Summary
Embryonic hematopoietic stem and progenitor cells (HSPCs) robustly proliferate while maintaining multilineage potential in vivo; however, an incomplete understanding of spatiotemporal cues governing their generation has impeded robust production from human induced pluripotent stem cells (iPSCs) in vitro. Using the zebrafish model, we demonstrate that NLRP3 inflammasome-mediated interleukin-1-beta (IL1β) signaling drives HSPC production in response to metabolic activity. Genetic induction of active IL1β or pharmacologic inflammasome stimulation increased HSPC number as assessed by in situ hybridization for runx1/cmyb and flow cytometry. Loss of inflammasome components, including il1b, reduced CD41+ HSPCs, and prevented their expansion in response to metabolic cues. Cell ablation studies indicated that macrophages were essential for initial inflammasome stimulation of Il1rl1+ HSPCs. Significantly, in human iPSC-derived hemogenic precursors, transient inflammasome stimulation increased multilineage hematopoietic colony-forming units and T-cell progenitors. This work establishes the inflammasome as a conserved metabolic sensor that expands HSPC production in vivo and in vitro.
Keywords: Inflammasome, Hematopoietic Stem Cell (HSC), IL1β, endothelial-to-hematopoietic transition (EHT), zebrafish, inflammation, iPSC, Nlrp3
eTOC:
Frame et al. identify an integral role for the inflammasome in stimulating de novo production of zebrafish and human hematopoietic stem and progenitor cells (HSPCs). Inflammasome-associated IL1β signaling is induced in vivo by developmental metabolic cues and relayed from primitive macrophages to hemogenic endothelium to promote embryonic HSPC formation.
Graphical Abstract
Introduction
Hematopoietic stem and progenitor cell (HSPC) transplantation can be used as a curative treatment in individuals with hematologic disorders or malignancies. However, these therapies depend on donor HSPC abundance and histocompatibility, thereby restricting their application. Improving HSPC generation from autologous pluripotent cell sources could overcome this limitation. Currently, attempts to derive bona fide human HSPCs from induced pluripotent stem cells (iPSCs) require genetic manipulation to induce robust expansion and achieve long-term multilineage engraftment in murine models (Daniel et al., 2016; Sugimura et al., 2017). Further elucidation of conserved spatiotemporal regulators of HSPC specification and expansion acting in vivo in model systems are necessary for the optimization of in vitro cultures for therapeutic use. Here, we describe a connection between metabolic state and sterile inflammatory signaling that regulates HSPC production through inflammasome activity in the zebrafish embryo. Furthermore, we demonstrate conservation of inflammasome activation in modulating expansion and multipotency of human iPSC-derived HSPCs.
The ontogeny of the vertebrate hematopoietic system is a complex yet tightly orchestrated process. Several highly conserved “waves” of hematopoietic cells emerge over developmental time, with each becoming increasingly diverse in terms of lineage potential and expansion capabilities (Dzierzak and Speck, 2008; Medvinsky et al., 2011; Clements and Traver, 2013). Initial waves of primitive erythroid and myeloid-restricted progenitors are closely followed by bipotent erythro-myeloid progenitors and lymphoid-restricted progenitors formed in the posterior blood island and caudal aortic endothelium of the zebrafish (Bertrand et al., 2007; Tian et al., 2017), and the yolk sac of murine and human embryos (McGrath et al., 2015; Böiers et al., 2013; Ivanovs et al., 2017). Finally, hematopoietic stem cells with extensive self-renewal and multilineage differentiation capacity arise from a subset of “hemogenic” endothelium lining the embryonic dorsal aorta in all vertebrates. In the zebrafish aorta, commitment of phenotypic endothelium to hemogenic fate is signified by the step-wise acquisition of gata2b expression, which in turn upregulates runx1 expression around 24 hours post fertilization (hpf) (Butko et al., 2015). Subsequently, individual Runx1+ cells acquire rounded, hematopoietic morphology, and egress from the ventral wall through a process termed “endothelial-to-hematopoietic transition” (EHT) (Bertrand et al., 2010; Kissa and Herbomel, 2010; Lam et al., 2010). The majority of Runx1-dependent HSPC “budding” initiates from 30–36hpf, followed by egress from the endothelium from 40–52hpf (Kissa and Herbomel, 2010). HSPCs subsequently migrate to the caudal hematopoietic tissue (CHT), and eventually, the thymus and kidney marrow to expand and differentiate. There is increasing evidence that the initial populations of embryonic hematopoietic cells provide instructive cues to trigger HSPC production. For example, sterile inflammatory cytokine signaling promotes formation of zebrafish and murine HSPCs during embryonic development, independently of infection or injury (Orelio et al., 2008, 2009; Li et al., 2014; Sawamiphak et al., 2014; Espín-Palazón et al., 2014; He et al., 2015). Both macrophages (Li et al., 2014; Mariani et al., 2019) and neutrophils (Espín-Palazón et al., 2014) have been identified as sources of inflammatory cues. However, it remains unclear how these accessory cell types initiate inflammatory cascades to specify and/or amplify embryonic HSPC production.
One of the master regulators of inflammation, IL1β, directs adult HSPCs to divide, and promotes emergency granulopoiesis and T cell activation through signaling of downstream cytokines (Dinarello, 2009, 2011; Pietras et al., 2016). Although the acute effects of IL1β in infection and immunity are typically beneficial, chronic inflammation can be detrimental to adult HSC maintenance, thus, inflammatory signals must be tightly modulated to maintain optimal physiologic responses (Essers et al., 2009; Baldridge et al., 2010; King and Goodell, 2011; Takizawa et al., 2011; Esplin et al., 2011). Typically sourced in large quantities by myeloid cells, especially macrophages, IL1β activity is controlled at the protein level by inflammasomes (Dinarello, 2009). These multimeric complexes are generally comprised of one member of a family of Nod-like pattern recognition receptors (NLR), the adapter protein Pycard, and Caspase1, which autoactivates and cleaves IL1β and a related cytokine IL18, in response to a variety of microbial and host-derived stimuli (He et al., 2016; Jo et al., 2016). Inflammasome-forming proteins are present in zebrafish (Masumoto et al., 2003; Kuri et al., 2017; Li et al., 2018), and zebrafish Caspase a (Caspa) has the ability to cleave IL1β (Vojtech et al., 2012). Inflammasome activation in vitro involves two sequential steps: a “priming” signal, usually mediated by Toll-like Receptor (TLR) activation, resulting in transcriptional activation of NFκB and synthesis of IL1β and NLR proteins, followed by a second “activation” signal, which is responsible for assembly and activation of caspase-1 to cleave IL1β into its active form (He et al., 2016; Jo et al., 2016). In vivo, exogenous induction of inflammasome priming is not usually required for pharmacologic activators to generate a response (Rock et al., 2010). Intriguingly, both TLR signaling and NFκB were previously shown to be active in developing HSPCs (Espín-Palazón et al., 2014; He et al., 2015), suggesting that sterile inflammasome “priming” occurs normally during embryogenesis.
The most well-studied inflammasome, NLRP3, is often associated with metabolic alterations in adult pathologies, such as type 2 diabetes and atherosclerosis (Lee et al., 2013; Patel et al., 2017; Hughes and O’Neill, 2018). In this context, inflammasome activation and subsequent IL1β production can be induced by heightened sterile “danger” signals, including elevations in the glycolytic enzyme hexokinase, reactive oxygen species (ROS), and cholesterol deposits (Moon et al., 2015; Duewell et al., 2010; Camell et al., 2015; Abderrazak et al., 2015). Priming of the NLRP3 inflammasome is reliant on glycolysis and HIF1α stabilization in adult macrophages (Masters et al., 2010; Tannahill et al., 2013), and is enhanced in response to high glucose concentrations or hypoxia in mesangial cells (Tseng et al., 2016). In mice, macrophages increase IL1β secretion after normal feeding (Dror et al., 2017). Adult HSPCs are also capable of inflammasome activation, as murine HSPCs with hyperactive Nlrp1a have compromised function (Masters et al., 2012). However, whether the inflammasome responds to metabolic cues to regulate embryonic HSPC production is undetermined.
We previously established that glucose metabolism expands HSPC formation in zebrafish embryos through elevated ROS generation and HIF1α-mediated induction of key hematovascular genes and signals (Harris et al., 2013; Lim et al., 2017). While HIF1α activation can induce il1b transcription in developing zebrafish macrophages (Ogryzko et al., 2019), a connection between energy metabolism and sterile inflammasome activity in the context of HSPC formation has not been explored. Here, we provide evidence that metabolic alterations promote inflammasome-induced IL1β signaling to enhance HSPC production, which has implications for future efforts to derive functional human HSPCs de novo for therapeutic application.
Results
Metabolic stimulation induces inflammasome priming in the embryo
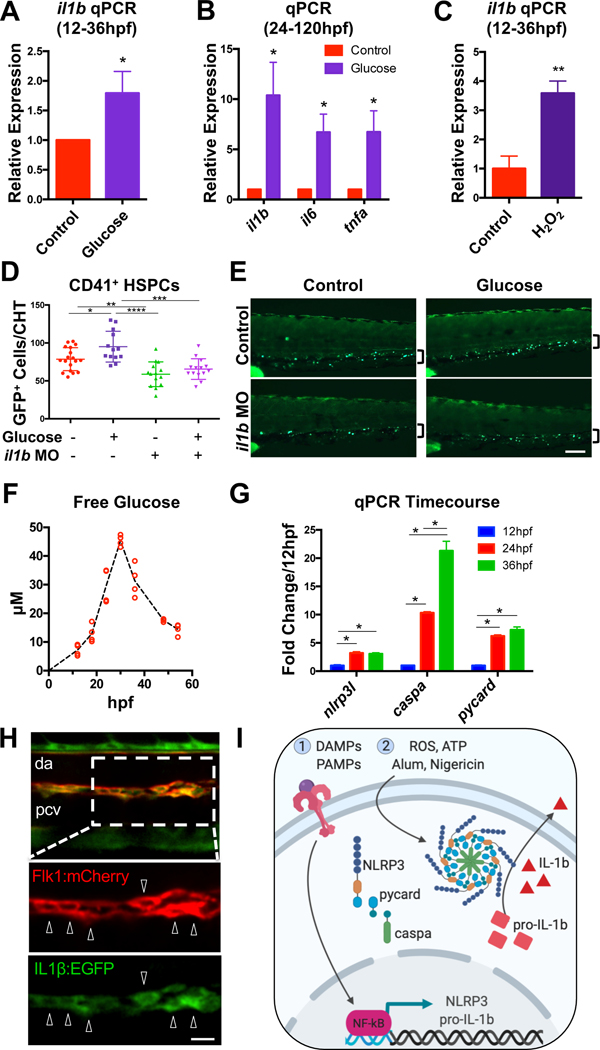
We previously demonstrated that metabolic stimulation promotes HSPC formation in zebrafish embryos via ROS-mediated HIF1α stabilization. In addition to a HIF1α signature, prior transcriptional analyses indicated that a strong inflammatory response was induced by glucose metabolism (Harris et al., 2013). Quantitative PCR (qPCR) analyses confirmed that exogenous glucose (12hpf) globally induced il1b at 24hpf and 36hpf, during the time of HSPC production (Fig. 1A, Fig. S1A). This induction was more pronounced after prolonged glucose exposure, and also resulted in substantial increases in expression of tnfa (Sieger et al., 2009) and il6, cytokines previously found to promote embryonic HSPC specification (Espín-Palazón et al., 2014; Lim et al., 2017)(Fig. 1B). Increased il1b expression was similarly observed following exposure to ROS during HSPC specification (12–36hpf) (Fig. 1C), or stabilization of HIF1α with CoCl2 treatment (24–120hpf) (Fig. S1B) (Harris et al., 2013). These data indicate that il1b transcripts are induced by activation of the glucose-ROS-HIF axis, and suggest that metabolic cues may prime embryos for HSPC formation and/or expansion through production of inflammatory cytokines.
