Abstract
Protein aggregation is a widespread process leading to deleterious consequences in the organism, with amyloid aggregates being important not only in biology but also for drug design and biomaterial production. Insulin is a protein largely used in diabetes treatment, and its amyloid aggregation is at the basis of the so-called insulin-derived amyloidosis. Here, we uncover the major role of zinc in both insulin dynamics and aggregation kinetics at low pH, in which the formation of different amyloid superstructures (fibrils and spherulites) can be thermally induced. Amyloid aggregation is accompanied by zinc release and the suppression of water-sustained insulin dynamics, as shown by particle-induced x-ray emission and x-ray absorption spectroscopy and by neutron spectroscopy, respectively. Our study shows that zinc binding stabilizes the native form of insulin by facilitating hydration of this hydrophobic protein and suggests that introducing new binding sites for zinc can improve insulin stability and tune its aggregation propensity.
Significance
Localized amyloidosis occurs at insulin injection sites for diabetes treatment, leading to deleterious inflammations known as insulin-derived amyloidosis. Amyloid superstructures are also promising candidates in the field of biomaterials. Here, we revealed that zinc, interacting with insulin in the native form, is released upon amyloid aggregation when insulin forms superstructures known as fibrils and spherulites. Zinc release leads to a full suppression of functionally essential protein dynamics through a modification of the protein’s hydration properties and completely modifies insulin amyloid kinetics. The results suggest that changes in protein hydration upon zinc binding and release modifies both stability and dynamics of insulin and might then be a general strategy to control protein stability and tune protein aggregation into amorphous and ordered superstructures.
Introduction
Under destabilizing conditions in vitro, a large number of proteins aggregate and form specific ordered superstructures (1). These can be amyloid fibrils, characterized by a typical cross-β pattern (2,3) or larger, micrometric assemblies of amyloid nature, like spherulites (4,5). The variety of amyloid species poses a challenge for pharmaceutical drug development, in which protein-based products have to be optimized while controlling and characterizing any by-product particles. Indeed, because of the simultaneous occurrence of different species, identifying and isolating each individual species is a highly demanding task, still remaining a conditio sine qua non for the quality control of the final product (6). On the other hand, having access to a number of markedly different protein self-assembled species offers a unique opportunity for the development of a rational platform for the design of bioinspired materials for applications ranging from biosensing and tissue engineering to drug delivery (7). Finally, yet importantly, a large-scale morphological variability of aggregates is also observed in the context of amyloid-related pathologies. This variability potentially determines different etiological subtypes of diseases (8, 9, 10), making it even more challenging to univocally establish a connection between aggregate structure and morphology and the onset and progression of the diseases. In all the above fields, it is then crucial to disentangle the concurring aggregation pathways and isolate the different species. Indeed, identifying the different aggregates would allow the mechanisms of formation of the different species to be mapped, as well as to unravel how the structure-function-dynamics relationship is perturbed by amyloid formation for each of the different aggregation states.
Insulin, a 5.8-kDa hormone consisting of two mainly α-helical chains (A and B) linked by disulfide bonds, can form amyloid fibrils (11,12) and spherulites (13). The two types of aggregate coexist at low pH when human insulin samples are thermally destabilized (14,15). Subcutaneous insulin injections are largely used during insulin therapy for diabetic patients. Near injection sites, localized amyloidosis has been found that can lead to deleterious repeated inflammations, known as the insulin-derived amyloidosis (IDA) (16, 17, 18, 19) or also “insulin ball” (20) or amyloidoma (21). The number of IDA reported cases has increased significantly in the last years (22). When IDA occurs, it leads to poor glycemic control and also to catheter occlusion in the case of continuous infusion. Recent in vitro studies on the stability of insulin formulations used to treat type 1 diabetes have shown that amyloid formation occurs either at neutral pH or below the insulin isoelectric point (pH <5.2) (23). An alternative way recently explored for insulin delivery to diabetic patients is the administration by the oral route that could potentially overcome the drawbacks of insulin injection, reducing the number of injections needed and the risk of side effects (24). However, when administered orally, insulin first arrives at the stomach, where pH is between 1.2 and 3.0, which is the range in which, in vitro, insulin forms both amyloid fibrils (11) and spherulites (13). It is also known that diabetes is commonly associated with neurodegenerative diseases (25) and insulin signaling impairment—for which insulin aggregation might play a role—was shown to promote neurodegeneration (26,27). Furthermore, insulin stability is a major concern in the pharmaceutical industry for production and storage (8, 9, 10). As a consequence, understanding the key factors involved in insulin self-assembly is highly desired for the optimization of downstream product processing, as well as for a more exhaustive mapping of the multiplicity of different structures occurring in its aggregation reaction. The insulin aggregation pathway starts from a mixture of oligomeric forms. The oligomerization state depends on the solution conditions, with mainly a dimer-monomer mixture occurring at a low pH (28) and a hexamer species at a higher pH. However, insulin amyloid species can be formed independent of the details of the early oligomerization process (29), and such aggregates are thioflavin T (ThT) positive, indicating the presence of a cross-β fibrillar structure as is found for toxic amyloid proteins such as τ- or α-synuclein (2,3). Several factors are reported to affect the aggregation process of insulin. Approaches involving sedimentation kinetics measurements (30), optical microscopy imaging (14), and small-angle x-ray scattering (31) have revealed the role of both insulin and salt concentration and of pH in the aggregation process. In the context of amyloid diseases, the pathological impact of metal ions in the alteration of amyloidogenesis is now accepted (32). In the specific case of insulin, zinc ions (Zn2+) are essential components for protein expression and activity (33). It has been shown that Zn2+ slows down fibrillation of monomeric insulin at physiological pH; this result has been used to propose that Zn2+ co-secreted from pancreatic β-cells protects the organism from the formation of non-native-insulin oligomers and aggregates (34). However, Zn2+ has been found to be coordinated in both functional and aggregated bovine insulin at physiological pH (35).
More generally, the significant influence of metal ions on aggregation kinetics, pathway, and aggregate morphology has been reported for several amyloid systems (36, 37, 38). As a matter of example, zinc and copper ions increase the stability of Aβ oligomers and reduce the stability of Aβ fibrils, the effect on fibrils being specific to the type of ion (39). Ions can also dramatically affect the balance between the numbers of native multimers and monomers (40), which are prone to form amyloid-like fibrils in systems containing GAPR-1 and heparin (41). Notwithstanding, pronounced metal ion-related effects are clearly detected in other amyloid-forming systems (37,42, 43, 44, 45, 46), the mechanism at play remains elusive. Metal coordination at the level of single protein molecules certainly changes the overall charge, protein structure, and/or hydration, eventually influencing the dynamics of the single protein molecule (47,48). How these changes modify the physicochemical properties of protein aggregates, however, remains unclear.
Recently, several pieces of evidence have been reported on the relationship between amyloid aggregation and the dynamical properties of aggregating proteins. Measurements of intramolecular reconfiguration dynamics in different pathological amyloid proteins suggested a direct correlation between internal dynamics and aggregation propensity and kinetics (49,50). Neutron-scattering results obtained on different proteins showed that upon amyloid aggregation, either the protein (51) or its hydration water (52) can show an increased mobility at temperatures above the so-called dynamical transition (53).
Here, we used neutron scattering to characterize the dynamics of human insulin in its native state and in both its fibril and spherulite amyloid forms. Controlled formation of the amyloid forms was achieved by formulating insulin in low pH solutions, which provide ideal in vitro experimental conditions for producing the above-mentioned amyloid structures. We first screened and selected the conditions for isolating samples containing either predominantly fibrils or predominantly spherulites. Neutron scattering revealed a complete suppression of functionally relevant internal protein dynamics in both insulin fibrils and spherulites. An analysis based on microbeam particle-induced x-ray emission (μPIXE) and x-ray absorption spectroscopy (XAS), combined with a protein conformational characterization, unexpectedly revealed that the protein rigidification is due to the release, upon amyloid aggregation, of the Zn2+ coordinated to insulin in the native state (33). A crucial role of Zn2+ in determining insulin aggregation kinetics was then unveiled by fluorescence spectroscopy.
Materials and methods
Sample preparation for neutron scattering, neutron, and x-ray diffraction and μPIXE: native, EDTA-treated, and “washed” insulin
Crystalline powder of human insulin was obtained from Novo Nordisk (Bagsværd, Denmark) and stored at −20°C before use. To obtain native-insulin samples, the powder was dissolved by adding 5-μL aliquots of 37% w/w HCl to a suspension of insulin crystals in H2O until the solution was clear. After flash cooling in liquid nitrogen and freeze drying, electron microscopy and x-ray powder diffraction experiments were used to check for the presence of any crystals—protein or salt—and for aggregates (see Fig. S5). To remove zinc, either partially or fully, from native insulin, we used washing steps (washed insulin) or EDTA addition followed by washing steps (EDTA-treated insulin), respectively. To prepare both washed and EDTA-treated insulin samples, the pH was set to the insulin isoelectric point (pI 5.2) to obtain a nonsoluble suspension of insulin in D2O (washed insulin) or in D2O containing EDTA at 1.5 EDTA/insulin molar ratio (EDTA-treated insulin). Washing steps were then performed for both samples using several cycles of centrifugation at 18,000 rpm for 20 min and resuspension in pure D2O (washed insulin) or D2O containing EDTA at 1.5 EDTA/insulin molar ratio (EDTA-treated insulin). These washing steps resulted in a complete removal of both EDTA and the chelated zinc from the EDTA-treated sample. At this stage, both washed and EDTA-treated samples still contain nonsoluble insulin, which is then solubilized by lowering the pH to 1.8. Finally, flash cooling and freeze drying were performed before D2O hydration for the neutron experiments.
