Abstract
The problem of resistance to therapy in cancer is multifaceted. Here we take a reductionist approach to define and separate the key determinants of drug resistance, which include tumour burden and growth kinetics; tumour heterogeneity; physical barriers; the immune system and the microenvironment; undruggable cancer drivers; and the many consequences of applying therapeutic pressures. We propose four general solutions to drug resistance that are based on earlier detection of tumours permitting cancer interception; adaptive monitoring during therapy; the addition of novel drugs and improved pharmacological principles that result in deeper responses; and the identification of cancer cell dependencies by high-throughput synthetic lethality screens, integration of clinico-genomic data and computational modelling. These different approaches could eventually be synthesized for each tumour at any decision point and used to inform the choice of therapy.
Drug resistance continues to be the principal limiting factor to achieving cures in patients with cancer. The problem of drug resistance in cancer has strong similarities to the field of infectious disease, in that both disciplines are challenged by highly proliferating intrinsic or extrinsic aggressors. As with antimicrobial therapy, the excitement that was brought about by initial successes of early chemotherapeutics (such as nitrogen mustard1 and aminopterin2) was quickly tempered by evidence showing that although tumours went into remission quickly, they developed resistance, resulting in disease relapse.
The initial solution to the problem of resistance to single-agent chemotherapy—the combined administration of agents with non-overlapping mechanisms of action, or polychemotherapy—was taken, unsurprisingly, from the rulebook of antimicrobial therapy3. This empirical approach worked remarkably well in some forms of lymphoma, breast cancer and testicular cancer4–6. Combination chemotherapy thus became a new paradigm for cancer therapy that led to the development of increasingly complex regimens. In addition, a number of different approaches to dose intensity7, including shorter-interval administrations of chemotherapy8,9 or higher doses of chemotherapy9 with growth factor support to prevent continued bone marrow suppression, resulted in improved success of these therapies by preventing early regrowth of tumours. At the turn of the century, almost 50 years after its introduction, the successes achieved with polychemotherapy had largely plateaued. Surgery, radiotherapy and polychemotherapy were clearly not enough to cure many tumour types.
As a result, new therapeutic strategies directed at targeting the key enabling characteristics and acquired capabilities that transform normal cells and tissues into malignancies began to be developed. The introduction of therapies that disrupted these hallmark features10,11, including targeted therapies, was an important leap forward. Indeed, the understanding of the biological determinants of cancer has resulted in highly efficacious therapies against tyrosine kinases, nuclear receptors and other molecular targets. The initial successes of oestrogen receptor (ER) and androgen receptor (AR) antagonists, as well as BCR-ABL, HER2 and EGFR inhibitors, led to a massive effort to develop agents that target oncogenes and other key cellular vulnerabilities. More recently, oncological therapy has advanced again by using immunological approaches to recognize and attack cancer. Anti-CTLA412 and anti-PD-1/PD-L113 monoclonal antibodies that disable negative regulators, or checkpoints, of the adaptive immune system have resulted in remarkable anti-tumour activity—and even cures—in multiple tumour types14. And yet, similarly to what was previously observed with conventional chemotherapy, eventual resistance to targeted and immunological therapies remains the norm. This is where the similarities between cancer and infectious diseases may dissociate: combination therapy often leads, for example, to disease becoming undetectable in HIV or cured in tuberculosis, but in metastatic cancers this is the exception rather than the rule15. Not unexpectedly, cancer appears to be a more-complex biological problem.
Here we attempt to present a framework for conceptualizing drug resistance in cancer by enumerating the basic determinants of resistance and considering their implications for the development of successful therapeutic strategies. These basic determinants of resistance, which are present in unique iterations during the history of a cancer, result in different clinical states that range from extreme sensitivity to complete resistance to therapy. We describe standard and emerging interventions that target these determinants, and consider how new technological and pharmacological advances can be integrated with these interventions to prevent, delay or revert resistance to therapy.
Biological determinants of resistance
In its simplest definition, cancer therapy operates as a three-component system: (i) a therapy; that targets (ii) a population of cancer cells; within (iii) a particular host environment. The pharmacological properties of the therapy, together with intrinsic and acquired physical and molecular parameters of cancer cells and extrinsic environmental factors, result in the spectrum of clinical responses. Many descriptions of drug resistance in cancer have focused on the binary differences between intrinsic and acquired resistance; however, in practice, many tumours are or become resistant owing to overlapping combinations of these factors. We propose that, in defining the fundamental biological principles of resistance, a framework can be created for understanding resistance both to existing and future therapies (Fig. 1a). We believe that by focusing both cancer and clinical science on addressing each determinant separately, the resistance problem can be managed (Fig. 1b). The biological determinants of resistance are delineated in the next sections.
Fig. 1 |. A framework for understanding drug resistance.
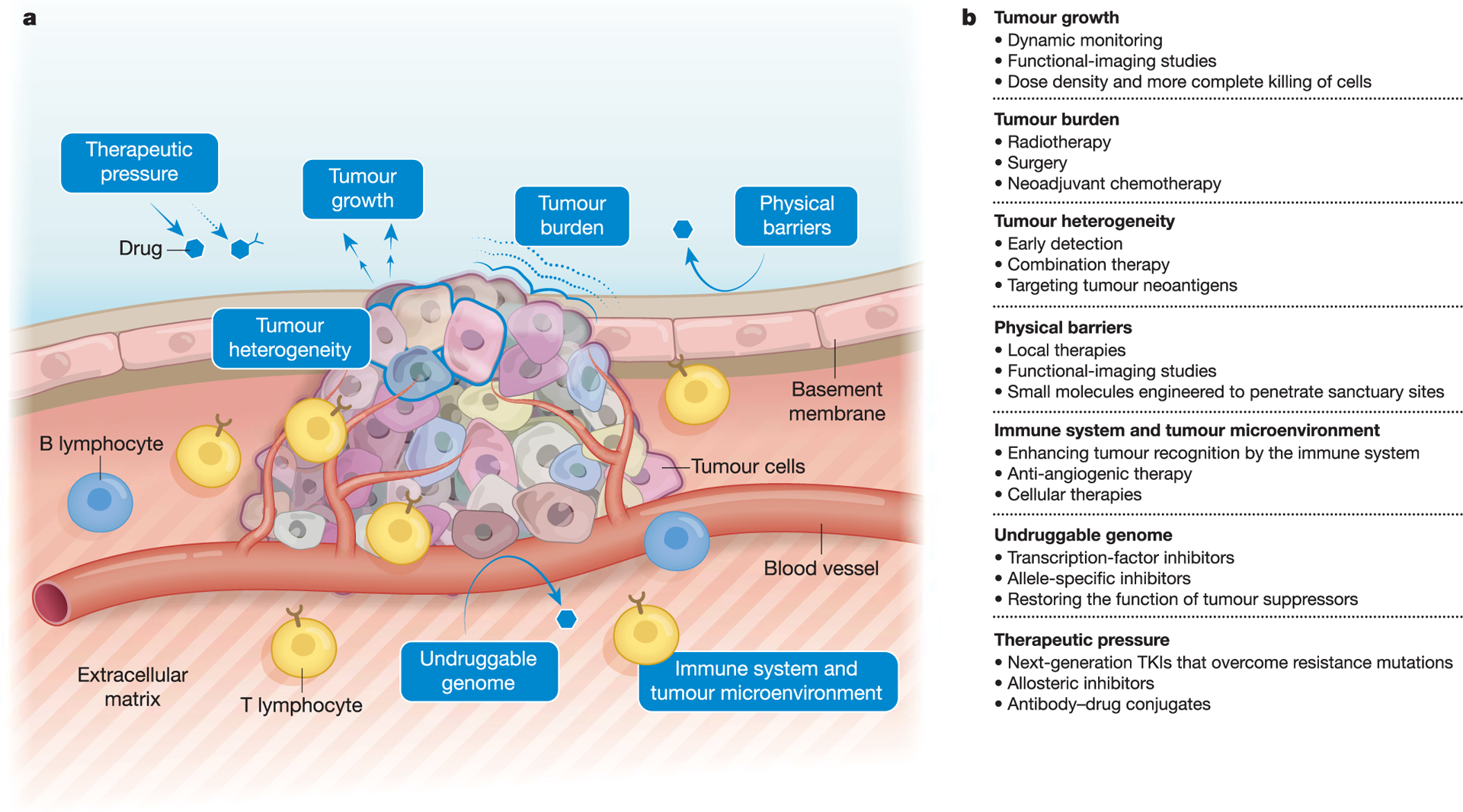
a, Biological determinants of drug resistance. Tumours are heterogeneous and are situated in a milieu that comprises the basement membrane, vasculature, immune cells and tumour microenvironment, among other components. Changes in the physical parameters, genome and surrounding environment of the tumour drive drug resistance. b, Standard of care and emerging approaches to managing the biological determinants of resistance. The determinants of resistance can be targeted by a number of clinical diagnostic and therapeutic strategies.
