Abstract
The genetic diversity of circulating HIV-1 strains poses a major barrier to the design, development and evaluation of HIV-1 vaccines. The assessment of both vaccine- and natural infection-elicited T cell responses is commonly done with multivalent peptides that are designed to maximally capture the diversity of potential T cell epitopes (PTEs) observed in natural circulating sequences. However, depending on the sequence diversity of viral subtypes and number of the HIV immunogens under investigation, PTE estimates, including HLA-guided computational methods, can easily generate enormous peptide libraries. Evaluation of T cell epitope specificity using such extensive peptide libraries is usually limited by sample availability, even for high-throughput and robust epitope mapping techniques like ELISpot assays. Here we describe a novel, two-step protocol for in-vitro polyclonal expansion of CD8 T cells from a single vial of frozen PBMC, which facilitated the screening 441 HIV-1 Gag peptides for immune responses among 32 HIV-1 positive subjects and 40 HIV-1 negative subjects for peptide qualification. Using a pooled-peptide mapping strategy, epitopes were mapped in two sequential ELISpot assays; the first ELISpot screened 33 large peptide pools using CD8 T cells expanded for 7 days, while the second step tested pool-matrix peptides to identify individual peptides using CD8 T cells expanded for 10 days. These comprehensive epitope screening established the breadth and magnitude of HIV-1 Gag-specific CD8 T cellsand further revealed the extent of immune responses to variable/polymorphic epitopes.
Keywords: ELISpot, CD8 T cells, Potential T-cell Epitopes, Epitope mapping, HIV-1 vaccines
1. Introduction
A successful and effective human immunodeficiency virus-1 (HIV-1) vaccine will likely have to stimulate both the cellular and humoral arms of the immune response. Both will need to address the genetic diversity and plasticity of the virus that permits the presentation of diverse variants, including escape mutations, of epitopes to the immune system. This requires the successful vaccine candidate to be one composed of epitopes able to elicit broadly reactive immune responses (1) (2). In T cell based vaccine development, the use of consensus sequences in the design and screening of epitopes has shown that some epitope variants in circulating strains can be missed (3). Therefore, for relevant HIV-1 T cell-based vaccine development, a broader assessment of vaccine-elicited T cell responses to circulating strains is vital. Multivalent-based vaccine immunogens have been considered as one approach to deal with the variability of HIV-1 strains (4) (5). This approach uses circulating viral sequences to generate the minimal set of multivalent sequences that include the maximal diversity of epitopes observed in the circulating viral strains.
T cells are activated upon recognition of peptide antigens on major histocompatibility complex (MHC) molecules. CD8 T cells are particularly responsive to peptide antigens that are 8–10 amino acids long with the majority being 9 amino acids (6) (7). Thus, any peptide of the appropriate length, embedded in HIV viral proteins can potentially function as a T cell epitope. However, depending on the number of circulating strains considered and the size and level of variability of the genomic region under consideration, the prediction of potential T-cell epitopes (PTE) covering circulating HIV-1 strains can easily result in peptide libraries of several hundred peptides. Predictive in-silico approaches have been used to economically and effectively predict the most relevant epitopes from sequences of circulating HIV-1 strains but even these approaches can result in extensive peptide libraries (8) (9) (10) (11). In clinical research, the use of the interferon gamma (IFNγ) ELISpot assay has proven to be an essential tool for screening of such extensive peptide libraries (12) (13), but even such a robust and high-throughput assay can still be limited by test sample availability. In particular, a key time point sample might not be sufficient for repeated measurements or for use in different experimental approaches. The classical approach of plating each peptide in its own well during ELISpot is precise but limited by the availability of test sample. The complexity of assessing epitope specific T cell responses in sensitive and specific T-cell assays, one peptide at a time, would make the assessment of extensive peptide libraries virtually impossible. This has been mitigated with the use of peptide pools and peptide matrix pools for single peptide deconvolution, which reduces the number of test wells without significantly compromising the specificity of the ELISpot procedure (12) (14). However, with these approaches at least two test sample allotments would be needed to screen the pools and then test deconvoluted individual peptides to identify specific T-cell epitopes (12) (14) (15).
To harness the high-throughput screening efficiency of the ELISpot assay and to further maximise the availability of limited clinical samples, we employed an in vitro polyclonal expansion procedure using a CD3/CD4 bi-specific monoclonal antibody (BSMAB) to generate CD8 T cells for testing of a large library of peptide in ELISpot (16, 17). In this approach, epitopes are mapped in two sequential ELISpot assays using two timepoints of polyclonal CD8 T cell expansion from a single vial of cryopreserved peripheral blood mononuclear cells (PBMC); the first assay, after seven days of expansion, screens large peptide pools and the second assay, after an additional three days of expansion, tests individual peptides contained in the pools that scored positive in the first assay (12, 14). We evaluated response specificities and magnitudes of in vitro polyclonal expanded CD8 T cell to HIV-1 Gag PTE 15-mer peptides designed by two different epitope mapping strategies: overlapping peptides designed from a subtype C consensus sequence (121 peptides) and peptides designed from worldwide circulating viral sequences that included variants of the same epitope (320 peptides). We show that polyclonal expanded CD8 T cells from PBMCs are sensitive and specific to antigenic stimulation and provide an effective and reliable option to maximize limited samples availability.
2. Materials and Methods
2.1. Study participants
All samples were collected under different study protocols at African Clinical Research Centres (CRCs) sponsored by the International AIDS Vaccine Initiative (IAVI). PBMC samples were isolated from blood samples collected with written informed donor consent. Protocols included sampling of healthy HIV-1 uninfected participants and sampling of 32 HIV-1-infected participants at approximately 365 days post Estimated Date of Infection (EDI) (Table 1). The HIV-1 positive participants were part of an IAVI-sponsored Protocol C study cohort where over 3,000 plasma and PBMC samples from 286 Zambian participants were collected at the Zambia-Emory HIV Research Project (ZEHRP) laboratories. Protocol C was a prospective, observational, multi-centre study conducted between February 2006 and December 2011 to evaluate laboratory, clinical, immunologic and viral markers of disease progression in recently HIV-infected volunteers (18, 19). Figure 1 shows the frequency of HLA-A, -B and -C of the participants selected for study. HLA genotyping was done as described previously on genomic DNA extracted from whole blood or buffy coats (Qama blood kit; Qiagen) using PCR with commercial sequence-specific primers for two-digit specificity (20). The study was reviewed and approved by the Emory University Institutional Review and University of Zambia Research Ethics Committee.
Table 1.
HIV-1 positive participants characteristics (n=32)
| Gender | Female | 19 |
| Male | 13 | |
| Age (years) | Median (Min-Max) | 33 (19–59) |
| Mean (SD) | 34±9 | |
| Day post EDI | Median (Min-Max) | 339 (324–393) |
| Mean± SD | 347±17 | |
| CD4 Count (cells/ul) | Median (Min-Max) | 580(241–1,010) |
| Mean± SD | 559±191 | |
| Viral load (copies/ml) | Median (Min-Max) | 8,640(156–18,500) |
| Mean± SD | 8,576±5,854 | |
| Mode of infection | Heterosexual discordant couples | |
| Infectious subtype | HIV-1 subtype C |
Figure 1. Distribution of HLA class I alleles in study participants.
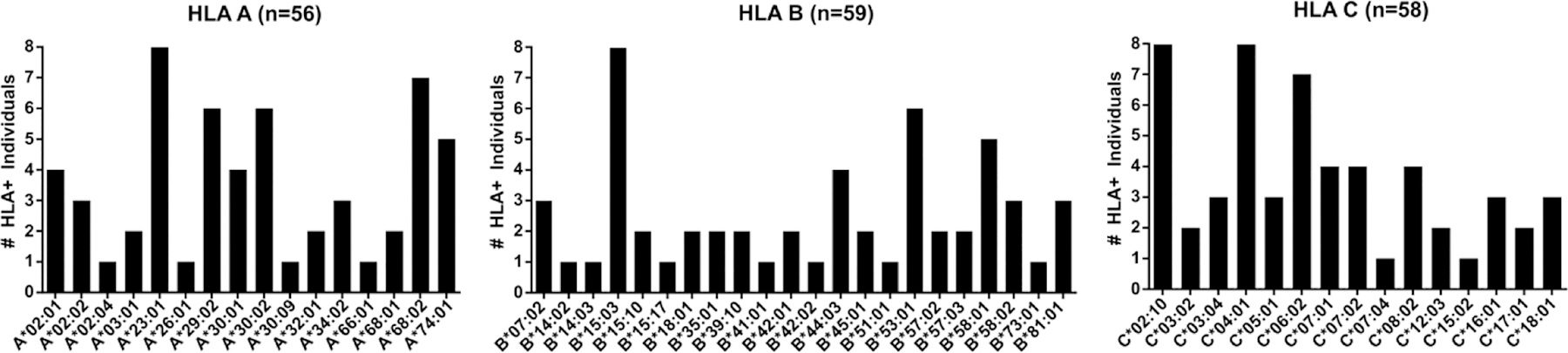
Allele frequencies of three HLA class I genes are based on 32 HIV-1 positive subjects who donated PBMC samples for analyses of CD8 T-cell responses to HIV-1 Gag protein peptides.