Sample preparation for neutron scattering, neutron, and x-ray diffraction and μPIXE: insulin fibrils and spherulites
The conditions for preparing pure fibril and pure spherulite samples are described in the main text as they were obtained and optimized in this work. Briefly, fibrils were produced by incubating a 5 mg/ml human insulin solution at 50°C for 24 h (pH 1.8), 0.25 mM NaCl under shaking at 600 rpm with an Eppendorf ThermoMixer (Hamburg, Germany). Spherulites were produced by incubating a 5 mg/ml human insulin solution at 60°C for 24 h (pH 1.8), 0.25 mM NaCl in quiescent conditions and then isolated by a postproduction treatment based on a series of vortexing, centrifugation, and supernatant-removal steps. Before neutron experiments, fibrils and spherulites were washed using several cycles of centrifugation at 18,000 rpm for 20 min and resuspension in pure D2O to remove any salt and contaminants. They were then flash cooled and freeze dried as for the monomeric samples.
Incoherent elastic neutron scattering
All data were recorded on the SPHERES backscattering spectrometer (proposal P14013, June 14–20, 2018; Jülich Centre for Neutron Science (JCNS), Jülich, Germany) (54,55) operated by JCNS at the Heinz Maier-Leibnitz Zentrum (MLZ) in Garching, Germany. Freeze-dried insulin powders were incubated with P2O5 for 24 h in a desiccator to further remove water. In the case of lysozyme, such a procedure resulted in a residual presence of four waters per protein molecule. The insulin powders were then weighed and rehydrated by vapor exchange with pure D2O to reach a hydration level h = 0.44 D2O/protein w/w. This water content was chosen to guarantee on one hand that the protein surface is fully hydrated (56) and on the other hand to avoid the presence of bulk water that would lead to crystalline water-ice formation at cryogenic temperatures (cryo-temperatures). Hydrated protein powders were then sealed in aluminum flat neutron cells (sample thickness = 0.3 mm) using 1-mm indium wire from Alfa Aesar (Ward Hill, MA). Samples were weighed before and after each neutron experiment to verify that they were correctly sealed and no water was lost. The cell was mounted in a cryostat at room temperature, and the temperature was subsequently lowered to 10 K within ∼2 h. The elastic scattering signal was recorded at an energy resolution of 0.66 μeV (FWHM), employing Si(111) monochromator and analyzer crystals in exact backscattering geometry (for large scattering angles) corresponding to an incident wavelength of λ = 6.27 Å. The elastic signal was recorded while continuously increasing the sample temperature from 10 to 300 K at a rate of 0.2 K/min and binned together to 0.05-K steps. Raw data were preprocessed—i.e., normalized to the detector efficiency and to the incident beam intensity recorded by a so-called monitor device—using the SLAW software available on facility computers (http://apps.jcns.fz-juelich.de/slaw). With the elastic signal arising mainly from the incoherent scattering of the hydrogen atoms, the mean-square displacements (MSDs) of hydrogen atoms can be obtained using the well-established Gaussian approximation (57,58). We performed this analysis using custom-made Python scripts (http://github.com/kpounot/nPDyn - v1.0.0), with the Gaussian approximation verified for the momentum transfer (q = 4πλ−1sin(θ/2), with θ being the scattering angle, range 0.6–1.2 Å−1. Using higher terms in cumulant expansion leading to a corrected Gaussian model (58) or using a gamma-distribution-based model (59) gave similar results but with higher numerical instability during the fitting procedure.
Neutron diffraction
To confirm the absence of ice formation at h = 0.44, neutron diffraction data were recorded on the D16 instrument at the Institut Laue-Langevin in Grenoble, France. The same samples within the sealed aluminum cells as used for measuring the incoherent elastic neutron scattering were mounted in an Orange cryostat to measure the data at two temperatures, 300 and 200 K. The diffracted beam was measured over an angular range 12–112.5°, which corresponds to a q-range of 0.05–2.5 Å−1. Neutron data were corrected for the empty cell scattering, the ambient room background, and the nonuniform detector response. The transmission and the thickness of the sample were also taken into account. The two-dimensional scattering intensities were normalized in absolute units with a standard calibration and radially integrated to obtain one-dimensional diffraction patterns. The neutron diffraction data (see Fig. S7) do not provide evidence for ice formation so that the presence of bulk water can be ruled out.
μPIXE
Zinc stoichiometries in the different insulin samples were measured using μPIXE analysis in combination with simultaneous Rutherford backscattering analysis to allow correction for sample matrix effects (60,61). For μPIXE measurements of proteins, sulfur acts as an internal standard. The measurements were carried out at the Ion Beam Centre, University of Surrey, UK (62). A 2.5-MeV proton beam of 1.5 μm in diameter was used to induce characteristic x-ray emission from dried insulin sample droplets (volume per droplet, ∼0.1 μL) under vacuum. The x-rays were detected using a solid-state lithium-drifted silicon detector, and backscattered protons were detected using a silicon particle detector. By scanning the proton beam in x and y over the dried sample, spatial maps were obtained of all elements heavier than magnesium present in the sample. Quantitative information was obtained by collecting three or four point spectra from each droplet. PIXE spectra were analyzed with GUPIX (63) using the matrix composition derived from the simultaneous RBS spectrum. Data processing is carried out using the data acquisition software OMDAQ-3 (Oxford Microbeams, Bicester, UK) to extract the relative amount of each element in the sample.
XAS
Data collection. X-ray absorption spectra were collected at the zinc K-edge (9660.75 eV) on the extended x-ray absorption fine structure (EXAFS) station (BM26A) of the Dutch-Belgian beamline (DUBBLE) (64) at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. The energy of the x-ray beam was tuned by a double-crystal monochromator operating in fixed-exit mode using an Si(111) crystal pair. Three different samples were put in separate glass capillaries and measured in fluorescence mode at ambient temperature and pressure: 1) a zinc solution prepared by dissolving 4 mg of ZnSO4 in 10 mL of 0.25 M NaCl solution at pH 1.8, 2) a native-insulin solution prepared by dissolving 10 mg of lyophilized insulin powder (Novo Nordisk) in 1 mL of 0.25 M NaCl solution at pH 1.8, and 3) the same solution as in 2 incubated at 60°C for ∼24 h to induce amyloid aggregation. The EXAFS spectra, three scans per sample with a new solution for each scan, were energy calibrated, averaged, and further analyzed using GNXAS (65,66).
Data analysis. In the GNXAS approach, the local atomic arrangement around the absorbing atom is decomposed into model atomic configurations containing 2, ..., n atoms. The theoretical EXAFS signal (k) is given by the sum of the n-body contributions γ2, γ3, …, γn, which take into account all the possible single- and multiple-scattering paths between the n atoms. The fitting of (k) to the experimental EXAFS signal allows refinement of the relevant structural parameters of the different coordination shells; the suitability of the model is also evaluated by comparison of the experimental EXAFS signal Fourier transform with the Fourier transform of the calculated (k) function. The coordination numbers and the global fit parameters that were allowed to vary during the fitting procedure were the distance R(Å), Debye-Waller factor (σ2), and the angles of the γn contributions, which were defined according to atomic structural models constructed using the VMD software (67). Zinc was solvated based on a published refined structure (68). For the model containing histidine, the residue was positioned by replacing one coordinating water molecule with it. Weak distance constraints were used so that water and histidine could compete to coordinate the zinc atom. Simulations were then performed using NAMD 2.13 (69) with the TIP3P model for water (70) and the CHARMM36 force field (71). The Nose-Hoover-Langevin piston algorithm maintained a constant pressure (72,73), and the stochastic velocity rescaling algorithm controlled the temperature (74). Bonds with hydrogen atoms were constrained using the SHAKE algorithm (75) with a force constant k set to 2, and the Verlet-I/r-RESPA multiple-time step scheme (75, 76, 77) integrated the equations of motion, with time steps of 2 fs for long-range nonbonded forces and 1 fs for short-range and bonded forces. Electrostatic interactions were computed using the smooth particle Mesh-Ewald (PME) sum (78), with a cutoff set to 10 Å, a switching function starting at 8 Å, and a pair list distance of 14 Å was used. A snapshot of the simulation run was then used to extract coordinates that were processed by GNXAS. According to the GNXAS approach, we used a single two-body configuration γ2 for the Zn-L (L = O or N) distance. To fit the higher shell contributions two η3 corresponding to the Zn-O-C and Zn-O-N three-body configurations were used (see inset in Fig. 5 c). Importantly, because of the high value of the vertex angle in the Zn-O-C and Zn-O-N configurations, which are connected to the aromatic ring present in the histidine, the signals of theses shells are enhanced because of the multiple-scattering effect, which is considered in the analysis. For these shells, whose Zn-O distance is the first shell, the only free parameter needed was the vertex angle θ.
