Summary
Genetic factors underlying coronary artery disease (CAD) have been widely studied using genome-wide association studies (GWASs). However, the functional understanding of the CAD loci has been limited by the fact that a majority of GWAS variants are located within non-coding regions with no functional role. High cholesterol and dysregulation of the liver metabolism such as non-alcoholic fatty liver disease confer an increased risk of CAD. Here, we studied the function of non-coding single-nucleotide polymorphisms in CAD GWAS loci located within liver-specific enhancer elements by identifying their potential target genes using liver cis-eQTL analysis and promoter Capture Hi-C in HepG2 cells. Altogether, 734 target genes were identified of which 121 exhibited correlations to liver-related traits. To identify potentially causal regulatory SNPs, the allele-specific enhancer activity was analyzed by (1) sequence-based computational predictions, (2) quantification of allele-specific transcription factor binding, and (3) STARR-seq massively parallel reporter assay. Altogether, our analysis identified 1,277 unique SNPs that display allele-specific regulatory activity. Among these, susceptibility enhancers near important cholesterol homeostasis genes (APOB, APOC1, APOE, and LIPA) were identified, suggesting that altered gene regulatory activity could represent another way by which genetic variation regulates serum lipoprotein levels. Using CRISPR-based perturbation, we demonstrate how the deletion/activation of a single enhancer leads to changes in the expression of many target genes located in a shared chromatin interaction domain. Our integrative genomics approach represents a comprehensive effort in identifying putative causal regulatory regions and target genes that could predispose to clinical manifestation of CAD by affecting liver function.
Keywords: coronary artery disease, GWAS, SNP, liver, hepatocyte, cholesterol, enhancer, CRISPR, STARR-seq, functional genomics
Introduction
Coronary artery disease (CAD) and its most important complication, myocardial infarction (MI), results primarily from atherosclerosis, an inflammatory disease of the large arteries characterized by lipid-rich lesions. Genome-wide association studies (GWASs) have identified ∼200 risk loci for CAD/MI.1,2 However, these loci correspond to thousands of common single-nucleotide polymorphisms (SNPs) in high linkage disequilibrium (LD) of which any could be causal. Furthermore, 90% of SNP-based heritability of CAD/MI is explained by variants located in intronic and intergenic regions with no known function, which complicates the functional interpretation.3,4 To translate the GWAS findings into therapeutic potential, we need to understand which of the risk variants are functional and what genes they regulate. It has been shown that around 30% of CAD variants5 can be explained by their association with traditional risk factors, including hypertension, obesity, diabetes, metabolic syndrome, dyslipidemia, insulin resistance, and non-alcoholic fatty liver disease (NAFLD), highlighting the key role of adipose tissue and the liver. Especially, NAFLD has been shown to promote both hyperglycemia and dyslipidemia that increases the risk of cardiovascular disease.6 However, little is known about the target genes and regulatory mechanisms by which these risk variants act.
Emerging evidence suggests that causal disease variants affecting gene expression are enriched in the enhancers of disease-relevant cell types.7 However, only a few such examples have been described for cardiovascular disease-associated variants, including SORT1,8 PHACTR1,9 ANRIL,10 LMOD1,11 and SMAD3,12 highlighting the pressing need for further studies. Enhancers act by looping with their target promoters where they bring in additional transcription factors and coactivators. Therefore, long-range interactions between regulatory elements and gene promoters play key roles in transcriptional regulation. Interacting regions are enriched with genetic variants linked to the altered expression of the genes they contact,13,14 which also highlights their important role in disease. Variants within enhancers have been shown to affect the binding of transcription factors (TFs),15 as well as the chromatin state16,17 and chromatin interactions.10
Here, we focused on the characterization of the GWAS CAD/MI risk loci that could act through the liver and in particular via hepatocyte-specific enhancers. First, we performed a promoter Capture Hi-C experiment in HepG2 cells to identify target genes for the risk SNPs that were further extended by a cis-eQTL analysis. Second, we used ChIP-seq intensity variation, DeepSEA computational predictions, and a massively parallel reporter assay to investigate the allele-specific enhancer activity and infer the causal regulatory SNPs. Finally, we characterized selected enhancer SNPs located in 3D regulatory hubs to demonstrate a functional link between enhancers and their target genes. Overall, we aim to provide deeper understanding of the genetic basis of CAD acting through the liver tissue.
Subjects and methods
Identification of susceptibility enhancers
GWAS lead SNPs associated with CAD and MI were obtained from the GWAS catalog18 by using the following terms: coronary artery disease, coronary heart disease, and myocardial infarction (Table S1). This corresponded to 262 cytogenic regions. As majority of the GWAS lead SNPs came from studies that were based on European ancestry, the co-inherited, proximal SNPs (dbSNP version 146) in tight LD (r2 > 0.80) with the GWAS lead SNPs (Table S2) were determined using the 1000 Genomes European samples phase 3, version 5a using PLINK v.1.90b5.3 with the following essential settings: ‘--extract <dbSNP v146 rsIDs>, --keep <EUR sample IDs>, --maf 0.01, --r2, --ld-snp-list <GWAS lead SNP rsIDs>, --ld-window 100000, --ld-window-kb 1000, --ld-window-r2 0.8’.19 Only SNPs were analyzed and other types of structural DNA variants were excluded. To study which of the susceptibility SNPs fall within with hepatocyte-specific enhancer marks, we checked for overlap of the enhancer peaks downloaded from ENCODE20 and Roadmap Epigenomics Mapping Consortium21 including GSM646355-6 (HepG2 H3K27ac), GSM646356 (HepG2 H3K4me1), and GSM1112808-9 (Liver H3K27ac) using the HOMER v4.1022 command ‘mergePeaks’ -cobound.
Prediction of pathogenic exonic SNPs using SIFT and PolyPhen-2
We used SIFT23 and PolyPhen-224 tools to predict deleterious effects of exonic SNPs as described below. Importantly, the CAD/MI risk loci that harbored exonic SNPs were included in the downstream analysis as we cannot rule out that both the coding and the noncoding SNPs could have a biological role. SIFT is a sequence homology-based tool that predicts whether an amino acid substitution in a protein will have a phenotypic effect. SIFT correlates protein function with evolution. We submitted the query in the form of SNP IDs. SIFT takes a query SNPs and uses multiple alignment information to predict tolerated and deleterious substitutions for every position of the query sequence. SIFT obtains alignment for SNP location with similar sequences that could share similar function to the query and normalized probabilities are calculated from the alignment. Normalized probabilities < 0.05 is considered to have deleterious effect while SNPs with value > 0.05 has tolerating effect. For a SNP, PolyPhen-2 calculates Naive Bayes posterior probability that a SNP is damaging, and reports estimates of false positive rate and true positive rate. A SNP is also evaluated as benign, possibly damaging, or probably damaging based on false positive rate (FPR) thresholds.
UK biobank GWAS association
The GWAS imputed V3 association files (Table S3) for cardiometabolic traits files were downloaded from nealelab including body mass index (BMI), basal metabolic rate (BMR), blood pressure, cholesterol (quantile), diseases of liver, HDL cholesterol (quantile), LDL direct (quantile), nonalcoholic fatty liver disease, non-cancer illness code, type 2 diabetes, and triglycerides (quantile). As described in nealelab (see web resources), the GWAS was based on linear regression model on all phenotypes including both sexes. Association for all phenotypes used a least-squares linear model predicting the phenotype with an additive genotype coding (0, 1, or 2 copies of the minor allele), with sex and the first 10 principal components from the UK Biobank sample QC file as covariates. p value < 5 × 10−5 were considered significantly associated. This lenient cut-off value was used to discover all suggestive candidate associations of the previously identified CAD/MI GWAS SNPs with liver-related traits.
Cell lines and culture reagents
HepG2 cells (ATCC, HB-8065) were cultured in Dulbecco’s modified Eagle medium (DMEM; 4.5 g/L glucose, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin; LONZA) supplemented with 10% fetal bovine serum (FBS; GIBCO). The cells were maintained at 37°C in a humidified atmosphere at 5% CO2.
Promoter Capture Hi-C library preparation and data processing
The promoter Capture Hi-C assay was performed using two replicates of 10 million HepG2 cells, as described previously.25 The libraries were sequenced on the Illumina HiSeq 4000 platform. On average, 114.4 M paired-end reads were obtained per sample. The reads were processed using HiCUP v0.7.226 software with the default settings except that the insert size restrictions for the filtering step were set to 200–600 bp. The reads were aligned to the GRCh37/hg19 human reference genome using bowtie2 v.2.2.9. Significant interactions were identified using the Capture Hi-C Analysis of Genome Organization (CHiCAGO) software v.1.1.8. which takes into account that the background levels in PCHi-C decrease as the genomic distance between the bait and other end increases.27 We used the default threshold of 5 for significant interactions. Gene set enrichment analysis for promoters within the interacting ends was performed using Enrichr.28
Feature enrichment at interaction endpoints
To associate interactions with SNPs or histone marks, study feature enrichment at interaction endpoints and connect features with HepG2 promoter capture interactions the HOMER v.4.1022 command ‘annotateInteractions.pl’ was used. The program uses positional overlaps to assign interaction endpoints to given genomic locations. GWAS SNPs18 (v.1.0, 2019-09-24) associated with HDL, LDL, schizophrenia, multiple sclerosis, Crohn disease, metabolite levels, prostate cancer, lung cancer, rheumatoid arthritis, psoriasis, ankylosing spondylitis, bipolar disorder, Alzheimer disease, Parkinson disease, migraine, autism spectrum disorder, and celiac disease were included in the analysis. To study the enrichment of chromatin marks, public data for HepG2 cell line was collected from ENCODE20 and Roadmap Epigenomics Mapping Consortium21 GSE26320 (H3K27ac/me3, H3K4me1/2/3), GSM1003519 (H3K9me3), GSM816662 (DNaseHS), and human liver from GSM1112808-9 (H3K27ac).
Identification of target genes from PCHi-C
First, we used HepG2 PCHi-C data to identify target genes of the CAD/MI SNP carrying regulatory regions. Ideally, each looping in PCHi-C represents an interaction between promoter of a gene and its regulatory region. If a promoter of a gene was found to have a looping interaction with a CAD/MI SNP carrying regulatory region, then we considered the gene to be a target gene. This was achieved by intersecting the SNP coordinates with the non-promoter end of the looping region. If there was a match, the promoter end of the coordinates was called by the gene name. This analysis was accomplished using a custom python script.
KOBS study cohort
A subset of 263 participants, with the RNA-sequencing data available from an ongoing KOBS study29,30 (Kuopio Obesity Surgery Study which includes severely obese individuals undergoing bariatric surgery), were included in our analysis. All participants provided informed consent and the study protocol was approved by the local ethics committee. Plasma glucose, insulin, and serum lipids and lipoproteins (total cholesterol, HDL cholesterol, and triglycerides) and free fatty acids (FFAs) were measured after fasting.