Figure 1. Metabolic stimulation promotes expression of inflammatory cytokines.
(A) Glucose (1% in E3 water, 12–36hpf) increases il1b expression during HSPC specification (n=8, mean ±SEM) and (B) expansion (1% glucose from 24–120hpf; n=4, mean ±SEM). (C) Reactive oxygen species (0.005% H2O2; 12–36hpf) increases il1b expression. (n=4, mean ±SEM). (D,E) Morpholino oligonucleotides (MO) targeting il1b prevents glucose (12–48hpf) from increasing CD41+ HSPCs in the CHT. Significance determined by ANOVA with Tukey’s multiple comparison test. Scale bar, 100μm. (F) Glucose levels measured in pooled embryo lysates from 2 independent clutches over developmental time. Each point represents the glucose concentration from 24 embryos. (G) Inflammasome component expression over developmental time (n=4, mean ±SEM). (H) IL1β:EGFP expression (bottom inset, arrowheads) in the Tg(il1b:EGFP;flk1:mCherry) dorsal aorta (da) at 36hpf. Pcv, posterior cardinal vein. Scale bar, 10μm. (I) NLRP3 inflammasome model. Image created with BioRender.com. *P<0.05, **P<0.01, ***P<0.001; ****P<0.0001. See also Figure S1.
To determine if IL1β function is required to mediate the effects of glucose metabolism on HSPC formation, morpholino-mediated knockdown of il1b was performed in the context of glucose exposure. Consistent with our prior observations (Li et al., 2014), il1b morphants (López-Muñoz et al., 2011) exhibited a reduction in expression of HSPC markers runx1/cmyb by whole-mount in situ hybridization (WISH), and generated fewer CD41+ HSPCs in Tg(−6.0itga2b:egfp) embryos (Harris et al., 2013; Li et al., 2015; Ma et al., 2011) when compared with controls at 48hpf (Fig. 1D–E, Fig. S1C–E). Furthermore, this deficit partially prevented glucose from inducing runx1/cmyb expression and HSPC number, suggesting that IL1β is required to promote HSPC expansion in the setting of increased glucose bioavailability.
To examine whether endogenous glucose fluctuations in the zebrafish embryo may promote sterile inflammatory signaling in the absence of exogenous metabolic stimulation, glucose levels were measured at frequent intervals throughout HSPC specification. Intriguingly, embryonic glucose levels tripled between 18 and 30hpf (Fig. 1F), consistent with prior observations, and coincident with the developmental onset of gluconeogenesis in zebrafish embryos (Dhillon et al., 2019; Jurczyk et al., 2011). To determine whether endogenous glucose production was temporally correlated with inflammasome priming, expression of inflammasome components in the embryo were assayed. Expression of nlrp3l, caspa, and pycard increased over the same developmental time (Fig. 1G), suggesting that inflammasome complexes become functional during the window of HSPC specification. Analysis of Tg(il1b:EGFP) embryos (Hasegawa et al., 2017) demonstrated EGFP co-expression with primitive macrophages in Tg(mfap4:tdtomato) embryos (Walton et al., 2015), both in circulation and closely associated with the aortic endothelium (Fig. S1F), as well as the endothelium itself in Tg(flk1:mCherry) embryos at 36hpf (Hogan et al., 2009) (Fig. 1H). Furthermore, FACS-isolated Il1β:EGFP+ cells expressed markers indicative of macrophage (mpeg1), neutrophil (mpx) and HSPC and/or thrombocyte (CD41) identity by PCR (Fig. S1G). Together, these data imply that inflammasome-derived IL1β signaling (Fig. 1I) is primed by metabolic cues, and is active in cell populations that are temporally and spatially associated with HSPC formation.
Inflammasome complexes are necessary for normal HSPC generation
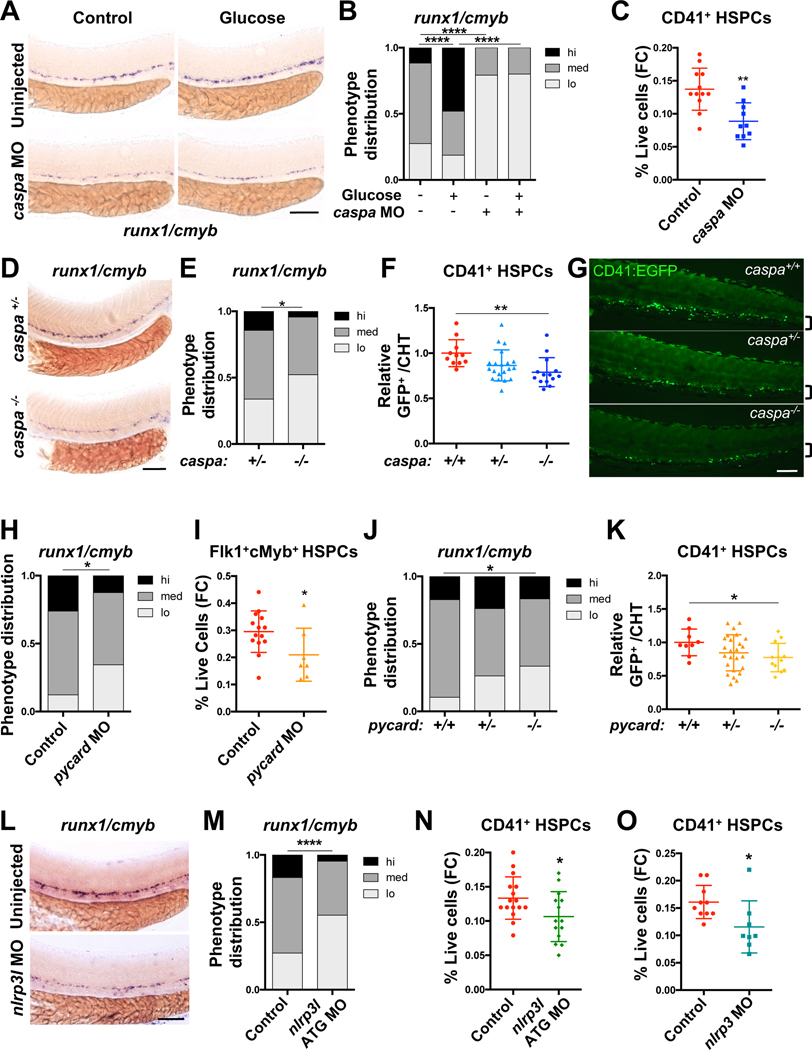
After transcriptional priming of il1b and inflammasome components, pro-IL1β protein is cleaved and activated by multimeric inflammasome complexes in response to sterile or microbial stimuli (Fig. 1I). To determine if inflammasome components are functionally required during HSPC formation, targeted knockdown studies were performed. Disruption of the effector caspa (Masumoto et al., 2003) substantially reduced runx1/cmyb expression by WISH (Fig. 2A–B; Fig. S2A–B), and glucose exposure failed to increase HSPC gene expression in morphant embryos. This decrease was not associated with alterations in arterial specification, as efnb2 expression was unchanged in morphants, but notably, expression of the hemogenic marker gata2b was reduced (Fig. S2C–D). A significant reduction of CD41+ HSPCs in caspa morphants was quantified by flow cytometry (Fig. 2C). While transient caspase inhibition with non-toxic doses of Ac-YVAD-CMK only caused a trend toward diminished runx1/cmyb expression (Fig. S2E), consistent with prior reports (Tyrkalska et al., 2019), analysis of caspa−/− embryos (Kuri et al., 2017) demonstrated a reduction of runx1/cmyb in the aorta (Fig. 2D–E), with mutants exhibiting a blunted response to glucose exposure (Fig. S2F). Fewer CD41+ HSPCs were also enumerated in the CHT of caspa−/− embryos compared with wild-type clutchmates (Fig. 2F–G).
Figure 2. Inflammasome action regulates HSPC formation.
(A-B) Expression of runx1/cmyb in embryos with or without morpholino (MO)-mediated caspa knockdown or 1% glucose treatment (12–36hpf) assessed by WISH. (C) caspa knockdown reduces frequency of CD41:EGFP+ HSPCs by flow cytometry (FC) at 48hpf. (D-E) runx1/cmyb WISH of caspa−/− aortas compared with caspa+/− embryos at 36hpf. (F-G) CD41:EGFP+ HSPCs in the CHT of embryos from caspa+/− incrosses at 48hpf. Counts were normalized relative to caspa+/+ embryos for each clutch. Significance was determined by Tukey’s multiple comparison test. (H-I) pycard knockdown reduces runx1/cmyb expression by WISH (36hpf), and the frequency of HSPCs in Tg(flk1:dsRed;cmyb:egfp) embryos by flow cytometry (48hpf). (J) runx1/cmyb WISH in pycard−/− embryos compared with stage-matched pycard+/+ and pycard+/− embryos. (K) CD41+ HSPCs in the CHT of embryos from pycard+/− incrosses. Counts were normalized relative to pycard+/+ embryos for each clutch. Significance was determined by Fisher’s LSD test. (L-M) nlrp3l knockdown reduces runx1/cmyb by WISH and (N) the frequency of HSPCs assayed by flow cytometry at 48hpf. (O) nlrp3 knockdown reduces the frequency of CD41+ HSPCs at 48hpf. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Scale bars, 100μm. See also Figure S2.