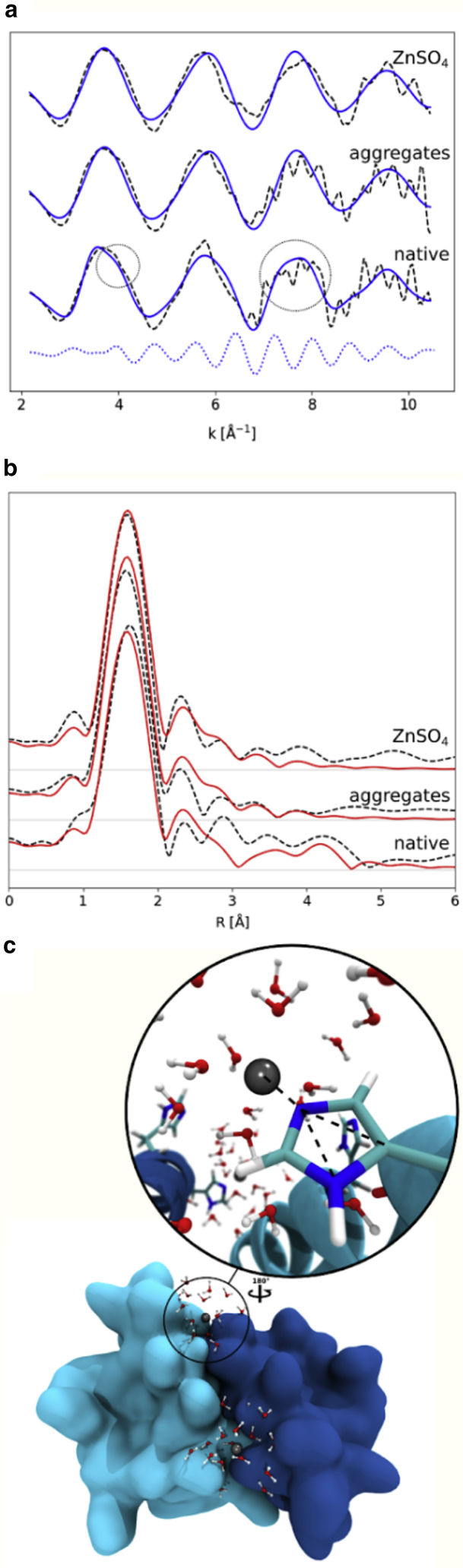
Figure 5.
XAS sheds light on the zinc environment in native insulin and insulin aggregates in solution. (a) Shown are k3-weighted EXAFS transmission spectra (dashed lines) of a native-insulin solution, an insulin solution treated at 60°C for ∼24 h (aggregates), and a ZnSO4 solution. Blue continuous lines represent the signals simulated on the basis of the structure of a fully solvated Zn2+ ion (for the ZnSO4 solution and for the aggregates) and of the structure of a solvated Zn2+ ion in the presence of a histidine residue (native insulin, see c). Details on the structural models are given in the Materials and methods. The blue dotted line represents the component of the simulated signal that is responsible for the features present only in native insulin and indicated by the circles (i.e., a shoulder at 4 Å−1 and a broadening of the peak around 7.6 Å−1). (b) k3-weighted Fourier-transformed EXAFS transmission spectra (dashed lines) of the same samples as in the top panel (a). Red continuous lines represent the signals simulated with the same structural models as in the top panel (a). (c) The structure of the dimeric human insulin (Protein Data Bank, PDB: 5CNY) is represented with a zoom on the structure of the zinc environment used to analyze the EXAFS data. Zinc is represented by a gray bead and is surrounded by water molecules and a histidine residue. Dashed black lines indicate the multiple-scattering contribution from the histidine to the EXAFS signal. To see this figure in color, go online.
Dynamic light scattering
Dynamic light scattering (DLS) measurements were performed using a DynaPro Nanostar detector (Wyatt Technology, Santa Barbara, CA) in Wyatt cuvettes with 10 μL of protein solution. Each measurement consisted of 5 s of integration 10 times with autoattenuation enabled. Insulin was dissolved at 5 mg/mL in a 0.25 M NaCl (pH 1.8), H2O solution without or with EDTA at the concentrations 0.25, 1, 5, and 20 mM, then centrifuged at 16,000 rpm for 30 min. Samples were measured three times, and data were analyzed using DYNAMICS software (Wyatt Technology).
Circular dichroism
Insulin was dissolved at 2 mg/mL in a 0.25 M NaCl (pH 1.8), H2O solution. The pH was set to 1.8 using sulfuric acid to avoid chlorine for the circular dichroism (CD) measurement. The insulin solution was filtered and measured with (5 mM) or without EDTA. Measurements were performed on a JASCO J-810 spectropolarimeter (Easton, MD) at 20 nm/s scanning speed with six accumulations.
X-ray powder diffraction
A fraction of the insulin powder samples prepared for neutron-scattering experiments was used for x-ray diffraction measurements. A small amount was sealed in a Mylar capillary, and diffraction patterns were recorded on beamline ID30B at the ESRF (Grenoble, France) using the following parameters: 20 images with oscillation step of 1° and 0.5 s exposure time for each image (100% transmission for 4 × 1012 ph/s at λ = 0.98 Å).
Aggregation kinetics
Insulin was dissolved in 0.25 M NaCl (pH 1.8), H2O solution, with or without EDTA at the concentrations 0.25, 1, and 5 mM, as indicated in Fig. 3, then filtered. 100 μL were pipetted into wells of a 96-well plate, and 2 μL of 1 mM ThT was added to each. Each well was sealed using vacuum grease and a glass slide to prevent any evaporation. One measurement was then performed every 5 min at 60°C for 24 h using 450 nm excitation light and a 480-nm detection filter for ThT. The measurements were performed on a BioTek Synergy H4 plate-reader (Winooski, VT) using the bottom configuration and a manual gain of 75.
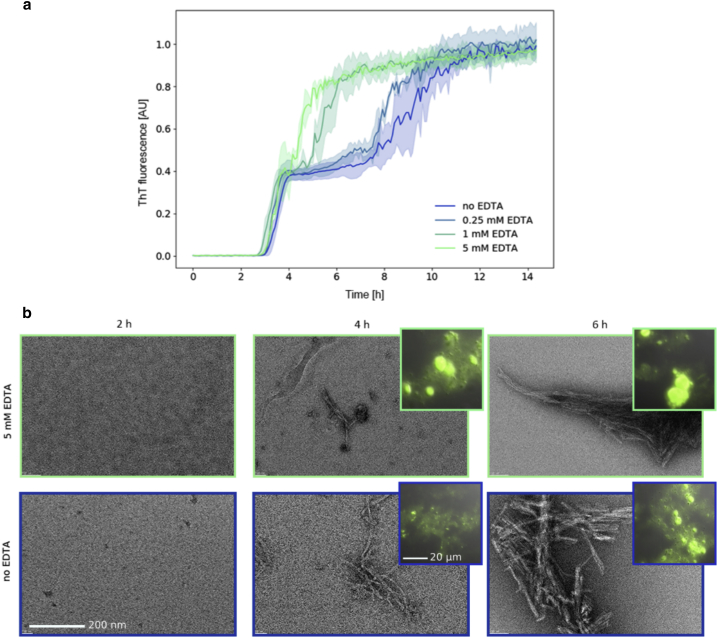
Figure 3.
EDTA modifies insulin aggregation kinetics. (a) Insulin aggregation kinetics at 60°C and pH 1.8 as probed by ThT fluorescence as a function of EDTA concentration. The color shaded areas indicate the standard deviation for three independent aggregation processes. (b) Transmission electron microscopy (fibrils) and fluorescence microscopy (spherulites, insets) images acquired during the kinetics without EDTA (blue curve in a, blue contoured images in b) and at 5 mM EDTA (light green in a, light green contoured images in b). Micrographs were acquired after 2 (left), 4 (middle), and 6 h (right) of incubation at 60°C and pH 1.8 and showed that fibrils and spherulites were formed roughly at the same time, with more numerous and larger spherulites appearing in the presence of EDTA. To see this figure in color, go online.
Electron microscopy
The same samples prepared for neutron-scattering experiments were absorbed on the clean side of a carbon film that was then deposited on mica, stained with 2% (w/v) uranyl acetate (pH 4.5), and transferred to a 400-mesh copper grid. The images were acquired under low dose conditions (<10 e−/Å2) with defocus values between 1.2 and 2.5 μm on a Tecnai 12 LaB6 electron microscope (Field Electron and Ion, Hillsboro, OR) at 120 kV accelerating voltage using a Gatan Orius 1000 CCD Camera (Pleasanton, CA).
Optical microscopy
For each sample prepared for neutron-scattering experiments, ThT was added to a final concentration of 20 μM. Subsequently, 8 μL of the sample was deposited between a glass slide and a coverslip. The samples were observed by an Olympus IX81 microscope (Tokyo, Japan) equipped with GFP filter cube set (Olympus) and differential interference contrast and an sCMOS Hamamatsu Orca Flash4 camera (Hamamatsu, Japan). The control of the microscope and the image acquisition were performed using the software Volocity.
Results
Screening and selection of homogeneous aggregated samples
The main approach followed for the isolation and identification of molecular mechanisms behind amyloid-related diseases (79) consists in changing the solution conditions (e.g., temperature, co-solvent, pH, and pressure) during in vitro experiments. The first step of this work was thus to screen and identify the experimental conditions for producing homogeneous samples, i.e., predominantly containing either fibrils or spherulites. This step was necessary to probe any potential difference in protein dynamics depending on the specific type of aggregates formed. The effect of salt, protein concentration, temperature (which has been shown to affect spherulite size and concentration (14)) and mechanical perturbation on insulin amyloid aggregation was investigated.