Tumour burden and growth kinetics
There is an almost universal correlation between tumour burden and curability16. In many tumour types, the size of the tumour (or number of cells, in the case of liquid tumours) at diagnosis is perhaps the most frequently used variable to estimate prognosis; larger tumours correlate with increased metastatic risk17. This inverse correlation between size and curability was not entirely anticipated in the infancy of chemotherapy. Early mathematical models, such as the ‘log kill’ hypothesis, proposed that combining multiple drugs that individually kill a logarithmic fraction of cells over multiple cycles would permit sequential decreases in tumour burden until the disease was fully eradicated18. This is true in tumours that are highly sensitive to chemotherapy, such as some lymphomas and germ cell tumours, but does not hold across many other cancer types.
To more accurately model cancer growth, the Goldie–Coldman hypothesis was proposed19. This model, which was informed by seminal microbiology experiments20, accounts for tumour size and also incorporates the emergence of resistance. According to this hypothesis, the probability that a cancer contains drug-resistant clones depends on the mutation rate and the size of the tumour19. In fact, given a certain mutation rate, size becomes the key determinant in predicting the presence of drug-resistant mutations. Multiple concepts have stemmed from this model, including the notion that alternating non-cross-resistant combinations of chemotherapy, rather than administering all therapies at once (which is often limited by toxicity), is superior in preventing drug resistance as compared to sequential therapies. Alternation of therapy sequences would allow the tumour to be exposed to a greater number of total drugs by an earlier time point. This hypothesis has not been uniformly borne out in clinical practice, suggesting that there are additional complexities that need to be considered21.
Moreover, although tumour size is a critical determinant of resistance, the rate of tumour growth and the changes in growth kinetics that are induced by therapy also have a critical role in responses to therapy and resistance. Tumour growth kinetics are highly variable, ranging from indolent to aggressive. Although tumours with low rates of growth are often associated with long survival, they are typically incurable with cytotoxic chemotherapy or even with targeted therapies. By contrast, tumours that grow at higher speeds can be exquisitely sensitive to chemotherapy. There is also a direct relationship between growth rate and tumour size, which explains, for example, the frequency of interval cancer cases in screening programs.
The model that perhaps best explains the growth of cancer and its regression after therapy is the Norton–Simon hypothesis22. This model, which applies to most solid tumours, is based on Gompertzian growth curves. According to the model, tumours grow in a sigmoidal manner—exponentially faster at low tumour burdens and subsequently approaching a plateau with slower growth rates as they reach a larger size23. Because drugs reduce the size of tumours, they affect growth kinetics. After a single administration of chemotherapy, the remaining tumour fractions may resume their early phase of exponential growth. Following this logic, the probability of eradication is maximized by preventing rapid regrowth of the tumour between treatments24. This led to the concept of dose-dense chemotherapy, an approach in which the most effective dose level of a drug is given over as short a time interval as possible. Clinical proof of concept of dose density has been demonstrated in early breast and ovarian cancer, for which chemotherapy that is administered more frequently has in select circumstances improved overall survival8,25,26. Dose-dense approaches have not been sufficient to convert ineffective therapies into effective ones, but rather have been used to improve the efficacy of established approaches. Less attention has been paid to the role of dose scheduling in targeted therapies than in chemotherapy, and we propose that this could be a strategy to deepen responses and increase cures.
Tumour heterogeneity
Cancer heterogeneity is perhaps the cause of drug resistance that is easiest to conceptualize27. Cancer cells acquire genomic alterations through a variety of mutational processes that generate spatial and temporal genetic diversity28. These processes occur at different evolutionary speeds—from the relatively slow rate of age-related mutations, to frequent editing of genes by APOBEC enzymes (a process that increases over the course of tumour evolution), to bursts of dramatic and catastrophic events that are induced by genomic instability29, chromothripsis30 and chromosomal instability31. Large chromosomal alterations can be envisioned as macro-evolutionary events and in some circumstances probably represent a point of no return in the development of resistance, illustrating the importance of early therapeutic intervention.
Together with ecosystem-selective pressures, mutational processes lead to parallel and convergent evolution, as well as spatial segregation of clones in primary and metastatic sites32. These pressures include exogenous exposures, internal environmental dynamics and cancer therapies themselves. The effects of selective therapeutic pressure have been well characterized and range from the disappearance of targeted cellular clones, to the acquisition of new resistance mutations, to adaptive responses in signalling and epigenetics and, finally, a complete change in tumour phenotype33. In some cases, the effects of therapies (mostly chemotherapy) can be profound and equivalent to inducing a state of genomic instability. For example, in low-grade gliomas, chemotherapy with temozolomide can result in hypermutated tumours at recurrence and in some cases bring about the transformation of tumours to highly aggressive glioblastoma multiforme34. Similarly, clonal haematopoiesis—as evidenced by recurrent somatic mutations in leukaemia-associated genes in haematopoietic stem cells—has been associated with prior exposure to radiotherapy and chemotherapy and carries an increased risk of developing leukaemia35. These observations should prompt us to weigh carefully the potential unintended consequences of administering chemotherapy or radiotherapy.
A critical clinical issue is how tumour heterogeneity is measured. Currently, heterogeneity is evaluated by genomic sequencing of either archived tumour samples at diagnosis or a subsequent biopsied tumour sample at recurrence. This approach—despite its usefulness in some cases for therapy selection36—has serious limitations, as it is unlikely to accurately capture tumour heterogeneity, with obvious implications for cancer therapy. For example, targeting an ‘actionable’ driver mutation may only prove effective if the mutation is truncal (that is, clonal and present in most subclones and regions of the tumour over its lifetime)37. In other cases, finding a given mutation may not be a guarantee of the mutation being clonal, and conversely, the paucity of a mutation does not guarantee that it is incidental. Indeed, subclonal driver mutations in ESR138 and in genes of the PI3K pathway39 are sufficient to drive resistance to targeted therapies. In this setting, we propose that a catalogue of ‘clonality’ of driver mutations may be informative.
Physical barriers
Cancer cells can create spatial gradients within tumours that prevent adequate blood flow, thereby creating a pro-tumorigenic hypoxic environment and decreasing the effective exposure of a tumour to drugs. Although attenuating blood flow to tumours is one known mechanism of action of anti-angiogenic agents, this is probably not the complete story. Alternative evidence suggests that anti-angiogenic agents may also normalize vascular structure and function40, facilitating the delivery of systemic agents such as chemotherapy or even targeted therapy41. Recently, the combination of anti-angiogenic tyrosine kinase inhibitors and anti-PD-1/PD-L1 antibodies has also demonstrated what appears to be synergistic activity42,43, although the precise mechanism underlying this clinical observation is uncertain.
Cancer cells may colonize and proliferate in ‘sanctuary sites’, or anatomical spaces in which systemically administered drugs do not reach therapeutic concentrations. The prototypical example of this is the central nervous system (CNS) and the physical boundary imposed by the blood–brain barrier. Additional sanctuary sites include the peritoneum, which can be addressed by intraperitoneal chemotherapy, and the testes, which has led to the administration of prophylactic radiation in children with acute lymphoblastic leukaemia.
Of these sanctuary sites, the CNS represents perhaps the highest unmet medical need. Some tumour types such as lung, HER2-positive breast, melanoma and kidney cancers have a particularly high level of CNS tropism. A variety of approaches have attempted to address this daunting clinical challenge, including improved radiotherapy techniques that allow a more selective targeting of tumours in the brain44, and targeted therapies that penetrate the blood–brain barrier45. Early investigations into the combination of immune checkpoint inhibitors and stereotactic radiosurgery have shown promise in inducing rapid and complete response of each lesion compared with stereotactic radiosurgery alone46. These data suggest that attacking a macroscopic brain metastasis with a combination of a local therapy and systemic checkpoint inhibition may be a promising approach for controlling CNS metastases. Ultimately, the solution to CNS invasion is not obvious but we are beginning to develop a deeper understanding of the underlying mechanisms that lead to migration across the blood–brain barrier, as well as CNS tropism and growth47–49, which may translate into therapeutic approaches.
Immune system and tumour microenvironment
The tumour microenvironment—the surrounding space composed of immune cells, stroma and vasculature—may mediate resistance by several mechanisms, including preventing immune clearance of tumour cells, hindering drug absorption and stimulating paracrine growth factors to signal cancer cell growth50.
The importance of immune evasion by tumours is underscored by the success of checkpoint blockade immunotherapy14, which has led to sustained long-term control of disease in advanced melanoma, renal cell carcinoma, non-small-cell lung cancer (NSCLC), urothelial cancers and microsatellite-unstable cancers, among others. Some immunotherapy-resistant tumours have a low mutational burden, which leads to a paucity of neoantigens that are available for presentation and ultimately prevents recognition51,52. Some mechanisms of resistance to checkpoint blockade have been elucidated, including the loss of β2-microglobulin (which impairs tumoral antigen presentation) and JAK1 or JAK2 mutations (which render tumour cells insensitive to interferon gamma)53.
