP interactions due to changes in actin polymerization
Differences in actin isoforms can be found with the acidic residues at the NH2-terminus between actin isoforms, which acts like a fishing rod to recruit ABPs (Fig. 1) [148], [149], [150]. Thus, variations in this region can modulate the binding affinities of different ABPs such as cofilin, utropin, formin and myosin, resulting in altered actin polymerization kinetics, cell migration, adhesion, cytokinesis, and cytoskeletal maintenance (Fig. 4) [151], [152], [153], [154], [155], [156]. For example, the greater affinity of cofilin to β and γ-actins compared to α-actin implies that smaller amounts of cofilin will cause those actin filaments to twist and sever [157]. Furthermore, the upregulation of TMs 1.1, 1.2 and 3.1, which is a consequence of aberrant actin isoform expression, can cause increased binding of gelsolin, which will also increase its rates of actin assembly and disassembly [158]. Taken together, this suggests that changes in the rates of actin assembly-disassembly, polymerization, elongation and severing which can have downstream effects in the intracellular trafficking, cell migration, and proliferation.
Clinical translation
An understanding of the diversity of actin subunit expression and function in multiple cancers may help to stratify patients into higher risk groups which may benefit from different treatment strategies. Furthermore, understanding how expression of subunits such as ACTA1, ACTA2, ACTB, ACTC1, and ACTG1 can promote chemoresistance against anti-microtubule agents in certain tumours could help to design new therapies to overcome such resistance. Drugs that disrupt actin polymerization or induce filament severing have been previously described; however, a major challenge in using these drugs in patients is that they would disrupt actin function in all cells in the body [159]. Recently, a more targeted approach aiming to disrupt specific tropomyosin interactions with actin has been presented. Small molecule inhibitors of TM 3.1 such as TR100 and ATM-3507 are used to destabilize TM 3.1-containing filaments with great effectiveness in vitro and in vivo. [160], [161], [162]. Another approach to target the actin cytoskeleton would be through the use of migrastatic agents that target actin polymerization.These include actin destabilizing compounds (i.e. cytochalasins, geodiamolides, and latrunculins), as well as stabilizing compounds that initiate dysregulated polymerization, monomer depletion, or formation of actin aggregates (i.e. jasplakinolide, chondramide, and cucurbitacin). The goal of this approach is to effectively inhibit cell invasion and metastasis [163]. Further understanding of the specific interactions of actin subunits that result in sustaining migration and cell survival may provide new avenues for rational design of drugs targeting aberrant actin signalling in cancer cells with less toxicity to normal cells.
Conclusion
In conclusion, actin has diverse biological function beyond just maintaining the structural integrity of cells or regulating cell motility. The actin composition of cells and subsequent altered F-actin properties can contribute to neoplastic transformation by driving tumour growth and promoting metastasis. Studying this fundamental change that takes place within cancer cells can further our understanding of oncogenesis. Furthermore, aberrant actin isoform expression could be used as a biomarker for the early onset of cancer, serve as a potential therapeutic target, and even act as an indicator for the efficacy of currently used chemotherapeutic agents.
Author contributions
Rahul Suresh: Methodology, Writing - original draft preparation, Visualization
Roberto Jose Diaz: Conceptualization, Writing - reviewing and editing, Supervision
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
Foundation of the Department of Neurosurgery and the Montreal Neurological Institute for funding the costs of publication.