Effect of salt and protein concentration
Insulin was incubated for 24 h at 60°C in different solutions at pH 1.8, with salt and protein concentration ranging from 0.1 to 0.5 M and from 0.5 up to 5 mg/mL, respectively. Whereas protein concentration has no relevant effect on the final aggregate formation (i.e., both spherulites and fibrils were found at all the concentrations explored), salt concentration strongly influences it: at high salt concentration (≥0.5 mM), neither spherulites nor fibrils were observed (Fig. S1).
Effect of temperature
Insulin was then incubated at pH 1.8 for 24 h at either 50, 60, or 70°C and at salt and protein concentrations of 0.25 M and 2 mg/mL, respectively. Amyloid aggregation in quiescent conditions (i.e., without shaking) occurs only at or above 60°C (Fig. S2).
Effect of mechanical perturbation
Having defined the conditions for maximizing amyloid aggregation in quiescent conditions, mechanical perturbation was used to select either fibrils or spherulites. It was found that shaking the sample at 600 or 900 rpm during incubation produces a sample in which a large majority of the initial native protein is converted into elongated amyloid-like fibrils, even at 50°C. However, even though shaking allows the suppression of spherulite formation, it has a noteworthy effect on the morphology of fibrils. Electron microscopy revealed the presence of well-defined fibrils at 600 rpm and strongly aggregated ThT-positive material at 900 rpm (Fig. S3). A pure spherulite sample could be obtained by heating the protein in 0.25 M NaCl solution at 60 or 70°C without shaking and then including a postproduction treatment based on a series of vortexing, centrifugations, and supernatant-removal steps. After this treatment, no fibrils could be found in the samples (Fig. S4).
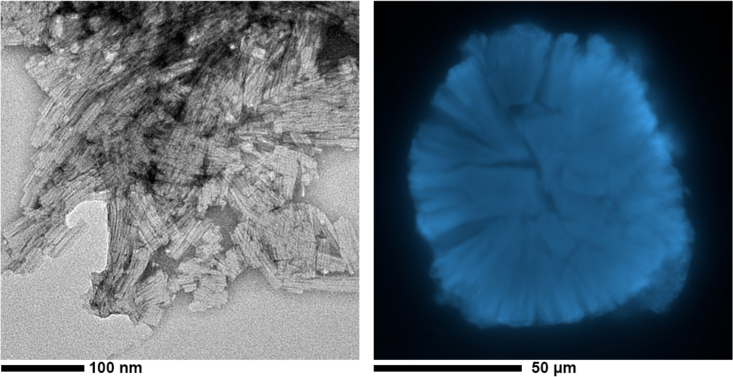
The screening described above enabled us to identify conditions for obtaining homogeneous fibril and spherulite samples. Fibrils were produced by incubating a human insulin solution at 50°C for 24 h at 5 mg/mL (pH 1.8), 0.25 mM NaCl under shaking at 600 rpm. Spherulites were produced by incubating a human insulin solution at 60°C for 24 h at 5 mg/mL (pH 1.8), 0.25 mM NaCl in quiescent conditions and then isolated by the postproduction treatment mentioned above and described in Materials and methods. Both fibril and spherulite samples were then washed in pure D2O (see Materials and methods for details) and subsequently freeze dried. As expected, the macroscopic morphology of both fibrils and spherulites appears slightly damaged after freeze drying, yet the main features of both fibrils and spherulites remain clearly visible (Fig. 1). Moreover, the presence of cross-β structure is revealed in both amyloid samples by x-ray powder diffraction, showing marked differences if compared with the α-helix-rich native state (Fig. S5).
Figure 1.
Morphology of insulin amyloid aggregates. (a) Shown is an electron microscopy image of freeze-dried insulin fibrils. (b) Shown is the confocal microscopy of a freeze-dried insulin spherulite. To see this figure in color, go online.
Protein dynamical transition is suppressed in insulin fibrils and spherulites
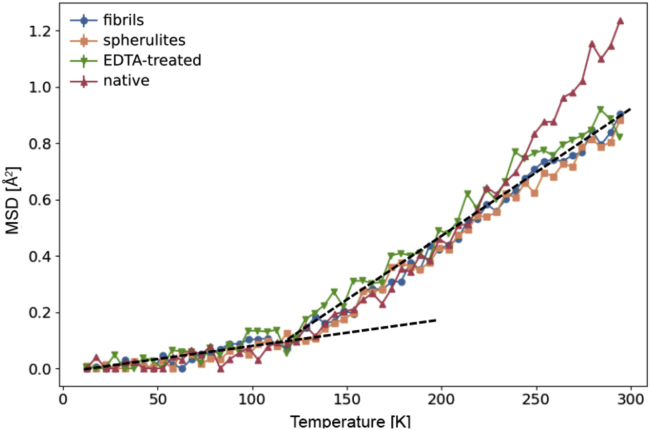
Incoherent neutron scattering of native insulin and of insulin fibrils and spherulites was then employed to monitor internal protein motions by measuring the elastic incoherent scattering signal as a function of temperature and the momentum transfer q. The dependence of the elastic incoherent signal on q provides information on the average motions of individual atoms in the system. From the q-dependence of incoherent elastic scattering from a D2O-hydrated protein powder, one can obtain the ensemble-averaged apparent MSDs of protein hydrogen atoms, reflecting the nanosecond signature of vibrations as well as backbone and side-chain relaxation (57). The scattering signal from hydrated insulin powders (hydration level h = 0.44 [D2O mass]/[protein mass]) was acquired during a heating scan at 0.2 K/min from 10 to 300 K, yielding MSDs as a function of temperature (Fig. 2). Native insulin shows the typical temperature-dependent MSDs of hydrated proteins (53), with harmonic behavior at cryo-temperature, a first anharmonic onset at ∼120 K, attributed to methyl group rotations entering the experimental window (80, 81, 82), and a second onset of water-sustained (56,83) anharmonic motions above ∼240 K (the so-called protein dynamical transition (53)). In both amyloid insulin species, i.e., fibrils and spherulites, the protein dynamical transition is completely suppressed, as in a dry protein (80), proving that the suppression is independent of the type of amyloid aggregates. Interestingly, the MSDs of native insulin treated with EDTA, known to be a highly efficient zinc-chelating agent, display a similar temperature dependence to that of aggregated samples and also lack the protein dynamical transition (Fig. 2; we note that EDTA was removed from the EDTA-treated sample before the neutron experiments (see Materials and methods)). Intriguingly, the dynamical transition can be partially suppressed by successive washing steps of native insulin without EDTA in pure D2O (Fig. S6). Because the formation of crystalline ice within the sample might also have led to dewetting of the protein at cryo-temperatures, and hence to the observed dry-like MSD temperature dependence, we measured the neutron diffraction signal of insulin fibrils and spherulites to determine if crystalline ice had formed at cryo-temperatures. As shown in Fig. S7, no Bragg peaks of hexagonal ice are present in the diffraction pattern of fibers at 200 K. A broad peak centered at ∼1.65 Å−1 and a peak at ∼1.35 Å−1 indicate the presence of amorphous ice and of amyloid cross-β structure, respectively.
Figure 2.
Amyloid aggregation suppresses the protein dynamical transition of hydrated insulin. Shown are mean-square displacements (MSDs) measured as a function of temperature by elastic incoherent neutron scattering on the backscattering spectrometer SPHERES (MLZ) for native insulin (red triangles), amyloid fibrils (blue circles), and spherulites (orange squares) and for insulin treated with EDTA (green triangles). To see this figure in color, go online.
Zinc release affects the kinetics of insulin amyloid aggregation
As revealed by neutron scattering, insulin amyloid species (both fibrils and spherulites) and EDTA-treated insulin (where zinc is expected to be chelated by EDTA and no aggregated material could be detected in x-ray diffraction data; Fig. S5) are characterized by the same dynamical behavior. This suggests that the presence or absence of zinc may affect the aggregation kinetics. To verify this, we monitored insulin aggregation kinetics at 60°C and pH 1.8 as a function of EDTA concentration. ThT (a probe whose fluorescence correlates with amyloid formation (84)) fluorescence was used as a probe of aggregate formation. As shown in Fig. 3, the kinetics in the absence of EDTA and without mechanical shaking is characterized by a double-sigmoidal curve as previously observed (85): a lag phase is followed by a first increase of the fluorescence signal, then by a plateau before a second increase. Adding EDTA clearly reduces the double-sigmoidal trend, which is essentially suppressed at high EDTA concentration. As expected, optical and electron microscopy observations at different time points along the aggregation kinetics showed that fibrils and spherulites are formed roughly at the same time, with more numerous and larger spherulites appearing in the presence of EDTA (Fig. 3). As shown above, shaking the insulin solution during incubation prevents spherulite formation. The kinetics experiments were then repeated as for those shown in Fig. 3 but in the presence of mechanical shaking, which inhibits the formation of spherulites and results in pure fibril samples. The results are shown in Fig. S8 and reveal that shaking suppresses the first plateau phase, whereas the addition of EDTA in the presence of mechanical perturbation does not produce any relevant effect. In addition, no spherulites were found with or without EDTA with mechanical shaking.
To explore any possible structural or conformational perturbation directly induced by EDTA, we also verified that in the range of EDTA concentration used both the oligomeric state and secondary structure of native insulin at room temperature are largely unaffected. In fact, a DLS analysis of size distribution showed a monodisperse signal for native insulin, with a single peak at a radius of ∼2.8 nm (suggestive of the presence of an insulin dimer) barely affected upon addition of EDTA up to a concentration of 5 mM (Fig. S9). CD in the 200–260-nm spectral region indicated that insulin secondary structure content is unchanged upon EDTA addition even at high protein/EDTA ratios (Fig. S10).
