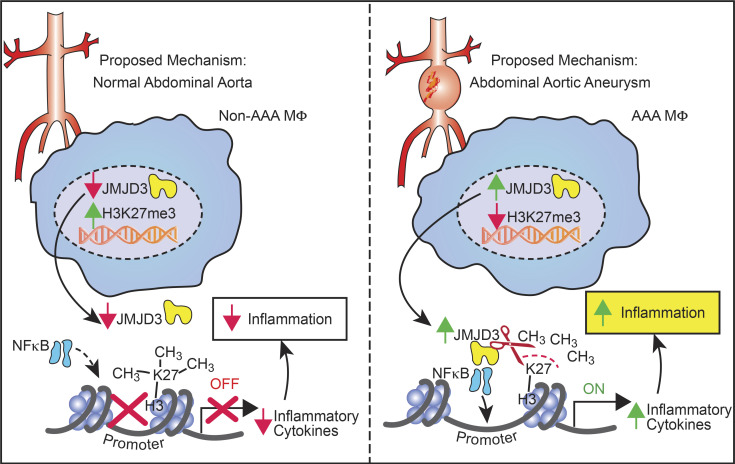
Abdominal aortic aneurysms (AAAs) are a life-threatening disease characterized by macrophage infiltration contributing to pathological vascular remodeling. Herein, we demonstrate that the histone demethylase JMJD3 is a critical regulator of inflammation during AAA development and cell-specific inhibition reduces AAA progression.
Abstract
Abdominal aortic aneurysms (AAAs) are a life-threatening disease for which there is a lack of effective therapy preventing aortic rupture. During AAA formation, pathological vascular remodeling is driven by macrophage infiltration, and the mechanisms regulating macrophage-mediated inflammation remain undefined. Recent evidence suggests that an epigenetic enzyme, JMJD3, plays a critical role in establishing macrophage phenotype. Using single-cell RNA sequencing of human AAA tissues, we identified increased JMJD3 in aortic monocyte/macrophages resulting in up-regulation of an inflammatory immune response. Mechanistically, we report that interferon-β regulates Jmjd3 expression via JAK/STAT and that JMJD3 induces NF-κB–mediated inflammatory gene transcription in infiltrating aortic macrophages. In vivo targeted inhibition of JMJD3 with myeloid-specific genetic depletion (JMJD3f/fLyz2Cre+) or pharmacological inhibition in the elastase or angiotensin II–induced AAA model preserved the repressive H3K27me3 on inflammatory gene promoters and markedly reduced AAA expansion and attenuated macrophage-mediated inflammation. Together, our findings suggest that cell-specific pharmacologic therapy targeting JMJD3 may be an effective intervention for AAA expansion.
Graphical Abstract
Introduction
Abdominal aortic aneurysms (AAAs) are a common vascular disease that can progress to the potentially fatal consequence of aortic rupture, which has a mortality of over 80% (Nordon et al., 2011). Recent studies suggest there may be greater than 1 million people in the United States living with an AAAs (Thom et al., 2006; Kent et al., 2010). Despite advances in the surgical and endovascular management of AAAs, to date there are no proven pharmacological interventions that slow AAA growth or prevent rupture (Baxter et al., 2008). Cumulative efforts to understand mechanisms that contribute to AAA dilation have consistently highlighted inflammation as a key feature of AAA onset as demonstrated by extensive medial and adventitial inflammatory cell infiltration into the vessel wall (Hellenthal et al., 2009). Within patient clinical samples and animal models, monocytes/macrophages play a critical and distinct role in AAA development and progression to rupture (Davis et al., 2014; Butt et al., 2016; Raffort et al., 2017; Wang et al., 2014a; Dale et al., 2015). Identifying the mechanisms that drive macrophage-mediated inflammation is a crucial step toward developing effective therapeutics.
Monocytes/macrophages are characterized by a remarkable degree of plasticity in tissues and are able to rapidly adapt to a wide range of environmental cues (Lawrence and Natoli, 2011). During vascular remodeling, monocytes are recruited to aortic tissue, where they differentiate into macrophages and represent the predominate myeloid population that participates in the inflammatory process together with specialized tissue-resident macrophages (Gordon and Plüddemann, 2017). The original in vitro definition of macrophage phenotypes presents an oversimplified M1/M2 dichotomy, where M1-like, or classically activated, macrophages secrete proinflammatory cytokines, including IL-6, IL-1β, and TNF. By contrast, M2-like, or alternatively activated, macrophages have an anti-inflammatory role and are involved in extracellular matrix remodeling. However, progress in the functional characterization of macrophages has revealed that the phenotypes are not limited to the in vitro M1/M2 extremes but rather in tissue exist along a continuous spectrum of phenotypes associated with differential cytokine production and functional characteristics (Xue et al., 2014; Colin et al., 2014). Within AAA development, proinflammatory macrophages arise from either hematopoietic progenitor cell proliferation/differentiation (Peshkova et al., 2019) or mobilization of splenic monocytes (Mellak et al., 2015) and have been shown to play a critical role in aortic expansion (Rao et al., 2015; Boytard et al., 2013; Qin et al., 2015). Further, proinflammatory monocyte/macrophage populations predominate in aortic tissue, resulting in increased proteolytic enzyme production, inflammatory cytokine expression, and pathological aortic dilation (Moran et al., 2013; Wang et al., 2014b; Juvonen et al., 1997; Karlsson et al., 2009b). In contrast, prevention of aortic expansion is associated with an anti-inflammatory macrophage phenotype (Martinez et al., 2009). These findings have been confirmed in human AAA tissue whereby an imbalance of the pro- and anti-inflammatory macrophage ratio occurs in the aneurysmal wall (Juvonen et al., 1997; Karlsson et al., 2009a). Despite the critical importance of macrophage phenotypic plasticity in AAA development and expansion, the molecular mechanisms that program and sustain inflammatory macrophage phenotypes in AAA disease have not been identified.
Macrophage functional plasticity is tightly regulated by transcriptional reprogramming, which is achieved by modifications of chromatin accessibility and the epigenetic landscape. Accumulating evidence suggests that epigenetic regulation of gene expression, via mechanisms such as histone modification, is a major influence on innate immune cell phenotypes in both normal and pathological conditions (Obata et al., 2015; Hoeksema and de Winther, 2016; Kuznetsova et al., 2020). Histone methylation is important, as it maintains active or suppressed gene expression, depending on the methylation site, and thereby controls downstream protein expression patterns. For example, methylation of lysine 27 (K27) of histone 3 (H3) maintains the chromatin in a confirmation such that the promoters for specific genes are unavailable for transcription and thus genes are epigenetically silenced (Schlesinger et al., 2007). In contrast, methylation of lysine 4 on H3 opens up the chromatin and allows for gene expression (Jaenisch and Bird, 2003). Prior work by our laboratory and others has identified that epigenetic modifications regulate immune-mediator expression in macrophages both in vitro and in vivo (Kroetz et al., 2015; Kuznetsova et al., 2020). Specifically, the chromatin-modifying enzyme Jumonji domain–containing protein D3 (JMJD3), a histone demethylase, acts to demethylate lysine residues on histones in a sequence-specific fashion. JMJD3 removes methyl groups from H3K27, effectively eliminating the repressive function of H3K27 trimethylation (H3K27me3), and thus allows for increased gene expression (De Santa et al., 2007; Lam and Lu, 2007). In addition, JMJD3 can influence macrophage polarization, as LPS-treated macrophages incubated with a JMJD3 inhibitor demonstrated a reduction in proinflammatory cytokine levels (Satoh et al., 2010; Gallagher et al., 2015; Ishii et al., 2009). From a cardiovascular standpoint, JMDJ3 has been shown to be an important regulator of macrophage inflammation (Neele et al., 2018, 2017; Krausgruber et al., 2011). Despite the importance of histone modification on macrophage function, there remains a paucity of data on epigenetic-based mechanisms that regulate macrophage inflammation in AAA formation (Davis and Gallagher, 2019). Additionally, little is known about the upstream mechanisms that regulate JMJD3 in in vivo macrophages. Current investigations on the role of epigenetic modifications in AAA development are extremely limited, and as such, epigenetic alterations have only been examined with histone acetylation in vascular smooth muscle cells (Lino Cardenas et al., 2018; Toghill et al., 2018) and with global methylation levels in peripheral blood mononuclear cells (Toghill et al., 2015, 2018; Ryer et al., 2015). Thereby, the mechanistic role of epigenetic modifications in the regulation of macrophage phenotype during AAA development remains undefined. Given JMJD3’s critical role in macrophage inflammation, we hypothesize that up-regulation of JMJD3 in macrophages induces aortic inflammation and AAA expansion.
Herein, we provide experimental evidence that JMDJ3 directs macrophage-mediated inflammation and aortic aneurysm formation in human tissue samples and well-established murine AAA models (elastase-induced AAAs and angiotensin II [AngII]–induced AAAs). Mechanistically, macrophage-specific JMJD3 induced inflammatory NF-κB–mediated gene transcription by reducing the repressive histone methylation mark H3K27me3 on inflammatory gene promoters in the murine AAA model. Jmjd3 expression in macrophages was regulated by IFNβ via the JAK/STAT1 pathway, as genetic depletion of the IFN receptor reduced JMJD3 expression and prevented AAA development. Further targeted inhibition of JMJD3 via myeloid-specific genetic depletion of JMJD3 (Jmjd3f/fLyz2Cre+) or pharmacological inhibition using a small-molecule inhibitor prevented AAA expansion and was associated with a reduction in the inflammatory macrophage phenotype. Lastly, using single-cell RNA transcriptome analysis, we demonstrate that the histone demethylase JMJD3 is primarily elevated in human AAA monocytes/macrophages, resulting in up-regulation of cellular activation, NF-κB–mediated inflammation, and T lymphocyte costimulatory pathways. Overall, these findings identify JMJD3 as a critical regulator of monocyte/macrophage-mediated inflammation during aneurysmal progression and demonstrate translational implications as cell-targeted manipulation of this pathway is feasible to reduce AAA development.
Results
JMJD3 is increased in human and murine AAA tissues and reduces the repressive H3K27 trimethylation on NF-κB–mediated inflammatory cytokine promoters in macrophages
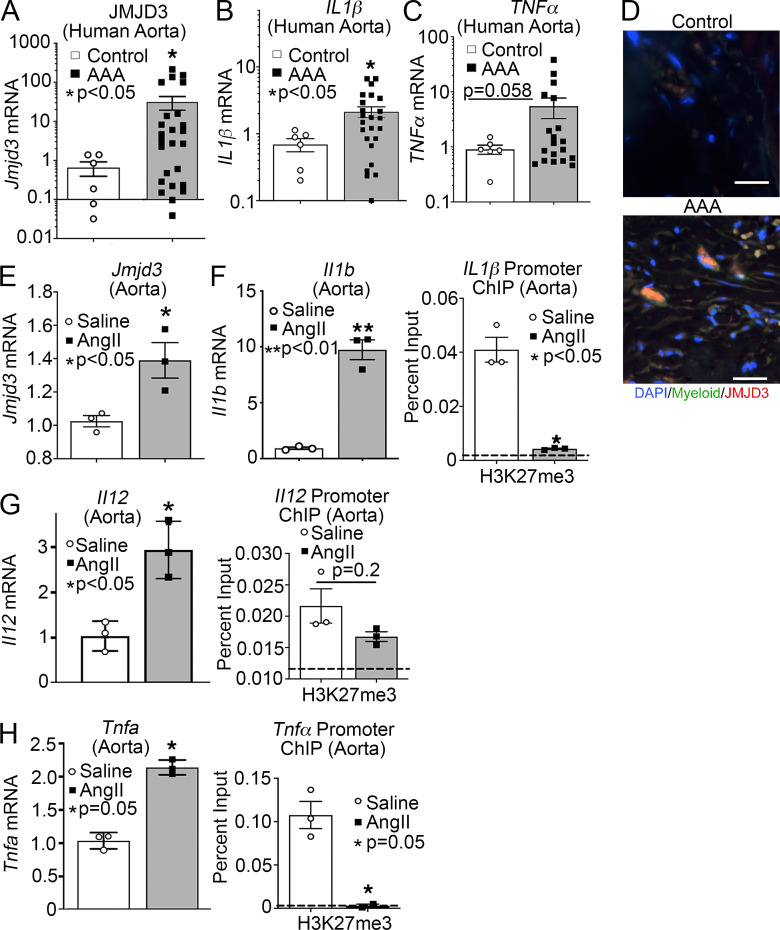
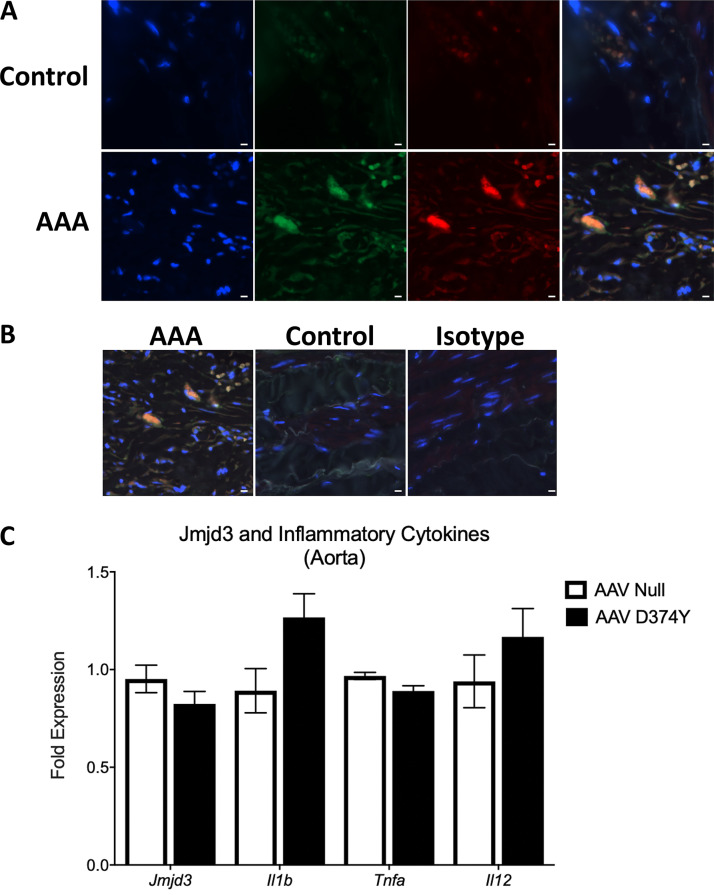
Since transmural macrophage inflammation is detrimental to vascular remodeling during AAA development, we first examined the presence of JMJD3, an epigenetic enzyme associated with inflammation, in human aortic tissue samples isolated from infrarenal AAAs and atherosclerotic aorto-iliac occlusive controls. We found expression of JMJD3 was markedly increased in AAA samples compared with controls (Fig. 1 A and Table S1). Additionally, proinflammatory cytokines (IL1β and TNFα), which are pathological for AAAs by eliciting vascular remodeling (Xiong et al., 2009; Johnston et al., 2013; Yan et al., 2019), were markedly elevated in human AAA samples (Fig. 1, B and C). Immunostaining showed that JMJD3 was highly expressed in the aortic media and adventitia of diseased aortas, particularly in infiltrating myeloid cells in the aortic wall (Fig. 1 D; and Fig. S1, A and B).