Immunosuppressive cancer microenvironments—so-called ‘immune deserts’—are now recognized as a major impediment to checkpoint inhibitors, owing to the presence of regulatory T cells, myeloid-derived suppressor cells, tumour-associated macrophages, cytokines and chemokines—all of which can inhibit immune-mediated anti-tumour effects50. This has led to the investigation of a range of techniques to turn immunologically ‘cold’ tumours into ‘hot’ ones by recruiting immune effectors. These techniques combine checkpoint blockade with anti-angiogenic agents42,43, targeted therapies54, metabolic modulators, oncolytic viruses, epigenetic therapies and other checkpoints.
Undruggable genomic drivers
Despite a growing number of successes in efforts to target oncogenic driver mutations, some of the most formidable oncogenes and tumour suppressor genes remain undruggable, including MYC, RAS and TP53. Several approaches are being explored to address these targets, including miniproteins that prevent MYC dimerization55, allele-specific inhibitors that trap and inactivate mutant KRAS(G12C)56 and small molecules that covalently bind to p53 to restore its normal (wild-type) function57.
Other oncogenic drivers, such as class 2 mutations in BRAF (which result in mutant BRAF proteins that signal as RAS-independent constitutive dimers58), are only partially inhibited by existing allosteric MEK inhibitors59, owing to an inadequate therapeutic index. New classes of RAF inhibitors that inhibit the formation of RAF homodimers and heterodimers may address these alterations. Furthermore, target indifference—in which the effects of targeting an oncogenic driver are attenuated by downstream or parallel alterations in the pathway—can drive resistance. This is exemplified by resistance to anti-EGFR therapies in colon cancer, which can be mediated by downstream KRAS- or NRAS-activating mutations60.
Selective therapeutic pressure
Cancer therapies are powerful inducers of changes in both the tumour and its ecosystem. Conventional chemotherapy and radiotherapy enhance genomic instability, with massive and widespread effects on surviving cells and non-cancer cells28,61, and can also induce immune responses in the host that attenuate anti-tumour responses62.
Under targeted therapies, changes are subtler and may be divided into early adaptive responses or—after prolonged exposures—acquired resistance. Adaptive responses can occur so rapidly that no response is ever clinically apparent, and may be responsible for short-lived durations of clinical response. Adaptive mechanisms are often the result of non-genetic relief of negative feedback of signalling pathways and/or epigenetic modulation, which causes the activation of parallel pathways, or reactivation of the initial pathway63,64. For example, BRAF-mutant colorectal cancers are insensitive to BRAF inhibitors, owing to reactivation of upstream receptor tyrosine kinases including EGFR65,66, but BRAF-mutant melanomas express only low levels of EGFR and are therefore not subject to this relief of negative feedback.
In acquired resistance, in which prolonged clinical response is followed by tumour regrowth, other mechanisms are involved that include the emergence of new activating mutations on the target itself, pathway alterations or even histological changes. In monogenic tumours, resistance to tyrosine kinase inhibitors (TKIs) is frequently driven by gatekeeper mutations67 and other driver mutations, which maintain oncogene dependence68 by preventing access to the site where ATP-competitive TKIs bind. Other acquired causes of TKI resistance include amplification of the gene and splice variants. Drug-discovery efforts have resulted in newer TKIs that can overcome on-target acquired resistance. Some of these drugs avoid the steric hindrance that is mediated by mutations in the kinase domain (such as the anti-BCR-ABL agent ponatinib69), bind more potently to the target (as exemplified by the anti-ALK agent alectinib70) and may even exhibit selectivity for mutant kinases by binding irreversibly to the mutated domains (such as osimertinib71). Characteristic mutations that are associated with acquired resistance can also develop in the ligand-binding domains of nuclear hormone receptors, resulting in ligand-independent transcription72.
Another mode of acquired resistance to kinase inhibition is the amplification of an upstream gene, which can bring about resistance to therapy that targets a downstream molecule; this is exemplified by MET amplification driving resistance to EGFR inhibitors73. Tumour suppressor mutations can also drive resistance to targeted therapies, including mutations in PTEN—which drive PI3Kβ signalling in response to PI3Kα inhibitors74—and reversion mutations in BRCA1 or BRCA2 in response to PARP inhibitors75.
Finally, phenotypic changes may occur that result in the evolution of treated tumours into new histological types. For example, transformation to aggressive clinical neuroendocrine phenotypes has been noted in prostate tumours that are initially responsive to antiandrogens, and in EGFR-mutant NSCLC that is initially responsive to TKIs. These tumours are treated, like newly diagnosed small-cell cancers, with platinum and etoposide chemotherapy, albeit with limited clinical benefit. Genomic studies have revealed that these neuroendocrine prostate and lung tumours that are induced by treatment acquire the loss of RB1 and TP53 and become resistant to antiandrogens76 or EGFR TKIs77. Investigations into the mechanisms of changes in cell plasticity have implicated the SOX2 transcription factor, which is upregulated in enzalutamide-resistant prostate cancer76. We speculate that additional transcriptional and epigenetic mechanisms drive lineage plasticity in cancers that are responsive to targeted therapies, leading to target independence.
Overcoming resistance
Although any single biological determinant of resistance can contribute to treatment becoming refractory, these factors frequently coexist in a cancer, arising in a time- and therapy-dependent manner. In the next sections we lay out four general solutions to drug resistance—earlier detection of disease; deepened responses; therapeutic monitoring and adaptive interventions; and exploitation of cancer dependencies— and illustrate how these interventions may increase the overall probability of cure (Fig. 2a). We also discuss how new diagnostic and therapeutic technologies are being used to disrupt the emergence of drug resistance.
Fig. 2 |. Proposed solutions to the problem of drug resistance in cancer.
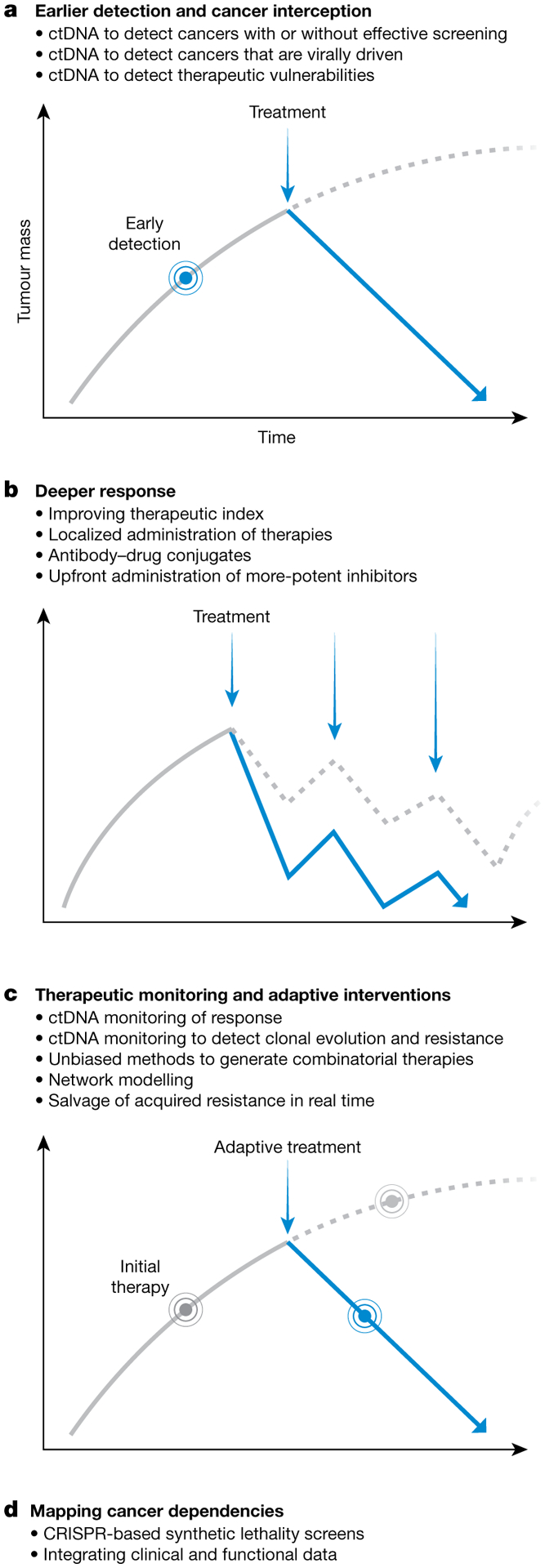
a–d, Proposed solutions. Many technologies—including plasma-based tumour diagnostics, more-potent pharmacological strategies, computational and systems biology modelling and genetic screening—may help to prevent cancer drug resistance through interceding earlier; enhancing therapeutic efficacy; ascertaining disease burden and changing therapies; and beginning to detail the behaviour of cancer cells under therapy. Early detection and treatment of cancer (a), deeper therapeutic responses (b), therapeutic monitoring with adaptive interventions (c) and mapping cancer dependencies (d) can alter the natural history of cancer or its initial therapeutic trajectory to increase the probability of cure (solid blue lines). Grey solid lines show the trajectory before the indicated intervention; grey dashed lines show the trajectory if the indicated intervention were not performed (a, c) or performed less effectively (b).