Insulin releases zinc upon aggregation
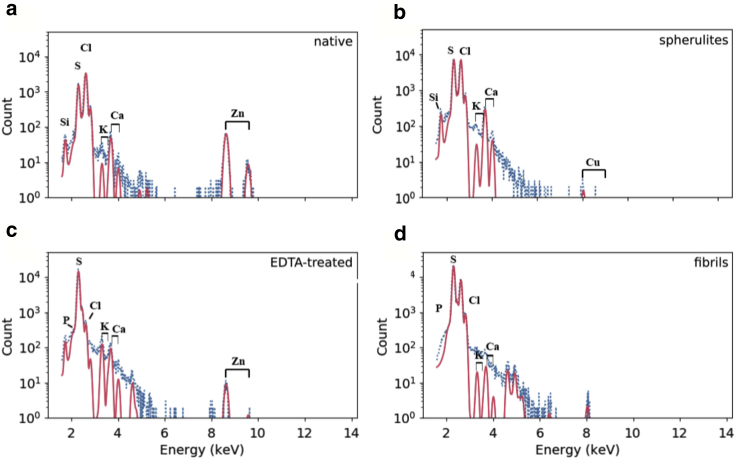
In the light of the effect of EDTA on both insulin dynamics and aggregation kinetics, our hypothesis then is that upon aggregation, insulin molecules release the zinc ions. To test this hypothesis, the presence of zinc in the different samples was explored. μPIXE (60,61) analysis was first used to determine the zinc concentration in the insulin samples investigated by neutron scattering. The results are shown in Fig. 4, and stoichiometric ratios are summarized in Table S1. μPIXE analysis revealed that native insulin contained 1.5 zinc atoms per insulin monomer, whereas zinc was below the detection limit (<0.001 atoms of zinc-monomer) in both fibrils and spherulites, and as expected, most of the zinc was removed in the EDTA-treated sample (0.02 zinc atoms per insulin monomer). However, the washed insulin sample still contains zinc with a stoichiometry of 0.116 zinc atoms per insulin monomer.
Figure 4.
Insulin loses coordinated zinc upon amyloid aggregation. Shown are the proton-induced x-ray emission spectra (blue points) and fit to the characteristic peaks (red lines) for (a) native insulin, (b) insulin spherulites, (c) EDTA-treated insulin, and (d) insulin fibrils. The theoretical positions for x-ray emission maxima are indicated in all cases. To see this figure in color, go online.
To get insights into the zinc-insulin interaction in native and amyloid conditions in the solution, we used XAS. XAS can be divided into x-ray absorption near-edge spectroscopy (XANES) and EXAFS, with XANES providing information on the oxidation state, geometry, and electronic configuration of the absorber atom. In contrast, EXAFS allows solving an atom’s local environment in terms of the number of neighboring atoms, their distance from the absorber atom, and their disorder on a short length scale. XAS experiments were performed at the zinc K-edge (9660.75 eV) on both a native-insulin solution at pH 1.8 and an insulin solution incubated at 60°C and pH 1.8 to induce the formation of amyloid species (because in quiescent conditions, both fibrils and spherulites are always formed, as described above, this sample was named “aggregates”) and on a ZnSO4 water solution. The XANES spectra in Fig. S11 do not show any significant difference between the various samples, indicating that Zn2+ ions are solvated in all the samples. Fig. 5 a shows k3-weighted EXAFS transmission spectra and their k3-weighted Fourier transforms (Fig. 5 b) for the three samples. The experimental curves in real space (Fig. 5 b) are comparable up to ∼3 Å, indicating that the first shell is very similar in all three samples. At larger distances, the curve for native insulin shows small but significant differences in the 3–5 Å range compared with the other two samples (in particular, the peak at ∼4.2 Å present only in native insulin). Small but significant differences are also evident in the reciprocal space (Fig. 5 a), namely a shoulder at 4 Å−1 and a broadening of the peak around 7.6 Å−1 visible only in native insulin. Fig. 5 a shows fitted curves in terms of structural models (see Materials and methods). The structural models contain either only a solvated Zn2+ ion, used to analyze data of both the ZnSO4 and the amyloid insulin solution samples, or a solvated Zn2+ ion in the vicinity of a histidine residue (Fig. 5 c), known to coordinate zinc in insulin, used to analyze data of the native-insulin solution. Both models were obtained from constrained molecular dynamics simulations based on refined geometrical parameters (86) and used to extract coordinates that were processed by GNXAS, as described in the Materials and methods. Structural parameters obtained from this analysis are reported in Table S2. A good agreement was found between experimental and calculated curves (Fig. 5 a). The Fourier-transformed, calculated curves also reproduced the peak positions of the Fourier-transformed experimental data up to ∼4.5 Å, in particular the differences in the 3–5-Å range (Fig. 5 b). XAS results indicate that Zn2+ interacts with a histidine residue in the native-insulin solution, whereas it is released in solution upon amyloid formation.
Discussion
Our study aimed at comparing the molecular dynamics of native and amyloid insulin species by neutron scattering and complementary biophysical methods. Zinc binding proved to be pivotal in controlling not only molecular dynamics at mesoscopic scale but also the aggregation kinetics. To enable the comparison of various insulin species and monitor potential differences in the protein dynamics depending on the type of aggregates, we first established protocols to produce batches containing either native insulin, insulin fibrils, or insulin spherulites at a high level of homogeneity. Formation of insulin fibrils and spherulites was triggered by reducing the pH value to 1.8 and increasing the temperature to at least 60°C. We found that although fine-tuning temperature and salt concentration allows amyloid aggregation into both fibrils and spherulites to be observed, shaking the vial during incubation leads to fibril formation and full inhibition of spherulite formation. We can argue that this inhibition of spherulite formation originates from mechanical perturbation preventing the formation of sufficiently stable spherulite-specific precursors. Spherulites are always produced along with fibrils in quiescent conditions. However, taking advantage of the much larger sedimentation rate of spherulites compared with the other species, they could be isolated from fibrils using gentle centrifugation.
Neutron backscattering spectroscopy showed a drastic change in the dynamics between native insulin and insulin in aggregated form above ∼240 K, but no significant difference between the two types of aggregates (spherulites and fibers) was observed (Fig. 2). Indeed large-amplitude motions associated with the protein dynamical transition are fully suppressed in insulin amyloid fibers and spherulites so that the temperature dependence of the MSDs resembles that of completely dry, nonfunctional proteins. In the native-insulin control measurement, a dynamical transition takes place (Fig. 2). These results are in contrast with our previous results on the τ-protein (52), in which no differences between native-state and fibril dynamics were observed on the same timescale, and on concanavalin A, in which internal dynamics were found to be larger in the amyloid than in the native species (51). The τ-results were suggestive of the protein internal energy, depending only on the amino acid composition (87) and not on the structure or aggregation state, as also supported by solution studies showing that the internal dynamics remain unchanged upon denaturation (88) and upon aggregation (89).
To rationalize the differential dynamical behavior of native and amyloid insulin, we suspected that interaction with Zn2+ ions might play a role because zinc is known to be essential for insulin expression and activity (33). We investigated how zinc binding affects the kinetics of insulin amyloid aggregation. Upon thermal treatment of the insulin solutions, ThT fluorescence profiles first showed an increase in the signal barely dependent on zinc presence, which has been suggested to be related to the formation of prefibrillar aggregates or early formation of fibrils (85) small enough to leave turbidity and temperature dependence unaffected. We attributed the intermediate plateau phase to spherulite formation as shown by fluorescence and electron microscopies (Fig. 3): both fibrils and spherulites are present, with spherulites being larger and more numerous in the presence of EDTA, i.e., when zinc is removed. After the first plateau, a second increase occurs, followed by a final plateau, where spherulites are densely present and significantly larger. Increasing EDTA concentration gradually suppresses the double-sigmoidal behavior and enhances spherulite formation. This indicates that the presence of zinc is the limiting step for the more efficient conversion of insulin native molecules into spherulites. When the zinc is fully removed by adding a sufficient amount of EDTA, the intermediate phase disappears. If the loss of zinc occurs at the level of single molecules before aggregation or happens once the molecules are embedded in high molecular weight (MW) species remains to be clarified. Importantly, shaking also suppresses the double-sigmoidal behavior, as well as the spherulite formation. From the kinetic results, we can conclude that 1) the first plateau can be attributed to spherulite formation, and 2) zinc binding inhibits spherulite formation, probably through electrostatic repulsion.