RNA-seq library preparation and mapping (KOBS cohort)
The KOBS RNA samples were isolated at the University of Eastern Finland using the miRNeasy (QIAGEN) kit according to the manufacturer’s protocol and subsequently sequenced at the UCLA sequencing core. The stranded RNA-seq libraries were prepared using Ribo-Zero gold. The RNA-seq libraries were sequenced as 50-bp paired-end reads on an Illumina HiSeq 2500 platform. On average 41.5 million uniquely mapped reads were obtained per sample.
The STAR31 2-pass method was used to align the reads to the GRCh38 reference genome (release 29). To remove lowly expressed genes, a gene had to have >10 reads in 80% of the samples. The Rsubread R package32 was used to count all the reads mapped within exon features. The gene-level quantification was estimated as the sums of the read counts and the TPM of all the transcripts of a given gene. We generated data quality statistics with Picard (see web resources). Hidden covariates were determined with the principal component analysis (PCA).
Expression quantitative trait loci (eQTL) analysis and overlap with CAD/MI GWAS SNPs
For the cis-eQTL analysis, we first carried out a surrogate variable analysis (SVA)33 on the KOBS gene expression data to identify latent factors representing unmeasured batch effects in RNA-seq. Then, KOBS liver gene expression TPM was adjusted for SVA factors, RNA integrity value (RIN), alignment rate, percent of mitochondria reads, 3' bias, BMI, age, and sex, as well as genetic principle components 1 and 2. We estimated the minor allele frequency (MAF) of SNP variants using VCFtools34 and only included SNPs with MAF > 5% in the cis-eQTL analysis. cis-eQTLs were identified with linear regression implemented in the R package Matrix eQTL v.2.1.1.35 The cis window was defined as 1 Mbp upstream or downstream of the TSS, and FDR of 5% was used to assess the significance. For this study, the list of significant SNP-eGene pairs were filtered for the CAD/MI GWAS (lead and proxy) SNPs described above. Together with the previously published liver cis-eQTL studies for the CAD/MI GWAS SNPs from GTEX v7,36 STARNET,37 and other publications38, 39, 40 (Table S4), a total of 138 eGenes were identified.
Gene association with non-alcoholic steatohepatitis (NASH) in KOBS
We used the edgeR’s41 negative binomial generalized linear model with quasi-likelihood F-test to test for differential expression, controlling for technical and 23 factors influencing gene expression identified with PCA, with selected gene expression levels as dependent and liver phenotypes (normal liver, simple steatosis, and non-alcoholic steatohepatitis) as independent variables. The following covariates were included in the analysis: uniquely aligned reads % and 3' bias, body mass index, sex, and age. The quantitative traits used in the correlation analysis (Pearson’s and partial correlation) were inverse normal transformed to avoid outlier effects.
HMDP study cohort
The Hybrid Mouse Diversity Panel (HMDP) is a resource for systems genetics studies consisting of about 100 diverse inbred strains of mice.42 The transcriptomic and clinical trait data examined here were generated as previously described.43
Visualization of KOBS and HMDP data gene-trait associations
The gene-trait associations as well as NASH differentially expressed (DE) genes data are presented as Circos (circular plots) using Circlize.44 circos.initializeWithIdeogram() function was used to create a track with chromosomal ideogram with hg19 annotation. The outer circle reflects the human chromosomes. circos.genomicLabels() function was used to add gene labels that are significantly associated with traits while circos.track() function was used to generate tracks for each trait. The inner circles in color dots reflects the genes that are nominally significant genes and the height of the dots represents the level of significance.
Processing of human liver single-cell RNA data
Human liver single-cell RNA-seq data, published by MacParland et al.,45 generated from fresh hepatic tissue obtained from five individuals during the transplantation surgery, was used to study the cell type specificity of the 714 candidate genes. The dataset consists of 8,444 parenchymal and non-parenchymal cells annotated by the original authors into 20 distinct cell populations including plasma cells, NK-like cells, B cells, cholangiocytes, erythroid cells, hepatic stellate cells, and multiple types of hepatocytes, T cells, endothelial cells, and intrahepatic macrophages. The UMI count data and cell type annotations were imported into the Seurat (v.3.1)46 software package and scaled to 10,000 counts per cell. The average expression for each gene was calculated in each cell type cluster as the number of counts per million. Genes with more than 1 count per million in at least one cell type were retained (492 genes). The cell type average expression levels were plotted on a gene-wise z-scored heatmap and the genes were clustered according to the Euclidean distance. The cell-type-specific genes (average TPM of hepatocyte clusters, endothelial cell clusters, macrophage clusters, and T/B cell clusters) were defined by TPM > 10 and a minimum fold change of > 2 in one cell type and < 0.5 in the other three cell types to be called cell-type-specific gene.
HepG2 IL1B treatment, ChIP-seq library preparation, and sequencing
HepG2 cells were stimulated with 10 ng/mL of IL-1β (PHC0814, GIBCO) at 2 h, 8 h, and 23 h time points. The ChIP-seq process was performed as previously described47 using an H3K27ac antibody (ab4729, Abcam). The data were mapped using the Bowtie software package allowing up to two mismatches and reporting only one alignment for each read. Poor quality reads were filtered out (minimum 97% of bp over a quality cutoff of 10). The peak calling was performed using HOMER 4.3,22 command ‘findPeaks’ and the settings for ‘-style histone’. The peaks were annotated to hg19 genes using the command ‘annotatePeaks’ and the settings ‘-log -size given -strand both’. For quantification, the tags from the two biological replicates were pooled. Average of 2 h, 8 h, and 23 h time points were calculated and H3K27ac peaks exhibiting more than 2-fold increase were considered inflammation-activated enhancers or promoters (H3K27ac peak ± 1 kb from an annotated promoter).
Identification of enhancer hubs
Enhancer hubs were defined as interacting domains composed of an enhancer-rich region connected through at least three interacting enhancers. Each end of the looping coordinates was taken as separate input files. Each line in these files were counted using sort | uniq -c command. The coordinates with less than three counts and trans looping coordinates (>1 MB) were removed from the analysis. Bedtools48 merge function was used to combine the selected overlapping interactions to constitute an enhancer hub. Further, looping coordinates were intersected with active H3K27ac ChIP-seq regions from untreated HepG2 cells described above, to identify enhancer-rich hubs. Gene coordinates were downloaded from UCSC table browser with assembly Feb. 2009 (GRCh37/hg19). The gene promoter coordinates harbored in each hub region were identified using homer command ‘mergePeaks’ -cobound function. The fold change for H3K27ac signal was derived for each promoter using the HOMER 4.322 using the command ‘annotatePeaks’ and the settings ‘-log -size given -strand both’ as described above. The Datamash package (see web resources) was used for mean and median calculation of the fold change across each pairwise comparison (control versus 2 h/8 h/23 h IL1β treatment) for each hub promoter. We ranked all enhancer hub gene promoters in descending order based on the median fold change in H3K27ac and plotted the max, median, and min values of as a running average of H3K27ac fold-change (window size 50). To identify how much the hepatocyte-specific genes are enriched in the enhancer hubs, a gene enrichment analysis was performed using the hypergeometric t test (see web resources). The parameters for the hypergeometric t test are as follows. Number of successes (k = 36) was the hepatocyte-specific genes significantly differentially expressed under cytokine treatment (p value < 0.05), sample size (s = 64) was the total hepatocyte-specific genes, number of successes in the population (M = 9,648) represented all the genes that were significantly differently expressed under cytokine treatment (p value < 0.05) while population size (N = 28,797) was the total number of genes included in the analysis. To understand the enrichment of the CAD/MI SNPs in enhancer hubs and super-enhancers compared to other random regions, RegioneR49 software package was used. The program utilizes permutation tests to assess the association between genomic region sets.
Analysis of RNA-seq data from cytokine-treated HepG2 cells
RNA-seq data for HepG2 cell line treated with cytokines were downloaded from GSE10200650 (GSM2720393–GSM2720400, GSM2720545–GSM2720554). GEO2R,51 an interactive web tool that allows users to compare two or more groups of samples, was used to generate differentially expressed genes for RNA-seq data. Nominal p value < 0.05 was considered significant. Lists of 50 super-enhancers, enhancer hubs, and random regions were generated using the command shuf -n 50. The genomic coordinates where there were limited interactions (i.e., regions not included as a hub) was considered as a random region. The genes that are present in the selected 50 super-enhancer, enhancer hubs, and random regions were identified by using mergePeaks command with -cobound options using the list of regions and gene coordinates as input files. Gene-gene correlations under each super-enhancer, enhancer hub, and random regions for GSE102006 were determined using the DGCA52 R package.
HepG2 nuclear protein extraction and EMSA
HepG2 cells were collected (5 × 106) in PBS by scraping them from culture flasks and were washed twice with cold PBS. The cells were re-suspended in 500 μL 1× hypotonic buffer (20 mM Tris-HCl [pH 7.4], 10 mM NaCl, and 3 mM MgCl2) with a cOmplete Protease Inhibitor Cocktail (Roche, Merck) by pipetting up and down several times and were then incubated on ice for 15 min. 25 μL of detergent (10% NP40) was added and vortexed for 10 s at the highest setting. Centrifugation was done with the homogenate for 10 min at 3,000 rpm at 4°C. The nuclear pellet was re-suspended in 50 μL complete Cell Extraction Buffer (10 mM Tris [pH 7.4], 2 mM Na3VO4, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 1 mM EGTA, 0.1% SDS, 1 mM NaF, 0.5% deoxycholate, and 20 mM Na4P2O7) for 30 min on ice by vortexing it at 10 min intervals followed by sonication and centrifugation for 30 min at 14,000 × g at 4°C. The supernatant (nuclear fraction) was saved. Quantitation of the protein concentration was performed using the Quant-iT Protein Quantitation Kit (Thermo Fisher Scientific) and the aliquoted supernatant was stored at −80°C. HepG2 nuclear extract lysate was also purchased from Abcam (ab14660, Lot: GR3223114-8).
Oligonucleotide probes (15 bp flanking SNP site for reference or alternate allele) with a biotin tag at the 5' end of the sequence (Integrated DNA Technologies) were incubated with the HepG2 nuclear protein and a working reagent from the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Catalog #20148). For competitor assays, an unlabeled probe of the same sequence was added to the reaction mixture at 100× excess. The reaction was incubated for 30 min at room temperature, and then loaded on a 6% retardation gel (Invitrogen, Catalog #EC6365BOX). The contents of the gel were transferred to a nylon membrane, cross-linked by UV, and visualized using a UV trans illuminator by Image Lab Software (Bio-rad).
Computational prediction of allele-specific binding using DeepSEA
The DeepSEA53 tool was used to predict the chromatin effects of sequence alterations with single nucleotide sensitivity. For the CAD/MI SNPs, DeepSEA predictions were obtained using the online tool with the SNPs in VCF files provided as input. Functional significance score represented a measure of the significance of magnitude of predicted chromatin effect and evolutionary conservation and these scores for SNPs at the single-nucleotide resolution were obtained. The SNP with functional significance score < 0.01 were considered significant for allele specific binding.
