Summary
Whether or not populations diverge with respect to the genetic contribution to risk of specific complex diseases is relevant to understanding the evolution of susceptibility and origins of health disparities. Here, we describe a large-scale whole-genome sequencing study of inflammatory bowel disease encompassing 1,774 affected individuals and 1,644 healthy control Americans with African ancestry (African Americans). Although no new loci for inflammatory bowel disease are discovered at genome-wide significance levels, we identify numerous instances of differential effect sizes in combination with divergent allele frequencies. For example, the major effect at PTGER4 fine maps to a single credible interval of 22 SNPs corresponding to one of four independent associations at the locus in European ancestry individuals but with an elevated odds ratio for Crohn disease in African Americans. A rare variant aggregate analysis implicates Ca2+-binding neuro-immunomodulator CALB2 in ulcerative colitis. Highly significant overall overlap of common variant risk for inflammatory bowel disease susceptibility between individuals with African and European ancestries was observed, with 41 of 241 previously known lead variants replicated and overall correlations in effect sizes of 0.68 for combined inflammatory bowel disease. Nevertheless, subtle differences influence the performance of polygenic risk scores, and we show that ancestry-appropriate weights significantly improve polygenic prediction in the highest percentiles of risk. The median amount of variance explained per locus remains the same in African and European cohorts, providing evidence for compensation of effect sizes as allele frequencies diverge, as expected under a highly polygenic model of disease.
Keywords: whole-genome sequencing of African Americans, rare variants, polygenic risk scores, differential genetic architecture, CALB2, PTGER4, understudied populations, trans-ethnic comparative analysis
Introduction
The inflammatory bowel diseases (IBDs [MIM: 604519, 266600, 191390]), Crohn disease (CD [MIM: 266600]), and ulcerative colitis (UC [MIM: 191390]) arise in the context of inappropriate activation of the intestinal immune system in response to an environmental trigger in individuals who are genetically predisposed. Genome-wide association studies (GWASs) of common and low frequency variants have so far identified 241 loci that confer significant risk for disease susceptibility.1, 2, 3 Although inflammatory bowel disease is one of the most successfully studied polygenic diseases with respect to identifying loci and risk alleles, four major challenges remain: (1) missing heritability—only a small fraction of disease liability is explained by the thus far known genetic risk factors (13% for Crohn disease and 8% for ulcerative colitis);2 (2) uncertainty as to the true causal genetic variants underlying GWAS associations—as most loci span several kb to tens of kb in length, containing credible sets that usually range from several to hundreds of variants in tight linkage disequilibrium (LD) having similar evidence of association; (3) lack of molecular insights into established genetic signals—as most association signals map to non-coding regions with their mechanistic effects largely unknown; and (4) lack of understanding on the different genetic architectures across populations.
Genetic discoveries of inflammatory bowel disease have been made primarily in populations of European ancestry and utilizing genome-wide genotype data.1, 2, 3 This predominance, combined with a focus on common alleles, has left our understanding of the role of rare variants among non-European populations incomplete. To this end, we have performed the whole-genome sequencing study of inflammatory bowel disease-affected individuals as compared to non-inflammatory bowel disease control individuals from over 3,600 Americans with African ancestry. Although most GWASs are performed with genotyping arrays, whole-genome sequencing offers advantages such as assessment of rare heterozygous effects—for example, for type 2 diabetes4 (T2D [MIM: 125853]) and on blood metabolites5—and comprehensive assessment of non-imputed common variants (for example, for COPD6 [MIM: 606963]), as well as mapping in the presence of high variability and short LD blocks in mixed ancestry populations (for example, plasma lipoproteins7 and serum peptides8).
Our goals were 4-fold: first, we hypothesized that genetic analysis of this understudied population would facilitate new locus discovery of common variants that have not been previously interrogated, including those that are specific to African-ancestry populations or shared across divergent populations. Second, given the high genetic diversity in African populations, we hypothesized that rare and potentially high-risk inflammatory bowel disease variants, within or near protein coding genes, have yet to be identified. Third, we evaluated the potential of our genetically diverse African American cohort to enhance fine-mapping of credible intervals. Fourth, we evaluated the degree to which effect sizes of GWAS variants differ across populations and assessed the impact on polygenic risk assessment based on established inflammatory bowel disease loci.
Subjects and methods
Study samples
This was a multi-center collaborative study involving self-identified African American subjects recruited from five primary sites and their collaborating centers across the US. These sample recruitment centers include Emory University (recruited as part of the GENESIS study and Emory African American Inflammatory Bowel Disease Consortium) and 12 other collaborating centers; Johns Hopkins/Rutgers (recruited as part of the Multicenter African American Inflammatory Bowel Disease Study) and 17 other collaborating centers; Cedars-Sinai Medical Center; Mount Sinai Medical Center; and Washington University (recruited as part of the Centers for Common Disease Genomics network). Sample breakdown, along with the proportion of affected individuals versus control individuals, per center is shown in Table S1. This study was approved by the institutional review boards at each of the participating sites and informed consent was obtained from all the participants. Deidentified datasets including genetic data are housed at Emory University with the approval of the local ethical board.
Library construction and whole-genome sequencing
All DNA samples investigated in this study (a total of 3,610 before quality control [QC]) were processed and sequenced at the Broad Institute of Harvard and MIT (Cambridge, MA) following the same protocol. Briefly, genomic DNA (350 ng in 50 μL) extracted from the blood of sampled participants was fragmented to a target size of 385 bp fragments via a Covaris Focused-ultrasonicator. Fragmented DNA was subjected to further size selection via a SPRI cleanup. Libraries were then constructed with kits commercially available from KAPA Biosystems (KAPA Hyper Prep without amplification module, product KK8505) and with palindromic forked adapters with unique 8-base index sequences embedded within the adaptor (purchased from Roche). Completed libraries were quantified with quantitative PCR (kit purchased from KAPA Biosystems), normalized to 2.2 nM, and were pooled into 24-plexes. Sample pools were combined with HiSeqX Cluster Amp Reagents EPX1, EPX2, and EPX3, and cluster generation was performed with the Illumina cBot according to the manufacturer’s protocol (Illumina). DNA libraries were sequenced with the HiSeqX sequencing system utilizing sequencing-by-synthesis kits to produce 151 bp paired-end reads. Because library amplification by PCR introduces substantial bias, we sequenced our samples by using the PCR-free protocol. Output from Illumina software was processed by the Picard data-processing pipeline to yield CRAM files containing demultiplexed, aggregated aligned reads.
Data processing and variant calling
The sequence reads from each sample were aligned to human reference genome build hg38 (GRCh38 assembly). Each sample was sequenced to an average depth of 30×. Variants were joint called across 3,610 samples via the Genome Analysis Toolkit (GATK) Best Practices for germline variants9 and were annotated via our in-house Bystro10 software. Our initial output, VCF file, had data for 3,610 samples and about 145 million variants, both single nucleotide polymorphisms (SNPs) and short insertions and deletions (INDELs), which was then subjected to both sample and variant QC as outlined below.
Sample QC and filtering
Using a subset of ∼1.4 million LD-independent (r2 < 0.1), high frequency (minor allele frequency [MAF] > 1%) variants, we performed sample QC to identify individuals with discordant sex information, low call rates (<95%), and unexpected relatedness. Briefly, we tested for agreement between X/Y genotypes and ascertained sex by using PLINK11 and excluded samples with discordant gender from subsequent analysis. Per-sample missingness rate was calculated via the –missing option, and individuals with more than 5% missing genotypes were removed from further analysis. To identify duplicated or related individuals, by using a subset of ∼1.4 million LD-independent (r2 < 0.1), high frequency (MAF > 1%) variants, we first calculated identity by state (IBS) for each pair of individuals, and then we estimated the degree of recent shared ancestry for each pair of individuals (identity by descent [IBD]; whose values range from 0 to 1) again by using PLINK.11 For duplicated samples (IBD > 0.5), we removed one of the samples either with low quality metrics at any of the QC steps or at random when both samples were of high quality. For pairs of individuals that appeared to be genetically related—first-degree relatives (IBD = 0.5) or second-degree relatives (IBD = 0.25)—one individual from each pair was removed from subsequent analyses. Similarly, two individuals from parent-offspring trios were dropped. While removing related samples, preference was given to keep the affected individual or the sample with high overall quality. Our final analytical set only included samples where the maximum relatedness between any pair of individuals is less than a second-degree relative. Collectively, these QC procedures resulted in the removal of 35 samples with sex discrepancies, 42 samples with missing variant data, and 122 duplicated samples or related individuals.
