Figure 4.
Structures of Fab 222 in complex with WT and mutant RBDs
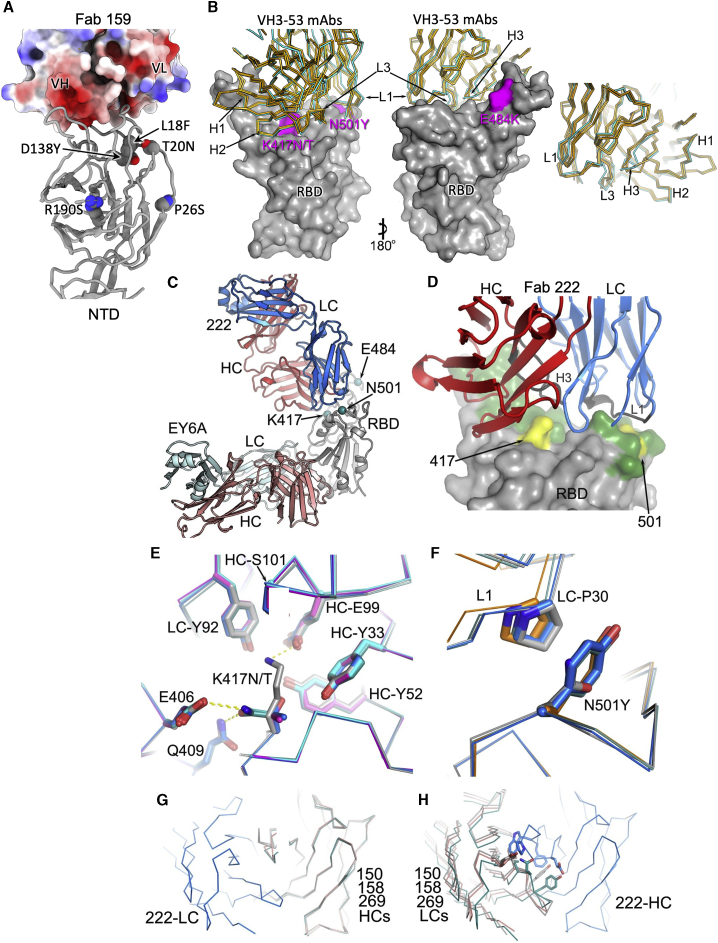
(A) Electrostatic surface depiction of Fab 159 in complex with the NTD depicted as a gray cartoon. Residues mutated in P.1 are shown as vdw radii representation for the original amino acid (oxygen, red; nitrogen, blue; carbon, gray).
(B) Left to right: back and front surfaces of the RBD (gray) bound to a number of typical VH3-53 Fabs (Cα trace with 222 shown in cyan and 150, 158, and 269 shown in gold). P1 mutations in the RBD are highlighted in magenta and labeled. In this group, mAb 222 has a slightly longer CDR-H3.
(C) Crystal structure of P1 RBD/222 Fab and EY6A Fabs (Zhou et al., 2020).
(D) Close-up of 222 CDRs interacting with the RBD (gray), mutations are highlighted in yellow on the green ACE2 interface.
(E and F) K417N/T interactions with Fab 222 (E) and N501Y interactions with Fab 222 (F) in the K417N (cyan), K417T (magenta), P.1 (blue), and P.1.351 (teal) RBD-Fab 222 complex structures compared with the WT RBD-Fab 222 (gray) complex by superimposing the RBD.
(G) Overlay of Vh domains of Fabs 150 (gray), 158 (teal), 269 (salmon), and 222 (blue) showing that the light chain of 222 does not clash with any of other three heavy chains, while (H) shows the light chains of 150, 158, and 269 clash with the heavy chain of 222. For clarity, only the light chain of 222 in (G) and the heavy chain of 222 in (H) are shown. Light-chain gene usage, RBD contacts, and somatic mutations are shown in Figure S3 and Table S3.