To obtain quantitative information on zinc content in our samples, we performed measurements with the μPIXE technique, the results of which unambiguously show that zinc was below the limit of detection (<0.001 zinc atom/monomer; Table S1) in amyloid aggregates, where the dynamical transition is suppressed. Furthermore, an EDTA-treated control sample is shown by μPIXE to have a zinc content reduced by a factor of 75 compared with the native protein (Fig. 4) and lacks the protein dynamical transition at 240 K in neutron-scattering experiments (Fig. 2). We note that the EDTA-treated sample and the native form present the same secondary structure according to CD data (Fig. S10) and are predominantly dimeric according to DLS (Fig. S9). This means that both the conformational and colloidal stability of the solution are not significantly influenced by the presence of EDTA and the loss of zinc is mainly affecting the dynamics of the single protein molecule. Indeed, an insulin sample with a zinc content intermediate between those of native and amyloid insulin (as confirmed by μPIXE; Table S1), obtained by successive washing steps of a native sample, shows a dynamical transition but with reduced MSD if compared with the native sample (Fig. S6). Zinc interaction with insulin thus modulates molecular dynamics, and its absence suppresses functionally relevant motions activated at the dynamical transition in amyloid insulin without modifying secondary, tertiary, or quaternary structure. XAS data in solution (Fig. 5) confirmed that zinc interacts with native insulin while it is released upon amyloid aggregation. Both experimental and computational studies have revealed weak and rather nonspecific interactions of divalent cations with insulin even at low pH (90,91). In the case of protein aggregation, a study showed that nonspecific divalent cation binding can occur for insulin and the SH3 domain of phosphatidyl-inositol 3-kinase at pH 2 during aggregation (92). Our results suggest that in our conditions, the zinc-insulin interaction is not compatible with a coordination mechanism because the zinc XANES spectra, and thus its electronic configuration, are not affected by the presence of insulin (Fig. S11), whereas the EXAFS signal can be analyzed in terms of an interaction with histidine residues (Fig. 5). Uncovering the exact zinc-insulin binding mechanism at low pH is beyond the scope of this article. The XAS results thus established that zinc loss is not an effect of the sample treatment (i.e., buffer exchange and freeze drying) preceding neutron-scattering experiments.
Translating our in vitro results to a physiological context, they imply that amyloid aggregation of insulin in vivo could induce an accumulation of free zinc ions at the injection sites, where IDA is observed. Zinc is anyway released from monomeric insulin during its function (33), yet our finding suggests that a substantially larger quantity of zinc may be released and could accompany IDA. Zinc overload is known to be a pro-oxidant condition leading to oxidative damage of biomolecules. When free zinc ion concentrations increase, zinc can bind to proteins that otherwise would not interact with zinc and affect their functions, thus inducing the production of free radicals by inhibiting antioxidant enzymes and the mitochondrial respiratory chain (93). On the basis of our results, we are not able to quantitatively assess the potential cytotoxicity of zinc release during IDA because the threshold at which zinc ion concentration becomes high enough to produce pro-oxidant and cytotoxic effects depends on the zinc buffering capacity of cells (33,93). In general, the direct implication of our results for physiological conditions is not straightforward, and only future in vivo investigations could clarify the physiological impact of the zinc accumulation revealed by our study.
An important information provided by the XAS results is that even in native insulin, which interacts with zinc, the metal ion environment is largely composed of water molecules. The presence of the high charge on the Zn2+ ion is known to perturb water molecules in the first and second solvation shells, which correspond to at least 20 water molecules interacting with one Zn2+ ion (94). Considering the 1.5 Zn/protein ratio indicated by the μPIXE analysis, the presence of zinc on the protein surface corresponds to a minimal hydration level h = [20 × 1.5 × water MW]/[insulin MW]∼0.1. In other words, the presence of Zn2+ favors the hydration of the highly hydrophobic insulin surface, thus influencing the dynamical behavior as revealed by neutron scattering.
Taken together, the results from neutron scattering, μPIXE, and XAS clearly show that interaction with zinc is the key parameter that allows hydrated insulin to undergo the protein dynamical transition, whereas the dramatic dynamical effect on fibrils and spherulites has to be attributed to loss of zinc. Removal of zinc results in a complete loss of water-sustained backbone and side-chain motions with the MSD profile corresponding to that of a dry protein, thus suggesting a dehydration effect upon zinc release. As previously suggested (52,95,96), the changes in hydration water entropy may be one of the “driving forces” leading proteins to form amyloid aggregates. Restructuring of hydration water is expected to be involved both in the initial steps of unfolding/cross-β-amyloid formation and in the growth of amyloid aggregates, thereby implying a possible change in the water configurational entropy (95). Protein dehydration has also been shown to modify amyloid aggregation kinetics (97). The presence of metal ions integrated within the protein structure, which generally influences protein solvation and electrostatics of the protein surface (98), can add a key parameter to the energetic balance between native and amyloid state. In the case of human insulin, which is highly hydrophobic (99), we postulate that zinc binding favors protein hydration, thus modifying both protein dynamics, as revealed by neutron scattering and water entropy. Indeed, dewetting of hydrophobic groups (which are known to reduce water mobility (100) and entropy (101)) during aggregation and subsequent release of water molecules in the bulk phase correspond to an increase of water entropy (96). The presence of zinc coordinated to the protein reduces the hydrophobicity of the insulin surface and then the propensity for aggregation, whereas zinc removal increases the entropy difference between native and amyloid state and then promotes amyloid aggregation.
It has been shown that a Thr to His mutation in the chain A of human insulin leads to a significant increase in the lag phase of amyloid aggregation (102). More generally, studies of protein rational design (48,103) have shown how the introduction of His binding sites for Zn2+ into hydrophobic peptides can modify both their stability and dynamics. Our results suggest that protein hydration changes upon Zn2+ binding/release can contribute to such modifications, as revealed by our finding that both dynamics and aggregation kinetics of human insulin change as an effect of Zn2+ release from His sites. Changes in metal-modulated protein hydration might be a general strategy to control protein stability and tune protein aggregation into amorphous and ordered superstructures. For this reason, our findings provide a novel perspective, to our knowledge, on the basic understanding of the insulin aggregation process as well as on strategies for the possible development of safer and more stable biopharmaceuticals.
Author contributions
K.P., V.F., M.W., and G.S. designed the research. K.P., G.S., D.N., and M.Z. performed neutron backscattering spectroscopy, and K.P. analyzed the data. K.P. and V.C. performed neutron diffraction and analyzed the data. K.P. and V.F. defined the sample preparation conditions, and K.P. prepared and characterized the samples. G.W.G. and E.F.G. performed μPIXE and analyzed the data. A.L., K.P., and G.S. performed XAS, and A.L. analyzed data based on structures extracted from the MD simulations performed by K.P. K.P. performed the CD, DLS, and fluorescence spectroscopy. K.P. and G.S. performed the x-ray powder diffraction. K.P., V.F., and G.S. wrote the article, with input from all authors.
Acknowledgments
We thank Daphna Fenel, Christine Moriscot, and Guy Schoehn from the Electron Microscopy platform; Caroline Mas and Marc Jamin for assistance and access to the Biophysical platform; and Aline Le Roy, Michel Thépaut, and Christine Ebel for assistance and access to the Protein Analysis Online platform. The access to Institut Laue-Langevin and ESRF beamlines to perform diffraction characterization is also acknowledged.
This work used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). V.F. acknowledges Villum Fonden for the Villum Young Investigator Grant “Protein Superstructures as Smart Biomaterials (ProSmart)” 2018–2023 (project number: 19175). The electron microscope facility is supported by the Rhône-Alpes Region, Fondation Recherche Médicale (FRM), and funds from FEDER, Centre National de la Recherche Scientifique (CNRS), CEA, University of Grenoble, EMBL, and GIS-Infrastructures en Biologie Santé et Agronomie (IBISA). The financial support provided by JCNS to perform neutron-scattering measurements at the Heinz Maier-Leibnitz Zentrum (MLZ), Garching, Germany, is gratefully acknowledged.
Editor: Michael Sattler.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2020.11.2280.
Contributor Information
Kevin Pounot, Email: pounot@ill.fr.
Vito Foderà, Email: vito.fodera@sund.ku.dk.
Giorgio Schirò, Email: giorgio.schiro@ibs.fr.