Footnotes
Conflict of Interest: On behalf of all authors, the corresponding author states that there is no conflict of interest
References
- 1.Dominguez R., Holmes K.C. Actin structure and function. Ann. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrin B.J., Ervasti J.M. The actin gene family: function follows isoform. Cytoskeleton (Hoboken) 2010;67(10):630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruddon R.W. Cancer Medicine. 6th edition ed. BC Decker; Hamilton (ON): 2003. What makes a cancer cell a cancer cell? [Google Scholar]
- 4.Franklin-Tong V.E., Gourlay C.W. A role for actin in regulating apoptosis/programmed cell death: evidence spanning yeast, plants and animals. Biochem. J. 2008;413(3):389–404. doi: 10.1042/BJ20080320. [DOI] [PubMed] [Google Scholar]
- 5.Stehn J.R. A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 2013;73(16):5169–5182. doi: 10.1158/0008-5472.CAN-12-4501. [DOI] [PubMed] [Google Scholar]
- 6.dos Remedios C.G. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83(2):433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 7.Courtemanche N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018;10(6):1553–1569. doi: 10.1007/s12551-018-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards M. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 2014;15(10):677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helfman D.M. Tropomyosin. Springer; 2008. Tropomyosin as a regulator of cancer cell transformation; pp. 124–131. [DOI] [PubMed] [Google Scholar]
- 10.Gunning P.W. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 2015;128(16):2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- 11.Lehman W. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 2000;302(3):593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 12.Belyantseva I.A. Gamma-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA, 2009;106(24):9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunnell T.M. beta-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell. 2011;22(21):4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunnell T.M., Ervasti J.M. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton (Hoboken) 2010;67(9):564–572. doi: 10.1002/cm.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford K. Mice lacking skeletal muscle actin show reduced muscle strength and growth deficits and die during the neonatal period. Mol. Cell. Biol. 2002;22(16):5887–5896. doi: 10.1128/MCB.22.16.5887-5896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc. Natl. Acad. Sci. USA. 1997;94(9):4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schildmeyer L.A. Impaired vascular contractility and blood pressure homeostasis in the smooth muscle alpha-actin null mouse. FASEB J. 2000;14(14):2213–2220. doi: 10.1096/fj.99-0927com. [DOI] [PubMed] [Google Scholar]
- 18.Tondeleir D. Cells lacking beta-actin are genetically reprogrammed and maintain conditional migratory capacity. Mol. Cell Proteomics. 2012;11(8):255–271. doi: 10.1074/mcp.M111.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vedula P., Kashina A. The makings of the 'actin code': regulation of actin's biological function at the amino acid and nucleotide level. J. Cell Sci. 2018;131(9) doi: 10.1242/jcs.215509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mounier N. Transfected muscle and non-muscle actins are differentially sorted by cultured smooth muscle and non-muscle cells. J. Cell Sci. 1997;110(Pt 7):839–846. doi: 10.1242/jcs.110.7.839. [DOI] [PubMed] [Google Scholar]
- 21.Vedula P. Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence. Elife. 2017;6 doi: 10.7554/eLife.31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329(5998):1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laing N.G. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30(9):1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang K. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int. J. Clin. Oncol. 2019;24(9):1030–1041. doi: 10.1007/s10147-019-01435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L. Screening and clinical significance of tumor markers in head and neck squamous cell carcinoma through bioinformatics analysis. Mol. Med. Rep. 2019;19(1):143–154. doi: 10.3892/mmr.2018.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell. Int. 2017;17:75. doi: 10.1186/s12935-017-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauvrak S.U. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer. 2013;109(8):2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omura N. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 2008;7(7):1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White-Al Habeeb N.M. Integrated analysis of epigenomic and genomic changes by DNA methylation dependent mechanisms provides potential novel biomarkers for prostate cancer. Oncotarget. 2014;5(17):7858–7869. doi: 10.18632/oncotarget.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J. Identification of key pathways and genes in PTEN mutation prostate cancer by bioinformatics analysis. BMC Med. Genet. 2019;20(1):191. doi: 10.1186/s12881-019-0923-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni W.D. Tenascin-C is a potential cancer-associated fibroblasts marker and predicts poor prognosis in prostate cancer. Biochem. Biophys. Res. Commun. 2017;486(3):607–612. doi: 10.1016/j.bbrc.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Dai Y. Identification of hub methylated-CpG sites and associated genes in oral squamous cell carcinoma. Cancer Med. 2020;9(9):3174–3187. doi: 10.1002/cam4.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu T. Identification of personalized chemoresistance genes in subtypes of basal-like breast cancer based on functional differences using pathway analysis. PLoS ONE. 2015;10(6) doi: 10.1371/journal.pone.0131183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gou R. Annexin A8 can serve as potential prognostic biomarker and therapeutic target for ovarian cancer: based on the comprehensive analysis of annexins. J. Transl. Med. 2019;17(1):275. doi: 10.1186/s12967-019-2023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evsyukova I., Plestant C., Anton E.S. Integrative mechanisms of oriented neuronal migration in the developing brain. Ann. Rev. Cell Dev. Biol. 2013;29:299–353. doi: 10.1146/annurev-cellbio-101512-122400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carson J.A., Wei L. Integrin signaling's potential for mediating gene expression in hypertrophying skeletal muscle. J. Appl. Physiol. 1985;88(1):337–343. doi: 10.1152/jappl.2000.88.1.337. [DOI] [PubMed] [Google Scholar]
- 37.Boppart M.D., Mahmassani Z.S. Integrin signaling: linking mechanical stimulation to skeletal muscle hypertrophy. Am. J. Physiol. Cell Physiol. 2019;317(4):C629–C641. doi: 10.1152/ajpcell.00009.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhagat T.D. Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. Elife. 2019;8 doi: 10.7554/eLife.50663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen N., Weingarten M., Peter Y. Very late antigen-5 facilitates stromal progenitor cell differentiation into myofibroblast. Stem Cells Transl. Med. 2014;3(11):1342–1353. doi: 10.5966/sctm.2014-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-Bueno G. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat. Protoc. 2009;4(11):1591–1613. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- 41.Yuan S.M. Alpha-smooth muscle actin and ACTA2 gene expressions in vasculopathies. Braz J. Cardiovasc. Surg. 2015;30(6):644–649. doi: 10.5935/1678-9741.20150081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X. Identification of key candidate genes and biological pathways in bladder cancer. PeerJ. 2018;6:e6036. doi: 10.7717/peerj.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeon M. Dimerization of EGFR and HER2 induces breast cancer cell motility through STAT1-dependent ACTA2 induction. Oncotarget. 2017;8(31):50570–50581. doi: 10.18632/oncotarget.10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee H.W. Alpha-smooth muscle actin (ACTA2) is required for metastatic potential of human lung adenocarcinoma. Clin. Cancer Res. 2013;19(21):5879–5889. doi: 10.1158/1078-0432.CCR-13-1181. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B. Identification of potential key genes and pathways in early-onset colorectal cancer through bioinformatics analysis. Cancer Control. 2019;26(1) doi: 10.1177/1073274819831260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang K. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30(6):1107–1119. doi: 10.1016/j.cmet.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Hanley C.J. Targeting the myofibroblastic cancer-associated fibroblast phenotype through inhibition of NOX4. J. Natl. Cancer Inst. 2018;110(1) doi: 10.1093/jnci/djx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlund D. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son G.M. Comparisons of cancer-associated fibroblasts in the intratumoral stroma and invasive front in colorectal cancer. Medicine (Baltimore) 2019;98(18):e15164. doi: 10.1097/MD.0000000000015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez-Outschoorn U.E. Tumor cells induce the cancer associated fibroblast phenotype via caveolin-1 degradation: implications for breast cancer and DCIS therapy with autophagy inhibitors. Cell Cycle. 2010;9(12):2423–2433. doi: 10.4161/cc.9.12.12048. [DOI] [PubMed] [Google Scholar]