In addition, using the observed genotypes across the entire genome, we estimated and removed samples with outlying (defined as ±3 standard deviations from the mean) heterozygotic/homozygotic changes (n = 44), theta (n = 12), exonic theta (n = 13), exonic transition/transversion (n = 1), silent/replacement (n = 1), silent transition/transversion (n = 1), and replacement transition/transversion (n = 1) ratios. We also removed three samples with missing phenotypes. Collectively, after all these sample QC procedures, a total of 192 unique samples were excluded from subsequent analyses.
Variant QC and filtering
After the removal of samples from above, we then filtered out variants genotyped in <95% of the samples (missingness > 5%) and those that showed a significant deviation from Hardy-Weinberg equilibrium in control individuals (p < 1 × 10−9). These procedures resulted in a final dataset of 3,418 samples and 93.4 million variants that include both SNPs and INDELs. In addition, we applied RepeatMasker12 to mask variants from repetitive and low complexity regions of the genome. For some common variant association analysis, we also included the pre-masked dataset in order to obtain summary statistics for some of the lead variants from the previously known disease loci (identified via populations of predominantly European ancestry) that were masked by the RepeatMasker program.
Principal-component analysis of sequence data
After excluding samples and variants with low quality, principal-component analysis of the whole-genome sequencing dataset was performed with EIGENSTRAT.13 Principal components were computed on the basis of a pruned version of the dataset consisting of ∼1.4 million LD-independent (r2 < 0.1), high frequency (MAF > 1%) variants. The first five principal components (based on the inspection of the scree plot) were included as covariates to control for population stratification within the whole-genomes dataset for all analyses (Figure S1).
Single-variant association testing of sequence data
We used a logistic regression model to test for association at each individual variant with the first five principal components of the genotype matrix included as covariates. Variants were tested separately for association with Crohn disease, ulcerative colitis, and inflammatory bowel disease (Crohn disease and ulcerative colitis together with inflammatory bowel disease-type unknown [IBDU]). We defined common variants as those that are present in at least 1% of the general African population from gnomAD and have an observed MAF > 1% in this dataset, yielding 14.9 million variants (pre-masked; ∼7 million variants in the post-masked dataset). The genomic control (λGC) values for these individual analyses of common variants ranged from 1.02 to 1.04, indicating little or no inflation or deflation due to population stratification. We defined rare variants as those that are either absent or present at an MAF of < 0.1% in the general African population from gnomAD. With these criteria, we observed 64.2 million rare variants in our dataset, and the genomic control (λGC) values for these individual analyses of rare variants ranged from 0.61 to 0.84, indicating deflation due to the limited number of rare alleles in the dataset.
Known association in ADCY7
Adenylate cyclase 7 (ADCY7 [MIM: 600385]) has recently been implicated in ulcerative colitis in European populations; using a low-pass whole-genome sequenced case-control cohort in conjunction with well-imputed newly genotyped and pre-existing GWAS case-control datasets of European ancestry and affected individuals and non-inflammatory bowel disease control individuals from the UK Biobank, Luo et al.14 identified a low-frequency (MAF = 0.006), missense variant, rs78534766, in ADCY7 in association with an increased risk of ulcerative colitis. In fact, rs78534766 is the largest effect allele that was identified to date for ulcerative colitis (odds ratio [OR] = 2.19; p = 9 × 10−12). However, although there is no evidence of association for ulcerative colitis in our study, we noticed an effect trending in the opposite direction (OR = 0.46; p = 0.47). The effect allele was seen in seven control individuals (MAF = 0.002) and one affected individual with ulcerative colitis (and four affected individuals with Crohn disease) in our whole-genome sequencing dataset: this could in part be due to the limited sample size of our dataset, fueled by the rarity of this allele, rs78534766, in general African populations (MAF in gnomAD v3 = 0.00098).
Power analysis
For power calculations, we assumed that disease prevalence (0.19% for Crohn disease and 0.20% for ulcerative colitis) and effect sizes are homogeneous across ancestries. We converted population frequencies (reported in African populations from gnomAD) and odds ratios (reported in the latest meta-analysis of European descent individuals3) to affected and control individual frequencies and calculated power of our whole-genomes cohort to detect previously known disease loci at p < 5 × 10−8 or p < 0.05 as a function of the MAF and the genotype relative risk of the variant under an additive model.
Aggregate rare-variant association testing
Using the optimal sequence kernel association test (SKAT-O),15 we performed gene-wide aggregation analyses to detect aggregate association of rare, likely deleterious (combined annotation dependent depletion (CADD) > 15) variants with the three traits. For aggregate tests, we selected all rare, likely deleterious (CADD > 15) variants (n = 1.5 million) across the genome and assigned them to the nearest gene. We then assessed the association of each gene with a collection of rare, likely deleterious variants in a SKAT-O model implemented in the R package “SKAT.” To interpret statistical significance, we applied experimental-wide, Bonferroni-corrected significance threshold of p < 2.2 × 10−6 = 0.05/22,521.
GWAS genotype data, QC, imputation, and association testing
Sample information, genotype data, and the application of QC procedures for the two existing GWAS cohorts considered in the current study were described extensively elsewhere.16 Briefly, genome-wide genotype data from non-overlapping African American affected individuals and matched control individuals generated with either the Illumina Omni (398 with Crohn disease, 238 with ulcerative colitis, and 1,551 control individuals) or the Affymetrix Axiom Genome-Wide AFR 1 World Array (451 with Crohn disease, 186 with ulcerative colitis, and 3,038 control individuals) SNP chips were considered for replicative evidence. Sample and variant QC, determination of principal components, and removal of outliers was done as described in the original paper.16 Both datasets were lifted from human reference build hg19 to hg38 via liftOver.17
Imputation
We phased the whole-genome sequences described above (n = 3,418; after QC) with Eagle v2.418 to create a reference panel. These pre-phased whole-genome sequences with MAF > 0.5% were imputed into each GWAS dataset, separately, via minimac3 software.19 By design, all the sequenced individuals have African descent and about half of these are inflammatory bowel disease-affected individuals, thereby enriching the reference panel for African-specific alleles that increase or decrease inflammatory bowel disease risk.
Common variant association testing for replicative evidence
After removing samples that were directly sequenced in the discovery phase, genotyped and imputed variants with INFO score > 0.6 were tested for association with Crohn disease, ulcerative colitis, and inflammatory bowel disease, separately, within each GWAS case-control dataset via SNPTEST 2.5.2,20 performing an additive frequentist association test conditioned on the first ten principal components. For sites that were present in both datasets and passed our QC filters, we performed meta-analysis by using META.21, 22, 23 For a common variant with p < 5 × 10−8 in the discovery cohort to be inferred to be associated with a trait, it had to have a directionally consistent effect and demonstrate at least a nominal evidence of association (p < 0.05) in the meta-analysis of the two GWAS datasets. At the 5p13.1 locus near Prostaglandin E Receptor 4 (PTGER4 [MIM: 601586]), the previously defined peak SNP rs7711427 was initially relegated to a “sub-PASS” tranche during GATK’s automated analysis, but closer inspection revealed that the reason for exclusion was an excess of heterozygotes in affected individuals. As this is a well-characterized disease-associated variant and strong disease alleles will give rise to excess heterozygosity in affected individuals, we elected to “rescue” this allele. Because this site is in complete LD (r2 = 1) with several other sites, no conclusions are altered, but we retain rs7711427 to facilitate comparison with published results.