Supporting material
References
- 1.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Fitzpatrick A.W.P., Falcon B., Scheres S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuttle M.D., Comellas G., Rienstra C.M. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetri V., Foderà V. The route to protein aggregate superstructures: particulates and amyloid-like spherulites. FEBS Lett. 2015;589:2448–2463. doi: 10.1016/j.febslet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Exley C., House E., Donald A.M. Spherulites of amyloid-β42 in vitro and in Alzheimer’s disease. J. Alzheimers Dis. 2010;20:1159–1165. doi: 10.3233/JAD-2010-091630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schack M.M., Møller E.H., Groenning M. Optimization of infrared microscopy to assess secondary structure of insulin molecules within individual subvisible particles in aqueous formulations. J. Pharm. Sci. 2019;108:1117–1129. doi: 10.1016/j.xphs.2018.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Knowles T.P.J., Mezzenga R. Amyloid fibrils as building blocks for natural and artificial functional materials. Adv. Mater. 2016;28:6546–6561. doi: 10.1002/adma.201505961. [DOI] [PubMed] [Google Scholar]
- 8.Fändrich M., Nyström S., Hammarström P. Amyloid fibril polymorphism: a challenge for molecular imaging and therapy. J. Intern. Med. 2018;283:218–237. doi: 10.1111/joim.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condello C., Lemmin T., Prusiner S.B. Structural heterogeneity and intersubject variability of Aβ in familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2018;115:E782–E791. doi: 10.1073/pnas.1714966115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen J., Mahler J., Jucker M. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2017;114:13018–13023. doi: 10.1073/pnas.1713215114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiménez J.L., Nettleton E.J., Saibil H.R. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brange J., Andersen L., Rasmussen E. Toward understanding insulin fibrillation. J. Pharm. Sci. 1997;86:517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 13.Krebs M.R.H., Bromley E.H.C., Donald A.M. The mechanism of amyloid spherulite formation by bovine insulin. Biophys. J. 2005;88:2013–2021. doi: 10.1529/biophysj.104.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith M.I., Foderà V., Donald A.M. Factors affecting the formation of insulin amyloid spherulites. Colloids Surf. B Biointerfaces. 2012;89:216–222. doi: 10.1016/j.colsurfb.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Vetri V., Piccirilli F., Foderà V. Ethanol controls the self-assembly and mesoscopic properties of human insulin amyloid spherulites. J. Phys. Chem. B. 2018;122:3101–3112. doi: 10.1021/acs.jpcb.8b01779. [DOI] [PubMed] [Google Scholar]
- 16.Shikama Y., Kitazawa J., Yagihashi S. Localized amyloidosis at the site of repeated insulin injection in a diabetic patient. Intern. Med. 2010;49:397–401. doi: 10.2169/internalmedicine.49.2633. [DOI] [PubMed] [Google Scholar]
- 17.Swift B. Examination of insulin injection sites: an unexpected finding of localized amyloidosis. Diabet. Med. 2002;19:881–882. doi: 10.1046/j.1464-5491.2002.07581.x. [DOI] [PubMed] [Google Scholar]
- 18.Gupta Y., Singla G., Singla R. Insulin-derived amyloidosis. Indian J. Endocrinol. Metab. 2015;19:174–177. doi: 10.4103/2230-8210.146879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dische F.E., Wernstedt C., Watkins P.J. Insulin as an amyloid-fibril protein at sites of repeated insulin injections in a diabetic patient. Diabetologia. 1988;31:158–161. doi: 10.1007/BF00276849. [DOI] [PubMed] [Google Scholar]
- 20.Nagase T., Katsura Y., Matsuoka T. The insulin ball. Lancet. 2009;373:184. doi: 10.1016/S0140-6736(09)60041-6. [DOI] [PubMed] [Google Scholar]
- 21.Samlaska C., Reber S., Murry T. Insulin-derived amyloidosis: the insulin ball, amyloidoma. JAAD Case Rep. 2020;6:351–353. doi: 10.1016/j.jdcr.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson M.R. Insulin amyloid at injection sites of patients with diabetes. Amyloid. 2016;23:139–147. doi: 10.1080/13506129.2016.1179183. [DOI] [PubMed] [Google Scholar]
- 23.Ohno Y., Seki T., Seki T. Investigation of factors that cause insulin precipitation and/or amyloid formation in insulin formulations. J. Pharm. Health Care Sci. 2019;5:22. doi: 10.1186/s40780-019-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J., Li A., Li J. Advances in pH-sensitive polymers for smart insulin delivery. Macromol. Rapid Commun. 2017;38:1700413. doi: 10.1002/marc.201700413. [DOI] [PubMed] [Google Scholar]
- 25.Ristow M. Neurodegenerative disorders associated with diabetes mellitus. J. Mol. Med. (Berl.) 2004;82:510–529. doi: 10.1007/s00109-004-0552-1. [DOI] [PubMed] [Google Scholar]
- 26.Craft S., Watson G.S. Insulin and neurodegenerative disease: shared and specific mechanisms. Lancet Neurol. 2004;3:169–178. doi: 10.1016/S1474-4422(04)00681-7. [DOI] [PubMed] [Google Scholar]
- 27.Schubert M., Gautam D., Brüning J.C. Role for neuronal insulin resistance in neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 2004;101:3100–3105. doi: 10.1073/pnas.0308724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupke D.W., Linderstrøm-Lang K. On the size of the monomer of insulin. Biochim. Biophys. Acta. 1954;13:153–154. doi: 10.1016/0006-3002(54)90293-0. [DOI] [PubMed] [Google Scholar]
- 29.Pease L.F., III, Sorci M., Belfort G. Probing the nucleus model for oligomer formation during insulin amyloid fibrillogenesis. Biophys. J. 2010;99:3979–3985. doi: 10.1016/j.bpj.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foderà V., Donald A.M. Tracking the heterogeneous distribution of amyloid spherulites and their population balance with free fibrils. Eur Phys J E Soft Matter. 2010;33:273–282. doi: 10.1140/epje/i2010-10665-4. [DOI] [PubMed] [Google Scholar]
- 31.Vestergaard B., Groenning M., Svergun D.I. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim A.C., Lim S., Kim Y.K. Metal ion effects on Aβ and tau aggregation. Int. J. Mol. Sci. 2018;19:128. doi: 10.3390/ijms19010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y.V. Zinc and insulin in pancreatic beta-cells. Endocrine. 2014;45:178–189. doi: 10.1007/s12020-013-0032-x. [DOI] [PubMed] [Google Scholar]
- 34.Noormägi A., Gavrilova J., Palumaa P. Zn(II) ions co-secreted with insulin suppress inherent amyloidogenic properties of monomeric insulin. Biochem. J. 2010;430:511–518. doi: 10.1042/BJ20100627. [DOI] [PubMed] [Google Scholar]
- 35.Frankær C.G., Sønderby P., Harris P. Insulin fibrillation: the influence and coordination of Zn2. J. Struct. Biol. 2017;199:27–38. doi: 10.1016/j.jsb.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Domínguez-Calva J.A., Pérez-Vázquez M.L., Quintanar L. Mercury-induced aggregation of human lens γ-crystallins reveals a potential role in cataract disease. J. Biol. Inorg. Chem. 2018;23:1105–1118. doi: 10.1007/s00775-018-1607-z. [DOI] [PubMed] [Google Scholar]
- 37.Ahmadi S., Zhu S., Kraatz H.-B. Interaction of metal ions with tau protein. The case for a metal-mediated tau aggregation. J. Inorg. Biochem. 2019;194:44–51. doi: 10.1016/j.jinorgbio.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Pan B.B., Yang Y., Su X.C. Coordination of platinum to α-synuclein inhibits filamentous aggregation in solution. ChemBioChem. 2019;20:1953–1958. doi: 10.1002/cbic.201900224. [DOI] [PubMed] [Google Scholar]
- 39.Lee M., Kim J.I., Eom K. Metal ions affect the formation and stability of amyloid β aggregates at multiple length scales. Phys. Chem. Chem. Phys. 2018;20:8951–8961. doi: 10.1039/C7CP05072K. [DOI] [PubMed] [Google Scholar]
- 40.Roosen-Runge F., Zhang F., Roth R. Ion-activated attractive patches as a mechanism for controlled protein interactions. Sci. Rep. 2014;4:7016. doi: 10.1038/srep07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng J., Olrichs N.K., Helms J.B. Metal ions and redox balance regulate distinct amyloid-like aggregation pathways of GAPR-1. Sci. Rep. 2019;9:15048. doi: 10.1038/s41598-019-51232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lautenschläger J., Stephens A.D., Schierle G.S.K. C-terminal calcium binding of α-synuclein modulates synaptic vesicle interaction. Nat. Commun. 2018;9:712. doi: 10.1038/s41467-018-03111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deas E., Cremades N., Abramov A.Y. Alpha-synuclein oligomers interact with metal ions to induce oxidative stress and neuronal death in Parkinson’s disease. Antioxid. Redox Signal. 2016;24:376–391. doi: 10.1089/ars.2015.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mantyh P.W., Ghilardi J.R., Maggio J.E. Aluminum, iron, and zinc ions promote aggregation of physiological concentrations of β-amyloid peptide. J. Neurochem. 1993;61:1171–1174. doi: 10.1111/j.1471-4159.1993.tb03639.x. [DOI] [PubMed] [Google Scholar]
- 45.Huang X., Atwood C.S., Bush A.I. Trace metal contamination initiates the apparent auto-aggregation, amyloidosis, and oligomerization of Alzheimer’s Abeta peptides. J. Biol. Inorg. Chem. 2004;9:954–960. doi: 10.1007/s00775-004-0602-8. [DOI] [PubMed] [Google Scholar]
- 46.Moir R.D., Atwood C.S., Bush A.I. Mounting evidence for the involvement of zinc and copper in Alzheimer’s disease. Eur. J. Clin. Invest. 1999;29:569–570. doi: 10.1046/j.1365-2362.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- 47.Ariöz C., Wittung-Stafshede P. Folding of copper proteins: role of the metal? Q. Rev. Biophys. 2018;51:e4. doi: 10.1017/S0033583518000021. [DOI] [PubMed] [Google Scholar]