Figure 1.
JMJD3 is increased in human and murine AAA tissues and reduces the repressive H3K27 trimethylation on NF-κB–mediated inflammatory cytokine promoters. (A–C) Aortic tissue from patients with AAA (n = 19) and atherosclerotic controls (n = 6) were collected. No statistical differences were found between groups with respect to sex, age, or comorbid conditions. JMJD3, Il1β, and TNFα gene expression was measured by quantitative PCR. *, P < 0.05, Welch’s t test replicated twice. (D) Immunofluorescence was performed on AAA and nonaneurysmal control tissue to analyze DAPI (blue), myeloid (green), and JMJD3 (red; n = 5 AAA and n = 3 controls with representative pictures shown). Representative images show 200× magnification, with scale bars representing 30 µm. (E–H) Male C57BL/6J mice were injected i.p. with an AAV containing mouse PCSK9D377Y and fed saturated fat diet for 6 wk. Mice were infused with saline or AngII (1,000 ng/min/kg) for 4 wk. Quantitative PCR analysis of Jmjd3, Il1b, Il12, and Tnfa isolated from aortas of mice exposed to saline or AngII for 28 d (n = 3/group run in triplicate). *, P < 0.05; **, P < 0.01, Welch’s t test. ChIP analysis for H3K27me3 at Il1b, Il12, and Tnfa promoter was performed (n = 3/group run in triplicate). For all ChIP experiments, isotype-matched IgG was run in parallel. Dotted line represents isotype control. *, P < 0.05 for Welch’s t test. Data are presented as the mean ± SEM.
Figure S1.
JMJD3 is increased in human AAA tissues. (A) Immunofluorescence was performed on AAA and nonaneurysmal control tissue to analyze DAPI (blue), myeloid (green), and JMJD3 (red). Representative images show 200× magnification, and scale bars represent 30 µm. (B) Immunofluorescence with DAPI, JMJD3, myeloid, and isotype control. Representative images show 200× magnification, and scale bars represent 30 µm. (C) Male C57BL/6 mice were injected i.p. with an either a null AAV or AAV containing mouse PCSK9D377Y and fed Western diet for 6 wk. Following a high-fat diet period, the aortic tissue was isolated, and expression of Jmjd3, Il1b, Tnfa, and Il12 was measured in aortic tissue on day 28 following viral vector injection (n = 3–4 mice/group run in triplicate). Data are presented as the mean ± SEM.
Given the elevated expression of JMJD3 in myeloid cells from human AAA tissue and its role in inflammation, we examined JMDJ3 in murine macrophages using an established AngII-induced murine model (Lu et al., 2016). Specifically, male C57BL/6J mice were injected i.p. with a single dose of an adeno-associated virus (AAV) vector expressing the mouse D377Y gain-of-function proprotein convertase subtilisin/kexin type 9 (PCSK9), which resulted in sustained hypercholesterolemia as described previously (Lu et al., 2016). Following this, mice were fed a saturated fat–enriched diet for 6 wk and received either saline or AngII infusion (1,000 ng/min/kg) for the last 4 wk. The use of a saturated fat diet and AngII infusion to induce AAAs recapitulates important clinical risk factors, including dyslipidemia and hypertension. Consistent with data from human aortic tissue, Jmjd3 expression was increased significantly in AngII-induced AAAs compared with nonaneurysmal controls (Fig. 1 E). This was associated with increased expression of NF-κB–mediated inflammatory genes (Il1b, Il12a, and Tnf), which are known to play a critical role in AAA inflammation (Fig. 1, F–H). Importantly, there was no alteration in Jmjd3 or inflammatory cytokine expression upon exposure to solely PCSK9 gain-of-function AAV compared with control AAV (Fig. S1 C).
JMJD3 has been shown to act via a H3K27me3-mediated mechanism on inflammatory gene promoters (Bosselut, 2016). As a demethylase, increased JMJD3 demethylates the H3K27 site, which normally acts to repress gene transcription (De Santa et al., 2007). Thus, increased JMJD3 results in decreased H3K27 methylation, which renders gene promoters accessible to transcription factor (NF-κB) binding, resulting in gene activation. To examine the transcriptional effects of JMJD3 up-regulation during murine AAA formation on NF-κB–mediated inflammatory gene expression in aortic tissue, chromatin immunoprecipitation (ChIP) was performed on aortic tissue from AngII-induced AAAs compared with those from saline-infused controls. Using primers for the NF-κB binding sites on the Il1b, Il12a, and Tnf promoters, we identified that H3K27me3 was reduced at the NF-κB binding sites in the AngII-induced AAA aortic tissue (Fig. 1, F–H). Further there was no difference in lysine-specific demethylase 6, a secondary histone demethylase involved in the regulation of H3K27me3 (data not shown). Collectively, these results suggested that the up-regulation of JMJD3 may play a causative role NF-κB–mediated inflammation in both human and murine AAAs.
IFNβ/JAK/STAT1 regulates Jmjd3 expression, resulting in decreased H3K27me3 at NF-κB binding sites on inflammatory gene promoters in AngII-induced AAA monocytes/macrophages
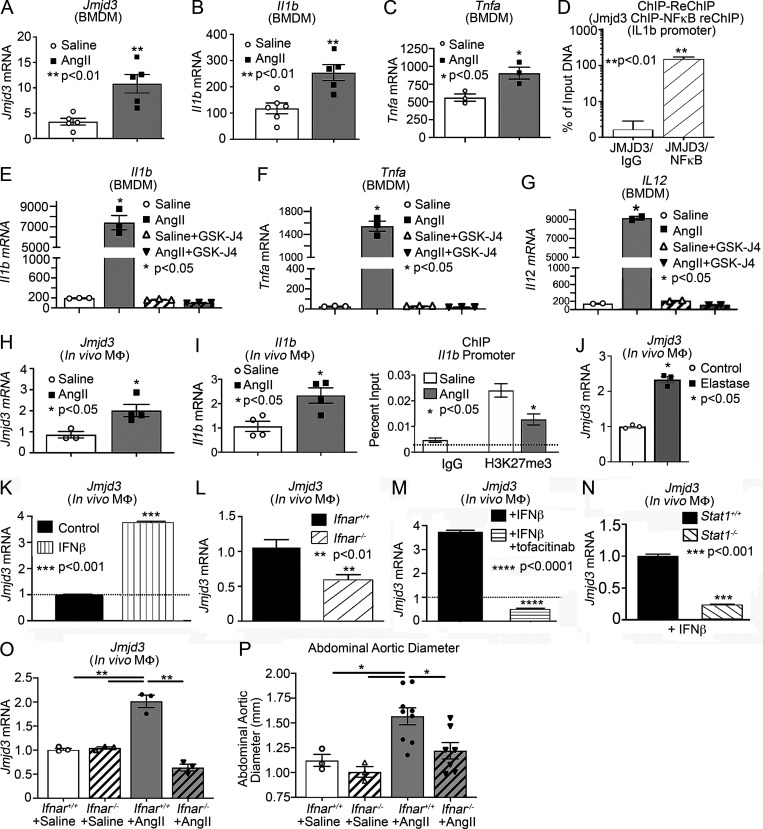
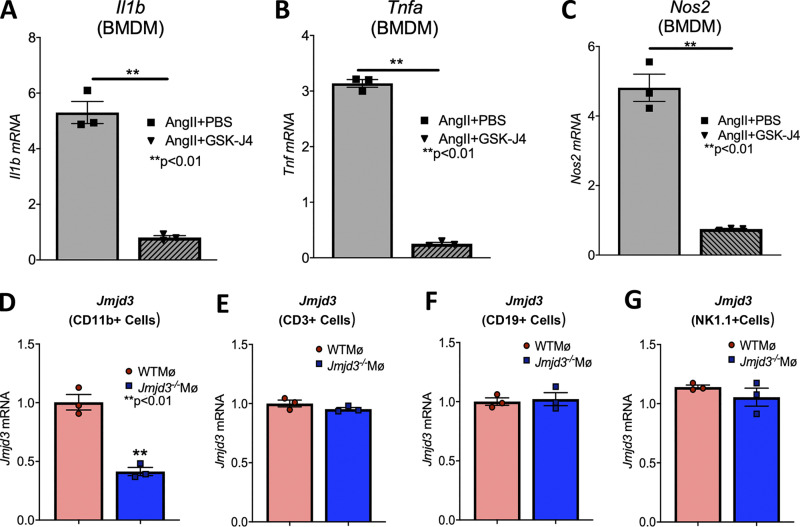
Since monocyte/macrophage inflammation is a pathological hallmark of AAA progression (Raffort et al., 2017), and we identified that JMJD3 is increased in human myeloid cells, we examined myeloid cells isolated from AngII-induced AAAs. It has been shown that although a minority of aortic macrophages are derived from embryonic precursors that populate the aorta before birth, the majority of monocytes/macrophages in AAA tissue originate from the hematopoietic lineage and hone from the blood into the aortic tissue following injury (Ginhoux and Guilliams, 2016). To more fully examine this, bone marrow (BM)–derived macrophages (BMDMs) were isolated from mice infused with saline or AngII. Jmjd3 and inflammatory cytokine expression (Il1b and Tnfa) were significantly elevated in BMDMs from AngII-induced AAA mice in comparison to controls (Fig. 2, A–C). To further dissect the mechanism for JMJD3 localization to specific NF-κB binding sites on inflammatory cytokine promoters, a sequential ChIP (ChIP-reChIP) was performed to identify colocalization of proteins interacting with a specific DNA sequence (i.e., NF-κB binding site of the Il1b promoter) using double and independent rounds of ChIP, as detailed previously (Beischlag et al., 2018; Kimball et al., 2019). In BMDMs, DNA bound to JMJD3 was isolated, followed by a second immunoprecipitation with antibody to NF-κB or IgG. The Il1b promoter was co-occupied by JMJD3 and NF-κB, which led to transcriptional activation of Il1b by removal of the repressive H3K27me3 mark in BMDMs (Fig. 2 D). These findings support that JMJD3 plays a role in regulating macrophage inflammatory cytokine production via direct interaction with NF-κB on promoters.
Figure 2.
Macrophages from AngII-induced AAA mice demonstrate increased Jmjd3 and decreased H3K27me3 at NF-κB binding sites on inflammatory gene promoters. (A–C) Jmjd3, Il1b, and Tnfa expression was measured by quantitative PCR in BMDMs isolated from either saline or AngII-infused mice on day 28 of infusion (n = 3/group run in triplicate). *, P < 0.05; **, P < 0.01 by Mann–Whitney U test. (D) Sequential ChIP was performed at the Il1b promoter on BMDMs treated for 6 h with IFNβ. DNA bound to Jmjd3 was isolated by ChIP, followed by a second immunoprecipitation with antibody to NF-κB or IgG (n = 6 mice/group run in triplicate). **, P < 0.01, Welch’s t test. (E–G) Il1b, Tnfa, and Il12 expression were measured by quantitative PCR in BMDMs isolated from either saline or AngII-infused mice administered with or without GSK-J4 (10 µM; JMJD3 inhibitor) for 6 h (n = 3/group run in triplicate). *, P < 0.05, ANOVA test with Dunnett multiple comparison. (H and I) Quantitative PCR analysis of Jmjd3and Il1b from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in mice infused with either saline or Ang II for 28 d (n = 3–4/group run in triplicate). *, P < 0.05 for Mann–Whitney U test. ChIP analysis for H3K27me3 at Il1b promoter was performed (n = 3/group run in triplicate). For all ChIP experiments, isotype-matched IgG was run in parallel. Dotted line represents isotype-matched control. *, P < 0.05 by ANOVA test with Newman–Keuls multiple comparison test. (J) Quantitative PCR analysis of Jmjd3 from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in control and elastase-treated mice (n = 3/group run in triplicate). *, P < 0.05, Mann–Whitney U test. (K) Quantitative PCR analysis of Jmjd3 was conducted in macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) treated with IFNβ (100 U) for 24 h (n = 3 mice/group run in triplicate). (L) Quantitative PCR analysis of Jmjd3 was conducted in macrophages isolated from Ifnar−/− and littermate controls (Ifnar+/+) and treated with IFNβ (100 U) for 24 h (n = 3 mice/group run in triplicate). **, P < 0.01 for Mann–Whitney U test. (M) Jmjd3 expression in macrophages treated with IFNβ ± tofacitinib (JAK inhibitor; n = 3 mice/group run in triplicate). ****, P < 0.0001 for Mann–Whitney U test. (N) Quantitative PCR analysis of Jmjd3 was conducted in macrophages isolated from Stat1−/− and littermate controls (Stat1+/+) and treated with IFNβ (100 U) for 24 h (n = 3 mice/group run in triplicate). ***, P < 0.001 for Mann–Whitney U test. (O) Quantitative PCR analysis of Jmjd3 from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in Ifnar+/+ and Ifnar−/− mice infused with either saline or Ang II for 28 d (n = 3 mice/group run in triplicated). **, P < 0.01 for Mann–Whitney U test. (P) Maximal abdominal aortic as determined by ultrasound in Ifnar+/+ and Ifnar−/− mice infused with either saline or AngII. *, P < 0.05 by ANOVA with Newman–Keuls multiple comparison test (n = 3–7/group). Data are presented as the mean ± SEM. MΦ, macrophage.
JMJD3 small-molecule inhibitors, such as GSK-J4, have been developed recently for inflammatory disease applications and oncological therapy with efficacious results both in vitro and in vivo (Doñas et al., 2016; Kruidenier et al., 2012). To determine if JMJD3 alters BMDM inflammatory cytokine expression from saline- and AngII-infused mice, BMDMs were isolated and stimulated with or without GSK-J4 (10 µM) for 6 h. BMDMs isolated from AngII-infused mice had significant up-regulation of Il1b, Tnfa, and Il12 inflammatory cytokine expression that was reversed with GSK-J4 (Fig. 2, E–G).