Genetic risk score calculation
Assessment of precision (equivalent to predicted prevalence in a percentile of risk) is sensitive to the ratio of affected individuals to control individuals, so it cannot be conducted directly on the whole-genome sequencing cohort. In order to directly contrast accuracy in a population with a maximal inflammatory bowel disease prevalence of ∼1%, we thus generated 150,000 pseudo-controls given expected genotype frequencies observed in our 1,644 African American whole-genome sequenced control individuals. Rather than using an external reference database of allele frequencies in African Americans, since these are known to be heterogeneous due to varying degrees of admixture, we used the control estimates from our whole-genome sequencing for internal consistency and simply generated random genotypes according to Hardy-Weinberg expectations. These were computed for 215 of the 241 known loci that were present in both the African American pseudo-controls cohort and UK Biobank. In the pseudo-controls, we combined genotypes between loci independently to minimize LD within the pseudo-control population. Note that we only use the lead SNPs and, as a result, make no attempt to capture independent secondary signals at each locus24 or to disentangle the combined effects of multiple interacting sites in high LD.25 Instead, we simply acknowledge that some of the observed differential effect sizes may be due to the combined influence of secondary sites in LD or epistasis. By ignoring neighboring sites with main effects, we are certainly under-representing the total amount of variance explained per locus25, 26, 27, 28 but eliminating any influence of these effects on the derivation of pseudo-controls. In the absence of a very large external dataset of African ancestry inflammatory bowel disease genotypes, this is currently the least-biased approach to estimating per-locus effects in polygenic risk score (PRS) estimation, but it should be recognized that there may be other systemic biases to the use of pseudo-controls that contribute to the ancestry-specific assessments. To confirm that long-range LD is also not biasing the analysis, we computed the pairwise matrix of LD among all lead SNPs in our whole-genome sequenced “real” control individuals. Reflecting the fact that the SNPs are dispersed across the genome, only 6 of 23,005 comparisons were significant at the Bonferroni adjusted threshold of 2 × 10−6 in this observed dataset, in each case generating squared genotype correlations (r2) between 2% and 7%, with an overall average r2 of just 0.07%. By design, there was no observed significant LD among the variants in the pseudo-controls. Consequently, bias introduced by LD among the lead SNPs appears to be measurably small in the African American pseudo-controls dataset, which has 1,774 inflammatory bowel disease affected individuals and 150,000 pseudo-controls. We similarly generated a UK Biobank dataset of the same size by down-sampling to the same case-control ratio; in this case, actual genotypes were used because UK Biobank is sufficiently large.
Next, to compute the genetic risk score, we divided each of the five sets of African American pseudo-controls and UK Biobank into a training set with 70% samples (1,242 affected individuals, 105,000 pseudo-controls) and test sets with the remaining 30% samples (532 affected individuals, 45,000 pseudo-controls). We used the 70% training datasets to estimate effect sizes both on the lnOR scale as well as the liability scale (these data were also used to estimate proportion of variance explained [PVE]). Using these estimated effect sizes at the most significant variants in the 215 of the 241 known loci that were present in both the African American pseudo-controls cohort and UK Biobank, we derived weighted PRSs in the withheld 30% datasets. Furthermore, for each PRS, the prevalence of inflammatory bowel disease, with standard error determined from the five test sets, was computed at each percentile bin on both the liability scale and lnOR scale. The proportion of variance explained by PRS was computed by determining the Negelkerke’s R-square via the “lrm” function from the “rms” package in R.29
Liability scale modeling
Liability effects were measured by assuming an underlying normal distribution (mean 0, variance 1) of liability representing disease status. Affected individuals are assumed to lie above a certain threshold of this continuous distribution. This threshold (Z score on x axis) is determined by computing the inverse of the cumulative normal function of the assumed prevalence of the disease. Assuming an upper limit for the prevalence of all inflammatory bowel diseases as 1%, odds ratios determined from logistic regression as the true odds ratios, and using the allele frequencies in control individuals within the study, we determined the additive effects on liability as follows:30,31 the displacement of each genotype threshold from the overall population threshold as [2.326-norminv(1-penetrance)] where the penetrance of the genotype is the overall prevalence (1%) multiplied by the ratio of the genotype between affected individuals and overall. The mean displacement was computed as the sum of the products of the three genotype displacements weighted by the genotype frequencies in the overall sample, which allows computation of the central displacement of each genotype by subtraction of the mean displacement. The liability alpha1 of the larger effect genotype is then [p∗central displacement_AA+q∗central displacement_Aa] where p is the frequency of the minor allele A and q is the frequency of the major allele a. Similarly, the liability alpha2 of the alternate allele is [p∗central displacement_Aa+q∗central displacement_aa]. The total variance explained by the locus on the liability scale is 2∗p∗alpha12 +2∗q∗alpha22. The polygenic risk score for an individual is the sum of the liabilities of the relevant genotypes, namely 2∗alpha1, alpha1+alpha2, or 2∗alpha2.
Results
Common variant associations in African Americans
After QC (see subjects and methods) and principal-component analysis (Figure S1) of our deeply sequenced whole genomes (median coverage of 30×), we present analyses of a total of 3,418 subjects: 1,774 affected individuals (1,335 with Crohn disease, 407 with ulcerative colitis, and 32 with IBDU) and 1,644 non-inflammatory bowel disease control individuals (Table S1) at 93 million variants that comprise both SNPs and short INDELs. These data include 14.9 million common variants (MAF > 1%), 13.9 million low-frequency variants (0.1% < MAF < 1%), and 64.2 million rare variants (MAF < 0.1%). First, we used single-variant analyses to test each variant, regardless of allele frequency, for association with Crohn disease, ulcerative colitis, and inflammatory bowel disease (Crohn disease and ulcerative colitis together with IBDU), separately, in a logistic regression framework conditioned on the first five principal components (see subjects and methods). Following these “discovery” analyses of whole-genome sequence data, we sought replication of the obtained results at common variants in an independent cohort of African American affected individuals and control individuals that were previously genotyped with Axiom or Omni genome-wide SNP arrays16 (subjects and methods; Table S1). We imputed our whole-genome sequences into these two (Axiom and Omni) existing GWAS datasets, thereby enriching the panel for inflammatory bowel disease risk alleles, and performed case-control association testing by using a logistic regression model, separately, within each dataset. Results from the meta-analysis of these two GWAS datasets served as our replicative evidence for common variation.
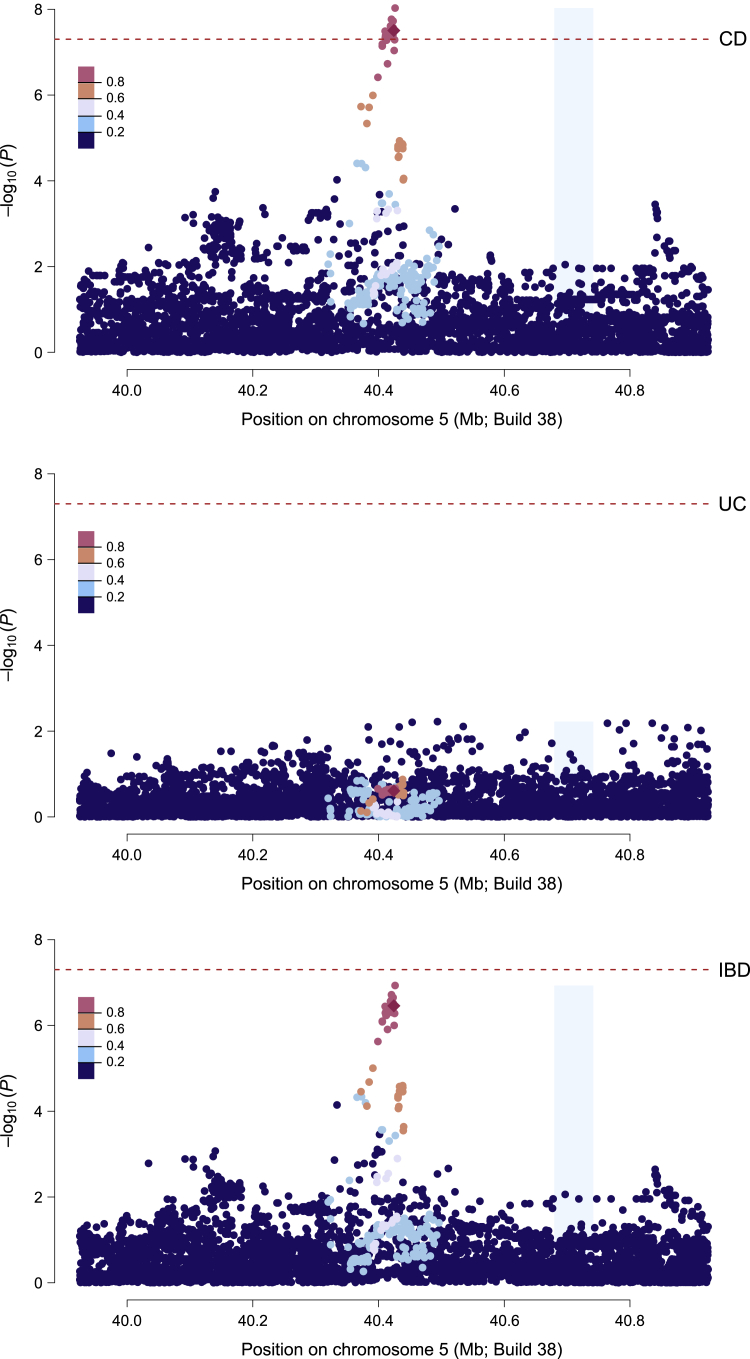
With a standard GWAS significance threshold of p < 5 × 10−8 in the discovery cohort and at least nominal (and directionally consistent) evidence of association (p < 0.05) in the replication cohort, we identified 22 common variants at a locus proximal to PTGER4 (260 kb) on chromosome 5p, previously discovered in cohorts of European descent, associated with a decreased risk of Crohn disease (Figure 1 and Table S2). All 22 variants were in complete LD with each other (r2 = 1). Following our previous report of suggestive evidence of association at this locus for Crohn disease in African Americans,16 here we present the first evidence of standard (GWAS) genome-wide significance. Consistent with being a Crohn disease-specific locus, the disease-associated alleles demonstrated strong effect sizes for Crohn disease (OR = 0.74; p < 5 × 10−8) but not ulcerative colitis (OR = 0.91; p = 0.25; Figure 1) despite being directionally consistent to both. Further, of all the ancestrally divergent populations studied thus far, the strongest effect on Crohn disease at these variants was observed in our African American cohort (Figure S2).