2.2. PBMC processing
Fresh PBMC were separated from whole blood by Ficoll-Hypaque (Histopaque®-1077 Hybri-Max™) density gradient centrifugation and frozen in liquid nitrogen using fetal bovine serum (FBS) supplemented with 10% dimethyl sulphoxide (DMSO) all from Sigma (St. Louis, Mo.) at a concentration of 10×106 cells/vial. PBMC were thawed with culture medium; RPMI 1640 medium supplemented with penicillin (100U/mL), streptomycin (100μg/mL), L-glutamine (2 mM), sodium pyruvate (1 mM), HEPES buffer solution (10mM) and 10% or 20% FBS (R10 or R20 media respectively), all from Sigma.
2.3. CD8 T cell expansion
On day 0, PBMC were thawed, washed with R20 medium and cultured at 1.0–1.5×106 cells/mL in R10 with IL-2 at 50U/mL (R10/50) and CD3/CD4 bi-specific antibody at 0.5μg/ml in an upright T25 culture flask, used for a starting culture volume of 2–5mL medium (16). The volume of culture medium was doubled on day 3 (same T25 flask placed horizontally) and again on day 5 of cell culture (when the culture was transferred to T75 flask) with R10/50. On day 7 of culture, cells were counted and approximately two-thirds of the cell population (approximately 18×106 cells from an initial culture of 10 × 106 PBMC) were removed from culture for the first ELISpot with peptide pools. Prior to the first ELISpot, cells were washed twice with 20ml PBS and once with 5ml R10. The cells were re-suspended in R20 without IL-2 and rested at 2.0×106 cells/mL in a fresh T75 flask at 37°C for 22–26hrs (day 8). The remaining cells were continued in culture, doubling the culture volume at days 7 and 9 with R10/50. On Day 10 cells were harvested, washed twice with 20mL 1x PBS and once with 5mL R10 cells and re-suspended in R20 at 2.0×106 cells/mL in a fresh T75 flask. Cells were rested at 37°C for 22–26hrs before proceeding with a second ELISpot (day 11). Typical expanded CD8 T cell culture purities at day 7 were at least 90% CD8+ T cells (17) (21).
2.4. Synthetic peptides
Two main sets of HIV-1 Gag protein peptides were used in this study. The first set consisted of 121 Consensus clade C Gag 15 amino acid (aa) peptides (15mer) with 11 aa overlap between peptides, covering the whole length of the Gag protein (NIH AIDS Reagents Program cat. 8118: HIV-1 Consensus C Gag Peptide Set, catalogue # 12756: HIV-1 Consensus C Gag Peptide Pool). The second set of 320 peptides were generated from sequences from worldwide circulating strains of HIV-1 in the LANL database (https://www.hiv.lanl.gov/content/index) using predictive algorithms to define potential T cell epitopes (PTE) (9) (NIH AIDS Reagents Program catalogue # 11554: HIV-1 PTE Gag Peptide Set and catalogue # 12437: HIV-1 PTE Gag Peptide Pool). This PTE set is commonly referred to as global PTE (22) and includes multiple variants of some peptides that are representative of circulating viruses, thereby overcoming the lack of variation in consensus peptides. The global PTE set is designed to cover the largest number of epitopes in the aa sequences of HIV-1 strains worldwide with a relatively small number of peptides, which would be practically manageable in laboratory assays.
Three additional large peptide pool sets were used for the evaluation of polyclonally expanded CD8 T cell specificities. These included; pooled 138 HCMV pp65 15-mer peptides (NIH AIDS Reagents Program cat. 11549), pooled 320 group M HIV-1 gag 15-mer peptides (NIH AIDS Reagents Program cat. 12437; peptide pool designed to represent the most frequent potential T cell epitopes (PTE) of HIV-1 Gag embedded in the sequences of circulating strains of HIV-1 worldwide), and a panel of 127 HIV-1 Nef peptides (NIH AIDS Reagents Program cat. 11553; peptide panel designed to permit expression of the most frequent potential T cell epitopes (PTE) of HIV-1 Nef embedded in the sequences of circulating strains of HIV-1 worldwide). These three proteins were selected as they typically elicit specific responses in most HIV-1 positive participants. All peptide concentrations were adjusted to be used at a final concentration of 1.5 – 2.0 µg/mL in an ELISpot well of 150µL assay medium volume.
2.5. Global Peptide design algorithm
The global peptide set is a publicly available reagent whose design is described by Li et al., (11) using a Potential T cell Epitopes (PTE) predicting algorithm. The algorithms work by selecting peptides to provide optimal coverage of an amino-acid sequence alignment. From the alignments, two sets of peptides are generated: (A) all 9mers representing potential epitopes and (B) all unique 15mers forming a pool of peptides to select from. Peptide selection begins by selecting a 15mer from (B) that provides maximal coverage of 9mers in (A), taking their frequency in the alignment into account. Naturally, 15mers covering conserved 9mers are selected first. Selection proceeds by removing the covered 9mers from (A) and picking the next best 15mer from (B) that provides maximal coverage. Selection continues until the desired number of PTEs is attained or until the marginal gain in coverage falls below a pre-specified threshold (i.e. stop selection when the next peptide provides less than 10% additional coverage of epitopes in the alignment). The peptide selection process is facilitated and guided by selected worldwide HLA-I molecules.
2.6. Design of peptide matrices
The two main sets of consensus and global peptides were organised into a 15-pool and 18-pool 3D-matrix for the consensus and global peptide sets respectively. The organisation was such that, within a given peptide matrix, each peptide was represented in three different peptide pools and the pools positioned one at each matric axis i.e. each axis pools contained all the peptides in a given set but differentially sorted among the pools. This allowed the identification of the respective peptide by responses in the three corresponding pools. The consensus pools composition ranged between 21–25 peptides per pool (median of 24 peptides) and the global PTE peptide sets ranged between 43–63 peptides per pool (median 52 peptides). Additionally, global PTE peptides were combined in families representing different variants (single aa substitutions) of the same epitope (see supplemental information on pool matrices). The final assay concentration of each peptide within a peptide pool or single deconvoluted peptide was 2.0μg/mL in 150μL of assay medium volume.
2.7. Two-step IFNγ ELISpot assays with in vivo expanded CD8 T cells
A two-step ELISpot assay approach was adapted to include a peptide pool matrix-screening step with a subsequent deconvoluted individual peptides analysis step (23) (Figure 2). The polyclonal CD8 T cell expansion extended the availability of limited, one time-point, clinical sample that allowed for a comprehensive characterization of T cell responses to evaluate a large set of 441 peptides covering the HIV-1 Gag protein using ELISPOT assay. The ELISpot assay procedures have been previously described (24). Briefly, day 7 polyclonal expanded and 24 hour rested (no IL2) CD8 T cells were recovered, viably counted (Beckman Coulter Vi-Cell XR) and plated in 96-well anti-IFNγ pre-coated flat bottom PVDF culture plates (25) (Mabtech, Nacka, Sweden) at 2X105 cells per well with stimuli. A no-cell well was used to access medium background. Non-specific IFNγ secretion by CD8 T cells was assessed by culture with 0.45% dimethyl sulphoxide (DMSO) (Sigma) in R10 (used as mock as peptides were dissolved on DMSO). Positive control stimuli were phytohemagglutinin (PHA, 10μg/mL) (Sigma) or cytomegalovirus (CMV) pp65 protein peptide pool. Plates were incubated overnight at 37°C, 5% CO2 and spots developed as described previously (24). ELISpots using single peptides deconvoluted from the day-7 CD8 T cells matrix pool ELISpots were performed on day 10 expanded CD8 T cells as above. The day 10 CD8 T-cell single peptide assay included a confirmatory re-testing of the day 7 CD8 T cell positive pools. The numbers of spot forming units (SFU) per well were counted using an automated ELISpot plate reader (AID ELISpot reader system; Autoimmune Diagnostika GmbH, Strassberg, Germany) and the number of specific T cells was calculated by subtracting the mock control values. SFU were expressed as mock corrected SFU per 106 cells i.e. the average mock SFU was subtracted from the observed pool or single peptide SFU values. Based on our previous work on assay validation and peptide qualification using IAVI sample PBMCs, a positive response was defined as a CD8 T cell ELISpot assay response to stimulant that averaged ≥38 SFU/106 CD8 T cells (mock-subtracted) and was > 4x the average mock back ground (or greater than 0 if the background was 0) (24, 25, 26).