- 51.Ning X., Deng Y. Identification of key pathways and genes influencing prognosis in bladder urothelial carcinoma. Onco. Targets Ther. 2017;10:1673–1686. doi: 10.2147/OTT.S131386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milewicz D.M. Genetic variants promoting smooth muscle cell proliferation can result in diffuse and diverse vascular diseases: evidence for a hyperplastic vasculomyopathy. Genet. Med. 2010;12(4):196–203. doi: 10.1097/GIM.0b013e3181cdd687. [DOI] [PubMed] [Google Scholar]
- 53.Nowak D. Beta-actin in human colon adenocarcinoma cell lines with different metastatic potential. Acta Biochim. Pol. 2005;52(2):461–468. [PubMed] [Google Scholar]
- 54.Popow A., Nowak D., Malicka-Blaszkiewicz M. Actin cytoskeleton and beta-actin expression in correlation with higher invasiveness of selected hepatoma Morris 5123 cells. J. Physiol. Pharmacol. 2006;57(Suppl 7):111–123. [PubMed] [Google Scholar]
- 55.Ruan W., Lai M. Actin, a reliable marker of internal control? Clin. Chim. Acta. 2007;385(1–2):1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Le P.U. Increased beta-actin expression in an invasive moloney sarcoma virus-transformed MDCK cell variant concentrates to the tips of multiple pseudopodia. Cancer Res. 1998;58(8):1631–1635. [PubMed] [Google Scholar]
- 57.Blanquicett C. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. Anal. Biochem. 2002;303(2):209–214. doi: 10.1006/abio.2001.5570. [DOI] [PubMed] [Google Scholar]
- 58.Goidin D. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001;295(1):17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 59.Liu Z. Proteomic identification of differentially-expressed proteins in esophageal cancer in three ethnic groups in Xinjiang. Mol. Biol. Rep. 2011;38(5):3261–3269. doi: 10.1007/s11033-010-0586-0. [DOI] [PubMed] [Google Scholar]
- 60.Morse D.L. Determining suitable internal standards for mRNA quantification of increasing cancer progression in human breast cells by real-time reverse transcriptase polymerase chain reaction. Anal. Biochem. 2005;342(1):69–77. doi: 10.1016/j.ab.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 61.Nguewa P.A. Identification of importin 8 (IPO8) as the most accurate reference gene for the clinicopathological analysis of lung specimens. BMC Mol. Biol. 2008;9:103. doi: 10.1186/1471-2199-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubie C. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol. Cell Probes. 2005;19(2):101–109. doi: 10.1016/j.mcp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Saviozzi S. Selection of suitable reference genes for accurate normalization of gene expression profile studies in non-small cell lung cancer. BMC Cancer. 2006;6:200. doi: 10.1186/1471-2407-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferguson R.E. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5(2):566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- 65.Shagieva G.S. Actin isoforms and reorganization of adhesion junctions in epithelial-to-mesenchymal transition of cervical carcinoma cells. Biochemistry (Moscow) 2012;77(11):1266–1276. doi: 10.1134/S0006297912110053. [DOI] [PubMed] [Google Scholar]
- 66.Weinkauf M. 2-D PAGE-based comparison of proteasome inhibitor bortezomib in sensitive and resistant mantle cell lymphoma. Electrophoresis. 2009;30(6):974–986. doi: 10.1002/elps.200800508. [DOI] [PubMed] [Google Scholar]
- 67.Ben-Ze'ev A. Cytoskeletal and adhesion proteins as tumor suppressors. Curr. Opin. Cell Biol. 1997;9(1):99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- 68.Decloitre F. Concomitant alterations of microfilaments and microtubules in human epithelial cells (HBL-100) in relation to their malignant conversion. Tumour Biol. 1991;12(2):111–119. doi: 10.1159/000217695. [DOI] [PubMed] [Google Scholar]
- 69.Frontelo P. Transforming growth factor beta 1 induces squamous carcinoma cell variants with increased metastatic abilities and a disorganized cytoskeleton. Exp. Cell Res. 1998;244(2):420–432. doi: 10.1006/excr.1998.4219. [DOI] [PubMed] [Google Scholar]