Earlier detection
Treatment of cancer when tumour burden and clonal diversity are low is perhaps the most obvious strategy to prevent drug resistance. Implementing this approach has two critical components: first, early disease detection; and second, treatment with the most effective upfront therapy to maximize tumour eradication (Fig. 2b). The latter concept is the basis for adjuvant therapy and has improved patient survival for many types of tumour. The most extreme realization of this approach would be cancer interception78—the detection of premalignant lesions followed by mechanistically based interventions to prevent the formation of a cancer. We will examine both components below.
Cancer screening, or efforts to detect pre-symptomatic cancer, is currently established in breast, cervical, colorectal and lung cancers and has generally led to improvements in disease-specific survival79. However, the tests that are currently available provide low sensitivity and specificity, often detect resistant and incurable disease and can also be tedious and invasive, which leads to poor compliance. A case in point is mammography, which even when optimally used in large populations of women who are at average risk for developing breast cancer reduces breast-cancer-specific mortality by only 20%80. This modest benefit is achieved at the expense of a high false-positive rate: over a 10-year period, almost a third of women screened will have had a benign biopsy after a positive radiographic finding81. Similarly, prostate-specific antigen (PSA) screening detects mostly low-grade disease but does not decrease mortality82.
Over the last two decades, it has become possible to detect DNA in the blood that originates from the fetus in pregnant women83,84 and from tumours in patients with cancer85. Circulating tumour DNA (ctDNA) testing offers many potential advantages, including non-invasive, dynamic and global detection of cancer and monitoring of clonal evolution86. The initial finding that ctDNA can be isolated from plasma87–89—coupled with methodological developments such as digital PCR90 to quantify mutant DNA, bead-based emulsions91 to enhance scalability and, most recently, next-generation sequencing using unique molecular barcoding techniques to correct errors92,93—has led to intensive research efforts to develop ctDNA-based ‘liquid biopsies’ for non-invasive detection94. We discuss ctDNA as a tool for cancer screening and surveillance in this section, and examine its role in response and resistance to therapy in later sections.
So far, ctDNA testing has primarily focused on the detection of actionable genomic alterations, including single nucleotide variants (SNVs), copy-number alterations and structural variants. However, for ctDNA to form the basis of an effective screening tool to identify cancer before it metastasizes, it must be able to detect a low burden of disease. Several studies have demonstrated that in early-stage disease, SNV-based ctDNA assays with analytical thresholds that are as low as 0.1% will miss an appreciable proportion of cancers95, although precise rates vary by cancer type96–98. These limitations are not easily overcome by increasing sequencing intensity or error-correction techniques, as the lower limit of SNV detection can be based on the total number of molecular templates available for sequencing—which itself is proportional to tumour burden. Several strategies to improve the ctDNA detection of low disease burden have recently emerged (Box 1). Another challenge for ctDNA detection is the delineation of the tumour tissue of origin. Some new strategies to overcome this hurdle capitalize on epigenetic modifications, combine multimodal methods to distinguish malignant from mutation-based non-malignant processes or focus on detecting specific DNA sequences (Box 1).
Box 1. Emerging methods to improve ctDNA-based screening of cancer.
Detection of low burden of disease
Increasing input DNA through phlebotomy of larger volumes of blood160.
Obtaining non-plasma tumour DNA (for example, stool161, Papanicolaou smear162).
Enriching for ctDNA-sized fragments163.
Characterizing the tissue of origin
Detecting ctDNA methylation164.
Assessing the footprints of nucleosome binding165.
Combining SNVs with epigenomic assays166 (to distinguish mutations from benign lesions or clonal haematopoiesis166).
Distinguishing cancers that are virally driven (for example, EBV-driven nasopharyngeal carcinoma167).
Evaluating oncogenic structural variants (for example, translocations168, amplifications169).
Noting these issues, we propose that as a single screening test, ctDNA may initially be used in select subtypes of monogenic cancer, especially those with high rates of cell turnover or in high-risk patient populations (for example, patients with germline BRCA mutations). In the case of NSCLC, computed tomography (CT)-based screening is effective in identifying small stage 1 tumours, whereas ctDNA is more effective for larger tumours95. Similarly, other cancers may require combined or risk-stratified screening modalities of conventional imaging or laboratory diagnostics (for example, mammography, PSA testing) with ctDNA. To maximize the benefits of early detection, these strategies must be coupled with a definitive therapy to prevent recurrence. Ultimately, population-based screening studies will be required to formally prove the efficacy of ctDNA screening in improving clinical outcomes.
Achieving deeper responses
As the persistence of even a few residual clones inevitably leads to tumour regrowth, the eradication of tumours is imperative (Fig. 2b). Strategies that can be used to deepen responses vary depending on the type of therapy but several guiding principles apply across therapeutic modalities, including optimizing dose, schedule and combination partners.
For chemotherapy, the ceiling for efficacy is often dictated not by a plateau in the dose–response effect, but rather by toxicity or therapeutic index. Nanoparticle platforms99—including liposomal or albumin-bound formulations—may mitigate chemotoxicity, but outside of selected cases of leukaemia100 they have generally not proven superior to the naked chemotherapeutic. In select haematological cancers and germ cell tumours, dose thresholds that are mandated by the limits of bone marrow tolerance have been overcome through the rescuing of marrow function after high-dose chemotherapy by reinfusion of autologous haematopoietic stem cells101. However, this approach has been notably unsuccessful in solid tumours102, suggesting that dose intensification alone will not always deepen responses—possibly owing to alternative mechanisms of promoting tumorigenesis62. Optimizing chemosensitivity has long been a goal of cancer therapy; however, ex vivo assays have been largely unsuccessful and patient-derived xenografts are costly, time-consuming and have not scaled to meet clinical demand. Newer approaches to determine chemosensitivity in vivo include implantable microdevices for real-time multiplex drug testing103, which could potentially increase the predictive value of chemotherapy benefit.
Another strategy to deepen therapeutic responses is the localized administration of chemotherapy through the liver vasculature, albeit with partial success104. A more elegant approach to deliver higher doses of chemotherapy, unconstrained by location or vascular supply, has been enabled by antibody–drug conjugates (ADCs), which consist of recombinant monoclonal antibodies that are covalently bound to cytotoxic agents through synthetic linkers105. ADCs enable the delivery of potent cytotoxic payloads directly to tumour cells, as the antibodies are engineered to bind with high affinity to antigens that are preferentially expressed on tumour cells.
Rather than simply increasing local amounts of a less-active chemotherapy, the net effect is to deepen response by localized delivery of a dose of chemotherapy that would be far too toxic if administered alone. Although we acknowledge that ADCs have so far not fully delivered on their initial promise to improve therapeutic index—despite the approval of several of these agents105—we believe that with modern ADC chemistry this is beginning to change. This is perhaps best exemplified by the high degree of durable efficacy that is observed with the latest generation of HER2-targeted ADC, DS-8201a, in so-called HER2-low (that is, defined by amplification of HER2 that is less than that necessary for response to current anti-HER2 therapies) breast cancer106. DS-8201a demonstrates the ability of a modern ADC to effectively deliver chemotherapy that is based on low levels of expression of a target protein, even when the target protein is not itself an oncogene for that tumour type. We anticipate that further engineering—such as optimizing the ratio of drug to antibody, or varying payloads to include immunomodulatory molecules—may improve the efficacy of ADCs.
With checkpoint blockade immunotherapy, the depth and durability of the response that is elicited have brought about a de facto cure for a fraction of patients with metastatic cancer. Because inhibition of CTLA4 primes and activates T cells at an earlier stage than inhibition of PD-1/PD-L1 (which drives recognition of cancer cells by T cells14), a combined anti-CTLA4 and anti-PD-1/PD-L1 blockade can exert a synergistic benefit and elicits a multicellular immune memory. This is distinct from combination targeted therapies, which almost always target multiple intracellular signalling factors and operate on a faster timescale. The notion that immune memory cannot be distilled to a single alteration may explain why PD-L1 is not a universal marker of response to checkpoint blockade across tumour histologies (for example, PD-L1-negative melanomas respond to anti-PD-L1 blockade107), and why global measures such as mismatch repair deficiencies in germline or somatic cells108 or tumour mutational burden109 can also be used to predict response to checkpoint blockade.