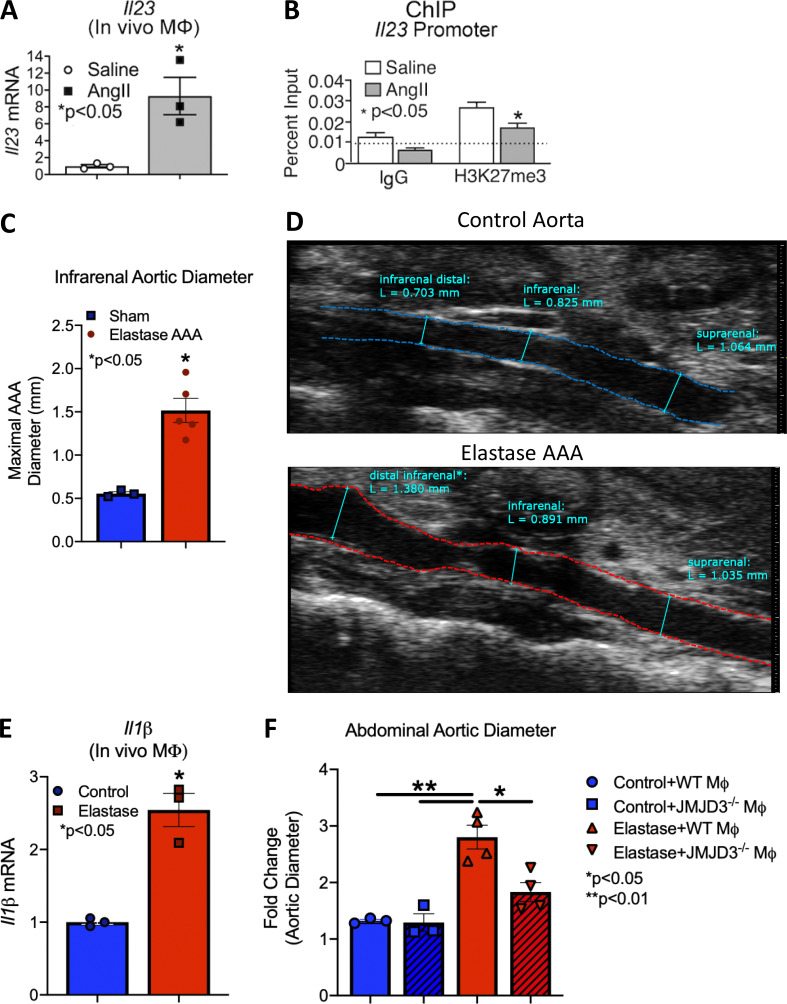
To further investigate the potential mechanisms underlying JMJD3-mediated macrophage inflammation in vivo during aortic aneurysm formation, we isolated in vivo macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) by cell sorting on day 28 from mice infused with AngII or saline. Consistent with the in vitro studies, Jmjd3 and inflammatory cytokine (Il1b and Il23) expression was increased in monocytes/macrophages from AngII-induced AAAs in comparison to controls (Fig. 2, H and I; and Fig. S2 A). Additionally, ChIP analysis was conducted for H3K27me3 on the NF-κB binding sites on inflammatory gene promoters, demonstrating significant reduction in repressive H3K27me3 on the Il1b and Il23 promoters (Fig. 2 I and Fig. S2 B). To further confirm the importance of Jmjd3 expression in AAA pathophysiology, we employed a second murine model of AAAs using topical elastase treatment, which results in nondissecting AAA formation (Sénémaud et al., 2017; Lysgaard Poulsen et al., 2016). At day 14, following elastase application, there was significant dilation of the infrarenal AAA (Fig. S2, C and D) as well as marked up-regulation of Jmjd3 and Il1b in in vivo macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) from elastase-treated mice compared with sham-operated control animals (Fig. 2 J and Fig. S2 E).
Figure S2.
Inflammatory cytokines and murine elastase AAA model. (A) Quantitative PCR analysis of IL-23 from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in mice infused with either saline or AngII for 28 d (n = 8–9 mice/group run in triplicate). *, P < 0.05 for Mann–Whitney U test. (B) ChIP analysis for H3K27me3 at Il23 promoter was performed (n = 5 mice/group run in triplicate). For all ChIP experiments, isotype-matched IgG was run in parallel. Dotted line represents isotype-matched control. *, P < 0.05 by ANOVA test with Newman–Keuls multiple comparison test. (C) Maximal infrarenal abdominal aortic diameter as determined by ultrasound in control and elastase-treated mice (n = 3–5 per group measured by two independent observers). *, P < 0.05; Mann–Whitney test. (D) Representative ultrasound images of the abdominal aorta at day 14 in control and elastase-treated mice. Dotted line represents aortic contour and arrows represent aortic wall diameter. Scale bar on the right side of image with each major axis representing 1 mm. (E) Quantitative PCR analysis of Il1b from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in control or elastase mice (n = 3 mice/group). *, P < 0.05; Mann–Whitney test. (F) Fold change abdominal aortic diameter was determined by ex vivo measurement in Jmjd3−/−MΦ or WTMΦ mice treated with or without elastase (n = 3 in control cohorts and n = 4 in elastase cohorts). *, P < 0.05; **, P < 0.001 by ANOVA with Newman–Keuls multiple comparison test.
Type 1 IFNs (IFN-I) serve as cell signaling molecules that bind to cell surface receptors and ultimately phosphorylate tyrosine kinases inducing specific transcription factors to activate inflammatory genes (Rayamajhi et al., 2010). Although IFN-I has been well studied in viral disorders and autoimmune diseases, very little is known about the role of IFN-I in both normal and pathological vascular remodeling (Ohno et al., 2018; Yan et al., 2016; Liao et al., 2012). Prior investigations have demonstrated that IFN-Is are increased in both human tissue samples and murine models of AAAs, and administration of an anti-IFN receptor 1 antibody attenuated AAA development (Ohno et al., 2018; Yan et al., 2016). Further, downstream IFN-I signaling pathways and the JAK1/STAT1 pathway have also been shown to be highly up-regulated in human AAA tissue samples (Liao et al., 2012). It has recently been suggested in microglial cells that transcriptional regulation of JMJD3 by IFNβ and STAT1 signaling may drive inflammatory responses (Sherry-Lynes et al., 2017; Przanowski et al., 2014). However, the mechanisms by which IFN-I and JAK/STAT signaling contribute to JMJD3 regulation in aneurysmal development remain undefined.
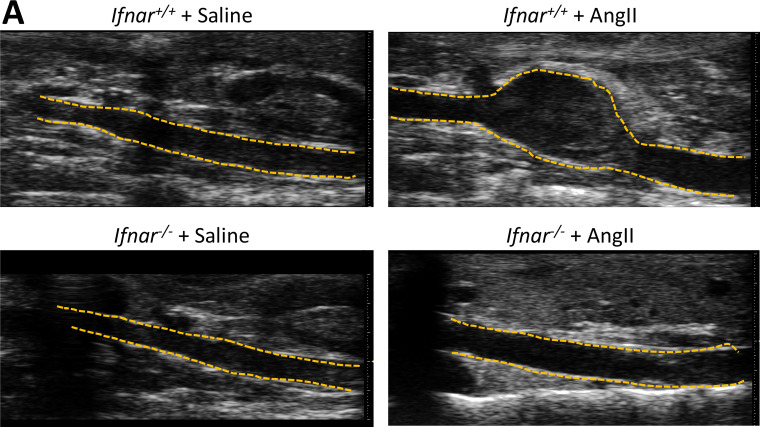
To determine if IFNβ stimulation regulated Jmjd3 expression in in vivo macrophages, we sorted macrophages, stimulated them ex vivo with IFNβ (100 U) for 6 h, and examined Jmjd3 gene expression. In macrophages, Jmjd3 was significantly increased following IFNβ stimulation (Fig. 2 K). To confirm that loss of IFNβ signaling decreased macrophage Jmjd3 expression, we isolated macrophages from Ifnar−/− mice and Ifnar+/+ controls. Following 6-h IFNβ stimulation, Jmjd3 was significantly reduced in the Ifnar−/− macrophages compared with matched controls, suggesting that IFNβ signaling alters JMJD3 expression in macrophages (Fig. 2 L). IFN-I signals through the JAK/STAT pathway to promote gene transcription during viral infection; however, the role of IFN-I/JAK/STAT in AAA pathogenesis remains undefined. We thereby isolated macrophages and stimulated them with IFNβ (100 U) with or without the JAK1 inhibitor tofacitinib (50 nM). Jmjd3 was significantly decreased in macrophages treated with the JAK1 inhibitor following stimulation with IFNβ (Fig. 2 M). Given the prior findings implicating STAT in AAA tissue, we further investigated JAK/STAT1 downstream signaling, and we analyzed Jmjd3 expression in macrophages from Stat1−/− mice and matched controls (Stat1+/+). Stat1−/− mice demonstrated decreased Jmjd3 expression in response to IFNβ stimulation (Fig. 2 N). Lastly, to determine the in vivo regulation of Jmjd3 expression, in the murine aneurysm model, Ifnar−/− mice and Ifnar+/+ controls were randomized to receive saline or AngII infusion for 28 d. Following the infusion, in vivo macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) were isolated by cell sorting and analyzed for Jmjd3 expression. Ifnar−/− mice had a significant decrease in Jmjd3 expression as well as a marked reduction in AAA development following AngII infusion (Fig. 2, O and P; and Fig. S3 A). Taken together, these results suggest that the increased JMJD3 responsible for the inflammatory macrophage phenotype during AAA development may be altered through upstream therapeutic manipulation of the IFNβ/JAK1/STAT1 pathway as well as via direct inhibition of JMJD3.
Figure S3.
Genetic deficiency of IFN-α receptor prevents AngII-induced AAAs. (A) Representative ultrasound images of the abdominal aorta at day 28 in WT Ifnar+/+ and Ifnar−/− mice that received either saline or AngII infusion. Dotted line represents aortic contour and arrows represent aortic wall diameter. Scale bar on the right side of the image with each major axis represents 1 mm.
Human aortic single-cell transcription profiling reveals elevated JMJD3 and inflammatory pathway expression in myeloid cells
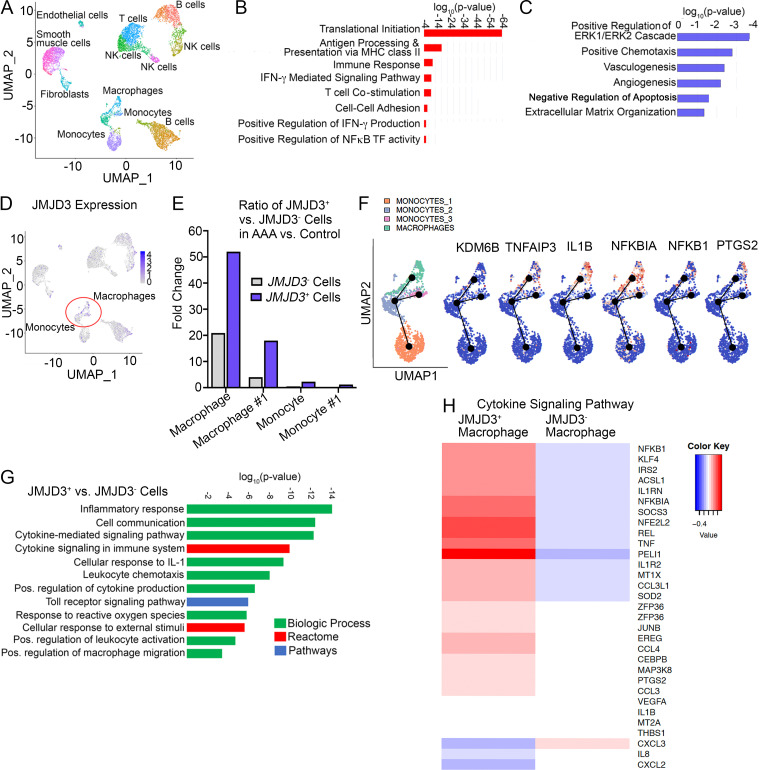
To translate our murine findings to human disease and further characterize the structural and infiltrating cells within the aortic wall, we performed single-cell RNA sequencing (scRNA-seq) analysis on a cohort of patients undergoing abdominal aortic surgery (Table S2). Cluster analysis using the uniform manifold approximation and projection technique identified 21 different cell clusters present in the aortic tissue. We attributed clusters to their putative identities and hierarchical similarities by differentially expressed gene signatures (Fig. 3 A). Analyzing the immune landscape in the aortic tissue demonstrated that there were five T lymphocyte clusters (CD160+, CD247+, CD3E+, and CD3D+), three monocyte clusters (CD14+, FCGR3A+, CD36+, and HLA-DRA+), three B cell clusters (CD19+, CD37+, and CD74+), and macrophage clusters (CD14+, FCGR3A+, CD68+, and TFRC+). Given that monocytes/macrophages are known to have a pivotal role in the inflammation associated with AAA progression (Raffort et al., 2017), we conducted pathway analysis on differentially expressed genes in the monocyte/macrophage populations. Gene Ontology demonstrated that monocytes from AAA samples were more activated in comparison to the same cellular subclusters in nonaneurysmal control samples, with up-regulation of inflammatory genes involved with regulation of cytokine-mediated signaling, NF-κB transcription factor activity, antigen processing, and T lymphocyte costimulation (Fig. 3 B). Further, the AAA monocyte clusters displayed a reduction in anti-inflammatory genes involved with regulation of the ERK cascade, angiogenesis, apoptosis, and response to hypoxia (Fig. 3 C). Given the pathway and phenotypic differences in monocytes from AAA and our murine findings that the epigenetic enzymes, including JMJD3, were up-regulated, we sought to determine if JMJD3 contributes to the monocyte/macrophage proinflammatory state. As such, we screened for alterations in known epigenetic enzymes in AAA macrophages/monocytes and found that JMDJ3 expression was markedly elevated in the macrophage and monocyte clusters in human AAA tissue in comparison to control samples (Fig. 3 D) with minimal expression in T and B lymphocyte subclusters, confirming our findings from the murine model. On further analysis, the ratio of JMJD3+ versus JMJD3− cells was markedly elevated in all macrophage/monocyte populations in AAA tissues in comparison to control samples with the macrophage population exhibiting the highest differential (Fig. 3 E).
Figure 3.
Human aortic single-cell transcription profiling reveals elevated JMJD3 and inflammatory pathway expression in infiltrating monocytes/macrophages. (A) Cluster analysis using the uniform manifold approximation and projection technique of single-cell sequencing from human AAA (n = 4) and nonaneurysmal (n = 2) samples revealed 21 distinct cell clusters (representative). Source data are provided as a source data file. (B and C) Gene Ontology biological pathway enrichment analysis of differentially expressed genes up-regulated (B) and down-regulated (C) in AAA samples. The combined score metric corresponds to the P value (two-tailed Fisher’s exact test) multiplied by the Z-score of the deviation from the expected rank, and q values determined by Benjamini–Hochberg correction. (D) Feature plots displaying the single-cell gene expression of JMJD3 across cell clusters. (E) Fold expression analysis of JMJD3+ vs. JMJD3− macrophages or monocytes (clusters 1–3) in AAA versus control tissue. (F) Focused Monocle pseudotime trajectory analysis including only the monocyte/macrophage defined clusters. Monocyte/macrophage clusters superimposed on pseudotime branches with gene expression plotted as a function of pseudotime. (G) Gene Ontology biological process, Reactome, or process enrichment analysis of differentially expressed genes JMJD3+ versus JMJD3− cells. The combined score metric corresponds to the P value (two-tailed Fisher’s exact test) multiplied by the Z-score of the deviation from the expected rank, and q values were determined by Benjamini–Hochberg correction. (H) Heatmap of differentially expressed genes in JMJD3+ versus JMJD3− macrophages from the cytokine-mediated signaling pathway.
To understand the hypothetical developmental relationships that might exist within the monocyte and macrophage clusters, we performed trajectory analysis on macrophage clusters and monocyte clusters 1–3 using the Monocle algorithm. Three branch points were determined based on changes in monocyte and macrophage gene expression, and this was plotted in pseudotime (Fig. 3 F). Clusters were superimposed on the Monocle pseudotime plot and revealed that monocyte cluster 1 fell toward the beginning of pseudotime, while monocyte clusters 2 and 3 and the macrophage cluster followed along the remaining trajectory. Gene expression plotted by cluster across pseudotime revealed that increased expression of JMJD3 was accompanied by an increased expression of TNFAIP3, IL1B, and NFKBIA.