Figure 1.
Regional plots of the PTGER4 region in the discovery whole-genome sequence data
Crohn disease (top); ulcerative colitis (middle); inflammatory bowel disease (bottom). Dashed red lines indicate genome-wide significance (p = 5 × 10−8). Variants are color coded to show linkage disequilibrium structure relative to a variant, rs6896969, that showed the strongest association with Crohn disease in African Americans in our previous GWAS.16
A protective role in Crohn disease of PTGER4 locus minor alleles was initially discovered in populations of European ancestry,32 and 2,819 common variants at the locus had genome-wide significant association. Using conditional stepwise regression to perform fine-mapping, Huang et al. further refined this region to a subset of 189 credible variants representing four independent signals that are more likely to be causal to Crohn disease24 (Figure S3). The 22-variant African American signal precisely captures the known primary peak (signal 1) of association in European populations (MAF in African American Crohn disease-affected individuals = 0.32 and control individuals = 0.39; MAF in European Crohn disease-affected individuals = 0.33 and control individuals = 0.39); there is no evidence for an association at signal 2 but nominal evidence at signals 3 and 4. The 22 Crohn disease-associated variants that we detected at this locus in African Americans were in high LD with the strongest signal (signal 1) comprising two potentially causal variants—rs7711427 and rs397897680—from the fine-mapping analysis of European populations32 (Figure S4). We note that rs397897680 has since been merged with rs5867512, while rs7711427 was excluded during our initial QC procedure (see subjects and methods). Despite a prior report30 of an expression quantitative trait effect on PTGER4 expression in lymphoblastoid cells in European individuals, the African American disease-associated alleles appear to be located distal to that signal as well as the major eQTL interval reported on the Blood eQTL browser. Experimental manipulation of epithelial cells supports a role for prostaglandin signaling promoting intestinal wound healing during inflammatory bowel disease,33 but it remains to be established whether PTGER4 is the target of the primary GWAS signal that lies in a gene desert 250 kb from a cluster of candidate genes and whether expression is affected in diseased epithelial cells.
Rare-variant associations in African Americans
With the sequencing data, we next assessed the contribution of rare variants (MAF < 0.1%) to inflammatory bowel disease. Our data was comprised of 64.2 million rare variants that include many alleles that were not genotyped or imputed in previous GWASs of inflammatory bowel diseases. Because our single-variant analyses at rare variants yielded deflated summary statistics (Figure S5), we performed aggregate analyses by selecting all rare, likely deleterious (CADD > 15) variants across the genome and assigning them to the nearest gene. In total, 1.5 million such variants were assigned to 22,521 genes with an average of 68 variants per gene (range = 1–3,593). Using the SKAT-O approach,15 we then tested whether any of these gene sets with a collection of rare, likely deleterious variants have an aggregate association with inflammatory bowel disease, Crohn disease, or ulcerative colitis. To interpret statistical significance, we applied a Bonferroni-corrected significance threshold of pSKAT < 2.2 × 10−6 (0.05 corrected for 22,521 tests).
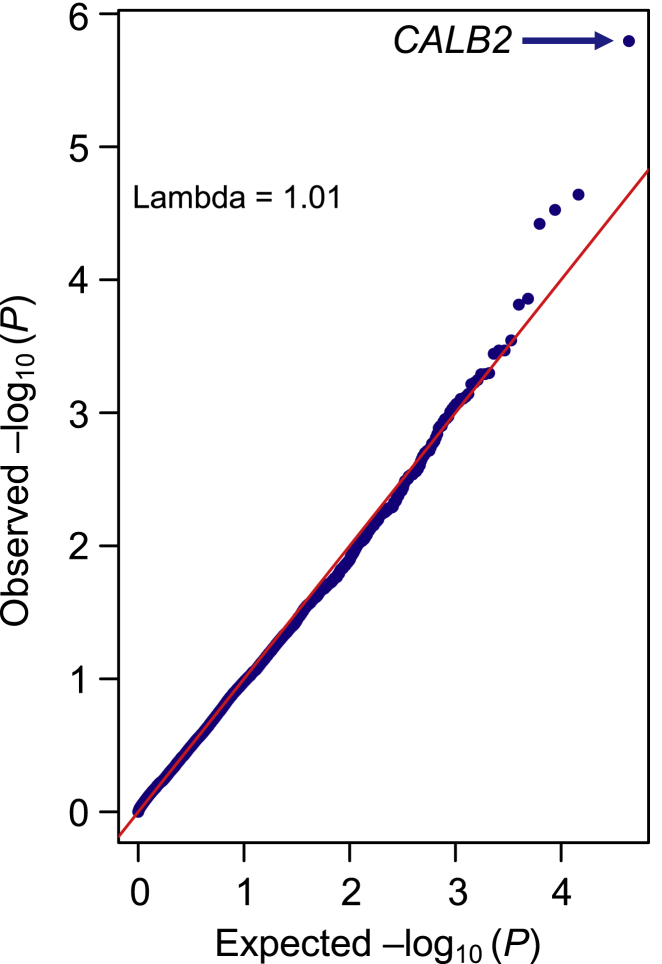
Using this strategy, we implicate variants in the vicinity of Calbindin 2 (CALB2 [MIM: 114051]) in ulcerative colitis. We detected an aggregate association of 35 rare, likely deleterious, heterozygous variants within or near CALB2 with ulcerative colitis (pSKAT = 1.61 × 10−6; Figure 2 and Table S3). Half of these variants were observed more frequently in affected individuals with ulcerative colitis compared to control individuals, while the other half were seen less frequently, representing a typical SKAT type of signal. Among the 35 variants that collectively contributed to this aggregate signal, many are absent in European (non-Finnish) populations in the gnomAD v3 release34 (15 as opposed to only 4 variants that were never seen before in African populations) despite a 0.6-fold larger European effective sample size, including an African-specific intronic variant, rs200083611, with a nominal evidence of association for increased risk of ulcerative colitis, showing an MAF of 0.009 in affected individuals and 0.0003 in control individuals (p = 0.001; OR = 30.5 from single variant association analysis). However, given the high-risk but weak evidence of association at rs200083611, it appears that the CALB2 gene-wide signal was driven by multiple additional rare variants.
Figure 2.