Thus, the nature of durable responses to immunotherapy may be due to the exertion of both on-target (cancer cell–T cell) and off-target (antigen-presenting cell–T cell) effects that contribute to a ‘bystander’ killing effect on neighbouring cells. Targeted therapies also cause bystander killing, as some targets (for example, the oestrogen receptor) need only be present on a subset of cells for the therapy to eradicate a population of tumour cells. A mechanistic understanding of immunological and non-immunological bystander effects may be relevant for deepening responses to targeted therapies.
Strategies to achieve deeper responses will necessarily be different with agents that target driver oncogenes. We divide targeted therapies into two classes on the basis of therapeutic index: the first (for example, HER2, ALK, NTRK, BRAF, mutant-selective EGFR) with high therapeutic indices and consequently high response rates; and the second (for example, anti-PI3K, MEK, FLT3, mTOR) with low therapeutic indices and lower response rates, owing to either off-target inhibition of other closely related mutant proteins or on-target inhibition of the wild-type protein (which can cause marked side effects as a result of the critical role of the protein in normal homeostasis).
In the case of drugs with a high therapeutic index, combination therapy with non-overlapping mechanisms of action, or more potent derivatives, may deepen or prolong responses. For monoclonal antibodies that target receptor tyrosine kinases, there is no additional dose effect once full target saturation has been achieved. Consistent with this, delivering higher doses of individual anti-HER2 monoclonal antibodies in HER2+ breast cancer does not result in additional benefit110. Simultaneously, the safety of naked antibodies affords the opportunity to use full doses of combination agents. In HER2+ breast cancer, the combination of synergistic anti-HER2 antibodies trastuzumab and pertuzumab has resulted in a marked improvement in survival111. Whereas most high-therapeutic-index drugs have such a high index because the kinase target has a limited role in normal biological function, other agents with a range of therapeutic indices can achieve efficacy because of their selectivity for mutant over wild-type kinases. We believe that this represents a particularly promising approach to drug development and may even allow targeting of previously intractable alterations such as KRAS(G12C)56.
For small-molecule inhibitors with a high therapeutic index, multiple agents of differing potency or robustness to resistance mechanisms are available, and upfront application of the best inhibitor or combination appears to result in improved outcomes compared to sequential use. For example, treatment with the more-potent EGFR-mutant-selective inhibitor osimertinib, or the ALK inhibitor alectinib, prolongs progression-free survival in NSCLC compared to first-generation EGFR or ALK inhibitors, respectively112,113. In chronic myeloid leukaemia, dual targeting of ABL by a combination of catalytic and allosteric inhibitors drives a potent response that is sustained even when treatment is discontinued114. Collectively, these studies demonstrate that using more-potent targeted therapies as a first-line treatment, which maintain activity in the presence of acquired resistant mutations or prevent their appearance altogether, may prolong survival compared to introducing these agents at the time of initial resistance. For example, patients with germline BRCA-mutant ovarian cancer who show an apparent partial or complete response after platinum-based chemotherapy can be driven into an even deeper response with two years of maintenance olaparib115. The resulting efficacy data, which demonstrate a remission rate of 60% at three years after the start of PARP inhibitor therapy, suggest that this approach is increasing the fraction of patients that are cured of their disease. By contrast, PARP maintenance therapy after the first relapse of ovarian cancer in an similar patient population prolongs progression-free survival but is not curative116. Together, these data suggest that early, aggressive interventions using the most-potent agents (or combination of agents) available may ultimately have more-dramatic effects on long-term patient outcome than sequentially pretreating patients with different drugs.
On the other hand, drugs with a low therapeutic index can be efficacious as monotherapy, but with substantial side effects. PI3K inhibitors yield low response rates as monotherapy, but when used in combination with anti-endocrine therapy in ER+ PIK3CA-mutant breast cancer they improve progression-free survival117. However, this comes at the cost of on-target side effects from insulin inhibition, including hyperglycaemia. Improving the depth of response to drugs with a low therapeutic index will require new approaches; for example, combining PI3K inhibitors with a ketogenic diet that blunts adaptive exogenous insulin signalling and resensitizes cells to PI3K inhibitors118. A unique example of an agent with a low therapeutic index is the BCL2 inhibitor venetoclax, which lowers the apoptotic threshold of cells and produces very deep responses in chronic lymphocytic leukaemia119. However, venetoclax can also lead to dramatic tumour lysis syndrome in patients with a high burden of disease before the onset of therapy; this on-target effect can be successfully managed with careful dose escalation.
Quantifying the depth of response to surgery or systemic therapy with ctDNA may provide opportunities to identify high-risk groups for additional therapeutic intervention, and predictive biomarkers in systemic therapy. After primary breast surgery, detection of minimal residual disease by ctDNA was possible eight months before detection by imaging, creating a lead time in which additional therapies could be tested to improve the depth of response120. The detection rate of minimal residual disease after surgery is improved in early-stage NSCLC by tracking five mutations by ctDNA rather than one121. In metastatic breast cancer122 and BRAF-mutant colorectal cancer123, decreased levels of ctDNA correlate better with therapeutic response than standard tumour markers. For immunotherapies, global ctDNA measurements—such as a high tumour mutational burden in the plasma—may correlate with response to checkpoint blockade124.
Monitoring of response and adaptive interventions
If we assume that our approach to cancer therapy is at best empirical, real-time monitoring of response to therapy would enable earlier modifications in dose, schedules and therapeutic regimens (Fig. 2b). Unfortunately, the conventional way of assessing response is based on measuring changes in tumour diameter using serial radiographic imaging. More recently, functional imaging during therapeutic intervention has enabled a faster and more-precise assessment of response in some tumour types. For example, using positron emission tomography–computed tomography (PET–CT) scan-based functional imaging to assess a patient’s interim response to ABVD chemotherapy (that is, a combination of adriamycin, bleomycin, vinblastine and dacarbazine) is now standard in Hodgkin’s lymphoma, after which treatment can be escalated or de-escalated accordingly125.
Acquired resistance is often ascertained by a single biopsy; however, the heterogeneity of resistance can manifest as multiple resistant subclones in multiple metastases74,126. In addition to functional-imaging techniques, there is growing evidence that ctDNA-based methods can be a valuable tool for monitoring tumour heterogeneity, evolution and response to therapy. As with HIV, for which viral load is used to monitor response and adherence to therapy, ‘tumour load’ (as measured by ctDNA) could be used in a similar fashion (Fig. 2b). Real-time monitoring by ctDNA may also provide a time window in which to identify patients who may relapse127–129, as well as providing information on clonal evolution under therapy and thereby helping to dictate the next therapy at an earlier time point, especially when a dominant resistance mutation is likely to arise130,131. Techniques including the barcoding of endogenous DNA132 may enable tracking of intratumoral heterogeneity. In EGFR-mutant NSCLC, detection of the most common resistance mutation, EGFRT790M, through ctDNA is now a standard of care for patients whose disease progresses after treatment with first-generation EGFR inhibitors. Moreover, detection of resistance mutations by ctDNA can enable therapy with second-line EGFR agents to begin before clinical resistance manifests (for example, in patients who received first-line erlotinib)—potentially further prolonging survival133. Early ctDNA detection of mutant KRAS alleles that arise in metastatic colorectal cancers as a result of acquired resistance to anti-EGFR antibody therapies potentially provides a therapeutic imperative to stop anti-EGFR treatments134.
At present, a more daunting challenge than the emergence of acquired resistance mechanisms is how to address responses that are initially adaptive but that result in outright resistance or short-lived clinical benefit. Robustness, a property that allows a system to function despite external and internal perturbations, is a defining feature of cancer. It is therefore not a surprise that activation of compensatory pathways is a prevalent mechanism of resistance to targeted therapies64. These compensatory adaptation processes are rapid and either result in reactivation of the targeted pathway itself (either upstream or downstream) or engage signalling nodes that bypass the oncogenic pathway. Although such compensatory processes are a sobering reality when attempting to target oncogenic drivers, we propose that identifying the ‘drivers’ that underlie the feedback mechanisms in each case could be therapeutically exploited.
Some oncogene targets—such as BCR–ABL in chronic myeloid leukaemia, gene fusions in members of the NTRK family in a variety of tumour types, or mutant EGFR in lung cancer—do not result in an adaptive response that is clinically meaningful. Classically, these tumours respond to their respective targeted therapies for prolonged periods of time until mechanisms of acquired resistance eventually take over. On the other side of the spectrum, adaptive responses are the norm when components of key cellular pathways such as the HER2–PI3K–AKT–mTOR and RAS–RAF–MEK–ERK pathways are targeted135–137. Similarly, targeting ER in breast cancer and AR in prostate cancer induces rich cellular responses that reduce the effects of internal or external perturbations. If we consider as a whole these targets that elicit powerful adaptive responses, they share some obvious commonalities: in particular, they belong to cellular pathways that are critical for both normal and cancer cells and that are highly regulated by complex feedback processes.