To confirm that these effects on HSPC development were due to loss of sterile inflammasome function, additional components of the complex were targeted. Knockdown of the adapter pycard (Progatzky et al., 2014) (Fig. S2G) significantly decreased runx1/cmyb expression by WISH and numbers of Flk1+cMyb+ HSPCs in Tg(flk1:dsRed;cmyb:EGFP) embryos (Bertrand et al., 2010; Kikuchi et al., 2011; North et al., 2007) assessed by flow cytometry (Fig. 2H–I). A reduction in runx1/cmyb, as well as numbers of CD41+ HSPCs, was also observed in pycard mutants (Matty et al., 2019) compared to pycard+/+ embryos (Fig. 2J–K). A large family of putative NLR sensor genes have been identified in zebrafish (Laing et al., 2008); knockdown of a predicted sensor gene nlrp3l phenocopied caspa and pycard morphants with reduced runx1/cmyb in the aorta at 36hpf (Fig. 2L–M, Fig. S2H–J). Rag1 expression, indicative of reduced or delayed HSPC colonization in the thymus, was also decreased in nlrp3l morphants (Fig. S2K–L). Fewer CD41+ HSPCs were detected in nlrp3l morphants, as well as in embryos in which a newly annotated and characterized nlrp3 gene was targeted (Li et al., 2020)(Fig. 2N–O). Together, these data suggest that Nlrp3 inflammasome activity is necessary for embryonic HSPC formation.
Overexpression of active IL1β enhances embryonic HSPC expansion
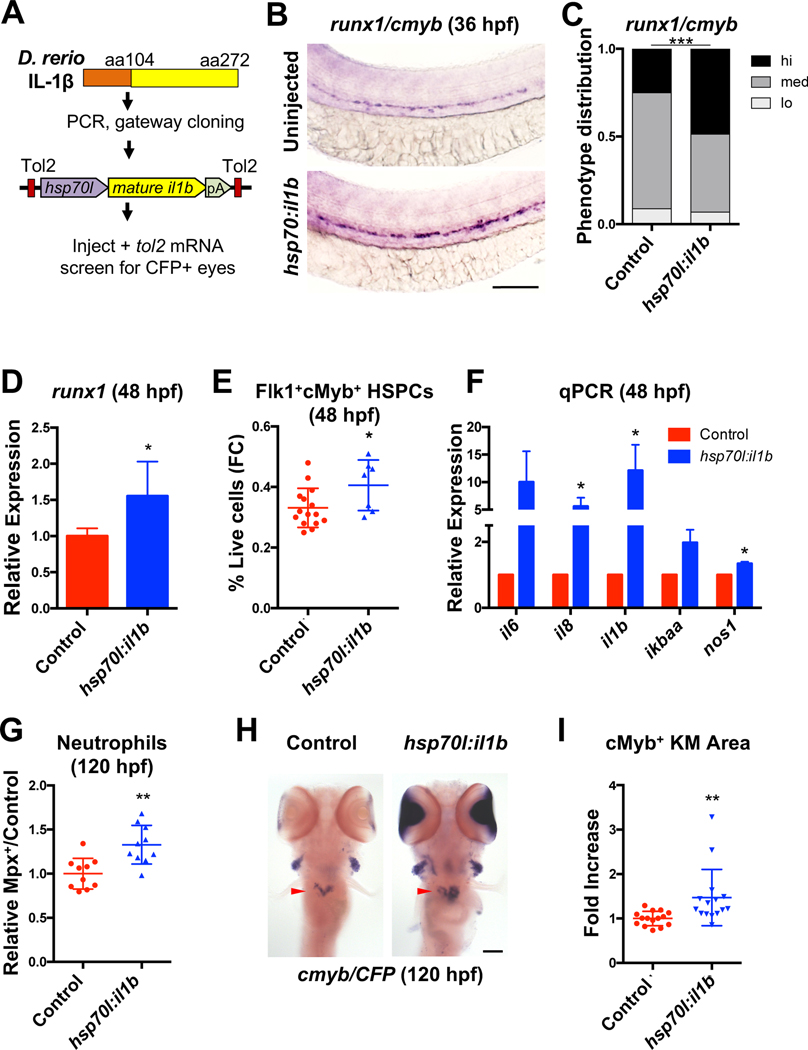
Previously reported effects of exogenous IL1β on embryonic HSPC formation relied on ex vivo culture (Orelio et al., 2008, 2009). To assess functional impact in vivo, an Hsp70-inducible transcript encoding the fully processed, active form of zebrafish IL1β was engineered (Vojtech et al., 2012) (Fig. 3A). Induction of active IL1β (actIL1β; 32hpf) increased runx1/cmyb expression in the developing aorta at 36hpf (Fig. 3B–C), which was confirmed by runx1 qPCR (Fig. 3D). Numbers of Flk1+cMyb+ HSPCs were likewise increased by flow cytometry (Fig. 3E). Consistent with the induction of an inflammatory response, expression of known target genes were increased with actIL1β induction (Fig. 3F)(Cortes et al., 2016; North et al., 2009) and numbers of neutrophils in Tg(mpx:gfp) embryos (Renshaw et al., 2006) were elevated by flow cytometry (Fig. 3G). Importantly, actIL1β overexpression at 24hpf was sufficient to recover wildtype levels of runx1 expression in il1b and pycard morphants by 36hpf (Fig. S3A–D), confirming that the active form bypasses a functional requirement for inflammasome assembly and Caspa activity. Finally, expression of cmyb was significantly increased in the kidney marrow at 120hpf with actIL1β overexpression, consistent with an acute expansion of the HSPC pool (Fig 3H–I).
Figure 3. IL1β mediates HSPC production.
(A) Generation of Tg(hsp70l:il1b) embryos. (B-C) il1b overexpression (actIL1β; 32hpf) promotes runx1/cmyb expression at 36hpf in chimeric injected animals. (D) actIL1β induces runx1 in whole Tg(hsp70l:il1b) embryos by qPCR (48hpf, n=5, mean ±SEM). (E) HSPCs quantified by flow cytometry (FC) in Tg(flk1:dsRed;cmyb:GFP;hsp70l:il1b) embryos compared with actIL1β− controls (48hpf). (F) actIL1β (32hpf) induces expression of il1b and common targets (n>3, 48hpf, mean ±SEM). (G) Relative frequency of neutrophils in 120hpf Tg(mpx:GFP;hsp70l:il1b) embryos heat-shocked at 32hpf. (H-I) Kidney marrow area (arrow), denoted by cmyb expression, is proportionally increased at 120hpf after actIL1β induction at 32hpf. CFP eye signal was used to identify presence of the hsp70l:il1b transgene. Scale bars, 100μm. *P<0.05, **P<0.01. See also Figure S3.
HSPC production is enhanced by inflammasome activation
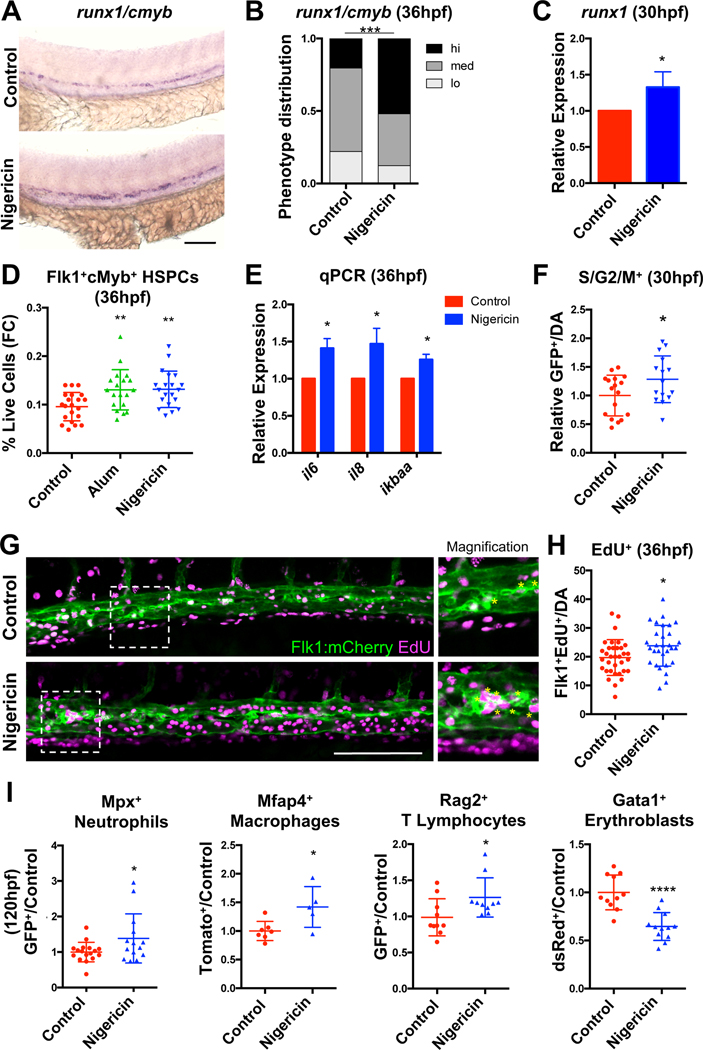
To determine whether pharmacologic inflammasome activation could be utilized to promote HSPC production through endogenous IL1β production, runx1/cmyb expression was assessed in embryos treated with established agonists of inflammasome assembly (Jo et al., 2016). Nigericin (0.1μM), which induces inflammasome action through alterations in intracellular potassium, increased runx1/cmyb expression in the dorsal aorta (Fig. 4A–B), and runx1 levels by qPCR (Fig. 4C). Acute inflammasome stimulation with nigericin or an alternative agonist, alum crystals (alum, 20 μg/mL), which prompts activation through lysosomal perturbations (Hornung et al., 2008), increased numbers of phenotypic Flk1+cMyb+ HSPCs (Fig. 4D). IL1β target expression was also increased by qPCR following inflammasome stimulation (Fig. 4E). These effects are likely due to enhanced proliferation, as numbers of dividing cells in 30hpf Tg(EF1a:mAG-zGem(1/100))rw0410h embryos (Sugiyama et al., 2009) as well as numbers of Flk1+EdU+ cells in the aortic floor at 36hpf were elevated in nigericin-treated embryos (Fig. 4F–H). Epistasis analyses with il6 morpholinos (Lim et al., 2017) suggest that il6 upregulation is at least partially required downstream of inflammasome action to promote runx1/cmyb expression (Fig. S4A–B). Furthermore, forced expression of a dominant negative IkB transgene, which suppresses il1b transcription (Espín-Palazón et al., 2014), prevented nigericin from increasing cmyb in the aorta of Tg(hsp70l:GAL4;UAS:dnnfkbiaa) embryos at 36hpf (Jeong et al., 2007), confirming the requirement for NFκB priming and subsequent IL1β signaling to boost numbers of HSPCs in vivo (Fig. S4C). Analyses of embryos at 120hpf revealed that inflammasome stimulation (24–120hpf) was correlated with increases in the proportion of Rag2+ T lymphocytes (Langenau et al., 2003), Mpx+ neutrophils, and Mfap4+ macrophages, while suppressing the frequency of Gata1+ erythroblasts (Traver et al., 2003), consistent with recent reports (Tyrkalska et al., 2019) (Figure 4I; Fig. S4D–I). To determine whether there were lasting effects of embryonic inflammatory stimulation on adult steady-state hematopoiesis, lineage distribution was assessed in kidney marrow from inflammasome-stimulated embryos. This analysis revealed no significant alterations in lymphoid, myeloid, or precursor compartments at 3 months of age (Fig. S4J). Taken together, these data indicate that transient inflammasome activation expands developing HSPCs, which may temporarily restrict erythroid commitment of embryonic progenitors in favor of lympho-myeloid potential, without compromising adult HSPC differentiation capacity or maintenance.