Figure 2. Two-step ELISpot assay with in vivo expanded CD8 T cells.
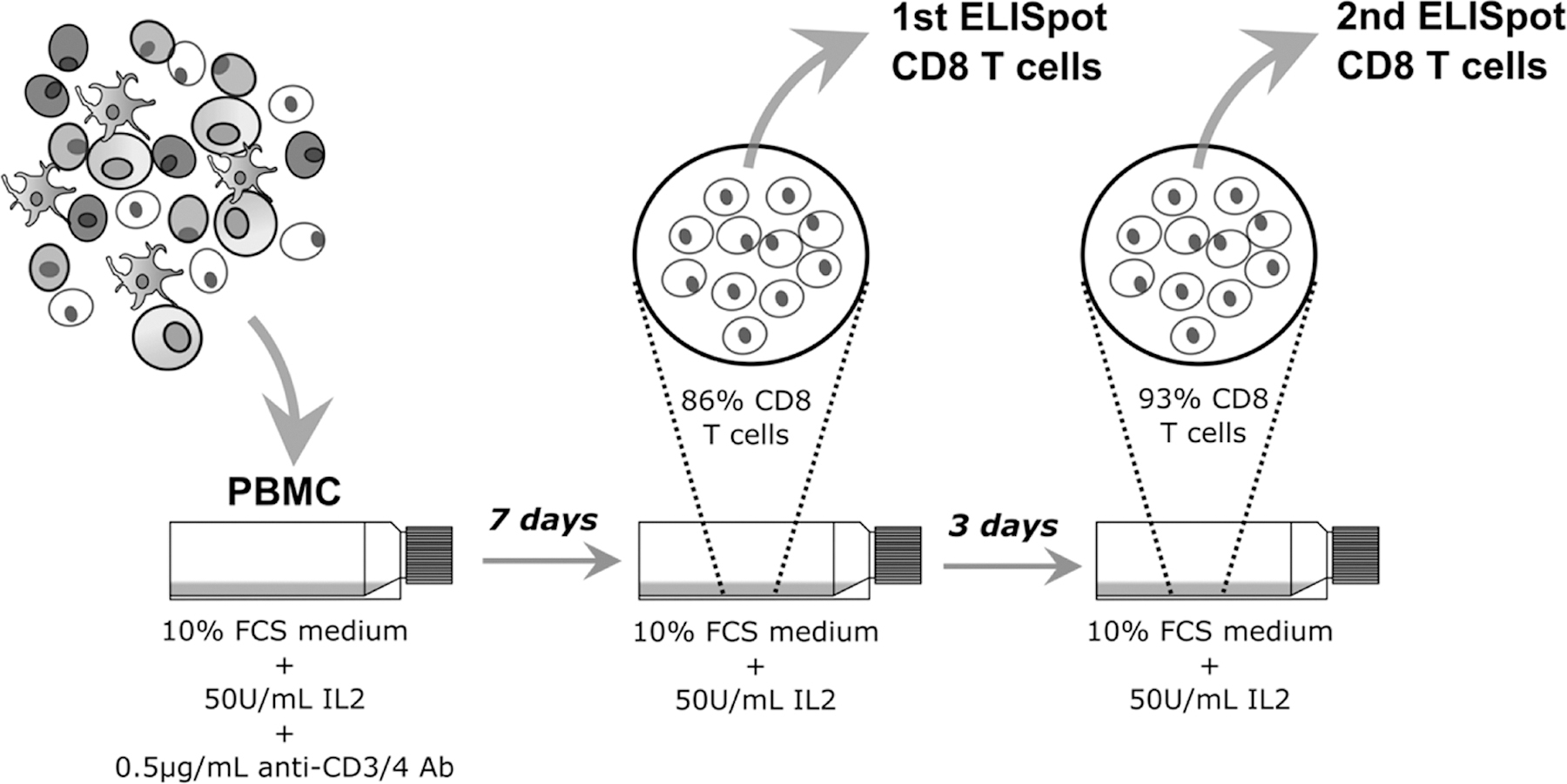
A two-step ELISpot assay approach was adapted to include a peptide pool matrix-screening step with a subsequent deconvoluted individual peptides analysis step for the screening of a large peptide library for potential T cell epitopes. Limited clinical sample was extended through an in vivo CD8 T cell expansion from PBMC; 7 day expanded CD8 T cells for the 1st ELISpot assays and an extra 3 days of expansion for 2nd ELISpot CD8 T cells.
2.8. Epitope predictions
The immune epitope database (IEDB) (https://www.iedb.org) and analysis resource used in the prediction of MHC-I binding of 9mer epitopes, identified epitopes in both consensus and global PTE 15mer peptides that were subsequently used as evaluation peptide sets (27). An IEDB percentile ranking score value of ≤ 1 % was used as a cut-off threshold for selecting high binding affinity 9mer epitopes as defined by our 32 HIV-1 positive samples HLA-I molecules (27, 28, 29, 30). The relative mapping of the predicted high affinity epitopes to the HBX2 HIV-1 gag protein sequence was compared to the mapping and coverage of the peptides determined from the ELISpot responses of polyclonal expanded CD8 T cells to the consensus and global peptide sets.
2.9. Comparison of IFNγ ELISpot responses of CD8 T cells isolated from PBMC or expanded CD8 T cells
IFNγ ELISpot responses of CD8 T cells isolated from PBMC or expanded CD8 T cells were assessed in a subset of samples from 6 HIV+ participants. CD8 T cells were expanded for 7 days, harvested, washed and rested as above. At this point additional vials of the same frozen PBMC samples were thawed and rested overnight in R20 medium. CD8 T cells were negatively selected from both expanded cells and PBMC in parallel using the MACS Miltenyi Biotec® CD8+ T Cell Isolation kit, Cat. 130–096-495 (Miltenyi Biotec, Bisley, UK), according to the package insert. The two-step ELISpot approach to deconvolute individual peptide specificities was not possible with CD8 T cells isolated directly from PBMC without polyclonal expansion due to limitations in PBMC number and number of negatively selected CD8 T cells derived from these PBMC. Comparisons of IFNγ ELISpot response specificities and magnitudes of either directly isolated or polyclonally expanded CD8 T cells were assessed by use of three peptide pools; CMVpp65, HIV-1 group M Gag and Nef peptide pools, along with mock and PHA controls.
2.10. Statistical analysis
Sequence alignment and analysis were performed using sequence analysis programs Jalview V.2 (31) and MEGA7 (32). Statistical analysis and graphical presentation were performed using GraphPad Prism 6 for Microsoft Windows v6.01. Group comparisons and effects/treatment correlations were performed using the Wilcoxon signed-rank test (data had right-skewed distributions) and the Spearman rank correlation test (linear regression used to determine the slope) respectively. Statistical results were given either as medians with 25% and 75% percentile ranges, sample group means or spearman’s rho (r) as required by the test. Significant was given as per test p-value (not-significant [ns] for p > 0.05).
3. Results
3.1. Comparison of bispecific antibody-expanded and directly negatively isolated CD8 T cells
To extend sample availability from the same time point for the evaluation of the extensive HIV-1 gag peptide library, we employed a T cell expansion protocol that non-specifically expanded polyclonal CD8 T cells. To ensure we did not significantly lose or gain CD8 T cell specificities during the expansion procedure we compared ELISpot responses of negatively selected CD8 T cells isolated from both PBMC and cells that had undergone 7 days of polyclonal CD8 T cell expansion (Figure 3). Correlation analysis showed a positive and moderate correlation in ELISpot SFU magnitudes between direct negatively isolated CD8 T cells from PBMC and CD8 T cells that had undergone polyclonal expansion and an additional CD8 T cell isolation step (spearman r = 0.67, p = 0.002), but with a result discrepancy of 0/18 paired ELISpot read outs (100% +ve/-ve result congruence). There was an instance of a difference between paired read outs in SFU magnitude were one value (1.7 Log10 SFU/106 PBMC) was closer to the set threshold (1.58 Log10 SFU/106 cells) defining a positive response compared to the respective pair value (2.7 Log10 SFU/106 CD8 T cells), howbeit, both responses qualified as positive responses.
Figure 3. Directly PBMC isolated and polyclonally expanded CD8 T cell IFNγ ELISpot responses.