To some extent, these adaptive cancer dependencies can be identified through experimental testing. For example, genes that confer resistance to inhibition of the MAP kinase pathway in BRAFV600E-mutant metastatic melanoma have been identified through gain-of-function genetic screens and genomic or transcriptomic sequencing of melanomas that are resistant to therapy. These large datasets of putative genes that confer drug resistance converge on ERK-dependent, ERK-independent and pathway-indifferent states that induce cellular pathway reactivation138–140. Similar examples have been observed in ER+ PIK3CA-mutant breast cancers, in which inhibition of PI3Kα elicits epigenetic reprogramming and increases ER transcription141,142.
Resistance to immunotherapy is necessarily different from resistance to targeted therapy. The direct targets of checkpoint blockade (PD-1 and CTLA-4) and their ligands are necessary to raise the threshold for activation of T cells and restrain the function of effector T cells, but are neither necessary nor sufficient for oncogenesis itself or response to checkpoint blockade. Moreover, antigen-presenting cells and T cells are genomically more stable relative to cancer cells. Thus, simple mutation or downregulation of PD-1, PD-L1 or CTLA-4 is not a common compensatory resistance mechanism; although mutation of PD-L1 would theoretically impair the recognition of checkpoint blockade antibodies, it would also impair the normal function of antigen-presenting cells and T cells. Rather, mechanisms that downregulate the presentation of major histocompatibility complex (MHC) proteins in cancer cells—such as mutation of B2M, JAK1 or JAK2—predominate among the known mechanisms of resistance to immunotherapy53. Adaptive responses that may hypothetically increase sensitivity to checkpoint blockade, such as DNA damage that is induced by chemotherapy, are under active clinical investigation. Serial multiplex testing of both the tumour and other cell types of the tumour microenvironment could enable the elucidation of adaptive responses to immunotherapy.
In addition to the type of experimental designs that we have described, adaptive responses can also be inferred from less-biased technologies that make few assumptions about the system, including network modelling. Logic-based models in which proteins are denoted as nodes can reconstruct cell signalling pathways143, and discrete dynamic models can be constructed that assess the node read-outs of inhibitory and activating states over time. The models can undergo computational simulations to determine the effects of an inhibitor and the relative fitness of different mechanisms of resistance in modulating apoptosis and proliferation. As an example, dynamic modelling of PI3K inhibition in ER+ breast cancer predicts that the combination of CDK4/6 inhibition together with PI3K inhibition can resensitize cells to PI3K inhibition144.
Another way to improve drug response would be to adapt the process of drug development. The traditional approach to developing drugs that target resistant mutations was to test the parent drug clinically, identify patients who develop resistance, molecularly characterize resistant tissue, model newly identified mutations preclinically and, finally, design new drugs for clinical testing. Unfortunately, this process takes many years, meaning that patients who experience a new first-in-class targeted therapy rarely live to receive a next-in-class agent. However, we are finally beginning to see examples of this bottleneck being overcome. Studies have shown that the selective TRK inhibitor larotrectinib is highly efficacious in an age- and tumour-agnostic manner in TRK-fusion-positive cancers145. Even before any acquired resistance mutations were detected in patients, preclinical and in silico modelling accurately predicted which mutations of the kinase domain of TRK would drive resistance to larotrectinib, and enabled the development of mutant-selective inhibitors. For those initial patients who received larotrectinib and developed acquired resistance, it was possible to move a next-generation TRK inhibitor into the clinic in a timeframe that enabled salvage of the therapeutic response146.
Mapping cancer dependencies
Some cancer dependencies have been discovered through classic methods of drug screening in cell lines—as in the case of the combination of ER and CDK4/6 antagonists in the treatment of ER+ breast cancer, which results in a synergistic control of disease compared to anti-ER therapy alone147. Exploring synthetic lethality, in which the concurrent perturbation of two genes leads to cell death, is another approach to identifying therapeutic vulnerabilities to a known driver mutation. The classical example of this phenomenon is PARP inhibition in BRCA-mutant tumours in which both DNA-repair genes are in parallel pathways148; however, this concept has now been expanded beyond tumour suppressors to discover new target genes that are synthetically lethal to selective oncogene inhibition or other specific drug therapy149.
Deciphering resistance to immunotherapy requires an understanding of the interplay between mechanisms that are intrinsic to cancer cells and extrinsic cellular and humoral factors, and these intrinsic and extrinsic components are likely to be different when comparing resistance to checkpoint blockade, adoptive cellular therapy, bispecific T cell engagers and cancer vaccines. Using small hairpin RNA (shRNA) to disrupt genes and performing high-throughput screening for loss of function can identify genes whose deletions cause resistance or enhance sensitivity to a specific driver or drug. This approach is limited, as gene knockdown may not be complete and off-target effects are common150. However, these problems can be overcome by CRISPR–Cas9 technology, which yields fewer off-target effects and increased efficiency given that the integration of a single-guide RNA can cleave both copies of a target DNA molecule151,152. One of the first genome-wide CRISPR-based knockout screens was used to identify potential mediators of resistance to RAF inhibition in BRAF-mutant melanoma that had not previously been defined by shRNA-based screens152. This has spurred a flurry of loss-of-function genome-wide CRISPR screens that have uncovered new genes associated with sensitivity and resistance153–155 to immune checkpoint blockade.
Integrating large clinico-genomic datasets with the outputs of synthetic lethality screening and computational modelling might one day facilitate a real-time reference guide of cancer dependencies to be produced for a diverse range of cancer types, with the ultimate goal of using predictive analytics to select combination drug candidates in a rational manner. For example, pan-cancer loss-of-function shRNA screens with deep coverage, analysed using subtraction methods to account for off-target effects, have revealed novel synthetic lethalities—including those that affect subunits in the SWI-SNF (switch/sucrose non-fermentable) chromatin remodelling complex, PRC2 repressor complex and Mediator complex156,157—which suggests that cancers driven by ‘undruggable’ targets may be targeted by drugging other enzymatic components within the same protein complex. Analysing these changes across therapy-naive and drug-resistant lines of cancer cells may reveal consensus dependencies that could eventually be exploited therapeutically.
In monogenic diseases such as chronic myeloid leukaemia, this level of analytics may not be required as the path to therapy is well defined. However, in most other cancers, which have a few dominant drivers but also a large number of known and unknown resistance mechanisms, the best treatments and orders of treatments to overcome drug resistance have not been established. In these cases, uncovering highly efficient combinations of therapies through a number of strategies including simultaneous co-targeting of multiple truncal alterations; targeting histology-specific synthetic lethal vulnerabilities; or identifying higher-order synthetic lethal interactions of perturbations that affect more than two genes may help to define new drug combination strategies158.
Conclusion
Resistance to therapy continues to be the biggest challenge in cancer today. There are as many underlying mechanisms of resistance as there are patients with cancer, because each tumour has its own defining set of characteristics that dictates tumour progression and that can eventually lead to death. Solving the resistance problem would therefore seem to be an unattainable goal. Here, we have proposed the creation of a framework that dissects and partitions resistance into its biological determinants. This enables the different problems to be tackled as separate working units at first, and then considered in a global manner.
Combining an assessment of the physical properties of the tumour (tumour burden, growth rate and localization) together with a deep analysis of tumour drivers and druggability, dependencies and vulnerabilities, early detection and precise monitoring and, finally, powerful analytics based on patient databases will no doubt be a challenging but formative approach to fighting the resistance of cancer to therapy. This combined knowledge should enable the development of tools that are capable of informing real-time clinical decisions, and which continuously improve through a feedback process that is reiterated by observed phenotypic outcomes. We recognize that many of the solutions proposed here would be costly at first, and further tax those systems of healthcare that are already strained159. These are important issues, and addressing them will require fundamental changes to economic models of oncology care.
Despite the caveats, it does finally seem possible—at least conceptually—to outline a roadmap for tackling the problem of resistance by using our understanding of its biological building blocks. Although we still need to fully understand key tumour drivers and their regulation (such as epigenetic and metabolic factors), cancer resistance has increasingly begun to resemble an engineering problem. We envision an amalgamation of engineering and cancer biology that will advance the field in unprecedented ways. The last example of a leading cause of death being eradicated by medicine was in the field of infectious diseases. As a next progressive step in the history of medicine, perhaps it is a reachable goal within our lifetimes to deliver cancer’s coup de grâce.
Acknowledgements
We thank K. Beal, K. Bolton, L. Diaz, A. Levine, L. Norton, J. Regan, D. Tsui and J. Wolchok for their critical reading of this manuscript, and S. Johnson for assistance with medical graphics. We acknowledge support from T32 CA009207 (N.V.), R01 CA207244 (D.M.H.), P30 CA008748 (D.M.H.) and R01 CA190642-01A1 (J.B.). We recognize that we could not cite many papers on this subject owing to space and editorial limits.