Figure 4. HSPC production is enhanced by inflammasome activation.
(A-B) Inflammasome stimulation (12–36hpf) promotes runx1/cmyb expression by WISH. (C) runx1 qPCR (n=4, mean ±SEM). (D) Nigericin or Alum (12–36hpf) increased HSPCs by flow cytometry (FC). Significance was determined by ANOVA with Dunnett’s post-hoc test. (E) qPCR of IL1β target genes (n=6, mean ±SEM). (F) Relative numbers of S/G2/M+ cells in Tg(EF1a:mAG-zGem(1/100)) aortas with nigericin stimulation (12–30hpf) (DA; dorsal aorta; Fold change calculated within each clutch). (G-H) Numbers of Flk1+EdU+ cells in the aortic floor in 36hpf Tg(flk1:mCherry) embryos labeled with EdU antibody. Asterisks in inset signify positive cells. (I) The proportion of Mpx+ neutrophils, Mfap4+ macrophages, Rag2+ lymphocytes, and Gata1+ erythroblasts in transgenic embryos was assessed with prolonged nigericin stimulation (24–120hpf) by flow cytometry. *P<0.05, **P<0.01, ****P<0.0001. Scale bars, 100μm. See also Figure S4.
Macrophages are an important source of IL1β during HSPC formation
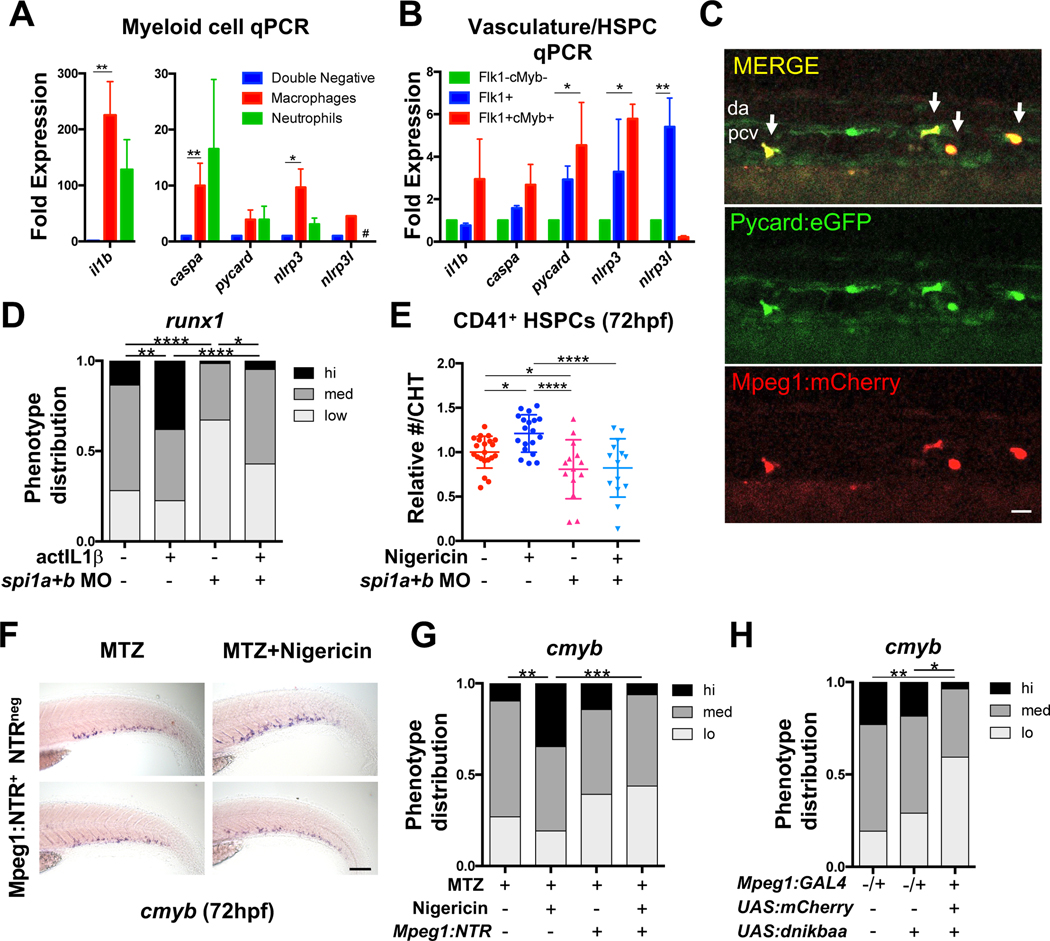
Inflammasome activation is thought to occur primarily in myeloid cells. To investigate the cell-type(s) involved in inflammasome regulation of embryonic HSPC production, primitive myeloid populations, vasculature, and developing HSPCs were profiled for expression of inflammasome components. Consistent with prior analyses (Nguyen-Chi et al., 2014) and our observations for Il1β:EGFP expression (Fig. S1F–G), il1b was significantly enriched (225-fold) in Mpeg1+ macrophages (Palha et al., 2013) and Mpx+ neutrophils (128-fold) isolated by FACS compared with the bulk negative (Mpeg1-Mpx-) population at 48 hpf; caspa and nlrp3 were also significantly enriched (10-fold) in macrophages (Fig. 5A). Inflammasome components were also detected at 48hpf in FACS-isolated Flk1+ vasculature and Flk1+cMyb+ HSPCs, with significant enrichments in nlrp3l (Flk1+) and pycard/nlrp3 (Flk1+cMyb+) compared with the bulk negative (Flk1-cMyb-) fraction (Fig. 5B). Expression of il1b in FACS-isolated Flk1+ and Mpeg1+ populations from Tg(mpeg1:gfp;flk1:mCherry) embryos (Ellett et al., 2011) indicated the strongest response to metabolic stimulation prior to HSPC specification (12–24hpf) was in macrophages (Fig. S5A). Imaging of Tg(pycard:pycard-EGFP) embryos (Kuri et al., 2017) (30 hpf) revealed that many pycardbright cells surrounding the aorta co-express the macrophage reporter Tg(mpeg1:mCherry) (Fig. 5C)(Ellett et al., 2011), together suggesting that macrophages serve as the major local source of inflammasome-processed IL1β during the onset of HSPC formation.
Figure 5. Myeloid cells are required to mediate the effects of the inflammasome on HSPCs.
(A) Inflammasome component qPCR in sorted Mpeg1+ macrophages and Mpx+ neutrophils relative to the Mpeg1-Mpx- fraction in 48hpf Tg(mpeg1:GAL4;UAS:NTR-mCherry;mpx:GFP) embryos (mean ±SEM, n>3, Holm-Sidak testing). Mpx+ v. Negative; P=0.056; #, below detection. (B) Inflammasome component qPCR in sorted Flk1+ vasculature and Flk+cMyb+ HSPCs relative to the Flk1-cMyb- fraction at 48hpf (mean ±SEM, n=4, Holm-Sidak testing). (C) Co-expression of EGFP and mCherry (arrows) beneath the dorsal aorta (DA) of 30hpf Tg(mpeg1:mCherry;pycard:pycard-EGFP) embryos. PCV, posterior cardinal vein. Scale bar, 20μm. (D) Knockdown of spi1a/spi1b reduces runx1 expression by WISH; il1b overexpression (24hpf) partially rescues runx1 expression (36hpf). (E) Loss of spi1a/spi1b blocks nigericin-induced expansion of CD41+ HSPCs in the CHT (24–72hpf). Significance was determined by ANOVA with Fisher’s LSD test. (F-G) Metronidazole-mediated macrophage ablation (MTZ; 30hpf) in Tg(mpeg1:GAL4;UAS:nfsB-mCherry) (Mpeg:NTR) embryos prevents nigericin (30hpf) from increasing cmyb CHT expression by WISH (72hpf). Scale bar, 100μm. (H) NFκB blockade in Tg(mpeg1:GAL4;UAS:nfsB-mCherry;UAS:dnikbaa) embryos reduces cmyb CHT expression by WISH (72hpf). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S5.