- 70.Liu C.R. Differential thymosin beta 10 expression levels and actin filament organization in tumor cell lines with different metastatic potential. Chin. Med. J. (Engl) 2004;117(2):213–218. [PubMed] [Google Scholar]
- 71.Guo C. ACTB in cancer. Clin Chim Acta. 2013;417:39–44. doi: 10.1016/j.cca.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 72.Kwiatkowski S. SETD3 protein is the actin-specific histidine N-methyltransferase. Elife. 2018;7 doi: 10.7554/eLife.37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu L. The Glycolytic Switch in Tumors: how Many Players Are Involved? J. Cancer. 2017;8(17):3430–3440. doi: 10.7150/jca.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pourcel L. Influence of cytoskeleton organization on recombinant protein expression by CHO cells. Biotechnol. Bioeng. 2020;117(4):1117–1126. doi: 10.1002/bit.27277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goggolidou P. A chronological expression profile of gene activity during embryonic mouse brain development. Mamm. Genome. 2013;24(11–12):459–472. doi: 10.1007/s00335-013-9486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.da Rocha R.G. Leptin impairs the therapeutic effect of ionizing radiation in oral squamous cell carcinoma cells. J. Oral Pathol. Med. 2019;48(1):17–23. doi: 10.1111/jop.12786. [DOI] [PubMed] [Google Scholar]
- 77.Ohtaki S. ACTC1 as an invasion and prognosis marker in glioma. J. Neurosurg. 2017;126(2):467–475. doi: 10.3171/2016.1.JNS152075. [DOI] [PubMed] [Google Scholar]
- 78.Huang H.C. Discovering disease-specific biomarker genes for cancer diagnosis and prognosis. Technol. Cancer Res. Treat. 2010;9(3):219–230. doi: 10.1177/153303461000900301. [DOI] [PubMed] [Google Scholar]
- 79.Zaravinos A. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE. 2011;6(4):e18135. doi: 10.1371/journal.pone.0018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Che C.L. DNA microarray reveals different pathways responding to paclitaxel and docetaxel in non-small cell lung cancer cell line. Int. J. Clin. Exp. Pathol. 2013;6(8):1538–1548. [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y., Rong G., Kang H. Taxotere-induced elevated expression of IL8 in carcinoma-associated fibroblasts of breast invasive ductal cancer. Oncol. Lett. 2017;13(3):1856–1860. doi: 10.3892/ol.2017.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang M. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018;17(5):6211–6226. doi: 10.3892/mmr.2018.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wanibuchi M. Actin, alpha, cardiac muscle 1 (ACTC1) knockdown inhibits the migration of glioblastoma cells in vitro. J. Neurol. Sci. 2018;392:117–121. doi: 10.1016/j.jns.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 84.Lee C.G., Jang J., Jin H.S. A novel missense mutation in the ACTG1 gene in a family with congenital autosomal dominant deafness: a case report. Mol. Med. Rep. 2018;17(6):7611–7617. doi: 10.3892/mmr.2018.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong X. Actin Gamma 1, a new skin cancer pathogenic gene, identified by the biological feature-based classification. J. Cell Biochem. 2018;119(2):1406–1419. doi: 10.1002/jcb.26301. [DOI] [PubMed] [Google Scholar]
- 86.Shum M.S. Gamma-Actin regulates cell migration and modulates the ROCK signaling pathway. FASEB J. 2011;25(12):4423–4433. doi: 10.1096/fj.11-185447. [DOI] [PubMed] [Google Scholar]
- 87.Yan Y. RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellular carcinoma. Onco. Targets Ther. 2019;12:1691–1703. doi: 10.2147/OTT.S197844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Durinck S. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat. Genet. 2015;47(1):13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang Y.C. Therapeutic targeting of aldolase a interactions inhibits lung cancer metastasis and prolongs survival. Cancer Res. 2019;79(18):4754–4766. doi: 10.1158/0008-5472.CAN-18-4080. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y. miR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017;8(4):e2739. doi: 10.1038/cddis.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J. Characterization of DNA hydroxymethylation profile in cervical cancer. Artif. Cells Nanomed. Biotechnol. 2019;47(1):2706–2714. doi: 10.1080/21691401.2019.1634578. [DOI] [PubMed] [Google Scholar]
- 92.Dugina V. Tumor promotion by gamma and suppression by beta non-muscle actin isoforms. Oncotarget. 2015;6(16):14556–14571. doi: 10.18632/oncotarget.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verrills N.M. Alterations in gamma-actin and tubulin-targeted drug resistance in childhood leukemia. J. Natl. Cancer Inst. 2006;98(19):1363–1374. doi: 10.1093/jnci/djj372. [DOI] [PubMed] [Google Scholar]