QQ plot of rare-variant gene-level association analysis of ulcerative colitis
Observed negative log p values are from the SKAT-O test.15
This CALB2 signal was approximately 3 Mb away from, and independent of, the nearby common variant, rs1728785 (intronic region of ZFP90 [MIM: 609451]), that has an established association for ulcerative colitis,2,14,35 indicating that these rare variant associations represent unique effects. CALB2 encodes an intracellular calcium-binding protein, calbindin 2 (also known as calretinin), that plays an important role in neuronal physiology and the maintenance of Ca2+ intracellular homeostasis. CALB2 has a common expression pattern in both the central and peripheral nervous systems. It has high expression in brain and intermediate expression in sigmoid and transverse colon. The absence of CALB2 in nerve fibers in colon is a widely used marker for Hirschsprung disease36,37 (HSCR1 [MIM: 142623]), whereas elevated expression of CALB2 has been reported as a hallmark of rapidly proliferating cancerous cell lines, including in colorectal cancer cell lines.38 Hirschsprung disease shares many of the clinical features with inflammatory bowel disease, where the latter is more commonly reported in affected individuals who had surgical treatment for Hirschsprung disease.39 Conversely, long-standing inflammatory bowel disease is an established risk factor for colorectal cancer.40,41 Given the intricate relationship of inflammatory bowel disease with these companion diseases, our implication of Hirschsprung disease- and colorectal cancer-associated CALB2 in ulcerative colitis makes this signal worthy of experimental validation.
Genetic landscape at the established disease loci between African Americans and Europeans
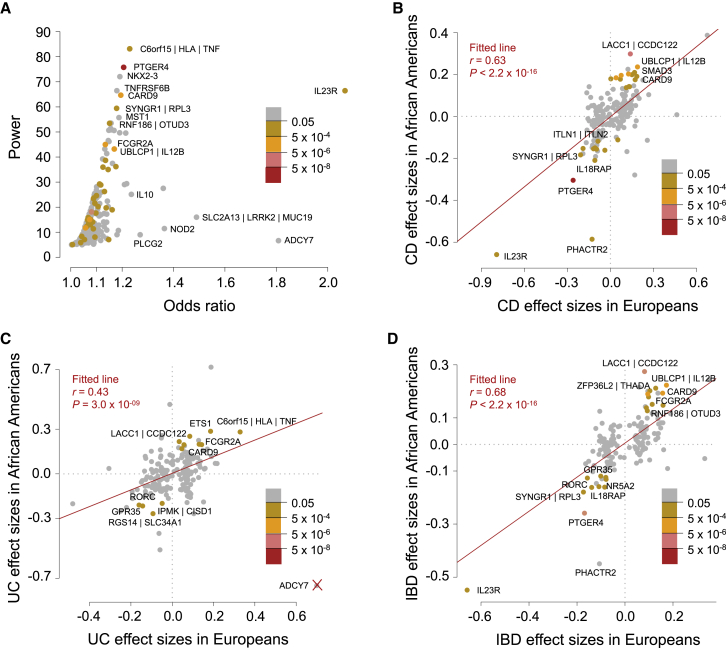
With our whole-genomes data, we next assessed whether the genetic landscape at established inflammatory bowel disease risk loci is shared between populations of European and African descent and whether trans-ethnic comparative analysis can be leveraged to further refine established GWAS signals. Of the 241 lead variants from the thus far established loci from the recent meta-analyses of cohorts of European descent3 (GWAS catalog), in addition to replicating the known PTGER4 locus at p < 5 × 10−8, we replicated 41 lead variants for association with inflammatory bowel disease, Crohn disease, or ulcerative colitis (see Table S4 for all lead variant data). Assuming Crohn disease and ulcerative colitis prevalence of 0.19% and 0.20%, respectively, and no ascertainment bias, our whole-genomes cohort was powered to replicate 42 of the known loci at p < 0.05 (see subjects and methods; Figure 3), matching the number of associations actually observed. Despite limited statistical evidence of association at many of the known loci as a result of the size of our African American cohort, we noted a strong concordance in the direction of effects (73%, p = 2.05 × 10−12 for inflammatory bowel disease; 69%, p = 1.26 × 10−8 for Crohn disease; and 64%, p = 2.48 × 10−5 for ulcerative colitis; sign test) and a strikingly positive correlation in effect sizes between European and African populations (R = 0.68, p < 2.2 × 10−16 for inflammatory bowel disease; R = 0.63, p < 2.2 × 10−16 for Crohn disease; and R = 0.43, p = 3.0 × 10−9 for ulcerative colitis), further supporting the previous notion that the genetic risk of inflammatory bowel diseases conferred by common variants is, to a great extent, shared across divergent populations (Figure 3).
Figure 3.
Comparison of effect sizes across populations
(A) Statistical power of the discovery whole-genome sequencing dataset to replicate previously known variants at p < 0.05. The known variants that showed an association (p < 0.05) with Crohn disease, ulcerative colitis, or inflammatory bowel disease in African Americans are colored to denote their p value of association.
(B–D) Comparison of effect sizes at known variants in African Americans and Europeans. Each variant is colored to denote the p value of association for Crohn disease (B), ulcerative colitis (C), or inflammatory bowel disease (D) in African Americans from the discovery whole-genome sequencing cohort. The red line shows the linear regression fit to indicate the general trend. Significance and goodness-of-fit are shown. ADCY7 was not included while computing goodness-of-fit in (C) (see subjects and methods).
Despite strong effect-size correlations, subtle differences in allele frequency or the magnitude of effects at the established disease loci may reveal differential genetic architectures underlying inflammatory bowel disease between the two populations. Given that our African American dataset is at least one order of magnitude smaller than the existing European datasets, in order to contrast effect estimates directly between the two populations, and assuming a maximal underlying inflammatory bowel disease prevalence of up to ∼1%, we generated 150,000 pseudo-controls from the observed genotypes in our whole-genome sequenced control individuals (n = 1,644) to contrast against the 1,774 inflammatory bowel disease-affected individuals. We refer to this from here on as African American pseudo-controls cohort (with 1,774 affected individuals and 150,000 pseudo-controls). For the European counterpart, we took advantage of the UK Biobank (all the observed genotypes were used because the UK Biobank is sufficiently large, 1,774 individuals with inflammatory bowel disease and 150,000 non-inflammatory bowel disease control individuals; see subjects and methods). For both the African American pseudo-controls cohort and UK Biobank, we generated five random subsets retaining 70% of the samples (1,242 affected individuals and 105,000 pseudo-controls) as discovery sets for estimating effect sizes. In a comparative analysis of effects (natural log transformed odds ratios [lnOR]) at the known loci, 69% of the African American estimates are within the 95% confidence interval of UK Biobank estimates (and 75% vice versa), implying general similarity of odds ratios. The distributions of the lnOR are significantly biased toward higher absolute values in African Americans (p = 0.01, two-tailed paired sample t test, n = 215 SNPs) and are highly correlated with those generated directly from the whole-genome sequencing study and shown in Figure 3 (mean r = 0.88). Scores generated directly from the discovery cohort may also bias the estimation (although note that we are only considering lead SNP effects previously identified in predominantly European ancestry studies), but considered together, the two approaches increase the robustness of our findings. The correlation between the odds ratio estimates from the de Lange et al. meta-analysis3 and the inflammatory bowel disease estimates in five UK Biobank iterations is just 0.77 on average, which is greater than those estimated between the African American pseudo-controls and UK Biobank (0.48) or the direct African American whole-genome sequencing GWAS and European meta-analysis (0.68). Thus, generation of pseudo-controls introduces minimal bias.
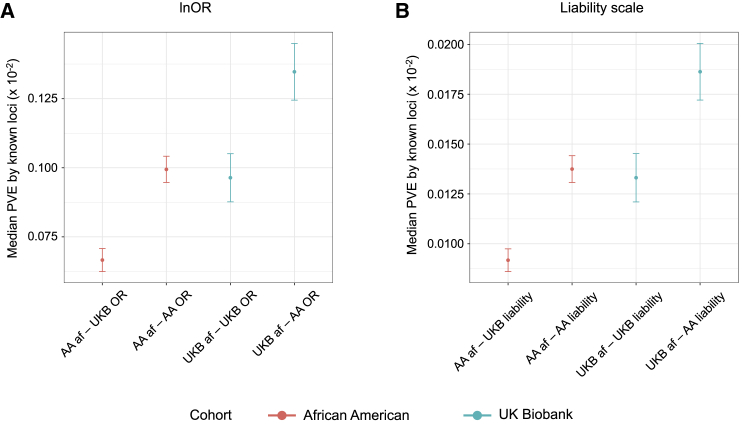
Since allele frequencies co-vary with odds ratios, explaining approximately 6.1% (mean R2 across five sets) of the difference in odds ratio between the populations (Figure S6), we also computed effect sizes on the liability scale (see subjects and methods), which is not a function of allele frequency. These estimates are also biased slightly upward in African Americans (mean R2 = 3.1%; Figure S6). The median percent variance explained per locus is very similar whether on the lnOR or liability scales in the two populations (Figure 4). Nevertheless, we estimate that given either the European or African allele frequencies, typically 40%–50% more variance would be explained with the observed African American effect sizes. Reciprocally, for either distribution of effect sizes, the European allele frequencies lead to 1.35–1.45 times greater variance explained. Collectively, these observations suggest that compensatory differences in both effect sizes and allele frequencies contribute to divergence in inflammatory bowel disease risk between European and African populations. The effect size differences may be due to local ancestry effects of LD with secondary associations at loci and to global interactions with the total genetic background.