- 48.Handel T.M., Williams S.A., DeGrado W.F. Metal ion-dependent modulation of the dynamics of a designed protein. Science. 1993;261:879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- 49.Acharya S., Ahmad B., Lapidus L. Aggregation of alpha-synuclein is kinetically controlled by intramolecular diffusion. Biophys. J. 2013;104:51a. doi: 10.1073/pnas.1109526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srivastava K.R., Lapidus L.J. Prion protein dynamics before aggregation. Proc. Natl. Acad. Sci. USA. 2017;114:3572–3577. doi: 10.1073/pnas.1620400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schirò G., Vetri V., Cupane A. Neutron scattering reveals enhanced protein dynamics in concanavalin A amyloid fibrils. J. Phys. Chem. Lett. 2012;3:992–996. doi: 10.1021/jz300082x. [DOI] [PubMed] [Google Scholar]
- 52.Fichou Y., Schirò G., Weik M. Hydration water mobility is enhanced around tau amyloid fibers. Proc. Natl. Acad. Sci. USA. 2015;112:6365–6370. doi: 10.1073/pnas.1422824112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doster W., Cusack S., Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989;337:754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- 54.Wuttke J., Budwig A., Staringer S. SPHERES, Jülich’s high-flux neutron backscattering spectrometer at FRM II. Rev. Sci. Instrum. 2012;83:075109. doi: 10.1063/1.4732806. [DOI] [PubMed] [Google Scholar]
- 55.Zamponi M., Khaneft M. SPHERES: Backscattering spectrometer. Journal of large-scale research facilities JLSRF. 2015;1:30. [Google Scholar]
- 56.Schirò G., Fichou Y., Weik M. Translational diffusion of hydration water correlates with functional motions in folded and intrinsically disordered proteins. Nat. Commun. 2015;6:6490. doi: 10.1038/ncomms7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabel F., Bicout D., Zaccai G. Protein dynamics studied by neutron scattering. Q. Rev. Biophys. 2002;35:327–367. doi: 10.1017/s0033583502003840. [DOI] [PubMed] [Google Scholar]
- 58.Yi Z., Miao Y., Smith J.C. Derivation of mean-square displacements for protein dynamics from elastic incoherent neutron scattering. J. Phys. Chem. B. 2012;116:5028–5036. doi: 10.1021/jp2102868. [DOI] [PubMed] [Google Scholar]
- 59.Kneller G.R., Hinsen K. Quantitative model for the heterogeneity of atomic position fluctuations in proteins: a simulation study. J. Chem. Phys. 2009;131:045104. doi: 10.1063/1.3170941. [DOI] [PubMed] [Google Scholar]
- 60.Garman E.F., Grime G.W. Elemental analysis of proteins by microPIXE. Prog. Biophys. Mol. Biol. 2005;89:173–205. doi: 10.1016/j.pbiomolbio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Grime G.W., Zeldin O.B., Garman E.F. High-throughput PIXE as an essential quantitative assay for accurate metalloprotein structural analysis: development and application. J. Am. Chem. Soc. 2020;142:185–197. doi: 10.1021/jacs.9b09186. [DOI] [PubMed] [Google Scholar]
- 62.Grime G.W., Dawson M., Watt F. The Oxford submicron nuclear microscopy facility. Nucl. Instrum. Methods Phys. Res. B. 1991;54:52–63. [Google Scholar]
- 63.Maxwell J.A., Teesdale W.J., Campbell J.L. The Guelph PIXE software package II. Nucl. Instrum. Methods Phys. Res. B. 1995;95:407–421. [Google Scholar]
- 64.Nikitenko S., Beale A.M., Bras W. Implementation of a combined SAXS/WAXS/QEXAFS set-up for time-resolved in situexperiments. J. Synchrotron Radiat. 2008;15:632–640. doi: 10.1107/S0909049508023327. [DOI] [PubMed] [Google Scholar]
- 65.Filipponi A., Natoli C.R., Natoli C.R. X-ray-absorption spectroscopy and n-body distribution functions in condensed matter. I. Theory. Phys. Rev. B Condens. Matter. 1995;52:15122–15134. doi: 10.1103/physrevb.52.15122. [DOI] [PubMed] [Google Scholar]
- 66.Filipponi A., Di Cicco A. X-ray-absorption spectroscopy and n-body distribution functions in condensed matter. II. Data analysis and applications. Phys. Rev. B Condens. Matter. 1995;52:15135–15149. doi: 10.1103/physrevb.52.15135. [DOI] [PubMed] [Google Scholar]
- 67.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 68.Obst S., Bradaczek H. Molecular dynamics simulations of zinc ions in water using CHARMM. J Mol Model. 1997;3:224–232. [Google Scholar]
- 69.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. [Google Scholar]
- 71.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 73.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 74.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 75.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 76.Grubmüller H., Heller H., Schulten K. Generalized verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991;6:121–142. [Google Scholar]
- 77.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 78.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 79.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 80.Roh J.H., Novikov V.N., Sokolov A.P. Onsets of anharmonicity in protein dynamics. Phys. Rev. Lett. 2005;95:038101. doi: 10.1103/PhysRevLett.95.038101. [DOI] [PubMed] [Google Scholar]
- 81.Schiró G., Caronna C., Cupane A. Direct evidence of the amino acid side chain and backbone contributions to protein anharmonicity. J. Am. Chem. Soc. 2010;132:1371–1376. doi: 10.1021/ja908611p. [DOI] [PubMed] [Google Scholar]
- 82.Wood K., Tobias D.J., Weik M. The low-temperature inflection observed in neutron scattering measurements of proteins is due to methyl rotation: direct evidence using isotope labeling and molecular dynamics simulations. J. Am. Chem. Soc. 2010;132:4990–4991. doi: 10.1021/ja910502g. [DOI] [PubMed] [Google Scholar]
- 83.Wood K., Frölich A., Weik M. Coincidence of dynamical transitions in a soluble protein and its hydration water: direct measurements by neutron scattering and MD simulations. J. Am. Chem. Soc. 2008;130:4586–4587. doi: 10.1021/ja710526r. [DOI] [PubMed] [Google Scholar]
- 84.LeVine H., III Quantification of beta-sheet amyloid fibril structures with thioflavin T. Meth. Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 85.Foderà V., Cataldo S., Leone M. Self-organization pathways and spatial heterogeneity in insulin amyloid fibril formation. J. Phys. Chem. B. 2009;113:10830–10837. doi: 10.1021/jp810972y. [DOI] [PubMed] [Google Scholar]
- 86.Cauët E., Bogatko S., Bylaska E.J. Structure and dynamics of the hydration shells of the Zn(2+) ion from ab initio molecular dynamics and combined ab initio and classical molecular dynamics simulations. J. Chem. Phys. 2010;132:194502. doi: 10.1063/1.3421542. [DOI] [PubMed] [Google Scholar]
- 87.Schirò G., Caronna C., Cupane A. The “protein dynamical transition” does not require the protein polypeptide chain. J. Phys. Chem. Lett. 2011;2:2275–2279. [Google Scholar]
- 88.Grimaldo M., Roosen-Runge F., Seydel T. High-resolution neutron spectroscopy on protein solution samples. EPJ Web of Conferences. 2015;83:02005. [Google Scholar]
- 89.Pounot K., Chaaban H., Seydel T. Tracking internal and global diffusive dynamics during protein aggregation by high-resolution neutron spectroscopy. J. Phys. Chem. Lett. 2020;11:6299–6304. doi: 10.1021/acs.jpclett.0c01530. [DOI] [PubMed] [Google Scholar]
- 90.Cunningham L.W., Fischer R.L., Vestling C.S. A study of the binding of zinc and cobalt by insulin. J. Am. Chem. Soc. 1955;77:5703–5707. [Google Scholar]
- 91.Duboué-Dijon E., Delcroix P., Jungwirth P. Binding of divalent cations to insulin: capillary electrophoresis and molecular simulations. J. Phys. Chem. B. 2018;122:5640–5648. doi: 10.1021/acs.jpcb.7b12097. [DOI] [PubMed] [Google Scholar]
- 92.Buell A.K., Hung P., Knowles T.P.J. Electrostatic effects in filamentous protein aggregation. Biophys. J. 2013;104:1116–1126. doi: 10.1016/j.bpj.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maret W. Zinc in pancreatic islet biology, insulin sensitivity, and diabetes. Prev. Nutr. Food Sci. 2017;22:1–8. doi: 10.3746/pnf.2017.22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fatmi M.Q., Hofer T.S., Rode B.M. An extended ab initio QM/MM MD approach to structure and dynamics of Zn(II) in aqueous solution. J. Chem. Phys. 2005;123:054514. doi: 10.1063/1.1996575. [DOI] [PubMed] [Google Scholar]
- 95.Thirumalai D., Reddy G., Straub J.E. Role of water in protein aggregation and amyloid polymorphism. Acc. Chem. Res. 2012;45:83–92. doi: 10.1021/ar2000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krone M.G., Hua L., Shea J.-E. Role of water in mediating the assembly of Alzheimer amyloid-β Abeta16-22 protofilaments. J. Am. Chem. Soc. 2008;130:11066–11072. doi: 10.1021/ja8017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mukherjee S., Chowdhury P., Gai F. Effect of dehydration on the aggregation kinetics of two amyloid peptides. J. Phys. Chem. B. 2009;113:531–535. doi: 10.1021/jp809817s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Capdevila D.A., Edmonds K.A., Giedroc D.P. Functional role of solvent entropy and conformational entropy of metal binding in a dynamically driven allosteric system. J. Am. Chem. Soc. 2018;140:9108–9119. doi: 10.1021/jacs.8b02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bigelow C.C. On the average hydrophobicity of proteins and the relation between it and protein structure. J. Theor. Biol. 1967;16:187–211. doi: 10.1016/0022-5193(67)90004-5. [DOI] [PubMed] [Google Scholar]
- 100.Rezus Y.L.A., Bakker H.J. Observation of immobilized water molecules around hydrophobic groups. Phys. Rev. Lett. 2007;99:148301. doi: 10.1103/PhysRevLett.99.148301. [DOI] [PubMed] [Google Scholar]
- 101.Dahanayake J.N., Mitchell-Koch K.R. Entropy connects water structure and dynamics in protein hydration layer. Phys. Chem. Chem. Phys. 2018;20:14765–14777. doi: 10.1039/c8cp01674g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nielsen L., Frokjaer S., Fink A.L. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry. 2001;40:8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
- 103.Pessi A., Bianchi E., Sollazzo M. A designed metal-binding protein with a novel fold. Nature. 1993;362:367–369. doi: 10.1038/362367a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.