- 94.Po'uha S.T., Kavallaris M. Gamma-actin is involved in regulating centrosome function and mitotic progression in cancer cells. Cell Cycle. 2015;14(24):3908–3919. doi: 10.1080/15384101.2015.1120920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Halim D. ACTG2 variants impair actin polymerization in sporadic Megacystis Microcolon Intestinal Hypoperistalsis Syndrome. Hum. Mol Genet. 2016;25(3):571–583. doi: 10.1093/hmg/ddv497. [DOI] [PubMed] [Google Scholar]
- 96.Benzoubir N. Gamma-smooth muscle actin expression is associated with epithelial-mesenchymal transition and stem-like properties in hepatocellular carcinoma. PLoS ONE. 2015;10(6) doi: 10.1371/journal.pone.0130559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu Y. Identification of ACTG2 functions as a promoter gene in hepatocellular carcinoma cells migration and tumor metastasis. Biochem. Biophys. Res. Commun. 2017;491(2):537–544. doi: 10.1016/j.bbrc.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 98.Gautreau A. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA. 2004;101(13):4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kunda P. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13(21):1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 100.Mendoza M.C. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 2011;41(6):661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mendoza M.C. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev. Biol. 2013;24(4):272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suria H. Cytoskeletal disruption induces T cell apoptosis by a caspase-3 mediated mechanism. Life Sci. 1999;65(25):2697–2707. doi: 10.1016/s0024-3205(99)00538-x. [DOI] [PubMed] [Google Scholar]
- 103.Puthalakath H. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293(5536):1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 104.Paul C. Hsp27 as a negative regulator of cytochrome C release. Mol. Cell Biol. 2002;22(3):816–834. doi: 10.1128/MCB.22.3.816-834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gourlay C.W. A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 2004;164(6):803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu X., Forbes J.G., Colombini M. Actin modulates the gating of Neurospora crassa VDAC. J. Membr. Biol. 2001;180(1):73–81. doi: 10.1007/s002320010060. [DOI] [PubMed] [Google Scholar]
- 107.Kusano H. Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene. 2000;19(42):4807–4814. doi: 10.1038/sj.onc.1203868. [DOI] [PubMed] [Google Scholar]
- 108.Gourlay C.W., Ayscough K.R. The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005;6(7):583–589. doi: 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- 109.Mashima T., Naito M., Tsuruo T. Caspase-mediated cleavage of cytoskeletal actin plays a positive role in the process of morphological apoptosis. Oncogene. 1999;18(15):2423–2430. doi: 10.1038/sj.onc.1202558. [DOI] [PubMed] [Google Scholar]
- 110.Kashina A.S. Regulation of actin isoforms in cellular and developmental processes. Semin. Cell Dev. Biol. 2020;102:113–121. doi: 10.1016/j.semcdb.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bergeron S.E. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J. Biol. Chem. 2010;285(21):16087–16095. doi: 10.1074/jbc.M110.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaech S. Isoform specificity in the relationship of actin to dendritic spines. J. Neurosci. 1997;17(24):9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khaitlina S.Y. Functional specificity of actin isoforms. Int. Rev. Cytol. 2001;202:35–98. doi: 10.1016/s0074-7696(01)02003-4. [DOI] [PubMed] [Google Scholar]
- 114.von Arx P. Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J. Cell Biol. 1995;131(6 Pt 2):1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Allen P.G. Phalloidin binding and rheological differences among actin isoforms. Biochemistry. 1996;35(45):14062–14069. doi: 10.1021/bi961326g. [DOI] [PubMed] [Google Scholar]
- 116.Fritzsche M. Analysis of turnover dynamics of the submembranous actin cortex. Mol. Biol. Cell. 2013;24(6):757–767. doi: 10.1091/mbc.E12-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schaks M., Giannone G., Rottner K. Actin dynamics in cell migration. Essays Biochem. 2019;63(5):483–495. doi: 10.1042/EBC20190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calzado-Martin A. Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy. ACS Nano. 2016;10(3):3365–3374. doi: 10.1021/acsnano.5b07162. [DOI] [PubMed] [Google Scholar]