Allele-specific binding analysis for HepG2 from ChIP-seq data
The BaalChIP54 tool was used for allele-specific measurements of transcription factor binding from the ChIP-seq data. A Bayesian statistical approach was used by the tool to correct for the effect of background allele frequency on the observed ChIP-seq read counts. We analyzed 210 TFs representing 580 samples obtained from ChIP-seq experiments in HepG2 cells by ENCODE20,55 (Table S5). The zygosity of the SNPs was identified using VCF file56 (dataset 1 and dataset 2 HepG2_phased_variants.vcf). The BaalChIP-analysis was corrected for altered karyotypes using HepG2 genomic DNA (gDNA) using ENCFF356NCL and ENCFF277OEP56 for the observed allelic imbalances and focused on the heterozygous loci (3,824 SNPs). The fold change was calculated by log2(Ref allele count/Alt allele count).
STARR-seq
A massively parallel reporter assay was performed for CAD/MI GWAS SNPs (lead+proxies) that overlapped the peaks extracted from the following data sources: GEO: GSE157306 (HepG2 H3K27ac inflammatory time course generated for this study), GEO: GSM646355-6 (HepG2 H3K27ac), GEO: GSM646356 (HepG2 H3K4me1), GEO: GSM1112808-9 (Liver H3K27ac), GEO: GSE98983 (HepG2 p65 ChIP-seq), GEO: GSM816662 (HepG2 DNaseHS), and GEO: GSM2400294 (Liver DnaseHS). In addition, TF binding peaks in HepG2 cells were downloaded from the ENCODE20 database under “Transcription Factor ChIP-seq Uniform Peaks from ENCODE/Analysis” and strong common DNase hypersensitive peaks assayed in a large collection of cell types were downloaded from source files found under “DNase I Hypersensitivity in 95 cell types” (hotspots) and “wgEncodeRegDnaseClusteredV3.bed.gz” (score equal or above 1,000). The overlap analysis was performed using the HOMER v.4.1022 command ‘mergePeaks’ -cobound.
The following computational pipeline was used to generate 198 bp sequences representing up to 5 haplotypes at each locus of interest (Table S6). HOMER-formatted peak files were generated using human genome reference coordinates in build hg19 demarcating the regions of interest for the STARR-seq library. Phased alleles within these regions were subset for European samples from the VCF files of the 1000 Genomes phase 3, version 5a using tabix with the options “--regions peak.file--print-header” using a custom R script. The HOMER program annotatePeaks.pl was used by inputting the peak file from step 1 along with the options “-vcf phased.vcf.file-size given” which output another HOMER-formatted peakfile with columns noting the bp positions within each peak and alleles of each haplotype. The sequence of the reference hg19 human genome was retrieved within each peak boundaries using the R package seqinr().57 A custom R script was then used to iterate through each peak to paste custom sequences together for each haplotype. Specifically, strings of non-polymorphic sequence were separated from polymorphic alleles using coordinates in the previous peak file, and then these were pasted together again for each haplotype. Resulting sequences were compared along each haplotype, and duplicated sequences were removed, which sometimes arose when peak sequences were identical between haplotypes.
The selected regions were cloned to hSTARR-seq_ORI plasmid (Addgene, #99296)58 backbone. 230-bp DNA inserts containing 198 base pairs of the SNP containing enhancer sequence, a 2-bp barcode at the 5′ end of the enhancer sequence, and 15-bp adapters at both ends matching the Illumina NGS sequencing primers were synthesized by Agilent. First round of emulsion PCR using Micellula DNA Emulsion & Purification Kit (Roboklon) was performed to complete the sequencing primers and to double-strand the oligos. The second round was used to amplify the material. The plasmid was linearized using AgeI and SalI restriction enzymes and inserts were cloned to the linearized plasmid in 17 reactions using the standard InFusion cloning (Clontech) protocol. The cloned DNA library was transformed to XL-10 gold ultra-competent bacteria (Agilent) in 15 reactions and the plasmid was purified using EndoFree Maxiprep kit (QIAGEN). The plasmid library was transfected following the manufacturer’s instructions in triplicates to 7 × 107 HepG2 cells using Lipofectamine-3000 transfection reagent (Invitrogen). Cells were harvested 24 h post-transfection and the total RNA was extracted using RNeasy midi kit (QIAGEN). Messenger-RNA was purified from the bulk RNA using Dynabeads Oligo(dT)25 beads (Invitrogen) with 2:1 beads to total RNA volume ratio. The purified mRNA was treated with Turbo DNaseI (Ambion) and purified using RNeasy MinElute clean up kit (QIAGEN). Reverse transcription was performed using UMI-primers. Unique molecular identifiers (UMIs) were added during cDNA synthesis to tag identifiable replicates of the constructs, which improves the data analysis by accounting for PCR duplicates.59 The samples were pooled and RNase A treated and cDNA was purified with AMPure XP beads using a 1.8:1 beads to cDNA ratio. The libraries were amplified using junction PCR. The junction PCR for the RNA library was implemented with junction_RNA_fwd and junction_RNA_rev primers,58 which allow the amplification of correctly inserted enhancer sequence cDNA. The jPCR products were purified using AMPure XPbeads with a beads to sample ratio of 0.8. A second PCR step was performed to add the index primers (NEBNext Multiplex Oligos for Illumina Dual Index Primers Set 1 and 2). PCR products were purified using SPRIselect beads (Beckman) (bead to sample ratio 0.8). Next generation sequencing was performed on the NextSeq 500 platform in paired end 75 cycle dual index runs.
The sequencing reads were mapped using Bowtie aligner60 using the set of synthesized oligo sequences as a reference genome. Then, UMI-tools61 was used to remove duplicates. To identify enhancers displaying allele-specific expression, QuASAR-MPRA62 and Fisher’s method63 were applied.
CRISPRa via VPR
To determine the effect of the enhancers on the transcription of hub genes, a guide RNA (gRNA) CRISPR-dCas9-derived activator system was used.64, 65, 66 The gRNAs oligos were designed with an online tool (IDTDNA) and cloned into a pSPgRNA plasmid (a gift from Charles Gersbach; Addgene plasmid # 47108) as previously described.66
Three gRNAs targeting different regions within each enhancer were cloned (Table S7) and the identity of final constructs was verified by sequencing. HepG2 cells were co-transfected with a gRNA plasmid and a CRISPR VPR activator plasmid (addgene ID: 63798) in a 1:1 mass ratio (ng) using Lipofectamine 3000 (Invitrogen). At 48 h post transfection, RNA was purified using RNeasy Mini Kit (QIAGEN) and the cDNA was prepared with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The mRNA level of the hub genes was measured by SYBR green chemistry qPCR using specific primers (Table S8) in StepOne real-time PCR system (Thermo Fisher Scientific). All gRNAs were tested in a pilot experiment and the two most effective ones per enhancer were selected for the replication. Three independent experiments with four technical replicates were performed. Data (ΔCt values) were checked for normal distribution before performing statistical tests. Paired Student’s t test (two-tailed) was used for data that followed normal distribution and equal variance. Otherwise, Mann-Whitney U test was used. p < 0.05 was used to define a significant difference between the groups.
CRISPR enhancer deletion
To delete the targeted enhancer regions, guides were designed using the custom Alt-R CRISPR-Cas9 guide RNA design tool (IDTDNA; Table S9). gRNAs predicted to produce the highest on target effect with the lowest off target risk were chosen. Positive (HPRT) and negative (non-targeting) crRNA (IDTDNA) were used as controls for the experiment. crRNA positive control are the ones that target HPRT in human and crRNA negative control are the one that contains a 20 nt “protospacer” sequence that is computationally designed to be non-targeting in human. Each RNA oligo Alt-R CRISPR-Cas9 crRNA and tracrRNA (IDTDNA) were resuspended in a duplex buffer at a final concentration of 100 μM. Two RNA oligos in equimolar concentrations were mixed in a sterile microcentrifuge tube to a final duplex concentration of 44 μM. The duplex was heated at 95°C for 5 min and allowed to cool on a bench to room temperature. For each electroporation, an Alt-R S.p. HiFi Cas9 enzyme (IDTDNA) was diluted to a working concentration of 36 μM in a resuspension buffer. An RNP complex was prepared by combining guide RNA (crRNA:tracrRNA duplex) with a Cas9 enzyme. The mixture was incubated at room temperature for 10–20 min. 1.2 × 105 HepG2 cells were electroporated with 1,300V for 30 ms with 1 pulse for each well and plated on 24-well plates. To avoid clonal heterogeneity that could cause significant genetic drifting and bias to the gene expression profiles,67,68 we opted on using pools of transfected cells for the analysis of the deletion effects. To achieve this, the cells were lysed after 48 h of transfection. The RNA extraction, cDNA synthesis and qPCR protocols were the same as those used for the CRIPSRa. Three independent experiments with 4 technical replicates were performed.
Results
A large fraction of CAD/MI GWAS SNPs are associated with liver-related traits
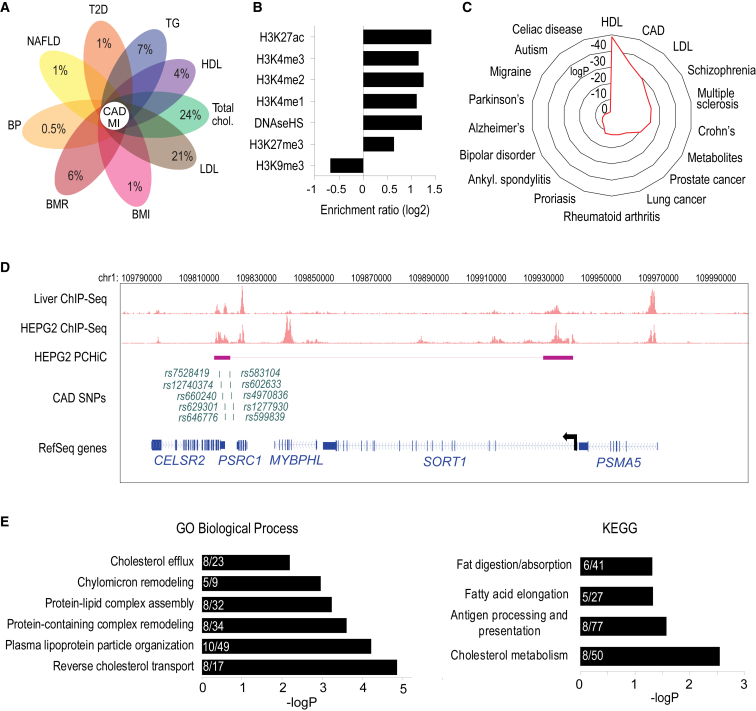
To investigate how a large fraction of the GWAS SNPs associated with CAD and its important clinical manifestation of MI could exert their function through liver-specific effects on cholesterol or lipid metabolism, we studied the genetic associations of the previously identified CAD/MI GWAS SNPs69 with liver-related traits, including cholesterol, triglycerides, and liver diseases using the UK Biobank data. The results demonstrated that of the 12,169 CAD/MI GWAS SNPs (lead and proxies in LD, r2 > 0.8), ∼37% (representing 99/262 risk loci) were associated with cholesterol or triglycerides and could thus act through liver tissue (Figure 1A) while 8% (30/262 loci) were further associated with blood pressure, T2D, BMR, and BMI possibly acting through other tissues. Altogether, this suggests that a significant fraction of risk loci exhibit pleiotropy between liver-related traits and CAD/MI and thus could provide a mechanistic explanation for these loci in disease development.