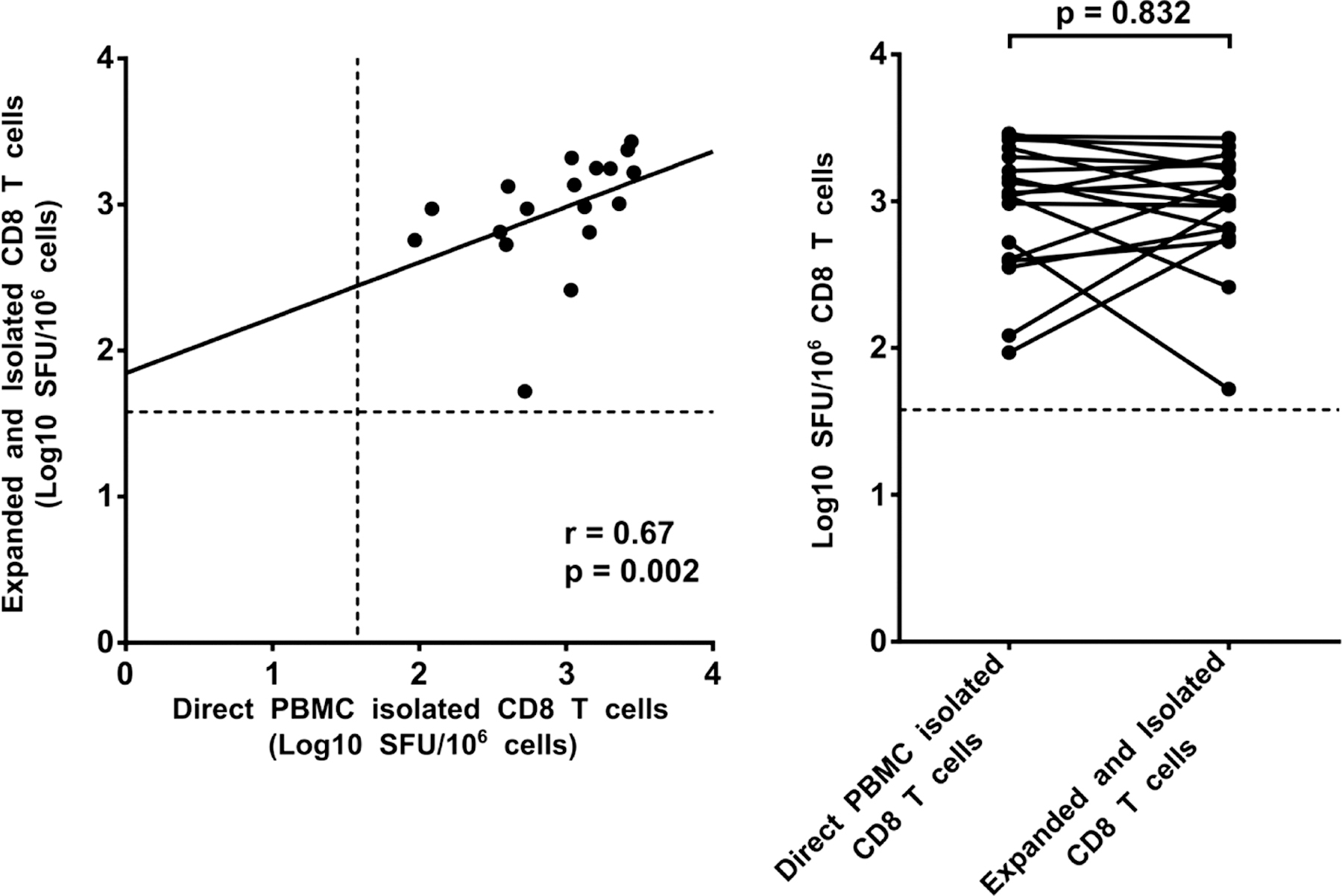
Responses to a set of 3 peptide pools, selected for their high probability to trigger a response in most of all HIV-1 positive participants, were compared between CD8 T cells negatively isolated from PBMC and from polyclonally expanded CD8 T cells. The peptide pools used included: HCMV pp65 (135 peptides), HIV-1 group M gag (320 peptides), and HIV-1 nef (127 peptides), all from the NIH AIDS reagent programme. Responses between direct PBMC isolated CD8 T cell and polyclonally expanded and isolated CD8 T cells from the same sample were correlated using spearman correlation rank test (left panel) and response magnitudes compared using wilcoxon matched pairs signed rank test (right panel). The dashed line indicates the positive response threshold of 1.58 Log10 SFC/106 CD8 T cells.
We further analysed T-cell response magnitudes between our different CD8 T-cell comparator groups using Wilcoxon matched-pairs signed rank test. There was no significant difference in SFU/106 CD8 T cells median values, p-value = 0.83 (Median [25%, 75% percentiles]); direct negatively isolated CD8 T-cell from PBMC (3.0 [2.6, 3.3]) Log10 SFU/106 CD8 T cells), and polyclonally expanded and isolated CD8 T cells (3.0 [2.7, 3.2] Log10 SFU/106 CD8 T cells). In-vitro polyclonal expanded CD8 T cells display similar antigen sensitivity as PBMC CD8 T cells even after several days of polyclonal expansion.
3.2. A two-step ELISpot-assay for epitope mapping
Our two-step ELISpot assay relied on the use of polyclonal day 7 and day 10 expanded CD8 T cells from the same PBMC sample to screen a large peptide set for PTEs. To assess consistency in expanded CD8 T cell responses, we compared the responses of day 7 and day 10 polyclonal expanded CD8 T cells to peptide pools. Correlation analysis of day 7 and 10 polyclonal expanded CD8 T cell responses to both consensus and global peptide pools showed a positive and significant correlation (consensus peptide pools spearman r = 0.87, p < 0.0001 and global peptide pools spearman r = 0.85, p < 0.0001) (Figure 4).
Figure 4. The comparison of 1st and 2nd ELISpot polyclonally expanded CD8 T-cell response specificities to HIV-1 gag peptide pools.
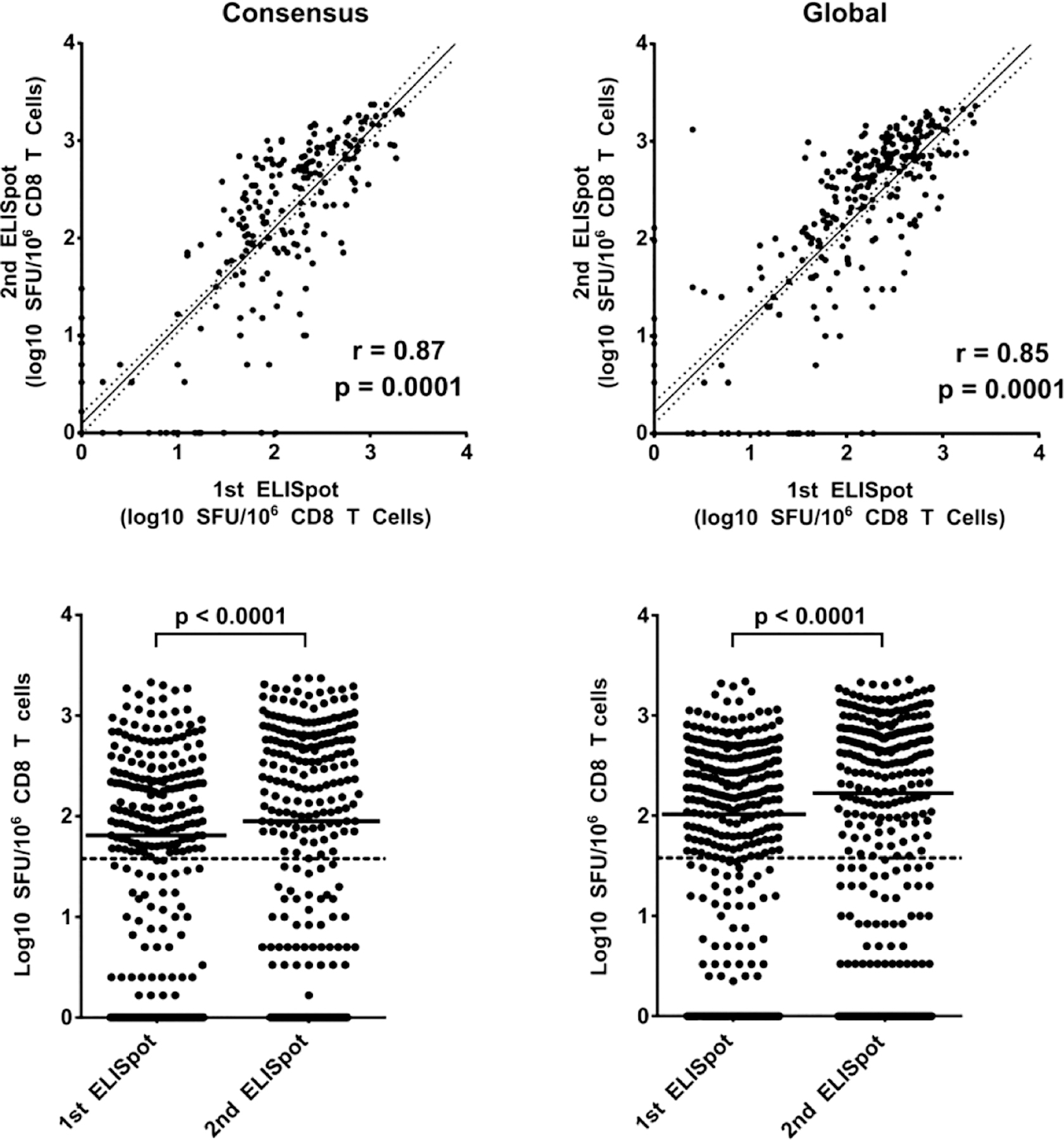
Panels of 121 consensus and 320 global 15-mer peptides were organised into 15-pool and 18-pool 3D-matrices respectively. The first ELISpot screened large peptide pool panels using day 7 polyclonal expanded CD8 T cells and the positive peptide pools were re-confirmed on the second ELISpot assay using day 10 polyclonal expanded CD8 T cells. Positive pool responses between day 7 polyclonal expanded CD8 T cells and day 10 polyclonally expanded CD8 T cells from the same samples were correlated using spearman correlation rank test (top panels) and response magnitudes compared using wilcoxon matched pairs signed rank test (bottom panels): consensus (left panels) and global peptides (right panels). Each spot indicates the SFU magnitude of CD8 T cell response to a pool of HIV-1 gag peptides. The dashed (lower panels) line indicates the positive response threshold of 1.58 Log10 SFC/10E6 CD8 T cells.