To determine whether the effects of inflammasome stimulation required myeloid cells, inflammasome activation was performed in embryos with or without knockdown of myeloid transcriptional regulators spi1a/1b (Bukrinsky et al., 2009). Morphant embryos exhibited a substantial reduction in macrophages, with a more modest effect on neutrophil number (Fig. S5B). As with prior studies, spi1a/1b morphants had reduced runx1 expression (Li et al., 2014) (Fig. 5D; Fig. S5C). While overexpression of actIL1β partially recovered runx1 expression in the aorta of spi1a/1b morphants at 36hpf, nigericin stimulation failed to expand HSPC numbers in morphant embryos at 72hpf (Fig. S5C–D, Fig. 5D–E). In further support of an essential role for macrophages, chemical ablation of macrophages to approximately 38% of untreated controls in Tg(mpeg1:GAL4;UAS:nfsB-mCherry) embryos (Palha et al., 2013) prevented an increase in cmyb expression in the CHT at 72hpf in response to inflammasome stimulation (Fig. 5F–G; Fig. S5E). Similarly, macrophage-specific blockade of NFκB activity, and thus inflammasome priming, in Tg(mpeg:GAL4;UAS:nfsB-mCherry;UAS:dnikbaa) embryos significantly impaired acquisition of cmyb expression in the CHT when compared to controls (Fig. 5H). Altogether, these data suggest that NFκB priming and inflammasome activation in macrophages positively influences HSPC number in the embryo.
IL1β signals are received by HSPCs to further prime inflammasome activity
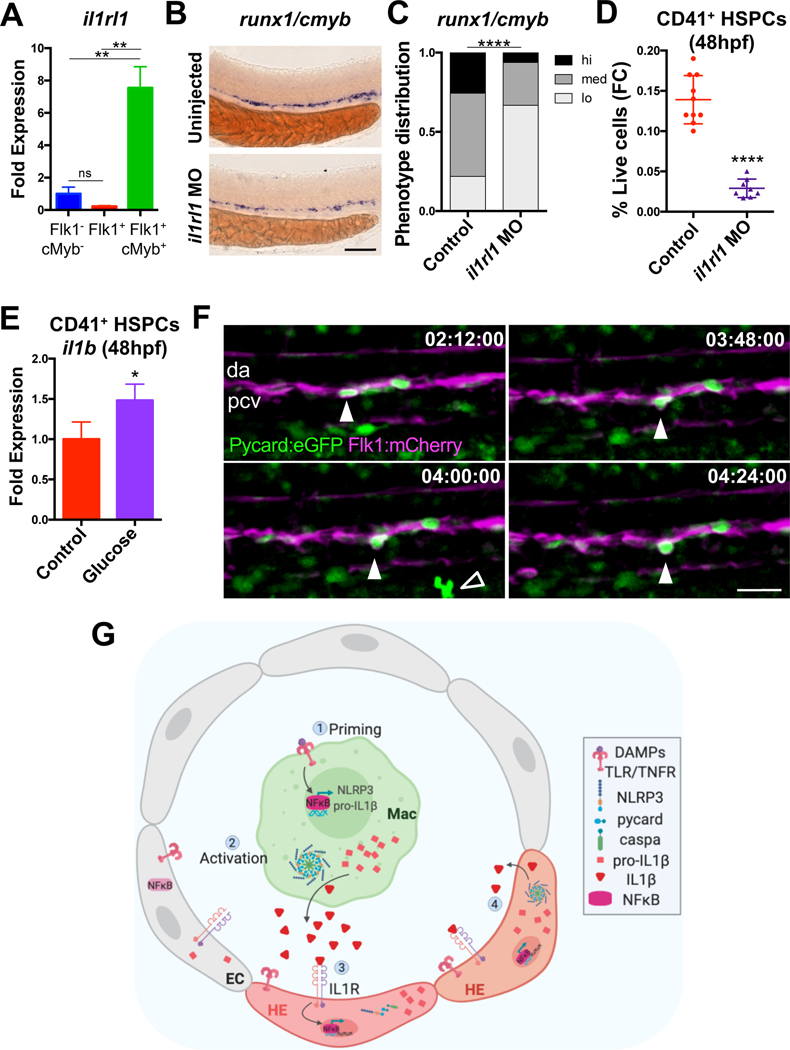
To assess whether developing HSPCs in the embryonic dorsal aorta can directly respond to IL1β stimulation provided by accessory macrophages, expression of a predicted IL-1 receptor 1 ortholog, il1rl1, was profiled and found to be significantly enriched in Flk1+cMyb+ HSPCs when compared with bulk vasculature and the non-endothelial fraction (Fig. 6A). Knockdown of il1rl1 (Fig. S6A) significantly reduced runx1/cmyb expression (Fig. 6B–C) and numbers of CD41+ HSPCs (Fig. 6D), confirming a functional role. As IL1β signaling is known to induce NFκB activation and il1b transcription (Dinarello, 2009), this raises the possibility that inflammasome priming and activation could subsequently be propagated within Il1rl1+ HSPCs. In support of this concept, CD41+ HSPCs were found to upregulate il1b at 48hpf in response to glucose treatment initiated at 12hpf (Fig. 6E). Furthermore, driving actil1b expression in Flk1+ cells was also sufficient to increase runx1 expression by WISH (Fig. S6B–C). Finally, in agreement with the observed enrichment of pycard by qPCR in Flk1+cMyb+ HSPCs (Fig. 5B), time-lapse microscopy confirmed the presence of budding Pycard+Flk1+ cells in the aorta (Fig. 6F, Movie S1, 34–36hpf), suggesting that developing HSPCs acquire competence to undergo inflammasome activation during the EHT process. Taken together, these data indicate that while macrophages provide critical inductive IL1β signals to promote embryonic HSPC development, this signal may be further relayed within nascent HSPCs to amplify their number (Fig. 6G).
Figure 6. IL1β signals are received by HSPCs to further prime inflammasome activity.
(A) il1rl1 qPCR in Flk1+ vasculature and Flk1+cMyb+ HSPCs relative to the Flk1-cMyb- fraction (48hpf). Mean ±SEM, n=3, Holm-Sidak testing. ns, not significant. (B-C) il1rl1 knockdown reduced runx1/cmyb expression in the aorta by WISH (36hpf). Scale bar, 100μm. (D) il1rl1 knockdown reduces CD41+ HSPCs (48hpf) by flow cytometry (FC). (E) il1b qPCR in sorted CD41+ HSPCs from 1% glucose-treated embryos (12–48hpf). (Mean ±SEM, n=6, 2 independent experiments; fold change was calculated from the average control in each experiment). (F) Sequential time-lapse images demonstrate budding of a Pycard+ cell from the dorsal aorta (DA) of a Tg(pycard:pycard-EGFP;flk1:mCherry) embryo (34–36hpf.) Open arrow, putative macrophage. PCV, posterior cardinal vein. Scale bar, 30μm. See also Movie S1. (G) Model of inflammasome regulation of HSPC production. EC, endothelial cell. HE, hemogenic endothelium. Image created from Biorender.com. *P<0.05, **P<0.01, ****P<0.0001. See also Figure S6.
Inflammasome stimulation promotes human HSPC expansion
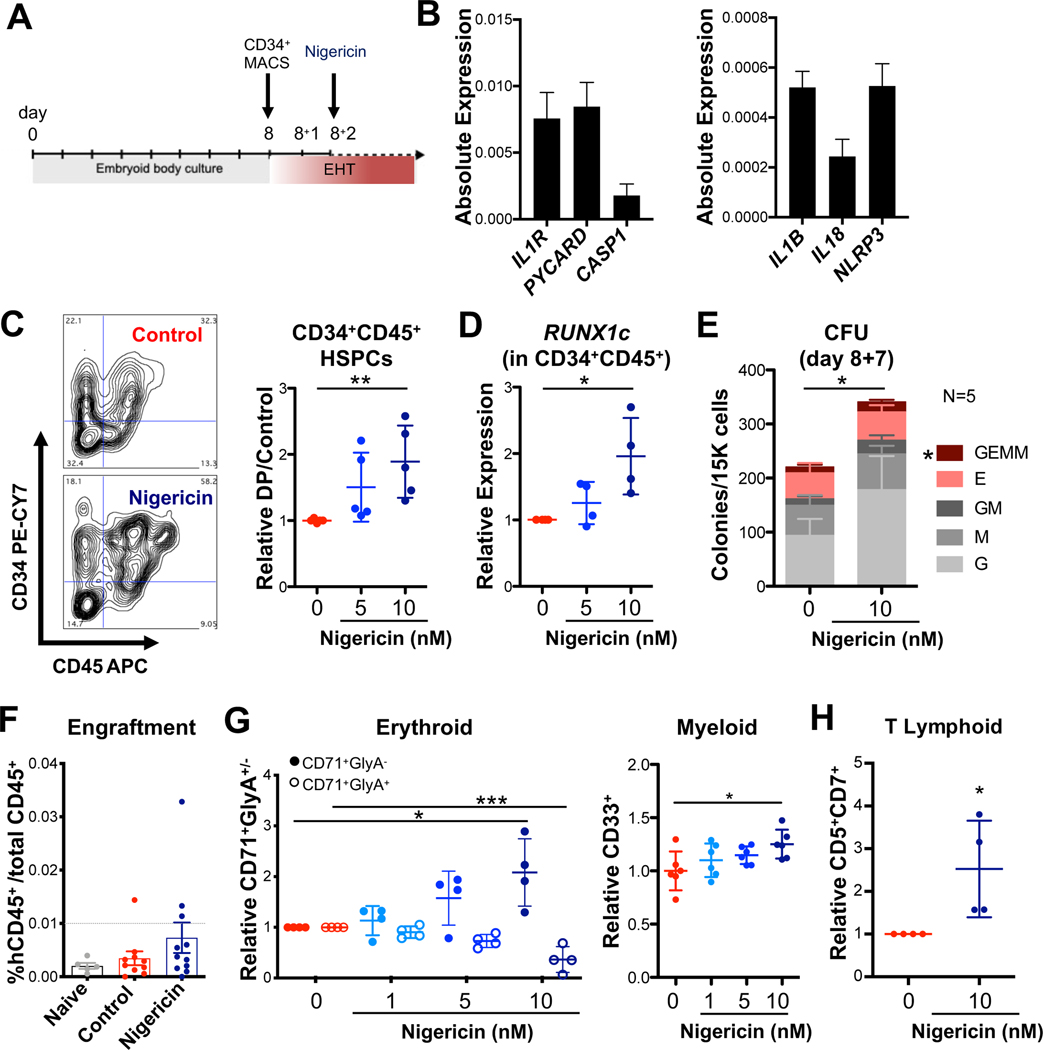
To build on the observations that inflammasome activity expands developing zebrafish HSPCs, a possible role for inflammasome stimulation in regulating human HSPC production from iPSC-derived CD34+ hemogenic endothelial cell cultures was next explored (Ditadi et al., 2015) (Fig. 7A). On day 2 post CD34 enrichment, cultures poised to undergo endothelial-to-hematopoietic transition (EHT) expressed IL1R, as well as NLRP3 inflammasome components (Gupta et al., 2019; Son et al., 2015; Spandidos et al., 2010), indicating that cells were competent to receive IL1β and further activate the inflammasome (Fig. 7B). Given the absence of CD34-CD45+ cells in EHT cultures on day 2 post CD34-enrichment (Fig. S7A), we postulated that addition of macrophages might be necessary to enhance the efficiency of IL1β production and inflammasome stimulation in vitro. To this end, THP-1 human monocytes were primed with lipopolysaccharide (LPS) to induce IL1B (Chanput et al., 2014; Sharif et al., 2007), seeded in transwell inserts over developing CD34+ EHT cultures, and subsequently activated with nigericin on day 2 of culture (Fig. S7B). Intriguingly, nigericin stimulation of co-cultures exerted a significant effect on phenotypic HSPC number (Fig. S7C). Co-culture with unstimulated or LPS-primed monocytes alone also improved the frequency of CD34+CD45+ phenotypic HSPCs over untreated controls, in the absence of nigericin-mediated inflammasome activation, suggesting that the monocytes supply both inflammasome-dependent and independent supportive factors to the HSPCs. As inflammasome stimulation with nigericin alone also significantly increased the frequency of CD34+/CD45+ cells in the absence of monocytes, we sought to further clarify the impact of inflammasome stimulation in the absence of other confounding paracrine signals.