- 119.Xuan B. Dysregulation in actin cytoskeletal organization drives increased stiffness and migratory persistence in polyploidal giant cancer cells. Sci. Rep. 2018;8(1):11935. doi: 10.1038/s41598-018-29817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muller M. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS ONE. 2013;8(7):e70636. doi: 10.1371/journal.pone.0070636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Khaitlina S. Polymerization of beta-like actin from scallop adductor muscle. FEBS Lett. 1986;198(2):221–224. doi: 10.1016/0014-5793(86)80409-4. [DOI] [PubMed] [Google Scholar]
- 122.Khaitlina S. Correlation between polymerizability and conformation in scallop beta-like actin and rabbit skeletal muscle alpha-actin. Arch. Biochem. Biophys. 1999;368(1):105–111. doi: 10.1006/abbi.1999.1303. [DOI] [PubMed] [Google Scholar]
- 123.Khaitlina S., Hinssen H. Difference in polymerization and steady-state dynamics of free and gelsolin-capped filaments formed by alpha- and beta-isoactins. Arch. Biochem. Biophys. 2008;477(2):279–284. doi: 10.1016/j.abb.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 124.Kunda P. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 2008;18(2):91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 125.Matthews H.K. Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev. Cell. 2012;23(2):371–383. doi: 10.1016/j.devcel.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raucher D., Sheetz M.P. Membrane expansion increases endocytosis rate during mitosis. J. Cell Biol. 1999;144(3):497–506. doi: 10.1083/jcb.144.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gunning P., O'Neill G., Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008;88(1):1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 128.Leavitt J. Expression of transfected mutant beta-actin genes: transitions toward the stable tumorigenic state. Mol. Cell Biol. 1987;7(7):2467–2476. doi: 10.1128/mcb.7.7.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leavitt J. Tropomyosin isoform switching in tumorigenic human fibroblasts. Mol. Cell Biol. 1986;6(7):2721–2726. doi: 10.1128/mcb.6.7.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsumura F., Yamashiro-Matsumura S. Purification and characterization of multiple isoforms of tropomyosin from rat cultured cells. J. Biol. Chem. 1985;260(25):13851–13859. [PubMed] [Google Scholar]
- 131.Gimona M., Kazzaz J.A., Helfman D.M. Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc. Natl. Acad. Sci. USA, 1996;93(18):9618–9623. doi: 10.1073/pnas.93.18.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mahadev K. Suppression of the transformed phenotype of breast cancer by tropomyosin-1. Exp. Cell Res. 2002;279(1):40–51. doi: 10.1006/excr.2002.5583. [DOI] [PubMed] [Google Scholar]
- 133.Prasad G.L., Fuldner R.A., Cooper H.L. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc. Natl. Acad. Sci. USA, 1993;90(15):7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Prasad G.L. Suppression of src-induced transformed phenotype by expression of tropomyosin-1. Oncogene. 1999;18(11):2027–2031. doi: 10.1038/sj.onc.1202264. [DOI] [PubMed] [Google Scholar]
- 135.Shah V. Cytoskeletal organization in tropomyosin-mediated reversion of ras-transformation: evidence for Rho kinase pathway. Oncogene. 2001;20(17):2112–2121. doi: 10.1038/sj.onc.1204291. [DOI] [PubMed] [Google Scholar]
- 136.Yager M.L. Functional analysis of the actin-binding protein, tropomyosin 1, in neuroblastoma. Br. J .Cancer. 2003;89(5):860–863. doi: 10.1038/sj.bjc.6601201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang C.L., Coluccio L.M. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int. Rev. Cell Mol. Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Viswanathan M.C. A role for actin flexibility in thin filament-mediated contractile regulation and myopathy. Nat. Commun. 2020;11(1):2417. doi: 10.1038/s41467-020-15922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hitchcock-DeGregori S.E., Sampath P., Pollard T.D. Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry. 1988;27(26):9182–9185. doi: 10.1021/bi00426a016. [DOI] [PubMed] [Google Scholar]