An analysis of the response agreement between day 7 and day 10 polyclonal expanded CD8 T-cell showed that responses to the global peptide pools had a +ve/-ve result congruence in 314/340 (92%) paired ELISpot read outs while response to the consensus peptide pools had a +ve/-ve result congruence in 262/288 (91%) paired ELISpot read outs. For both peptide sets, the discrepant results were mostly in sample wells that registered SFU read values of below 2.0 log10 SFU/106 CD8 T cells. Response magnitude comparison using the Wilcoxon signed rank test showed a significant difference in response magnitudes with the day 10 CD8 T-cell registering higher median responses to both global and consensus peptide pools (Median [25%, 75% percentiles]); consensus peptide pools, day 7 vs day 10 CD8 T cells, 2.0 [0.5, 2.5] Log10 SFU/106 CD8 T cells vs 2.2 [0.5, 2.8] Log10 SFU/106 CD8 T cells p < 0.0001 and global peptide pools, day 7 vs day 10 CD8 T-cell, 1.8 [0.2, 2.4] Log10 SFU/106 CD8 T cells vs 1.9 [0.0, 2.7] Log10 SFU/106 CD8 T cells p < 0.0001. Day-7 and day-10 polyclonal expanded CD8 T cells had similar antigenic pool specificity and sensitivity even though the latter exhibited higher responsivity in SFU magnitudes.
3.3. Qualification of HIV-1 gag peptides using HIV negative samples
Previously, we defined a positive IFNγ ELISpot response as one having a mock subtracted response to stimulation of ≥38 SFU/106 (1.58 Log10 SFU/106) cells and ≥4-fold SFU value over mock, a criteria aimed at a false positive rate of < 2% (24) (25). For the qualification of our HIV-1 gag peptides, we used 7 days polyclonal in vitro expanded CD8 T cells from 32 HIV-1 positive and 40 HIV-1 negative participant PBMCs to demonstrate expected responses from pathogen infected and uninfected participants, respectively, stimulated with peptide pools. Responses from 64 triplicate medium control (mock) wells using HIV-1 positive 7 day in vitro expanded CD8 T cells were used to estimate the limit of detection of our experimental set-up. The means of these triplicates ranged from 0 to 6.67 SFU per well of CD8 T cells (mean 0.8, SD 1.5 SFU per well). The median of the triplicate background means was 0.3 SFU per well with the 25th and 75th percentiles of 0.0 and 1.0 SFU per well, respectively (Figure 5A). Subsequent stimulation of our CD8 T cells for antigen specific responses was done using pooled peptides. The range of SFU detected in CD8 T cells from pathogen un-exposed participants would validate the cut-off for true positive sample responses observed with HIV-1 positive samples. The 121 consensus peptides were arranged into a 3D matrix of peptide pools with each axis having 5 peptide pools that contained all the consensus peptides while the 320 global peptides were designed into a 6-pool per axis 3D matrix with each axis containing all the global peptides; the peptides allocated to different pools to allow individual peptide deconvolution (12). Each matrix axis set of peptide pools was used to stimulate day 7 polyclonal expanded CD8 T cells from different sets of HIV-1 negative samples (n=11–15). Responses to mock stimulation of the HIV-1 negative polyclonal expanded CD8 T cells averaged a low 1.45 SFU/106 CD8 T cells, with a range of 0 – 8.75 SFU (min-max). Mock subtracted SFU responses by the HIV-1 negative samples to HIV-1 gag peptide pools were also low (mean [consensus] = 0.97 SFU/106 CD8 T cells range: 0–10 SFU (Figure 5B) and mean [global] = 3.34 SFU/106 CD8 T cells range: 0–428.8 SFU/106 CD8 T cells (data not shown). HIV-1 negative sample cumulative distribution of SFU/106 cells ELISpot responses to both consensus and global peptide pools determined a 97.5 percentile upper response limit of 5 and 10 SFU/106 cells for consensus and global peptide pools respectively. However, since the responses to the global included two unusually high SFU values, 53.75 SFU/106 CD8 T cells and 428.80 SFU/106 CD8 T cells, we used the interquartile range (IQR) approach to create outlier fences that determined several SFU values as outliers, including the two high values: 13.13, 15.00,16.25, 53.75 and 428.8 SFU/106 CD8 T cells (data not shown). The exclusion of the outliers resulted in mock subtracted SFU response data set with a low mean SFU value of 1.23 SFU/106 CD8 T cells (range: 0–10 SFU SFU/106 CD8 T cells) and a 97.5 percentile upper response limit of 7.5 SFU/106 cells (Figure 5C).
Figure 5. Cumulative distribution of ELISpot responses.
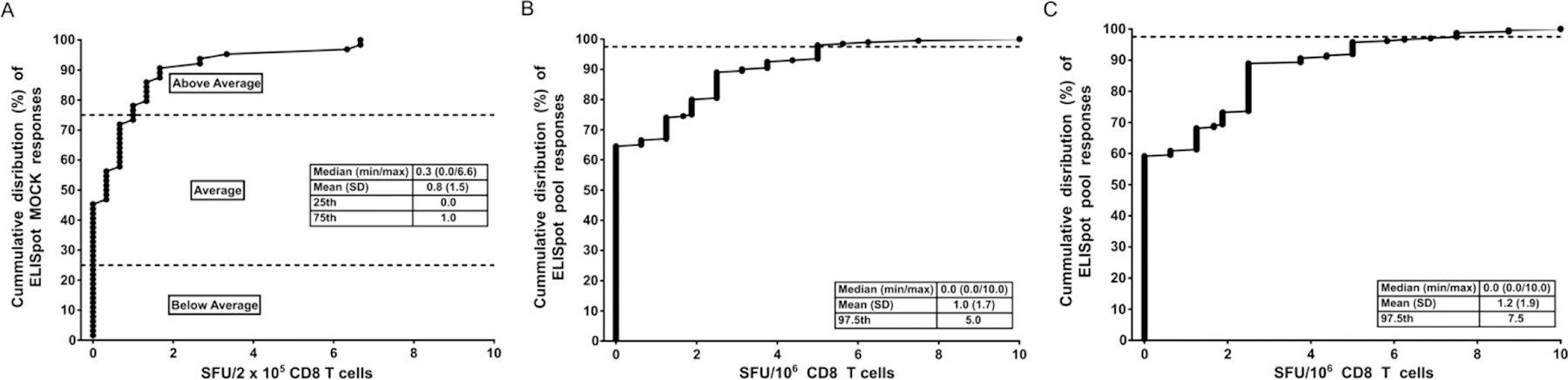
(A) The ELISpot assay limit of detection was defined as the lowest number of SFU that would distinguish a stimulated from unstimulated control well. The limited of detection was estimated using the means of triplicate medium control (mock) responses using polyclonal in vitro expanded CD8 T cell derived from HIV-1 positive participant samples PBMCs used at 2 × 105 cells per well. (B and C) Peptide pools were used to stimulate polyclonal in vitro expanded CD8 T cells from HIV-1 negative participants to assess the SFU distribution of negative responses that can be used to determine the cut-off for true positive sample responses observed with HIV-1 positive samples. Cumulative distribution of 40 HIV-1 negative sample SFU/106 CD8 T-cell ELISpot responses to both consensus (B) and global (C) peptide pools with inserts showing the medium, mean, SD and the 97.5 percentile of the HIV-1 negative samples’ responses.