Footnotes
Competing interests N.V. reports advisory board activities for Novartis and consulting activities for Petra Pharmaceuticals. D.M.H. reports receipt of personal fees from Atara Biotherapeutics, Chugai Pharma, CytomX Therapeutics, Boehringer Ingelheim and AstraZeneca, and research funding from Puma Biotechnology, AstraZeneca, Loxo Oncology and Kinnate/Fount Therapeutics. J.B. is an employee of AstraZeneca. He is on the Board of Directors of Foghorn and is a past board member of Varian Medical Systems, Bristol-Myers Squibb, Grail, Aura Biosciences and Infinity Pharmaceuticals. He has performed consulting and/or advisory work for Grail, PMV Pharma, ApoGen, Juno, Lilly, Seragon, Novartis and Northern Biologics. He has stock or other ownership interests in PMV Pharma, Grail, Juno, Varian, Foghorn, Aura, Infinity Pharmaceuticals and ApoGen, as well as Tango and Venthera, for which he is a co-founder. He has previously received honoraria or travel expenses from Roche, Novartis and Eli Lilly.
Peer review information Nature thanks Robert Kerbel, Arjun Raj and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Goodman LS et al. Nitrogen mustard therapy; use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. J. Am. Med. Assoc 132, 126–132 (1946). [DOI] [PubMed] [Google Scholar]; This study and the next one (reference 2) were the first clinical reports of chemotherapy in advanced cancers.
- 2.Farber S & Diamond LK Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med 238, 787–793 (1948). [DOI] [PubMed] [Google Scholar]
- 3.Crofton J Chemotherapy of pulmonary tuberculosis. BMJ 1, 1610–1614 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeVita VT Jr et al. Curability of advanced Hodgkin’s disease with chemotherapy. Long-term follow-up of MOPP-treated patients at the National Cancer Institute. Ann. Intern. Med 92, 587–595 (1980). [DOI] [PubMed] [Google Scholar]; This study and the next one (reference 5) were the first clinical trials of combination chemotherapy, in advanced Hodgkin’s lymphoma and localized breast cancer, respectively.
- 5.Bonadonna G et al. Combination chemotherapy as an adjuvant treatment in operable breast cancer. N. Engl. J. Med 294, 405–410 (1976). [DOI] [PubMed] [Google Scholar]
- 6.Bosl GJ et al. VAB-6: an effective chemotherapy regimen for patients with germ-cell tumors. J. Clin. Oncol 4, 1493–1499 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Hryniuk W & Bush H The importance of dose intensity in chemotherapy of metastatic breast cancer. J. Clin. Oncol 2, 1281–1288 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Citron ML et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J. Clin. Oncol 21, 1431–1439 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Sternberg CN et al. Randomized phase III trial of high-dose-intensity methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) chemotherapy and recombinant human granulocyte colony-stimulating factor versus classic MVAC in advanced urothelial tract tumors: European Organization for Research and Treatment of Cancer protocol no. 30924. J. Clin. Oncol 19, 2638–2646 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D & Weinberg RA The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Leach DR, Krummel MF & Allison JP Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Iwai Y et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl Acad. Sci. USA 99, 12293–12297 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribas A & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glickman MS & Sawyers CL Converting cancer therapies into cures: lessons from infectious diseases. Cell 148, 1089–1098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldie JH & Coldman AJ The genetic origin of drug resistance in neoplasms: implications for systemic therapy. Cancer Res. 44, 3643–3653 (1984). [PubMed] [Google Scholar]
- 17.Fisher B, Slack NH & Bross ID Cancer of the breast: size of neoplasm and prognosis. Cancer 24, 1071–1080 (1969). [DOI] [PubMed] [Google Scholar]
- 18.Skipper HE, Schabel FM Jr & Wilcox WS Experimental evaluation of potential anticancer agents. XIII. On the criteria and kinetics associated with “curability” of experimental leukemia. Cancer Chemother. Rep 35, 1–111 (1964). [PubMed] [Google Scholar]
- 19.Goldie JH & Coldman AJ A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat. Rep 63, 1727–1733 (1979). [PubMed] [Google Scholar]
- 20.Luria SE & Delbrück M Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tannock IF Cancer: Resistance through repopulation. Nature 517, 152–153 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Norton L, Simon R, Brereton HD & Bogden AE Predicting the course of Gompertzian growth. Nature 264, 542–545 (1976). [DOI] [PubMed] [Google Scholar]; This study, references 23 and 24 are among the first applications of mathematical models of Gompertzian growth kinetics to tumour growth and response to therapy.
- 23.Laird AK Dynamics of tumor growth. ANL-6723. ANL Rep 1963, 216–222 (1963). [PubMed] [Google Scholar]
- 24.Norton L & Simon R Growth curve of an experimental solid tumor following radiotherapy. J. Natl Cancer Inst 58, 1735–1741 (1977). [DOI] [PubMed] [Google Scholar]
- 25.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Increasing the dose intensity of chemotherapy by more frequent administration or sequential scheduling: a patient-level meta-analysis of 37 298 women with early breast cancer in 26 randomised trials. Lancet 393, 1440–1452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This meta-analysis of over 37,000 patients from 26 randomized clinical trials of early-stage breast cancer shows that dose-dense chemotherapy increases overall survival compared to chemotherapy of standard dose intensity.
- 26.Katsumata N et al. Long-term results of dose-dense paclitaxel and carboplatin versus conventional paclitaxel and carboplatin for treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer (JGOG 3016): a randomised, controlled, open-label trial. Lancet Oncol. 14, 1020–1026 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Nowell PC The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 28.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study analysed about 5 million mutations from around 7,000 cancers to derive 21 distinct mutational signatures, including signatures of APOBEC enzymes and kataegis.
- 29.Lengauer C, Kinzler KW & Vogelstein B Genetic instabilities in human cancers. Nature 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Stephens PJ et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sansregret L, Vanhaesebroeck B & Swanton C Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol 15, 139–150 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Greaves M Evolutionary determinants of cancer. Cancer Discov. 5, 806–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGranahan N & Swanton C Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Johnson BE et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coombs CC et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap TA, Gerlinger M, Futreal PA, Pusztai L & Swanton C Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med 4, 127ps10 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Schiavon G et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med 7, 313ra182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGranahan N et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med 7, 283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain RK Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science 307, 58–62 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Minchinton AI & Tannock IF Drug penetration in solid tumours. Nat. Rev. Cancer 6, 583–592 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Makker V et al. Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer: an interim analysis of a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 20, 711–718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rini BI et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med 380, 1116–1127 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Andrews DW et al. Whole brain radiation therapy with or without stereotactic radiosurgery boost for patients with one to three brain metastases: phase III results of the RTOG 9508 randomised trial. Lancet 363, 1665–1672 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Valiente M et al. The evolving landscape of brain metastasis. Trends Cancer 4, 176–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson ES et al. Melanoma brain metastases treated with stereotactic radiosurgery and concurrent pembrolizumab display marked regression; efficacy and safety of combined treatment. J. Immunother. Cancer 5, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Q et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valiente M et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boire A et al. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell 168, 1101–1113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaretsky JM et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study was one of the first to discover mechanisms of acquired resistance to checkpoint blockade in metastatic melanoma, including JAK mutations and loss of β2-microglobulin.
- 54.De Henau O et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature 539, 443–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soucek L et al. Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ostrem JM, Peters U, Sos ML, Wells JA & Shokat KM K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lambert JM et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Yao Z et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28, 370–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan RJ et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase I dose-escalation and expansion study. Cancer Discov. 8, 184–195 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Karapetis CS et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med 359, 1757–1765 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Glen CD & Dubrova YE Exposure to anticancer drugs can result in transgenerational genomic instability in mice. Proc. Natl Acad. Sci. USA 109, 2984–2988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaked Y Balancing efficacy of and host immune responses to cancer therapy: the yin and yang effects. Nat. Rev. Clin. Oncol 13, 611–626 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Lito P, Rosen N & Solit DB Tumor adaptation and resistance to RAF inhibitors. Nat. Med 19, 1401–1409 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Sun C & Bernards R Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem. Sci 39, 465–474 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Prahallad A et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Corcoran RB et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2, 227–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azam M, Seeliger MA, Gray NS, Kuriyan J & Daley GQ Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol 15, 1109–1118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, Yang PL & Gray NS Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cortes JE et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N. Engl. J. Med 369, 1783–1796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw AT et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 17, 234–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mok TS et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med 376, 629–640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toy W et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet 45, 1439–1445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engelman JA et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Juric D et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patch AM et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Mu P et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oser MG, Niederst MJ, Sequist LV & Engelman JA Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blackburn EH Cancer interception. Cancer Prev. Res 4, 787–792 (2011). [DOI] [PubMed] [Google Scholar]; This paper, to our knowledge, was the first to codify the concept of cancer interception.