Figure 1.
CAD/MI SNPs are enriched in regulatory regions of hepatocytes
(A) Flower plot depicting the percentage of CAD/MI GWAS SNPs that are also associated with type 2 diabetes (T2D), triglycerides (TG), high-density lipoproteins (HDL), total cholesterol, low-density lipoprotein (LDL), body mass index (BMI), basal metabolic rate (BMR), blood pressure (BP), and nonalcoholic fatty liver disease (NAFLD) in the UK Biobank.
(B) Enrichment analysis of non-promoter regions in hepatocyte chromosomal interactions for enhancer- (H3K4me1-3, and H3K27ac) and repressor- (H3K27me3, H3K9me3) associated histone marks, and DNase I hypersensitive sites (DNaseHS).
(C) Radar chart showing the enrichment of GWAS variants within HepG2 chromatin interactions.
(D) Washu genome browser shot showing the location of SORT1 (chr1:109782257–109979272), H3K27ac ChIP-seq track for liver and HepG2, CAD-risk SNPs that fall within the looping ends, and PCHi-C interactions in HepG2 cells. Interacting restriction fragments are represented as boxes connected by a line on the HEPG2 PCHi-C track.
(E) Gene ontology analysis of the target genes identified from PCHi-C data.
Characterization of the hepatocyte chromosomal interactions harboring CAD/MI GWAS SNPs
Among the 12,321 CAD/MI GWAS SNPs, 152 SNPs (48 loci) were located within exons, and only 26 SNPs (24 loci) were further predicted to produce altered proteins (Figure S1; Table S10). Of the remaining SNPs, 20% (2,426/12,169 in 262 loci) were localized to hepatocyte-specific enhancers and 12% (1,446/12,169 in 262 loci) were localized to liver enhancers and could thus act by regulating gene expression in the liver. To pinpoint potential target genes for these enhancers, we prepared HindIII-digested PCHi-C libraries from HepG2 hepatocellular carcinoma cell line as described in Garske et al.25 (Table S11). In total, 91,498 promoter interactions were detected (Table S12). As expected, the majority of the interactions were enriched for active chromatin marks (H3K4me1/2/3 and H3K27ac) and transcription factor bindings and depleted for H3K9me3-marked heterochromatin, based on ChIP-seq from HepG2 cells (Figure 1B; Table S13). This suggests that we were mainly detecting interactions involving active regulatory regions, such as enhancers. Furthermore, risk SNPs associated with CAD/MI, HDL, and LDL were significantly enriched within promoter-interacting fragments compared to SNPs associated with non-liver-related diseases (Figure 1C). Altogether the CAD/MI GWAS SNPs interacted with 621 genes with a median distance of 165 kb from the promoter. Lower-resolution Hi-C data from the liver70 confirmed 101 of these candidates (Table S14). This included SORT1 for which causal SNPs were traced to an enhancer located 33 kb from the promoter8 (Figure 1D). Finally, genes interacting with fragments containing CAD/MI SNPs were more likely to be enriched for disease-relevant functional annotations such as metabolic process, chylomicron, and triglyceride-rich lipoprotein (Figure 1E), supporting the importance of the identified genes in disease etiology.
Cell type-specific expression of liver cis-eQTLs and PCHi-C target genes
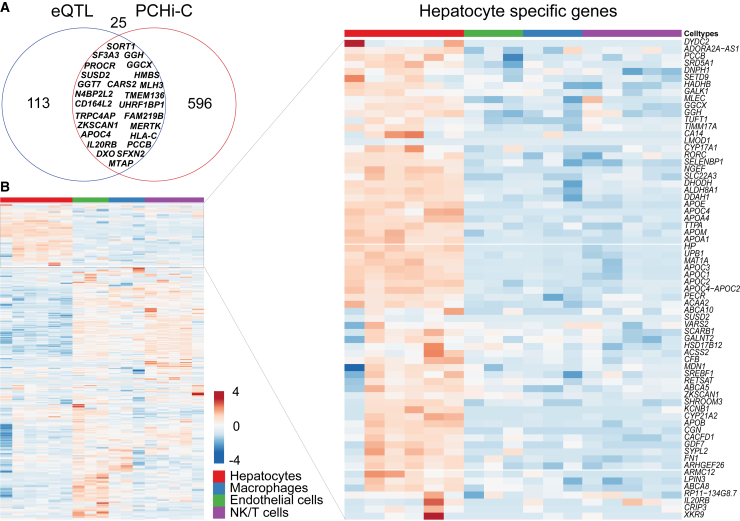
To complement the identification of liver-specific target genes of CAD/MI GWAS SNPs, we performed a cis-eQTL analysis using 263 individuals from the Kuopio Obesity Surgery (KOBS) cohort29,30 and collected published cis-eQTL information from GTEX v7,36 STARNET,37 and other publications.38, 39, 40 Altogether, 138 cis-eGenes were identified for CAD/MI risk SNPs (lead+proxies) corresponding to 62 risk loci. Intersection of the HepG2 target genes defined by PCHi-C and cis-eQTL analysis identified 25 genes common to both analysis (Figures 2A and S2). Importantly, 23 of these genes corresponded to the same SNP-eGene pair. These included SORT1 and APOC48 that have been functionally associated with lipid metabolism. No studies reporting a role in CAD or risk factor-related traits exist for other identified genes, including N4BP2L2, CD164L2, and ZKSCAN1. Importantly, only 19.7% of the genes identified by PCHi-C and cis-eQTL analysis corresponded to the GWAS annotated nearest gene.
Figure 2.
Identification of target genes regulated by CAD SNP harboring enhancers using PCHi-C and cis-eQTL analysis
(A) Venn diagram showing the intersection of cis-eQTL genes and PCHi-C target genes. The names of the 25 most common genes are shown.
(B) Expression of CAD GWAS SNP target genes (cis-eQTL and PCHi-C) in different cell types of the human liver based on scRNA-seq.45 A zoomed-in heatmap is shown for hepatocyte-specific genes.
The low overlap of PCHi-C identified target genes in HepG2 cells and cis-eQTL analysis from the liver could be due to a heterogeneous tissue composition that does not capture cell-type-specific regulatory landscapes. To better understand the cell-type-specific gene expression in the liver, we examined the expression of the 734 SNP target genes (PCHi-C+cis-eQTL) using published liver scRNA-seq data.45 Altogether, 492 SNP target genes were found expressed in any given cell type in the liver scRNA-seq data of which 14% (69) were specific to hepatocytes. These were exemplified by the 3- to 50-fold higher expression of apolipoproteins (APOA1/A4/B/C1/C2/C4/E/M) and cholesterol efflux regulatory proteins (ABCA1/5/8) (Figure 2B) compared to other cell types of the liver. We further discovered that 15% (76/492) of the genes were expressed specifically in endothelial cells (e.g., PROCR, KDM3A, and CD164L2) and 26% (127/492) mainly in inflammatory cells (e.g., MERTK in macrophages and FAS in T cells), whereas the rest were shared among many cell types (Figures S2A and S2B, Table S15). Importantly, 23/138 (20/62 loci) of the eGenes for CAD-SNPs were specific to hepatocytes, whereas the rest of them were predominantly expressed in macrophages. This could suggest that the eQTL information obtained from the KOBS cohort with participants displaying extreme obesity and/or NASH/NAFLD could reflect potential macrophage accumulation that has been associated these conditions in recent single-cell studies.71,72
Hepatocyte-specific enhancer hubs
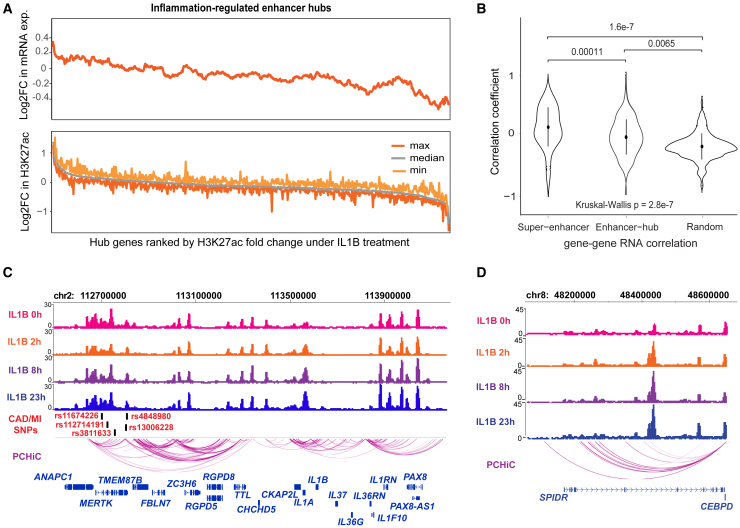
The finding that more than half of the target genes defined by PCHi-C were not specific to hepatocytes also suggests that many of the interactions are likely to be shared by other cell types as shown for MERTK and LRRC16A (Figures S3A and S3B). However, recent studies have demonstrated that genes which are important for cell identify and tissue-relevant disease SNPs are often enriched in cell-type-specific superenhancers73 and enhancer hubs74 that exhibit a significantly higher frequency of 3D chromatin interactions.75 We therefore sought to investigate whether the hepatocyte-specific genes (genes primarily expressed in hepatocytes) and CAD/MI GWAS SNPs were found near hepatocyte super enhancers or enhancer hubs. Altogether, we identified 497 super enhancers defined by linear clusters of the H3K27ac-marked regulatory regions and 1,028 enhancer hubs that clustered in 3D space (see subjects and methods). In line with the higher connectivity of super enhancers,76 these two measures exhibited similarity, with 27% (277/1,028) of enhancer clusters overlapping super enhancers. Importantly, the hepatocyte-specific genes were 1.6 times more enriched in the enhancer hubs (hypergeometric t test p = 1.52 × 10−4) compared to all expressed genes. Supporting this, a large majority (58/68) of the hepatocyte-specific genes fell within a subset of 41 enhancers hubs suggesting they could exhibit features of regulatory domains that control genes which are important for hepatocyte function. Finally, we demonstrated that the CAD/MI GWAS SNPs were significantly enriched in the enhancer hubs and super enhancers compared to other random regions (Figure S3C), supporting their functional importance in the disease.
To explore the behavior of hepatocyte-specific hubs as functional domains, we exposed HepG2 cells to IL1β stimulus time course (0 h, 2 h, 8 h, and 23 h) to simulate the low-grade systemic inflammation prevalent in CAD. Inflammation induced the H3K27ac signal in 15,266 enhancers and 700 promoters by at least 2-fold. Of the 700 promoters that showed inflammation-induced H3K27ac in the hubs, 28.8% also exhibited an increase in mRNA expression (p < 0.05; Figure 3A). Importantly, super enhancer and enhancer hub associated H3K27ac regions (enhancers/promoters) demonstrated coordinated changes during the inflammatory time course that were less evident in size-matched control regions within non-hub interactions carrying H3K27ac (Figure 3B). Altogether, 48 hepatocyte hubs (0.5 < log2FC < −0.5) were enriched for inflammation-regulated regulatory elements as exemplified by IL1A, IL1B, MERTK, CEBPD (Figures 3C and 3D), CXCL8, MTHFD2L, CXCL3, CXCL2, and CXCL5 (data not shown) which contained altogether 97 CAD/MI-associated SNPs (42 loci, Table S16). This information could be used to prioritize risk loci that are affected by the inflammatory burden associated with CAD and play an important role in hepatocyte function.