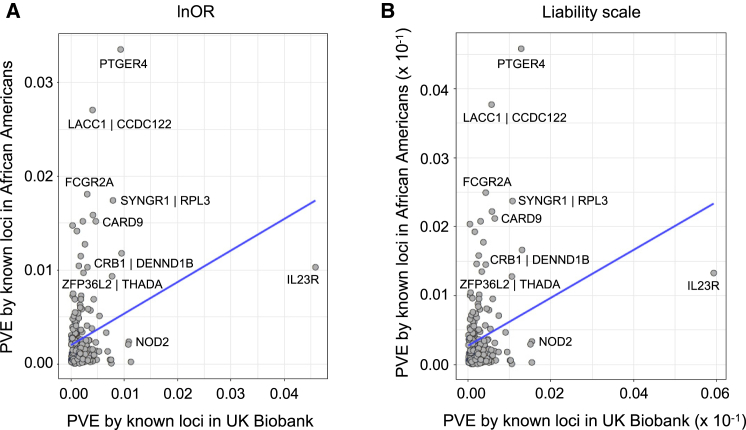
Figure 4.
Median proportion of variance explained by inflammatory bowel disease loci in African Americans and Europeans via effects estimated from the two populations
(A and B) Median proportion of variance explained (PVE) by the known loci (n = 215 SNPs) assessed via allele frequencies and effect sizes estimated in African American pseudo-controls and UK Biobank datasets measured on lnOR and liability scales across five discovery sets. PVE is computed as 2p.q.ln(OR)2 on lnOR scale (A); 2.p.alpha12 + 2.q.alpha22, where alpha1 and alpha2 are liability estimates on liability scale (B).
Variance in disease liability in African Americans and Europeans
For 215 of the 241 known loci that we had data for in both the African American pseudo-controls cohort and UK Biobank, we estimated the mean PVE by each locus from five discovery subsets in both lnOR and liability scale, and this is illustrated in Figure 5. While the two large effect European Crohn disease-associated alleles at IL23R and NOD2 explained 4.58% and 1.08% PVE on lnOR scale (0.59% and 0.15% on liability scale) in the UK Biobank cohort, they only accounted for 1.02% and 0.24% (0.13% and 0.03% on liability scale) in the African American pseudo-controls cohort, respectively. Conversely, the risk allele at rs3764147 (genes in the region include LACC1 [MIM: 613409] and CCDC122 [MIM: 613408]) that depicted suggestive evidence of association (p = 1.2 × 10−7) in our discovery African American cohort accounted for 2.71% PVE in the African American pseudo-controls cohort, while it only explained 0.40% in Europeans (0.37% and 0.05% on liability scale, respectively; Figure 5). Similarly, the PTGER4 signal that we detected at p < 5 × 10−8 in our whole-genome sequencing analysis explained 3.35% of PVE in African Americans versus 0.93% in Europeans (0.45% and 0.12% on liability scale, respectively).
Figure 5.
Comparison of proportion of variance explained (PVE) per locus in African Americans and Europeans at known loci (n = 215 SNPs) via effects estimated on the lnOR and liability scales
(A) PVE is computed as 2p.q.ln(OR)2.
(B) PVE is computed as 2.p.alpha12 + 2.q.alpha22, where alpha1 and alpha2 are liability scale estimates.
Transferability of polygenic risk scores across populations
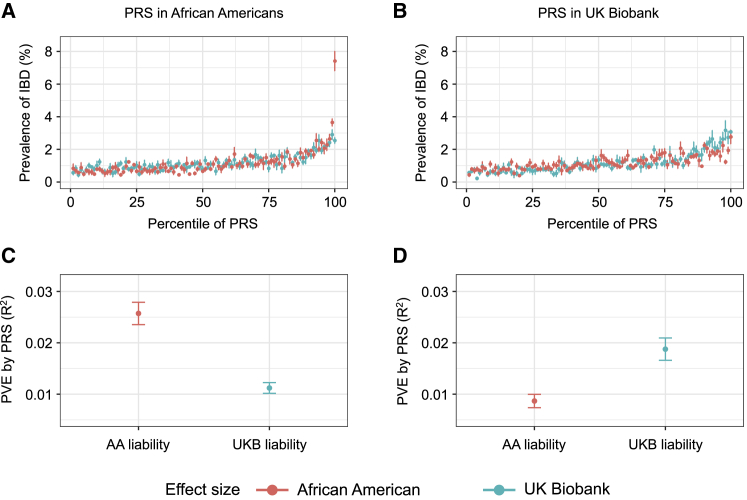
These subtle differences at each individual locus when combined across a genome-wide feature set, for example while deriving polygenic risk predictors for a common complex disease, lead to significant biases in populations distinct from the discovery samples.42,43 In order to directly contrast accuracy in a population with inflammatory bowel disease prevalence of up to ∼1%, we utilized the effects estimated from our five discovery sets generated from 70% of 1,774 affected individuals and 150,000 pseudo-controls and used the remaining 30% of the withheld samples from each of the five sets for computing PRSs at 215 of the 241 known loci that we had data for in both the African American pseudo-controls cohort and UK Biobank. Strikingly, after 5-fold cross validation, the African American-estimated betas gave a 7-fold elevation of the top percentile in African Americans but underestimated risk in the UK Biobank. Similar results were observed with liability scale estimates (Figure 6) and lnOR’s (Figure S7). On the other hand, the UK Biobank-estimated betas applied to UK Biobank samples yielded 3-fold elevation in the top percentile and performed better than the African American beta estimates applied to the UK Biobank. Specifically, for PRSs computed in African Americans, the prevalence in the top percentile was 7.4% (mean PVE by PRS from five test sets: R2 = 0.026) with African American summary statistics, 2.5% (mean PVE: R2 = 0.011) with UK Biobank summary statistics, and 2.9% (mean PVE: R2 = 0.010) with summary statistics obtained from the largest GWAS meta-analysis of European individuals.3 For PRSs computed in UK Biobank individuals, the prevalence in the top percentile was 2.8% (mean PVE: R2 = 0.009) with African American summary statistics, 3.0% (mean PVE: R2 = 0.019) with UK Biobank summary statistics, and 3.8% (mean R2 = 0.027) with summary statistics from GWAS meta-analysis of European individuals (Figure S7). Although there may be systemic biases in the use of pseudo-controls (see subjects and methods), these results confirm and extend recent observations concerning the propensity for PRS distributions to differ between Europeans and Africans based on discovery ancestry group.42,44,45
Figure 6.
Polygenic risk scores (PRSs) in African Americans and Europeans as a function of different discovery ancestry groups
PRSs in African Americans and UK Biobank individuals derived via 215 of the known disease SNPs and liability scale effects estimated from the African American discovery cohort (AA_liability, red) or the UK Biobank discovery cohort (UKBB_liability, green).
(A and B) Prevalence of IBD (%) versus percentile of PRS in each cohort.
(C and D) Mean proportion of variance explained (PVE) by each PRS. Standard error bars are from five test sets.
In order to further validate these conclusions, we also stress-tested the PRS estimation by a series of perturbations summarized in Figure S8, all of which retain the core result that using African American-estimated weights improves PRS discrimination. Substituting lnOR estimated directly from the whole-genome sequencing (Figures S8A–S8D), the overall PVE actually increases to 4.5% in the African ancestry samples, although the prevalence in the upper percentile is reduced a percentage point, while there is little impact on the European ancestry evaluation. Substitution of European meta-analysis effect-size estimates for ones generated from the UK Biobank also improves the PVE in Europeans (but not Africans), most likely because the down-sampling procedure—like the pseudo-control generation—uses a reduced sample size in the estimation. Furthermore, we removed various large-effect SNPs to evaluate whether they may be driving the improved performance. Figures S8E and S8F show that removing four variants with MAF < 0.02, three with OR > 1.2, has just a small impact on the prevalence-risk score relationship, as does removing the five variants with the largest absolute value of the OR, three of which have protective minor alleles.