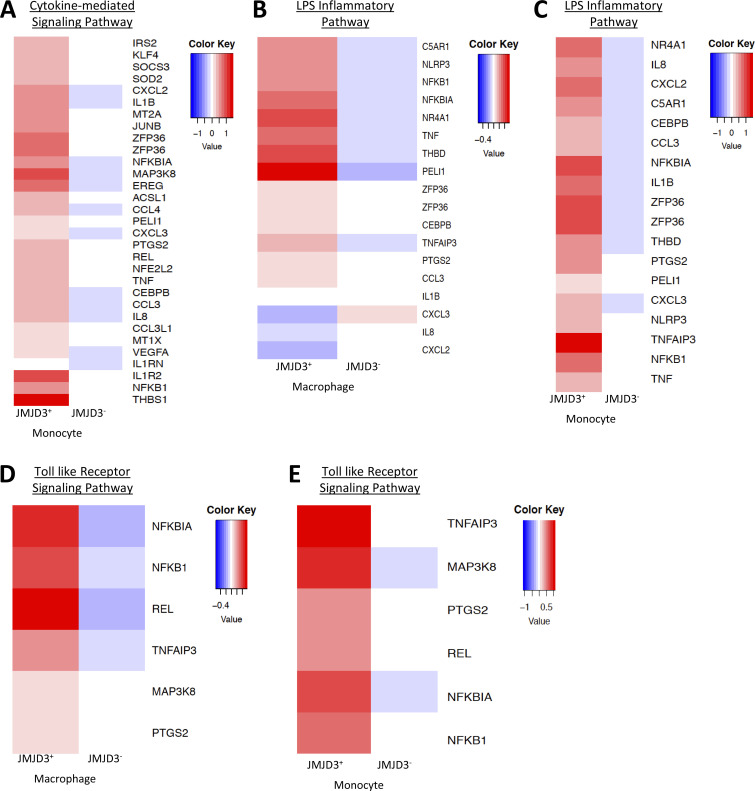
On further analysis of JMJD3+ macrophages/monocytes compared with JMJD3− macrophages/monocytes from human AAA samples, Gene Ontology analysis and differential gene expression analysis demonstrated that multiple inflammatory pathways and cytokines were elevated in JMJD3+ monocytes in comparison to JMJD3− monocytes, further confirming the associated between JMJD3 and inflammatory myeloid state (Fig. 3, G and H; and Fig. S4). Taken together, these results suggest human AAAs are characterized by a significant up-regulation of the histone demethylase JMJD3 in monocytes that likely leads to inflammatory pathway activation.
Figure S4.
Differential expressed gene heatmaps. (A–E) Heatmaps of differentially expressed genes in JMJD3− versus JMJD3+ macrophage/monocytes in human aortic tissue samples by scRNA-seq.
Pharmacological inhibition of JMJD3 via GSK-J4 prevents AAA formation and decreases macrophage inflammation
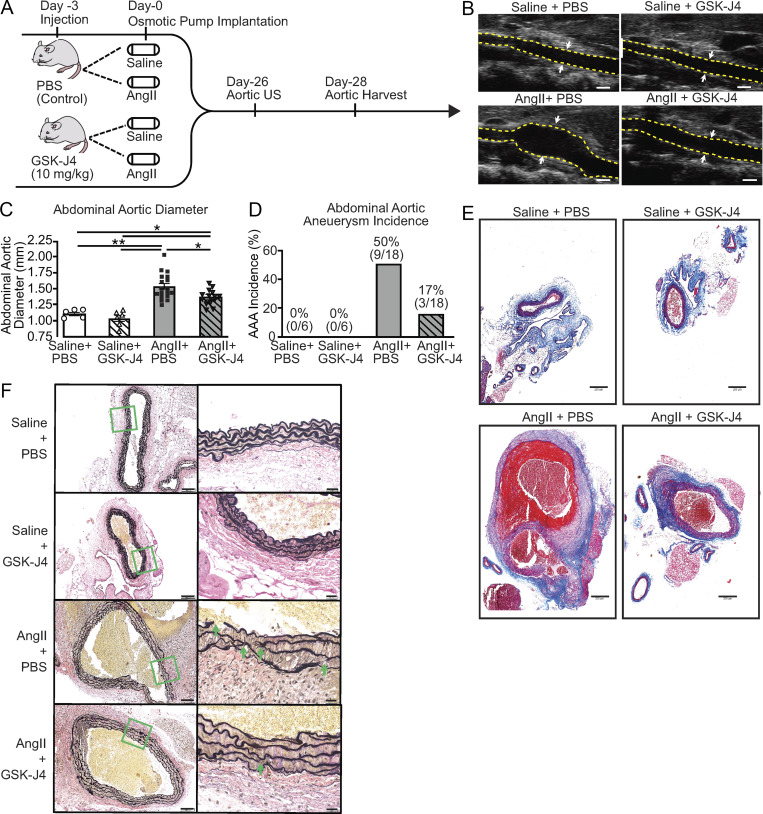
JMJD3-mediated epigenetic modifications increased inflammatory cytokine expression in infiltrating aortic monocytes/macrophages; hence, we investigated the translational potential of JMJD3 inhibition in regulating AAA formation and aortic inflammation. As shown in the schematic in Fig. 4 A, C57BL/6 mice were randomized to receive every-other-day injection of GSK-J4 (10 mg/kg) or PBS control starting 3 d before AngII or saline pump implantation, with injections continuing throughout the 28-d infusion period. JMJD3 inhibition resulted in a marked reduction in AAA incidence and diameter (Fig. 4, B–D). Histological analyses showed that aortas from AngII-infused + GSK-J4–treated mice maintained a normal aortic architecture with preserved smooth muscle cell layer and reduced elastic fiber fragmentation (Fig. 4, E and F).
Figure 4.
Pharmacological inhibition of JMJD3 via GSK-J4 prevents AAA formation. (A) Experimental design of JMJD3 inhibition (GSK-J4) in murine AAA model. WT mice were fed a high-fat diet for 6 wk and infused with saline or AngII (1,000 ng/min/kg) for 4 wk. During this period, mice were randomized to receive either PBS or GSK-J4 (10 mg/kg) injection thrice weekly. (B) Representative ultrasound images of the abdominal aorta at day 28 in WT mice that received either saline or AngII infusion with or without GSK-J4 treatment. Dotted line represents aortic contour, and arrows represent aortic wall diameter. Scale bar represents 1-mm distance. (C and D) Maximal abdominal aortic diameter and aneurysm incidence as determined by ultrasound measured by two observers in WT mice infused with either saline or AngII with or without GSK-J4 administration (n = 6 in saline-infused cohorts and 18 in AngII-infused cohorts). *, P < 0.05; **, P < 0.001, ANOVA with Newman–Keuls multiple comparison test. Data are presented as the mean ± SEM. (E and F) Representative Masson’s trichrome staining and Verhoeff–van Gieson elastin staining of abdominal aortic sections showing preserved aortic structure in AngII + GSK-J4 mice compared with AngII + PBS mice; scale bars represent 200 µm in Masson’s trichrome and 100 µm or 50 µm in Verhoeff–van Gieson stain; arrows represent elastin fragmentation.
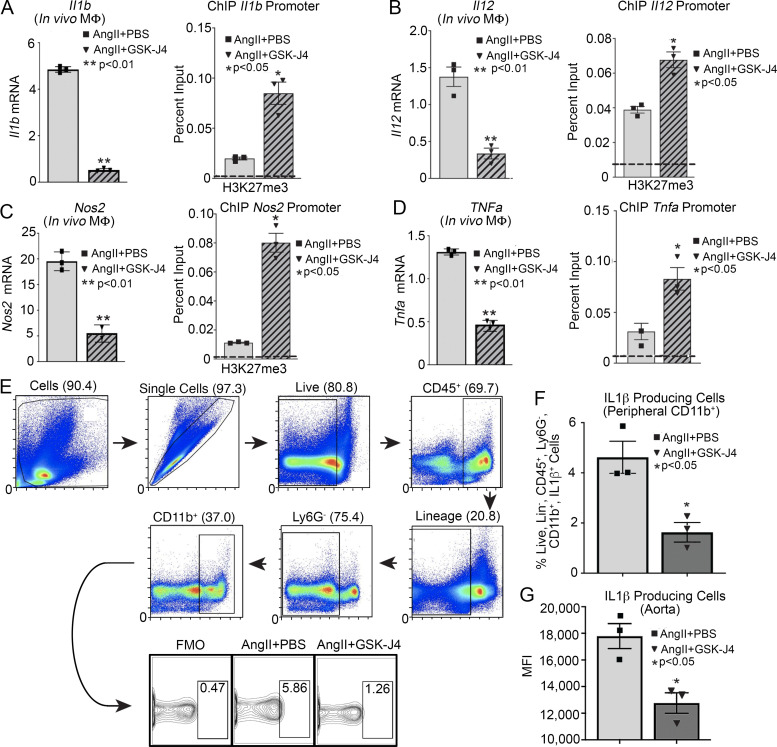
To further determine the involvement of JMJD3 inhibition on macrophage-mediated inflammation and AAA development, macrophages were sorted on day 28 to analyze inflammatory cytokine profiles and histone methylation marks on NF-κB binding sites of inflammatory gene promoters. Mice that received AngII + GSK-J4 treatment displayed a significant reduction in inflammatory cytokine expression in macrophages as well as a corresponding increase in H3K27me3 on NF-κB binding sites of inflammatory gene promoters (Fig. 5, A–D). Inflammatory cytokine expression was also noted to be reduced in BMDMs isolated from AngII + GSK-J4 mice compared with controls (Fig. S5, A–C). Additionally, intracellular flow cytometry analyses revealed reduction in IL-1β protein in both peripheral monocytes (CD11b+Ly6CHi [CD3−CD19−Nk1.1−Ly6G−]) and aortic tissue macrophages in AngII + GSK-J4–treated mice compared with controls (Fig. 5, E–G). Taken together, these findings suggest that JMJD3-mediated histone modification in monocytes/macrophages may contribute to AAA development and that JMJD3 inhibition abrogates aneurysm formation by modulating macrophage inflammation.
Figure 5.
Pharmacological inhibition of JMJD3 via GSK-J4 decreases macrophage inflammatory cytokine production. (A–D) Quantitative PCR analysis of Il1b, Il12, Nos2, and Tnfa mRNA from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]) in mice exposed to saline + PBS, saline + GSK-J4, AngII + PBS, or AngII + GSK-J4 infusion for 28 d (n = 5 mice/group pooled and run in triplicate). ChIP analysis for H3K27me3 at Il1b, Il12, Nos2, and Tnfa promoter was performed (n = 5 mice/group pooled and run in triplicate). For all ChIP experiments, isotype-matched IgG was run in parallel. Dotted line represents isotype-matched control. *, P < 0.05; **, P < 0.01, ANOVA test with Newman–Keuls multiple comparison test. (E) Gating strategy to select single, live, lineage− [CD3, CD19, NK1.1, Ter-119]−, Ly6G−, CD11b+ by flow cytometry at day 28 peripheral blood and aortic tissue. (F and G) Percentage of CD11b+ cells staining positive for IL-1β in monocytes and aortic tissue macrophages (n = 6 mice/group pooled and run in triplicate). Tissues of two mice per mouse were pooled for a single biological replicate. *, P < 0.05, Welch’s t test. Data are presented as the mean ± SEM. FMO, fluorescence minus one; MΦ, macrophage.
Figure S5.
JMJD3 inhibition reduced BMDM inflammatory cytokine expression. (A–C) Expression of Il1b, Tnfa, and Nos2 was measured in BMDMs isolated from mice undergoing AngII + PBS or AngII + GSK-J4 infusion for 28 d (n = 3–4 mice/group run in triplicate). *, P < 0.05; **, P < 0.01 by Mann–Whitney U test. Data are presented as the mean ± SEM. (D–G) Jmjd3 expression was measured by quantitative PCR in CD11b+[CD3−CD19−Ly6G−], CD3+[CD11b−CD19−Ly6G−], CD19+[CD3−CD11b−Ly6G−], or Nk1.1+[CD3−CD11b−CD19−Ly6G−] cells isolated from Jmjd3flox/floxLys2cre+/− (Jmjd3−/−MΦ) and Jmjd3flox/floxLys2cre−/− (WTMΦ) control mice. **, P < 0.01, t test. Data are presented as the mean ± SEM.
AAA formation is inhibited in macrophage-specific JMJD3-deficient mice
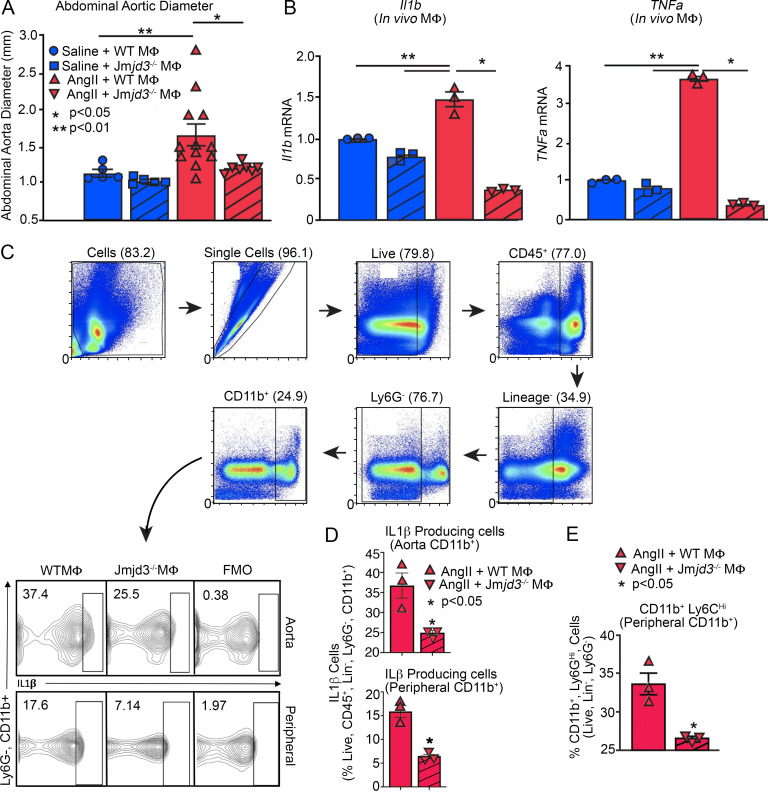
Since global pharmacological inhibition of JMDJ3 prevented AAA formation, we examined the effects of macrophage-specific genetic deletion of JMJD3 on AAA development in a murine model. To define the cell-specific function of JMJD3 in macrophages during AAA development, we uniquely generated a myeloid-specific Jmjd3-deficient mouse. Specifically, using targeted deletion of Jmjd3 in murine embryonic stem cells (ESC), we created mice with the Jmjd3 gene flanked by Lox-P sequences, which were then bred with mice expressing the Cre recombinase in mature macrophage lineage within the lysozyme C-2 gene Lyz2 (LysMcre) to generate Jmjd3flox/floxLys2Cre+ (Jmjd3−/−MΦ) and Jmjd3flox/floxLys2Cre−/− (WTMΦ) control mice. No phenotypic defects were observed in these mice, and confirmation of reduced Jmjd3 expression in macrophages was verified (Fig. S5, D–G). Jmjd3−/−MΦ and WTMΦ mice underwent AngII-induced AAA induction with temporal monitoring of aortic dilation. Jmjd3−/−MΦ mice were less susceptible to AngII-induced AAA formation, with significantly decreased abdominal aortic diameter (Fig. 6 A). We also conducted a second model of AAA development using the topical elastase application, which also demonstrated a significant reduction in AAA development in Jmjd3−/−MΦ mice compared with controls (Fig. S2 F).