Thus, our use of ≥1.58 Log10 SFU/106 CD8 T cells as the threshold defining positive pool or peptide responses in the 32 HIV-1 positive samples ensured a higher degree of confidence in the observed positive results as true positives (Figure 6).
Figure 6. IFN-γ ELISpot mock corrected SFU/106 cell responses for polyclonal expanded CD8 T cells from 40 HIV seronegative (A) and 32 HIV seropositive samples (B).
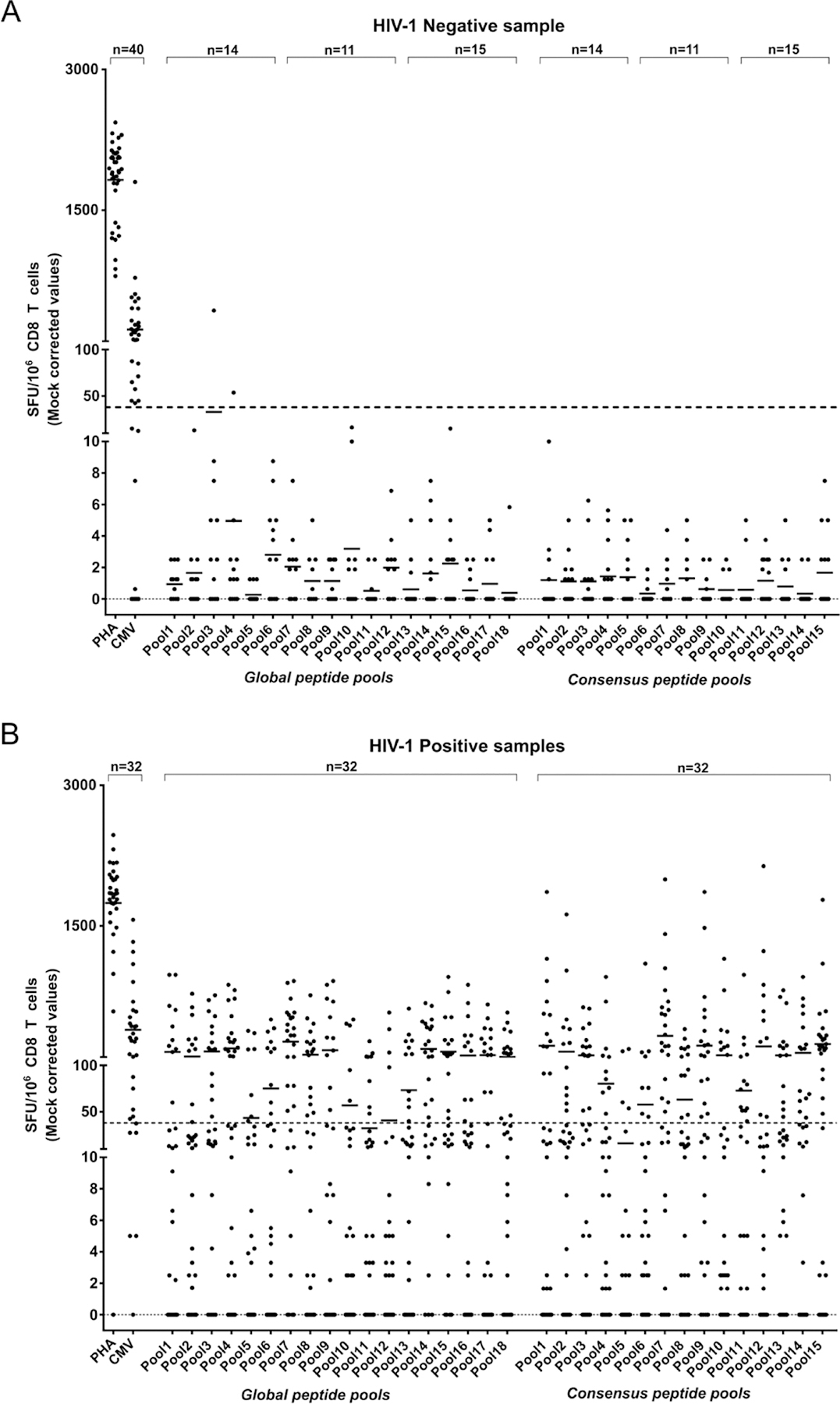
HIV-1 gag peptide pools were used to stimulate polyclonal in-vitro expanded CD8 T cells to compare responses to a previously determined response threshold (dotted line = 38SFU/106 cells) defining a positive response to stimulation in the ELISpot assay. Each spot indicates the SFU magnitude of CD8 T cell response to a pool of HIV-1 gag peptides and the bar associated with each pool represents the average CD8 T cell SFU response to the pool peptides.
3.4. Consensus and Global response peptide mapping
Epitopes were mapped in two sequential ELISpot assays; the first ELISpot screened matrix peptide pools, while the second tested individual peptides. Positive peptide pools from the ELISpot assay with day 7 polyclonal expanded CD8 T cells dictated the deconvolution of single peptides using the 3D pool matrices. These individual peptides were analysed using day 10 expanded polyclonal CD8 T cells from the same samples as the matrix peptide pools. ELISpot on deconvoluted peptides identified 38/121 consensus and 71/320 global unique reactive peptides from polyclonal expanded CD8 T cells from the HIV-1 positive participant samples. We also developed additional comparator sets of peptides by using the immune epitope database (IEDB) (https://www.iedb.org) to predict within the consensus or global peptides sets 9mers that could bind to HLA-I molecules expressed by the expanded CD8 T cells (27, 30, 33). The IEDB predicted 115 and 419 peptides, within the consensus and global peptides sets respectively, using the specified percentile ranking score value of <=1% as a cut-off threshold for the selection of high binding affinity epitopes (27, 28, 29, 30).
Comparatively aligning of all the consensus and global ELISpot positive peptides to the HBX2 HIV-1 gag 500 amino acid sequence, we counted and plotted the number of peptides aligned at each position (Figure 7 upper panel spectrum). The count of peptides aligned at each HBX2 HIV-1 gag protein position showed the global peptides averaged a count of 2 peptides compared to a consensus peptides’ average of 1 peptide covering the HBX2 HIV-1 gag protein positions, with a maximum count of 9 peptides and 4 peptides respectively. Using the HBX2 HIV-1 gag protein sequence for reference, all the identified ELISpot positive peptides from both the consensus and global peptide sets were mapped back to determine their relative positions within the gag protein. All peptides from the consensus and global sets mapped to relatively the same regions suggesting similar immunodominant hotspot targets underlying observed response peptides in both the consensus and global peptide sets (34) (Figure 7 middle panels). The alignments also showed that the global peptide covered 371/500 while the consensus peptides covered 305/500 of the HBX2 HIV-1 gag protein positions representing a 71% and 61% coverage respectively. The predicted epitope peptides alignment translated to higher HBX2 HIV-1 gag protein coverage of 86% and 93% for the consensus and global predicted epitopes respectively. However, the covered region of the HBX2 HIV-1 gag protein by the observed ELISpot positive consensus (61%) and global (71%) peptides fell within the regions covered by the predicted high binding affinity epitopes (not shown).
Figure 7. Mapping of ELISpot positive peptides to the 500aa of the HBX2 HIV-1 gag protein sequence.