Potential of population diversity to improve fine-mapping
Analysis of genetic data from divergent populations is thought to improve fine-mapping resolution of causal variants by leveraging trans-ethnic differences in LD, effect sizes, and allele frequencies. The fixed-effects meta-analysis of our African American whole-genome sequence data with summary statistics from European individuals2 at the previously fine-mapped (to 95% credible sets) loci31 showed marked improvement in fine-mapping resolution. Of the 3,529 credible variants from 94 loci that we meta-analyzed, 552 variants (16%) from 40 signals showed a change in p value of association with the previously assigned phenotype (inflammatory bowel disease, Crohn disease, or ulcerative colitis) by a factor of at least 10 (up to 100,000; Table S5). Of these, we noted an improvement in the strength of the association for 320 variants, while weaker association was observed for 232 variants. For instance, of the three credible variants that constituted a 95% credible set near Interleukin 23 Receptor (IL23R [MIM: 607562]; HD 7, signal 1 in Libioulle et al.32), we noted an improvement in association at the variant with the highest posterior probability (PP) of being causal (PP = 0.49). Conversely, a weaker association was observed for one (PP = 0.26) of the three 95% credible set-variants near SBNO2 (MIM: 615729). Similarly, 98 of the analyzed 120 credible variants near IRGM (MIM: 608212) (HD 67, signal 1, 95% credible size = 145 variants) and 102 (including a variant with PP = 0.58) of the analyzed 355 credible variants near DOCK3 (MIM: 603123) (HD 50, signal 1, 95% credible size = 437 variants) demonstrated weaker evidence of association, supporting the potential of trans-ethnic meta-analysis as a means to further improve the resolution of credible sets that had been constructed with a homogeneous population.
Discussion
To further resolve the genetic architecture of inflammatory bowel disease and better define the differential genetic structure of the disease across divergent ancestries, we have performed the whole-genome sequencing analyses that include many alleles that were not previously examined in a population that remains very significantly understudied. Similar to the findings of a whole-genome sequencing association study of inflammatory bowel disease in 4,280 European affected individuals,14 we find little evidence for large effect rare variants explaining much of the heritability. We did observe an aggregate association of rare, likely deleterious variants at CALB2 with ulcerative colitis, and many variants were specific to African populations (absent in European [non-Finnish] populations in gnomAD). However, though it exceeded exome-wide significance, this finding needs to be interpreted with caution because of the lack of replication in an independent cohort or functional validation. Our study assesses rare variant contributions to inflammatory bowel diseases in African Americans and will need evidence from future studies to further support the association at CALB2 with ulcerative colitis. Nevertheless, our study highlights that multiple rare variants with small to moderate effects exist, and at least, when clustered in a small number of sets (genes, windows etc.), are likely to account for some of the missing heritability. However, much larger-scale, deep-sequencing studies will be needed to precisely estimate the variance in disease liability explained by such variants.
Besides providing further evidence for the emerging notion that the overall genetic risk of inflammatory bowel disease conferred by common alleles is shared across populations, our data highlight the impact that subtle differences in the effect size and/or allele frequency can have on the phenotype and the likelihood that rare variant contributions exert population-specific effects. Our effect size estimates were based on univariate associations at lead SNPs, an approach that most likely under-represents the variance explained at each locus because it does not capture the effect of secondary associations that are modeled by methods such as LD score regression.25 Since the apparent proportion of the variance explained by a particular SNP is the sum of the effects of that SNP plus those in LD (weighted by the extent of LD captured by the squared genotype correlation), local differences in LD and rare variant contributions will contribute to observed effect size differences. However, interactions with the genetic background as well as environmental/cultural differences, including access to healthcare and the age distribution of disease, all may also subtly contribute to variable allelic effects across populations. From the point of view of using PRSs in practice, whether the cause of the difference is due to main effects at the lead SNP or combined effects of LD at neighboring SNPs is largely immaterial. It remains to be seen whether such population-specific genetic contributions may provide insights into differences in diseases incidence and progression across populations.
Our analyses also provide an example of how polygenic analysis needs to be adjusted for ancestry when considering ethnic disparities in health care. It is well established that frequency distributions of PRSs can differ markedly across populations, mostly because of deviations in allele frequencies,42, 43, 44 although ascertainment biases in discovery of common variant associations are also a concern.45 Fine-scale mapping of diverse clinical, behavioral, and hematological traits in large multi-ethnic cohorts such as PAGE and BioME has been shown to identify new loci, resolve secondary signals, and quantify divergent allelic effects46,47 despite high overall levels of repeatability, consistent with our observations. Admixture mapping has also been used to refine PRS estimation, for example producing better discrimination in high risk percentiles for African Americans with multiple myeloma.48 New methods for incorporating such findings into polygenic prediction algorithms are rapidly emerging, notably those incorporating both local and global ancestry adjustments.49, 50, 51 Our findings similarly illustrate how observed risk distributions that differ across populations and as a function of the discovery ancestry group can be used to improve risk assessment. Optimal risk scores should combine genetic and clinical features and will require better estimates based on expanded sample collection within each ethnic background and in admixed populations.
It is not known to what extent the rank order of risk is conserved in trans-ethnic risk assessment: if it is, then a simple rescaling of the PRS distribution may suffice. However, we show, similar to a multi-ethnic comparison of diabetes genetic risk,52 that incorporation of ancestry-specific weights significantly increases the inferred risk at the upper tail of the polygenic risk distribution. This occurs despite overall conservation of the average variance explained per locus accounting for compensation of changes in allele frequency and allelic effect size. It is striking that most of the gains in risk assessment occur at the extreme tails of the PRS distribution. Our intuition about this is that for most people, increases in effects at some loci are offset by decreases at others, since the magnitude and sign of the effects are generally uncorrelated. However, in the top and bottom percentiles, there is an excess of alleles with the same sign of effect and necessarily those individuals share more genotypes, which establishes a large enough correlation of ancestry-specific estimates to enhance the PRS discrimination in the correct population and add to the error in the incorrect one. Further research into the mechanisms responsible for ancestry-specific effects is warranted, including evaluation of the influences of linked rare variants, cumulative genetic background modifiers, and genotype-by-environment interactions.
Data and code availability
Individual-level whole-genome sequencing data relating to this study are available at dbGaP: phs001642.v1.p1. All other data are contained in the paper and its supplementary information or are available upon request.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
The National Institutes of Health (NIH) grants DK062431 (S.R.B.); DK087694 (S.K.); DK062413 (D.P.B.M. and K.T.); DK046763-19, AI067068, and U54DE023789-01 (D.P.B.M.); DK062429 (J.H.C. and P.S.); DK062422 (J.H.C.); DK062420 (R.H.D.); DK062432 (J.D.R.); and DK062423 (M.S.S.) supported this study. Sequencing at the Broad Institute was supported by NHGRI grants 5U54HG003067-13 and UM1HG008895. S.R.B. was also supported in part by funding from Rutgers Crohns and Colitis Center of New Jersey. We acknowledge the clinicians and organizations that contributed to samples used in this study. Finally, we are grateful to the many families whose participation made this study possible.
Published: February 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.02.001.