- 140.Vindin H., Gunning P. Cytoskeletal tropomyosins: choreographers of actin filament functional diversity. J. Muscle Res. Cell Motil. 2013;34(3–4):261–274. doi: 10.1007/s10974-013-9355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blanchoin L., Pollard T.D., Hitchcock-DeGregori S.E. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 2001;11(16):1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 142.Eppinga R.D. Tropomyosin and caldesmon regulate cytokinesis speed and membrane stability during cell division. Arch. Biochem. Biophys. 2006;456(2):161–174. doi: 10.1016/j.abb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 143.Hughes J.A. High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia. 2003;42(1):25–35. doi: 10.1002/glia.10174. [DOI] [PubMed] [Google Scholar]
- 144.Kee A.J. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic. 2015;16(7):691–711. doi: 10.1111/tra.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McIntosh B.B., Holzbaur E.L., Ostap E.M. Control of the initiation and termination of kinesin-1-driven transport by myosin-Ic and nonmuscle tropomyosin. Curr. Biol. 2015;25(4):523–529. doi: 10.1016/j.cub.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang N., Ostap E.M. Motor domain-dependent localization of myo1b (myr-1) Curr. Biol. 2001;11(14):1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 147.Yamashiro S. Differential actin-regulatory activities of Tropomodulin1 and Tropomodulin3 with diverse tropomyosin and actin isoforms. J. Biol. Chem. 2014;289(17):11616–11629. doi: 10.1074/jbc.M114.555128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vandekerckhove J., Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 1978;126(4):783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- 149.Vandekerckhove J., Weber K. Actin typing on total cellular extracts: a highly sensitive protein-chemical procedure able to distinguish different actins. Eur. J. Biochem. 1981;113(3):595–603. doi: 10.1111/j.1432-1033.1981.tb05104.x. [DOI] [PubMed] [Google Scholar]
- 150.Van Troys M., Vandekerckhove J., Ampe C. Structural modules in actin-binding proteins: towards a new classification. Biochim. Biophys. Acta. 1999;1448(3):323–348. doi: 10.1016/s0167-4889(98)00152-9. [DOI] [PubMed] [Google Scholar]
- 151.Yonezawa N. An actin-interacting heptapeptide in the cofilin sequence. Eur. J. Biochem. 1989;183(1):235–238. doi: 10.1111/j.1432-1033.1989.tb14918.x. [DOI] [PubMed] [Google Scholar]
- 152.Cook R.K. Enhanced stimulation of myosin subfragment 1 ATPase activity by addition of negatively charged residues to the yeast actin NH2 terminus. J. Biol. Chem. 1993;268(4):2410–2415. [PubMed] [Google Scholar]
- 153.Dugina V. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J. Cell Sci. 2009;122(Pt 16):2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- 154.Heissler S.M., Manstein D.J. Nonmuscle myosin-2: mix and match. Cell Mol. Life Sci. 2013;70(1):1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harris, A.R., et al., Biased localization of actin binding proteins by actin filament conformation. bioRxiv, 2020: p. 2020.02.21.959791. [DOI] [PMC free article] [PubMed]
- 156.Shekhar S., Pernier J., Carlier M.F. Regulators of actin filament barbed ends at a glance. J. Cell Sci. 2016;129(6):1085–1091. doi: 10.1242/jcs.179994. [DOI] [PubMed] [Google Scholar]
- 157.De La Cruz E.M. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol Biol. 2005;346(2):557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 158.Kis-Bicskei N. Tropomyosins regulate the severing activity of gelsolin in isoform-dependent and independent manners. Biophys. J. 2018;114(4):777–787. doi: 10.1016/j.bpj.2017.11.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jordan M.A., Wilson L. Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr. Opin Cell Biol. 1998;10(1):123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 160.Bonello T.T. A small molecule inhibitor of tropomyosin dissociates actin binding from tropomyosin-directed regulation of actin dynamics. Sci. Rep. 2016;6:19816. doi: 10.1038/srep19816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Currier M.A. Identification of cancer-targeted tropomyosin inhibitors and their synergy with microtubule drugs. Mol. Cancer Ther. 2017;16(8):1555–1565. doi: 10.1158/1535-7163.MCT-16-0873. [DOI] [PubMed] [Google Scholar]
- 162.Wang Y. Drug targeting the actin cytoskeleton potentiates the cytotoxicity of low dose vincristine by abrogating actin-mediated repair of spindle defects. Mol. Cancer Res. 2020;18(7):1074–1087. doi: 10.1158/1541-7786.MCR-19-1122. [DOI] [PubMed] [Google Scholar]
- 163.Gandalovicova A. Migrastatics-anti-metastatic and anti-invasion drugs: promises and challenges. Trends Cancer. 2017;3(6):391–406. doi: 10.1016/j.trecan.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]