Figure 6.
AAA formation is inhibited in macrophage-specific JMJD3-deficient mice. (A) Maximal abdominal aortic diameter was determined by ultrasound in Jmjd3−/−MΦ or WTMΦ mice infused with either saline or AngII (n = 5 in saline-infused cohorts and 7–10 in AngII-infused cohort). *, P < 0.05; **, P < 0.001, ANOVA with Newman–Keuls multiple comparison test. (B) Quantitative PCR analysis of Il1b and Tnfa from macrophages (CD11b+[CD3−CD19−Nk1.1−Ly6G−]; n = 3 mice/group run in triplicate). *, P < 0.05; **, P < 0.01, ANOVA test with Tukey's multiple comparison test. (C) Gating strategy to select single, live, lineage− [CD3, CD19, NK1.1, Ter-119]−, Ly6G−, CD11b+ by flow cytometry at day 28 peripheral blood monocytes and aortic tissue macrophages. (D) Percentage of CD11b+ cells from monocytes and aortic tissue macrophages immunostained positive for IL-1β in Jmjd3−/−MΦ or WTMΦ mice (n = 3/group run in triplicate). Tissues of two mice per mouse were pooled for a single biological replicate. *, P < 0.05 by Welch’s t test. (E) Percentage of CD11b+Ly6CHi cells in Jmjd3−/−MΦ or WTMΦ mice (n = 3 mice/group). *, P < 0.05 by Welch’s t test. Data are presented as the mean ± SEM. MΦ, macrophage.
To determine the transcriptional effects of macrophage-specific JMJD3 deficiency on NF-κB–mediated inflammatory gene expression, in vivo macrophages were sorted from our Jmjd3−/−MΦ and WTMΦ mice after 28 days of saline or AngII infusion. We found a significant reduction in Il1b and Tnfa inflammatory gene expression in Jmjd3−/−MΦ mice following AngII infusion (Fig. 6 B). Similarly, we performed flow cytometry and found decreased IL-1β protein in peripheral blood monocytes and aortic tissue macrophages in Jmjd3−/−MΦ mice (Fig. 6, C and D). Jmjd3−/−MΦs mice had a significant reduction in the proinflammatory CD11b+Ly6CHi monocyte population compared with controls (Fig. 6 E). These data identify that JMJD3 is important in myeloid cells for AAA development and that in vivo macrophage NF-κB–mediated inflammatory gene expression is controlled, at least partly, by JMJD3.
Discussion
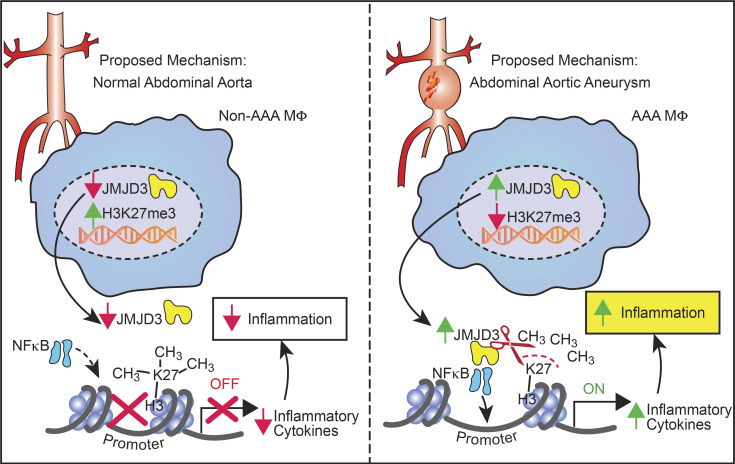
Our results have uncovered dynamic mechanisms that bridge sustained inflammation and AAA expansion and have identified the epigenetic chromatin-modifying enzyme JMJD3 as an instrumental signal in perpetuating the aortic inflammatory response (Fig. 7). Chronic macrophage-mediated inflammation is a key driver for AAA pathogenesis, as prior work has demonstrated that distinct monocyte/macrophage phenotypes have differential roles in initiation, progression, and healing of aneurysmal disease. However, the etiology behind this resilient proinflammatory macrophage population responsible for AAA development remains unclear. Herein, using human AAA tissue samples and two murine models (AngII-induced AAAs and elastase AAA model), we identified that the epigenetic enzyme JMJD3 is a crucial regulator of macrophage phenotype during AAA development. Mechanistically, IFNβ signals through the JAK/STAT1 pathway to increase JMJD3, which colocalizes with NF-κB to binding sites on inflammatory gene promoters, where it removes the repressive histone methylation mark H3K27me3, allowing for transcriptional activation of inflammatory genes. Additionally, single-cell transcriptomics demonstrated human monocytes and tissue macrophages have increased JMJD3 expression, resulting in the activation of multiple immune stimulatory pathways. Ultimately, manipulation of this pathway, either by the use of a macrophage-specific genetic model (Jmjd3flox/floxLys2Cre+) or with pharmacological inhibition, decreased proinflammatory monocyte infiltration into the aortic wall, inhibited inflammatory cytokine production, and decreased AAA formation, offering promise as a translational therapy.
Figure 7.
Schematic of JMJD3-mediated regulation of macrophage inflammation during AAA development.
To date, there is limited literature demonstrating the role of epigenetic histone modifications in regulation of cellular function, particularly monocyte/macrophage phenotype, during AAA development (Krishna et al., 2010; Toghill et al., 2015). Initial investigations found histone deacetylases were increased in AAA tissues from humans and AngII-infused mice. Further, inhibition of class I or class IIa histone deacetylases improved survival and decreased AAA formation in mice; however, the cell-specific mechanisms were not fully investigated, and the genes these deacetylases regulated were unknown (Han et al., 2016; Galán et al., 2016; Vinh et al., 2008). More recent studies have attempted to provide cell-specific information by analyzing immune cell subsets within AAA disease. Specially, isolation of regulatory T lymphocytes (FOXP3+ CD4+ CD25+) from human AAA tissue had reduced acetylation on H3 and increased histone deacetylase enzymes compared with healthy controls but failed to correlate changes in histone acetylation to regulatory T lymphocyte function (Xia et al., 2019; Jiang et al., 2018).
In contrast to histone acetylation, the role of histone methylation has not been investigated in AAA disease. In this study, we found that the histone demethylase JMJD3 stimulates a proinflammatory monocyte/macrophage phenotype in AAA tissue via selective removal of the repressive H3K27me3 mark on inflammatory gene promoters. In addition, we identified via sequential ChIP that JMDJ3 and NF-κB colocalized to inflammatory gene promoters thereby driving gene transcription. These changes are seen both at the BM and peripheral myeloid levels, suggesting epigenetic modifications in macrophages may occur before cell arrival within the aneurysmal tissue. Prior investigations have demonstrated that epigenetic changes can contribute to a phenomenon known as innate immune memory, where chromatin modifications persist even after a normal physiological environment is restored. Considering the nature of monocyte/macrophage differentiation, as well as their relatively short lifespan, innate immune memory arises from epigenetic imprinting of hematopoietic stem cells (Kaufmann et al., 2018; Mitroulis et al., 2018). This response is considered to be independent of adaptive immunity and driven by epigenetic modifications that influence inflammatory gene expression in mononuclear phagocytes (Novakovic et al., 2016; Kleinnijenhuis et al., 2014). This is of important mechanistic relevance, as increased inflammatory cytokine production, particularly expression of IL-1β, TNFα, IL-12, and IL-23, have all been demonstrated to be pathological in AAA disease development, with inhibition of these cytokines preventing aneurysm formation (Xiong et al., 2009; Johnston et al., 2013; Yan et al., 2019). JMJD3 has been shown to be a key driver in NF-κB–dependent inflammation, as in vitro, 70% of proinflammatory LPS-responsive genes are regulated by JMJD3 (De Santa et al., 2009, 2007; Kruidenier et al., 2012). More recently, JMJD3 has also been shown to be highly inducible by serum amyloid A (Yan et al., 2014), an acute-phase protein that triggers an inflammatory response and has also be linked to AAA development in human and murine models (Rohde et al., 1999; Webb et al., 2015). JMJD3 depletion attenuated serum amyloid A–induced expression of proinflammatory genes in vitro along with enrichment of H3K27me3 onto target gene promoters (Yan et al., 2014).
Given the importance of JMJD3, we investigated if inhibition of JMJD3 altered macrophage function and reduced AAA development. Administration of a GSK-J4 led to reduced AAA formation, improved vascular wall remodeling, and reduced macrophage-mediated inflammation. Although pharmacological inhibition of JMJD3 with GSK-J4 provides important mechanistic insight, GSK-J4 can have off-target effects on other histone demethylases when administered at high concentrations (Heinemann et al., 2014). Our pharmacological observations were confirmed with a novel myeloid-specific JMJD3 knockout animal, which also demonstrated a reduction in AAA formation and decreased macrophage inflammation. This is in agreement with a recent study where in vitro treatment of macrophages with a selective JMJD3 inhibitor led to alterations in proinflammatory cytokine production (Kruidenier et al., 2012). As macrophages exhibit different functional phenotypes during tissue repair (Watanabe et al., 2019), the ability to modulate macrophage phenotype at a particular time after injury is an attractive therapeutic strategy. Indeed, epigenetic therapies have been shown to be effective in the treatment of cancer with drugs, resulting in dose-dependent inhibition of cell proliferation, invasion, and cell migration (Verma et al., 2012; Mottamal et al., 2015; Shukeir et al., 2015). More recently, epigenetic therapies are beginning to be examined in cardiovascular disease (Kuznetsova et al., 2020; Luque-Martin et al., 2019). Inflammation is a chronic hallmark of atherogenesis, which in turn manifests as genome-wide and gene-specific DNA hypermethylation and histone modification (Zaina et al., 2014; Cao et al., 2014). Cao et al. (2014) demonstrated that administration of 5-aza-2′-deoxycytidine, a demethylating compound, reduced atherosclerosis development and induced a phenotypic shift in macrophages associated with a down-regulation of proinflammatory genes. Histone deacetylase inhibition has also been shown to prevent atherosclerosis development in murine models (Xu et al., 2017; Huang et al., 2017). Lastly, epigenetic therapies for atherosclerosis are currently undergoing phase 3 clinical trials, including apabetalone, a bromodomain and extra-terminal protein inhibitor, which has demonstrated mixed results (NCT02586155; Ray et al., 2019). Despite these preclinical and ongoing trials for cardiovascular disease, the role of JMJD3 inhibition in AAA development has not been vigorously investigated. Our results demonstrate JMJD3 may be an attractive therapeutic target, either in peripheral monocytes or aortic tissue macrophages, due to its capacity to alleviate inflammation and decrease vascular remodeling.
Although this study provides valuable insight into the mechanisms behind dysregulated macrophage inflammation in AAA development, some limitations must be addressed. First, the human AAA samples are taken from end-stage disease and therefore may not represent the entire etiology of AAA formation. Second, GSK-J4 is a potent inhibitor of JMJD3 but has also been shown to inhibit the epigenetic enzyme UTX at high concentrations (Yin et al., 2019). Within our investigation, we did not see a difference in UTX expression in AAA and control samples suggesting minimal contribution of UTX to AAA disease development. Further, the importance of JMJD3 to macrophage inflammation demonstrated in our GSK-J4 pharmacological observations were confirmed with a novel myeloid-specific JMJD3 knockout animal, which also demonstrated a reduction in AAA formation and decreased macrophage inflammation. Third, within our novel myeloid-specific JMJD3 murine model, we used the Lyz2Cre system. We acknowledge that gene expression profiles between monocytes/macrophages, neutrophils, and dendritic cells overlap due to their close lineage relationship. As such, there is no Cre-transgenic line that is perfectly specific for macrophages (Shi et al., 2018). However, the contributions of neutrophils and dendritic cells for AAA development have been shown to be minor in comparison to macrophage pathophysiology (Raffort et al., 2017; Davis et al., 2014, 2015). Lastly, we recognize that other epigenetic enzymes or vascular cell types may regulate aberrant macrophage function during AAA development as well (Kruidenier et al., 2012).
In conclusion, our study provides important mechanistic evidence that JMJD3 is a regulator of macrophage polarity and inflammation during human and murine AAA development. Increased JMJD3 in macrophages removed the repressive histone methylation mark H3K27me3 on NF-κB inflammatory gene promoters, leading to adverse vascular remodeling and aortic dilation. Targeting the JMJD3 pathway in a cell-specific manner affords the opportunity to intervene in pathological macrophage inflammation and potentially limit AAA progression and rupture.
Materials and methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts, Frank Davis (davisfr@med.umich.edu) and Katherine Gallagher (kgallag@med.umich.edu).
Experimental model and subject details
Mice
Mice were maintained in the University of Michigan pathogen-free animal facility, and all protocols were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee. Male mice were used for AAA experiments as detailed in the American Heart Association Council statement (Robinet et al., 2018). Only male mice were used for these studies, as female mice do not adequately develop AAAs (Robinet et al., 2018). Mouse strains include C57BL/6J and Ifnar−/− mice maintained on a normal diet (13.5% kcal fat; LabDiet 5001) purchased at 8–10 wk from The Jackson Laboratory. For creation of Jmjd3f/fLyz2Cre+ and Jmjd3f/fLyz2Cre- strains, Jmjd3 gene–targeted ESC clones EPD0164_4-B10, -E09, and -E12 were obtained from the trans-NIH Knockout Mouse Project (Knockout Mouse Project Repository). The JM8.N4 C57BL/6N ESC clones (Pettitt et al., 2009) carried the knockout first tm1a(KOMP)Wtsi Jmjd3 allele (Ryder et al., 2013). ESCs were expanded in cell culture, and chromosome counts were performed. Correct targeting of the Jmjd3 gene was confirmed by genetic analysis of DNA from the ESC clones. Germline transmission of the Jmjd3tm1a allele was obtained by breeding ESC-mouse chimeras produced by the microinjection of C57BL/6/BrdCrHsd-Tyrc albino C57BL/6 blastocysts with the ESC clones. Chimeras were mated with FLPo recombinase mice to remove the drug selection neomycin cassette to produce mice carrying the conditional floxed Jmjd3tm1c allele. C57BL/6-Tg(CAG-Flpo)1Afst/Mmucd FLPo recombinase mice (Kranz et al., 2010) were obtained from the Mutant Mouse Resource and Research Centers (stock no. 032247-UCD). FLPo mice were backcrossed onto albino C57BL/6 mice so that coat-color selection could be used to chimeras to identify germline transmission and maintain an inbred C57BL/6 genetic background. The resulting progeny with a floxed Jmjd3 allele were bred with Lyz2cre (The Jackson Laboratory) mice to generate control and mice deficient for Jmjd3 in monocytes, macrophages, and granulocytes. Lyz2cre was chosen for the cell-specific line, as this affects myelomonocytic cells. However, this Cre line can also result in deletion of JMJD3 in neutrophils and dendritic cells. Given macrophages are highly heterogenous, their gene expression patterns dynamically change during prenatal development and adult tissue homeostasis and in various inflammatory diseases. Moreover, such gene expression profiles are largely overlapped among monocytes/macrophages, granulocytes, and dendritic cells due to their close lineage relationship. Therefore, there is no Cre-transgenic line that is perfectly specific for macrophages (Shi et al., 2018). IFN-α/β receptor−/− (Ifnar−/−), Stat1−/−, and BALB/c control mice were obtained from Christiane Wobus (University of Michigan, Ann Arbor, MI) and maintained in breeding pairs at the Unit of Laboratory Animal Medicine facilities. Animals were housed in a barrier facility on a light/dark cycle of 14:10 h (ambient temperature of 22°C) with free access to water, food (LabDiet 5001), and bedding (Andersons Lab Bedding; Bed o’Cobs combo). Animals underwent all procedures at 8–10 wk of age.