- 79.Shieh Y et al. Population-based screening for cancer: hope and hype. Nat. Rev. Clin. Oncol 13, 550–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Myers ER et al. Benefits and harms of breast cancer screening: a systematic review. J. Am. Med. Assoc 314, 1615–1634 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Elmore JG et al. Ten-year risk of false positive screening mammograms and clinical breast examinations. N. Engl. J. Med 338, 1089–1096 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Martin RM et al. Effect of a low-intensity PSA-based screening intervention on prostate cancer mortality: the CAP randomized clinical trial. J. Am. Med. Assoc 319, 883–895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lo YM et al. Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997). [DOI] [PubMed] [Google Scholar]
- 84.Lo YM et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med 2, 61ra91 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Leon SA, Shapiro B, Sklaroff DM & Yaros MJ Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650 (1977). [PubMed] [Google Scholar]; This clinical report was the first, to our knowledge, to detect circulating DNA in the serum of patients with cancer and show that the levels of DNA correlate with therapeutic intervention.
- 86.Wan JCM et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Stroun M, Anker P, Lyautey J, Lederrey C & Maurice PA Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol 23, 707–712 (1987). [DOI] [PubMed] [Google Scholar]
- 88.Chen XQ et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat. Med 2, 1033–1035 (1996). [DOI] [PubMed] [Google Scholar]
- 89.Nawroz H, Koch W, Anker P, Stroun M & Sidransky D Microsatellite alterations in serum DNA of head and neck cancer patients. Nat. Med 2, 1035–1037 (1996). [DOI] [PubMed] [Google Scholar]
- 90.Vogelstein B & Kinzler KW Digital PCR. Proc. Natl Acad. Sci. USA 96, 9236–9241 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dressman D, Yan H, Traverso G, Kinzler KW & Vogelstein B Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newman AM et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kinde I, Wu J, Papadopoulos N, Kinzler KW & Vogelstein B Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siravegna G, Marsoni S, Siena S & Bardelli A Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol 14, 531–548 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Abbosh C et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen JD et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein EA et al. Development of a comprehensive cell-free DNA (cfDNA) assay for early detection of multiple tumor types: the Circulating Cell-free Genome Atlas (CCGA) study. J. Clin. Oncol 36, 12021 (2018). [Google Scholar]
- 98.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang AZ, Langer R & Farokhzad OC Nanoparticle delivery of cancer drugs. Annu. Rev. Med 63, 185–198 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Lancet JE et al. CPX-351 (cytarabine and daunorubicin) liposome for injection versus vonventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J. Clin. Oncol 36, 2684–2692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thomas ED et al. Marrow transplantation for acute nonlymphoblastic leukemia in first remission. N. Engl. J. Med 301, 597–599 (1979). [DOI] [PubMed] [Google Scholar]
- 102.Peters WP et al. Prospective, randomized comparison of high-dose chemotherapy with stem-cell support versus intermediate-dose chemotherapy after surgery and adjuvant chemotherapy in women with high-risk primary breast cancer: a report of CALGB 9082, SWOG 9114, and NCIC MA-13. J. Clin. Oncol 23, 2191–2200 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Jonas O et al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci. Transl. Med 7, 284ra57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kemeny NE & Gonen M Hepatic arterial infusion after liver resection. N. Engl. J. Med 352, 734–735 (2005). [DOI] [PubMed] [Google Scholar]
- 105.Beck A, Goetsch L, Dumontet C & Corvaïa N Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov 16, 315–337 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Modi S et al. Trastuzumab deruxtecan (DS-8201a) in subjects with HER2-low expressing breast cancer: updated results of a large phase 1 study. Cancer Res. 79, abstr. P6–17–02 (2019). [Google Scholar]
- 107.Robert C et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 108.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hellmann MD et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med 378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vogel CL et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol 20, 719–726 (2002). [DOI] [PubMed] [Google Scholar]
- 111.Baselga J et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med 366, 109–119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Soria JC et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med 378, 113–125 (2018). [DOI] [PubMed] [Google Scholar]; This clinical trial and the next one (reference 113) both demonstrated the superiority of more-potent and mutant-specific TKIs as a first-line treatment compared to first-generation TKIs in EGFR-mutated and ALK-translocated NSCLC.
- 113.Peters S et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med 377, 829–838 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Wylie AA et al. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543, 733–737 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Moore K et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med 379, 2495–2505 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Mirza MR et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med 375, 2154–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 117.André F et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Hopkins BD et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature 560, 499–503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roberts AW et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N. Engl. J. Med 374, 311–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garcia-Murillas I et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med 7, 302ra133 (2015). [DOI] [PubMed] [Google Scholar]
- 121.Abbosh C, Birkbak NJ & Swanton C Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol 15, 577–586 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Dawson SJ et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med 368, 1199–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 123.Corcoran RB et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAFV600-mutant colorectal cancer. J. Clin. Oncol 33, 4023–4031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rizvi NA et al. Durvalumab with or without tremelimumab vs platinum-based chemotherapy as first-line treatment for metastatic non-small cell lung cancer: MYSTIC. Ann. Oncol 29, abstr. LBA6 (2018). [Google Scholar]
- 125.Johnson P et al. Adapted treatment guided by interim PET-CT scan in advanced Hodgkin’s lymphoma. N. Engl. J. Med 374, 2419–2429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murtaza M et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun 6, 8760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Diehl F et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med 14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Murtaza M et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Forshew T et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med 4, 136ra68 (2012). [DOI] [PubMed] [Google Scholar]
- 130.Misale S et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuang Y et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin. Cancer Res 15, 2630–2636 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; This translational study showed the feasibility of using plasma ctDNA to detect EGFRT790M mutations in patients with EGFR-mutated NSCLC who progressed on gefitinib or erlotinib.
- 132.Guernet A et al. CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol. Cell 63, 526–538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu YL et al. Osimertinib vs platinum-pemetrexed for T790M-mutation positive advanced NSCLC (AURA3): plasma ctDNA analysis. J. Thorac. Oncol 12, S386 (2017). [Google Scholar]
- 134.Diaz LA Jr et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rodrik-Outmezguine VS et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 1, 248–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chandarlapaty S et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lito P et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Johannessen CM et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wagle N et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 4, 61–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van Allen EM et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bosch A et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci. Transl. Med 7, 283ra51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Toska E et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 355, 1324–1330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pe’er D & Hacohen N Principles and strategies for developing network models in cancer. Cell 144, 864–873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gómez Tejeda Zañudo J, Scaltriti M & Albert R A network modeling approach to elucidate drug resistance mechanisms and predict combinatorial drug treatments in breast cancer. Cancer Converg. 1, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drilon A et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med 378, 731–739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Drilon A et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 7, 963–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This clinical trial demonstrates the activity of LOXO-195, a next generation TRK inhibitor that targets solvent-front and kinase-domain TRK mutants, and shows the feasibility of designing next-generation inhibitors in real time to combat acquired resistance.
- 147.Finn RS et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med 375, 1925–1936 (2016). [DOI] [PubMed] [Google Scholar]
- 148.Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]; This was the one of the first reports to exploit synthetic lethality in cancer through PARP inhibition in BRCA-mutant cells.
- 149.O’Neil NJ, Bailey ML & Hieter P Synthetic lethality and cancer. Nat. Rev. Genet 18, 613–623 (2017). [DOI] [PubMed] [Google Scholar]
- 150.Mohr SE, Smith JA, Shamu CE, Neumüller RA & Perrimon N RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol 15, 591–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang T, Wei JJ, Sabatini DM & Lander ES Genetic screens in human cells using the CRISPR–Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; To our knowledge, this and the next study (reference 152) are the first CRISPR–Cas9-based synthetic lethality screens reported that screened for resistance to chemotherapy and targeted therapy.
- 152.Shalem O et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Patel SJ et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Burr ML et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pan D et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McDonald ER III et al. Project DRIVE: a compendium of cancer dependencies and synthetic-lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592 (2017). [DOI] [PubMed] [Google Scholar]
- 157.Tsherniak A et al. Defining a cancer dependency map. Cell 170, 564–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ashworth A & Lord CJ Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol 15, 564–576 (2018). [DOI] [PubMed] [Google Scholar]
- 159.Tannock IF & Hickman JA Limits to personalized cancer medicine. N. Engl. J. Med 375, 1289–1294 (2016). [DOI] [PubMed] [Google Scholar]
- 160.Haque IS & Elemento O Challenges in using ctDNA to achieve early detection of cancer. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/237578v1 (2017). [Google Scholar]
- 161.Imperiale TF et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med 370, 1287–1297 (2014). [DOI] [PubMed] [Google Scholar]
- 162.Kinde I et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med 5, 167ra4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Underhill HR et al. Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lehmann-Werman R et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Snyder MW, Kircher M, Hill AJ, Daza RM & Shendure J Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bowman RL, Busque L & Levine RL Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chan KCA et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med 377, 513–522 (2017). [DOI] [PubMed] [Google Scholar]
- 168.Heitzer E et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 5, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Romanel A et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med 7, 312re10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