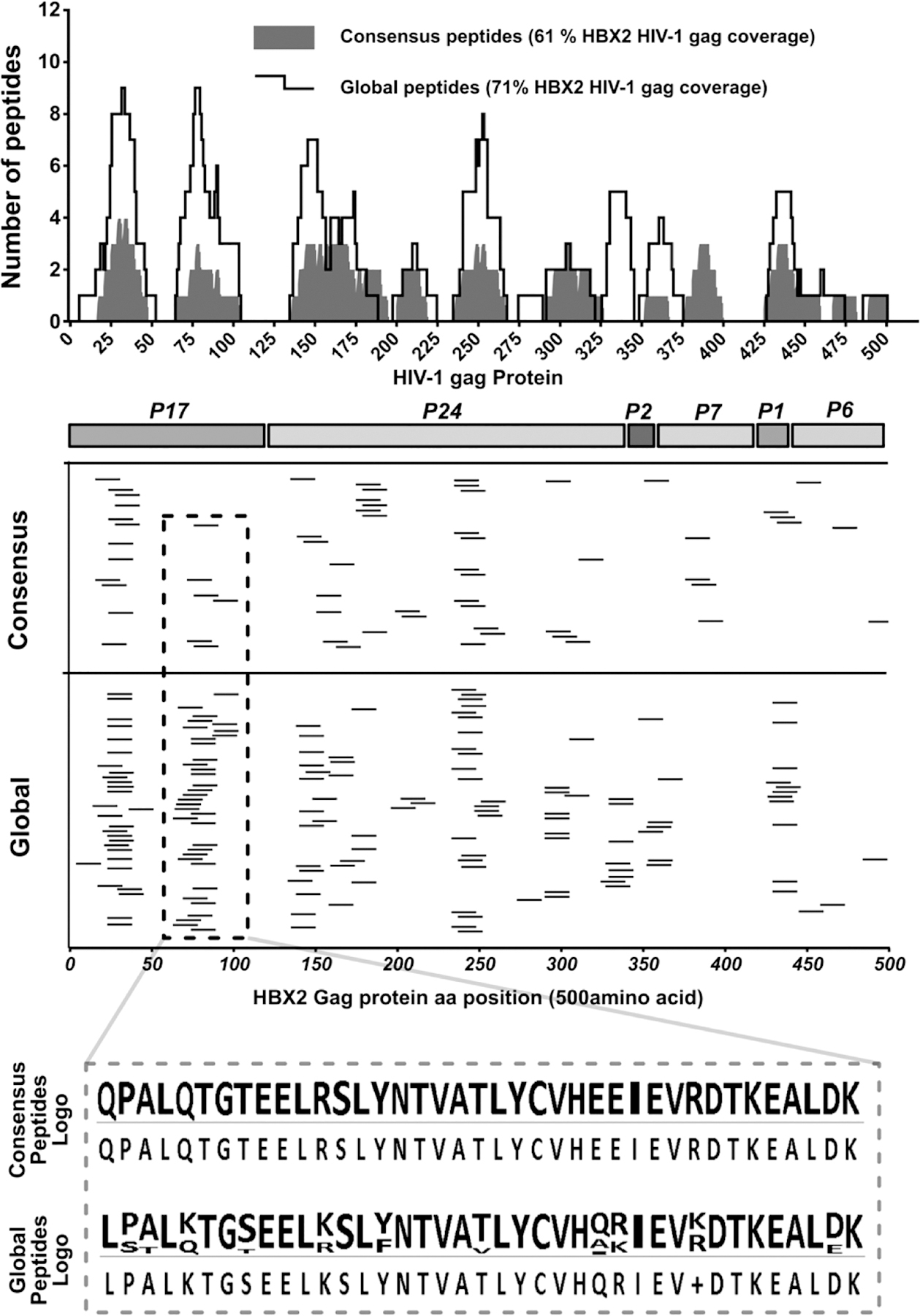
Epitopes were mapped in two subsequent ELISpot assays; the first ELISpot screened large peptide pools, while the second tested individual peptides. Determined response peptides were mapped to the HBX2 HIV-1 gag protein reference sequence to determine their relative locations. (Upper panel spectrum) Consensus and global peptides that elicited a positive ELISpot response with polyclonal expanded CD8 T cells were comparatively aligned to the HBX2 HIV-1 gag proteins. The number of peptides covering each gag protein sequence position was counted and plotted for each peptide set and the total coverage of the entire gag protein sequence determined. (Middle panels) Mapping of the positive ELISpot peptides from the consensus and global peptide sets to the HXB2 HIV-1 gag protein sequence positions and (lower panel) sequence logo of positive peptides from a select region on the Gag protein showing response global peptide sequence variations.
Unlike the consensus peptide set, the global peptide set included multiple variants of the same peptide. Thus, the higher number of ELISpot positive peptides within the global peptide set yet with a comparable coverage and similarity in the mapped peptide target regions to consensus peptides can only be explained by the polyclonal expanded CD8 T cells being able to detect variants of the same target region within the global peptide sets (figure 7 lower panel logo). Taken together, these results suggest the ability of the polyclonal expanded CD8 T cells to not only detect and respond to high binding affinity targets but also exhibit target variance response plasticity.
4. Discussion
In this study, we evaluated an immunological assays using in-vitro polyclonally expanded CD8 T cells and HIV-1 gag protein peptides designed with two different strategies: peptides corresponding to either consensus sequences or worldwide circulating viral sequences that included known variants. We employed a two-step ELISpot assay for the identification of CD8 T-cell responses in an extensive HIV-1 gag peptide library by first screening peptides pools to deconvolute single peptides for testing. CD8 T cells were expanded from a single vial of frozen PBMC by a 7-day and 10-day in-vitro polyclonal expansion of CD8 T cells for the first and second ELISpot assays respectively. Compared with CD8 T cells isolated directly from PBMC, we show that 7- and 10-day polyclonal expanded CD8 T cells were both sensitive and specific to stimulation using HIV-1 gag antigenic peptides.
The in-vitro expansion of CD8 T cells increased the application of the ELISpot assay by extending the availability of samples. The in-vitro expanded CD8 T cells have been shown to have a predominantly central memory phenotype (CD45RO+/CD27+) and some effector memory (CD45RO+/CD27-), with the expansion process resulting in a 4.9 fold (minimum 3 fold) increase from starting PBMC to >83% CD8+ cells by day 7 of cell expansion (17). Central memory T cells are thought to be the main effector cells in the cultured ELISpot assay and have been argued to have increased sensitivity compared to the standard ELISpot assay (35). In a viral inhibition assay, assessing the ability of CD8 T cells to inhibit HIV-1 replication, it was also shown that bispecific antibody expanded CD8 T cells demonstrated similar HIV-1 inhibition capacity compared with CD8 T cells directly isolated from PBMC (17).
Comparing day 7 and day 10 polyclonal expanded CD8 T cells we observed mean higher response magnitudes in day 10 CD8 T cells and some discrepancy in the congruence of positive peptide pool responses. The protocol we used for the in-vitro expansion of CD8 T cells from PBMC progresses with a selective proliferation of CD8+ T cells with purities of about 83% CD8 T cells by day 7 of expansion and reaching over 90% CD8 T cells by day 10 (17, 21, 36). Serial dilution experiments of ELISpot samples have shown a linear relationship in the response magnitude and plated cells in the wells which has an effect of increasing the target cell population with increased PBMC plated per well (37). As we controlled for cell numbers, 2×105 cell per well, and cell viability of samples used in the ELISpot assay, the only obvious variability was a difference in CD8 T cells in day 7 vs day 10 expanded cell populations; the superior CD8 T-cell response in SFU magnitude could be explained the slightly higher proportion of CD8 T cells by day 10 of cell expansion.
The IFNγ ELISpot assay has proven a versatile and robust method for immune monitoring that has allowed the detection of both immunodominant and subdominant T-cell specific antigens at the single-cell level. However, currently no consensus method is agreed on what defines a positive and relevant ELISpot result. The empirical approaches for ELISpot response determination have proven sufficient for clear-cut results that are obtained with a low background and high signal-to-noise resolution, with SFU values of over 4 fold over negative controls (38) (39) (40). However, more sensitivity enhanced approaches including the statistical definition of positive ELISpot results have been demonstrated to be required for subdominant and weak T-cell responses (37). Most of the response discrepancies between 7-day and 10-day expanded CD8 T cells from the same sample were in low SFU responses that probably would have needed a higher replicate well set-up and a statistical determination of the results to be certain of the discrepancy (38, 39, 40, 37).
Bioinformatic approaches are also currently being employed to contribute to the design of epitope-based vaccines with a goal to streamline and effectively interpret proteome data (41). The Li et al., study describes a bioinformatics approach in the global peptide set design (9) that resulted in a large peptide library of 1708 15-mer peptides covering HIV-1 Gag, Pol, Nef and Env HIV-1 proteins. Hence, for epitope mapping of the whole HIV-1 proteome, the use of peptide pools and matrices with the ELISpot assay has proved to be a more convenient and efficient approach to flow-cytometry approaches as it allows assessing of large peptide sets easily without compromising the detection of immunologically relevant epitopes. In our set up, the largest pool comprised of 63 peptides which is far less than larger pools of up to 134 15-mer peptides that were shown not to be inhibitive to specific responses of HIV-1 specific T-cell to known epitopes in the pools (23). Another bioinformatics approach we used in this study is the prediction of peptide-MHC binding as the basis of anticipating T cell epitopes (42). We used the IEDB to predict, within the consensus and global peptides sets, high binding affinity epitopes whose coverage of the HBX2 HIV-1 gag protein encompassed the target regions of our polyclonal expanded CD8 T cells responses.
In this set of experiments, we set out to evaluate the potential of mapping, from a single cryopreserved vial of ~ 10 million PBMC, responses of in-vitro polyclonal expanded CD8 T cells to an extensive peptide library using the gamma-interferon ELISPOT assay. We show that an in-vitro polyclonal expansion of CD8 T cells from PBMCs to extend clinical sample availability is both a feasible and strategic option to augment the already robust potential of the IFNγ ELISpot assay using expanded CD8 T cells that are both sensitive and specific to antigen stimulation.