Figure 3.
Identification of regulatory regions influenced by inflammatory conditions in hepatocytes
(A) Changes in H3K27ac signal and gene expression within enhancer hubs upon inflammatory stimulus. Hub promoters were ranked by their median fold change (FC) in H3K27ac upon cytokine treatment (2–23 h), so that inflammatory-induced promoters are on the left of the x axis. Similarly, median fold change in mRNA expression is shown for genes associated with each hub.
(B) Gene pairs located within the same super enhancer or enhancer hub region show higher expression correlation across inflammatory treatment conditions than gene pairs from the random regions. p values were derived using the Kruskall-Wallis analysis of variance.
(C and D) Examples of coordinated inflammation-induced H3K27ac at chromatin hubs encompassing IL1A, IL1B, MERTK, and CEBPD.
Understanding disease association through gene function and expression
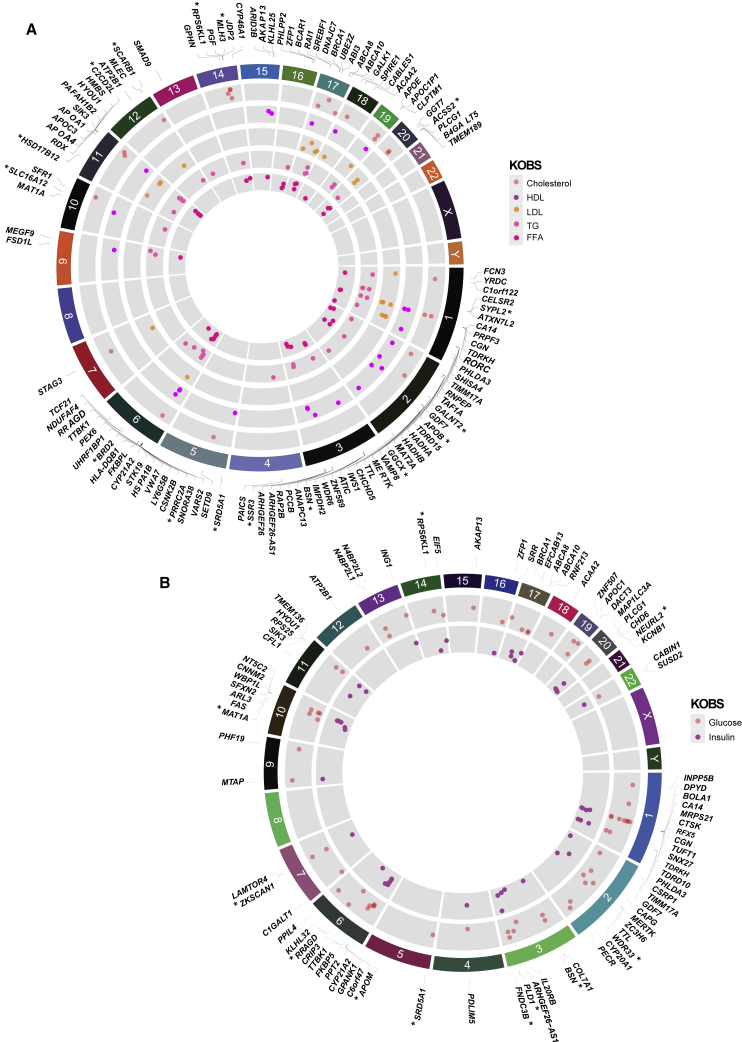
Next, we sought to understand the association of hepatocyte-specific genes with CAD by analyzing the target gene function and expression. First, we studied whether the expression of the gene itself was correlated with the liver-related traits in the KOBS cohort. Among the 714 identified target genes, 420 genes were found expressed in the liver and 113 of them were nominally associated with cholesterol-related traits such as HDL, LDL, and triglycerides (Figure 4A), whereas 84 genes were associated with glucose and insulin (Figure 4B; Table S17). These associations (cholesterol- and glucose-related traits) were confirmed for 96 genes in hyperlipidemic mice from the Hybrid Mouse Diversity Panel77 (Figure S4).
Figure 4.
Association of target gene expression with liver-related traits
Circos44 plot (circular manhattan plot) showing the association of target gene expressions with the KOBS data. Shown are (A) lipid traits (cholesterol, HDL, TG, LDL, and FFA) and (B) glucose and insulin levels in the KOBS cohort.29 Asterisk (∗) denotes genes with a significant association also in the Hybrid Mouse Diversity Panel (HMDP). The height of the plot indicates the significance of the association (p value).
Inflammation also links CAD to NAFLD, which covers a spectrum of diseases from simple steatosis to non-alcoholic steatohepatitis (NASH). NAFLD has been associated with an increased risk of CAD, independently of classical CAD risk factors.78 Therefore, we also studied the liver expression of the candidate genes in the KOBS cohort and tested whether their expression was associated with NASH by comparing diseased and the non-diseased samples. Out of the 420 genes expressed in liver, the expression of 105 genes was significantly associated (FDR < 0.05) with NASH compared to controls (Figures S5A and S5B, Table S18). Among them, the expression of 70 genes was also correlated with the levels of inflammatory cytokines such as IL6, TNFa, MIP-1b, MCP1, and IL1A in the atherosclerosis mouse model in the Hybrid Mouse Diversity Panel43,77 (Figure S6). These included known genes such as APOM,79 GALNT2,80 and MTAP81 but also candidate genes N4BP2L1 and ZKSCAN1 with no previous association with cardiometabolic traits. Altogether, our analysis provides a list of target genes whose liver expression changes could explain the SNP association with the risk of CAD through lipid traits or inflammation.
Identification of susceptibility enhancers associated with allele-specific enhancer activity
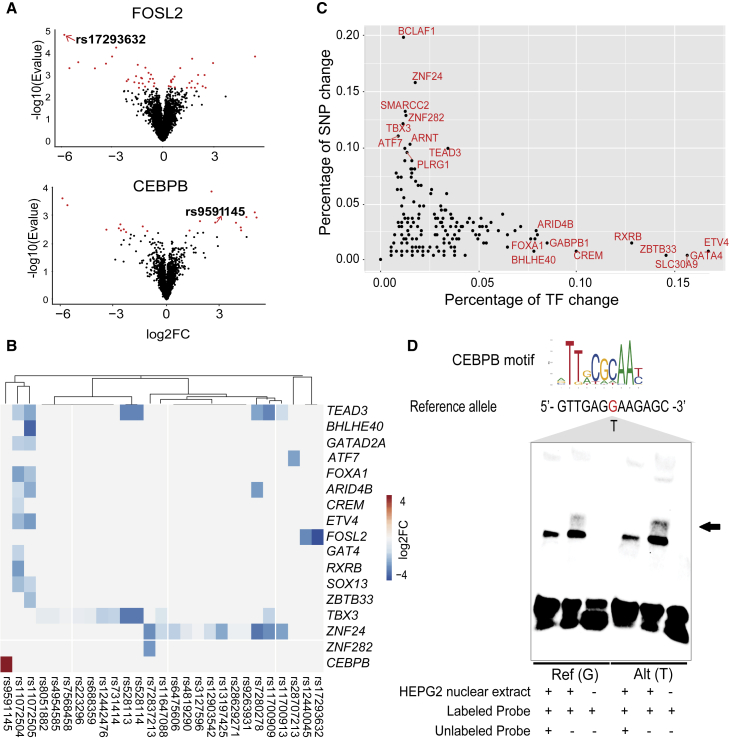
There is mounting evidence showing that SNPs within enhancers affect gene regulation mainly by altering the transcription factor (TF) binding.15 To provide a comprehensive characterization of CAD/MI susceptibility SNPs located in liver-specific enhancer regions, we used a combination of computational and experimental approaches. First, we used the DeepSEA deep learning-based algorithmic framework to predict the differential allele-specific binding of TFs. Altogether 225/12,169 SNPs passed the functional significance score < 0.01 (Table S19, Figure S1). As an example, rs17293632, previously validated eQTL for SMAD3,12 demonstrates that the “T” allele greatly reduces the FOSL2 binding (Figure 5A).
Figure 5.
Analysis of allele-specific binding activity of CAD SNPs
(A) Volcano plot of DeepSEA-predicted probability differences between the reference and alternative alleles for FOSL2 and CEBPB motifs that overlap CAD/MI GWAS alleles (lead+proxies). Two example SNPs, rs17293632 and rs9591145, predicted to result in high-significance allele-specific binding of the TF are shown. E-value is defined as the expected proportion of SNPs with larger predicted effect (from reference allele to alternative allele) for a given chromatin feature.
(B) Heatmap82 of selected SNPs demonstrating allele-specific TF binding based on BaalChIP-analysis. See Figure S5 for a complete list.
(C) A dot plot demonstrating negative correlation between the percentage of SNPs and the percentage of TFs demonstrating a change in TF binding. The x axis represents percent of SNPs that demonstrate significant allele-specific binding (>2-fold) for a given TF among all studied CAD/MI SNPs. The higher value indicates that among all SNPs demonstrating an allele-specific binding in BaalChIP, a specific TF is more likely to exhibit allele-specific binding. The y axis represents the percent from all studied TFs exhibiting an allele-specific binding across CAD/MI SNPs that exhibit allele-specific binding by at least one TF. The higher value indicates that the allele-specific binding of a specific TF could be more likely to translate to an allele-specific binding by the other studied TFs.
(D) EMSA for the rs9591145 SNP showing that the “T” allele significantly gains binding affinity of CEBPB compared to the “G” allele.
Second, we took advantage of the extensive ChIP-seq data resource from ENCODE evaluating the allele-specific TF binding in HepG2 cells comprising of 210 TFs and 580 samples. Altogether, 3,824 heterozygous loci were identified in HepG2, and 23.6% (908) of them exhibited an allele-specific ChIP-seq signal (Figures 5B and S1; Table S20). To investigate the possibility that certain TFs are frequently mutated, or have a large effect on the binding of other TFs (as has been shown for pioneering factors15,83), we calculated the percentage of SNPs that demonstrated a change in the binding and the percentage of other TFs affected by the same SNP at least 2-fold (Figures 5C and S7). Interestingly, a clear negative correlation was observed in which frequently observed SNPs were less likely to affect the binding of other TFs. This was exemplified by the high frequency of SNPs affecting the binding of TBX3 and ZNF24 and the low frequency of SNPs affecting known pioneering factors, such as FOXA1 and GATA4. This is in line with studies from us and others demonstrating that SNPs affecting the binding of lineage determining TFs are likely to affect other TFs that are considered lower in hierarchy.15,83 Interestingly, our data also show that FOXA1 and GATA4 mutations were less frequent among all the SNPs that demonstrated allele-specific binding.