Web resources
Multicenter African American Inflammatory Bowel Disease Study, https://www.clinicaltrials.gov/ct2/show/NCT01169194
RepeatMasker, http://www.repeatmasker.org/
UK Biobank, https://www.ukbiobank.ac.uk/
Washington University Center for Common Disease Genomics network, https://www.genome.wustl.edu/items/center-for-common-disease-genomics/
Supplemental information
References
- 1.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.G. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long T., Hicks M., Yu H.C., Biggs W.H., Kirkness E.F., Menni C., Zierer J., Small K.S., Mangino M., Messier H. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 6.Prokopenko D., Sakornsakolpat P., Fier H.L., Qiao D., Parker M.M., McDonald M.N., Manichaikul A., Rich S.S., Barr R.G., Williams C.J. Whole-genome sequencing in severe chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2018;59:614–622. doi: 10.1165/rcmb.2018-0088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zekavat S.M., Ruotsalainen S., Handsaker R.E., Alver M., Bloom J., Poterba T., Seed C., Ernst J., Chaffin M., Engreitz J., NHLBI TOPMed Lipids Working Group Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. Nat. Commun. 2018;9:2606. doi: 10.1038/s41467-018-04668-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries P.S., Yu B., Feofanova E.V., Metcalf G.A., Brown M.R., Zeighami A.L., Liu X., Muzny D.M., Gibbs R.A., Boerwinkle E., Morrison A.C. Whole-genome sequencing study of serum peptide levels: the Atherosclerosis Risk in Communities study. Hum. Mol. Genet. 2017;26:3442–3450. doi: 10.1093/hmg/ddx266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotlar A.V., Trevino C.E., Zwick M.E., Cutler D.J., Wingo T.S. Bystro: rapid online variant annotation and natural-language filtering at whole-genome scale. Genome Biol. 2018;19:14. doi: 10.1186/s13059-018-1387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smit A.F.A., Hubley R., Green P. 2013–2015. RepeatMasker Open-4.0. [Google Scholar]
- 13.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y., de Lange K.M., Jostins L., Moutsianas L., Randall J., Kennedy N.A., Lamb C.A., McCarthy S., Ahmad T., Edwards C. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 2017;49:186–192. doi: 10.1038/ng.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., Christiani D.C., Wurfel M.M., Lin X., NHLBI GO Exome Sequencing Project—ESP Lung Project Team Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brant S.R., Okou D.T., Simpson C.L., Cutler D.J., Haritunians T., Bradfield J.P., Chopra P., Prince J., Begum F., Kumar A. Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology. 2017;152:206–217.e2. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bioconductor Package Maintainer . R package version 1.12.0; 2020. liftOver: Changing genomic coordinate systems with rtracklayer::liftOver. [Google Scholar]
- 18.Loh P.R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 21.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17(R2):R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J.Z., Tozzi F., Waterworth D.M., Pillai S.G., Muglia P., Middleton L., Berrettini W., Knouff C.W., Yuan X., Waeber G., Wellcome Trust Case Control Consortium Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 24.Huang H., Fang M., Jostins L., Umićević Mirkov M., Boucher G., Anderson C.A., Andersen V., Cleynen I., Cortes A., Crins F., International Inflammatory Bowel Disease Genetics Consortium Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rio S., Mary-Huard T., Moreau L., Bauland C., Palaffre C., Madur D., Combes V., Charcosset A. Disentangling group specific QTL allele effects from genetic background epistasis using admixed individuals in GWAS: An application to maize flowering. PLoS Genet. 2020;16:e1008241. doi: 10.1371/journal.pgen.1008241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wientjes Y.C.J., Bijma P., Vandenplas J., Calus M.P.L. Multi-population genomic relationships for estimating current genetic variances within and genetic correlations between populations. Genetics. 2017;207:503–515. doi: 10.1534/genetics.117.300152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker E., Schmidt K.M., Sims R., O’Donovan M.C., Williams J., Holmans P., Escott-Price V., Consortium W.T.G. POLARIS: Polygenic LD-adjusted risk score approach for set-based analysis of GWAS data. Genet. Epidemiol. 2018;42:366–377. doi: 10.1002/gepi.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrell F.E., Jr. R package version 5.1-3.1; 2019. rms: Regression Modeling Strategies. [Google Scholar]
- 30.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cutler D.J., Zwick M.E., Okou D.T., Prahalad S., Walters T., Guthery S.L., Dubinsky M., Baldassano R., Crandall W.V., Rosh J., PRO-KIIDS Research Group Dissecting allele architecture of early onset IBD using high-density genotyping. PLoS ONE. 2015;10:e0128074. doi: 10.1371/journal.pone.0128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libioulle C., Louis E., Hansoul S., Sandor C., Farnir F., Franchimont D., Vermeire S., Dewit O., de Vos M., Dixon A. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi H., VanDussen K.L., Malvin N.P., Ryu S.H., Wang Y., Sonnek N.M., Lai C.W., Stappenbeck T.S. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017;36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett J.C., Lee J.C., Lees C.W., Prescott N.J., Anderson C.A., Phillips A., Wesley E., Parnell K., Zhang H., Drummond H., UK IBD Genetics Consortium. Wellcome Trust Case Control Consortium 2 Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakhshani N., Araste M., Imanzade F., Panahi M., Safarnezhad Tameshkel F., Sohrabi M.R., Karbalaie Niya M.H., Zamani F. Hirschsprung disease diagnosis: Calretinin marker role in determining the presence or absence of ganglion cells. Iran. J. Pathol. 2016;11:409–415. [PMC free article] [PubMed] [Google Scholar]
- 37.Anbardar M.H., Geramizadeh B., Foroutan H.R. Evaluation of Calretinin as a new marker in the diagnosis of Hirschsprung disease. Iran. J. Pediatr. 2015;25:e367. doi: 10.5812/ijp.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marilley D., Schwaller B. Association between the calcium-binding protein calretinin and cytoskeletal components in the human colon adenocarcinoma cell line WiDr. Exp. Cell Res. 2000;259:12–22. doi: 10.1006/excr.2000.4942. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura H., Lim T., Puri P. Inflammatory bowel disease in patients with Hirschsprung’s disease: a systematic review and meta-analysis. Pediatr. Surg. Int. 2018;34:149–154. doi: 10.1007/s00383-017-4182-4. [DOI] [PubMed] [Google Scholar]
- 40.Scharl S., Barthel C., Rossel J.B., Biedermann L., Misselwitz B., Schoepfer A.M., Straumann A., Vavricka S.R., Rogler G., Scharl M., Greuter T. Malignancies in inflammatory bowel disease: frequency, incidence and risk factors-Results from the Swiss IBD cohort study. Am. J. Gastroenterol. 2019;114:116–126. doi: 10.1038/s41395-018-0360-9. [DOI] [PubMed] [Google Scholar]
- 41.Peneau A., Savoye G., Turck D., Dauchet L., Fumery M., Salleron J., Lerebours E., Ligier K., Vasseur F., Dupas J.L. Mortality and cancer in pediatric-onset inflammatory bowel disease: a population-based study. Am. J. Gastroenterol. 2013;108:1647–1653. doi: 10.1038/ajg.2013.242. [DOI] [PubMed] [Google Scholar]
- 42.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson R.E., Kuchenbaecker K., Walters R.K., Chen C.Y., Popejoy A.B., Periyasamy S., Lam M., Iyegbe C., Strawbridge R.J., Brick L. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589–603. doi: 10.1016/j.cell.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M.S., Patel K.P., Teng A.K., Berens A.J., Lachance J. Genetic disease risks can be misestimated across global populations. Genome Biol. 2018;19:179. doi: 10.1186/s13059-018-1561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen M.H., Raffield L.M., Mousas A., Sakaue S., Huffman J.E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., VA Million Veteran Program Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Z., Weinhold N., Song G.C., Rand K.A., Van Den Berg D.J., Hwang A.E., Sheng X., Hom V., Ailawadhi S., Nooka A.K. A meta-analysis of genome-wide association studies of multiple myeloma among men and women of African ancestry. Blood Adv. 2020;4:181–190. doi: 10.1182/bloodadvances.2019000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin E.R., Tunc I., Liu Z., Slifer S.H., Beecham A.H., Beecham G.W. Properties of global- and local-ancestry adjustments in genetic association tests in admixed populations. Genet. Epidemiol. 2018;42:214–229. doi: 10.1002/gepi.22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Y., Perera M.A., Gamazon E.R. On using local ancestry to characterize the genetic architecture of human traits: genetic regulation of gene expression in multiethnic or admixed populations. Am. J. Hum. Genet. 2019;104:1097–1115. doi: 10.1016/j.ajhg.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H., Burch K.S., Johnson R., Freund M.K., Kichaev G., Mancuso N., Manuel A.M., Dong N., Pasaniuc B. Localizing components of shared transethnic genetic architecture of complex traits from GWAS summary data. Am. J. Hum. Genet. 2020;106:805–817. doi: 10.1016/j.ajhg.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Márquez-Luna C., Loh P.R., Price A.L., South Asian Type 2 Diabetes (SAT2D) Consortium. SIGMA Type 2 Diabetes Consortium Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 2017;41:811–823. doi: 10.1002/gepi.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level whole-genome sequencing data relating to this study are available at dbGaP: phs001642.v1.p1. All other data are contained in the paper and its supplementary information or are available upon request.