Figure 7. Inflammasome stimulation boosts iPS-derived human HSPC activity in vitro.
(A) Schematic of human iPS cell hematopoietic differentiation cultures. Embryoid body-derived cells are enriched for CD34+ hemogenic endothelium (HE) on day 8 of differentiation and subjected to 7 more days of EHT culture. Nigericin (or EtOH to controls) was added on day 2 of EHT. (B) Expression of inflammasome components (relative to GAPDH) in day 2 EHT cultures (mean ±SEM, n=3 independent cultures, 3 samples/ culture). (C) Nigericin increases CD34+CD45+ cells on day 7, representative FACS plots are shown; n=5 independent differentiation cultures. (D) RUNX1c qPCR in CD34+CD45+ cells from nigericin-treated cultures relative to untreated controls. (E) CFU assays of floating cells harvested on day 7 of EHT culture from 5 differentiations. Nigericin significantly increases both total colonies and GEMM colonies (*P<0.05). (F) Engraftment potential of iPSCs transplanted on day 4 of EHT culture. % of human CD45+ cells was assessed from the total CD45+ population in the bone marrow at 8 weeks post-transplant (mean ±SEM). (G) Nigericin-stimulated cultures yield proportionally fewer CD71+GlyA+ cells and more CD33+ myeloid cells by day 8. (H) Nigericin stimulation on day 2 of EHT increases pro-T progenitors after 2 weeks. Each point represents the average fold change across multiple wells within a differentiation culture; n=4 independent differentiation cultures. *P<0.05, ***P<0.001. See also Figure S7.
As our zebrafish studies suggested that developing HSPCs are competent to not only respond to IL1β, but later activate the inflammasome, the impact of direct nigericin stimulation was further assessed. In addition to increasing the proportion of CD34+CD45+ phenotypic hematopoietic progenitor cells assayed on day 7 in a dose-dependent manner, nigericin treatment of EHT cultures also elevated the expression of RUNX1c within this population (Challen and Goodell, 2010; Navarro-Montero et al., 2017) (Fig. 7C–D), suggestive of improved commitment and/or expansion. Importantly, inflammasome stimulation increased the total numbers of functional hematopoietic colony-forming units as well as the number of CFU-GEMM colonies harvested on day 7 of EHT cultures (Fig. 7E). To determine whether inflammasome stimulation improved in vivo engraftment and differentiation capacity of iPSC-derived HSPCs, transplantation studies were performed. While transient inflammasome stimulation showed a trend toward enhancing human CD45+ engraftment of transplanted mice after 8 weeks, the overall lack of robust engraftment in the absence of additional genetic modification made it difficult to discern a selective advantage in multi-potent repopulating potential (Fig. 7F; Fig. S7D). Intriguingly, consistent with in vivo observations in the zebrafish embryo, continued differentiation in suspension cultures demonstrated that nigericin stimulation induced a strong bias toward myeloid over erythroid fate (Fig. 7G; Fig. S7E–F). Importantly, however, inflammasome-expanded progenitors were equally able to undergo lymphoid commitment in vitro, resulting in a net increase in iPSC-derived CD5+CD7+ pro-T cell formation (Fig. 7H; Fig. S7G). Together, these data indicate that inflammasome activation promotes short-term expansion of human HSPCs, and restrains premature erythroid differentiation in vitro.
Discussion
In the vertebrate embryo, there is emerging evidence that sterile inflammation is an endogenous regulator of HSPC number and lineage output. However, it has remained unclear how these signals are initiated and relayed to regulate the specification and differentiation of HSPCs. Here, we show that metabolic alterations associated with embryonic growth trigger the production of inflammatory signals through activation of the NLRP3 inflammasome to expand HSPC number. Our time course analyses demonstrate a precise temporal correlation between the peak of glucose bioavailability and the onset of HSPC budding from the aortic floor (Kissa and Herbomel, 2010). This energy-intensive budding process is also contemporaneous with the ability of embryos to utilize oxidative phosphorylation, which results in accumulation of ROS, a known NLRP3 inflammasome agonist, and HIF1α stabilization, which promotes il1b transcription in the embryo (Harris et al., 2013; Guo et al., 2015; Jo et al., 2016; Ogryzko et al., 2019). Coincidentally, global expression of inflammasome components increased during the time of HSPC formation, while il1b was observed to accumulate in developing myeloid populations by qPCR and in aortic endothelium of Tg(il1b:EGFP) embryos. Exogenous glucose stimulation further induced il1b expression, consistent with established connections between heightened metabolism and ROS formation in macrophages and subsequent IL1β production (Tannahill et al., 2013; Hall et al., 2018). Importantly, our observations that the endogenous peak of embryonic glucose concentration subsides after HSPC formation also suggests that the metabolic inflammasome stimulation is likely transient in nature, minimizing the potential for HSPC exhaustion and/or damage. Together, our data indicate that dynamic developmental fluctuations of energy sources coordinate the timing of sterile inflammatory signaling and the subsequent production of embryonic HSPCs, which may be suboptimal in current in vitro culture approaches.
IL1β was previously reported to impact murine HSPC formation (Orelio et al., 2008, 2009), however, its upstream developmental regulator was not fully defined. Consistent with a role for the NLRP3 inflammasome in regulating sterile IL1β activity in adult mammals (Haneklaus and O’Neill, 2015), morphants and mutants of zebrafish NLRP3 inflammasome components demonstrated a reduction in robust HSPC formation. These data reveal a previously undescribed role for NLRP3 activity as a homeostatic regulator of developmental hematopoiesis. Furthermore, epistasis analyses suggest that energy metabolism promotes inflammasome activity to increase IL1β production and HSPC marker expression. Our data also indicate that NFκB activity is a key requirement for inflammasome-mediated regulation of HSPC number in vivo, and is required in macrophages to initiate normal levels of cmyb expression. Intriguingly, NFκB also regulates Notch signaling within hemogenic endothelium (Espín-Palazón et al., 2014; He et al., 2015), and can regulate Hif1α, which was previously shown by our laboratory to promote HSPC development (Tirziu et al., 2012; D’Ignazio et al., 2016; Harris et al., 2013). Given that IkB itself is also an ΝFκΒ target (Sun et al., 1993), it will be interesting to further dissect how these transcriptional networks converge to establish and subsequently modulate inflammatory activity during HSPC formation. Although prior in vitro work has indicated that inflammasome priming is necessary to boost NLRP3 expression to induce robust pro-IL1β cleavage in macrophages (He et al., 2016), we find that human iPSC-derived CD34+ cultures basally express low levels of inflammasome components and respond to low concentrations of nigericin. Thus, it is possible that combinatorial use of agonists to further prime or stimulate the inflammasome may elicit a potent impact on HSPC production.
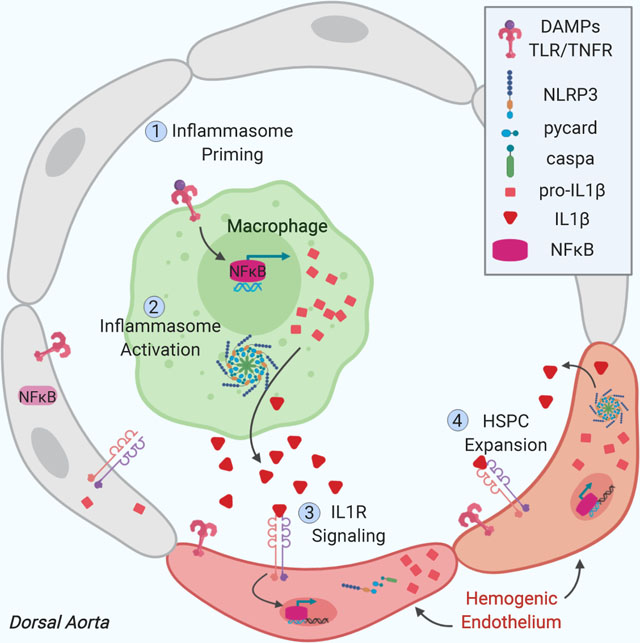
Our studies support the concept that macrophages are a major source of IL1β and play an important role in initiating inflammasome activation during zebrafish HSPC formation. Based on our in vivo time-lapse imaging data and inflammasome expression profiling, we propose that macrophage-derived IL1β might propagate inflammasome activity within Il1rl1+ HSPCs through downstream NFκB activation (Bauernfeind et al., 2009; Dinarello, 2009). Recent transcriptome analyses of aortic-associated macrophages from the mouse embryo similarly indicate expression of a multitude of cytokines and growth factors which may support HSPC production, including inflammasome-derived IL18 and IL1β (Mariani et al., 2019), both of which result in downstream NFκB activity and can generate similar cellular responses (Dinarello, 1999). Although not specifically enriched in murine CD206+ macrophages, regulation of IL1β at the protein level suggests that IL1β is likely an influential paracrine signal (Orelio et al., 2008). However, our data do not diminish the possibility that other macrophage-derived factors, or additional cell populations, including neutrophils, can “prime” inflammasome activation or cooperate with downstream IL1β signaling to further promote HSPC expansion. Intriguingly, macrophages are rare in human EHT cultures during the window of inflammasome stimulation; addition of monocytes alone positively influenced the frequency of CD34+CD45+ phenotypic HSPCs formed in human EHT cultures. Thus, absence of these additional cues from iPSC cultures might explain the suboptimal numbers of HSPCs generated with multipotent engraftment activity to date. In future work, it will be of interest to evaluate whether the presence of ontogenically distinct myeloid populations differentially influence developing human EHT cultures in vitro, and the precise temporal windows needed for their interaction, to maximize HSPC production and maintenance.