Altogether, 31 of the candidates investigated in this study, representing 14% of the DeepSEA-predicted enhancers and 3.4% of the enhancers exhibiting ChIP-seq intensity variation, showed an allele bias with both methods. Among them, we selected the looping candidate of N4BP2L2 (rs9591145) (Figure 5D, Table S21) for functional validation using an electrophoretic mobility shift assay (EMSAs) from nuclear extracts of HepG2. As shown in Figure S8, rs9591145 was predicted to have a higher affinity to the reference allele “T” while binding to CEBPB. The EMSA results confirmed the reduced protein binding of the alternate allele (G) compared to the reference allele (T) of rs9591145, validating the computational predictions and allele-specific ChIP-seq data for rs9591145, i.e., that the “T” allele increases the binding affinity of CEBPB.
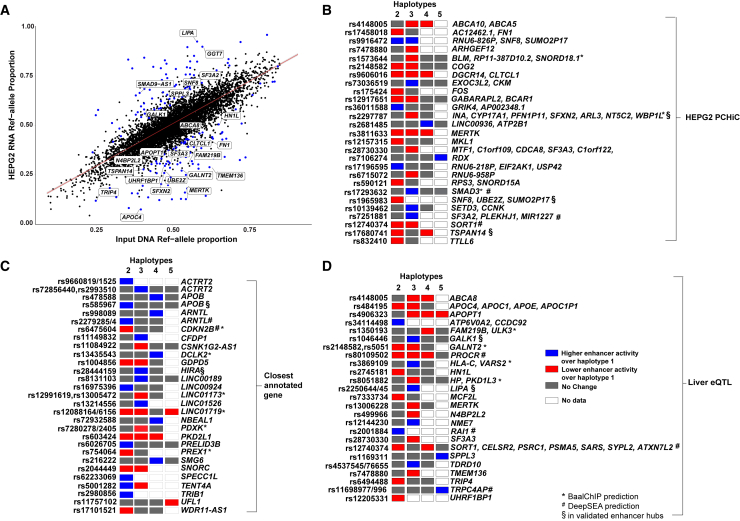
Finally, to more broadly validate the allele-specific enhancer activity of the susceptibility loci, we investigated the functional effects of CAD/MI GWAS SNPs on the enhancer activity using the STARR-seq84-based massively parallel reporter assay. In contrast to previous single SNP-centric studies, we took advantage of haplotype-specific information to design a reporter library that incorporated the most common combinations of SNPs in the population within a given 200 bp region. Altogether 3,661 SNPs located within enhancer regions were included (see subjects and methods). The plot between the input DNA ref-allele proportions compared to the HEPG2 RNA-allele proportion is shown in Figure 6A. The results demonstrated that 212 susceptibility enhancers, corresponding to 42/262 CAD/MI GWAS loci, exhibited a significant allele-specific activity in STARR-seq. These include the rs17293632 located in intron 1 of SMAD3, a key contributor to transforming growth factor-β pathway signaling, which in addition to CAD is found to be associated with inflammatory conditions such as Crohn disease and ulcerative colitis in GWAS.18 Recent fine-mapping efforts have demonstrated causality for the rs17293632 SNP in mediating an anti-proliferative effect on vascular smooth muscle cells.12 Our HepG2 PCHi-C further showed that rs17293632 (Figure S9) indeed interacted with the promoter of SMAD3 although it was not a liver cis-eQTL. In addition, allele-specific enhancer activity was also detected for an enhancer harboring rs6475604, a SNP located in intron 3 of CDKN2B-AS1 that was recently found to be associated with atherosclerosis using fine mapping of 9q21.85 Altogether, 79 SNPs (38 loci) identified in STARR-seq had a predicted target gene identified by PCHi-C analysis, closely annotated genes or cis-eQTL (Figures 6B–6D), including important players in lipoprotein metabolism such as APOC1/4, APOE, and LIPA. Altogether, our analysis summarized in Table S22 provides a resource for further prioritization and functional characterization of causal enhancer SNPs and their target genes associated with the risk of CAD in the liver.
Figure 6.
Allele-specific activity of enhancers investigated by STARR-seq
(A) Dot plot depicting the STARR-seq input DNA library ref-allele proportions in relation to the experimentally quantified RNA library ref-allele proportions in HepG2 cells. SNPs with significant allele-specific effects are highlighted in blue. Selected genes associated with the studied enhancer regions are shown.
(B–D) Heatmap of selected enhancers showing a significant allele-specific difference compared to the most common haplotype (1) observed in the European population for which the target genes were defined by (B) PCHi-C, (C) proximity, and (D) cis-eQTL analysis. Asterisk (∗) represents the SNP that demonstrated allele-specific binding in BaalChIP while hatch mark (#) denotes SNPs that were predicted to disrupt TF binding by DeepSEA and double s (§) is an experimentally validated location from Figure 7.
Genetic perturbations of hepatocyte-specific enhancer hubs
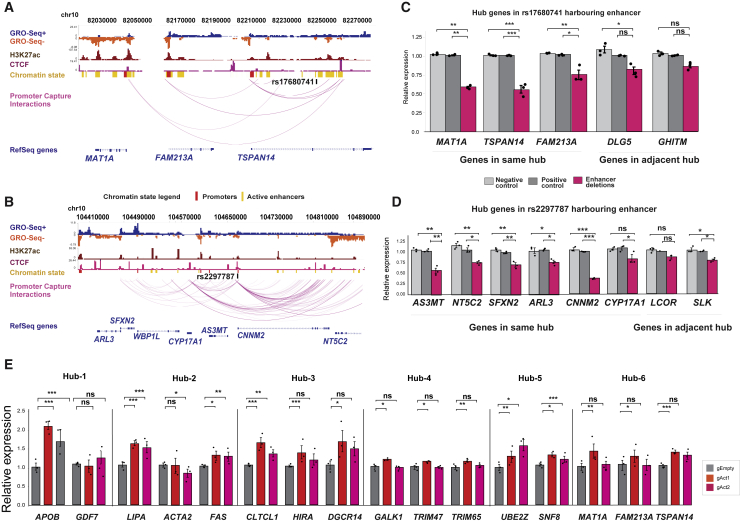
Among the STARR-seq-validated susceptibility enhancers with a high probability of causality, we selected seven targets for genetic perturbations. We prioritized enhancers located within hepatocyte-specific hubs (Figures 6B–6D, marked with §) for their potential to perturb several genes within the same regulatory environment.74 First, we selected two loci for CRISPR-Cas9-mediated deletion of the susceptibility enhancer (Figures 7A and 7B). We demonstrate that a 300 bp deletion of an enhancer harboring rs17680741 led to downregulation of all three genes MAT1A, FAM23A, and TSPAN14 in the hub, while deletion of rs2297787-enhancer led to the downregulation of AS3MT, NT5C2, SFXN2, ARL3, and CNNM2 (Figures 7C and 7D). Importantly, the deletion effect on the non-target genes on adjacent hubs was often not significant, supporting a model where variant carrying enhancers primarily regulate the genes within the same chromatin interaction space. To further confirm these findings with a different approach, we employed CRISPRa-mediated activation of six loci (Figures 7A, 7B, S10, and S11). Importantly, this validated the effect of the enhancer containing rs17680741 in regulating the expression of MAT1A, FAM23A, and TSPAN14 (Figure 7E, hub-6). In addition, activation of five other susceptibility enhancers led to the activation of 12 genes including the well-established disease associated genes APOB, LIPA, and FAS (Figure 7E, Table S23). Altogether, our findings demonstrate how a single enhancer carrying a risk variant in a large 3D hub is able to regulate many genes, which supports the concept of enhancer interaction hubs acting as functional regulatory domains.
Figure 7.
CRISPR-mediated genetic perturbations of enhancer hubs
(A and B) Washu genome browser shots of two enhancer hubs containing (A) TSPAN14 and (B) SFXN2 where the CRISPR-Cas9 system was used to delete the enhancer region harboring a CAD GWAS variant. GRO-seq showing enhancer RNA (eRNA) for HEPG2 comes from GSE92375 (GSM2428726).
(C and D) Analysis of the effect of enhancer deletion on gene expression within the TSPAN14 and SFXN2 hubs in HepG2 cells. qPCR was performed for genes located in the same hub as well as for genes in adjacent hubs.
(E) Analysis of the effect of CRISPRa-mediated activation of enhancer variants in six selected chromatin hubs. For locus information, see Figure S9. Gene expression data are presented as the mean ± SEM of three independent experiments. The statistical significance was evaluated using a two-tailed Student’s t test or Mann-Whitney U test. For all bar plots, significance is denoted with asterisk. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005.
Discussion
The involvement of the liver in the progression of coronary artery disease is incompletely understood. Especially, the causality of the associations and the mechanisms behind the SNP-disease interactions outside the well-characterized lipid associations86 have remained largely unknown. This is mostly due to the fact that a majority of the CAD GWAS variants are located in non-coding regions.3,4 In this study, we performed a comprehensive identification of target genes in hepatocytes using eQTL analysis and PCHi-C while providing prioritization of regulatory SNPs using computational predictions, allele-specific ChIP-seq analysis, and STARR-seq. Among the susceptibility loci, we were able to identify hundreds of potentially causal enhancer SNPs and target genes. Importantly, we add to the growing evidence that the nearest gene defined by GWAS studies does not always represent the causal gene for disease association.87
Chromatin interactions are important for gene regulation and they have been recently used to identify target genes of variants in adipocytes,13 endothelial cells,88 pluripotent stem cells (iPSCs),89 iPSC-derived cardiomyocytes (CMs),90 and even hepatocytes.91 In line with our data and others, PCHi-C data predict significantly more target genes compared to cis-eQTL analysis. This could be partially due to underpowered eQTL studies but also by the fact that physical chromatin interaction does not necessarily mean functional interaction between an enhancer and a gene. Instead of a direct functional interaction, three additional types of interactions have been proposed, namely random interactions, bystander interactions (i.e., DNA close to a direct interaction will be also close as a consequence of the former), and interactions due to sharing of the same nuclear structure.92 This limitation was also evident in our data where chromatin interactions were shared between hepatocytes, macrophages, and endothelial cells, despite the cell-type-specific expression of a gene. One advantage of chromatin interactions, however, is the limited effect of the environment on the interactions,93 which largely impacts an eQTL analysis.94 In addition, a cis-eQTL analysis relies on RNA-seq data that represents a sum of transcriptional and post-transcriptional gene regulation, making it less ideal for the analysis of regulatory variants. Future studies based on the analysis of nascent RNAs are hoped to overcome this limitation. On the other hand, limited overlap of the PCHi-C and cis-eQTL based target gene identification can also be explained by the PCHi-C being limited to a HepG2 hepatocellular carcinoma cell line solely representing hepatocyte-like cells that represents about ∼60% overlap with bulk liver DNaseHS-sites.20 Still, we believe that a more robust target gene identification can be achieved by applying both PCHi-C and cis-eQTL analysis approaches and that further functional validations should be conducted to verify the effect of regulatory variants on gene expression. These data could be further stratified by deconvolution of the cellular composition of bulk RNA-seq based on scRNA-seq to shed light on the functional impacts of cell-type-specific genetic variation.95
Our study represents one step forward in the path to comprehensively understand the function and regulation of CAD/MI risk genes in the liver. Previous efforts to annotate the genes mapped to CAD loci have identified 32 genes which are likely to regulate lipid metabolism and inflammation. However, we provide evidence that the number of susceptibility genes acting through similar mechanisms could be higher. This was supported by the observation that ∼37% of the CAD/MI GWAS SNPs (99/262 loci) were associated with cholesterol and triglyceride levels and the expression of more than a hundred target genes (PCHi-C/cis-eQTL) exhibited correlations with lipid traits, NASH, and inflammatory molecule levels. Interestingly, we also identified interaction hubs where several genes were shown to be similarly regulated in response to an inflammatory stimulus and where the perturbation of a single enhancer was able to modulate the expression of most genes within the hub. These are highly similar to the 3D hubs that were previously described in pancreatic islets to exhibit coordinated glucose-dependent activity and which have the ability to predict a T2D risk driven by islet regulatory variants.74 Future studies further focusing on hepatocyte-specific enhancer hubs should thus be evaluated for the quantification of genetic risks acting through inflammatory pathways. Previous work have demonstrated that with increasing dietary cholesterol intake, the liver switches from a resilient, adaptive state to an inflammatory, pro-atherosclerotic state.96 Therefore, future studies investigating the effect of the genotype and dietary saturated fat intake on CAD could improve our understanding of gene-environment interactions acting to shape the regulatory wiring at inflammatory enhancer hubs in the liver.