Production and injection of AAV vectors
AAV vectors (serotype 8) were produced by the Viral Vector Core at the University of Pennsylvania (https://gtp.med.upenn.edu/core-laboratories-public/vector-core). These AAV vectors contained inserts expressing mouse PCSK9D377Y mutation (equivalent to human PCSK9D374Y gain-of-function mutation). Empty AAV vector (null AAV) was used as control. AAV vectors were diluted in sterile PBS (200 µl per mouse) and injected i.p. as reported previously (Lu et al., 2016; Wu et al., 2015). Mice inclusion and randomization were conducted as previously specified. Further, mice had a predefined exclusion from the data analysis if plasma total cholesterol concentrations were <250 mg/dl 3 wk and <500 mg/dl 6 wk after PCSK9D377Y.AAV injection (Lu et al., 2016). Briefly, mice received injections of AAVs containing either a null insert or a mouse PCKS9 insert expressing D377Y mutation. Immediately after AAV injections, normolipidemic mice were fed a diet containing saturated fat (Envigo; milk fat 21% wt/wt; Diet #TD.88137) for 2 wk, at which time they underwent implantation of mini-osmotic pumps as described below. Body weights were determined before experimentation.
Osmotic mini-pump implantation and AngII infusion
To induce AAAs, 8–10-wk-old male C57BL/6J mice were injected with AAV and started on a saturated fat diet as detailed above. Following 2 wk of saturated fat diet feeding, mice were randomized to receive mini-osmotic pumps (Alzet; Model 2004) containing AngII (Bachem; 1,000 ng/min/kg, catalog no. H-1706) or saline implanted subcutaneously in the neck region of anesthetized mice following a protocol described previously (Lu et al., 2015). Briefly, mice were anesthetized in a closed chamber with isoflurane (3%) in oxygen for 2–5 min until immobile. Each mouse was then removed and taped on a heated (35–37°C) procedure board with isoflurane (1.0–1.5%) administered via nosecone during minor surgery. Pumps were implanted subcutaneously on the right flank of each mouse, which provided AngII or saline infusion for 28 d. Incisions were closed with surgical staples, and postoperative analgesia (buprenorphine, 0.05 mg/kg/12 h, i.p.) was administered. For inhibitor studies, mice were randomized to receive every-other-day i.p. injection of GSK-J4 (Tocris; 10 mg/kg; 4594) in 0.1% DMSO in PBS or of 0.1% DMSO in PBS alone. These injections began 3 d before mini-osmotic pump implantation and continued for the 28-d duration. Animal experiments were conducted following the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of the University of Michigan.
Elastase treatment model of aneurysm formation
A murine elastase treatment model of AAA formation was used as described by Laser et al. (2012). In brief, the infrarenal aorta was treated topically with 30 µl elastase reconstituted with normal saline (5 U/mg protein) or 30 µl heat-inactivated elastase (at 90°C for 30 min) as a control group. The topical application was accomplished by dropping the elastase on the anterior aorta from a 2 cm height for 5 min. Video micrometric measurements of aortic diameters were made in situ before perfusion, after perfusion, and before harvesting the aorta on day 14. Maximum infrarenal aortic diameter and ratio of treated versus untreated section of the aorta was calculated.
Quantification of aortic pathologies
For in vivo imaging of the abdominal aorta in mice, 2D (B-mode) ultrasound images were obtained 27 d after the implantation of osmotic pumps using a VisualSonics Vevo2100 imaging system with a mechanical transducer (MS400) from the University of Michigan Frankel Center for Physiology. Two independent investigators measured aortic diameters at systole, with no significant interobserver or intraobserver variability.
At the completion of each murine aneurysm experiment (day 28), mice were deep anesthetized with ketamine (100 mg/kg) and xylazine (20 mg/kg). At termination after blood collection, the right atrium was cut open, and saline was perfused through the left ventricle to remove blood in aortas. Subsequently, aortas were dissected and placed in either RNA later or 10% neutrally buffered formalin overnight at room temperature. After fixation, periaortic adventitia were removed thoroughly. Maximal outer diameters of suprarenal aortas were measured ex vivo as a parameter for AAA quantification using ImageJ software (National Institutes of Health).
Necropsy was performed for mice that died during AngII infusion. Aortic rupture was defined as observation of blood clots in either the thoracic cavity (thoracic aortic rupture) or retroperitoneal cavity (abdominal aortic rupture). There was no difference in early rupture between treatment groups and genotypes.
Histology/immunofluorescence
For aortic histology, aortas were excised as above and fixed in formalin (10%) overnight before embedding in paraffin. Sections (5 µm) were stained with hematoxylin and eosin, Masson’s trichrome stain for collagen deposition (Newcomers Supply; 9179B), or Verhoeff–Van Gieson stain for elastin (Sigma; HT25A). Images were captured using Olympus BX43 microscope and Olympus cell Sens Dimension software.
For immunofluorescence, formalin-fixed, paraffin-embedded tissue slides obtained from patients with AAA and aortic healthy control were heated for 30 min at 60°C, deparaffinized, and rehydrated. Slides were placed in pH 9 antigen retrieval buffer and heated at 125°C for 3 s in a pressure-cooker water bath. After cooling, slides were blocked using 10% donkey serum (30 min). Overnight coincubation (4°C) was then performed using anti-human JMJD3 (Abcam; catalog no. ab3811), anti-human CD11c (Abcam; ab216028), and isotype control at a concentration of 1 µg/ml. Slides where then washed and treated with relative fluorescence-conjugated secondary antibodies (30 min). Slides were prepared in mounting medium with DAPI (Vector; VectaShield, Antifade Mounting Medium with DAPI, H-1200). Images were acquired using a Zeiss Axioskop 2 microscopy. Images presented are representative of at least three biological replicates.
Flow cytometry
For surface staining, aortas were collected following saline perfusion. Aortas were minced, digested with (elastase, 1 mg/ml [Worthington; LS002292]; collagenase D, 0.2 mg/ml [Roche Diagnostics; 11088866001]; DNase I, and 0.2 mg/ml [Roche Diagnostics; 90083] in RPMI 1640) for 45 min at 37°C. Enzymatic activity was stopped by washing with PBS at 4°C. Spleen suspensions were prepared after lysing red blood cells. Single-cell suspensions were meshed on a 100-µm nylon filter to yield a single-cell suspension. Cells were stained with a Fixable LIVE/DEAD viability dye (BioLegend; 564406; 1:1,000 dilution). Fc receptors were then blocked with anti-CD16/32 (BioXCell; catalog no. CUS-HB-197, 1:200 dilution) for 10 min. Monoclonal antibodies for surface staining included anti-F4/80 (BioLegend, catalog no. 123149, 1:400 dilution), anti-CD11b (BioLegend; catalog no. 101206, 1:400 dilution), anti-CD45 (BioLegend, catalog no. 103106, 1:400 dilution), anti-Ly6C (BioLegend, catalog no. 128028, 1:400 dilution), anti-Ly6G (BioLegend, catalog no. 127624, 1:400 dilution), anti-CD3 (BioLegend; catalog no. 100304, 1:400 dilution), anti-CD19 (BioLegend, catalog no. 116204, 1:400 dilution), and anti-Ter119 (BioLegend; catalog no. 115504, 1:400 dilution). After surface staining, cells were washed twice, and biotinylated antibodies were labeled with streptavidin-fluorophore (BioLegend; 405249, 1:1,000 dilution). Next, cells were either washed and acquired for surface-only flow cytometry or fixed with 2% formaldehyde and then washed/permeabilized with BD Perm/wash buffer (BD Biosciences; reference no. 00–8333-56) for intracellular flow cytometry. After permeabilization, intracellular stains included: anti-IL-1β-Pro-PE Cy7 (eBioscience; reference no. 25–7114-82, 1:200 dilution), and anti-TNF-α-APC (BioLegend; catalog no. 506308, 1:200 dilution). Samples were acquired on a three-laser Novocyte Flow Cytometer (Acea Biosciences). FACS was performed with FACsDiva Software (BD Biosciences), analysis was performed using FlowJo software version 10.0 (Tree Star), and data were compiled using Prism software (GraphPad). All populations were routinely backgated to verify gating and purity.
Peripheral macrophage isolation and magnetic-activated cell sorting
Peripheral circulating macrophages were isolated from splenic tissue. Single-cell suspensions were incubated with fluorescein isothiocyanate–labeled anti-CD3, anti-CD19, anti-NK1.1, and anti-Ly6G (BioLegend) followed by EasySep Mouse Streptavidin RapidSpheres (Stem Cell Technologies; 19860). Flow-through was then incubated with EasySep Mouse CD11b Positive Selection Kit (Stem Cell Technologies; 18970) to isolate the nonneutrophil, non–natural killer cells, nonlymphocyte, CD11b+ cells. When indicated, peripheral macrophages were stimulated with/without IFNβ (PBL Assay Science; catalog no. 12400–01; 100 U/ml). For JAK1 and JAK3 inhibition, cells were treated with 50 nM tofacitinib (Cayman Chemicals) at the time of stimulation with IFNβ. Cells were saved in Trizol (Invitrogen) for quantitative RT-PCR analyses or processed for chromatin immunoprecipitation as described below.
Cell culture and cytokine analysis
BM cells were collected by flushing mouse femurs and tibias at day 28 following AngII or saline infusion with RPMI. BMDMs were cultured as previously detailed (Ishii et al., 2009). On day 6, cells were replated, and after resting for 24 h, they were incubated with or without GSK-J4 (10 µM) for 2–6 h, after which cells were fixed in paraformaldehyde for ChIP analysis or placed in Trizol (Invitrogen) for RNA analysis.
ChIP assay
ChIP assay was performed as described previously (Ishii et al., 2009). Briefly, cells fixed in paraformaldehyde were lysed and sonicated to generate 100–300-bp fragments. Immunoprecipitated samples were incubated in anti-H3K27trimethyl antibody (Active Motif; 39155) or isotype control (Millipore; rabbit polyclonal IgG) in parallel samples overnight followed by addition of protein A Sepharose beads (Thermo Fisher). Bound DNA was eluted and purified using phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. Primers were designed using the Ensembl genome browser to search the IL-1β, IL-12, and IL-23 promoter for NF-κB within the promoter region, and then NCBI Primer-BLAST was used to design primers that flank this site. Data are representative of two or three independent experiments. Primer sequences are available in Table S3.
RNA analysis
Total RNA extraction was performed using Trizol (Invitrogen) or Trizol LS (for human samples) according to the manufacturer’s instructions. RNA was then reversed transcribed to cDNA using iScript (Bio-Rad). PCR was performed with 2X TaqMan PCR mix using the 7500 Real-Time PCR System. Primers for Jmjd3 (Mm01332680_m1), Il-1β (Mm00434228_m1), Tnf⍺ (Mm00443258_m1), Il-12 (Mm00434165_m1, Mm01288992_m1), Il-23 (Mm00518984_m1), Nos2 (Mm00440502_m1), human IL-1β (Hs00233688_m1), human IL-23 (Hs00372324_m1), and human JMJD3 (HS00389738_m1) were purchased from Applied Biosystems. 18S or GAPDH was used as the internal control. Data were then analyzed relative to 18s ribosomal RNA or GAPDH (2ΔCt). All samples were assayed in triplicate. The threshold cycle values were used to plot a standard curve. Data are representative of two or three independent experiments were compiled in Microsoft Excel and presented using Prism software (GraphPad).
Human tissue
Full-thickness aortic wall tissue specimens were collected from the infrarenal abdominal aorta from patients undergoing open aortic aneurysm repair (n = 19) or open aortobifemoral bypass (n = 6). Aneurysmal samples were taken from the midportion of the aneurysmal sac. For control samples, aortic tissue was isolated from patients with atherosclerotic occlusive disease but no history of aneurysmal disease at the time of open aortobifemoral bypass. Patient medical comorbidities are shown in Table S1. All aortic samples were processed for both histology and protein/RNA analyses. For histology, human aortas were placed in formalin (10%) for 24 h before paraffin embedding. Sections (5 µm) were stained with hematoxylin and eosin or Masson’s trichrome stain for collagen deposition. Images were captured using an Olympus BX43 microscope and Olympus cell Sens Dimension software. For protein/RNA analysis, specimens were stored at −80°C for future protein and RNA analyses. For scRNA-seq, a second cohort of samples was retrieved from the infrarenal abdominal aorta of patients undergoing open aortic aneurysm repair (n = 4) or open aortobifemoral bypass (n = 2). Patient medical comorbidities can be seen in Table S2. These samples were immediately processed as described below. This study was approved by the University of Michigan Institutional Review Board (HUM00098915).
scRNA-seq and bioinformatics analysis
Generation of single-cell suspensions for scRNA-seq was performed as follows. Aortic tissue from aneurysmal and nonaneurysmal controls was harvested at the time of surgical intervention (Gene Expression Omnibus accession no. GSE166676). Samples were minced, digested in 0.2% Collagenase II (Life Technologies) and 0.2% Collagenase V (Sigma) in plain medium for 1 h at 37°C, and strained through a 70-µM mesh. The scRNA-seq samples were analyzed by the University of Michigan Advanced Genomics Core on the 10X Chromium system. Libraries were sequencing on the Illumina NovaSeq 6000 sequencer to generate 151-bp paired-end reads. Data processing, including quality control, read alignment, and gene quantification, was conducted using 10X Cell Ranger software. Seurat was used for normalization, data integration, and clustering analysis (Butler et al., 2018). The 10X Cell Ranger aggr function was used to integrate the scRNA-seq datasets, with subsampling used for normalizing the data. Postnormalization resulted in >100 million reads. Human reference hg19 was used for alignment, and Seurat was employed for cross-sample adjustment, processing, and quality control, including removal of cells with >25% of reads from mitochondria genes and removal of cells with <200 or >2,500 genes detected, resulting in ∼8,000 cells. Clustered cells were mapped to corresponding cell types by matching cell cluster gene signatures with putative cell-type–specific markers.
Statistical analysis
Data were analyzed using GraphPad Prism software version 6. Data are represented as means ± SEM. Shapiro–Wilk test was used to determine normality of data, Brown–Forsythe or F test to determine variances, or ROUT test to identify outliers in given datasets, and then a parametric or nonparametric test was performed accordingly. Parametric statistical analysis was performed using unpaired Student’s t test (two tailed) between two groups and one-way ANOVA followed by post hoc analysis (Bonferroni, Dunnett, or Newman–Keuls multiple comparison test) for analysis of differences between >2 groups. Nonparametric statistical analysis was performed using Mann–Whitney U test. P values <0.05 were considered significant.