Supplementary Material
Highlights.
In-vitro polyclonal expansion of CD8 T-cells facilitated the screening of hundreds of HIV-1 epitopes per individual.
The efficiency of ELISpot assays was improved through peptide pool matrices that allowed single peptide deconvolution.
5. Acknowledgments
This work was supported grant AI −1064060 from the National Institute for Allergy and Infectious Diseases, the National Institutes of Health, and through the DELTAS Africa Initiative [grant # DEL-15–006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Science (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (grant # 107752/Z/15/Z) and the UK government. This work was funded in part by IAVI and made possible by the support of many donors, including United States Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org. The contents of this manuscript are the responsibility of the authors and do not necessarily reflect the views of USAID, the US Government, AAS, NEPAD Agency, Wellcome Trust or the UK government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6 References
- 1.Kwong PD, Mascola JR, Nabel GJ. The changing face of HIV vaccine research. J Int AIDS Soc. 2012;15(2):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nabel GJ, Kwong PD, Mascola JR. Progress in the rational design of an AIDS vaccine. Philos Trans R Soc Lond B Biol Sci [Internet]. 2011;366(1579):2759–65. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3146778&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol [Internet]. 2003. July;77(13):7330–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/164796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santra S, Liao H, Zhang R, Muldoon M, Watson S, Fischer W, et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med [Internet]. 2010. March;16(3):324–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20173754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakari M, Aboud S, Nilsson C, Francis J, Buma D, Moshiro C, et al. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine [Internet]. 2011. October 26;29(46):8417–28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21864626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llano A, Williams A, Olvera A, Silva-Arrieta S, Brander C. Best-Characterized HIV-1 CTL Epitopes: The 2013 Update. HIV Mol Immunol 2013. 2013;3–25.
- 7.Trolle T, McMurtrey CP, Sidney J, Bardet W, Osborn SC, Kaever T, et al. The Length Distribution of Class I–Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele–Specific Binding Preference. J Immunol [Internet]. 2016;196(4):1480–7. Available from: 10.4049/jimmunol.1501721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Zhang M, Cong H. Advances in the study of HLA-restricted epitope vaccines. Hum Vaccin Immunother [Internet]. 2013. December;9(12):2566–77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24439530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24(47–48):6893–904. [DOI] [PubMed] [Google Scholar]
- 10.Malhotra U, Li F, Nolin J, Allison M, Zhao H, Mullins JI, et al. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J Virol [Internet]. 2007;81(10):5225–37. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1900243&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asbach B, Meier JP, Pfeifer M, Köstler J, Wagner R. Computational Design of Epitope-Enriched HIV-1 Gag Antigens with Preserved Structure and Function for Induction of Broad CD8+ T Cell Responses. Sci Rep. 2018;8(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anthony DD, Lehmann P V. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29(3):260–9. [DOI] [PubMed] [Google Scholar]
- 13.Draenert R, Altfeld M, Brander C, Basgoz N, Corcoran C, Wurcel AG, et al. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J Immunol Methods. 2003;275(1–2):19–29. [DOI] [PubMed] [Google Scholar]
- 14.Tobery TW, Wang S, Wang X-M, Neeper MP, Jansen KU, McClements WL, et al. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J Immunol Methods [Internet]. 2001;254(1–2):59–66. Available from: http://www.sciencedirect.com/science/article/pii/S0022175901003970 [DOI] [PubMed] [Google Scholar]
- 15.Lehmann PV, Suwansaard M, Zhang T, Roen DR, Kirchenbaum GA, Karulin AY, et al. Comprehensive Evaluation of the Expressed CD8+ T Cell Epitope Space Using High-Throughput Epitope Mapping. Front Immunol. 2019;10(April):655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong JT, Colvin RB. Selective reduction and proliferation of the CD4+ and CD8+ T cell subsets with bispecific monoclonal antibodies: Evidence for inter-T cell-mediated cytolysis. Clin Immunol Immunopathol. 1991;58(2):236–50. [DOI] [PubMed] [Google Scholar]
- 17.Hayes PJ, Cox JH, Coleman AR, Fernandez N, Bergin PJ, Kopycinski JT, et al. Adenovirus-based HIV-1 vaccine candidates tested in efficacy trials elicit CD8+ T cells with limited breadth of HIV-1 inhibition. Aids [Internet]. 2016;30(11):1703–12. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=0000203020-1607170-00005 [DOI] [PubMed] [Google Scholar]
- 18.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, et al. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 2016;12(1):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amornkul PN, Karita E, Kamali A, Rida WN, Sanders EJ, Lakhi S, et al. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. Aids. 2013;27(17):2775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol [Internet]. 2002;76(16):8276–84. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12134033%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC155130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, Kaltsidis H, et al. Viral inhibition assay: a CD8 T cell neutralization assay for use in clinical trials of HIV-1 vaccine candidates. J Infect Dis [Internet]. 2010;201(5):720–9. Available from: http://jid.oxfordjournals.org/cgi/content/long/201/5/720 [DOI] [PubMed] [Google Scholar]
- 22.Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24(47–48):6893–904. [DOI] [PubMed] [Google Scholar]
- 23.Fiore-Gartland A, Manso BA, Friedrich DP, Gabriel EE, Finak G, Moodie Z, et al. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: An evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PLoS One. 2016;11(2):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill DK, Huang Y, Levine GL, Sambor A, Carter DK, Sato A, et al. Equivalence of ELISpot assays demonstrated between major HIV network laboratories. PLoS One. 2010;5(12):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boaz MJ, Hayes P, Tarragona T, Seamons L, Cooper A, Birungi J, et al. Concordant proficiency in measurement of T-cell immunity in human immunodeficiency virus vaccine clinical trials by peripheral blood mononuclear cell and enzyme-linked immunospot assays in laboratories from three continents. Clin Vaccine Immunol. 2009;16(2):147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubey S, Clair J, Fu T-M, Guan L, Long R, Mogg R, et al. Detection of HIV vaccine-induced cell-mediated immunity in HIV-seronegative clinical trial participants using an optimized and validated enzyme-linked immunospot assay. J Acquir Immune Defic Syndr [Internet]. 2007. May 1;45(1):20–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17310936 [DOI] [PubMed] [Google Scholar]
- 27.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(D1):D405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui H-H, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol [Internet]. 2006. July;24(7):817–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16767078 [DOI] [PubMed] [Google Scholar]
- 29.Kotturi MF, Peters B, Buendia-Laysa F, Sidney J, Oseroff C, Botten J, et al. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol [Internet]. 2007. May;81(10):4928–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17329346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8(March):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci [Internet]. 2013;110(22):E2046–53. Available from: 10.1073/pnas.1305227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertz T, Ahmed H, Friedrich DP, Casimiro DR, Self SG, Corey L, et al. HIV-1 Vaccine-Induced T-Cell Reponses Cluster in Epitope Hotspots that Differ from Those Induced in Natural Infection with HIV-1. PLoS Pathog. 2013;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calarota SA, Baldanti F. Enumeration and characterization of human memory t cells by enzyme-linked immunospot assays. Clin Dev Immunol. 2013;2013(Il). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, Kaltsidis H, et al. Viral Inhibition Assay: A CD8 T Cell Neutralization Assay for Use in Clinical Trials of HIV-1 Vaccine Candidates. J Infect Dis [Internet]. 2010;201(5):720–9. Available from: 10.1086/650492 [DOI] [PubMed] [Google Scholar]
- 37.Karulin A, Caspell R, Dittrich M, Lehmann P. Normal Distribution of CD8+ T-Cell-Derived ELISPOT Counts within Replicates Justifies the Reliance on Parametric Statistics for Identifying Positive Responses. Cells. 2015;4(1):96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moodie Z, Price L, Gouttefangeas C, Mander A, Janetzki S, Löwer M, et al. Response definition criteria for ELISPOT assays revisited. Cancer Immunol Immunother. 2010;59(10):1489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCutcheon M, Wehner N, Wensky A, Kushner M, Doan S, Hsiao L, et al. A sensitive ELISPOT assay to detect low-frequency human T lymphocytes. J Immunol Methods. 1997;210(2):149–66. [DOI] [PubMed] [Google Scholar]
- 40.Hudgens MG, Self SG, Chiu YL, Russell ND, Horton H, McElrath MJ. Statistical considerations for the design and analysis of the ELISpot assay in HIV-1 vaccine trials. J Immunol Methods. 2004;288(1–2):19–34. [DOI] [PubMed] [Google Scholar]
- 41.Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J Biomed Inform [Internet]. 2015;53:405–14. Available from: doi: 10.1016/j.jbi.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J Immunol Res. 2017;2017. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