We also provide a comprehensive analysis of the effect of the regulatory SNPs on transcription factor binding. Altogether, 9% of the SNPs exhibited allele-specific TF binding using computational predictions or ChIP-seq data. To provide the most comprehensive characterization of CAD/MI regulatory variants in hepatocytes, we also performed the first massively parallel STARR-seq reporter assay with all hepatocyte-specific enhancers carrying risk SNPs. To this end, we studied common haplotype combinations for each of 200 bp regions. This identified 212 enhancer SNPs (6% of the studied regions) with allele-specific enhancer activity. While STARR-seq does not prove causality, it does substantially reduce the test space of alleles linked to a trait locus and provides a concise list of high-priority targets for follow-up. Interestingly, STARR-seq identified such enhancer SNP variants for APOC1/4, APOE, APOB, and LIPA. All of these genes have established roles in lipoprotein metabolism, and common exonic variants for APOE (rs429358, rs7412) have been associated with LDL cholesterol, apoB levels, and Alzheimer disease,97,98 and common exonic variants for LIPA (rs1051338) have been shown to be associated with the risk of CAD, obesity-related metabolic complications, and blood pressure.99,100 Our results suggest that common enhancer variants could represent an additional layer of regulation that together with exonic variants affect gene expression and disease susceptibility. However, despite the demonstration of potential functional roles of the enhancers (CRISPR-perturbation) and the SNP variants (STARR-seq) in our study, we recognize that in the case of coexisting pathogenic exonic variants, further follow-up studies are needed to confirm whether indeed multiple SNP(s) are responsible for the GWAS associations. As a preliminary evidence of such mechanism, we demonstrate that the activation of the enhancer variant locus for APOB and LIPA results in a significant effect on gene expression. Therefore, similar to SORT1,8 common variation within the enhancers of lipoprotein genes could contribute to clinical liver phenotypes.
Since the increased understanding of the importance of enhancers as the key regulators of gene expression, it has become increasingly important to understand the effects of genetic variation on enhancer function and target gene expression. Here, we studied all known CAD/MI-associated GWAS SNPs located within the liver and hepatocyte-specific regulatory regions in an effort to understand the possible mechanisms through which they contribute to disease. Overall, our findings expand the repertoire of genes and regulatory mechanisms acting in the liver and governing the risk of CAD development.
Data and code availability
All HMDP microarray data are deposited in the NCBI Gene Expression Omnibus (GEO) under the accession number GSE66570. The promoter capture Hi-C and H3K27ac ChIP-seq experiments reported in this study are deposited in the GEO database under the accession number GSE157306. The published article includes all other analysis results generated during this study.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
The authors would like to thank pathologist Vesa Kärjä, gastro-surgeons Pirjo Käkelä and Sari Venesmaa (Kuopio University Hospital), and study nurses Päivi Turunen and Matti Laitinen (University of Eastern Finland) for their assistance with the tissue biopsies and laboratory analyses. The authors also wish to acknowledge CSC – IT Center for Science, Finland and Bioinformatics center of University of Eastern Finland for the computational resources. This study was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant no. 802825 to M.U.K.), National Institutes of Health (NIH) grants HL-095056, HL-28481, and U01DK105561 (to P.P.) and R01 DK117850 and HL147883 (to A.J.L.), and Sigrid Juselius Foundation (to M.U.K. and A.-L.L.). I.S., A.T., P.R.M., and O.H.L. were supported by the University of Eastern Finland Doctoral Program in Molecular Medicine. K.M.G. was supported by NIH/NHLBI grant F31HL142180. A.K. was supported by American Heart Association grant 19CDA34769186. D.K. was supported by the Academy of Finland (contract no. 316458). Kuopio Obesity Surgery Study (PI J.P.) was supported by the Academy of Finland grant (contract no. 138006), the Finnish Diabetes Research Foundation, and Kuopio University Hospital Project grants (EVO/VTR grants 2005-2019). M.U.K. was further supported by the Academy of Finland (grants nos. 287478 and 294073), the Finnish Foundation for Cardiovascular Research, and the Jane and Aatos Erkko Foundation.
Published: February 23, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ajhg.2021.02.006.
Web Resources
Datamash, https://www.gnu.org/software/datamash/
HOMER, http://homer.ucsd.edu/homer/
Hybrid Mouse Diversity Panel web interface, https://systems.genetics.ucla.edu
Hypergeometric test, https://systems.crump.ucla.edu/hypergeometric/
UK Biobank — Neale lab, http://www.nealelab.is/uk-biobank
Supplemental Information
References
- 1.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Won H.-H., Natarajan P., Dobbyn A., Jordan D.M., Roussos P., Lage K., Raychaudhuri S., Stahl E., Do R. Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease. PLoS Genet. 2015;11:e1005622. doi: 10.1371/journal.pgen.1005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khera A.V., Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat. Rev. Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasper P., Martin A., Lang S., Kütting F., Goeser T., Demir M., Steffen H.M. NAFLD and cardiovascular diseases: a clinical review. Clin. Res. Cardiol. 2020 doi: 10.1007/s00392-020-01709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauberg M.E., Zhang W., Giambartolomei C., Franzén O., Morris D.L., Vyse T.J., Ruusalepp A., Sklar P., Schadt E.E., Björkegren J.L.M., Roussos P., CommonMind Consortium Large-Scale Identification of Common Trait and Disease Variants Affecting Gene Expression. Am. J. Hum. Genet. 2017;100:885–894. doi: 10.1016/j.ajhg.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudoin M., Gupta R.M., Won H.H., Lo K.S., Do R., Henderson C.A., Lavoie-St-Amour C., Langlois S., Rivas D., Lehoux S. Myocardial Infarction-Associated SNP at 6p24 Interferes With MEF2 Binding and Associates With PHACTR1 Expression Levels in Human Coronary Arteries. Arterioscler. Thromb. Vasc. Biol. 2015;35:1472–1479. doi: 10.1161/ATVBAHA.115.305534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakaoka H., Gurumurthy A., Hayano T., Ahmadloo S., Omer W.H., Yoshihara K., Yamamoto A., Kurose K., Enomoto T., Akira S. Allelic Imbalance in Regulation of ANRIL through Chromatin Interaction at 9p21 Endometriosis Risk Locus. PLoS Genet. 2016;12:e1005893. doi: 10.1371/journal.pgen.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanda V., Wang T., Pjanic M., Liu B., Nguyen T., Matic L.P., Hedin U., Koplev S., Ma L., Franzén O. Functional regulatory mechanism of smooth muscle cell-restricted LMOD1 coronary artery disease locus. PLoS Genet. 2018;14:e1007755. doi: 10.1371/journal.pgen.1007755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner A.W., Martinuk A., Silva A., Lau P., Nikpay M., Eriksson P., Folkersen L., Perisic L., Hedin U., Soubeyrand S., McPherson R. Functional analysis of a novel genome-wide association study signal in SMAD3 that confers protection from coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2016;36:972–983. doi: 10.1161/ATVBAHA.116.307294. [DOI] [PubMed] [Google Scholar]
- 13.Pan D.Z., Garske K.M., Alvarez M., Bhagat Y.V., Boocock J., Nikkola E., Miao Z., Raulerson C.K., Cantor R.M., Civelek M. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 2018;9:1512. doi: 10.1038/s41467-018-03554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., BLUEPRINT Consortium Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilpinen H., Waszak S.M., Gschwind A.R., Raghav S.K., Witwicki R.M., Orioli A., Migliavacca E., Wiederkehr M., Gutierrez-Arcelus M., Panousis N.I. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasowski M., Kyriazopoulou-Panagiotopoulou S., Grubert F., Zaugg J.B., Kundaje A., Liu Y., Boyle A.P., Zhang Q.C., Zakharia F., Spacek D.V. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei I., Jordan D.M., Sunyaev S.R. Curr. Protoc. Hum. Genet; 2013. Predicting functional effect of human missense mutations using PolyPhen-2, Chapter 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garske K.M., Pan D.Z., Miao Z., Bhagat Y.V., Comenho C., Robles C.R., Benhammou J.N., Alvarez M., Ko A., Ye C.J. Reverse gene-environment interaction approach to identify variants influencing body-mass index in humans. Nat Metab. 2019;1:630–642. doi: 10.1038/s42255-019-0071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingett S., Ewels P., Furlan-Magaril M., Nagano T., Schoenfelder S., Fraser P., Andrews S. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.M., Osborne C. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90-7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonen M., Männistö V., Leppänen J., Kaminska D., Kärjä V., Venesmaa S., Käkelä P., Kuusisto J., Gylling H., Laakso M., Pihlajamäki J. Desmosterol in human nonalcoholic steatohepatitis. Hepatology. 2013;58:976–982. doi: 10.1002/hep.26342. [DOI] [PubMed] [Google Scholar]
- 30.Benhammou J.N., Ko A., Alvarez M., Kaikkonen M.U., Rankin C., Garske K.M., Padua D., Bhagat Y., Kaminska D., Kärjä V. Novel Lipid Long Intervening Noncoding RNA, Oligodendrocyte Maturation-Associated Long Intergenic Noncoding RNA, Regulates the Liver Steatosis Gene Stearoyl-Coenzyme A Desaturase As an Enhancer RNA. Hepatol Commun. 2019;3:1356–1372. doi: 10.1002/hep4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leek J.T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42:e161. doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franzén O., Ermel R., Cohain A., Akers N.K., Di Narzo A., Talukdar H.A., Foroughi-Asl H., Giambartolomei C., Fullard J.F., Sukhavasi K. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strunz T., Grassmann F., Gayán J., Nahkuri S., Souza-Costa D., Maugeais C., Fauser S., Nogoceke E., Weber B.H.F. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci. Rep. 2018;8:5865. doi: 10.1038/s41598-018-24219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu C.-H., Pal L.R., Moult J. Consensus Genome-Wide Expression Quantitative Trait Loci and Their Relationship with Human Complex Trait Disease. OMICS. 2016;20:400–414. doi: 10.1089/omi.2016.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lusis A.J., Seldin M.M., Allayee H., Bennett B.J., Civelek M., Davis R.C., Eskin E., Farber C.R., Hui S., Mehrabian M. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 2016;57:925–942. doi: 10.1194/jlr.R066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett B.J., Davis R.C., Civelek M., Orozco L., Wu J., Qi H., Pan C., Sevag Packard R.R., Eskin E., Yan M. Correction: Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS Genet. 