Finally, our findings indicate that inflammasome activation appears to robustly expand the HSPC pool in vivo and in vitro. A frequent concern with stimulation of proliferative expansion in HSPCs is the potential for exhaustion or premature differentiation (Mirantes et al., 2014). While we found no evidence for exhaustion in vivo, a reduction in erythroid differentiation was observed, suggesting that inflammasome activation may also restrain erythroid commitment within HSPCs. This would effectively counter the effects of embryonic hypoxia, which strongly induces erythroid differentiation signals in the embryo (Imtiyaz and Simon, 2010). In support of this idea, a recent report demonstrates that embryonic HSPCs cell-autonomously activate low levels of Caspase-1 to inactivate Gata1 and prevent erythroid differentiation in favor of myeloid differentiation (Tyrkalska et al., 2019). Although our data also indicate a reduction in erythroid differentiation in the context of inflammasome activity, this may be a secondary signaling mechanism that is initiated after macrophage-derived IL1β drives HSPC expansion. This latter hypothesis is supported by our observation that similar proportions of human erythroid colonies and lymphoid cells are formed from inflammasome-stimulated progenitors in vitro. Conversely, when HSPCs are left to differentiate in nigericin-treated suspension cultures, or when zebrafish embryos undergo prolonged inflammasome stimulation, both eventually exhibit lineage bias toward lymphoid and myeloid fates, mimicking inflammation-associated anemia. This effect may be due to amplified production of downstream cytokines, including IL6 (Nemeth et al., 2004; Zhao et al., 2014). These findings also underscore the importance of the timing and duration of inflammasome action in providing the optimal HSPC amplification response in vitro, and may be one reason why a prior study reported no benefit of exogenous inflammatory cytokines on the numbers of phenotypic human HSPCs specified in vitro (Giorgetti et al., 2017). It is likely that the acute stimulation of inflammasome activation in our cultures provides a more physiological inflammatory signaling cascade to better support HSPC expansion, as opposed to a continuous supply of recombinant cytokines, which may damage self-renewal and/or differentiation potential. As such, we favor a model in which endogenous, acute IL1β production serves to primarily expand multipotent HSPCs and secondarily restrain erythroid potential in vitro and in vivo.
Together, these studies reveal that inflammasome activation serves as a developmental metabolic sensor to trigger IL1β production and expand developing HSPCs. In addition, transient inflammasome activation in vitro provides a physiological burst of IL1β that expands functional human HSPCs. Inflammasome activity is initiated by macrophages in vivo, which, alongside other work, highlights the importance of supporting cell types in the development and function of HSPCs. It will be of interest to further explore and optimize potential co-culture conditions of human hemogenic endothelium alongside myeloid populations to better recapitulate the metabolic cues they interpret and relay to developing HSPCs to promote their expansion and maintenance.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for resources should be directed to and will be fulfilled by the lead contact, Trista E. North (Trista.north@childrens.harvard.edu).
Materials Availability
Materials generated in this study are available upon request.
Data and Code Availability
This study did not generate datasets or code requiring public database deposition.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
Zebrafish maintenance and experiments were performed according to guidelines set forth by the Beth Israel Deaconess Medical Center and Boston Children’s Hospital Institutional Animal Care and Use Committees. Adult zebrafish were housed in a standard circulating water system at 27°C and fed a brine shrimp diet. AB or TU strains were used for wild-type studies and outcrosses. Transgenic and mutant lines are noted in the Key Resources Table. Mutants were genotyped by PCR followed by Sanger sequencing. Transgenics were identified by fluorescence microscopy or transgene expression by WISH. For embryonic studies, males and females were separated off-system overnight, and barriers removed the following morning to allow for timed embryo generation. After collection, embryos were maintained under static conditions in E3 embryo buffer in incubators at 28.2°C, 25°C, or 22°C to desired developmental stages reported in figure legends. Embryos exhibiting developmental abnormalities or delay were discarded. Sex is not determined in zebrafish embryos and therefore was not assessed. For adult kidney marrow analyses, male zebrafish were maintained at equivalent tank densities until the experimental endpoint (3 months).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human CD34 PE-CY7 | Thermo Fisher Scientific | Cat# BDB560710 |
| Human CD45 APC | Beckman Coulter | Cat# IM2473U |
| Human GLYA/CD235a FITC | Beckman Coulter | Cat# IM2212U |
| Human CD71 PE | BD Biosciences | Cat# 555537 |
| Human CD19 PE | BioLegend | Cat# 302208 |
| Human CD3 PE-CY7 | BioLegend | Cat# 344816 |
| Human CD33 APC | BioLegend | Cat# 303407 |
| Mouse CD45.1 APC-CY7 | BioLegend | Cat# 110716 |
| TruStain FcX | BioLegend | Cat# 101320 |
| Human CD34 Microbeads | Miltenyi Biotec | Cat# 130-046-702 |
| Human CD5 PE (M-T701) | BD Biosciences | Cat# 555361 |
| Human CD7 BV510 (UCHT2) | BD Biosciences | Cat# 563381 |
| Anti-digoxigenin-AP Fab Fragments | Millipore Sigma | Cat# 11093274910 |
| Biological Samples | ||
| Human cord blood mononuclear cells (for gating controls) | Stem Cell Technologies | Cat# 70007.1 |
| Corning™ Matrigel™ hESC-Qualified Matrix | ThermoFisher Scientific | Cat# 08774552 |
| CF-1 MEF 4M IRR | ThermoFisher Scientific | Cat# A34181 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CoCl2 | Sigma | Cat# C8661 |
| Alum Crystals | Invivogen | Cat# tlrl-alk |
| Nigericin | Cayman Chemical | Cat# 11437 |
| Metronidazole | Millipore Sigma | Cat# M3761 |
| Ac-YVAD-CMK | Cayman Chemical | Cat#10014 |
| LPS | Invivogen | Cat# tlrl-eklps |
| CHIR99021 | Stem Cell Technologies | Cat# 72052 |
| SB431542 | Stem Cell Technologies | Cat# 72232 |
| Recombinant Human BMP4 | PeproTech | Cat# 10779-138 |
| Recombinant Human bFGF | Gibco | Cat# PHG0266 |
| Recombinant Human VEGF | R&D Systems | Cat# 293-VE |
| Recombinant Human IL6 | PeproTech | Cat# 200-06 |
| Recombinant Human IL11 | PeproTech | Cat# 200-11 |
| Recombinant Human IGF1 | PeproTech | Cat# 100-11 |
| Recombinant Human SCF | PeproTech | Cat# 300-07 |
| Recombinant Human EPO | Epogen | Cat# 55513-144-01 |
| Recombinant Human TPO | PeproTech | Cat# 300-18 |
| Recombinant Human Flt3L | PeproTech | Cat# 300-19 |
| Recombinant Human IL3 | PeproTech | Cat# 200-03 |
| Recombinant human Sonic Hedgehog | PeproTech | Cat# 100-45 |
| Angiotensin II | Millipore Sigma | Cat# A9525-5MG |
| Losartan Potassium | ThermoFisher Scientific | Cat# 37-985-0 |
| Liberase TM | Millipore Sigma | Cat# 5401127001 |
| Proteinase K | Millipore Sigma | Cat# 3115828001 |
| BCIP | Millipore Sigma | Cat# 11383221001 |
| NBT | Millipore Sigma | Cat# 11383213001 |
| Critical Commercial Assays | ||
| MethoCult™ SF H4636 | Stem Cell Technologies | Cat# 04636 |
| StemFlex™ Medium | ThermoFisher Scientific | Cat# A3349401 |
| StemSpan T cell generation kit | Stem Cell Technologies | Cat# 09940 |
| Amplex Red Glucose Assay kit | ThermoFisher Scientific | Cat# A22189 |
| RNEasy Micro Kit | Qiagen | Cat# 74004 |
| RNEasy Plus Micro Kit | Qiagen | Cat# 74034 |
| RNAqueous Kit | ThermoFisher Scientific | Cat# AM1912 |
| Superscript III cDNA Synthesis Supermix | Invitrogen | Cat# 11752050 |
| Superscript VILO cDNA Synthesis kit | Invitrogen | Cat# 11754050 |
| Maxima Reverse Transcriptase kit | ThermoFisher Scientific | Cat# EP0741 |
| Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647 | Invitrogen | Cat# C10340 |
| LS Columns | Miltenyi Biotec | Cat# 130-042-401 |
| Experimental Models: Cell Lines | ||
| hiPS cells | hESC core facility at Boston Children’s Hospital | 1157.2 |
| THP-1 monocytes | Invivogen | Cat# thp-null |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: Tg(−6.0itga2b:egfp)la2 (CD41:eGFP) | (Lin et al., 2005) | ZFIN: ZDB-ALT-051223-4 |
| Zebrafish: TgBAC(il1b:NTR-EGFP,cryaa:EGFP) | (Hasegawa et al., 2017) | ZFIN: ZDB-ALT-170531-2 |