Online supplemental material
Fig. S1 describes JMJD3 expression in human control and AAA tissue samples. Fig. S2 describes the elastase-induced AAA murine model and changes in JMJD2 expression. Fig. S3 demonstrates ultrasound analysis of the AngII-induced AAAs in Ifnar+/+ and Ifnar−/− mice. Fig. S4 shows heatmaps of differentially expressed genes in JMJD3− versus JMJD3+ macrophage/monocytes in human aortic tissue samples. Fig. S5 shows JMJD3 expression across different immune cell populations. Table S1 shows the human AAA and atherosclerotic tissue cohort. Table S2 shows the human AAA and atherosclerotic scRNA-sequencing cohort. Table S3 lists the primers used in this study.
Supplementary Material
lists the human AAA and atherosclerotic tissue cohort.
lists the human AAA and atherosclerotic scRNA-seq cohort.
lists the primers used in this study.
Acknowledgments
We thank Robin Kunkel for her assistance with the graphical illustrations.
This work is supported in part by National Institutes of Health grants R01-HL137919 (K.A. Gallagher) and F32-DK117545 (F.M. Davis), an American College of Surgeons resident fellowship (F.M. Davis), a Vascular and Endovascular Surgery Society resident research award (F.M. Davis), National Institutes of Health grants P30 AR075043 (J.E. Gudjonsson and L.C. Tsoi) and R01-AR069071 (J.E. Gudjonsson), and the Doris Duke Foundation (K.A. Gallagher).
Author contributions: F.M. Davis, L.C. Tsoi, J.E. Gudjonsson, B.B. Moore, and K.A. Gallagher designed the experiments. F.M. Davis, A. denDekker, A.D. Joshi, R. Wasikowski, S. Wolf, A.T. Obi, A.C. Billi, X. Xing, C. Audu, W.J. Melvin, and J.E. Gudjonsson performed experiments. F.M. Davis, L.C. Tsoi, A. denDekker, A.D. Joshi, R. Wasikowski, S. Wolf, A.T. Obi, C. Audu, W.J. Melvin, B.B. Moore, S.L. Kunkel, A. Daugherty, H.S. Lu, J.E. Gudjonsson, and K.A. Gallagher prepared the manuscript and provided key edits.
References
- Baxter, B.T., Terrin M.C., and Dalman R.L.. 2008. Medical management of small abdominal aortic aneurysms. Circulation. 117:1883–1889. 10.1161/CIRCULATIONAHA.107.735274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag, T.V., Prefontaine G.G., and Hankinson O.. 2018. ChIP-re-ChIP: Co-occupancy Analysis by Sequential Chromatin Immunoprecipitation. Methods Mol. Biol. 1689:103–112. 10.1007/978-1-4939-7380-4_9 [DOI] [PubMed] [Google Scholar]
- Bosselut, R. 2016. Pleiotropic Functions of H3K27Me3 Demethylases in Immune Cell Differentiation. Trends Immunol. 37:102–113. 10.1016/j.it.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boytard, L., Spear R., Chinetti-Gbaguidi G., Acosta-Martin A.E., Vanhoutte J., Lamblin N., Staels B., Amouyel P., Haulon S., and Pinet F.. 2013. Role of proinflammatory CD68(+) mannose receptor(-) macrophages in peroxiredoxin-1 expression and in abdominal aortic aneurysms in humans. Arterioscler. Thromb. Vasc. Biol. 33:431–438. 10.1161/ATVBAHA.112.300663 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H.Z., Sylvius N., Salem M.K., Wild J.B., Dattani N., Sayers R.D., and Bown M.J.. 2016. Microarray-based Gene Expression Profiling of Abdominal Aortic Aneurysm. Eur. J. Vasc. Endovasc. Surg. 52:47–55. 10.1016/j.ejvs.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Cao, Q., Wang X., Jia L., Mondal A.K., Diallo A., Hawkins G.A., Das S.K., Parks J.S., Yu L., Shi H., et al. 2014. Inhibiting DNA Methylation by 5-Aza-2′-deoxycytidine ameliorates atherosclerosis Cao, Q., Wang, X., Jia, L., Mondal, A. K., Diallo, A., Hawkins, G. A., … Xue, B. (2014). Inhibiting DNA Methylation by 5-Aza-2′-deoxycytidine ameliorates atherosclerosis thro. Endocrinology. 155:4925–4938. 10.1210/en.2014-1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin, S., Chinetti-Gbaguidi G., and Staels B.. 2014. Macrophage phenotypes in atherosclerosis. Immunol. Rev. 262:153–166. 10.1111/imr.12218 [DOI] [PubMed] [Google Scholar]
- Dale, M.A., Ruhlman M.K., and Baxter B.T.. 2015. Inflammatory cell phenotypes in AAAs: their role and potential as targets for therapy. Arterioscler. Thromb. Vasc. Biol. 35:1746–1755. 10.1161/ATVBAHA.115.305269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, F.M., and Gallagher K.A.. 2019. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler. Thromb. Vasc. Biol. 39:623–634. 10.1161/ATVBAHA.118.312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, F.M., Rateri D.L., and Daugherty A.. 2014. Mechanisms of aortic aneurysm formation: translating preclinical studies into clinical therapies. Heart. 100:1498–1505. 10.1136/heartjnl-2014-305648 [DOI] [PubMed] [Google Scholar]
- Davis, F.M., Rateri D.L., and Daugherty A.. 2015. Abdominal aortic aneurysm: novel mechanisms and therapies. Curr. Opin. Cardiol. 30:566–573. 10.1097/HCO.0000000000000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa, F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., and Natoli G.. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 130:1083–1094. 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- De Santa, F., Narang V., Yap Z.H., Tusi B.K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S., et al. 2009. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28:3341–3352. 10.1038/emboj.2009.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doñas, C., Carrasco M., Fritz M., Prado C., Tejón G., Osorio-Barrios F., Manríquez V., Reyes P., Pacheco R., Bono M.R., et al. 2016. The histone demethylase inhibitor GSK-J4 limits inflammation through the induction of a tolerogenic phenotype on DCs. J. Autoimmun. 75:105–117. 10.1016/j.jaut.2016.07.011 [DOI] [PubMed] [Google Scholar]
- Galán, M., Varona S., Orriols M., Rodríguez J.A., Aguiló S., Dilmé J., Camacho M., Martínez-González J., and Rodriguez C.. 2016. Induction of histone deacetylases (HDACs) in human abdominal aortic aneurysm: therapeutic potential of HDAC inhibitors. Dis. Model. Mech. 9:541–552. 10.1242/dmm.024513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, K.A., Joshi A., Carson W.F., Schaller M., Allen R., Mukerjee S., Kittan N., Feldman E.L., Henke P.K., Hogaboam C., et al. 2015. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes. 64:1420–1430. 10.2337/db14-0872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F., and Guilliams M.. 2016. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 44:439–449. 10.1016/j.immuni.2016.02.024 [DOI] [PubMed] [Google Scholar]
- Gordon, S., and Plüddemann A.. 2017. Tissue macrophages: heterogeneity and functions. BMC Biol. 15:53. 10.1186/s12915-017-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y., Tanios F., Reeps C., Zhang J., Schwamborn K., Eckstein H.-H., Zernecke A., and Pelisek J.. 2016. Histone acetylation and histone acetyltransferases show significant alterations in human abdominal aortic aneurysm. Clin. Epigenetics. 8:3. 10.1186/s13148-016-0169-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, B., Nielsen J.M., Hudlebusch H.R., Lees M.J., Larsen D.V., Boesen T., Labelle M., Gerlach L.O., Birk P., and Helin K.. 2014. Inhibition of demethylases by GSK-J1/J4. Nature. 514:E1–E2. 10.1038/nature13688 [DOI] [PubMed] [Google Scholar]
- Hellenthal, F.A.M.V.I., Buurman W.A., Wodzig W.K.W.H., and Schurink G.W.H.. 2009. Biomarkers of abdominal aortic aneurysm progression. Part 2: inflammation. Nat. Rev. Cardiol. 6:543–552. 10.1038/nrcardio.2009.102 [DOI] [PubMed] [Google Scholar]
- Hoeksema, M.A., and de Winther M.P.J.. 2016. Epigenetic Regulation of Monocyte and Macrophage Function. Antioxid. Redox Signal. 25:758–774. 10.1089/ars.2016.6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A., Young T.L., Dang V.T., Shi Y., McAlpine C.S., and Werstuck G.H.. 2017. 4-phenylbutyrate and valproate treatment attenuates the progression of atherosclerosis and stabilizes existing plaques. Atherosclerosis. 266:103–112. 10.1016/j.atherosclerosis.2017.09.034 [DOI] [PubMed] [Google Scholar]
- Ishii, M., Wen H., Corsa C.A.S., Liu T., Coelho A.L., Allen R.M., Carson W.F. IV, Cavassani K.A., Li X., Lukacs N.W., et al. 2009. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 114:3244–3254. 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch, R., and Bird A.. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(S3, Suppl):245–254. 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Xin S., Yan Y., Lun Y., Yang X., and Zhang J.. 2018. Abnormal acetylation of FOXP3 regulated by SIRT-1 induces Treg functional deficiency in patients with abdominal aortic aneurysms. Atherosclerosis. 271:182–192. 10.1016/j.atherosclerosis.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Johnston, W.F., Salmon M., Su G., Lu G., Stone M.L., Zhao Y., Owens G.K., Upchurch G.R. Jr., and Ailawadi G.. 2013. Genetic and pharmacologic disruption of interleukin-1β signaling inhibits experimental aortic aneurysm formation. Arterioscler. Thromb. Vasc. Biol. 33:294–304. 10.1161/ATVBAHA.112.300432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvonen, J., Surcel H.M., Satta J., Teppo A.M., Bloigu A., Syrjälä H., Airaksinen J., Leinonen M., Saikku P., and Juvonen T.. 1997. Elevated circulating levels of inflammatory cytokines in patients with abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 17:2843–2847. 10.1161/01.ATV.17.11.2843 [DOI] [PubMed] [Google Scholar]
- Karlsson, L., Bergqvist D., Lindbäck J., and Pärsson H.. 2009a. Expansion of small-diameter abdominal aortic aneurysms is not reflected by the release of inflammatory mediators IL-6, MMP-9 and CRP in plasma. Eur. J. Vasc. Endovasc. Surg. 37:420–424. 10.1016/j.ejvs.2008.11.027 [DOI] [PubMed] [Google Scholar]
- Karlsson, L., Gnarpe J., Bergqvist D., Lindbäck J., and Pärsson H.. 2009b. The effect of azithromycin and Chlamydophilia pneumonia infection on expansion of small abdominal aortic aneurysms--a prospective randomized double-blind trial. J. Vasc. Surg. 50:23–29. 10.1016/j.jvs.2008.12.048 [DOI] [PubMed] [Google Scholar]
- Kaufmann, E., Sanz J., Dunn J.L., Khan N., Mendonça L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.-C., et al. 2018. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell. 172:176–190.e19. 10.1016/j.cell.2017.12.031 [DOI] [PubMed] [Google Scholar]
- Kent, K.C., Zwolak R.M., Egorova N.N., Riles T.S., Manganaro A., Moskowitz A.J., Gelijns A.C., and Greco G.. 2010. Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J. Vasc. Surg. 52:539–548. 10.1016/j.jvs.2010.05.090 [DOI] [PubMed] [Google Scholar]
- Kimball, A.S., Davis F.M., denDekker A., Joshi A.D., Schaller M.A., Bermick J., Xing X., Burant C.F., Obi A.T., Nysz D., et al. 2019. The Histone Methyltransferase Setdb2 Modulates Macrophage Phenotype and Uric Acid Production in Diabetic Wound Repair. Immunity. 51:258–271.e5. 10.1016/j.immuni.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis, J., Quintin J., Preijers F., Joosten L.A.B., Jacobs C., Xavier R.J., van der Meer J.W.M., van Crevel R., and Netea M.G.. 2014. BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin. Immunol. 155:213–219. 10.1016/j.clim.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, A., Fu J., Duerschke K., Weidlich S., Naumann R., Stewart A.F., and Anastassiadis K.. 2010. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 48:512–520. 10.1002/dvg.20641 [DOI] [PubMed] [Google Scholar]
- Krausgruber, T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., and Udalova I.A.. 2011. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12:231–238. 10.1038/ni.1990 [DOI] [PubMed] [Google Scholar]
- Krishna, S.M., Dear A.E., Norman P.E., and Golledge J.. 2010. Genetic and epigenetic mechanisms and their possible role in abdominal aortic aneurysm. Atherosclerosis. 212:16–29. 10.1016/j.atherosclerosis.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Kroetz, D.N., Allen R.M., Schaller M.A., Cavallaro C., Ito T., and Kunkel S.L.. 2015. Type I Interferon Induced Epigenetic Regulation of Macrophages Suppresses Innate and Adaptive Immunity in Acute Respiratory Viral Infection. PLoS Pathog. 11:e1005338. 10.1371/journal.ppat.1005338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier, L., Chung C.W., Cheng Z., Liddle J., Che K., Joberty G., Bantscheff M., Bountra C., Bridges A., Diallo H., et al. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 488:404–408. 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova, T., Prange K.H.M., Glass C.K., and de Winther M.P.J.. 2020. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat. Rev. Cardiol. 17:216–228. 10.1038/s41569-019-0265-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, Q.L.K., and Lu L.. 2007. Role of leptin in immunity. Cell. Mol. Immunol. 4:1–13. [PubMed] [Google Scholar]
- Laser, A., Lu G., Ghosh A., Roelofs K., McEvoy B., DiMusto P., Bhamidipati C.M., Su G., Zhao Y., Lau C.L., et al. 2012. Differential gender- and species-specific formation of aneurysms using a novel method of inducing abdominal aortic aneurysms. J. Surg. Res. 178:1038–1045. 10.1016/j.jss.2012.04.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, T., and Natoli G.. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11:750–761. 10.1038/nri3088 [DOI] [PubMed] [Google Scholar]
- Liao, M., Xu J., Clair A.J., Ehrman B., Graham L.M., and Eagleton M.J.. 2012. Local and systemic alterations in signal transducers and activators of transcription (STAT) associated with human abdominal aortic aneurysms. J. Surg. Res. 176:321–328. 10.1016/j.jss.2011.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino Cardenas, C.L., Kessinger C.W., Cheng Y., MacDonald C., MacGillivray T., Ghoshhajra B., Huleihel L., Nuri S., Yeri A.S., Jaffer F.A., et al. 2018. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun. 9:1009. 10.1038/s41467-018-03394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Howatt D.A., Balakrishnan A., Moorleghen J.J., Rateri D.L., Cassis L.A., and Daugherty A.. 2015. Subcutaneous Angiotensin II Infusion using Osmotic Pumps Induces Aortic Aneurysms in Mice. J. Vis. Exp. (103). 10.3791/53191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Howatt D.A., Balakrishnan A., Graham M.J., Mullick A.E., and Daugherty A.. 2016. Hypercholesterolemia induced by a PCSK9 gain-of-function mutation augments angiotensin II-induced abdominal aortic aneurysms in C57BL/6 mice-brief report. Arterioscler. Thromb. Vasc. Biol. 36:1753–1757. 10.1161/ATVBAHA.116.307613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Martin, R., Van den Bossche J., Furze R.C., Neele A.E., van der Velden S., Gijbels M.J.J., van Roomen C.P.P.A., Bernard S.G., de Jonge W.J., Rioja I., et al. 2019. Targeting histone deacetylases in myeloid cells inhibits their maturation and inflammatory function with limited effects on atherosclerosis. Front. Pharmacol. 10:1242. 10.3389/fphar.2019.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysgaard Poulsen, J., Stubbe J., and Lindholt J.S.. 2016. Animal Models Used to Explore Abdominal Aortic Aneurysms: A Systematic Review. Eur. J. Vasc. Endovasc. Surg. 52:487–499. 10.1016/j.ejvs.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Martinez, F.O., Helming L., and Gordon S.. 2009. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27:451–483. 10.1146/annurev.immunol.021908.132532 [DOI] [PubMed] [Google Scholar]