2016;12:e1005913. doi: 10.1371/journal.pgen.1005913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 45.MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaikkonen M.U., Niskanen H., Romanoski C.E., Kansanen E., Kivelä A.M., Laitalainen J., Heinz S., Benner C., Glass C.K., Ylä-Herttuala S. Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization. Nucleic Acids Res. 2014;42:12570–12584. doi: 10.1093/nar/gku1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gel B., Díez-Villanueva A., Serra E., Buschbeck M., Peinado M.A., Malinverni R. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016;32:289–291. doi: 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang J., Mathijs K., Timmermans L., Claessen S.M., Hecka A., Weusten J., Peters R., van Delft J.H., Kleinjans J.C.S., Jennen D.G.J., de Kok T.M. The idiosyncratic drug-induced gene expression changes in HepG2 cells. Data Brief. 2017;14:462–468. doi: 10.1016/j.dib.2017.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenzie A.T., Katsyv I., Song W.M., Wang M., Zhang B. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst. Biol. 2016;10:106. doi: 10.1186/s12918-016-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Santiago I., Liu W., Yuan K., O’Reilly M., Chilamakuri C.S., Ponder B.A., Meyer K.B., Markowetz F. BaalChIP: Bayesian analysis of allele-specific transcription factor binding in cancer genomes. Genome Biol. 2017;18:39. doi: 10.1186/s13059-017-1165-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou B., Ho S.S., Greer S.U., Spies N., Bell J.M., Zhang X., Zhu X., Arthur J.G., Byeon S., Pattni R. Haplotype-resolved and integrated genome analysis of the cancer cell line HepG2. Nucleic Acids Res. 2019;47:3846–3861. doi: 10.1093/nar/gkz169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charif D., Lobry J.R. Springer; Berlin, Heidelberg: 2007. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. in 207–232. [DOI] [Google Scholar]
- 58.Muerdter F., Boryń Ł.M., Woodfin A.R., Neumayr C., Rath M., Zabidi M.A., Pagani M., Haberle V., Kazmar T., Catarino R.R. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat. Methods. 2018;15:141–149. doi: 10.1038/nmeth.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalita C.A., Brown C.D., Freiman A., Isherwood J., Wen X., Pique-Regi R., Luca F. High-throughput characterization of genetic effects on DNA-protein binding and gene transcription. Genome Res. 2018;28:1701–1708. doi: 10.1101/gr.237354.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalita C.A., Moyerbrailean G.A., Brown C., Wen X., Luca F., Pique-Regi R. QuASAR-MPRA: accurate allele-specific analysis for massively parallel reporter assays. Bioinformatics. 2018;34:787–794. doi: 10.1093/bioinformatics/btx598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López Rodríguez M., Kaminska D., Lappalainen K., Pihlajamäki J., Kaikkonen M.U., Laakso M. Identification and characterization of a FOXA2-regulated transcriptional enhancer at a type 2 diabetes intronic locus that controls GCKR expression in liver cells. Genome Med. 2017;9:63. doi: 10.1186/s13073-017-0453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stojic L., Lun A.T.L., Mangei J., Mascalchi P., Quarantotti V., Barr A.R., Bakal C., Marioni J.C., Gergely F., Odom D.T. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 2018;46:5950–5966. doi: 10.1093/nar/gky437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verweij N., Eppinga R.N., Hagemeijer Y., van der Harst P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci. Rep. 2017;7:2761. doi: 10.1038/s41598-017-03062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung D., Jung I., Rajagopal N., Schmitt A., Selvaraj S., Lee A.Y., Yen C.A., Lin S., Lin Y., Qiu Y. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., Zhao X.Y., Ji Y., Li C., Guo L. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol. Cell. 2019;75:644–660.e5. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krenkel O., Hundertmark J., Abdallah A.T., Kohlhepp M., Puengel T., Roth T., Branco D.P.P., Mossanen J.C., Luedde T., Trautwein C. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut. 2020;69:551–563. doi: 10.1136/gutjnl-2019-318382. [DOI] [PubMed] [Google Scholar]
- 73.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miguel-Escalada I., Bonàs-Guarch S., Cebola I., Ponsa-Cobas J., Mendieta-Esteban J., Atla G., Javierre B.M., Rolando D.M.Y., Farabella I., Morgan C.C. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat. Genet. 2019;51:1137–1148. doi: 10.1038/s41588-019-0457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J., Li K., Cai W., Liu X., Zhang Y., Orkin S.H., Xu J., Yuan G.C. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun. 2018;9:943. doi: 10.1038/s41467-018-03279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thibodeau A., Márquez E.J., Shin D.G., Vera-Licona P., Ucar D. Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin. Sci. Rep. 2017;7:14466. doi: 10.1038/s41598-017-14389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bennett B.J., Davis R.C., Civelek M., Orozco L., Wu J., Qi H., Pan C., Packard R.R., Eskin E., Yan M. Genetic Architecture of Atherosclerosis in Mice: A Systems Genetics Analysis of Common Inbred Strains. PLoS Genet. 2015;11:e1005711. doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francque S.M. The role of non-alcoholic fatty liver disease in cardiovascular disease. Eur. Cardiol. 2014;9:10–15. doi: 10.15420/ecr.2014.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Huang L.Z., Yang Q.L., Liu Y., Zhou X. Correlation analysis between ApoM gene-promoter polymorphisms and coronary heart disease. Cardiovasc. J. Afr. 2016;27:228–237. doi: 10.5830/CVJA-2016-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng P., Wang L., Yang X., Huang X., Ba Y., Chen X., Guo J., Lian J., Zhou J. A preliminary study of the relationship between promoter methylation of the ABCG1, GALNT2 and HMGCR genes and coronary heart disease. PLoS ONE. 2014;9:e102265. doi: 10.1371/journal.pone.0102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J.B., Deluna A., Mungrue I.N., Vu C., Pouldar D., Civelek M., Orozco L., Wu J., Wang X., Charugundla S. Effect of 9p21.3 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation. 2012;126:1896–1906. doi: 10.1161/CIRCULATIONAHA.111.064881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566-70. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A.J., Mann A.L., Kundu K., Hale C., Dougan G., Gaffney D.J., HIPSCI Consortium Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arnold C.D., Gerlach D., Stelzer C., Boryń Ł.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 85.Vargas J.D., Manichaikul A., Wang X.Q., Rich S.S., Rotter J.I., Post W.S., Polak J.F., Budoff M.J., Bluemke D.A. Detailed analysis of association between common single nucleotide polymorphisms and subclinical atherosclerosis: The Multi-ethnic Study of Atherosclerosis. Data Brief. 2016;7:229–242. doi: 10.1016/j.dib.2016.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erdmann J., Kessler T., Munoz Venegas L., Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc. Res. 2018;114:1241–1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 87.Anderson C.A., Soranzo N., Zeggini E., Barrett J.C. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lalonde S., Codina-Fauteux V.A., de Bellefon S.M., Leblanc F., Beaudoin M., Simon M.M., Dali R., Kwan T., Lo K.S., Pastinen T., Lettre G. Integrative analysis of vascular endothelial cell genomic features identifies AIDA as a coronary artery disease candidate gene. Genome Biol. 2019;20:133. doi: 10.1186/s13059-019-1749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ikeda H., Sone M., Yamanaka S., Yamamoto T. Structural and spatial chromatin features at developmental gene loci in human pluripotent stem cells. Nat. Commun. 2017;8:1616. doi: 10.1038/s41467-017-01679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Montefiori L.E., Sobreira D.R., Sakabe N.J., Aneas I., Joslin A.C., Hansen G.T., Bozek G., Moskowitz I.P., McNally E.M., Nóbrega M.A. A promoter interaction map for cardiovascular disease genetics. eLife. 2018;7 doi: 10.7554/eLife.35788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Çalışkan M., Manduchi E., Rao H.S., Segert J.A., Beltrame M.H., Trizzino M., Park Y., Baker S.W., Chesi A., Johnson M.E. Genetic and Epigenetic Fine Mapping of Complex Trait Associated Loci in the Human Liver. Am. J. Hum. Genet. 2019;105:89–107. doi: 10.1016/j.ajhg.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niskanen H., Tuszynska I., Zaborowski R., Heinäniemi M., Ylä-Herttuala S., Wilczynski B., Kaikkonen M.U. Endothelial cell differentiation is encompassed by changes in long range interactions between inactive chromatin regions. Nucleic Acids Res. 2018;46:1724–1740. doi: 10.1093/nar/gkx1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Romanoski C.E., Lee S., Kim M.J., Ingram-Drake L., Plaisier C.L., Yordanova R., Tilford C., Guan B., He A., Gargalovic P.S. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donovan M.K.R., D’Antonio-Chronowska A., D’Antonio M., Frazer K.A. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat. Commun. 2020;11:955. doi: 10.1038/s41467-020-14561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kleemann R., Verschuren L., van Erk M.J., Nikolsky Y., Cnubben N.H., Verheij E.R., Smilde A.K., Hendriks H.F., Zadelaar S., Smith G.J. Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: a combined transcriptomics and metabolomics analysis. Genome Biol. 2007;8:R200. doi: 10.1186/gb-2007-8-9-r200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smolková B., Bonassi S., Buociková V., Dušinská M., Horská A., Kuba D., Džupinková Z., Rašlová K., Gašparovič J., Slíž I. Genetic determinants of quantitative traits associated with cardiovascular disease risk. Mutat. Res. 2015;778:18–25. doi: 10.1016/j.mrfmmm.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 98.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease. Alzheimer’s Disease Genetic Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morris G.E., Braund P.S., Moore J.S., Samani N.J., Codd V., Webb T.R. Coronary Artery Disease-Associated LIPA Coding Variant rs1051338 Reduces Lysosomal Acid Lipase Levels and Activity in Lysosomes. Arterioscler. Thromb. Vasc. Biol. 2017;37:1050–1057. doi: 10.1161/ATVBAHA.116.308734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guénard F., Houde A., Bouchard L., Tchernof A., Deshaies Y., Biron S., Lescelleur O., Biertho L., Marceau S., Pérusse L., Vohl M.C. Association of LIPA gene polymorphisms with obesity-related metabolic complications among severely obese patients. Obesity (Silver Spring) 2012;20:2075–2082. doi: 10.1038/oby.2012.52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All HMDP microarray data are deposited in the NCBI Gene Expression Omnibus (GEO) under the accession number GSE66570. The promoter capture Hi-C and H3K27ac ChIP-seq experiments reported in this study are deposited in the GEO database under the accession number GSE157306. The published article includes all other analysis results generated during this study.