- Mellak, S., Ait-Oufella H., Esposito B., Loyer X., Poirier M., Tedder T.F., Tedgui A., Mallat Z., and Potteaux S.. 2015. Angiotensin II mobilizes spleen monocytes to promote the development of abdominal aortic aneurysm in Apoe-/- mice. Arterioscler. Thromb. Vasc. Biol. 35:378–388. 10.1161/ATVBAHA.114.304389 [DOI] [PubMed] [Google Scholar]
- Mitroulis, I., Ruppova K., Wang B., Chen L.-S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I., et al. 2018. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 172:147–161.e12. 10.1016/j.cell.2017.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, C.S., Jose R.J., Moxon J.V., Roomberg A., Norman P.E., Rush C., Körner H., and Golledge J.. 2013. Everolimus limits aortic aneurysm in the apolipoprotein E-deficient mouse by downregulating C-C chemokine receptor 2 positive monocytes. Arterioscler. Thromb. Vasc. Biol. 33:814–821. 10.1161/ATVBAHA.112.301006 [DOI] [PubMed] [Google Scholar]
- Mottamal, M., Zheng S., Huang T.L., and Wang G.. 2015. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 20:3898–3941. 10.3390/molecules20033898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neele, A.E., Prange K.H., Hoeksema M.A., van der Velden S., Lucas T., Dimmeler S., Lutgens E., Van den Bossche J., and de Winther M.P.. 2017. Macrophage Kdm6b controls the pro-fibrotic transcriptome signature of foam cells. Epigenomics. 9:383–391. 10.2217/epi-2016-0152 [DOI] [PubMed] [Google Scholar]
- Neele, A.E., Gijbels M.J.J., van der Velden S., Hoeksema M.A., Boshuizen M.C.S., Prange K.H.M., Chen H.J., Van den Bossche J., van Roomen C.P.P.A., Shami A., et al. 2018. Myeloid Kdm6b deficiency results in advanced atherosclerosis. Atherosclerosis. 275:156–165. 10.1016/j.atherosclerosis.2018.05.052 [DOI] [PubMed] [Google Scholar]
- Nordon, I.M., Hinchliffe R.J., Loftus I.M., and Thompson M.M.. 2011. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat. Rev. Cardiol. 8:92–102. 10.1038/nrcardio.2010.180 [DOI] [PubMed] [Google Scholar]
- Novakovic, B., Habibi E., Wang S.-Y., Arts R.J.W., Davar R., Megchelenbrink W., Kim B., Kuznetsova T., Kox M., Zwaag J., et al. 2016. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 167:1354–1368.e14. 10.1016/j.cell.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata, Y., Furusawa Y., and Hase K.. 2015. Epigenetic modifications of the immune system in health and disease. Immunol. Cell Biol. 93:226–232. 10.1038/icb.2014.114 [DOI] [PubMed] [Google Scholar]
- Ohno, T., Aoki H., Ohno S., Nishihara M., Furusho A., Hiromatsu S., Akashi H., Fukumoto Y., and Tanaka H.. 2018. Cytokine Profile of Human Abdominal Aortic Aneurysm: Involvement of JAK/STAT Pathway. Ann. Vasc. Dis. 11:84–90. 10.3400/avd.oa.17-00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkova, I.O., Aghayev T., Fatkhullina A.R., Makhov P., Titerina E.K., Eguchi S., Tan Y.F., Kossenkov A.V., Khoreva M.V., Gankovskaya L.V., et al. 2019. IL-27 receptor-regulated stress myelopoiesis drives abdominal aortic aneurysm development. Nat. Commun. 10:5046. 10.1038/s41467-019-13017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt, S.J., Liang Q., Rairdan X.Y., Moran J.L., Prosser H.M., Beier D.R., Lloyd K.C., Bradley A., and Skarnes W.C.. 2009. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 6:493–495. 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przanowski, P., Dabrowski M., Ellert-Miklaszewska A., Kloss M., Mieczkowski J., Kaza B., Ronowicz A., Hu F., Piotrowski A., Kettenmann H., et al. 2014. The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia. J. Mol. Med. (Berl.). 92:239–254. 10.1007/s00109-013-1090-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Z., Bagley J., Sukhova G., Baur W.E., Park H.J., Beasley D., Libby P., Zhang Y., and Galper J.B.. 2015. Angiotensin II-induced TLR4 mediated abdominal aortic aneurysm in apolipoprotein E knockout mice is dependent on STAT3. J. Mol. Cell. Cardiol. 87:160–170. 10.1016/j.yjmcc.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Raffort, J., Lareyre F., Clément M., Hassen-Khodja R., Chinetti G., and Mallat Z.. 2017. Monocytes and macrophages in abdominal aortic aneurysm. Nat. Rev. Cardiol. 14:457–471. 10.1038/nrcardio.2017.52 [DOI] [PubMed] [Google Scholar]
- Rao, J., Brown B.N., Weinbaum J.S., Ofstun E.L., Makaroun M.S., Humphrey J.D., and Vorp D.A.. 2015. Distinct macrophage phenotype and collagen organization within the intraluminal thrombus of abdominal aortic aneurysm. J. Vasc. Surg. 62:585–593. 10.1016/j.jvs.2014.11.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K.K., Nicholls S.J., Ginsberg H.D., Johansson J.O., Kalantar-Zadeh K., Kulikowski E., Toth P.P., Wong N., Cummings J.L., Sweeney M., and Schwartz G.G.. 2019. Effect of selective BET protein inhibitor apabetalone on cardiovascular outcomes in patients with acute coronary syndrome and diabetes: Rationale, design, and baseline characteristics of the BETonMACE trial. Am. Heart J. 217:72–83. 10.1016/j.ahj.2019.08.001 [DOI] [PubMed] [Google Scholar]
- Rayamajhi, M., Humann J., Kearney S., Hill K.K., and Lenz L.L.. 2010. Antagonistic crosstalk between type I and II interferons and increased host susceptibility to bacterial infections. Virulence. 1:418–422. 10.4161/viru.1.5.12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinet, P., Milewicz D.M., Cassis L.A., Leeper N.J., Lu H.S., and Smith J.D.. 2018. Consideration of Sex Differences in Design and Reporting of Experimental Arterial Pathology Studies-Statement From ATVB Council. Arterioscler. Thromb. Vasc. Biol. 38:292–303. 10.1161/ATVBAHA.117.309524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, L.E.P., Arroyo L.H., Rifai N., Creager M.A., Libby P., Ridker P.M., and Lee R.T.. 1999. Plasma concentrations of interleukin-6 and abdominal aortic diameter among subjects without aortic dilatation. Arterioscler. Thromb. Vasc. Biol. 19:1695–1699. 10.1161/01.ATV.19.7.1695 [DOI] [PubMed] [Google Scholar]
- Ryder, E., Gleeson D., Sethi D., Vyas S., Miklejewska E., Dalvi P., Habib B., Cook R., Hardy M., Jhaveri K., et al. Sanger Mouse Genetics Project . 2013. Molecular characterization of mutant mouse strains generated from the EUCOMM/KOMP-CSD ES cell resource. Mamm. Genome. 24:286–294. 10.1007/s00335-013-9467-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryer, E.J., Ronning K.E., Erdman R., Schworer C.M., Elmore J.R., Peeler T.C., Nevius C.D., Lillvis J.H., Garvin R.P., Franklin D.P., et al. 2015. The potential role of DNA methylation in abdominal aortic aneurysms. Int. J. Mol. Sci. 16:11259–11275. 10.3390/ijms160511259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11:936–944. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Schlesinger, Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39:232–236. 10.1038/ng1950 [DOI] [PubMed] [Google Scholar]
- Sénémaud, J., Caligiuri G., Etienne H., Delbosc S., Michel J.-B., and Coscas R.. 2017. Translational Relevance and Recent Advances of Animal Models of Abdominal Aortic Aneurysm. Arterioscler. Thromb. Vasc. Biol. 37:401–410. 10.1161/ATVBAHA.116.308534 [DOI] [PubMed] [Google Scholar]
- Sherry-Lynes, M.M., Sengupta S., Kulkarni S., and Cochran B.H.. 2017. Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS One. 12:e0174775. 10.1371/journal.pone.0174775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Hua L., Harmer D., Li P., and Ren G.. 2018. Cre driver mice targeting macrophages. In Methods in Molecular Biology. Humana Press Inc. pp. 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukeir, N., Stefanska B., Parashar S., Chik F., Arakelian A., Szyf M., and Rabbani S.A.. 2015. Pharmacological methyl group donors block skeletal metastasis in vitro and in vivo. Br. J. Pharmacol. 172:2769–2781. 10.1111/bph.13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom, T., Haase N., Rosamond W., Howard V.J., Rumsfeld J., Manolio T., Zheng Z.-J., Flegal K., O’Donnell C., Kittner S., et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee . 2006. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 113:e85–e151. 10.1161/CIRCULATIONAHA.105.171600 [DOI] [PubMed] [Google Scholar]
- Toghill, B.J., Saratzis A., Harrison S.C., Verissimo A.R., Mallon E.B., and Bown M.J.. 2015. The potential role of DNA methylation in the pathogenesis of abdominal aortic aneurysm. Atherosclerosis. 241:121–129. 10.1016/j.atherosclerosis.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Toghill, B.J., Saratzis A., Freeman P.J., Sylvius N., and Bown M.J.. UKAGS collaborators . 2018. SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin. Epigenetics. 10:29. 10.1186/s13148-018-0460-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S.K., Tian X., Lafrance L.V., Duquenne C., Suarez D.P., Newlander K.A., Romeril S.P., Burgess J.L., Grant S.W., Brackley J.A., et al. 2012. Experimental Methods (Chemistry). ACS Med. Chem. Lett. 3:1091–1096. 10.1021/ml3003346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh, A., Gaspari T.A., Liu H.B., Dousha L.F., Widdop R.E., and Dear A.E.. 2008. A novel histone deacetylase inhibitor reduces abdominal aortic aneurysm formation in angiotensin II-infused apolipoprotein E-deficient mice. J. Vasc. Res. 45:143–152. 10.1159/000110041 [DOI] [PubMed] [Google Scholar]
- Wang, J., Lindholt J.S., Sukhova G.K., Shi M.A., Xia M., Chen H., Xiang M., He A., Wang Y., Xiong N., et al. 2014a. IgE actions on CD4+ T cells, mast cells, and macrophages participate in the pathogenesis of experimental abdominal aortic aneurysms. EMBO Mol. Med. 6:952–969. 10.15252/emmm.201303811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Ren J., Morgan S., Liu Z., Dou C., and Liu B.. 2014b. Monocyte chemoattractant protein-1 (MCP-1) regulates macrophage cytotoxicity in abdominal aortic aneurysm. PLoS One. 9:e92053. 10.1371/journal.pone.0092053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Alexander M., Misharin A.V., and Budinger G.R.S.. 2019. The role of macrophages in the resolution of inflammation. J. Clin. Invest. 129:2619–2628. 10.1172/JCI124615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, N.R., De Beer M.C., Wroblewski J.M., Ji A., Bailey W., Shridas P., Charnigo R.J., Noffsinger V.P., Witta J., Howatt D.A., et al. 2015. Deficiency of Endogenous Acute-Phase Serum Amyloid A Protects apoE-/- Mice From Angiotensin II-Induced Abdominal Aortic Aneurysm Formation. Arterioscler. Thromb. Vasc. Biol. 35:1156–1165. 10.1161/ATVBAHA.114.304776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Xu Y., Lu H., Howatt D.A., Balakrishnan A., Moorleghen J.J., Vander Kooi C.W., Cassis L.A., Wang J.A., and Daugherty A.. 2015. Cys18-Cys137 disulfide bond in mouse angiotensinogen does not affect AngII-dependent functions in vivo. Hypertension. 65:800–805. 10.1161/HYPERTENSIONAHA.115.05166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Q., Zhang J., Han Y., Zhang X., Jiang H., Lun Y., Wu X., Gang Q., Liu Z., Böckler D., et al. 2019. Epigenetic regulation of regulatory T cells in patients with abdominal aortic aneurysm. FEBS Open Bio. 9:1137–1143. 10.1002/2211-5463.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, W., MacTaggart J., Knispel R., Worth J., Persidsky Y., and Baxter B.T.. 2009. Blocking TNF-α attenuates aneurysm formation in a murine model. J. Immunol. 183:2741–2746. 10.4049/jimmunol.0803164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Xu S., Liu P., Koroleva M., Zhang S., Si S., and Jin Z.G.. 2017. Suberanilohydroxamic acid as a pharmacological Kruppel-like factor 2 activator that represses vascular inflammation and atherosclerosis. J. Am. Heart Assoc. 6:e007134. 10.1161/JAHA.117.007134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L., et al. 2014. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 40:274–288. 10.1016/j.immuni.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q., Sun L., Zhu Z., Wang L., Li S., and Ye R.D.. 2014. Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression in serum amyloid A-stimulated macrophages. Cell. Signal. 26:1783–1791. 10.1016/j.cellsig.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H., Zhou H.-F., Akk A., Hu Y., Springer L.E., Ennis T.L., and Pham C.T.N.. 2016. Neutrophil Proteases Promote Experimental Abdominal Aortic Aneurysm via Extracellular Trap Release and Plasmacytoid Dendritic Cell Activation. Arterioscler. Thromb. Vasc. Biol. 36:1660–1669. 10.1161/ATVBAHA.116.307786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H., Hu Y., Akk A., Ye K., Bacon J., and Pham C.T.N.. 2019. Interleukin-12 and -23 blockade mitigates elastase-induced abdominal aortic aneurysm. Sci. Rep. 9:10447. 10.1038/s41598-019-46909-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X., Yang S., Zhang M., and Yue Y.. 2019. The role and prospect of JMJD3 in stem cells and cancer. Biomed. Pharmacother. 118:109384. 10.1016/j.biopha.2019.109384 [DOI] [PubMed] [Google Scholar]
- Zaina, S., Heyn H., Carmona F.J., Varol N., Sayols S., Condom E., Ramírez-Ruz J., Gomez A., Gonçalves I., Moran S., and Esteller M.. 2014. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 7:692–700. 10.1161/CIRCGENETICS.113.000441 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the human AAA and atherosclerotic tissue cohort.
lists the human AAA and atherosclerotic scRNA-seq cohort.
lists the primers used in this study.