| Zebrafish: Tg(kdrl:Hsa.HRAS-mCherry)s916 | (Hogan et al., 2009) | ZFIN: ZDB-ALT-090506-2 |
| Zebrafish: Tg(kdrl:dsred2) (commonly Flk1:dsred) | (Kikuchi et al., 2011) | ZFIN: ZDB-ALT-110408-4 |
| Zebrafish: Tg(cmyb:EGFP) | (North et al., 2007) | ZFIN: ZDB-ALT-071017-1 |
| Zebrafish: caspahdb12 | (Kuri et al., 2017) | ZFIN: ZDB-ALT-180207-12 |
| Zebrafish: pycardzf2174/zf2174 (AB) | (Matty et al., 2019) | ZFIN: ZDB-GENO200316-6 |
| Zebrafish: Tg(hsp70l:il1b;cryaa:CFP) | This paper | N/A |
| Zebrafish: Tg(mpx:GFP) | (Renshaw et al., 2006) | ZFIN: ZDB-ALT-070118-2 |
| Zebrafish: Tg(mfap4:tdTomato-CAAX)xt6 | (Walton et al., 2015) | ZFIN: ZDB-ALT-160122-3 |
| Zebrafish: Tg(gata1a:dsRed) | (Traver et al., 2003) | ZFIN: ZDB-ALT-051223-6 |
| Zebrafish: Tg(UAS:dnnfkbiaa)sd35 | (Espín-Palazón et al., 2014) | ZFIN: ZDB-ALT-151228-5 |
| Zebrafish: Tg(hsp70l:GAL4) | (Jeong et al., 2007) | ZFIN: ZDB-ALT-070706-2 |
| Zebrafish: Tg(pycard:pycard-EGFP); pycardhdb10Tg | (Kuri et al., 2017) | ZFIN: ZDB-ALT-180207-11 |
| Zebrafish: Tg(mpeg1:mCherry)gl23 | (Ellett et al., 2011) | ZFIN: ZDB-ALT-120117-2 |
| Zebrafish: Tg(mpeg1:EGFP)gl22 | (Ellett et al., 2011) | ZFIN: ZDB-ALT-120117-1 |
| Zebrafish: Tg(mpeg1:GAL4;UAS:NTR-mCherry) | (Palha et al., 2013) | ZFIN: ZDB-ALT-120117-3; ZDB-ALT-070316-1 |
| Zebrafish: Tg(EF1a:mAG-zGem(1/100))rw0410h) | (Sugiyama et al., 2009) | ZFIN: ZDB-ALT-101018-2 |
| Zebrafish: Tg(rag2:gfp) | (Langenau et al., 2003) | ZFIN: ZDB-ALT-051111-6 |
| Mouse: NBSGW (NOD.Cg-KitW−41J Tyr + Prkdcscid Il2rgtm1Wjl/ThomJ) | Jackson Laboratory | JAX: 026622 |
| Oligonucleotides | ||
| See Table S1 | See Table S1 | N/A |
| Recombinant DNA | ||
| Plasmid: pME-MCS | (Kwan et al., 2007) | Tol2kit v1.2 #237 |
| Plasmid: p3E-polyA | (Kwan et al., 2007) | Tol2kit v1.2 #302 |
| Plasmid: p5E-Hsp70l | (Kwan et al., 2007) | Tol2kit v1.2 #222 |
| Plasmid: p5E-kdrl | Addgene | Cat# 78687 |
| Plasmid: pDestTol2pA2 | (Kwan et al., 2007) | Tol2kit v1.2 #394 |
| Plasmid: pCS2FA-Transposase | (Kwan et al., 2007) | Tol2kit v1.2 #396 |
| Plasmid: pDestTol2pA2CryCFP-hsp70l:il1b | This paper | N/A |
| Plasmid: pDestTol2pA2CryCFP-flk1:il1b | This paper | N/A |
| Software and Algorithms | ||
| Fiji | (Schindelin et al., 2012) | https://imagej.net/Fiji |
| Prism (Version 5, 6, 8) | GraphPad | https://www.graphpad.com/ |
| FlowJo (Version 10) | BD Biosciences | https://www.flowjo.com/ |
| PCRMiner | (Zhao and Fernald, 2005) | http://ewindup.info/miner/home.htm |
Mice
Murine housing, maintenance, and transplantation studies were performed in accordance with guidelines set forth by the Boston Children’s Hospital Institutional Animal Care and Use Committee. Female NBSGW (NOD.Cg-KitW−41J Tyr + Prkdcscid Il2rgtm1Wjl/ThomJ) immunocompromised mice were purchased from Jackson Laboratory and maintained in a standard barrier facility. 8–12 week old mice were used for transplantation. Antibiotic water (0.5mg/mL Sulfatrim) was given for 2 weeks post-transplant.
Primary Cells
Irradiated murine embryonic fibroblasts (MEF; mixed/unknown sex) used to passage iPSCs were purchased directly from ThermoFisher Scientific. Human umbilical cord blood mononuclear cells isolated from unknown donors were purchased (Stem Cell Technologies) and used for gating controls in transplantation analyses.
Cell lines
1157.2-iPS cells were generated by the Boston Children’s Hospital hESC (human embryonic stem cell) Core Facility from a healthy male donor under Institutional Review Board approved protocols. iPSCs were maintained on hESC-qualified Matrigel-coated dishes in Stemflex medium (ThermoFisher Scientific) at 37°C /20% O2/5% CO2 and routinely karyotyped and tested for mycoplasma contamination. THP-1 monocytes (from a male with monocytic leukemia (Tsuchiya et al., 1980)) were purchased for use (Invivogen) and maintained in RPMI medium with 2mM glutamine, 25mM HEPES, and 10% fetal bovine serum (FBS) at 37°C /20% O2/5% CO2. Authenticity, purity and functionality to activate the inflammasome was verified by the vendor.
Human iPSC differentiation
1157.2-iPSCs were differentiated into hemogenic endothelium essentially as described (Sugimura et al., 2017). In brief, iPSCs were cultured for one passage on CF-1 irradiated MEFs prior to embryoid body (EB) formation. Colonies were detached using Collagenase IV and seeded into low attachment dishes (Corning) in aggregation media (Sturgeon et al., 2014). 24 hours later bFGF (5 ng/ml) was added. On day 2 aggregation media was supplemented with bFGF (5 ng/ml), BMP4 (10 ng/ml), CHIR99021 (3 μM) and SB431542 (6 μM). For the following 3 days, EB media (Stempro-34, Thermo Fisher Scientific) was supplemented with ascorbic acid (1 mM), holo-Transferrin (150 μg/ml) and α-Monothioglycerol (MTG, 0.4 mM)). On day 6 EB media was supplemented with bFGF (5 ng/ml), VEGF (15 ng/ml), IL-6 (10 ng/ml), IL-11 (5 ng/ml), IGF-1 (25 ng/ml), SCF (50 ng/ml) and EPO (2U/ml). EBs were harvested on day 8 and dissociated with trypsin/EDTA followed by collagenase IV. CD34+ hemogenic endothelium was enriched by Magnetic-activated cell sorting (MACS) (Miltenyi). For MACS, freshly dissociated EBs were filtered through a 40 μm cell strainer and incubated with anti-CD34 magnetic beads for 30 min on ice per the manufacturer’s recommendation. Unbound beads were washed off with Phosphate Buffered Saline (PBS)/2%FBS and cells separated using LS columns. CD34+ cells were counted and 50k cells per well of a 24 well plate were plated onto Matrigel (ThermoFisher Scientific) coated dishes in EHT media (EB media + BMP4 (10 ng/ml), bFGF (5 ng/ml), IL-3 (30 ng/ml), IL-6 (10 ng/ml), IL-11 (5 ng/ml), IGF-1 (25 ng/ml), VEGF (5 ng/ml), SCF (100 ng/ml), EPO (2U/ml), TPO (30 ng/ml), FLT-3L (10 ng/ml), SHH (20 ng/ml), Angiotensin II (10 μg/ml ), Losartan (100 μM). Nigericin (1–10 nM) or EtOH treatment (for controls) was performed on day 2 prior to emergence of round floating cells. Once EB differentiation was started, cultures were incubated at 37°C in a 5% CO2/5%O2/90% N2 environment.
METHOD DETAILS
Generation of inducible IL1β transgenic zebrafish
The Tg(hsp70l:il1b) line was generated by PCR amplification of the portion of il1b transcript corresponding to amino acids 104–272 (following the Caspa cleavage site)(Vojtech et al., 2012), insertion into a Gateway middle entry vector (pME-MCS), and subsequent multi-site Gateway recombination (Invitrogen) into a destination vector carrying the cryaa:cerulean transgene using the Tol2Kit workflow (plasmids listed in Key Resources Table) (Kwan et al., 2007). Germline transgenesis was achieved in 7 founders via co-injection of the construct with Tol2 transposase mRNA. Overexpression of il1b was confirmed in embryos by WISH after incubation at 38°C for 30 minutes. For endothelial-specific induction of active il1b, a p5E-flk1 promoter construct (Addgene) was utilized in place of the Hsp70l promoter construct, and chimeric injected embryos were screened for expression of cryaa:cerulean by WISH alongside runx1 and il1b.
Zebrafish embryo injections
Morpholinos (Gene Tools) were loaded into a needle pulled from 1.0mm borosilicate glass capillary tubes, and injected into the yolk of fertilized eggs immobilized in agarose molds at the 1-cell stage using a microinjector (Narishige). Morpholinos are listed in Table S1. For transgenesis, constructs were injected directly into the fertilized egg cell.
Zebrafish Chemical Exposures and WISH
Groups of 20 stage-matched embryos were arranged in 6 well plates in 5mL E3 water with or without chemical treatments of interest. Embryos were manually dechorionated and fixed in 4% paraformaldehyde for WISH analyses. Pigment was subsequently removed with 0.8%KOH/0.9%H2O2 treatment; embryos were then dehydrated into 100% methanol prior to WISH using standard published protocols and probes (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Briefly, embryos were permeabilized with 10μg/mL Proteinase K (Millipore Sigma), re-fixed in 4% paraformaldehyde, washed in 1xPBS containing 0.1% Tween (PBST) and hybridized with digoxigenin-UTP-labeled riboprobes in hybridization buffer containing 50% formamide at 70°C overnight. Embryos were washed in sodium citrate buffer/0.1% Tween and incubated overnight with a digoxigenin antibody conjugated to alkaline phosphatase in PBST. After several PBST washes, antibody staining was revealed by incubation in Tris buffer (pH 9.5) with BCIP/NBT substrate (Millipore Sigma). Stained embryos were directly compared with stage-matched sibling controls across multiple clutches, with the exception of mutant clutches, which were compared alongside matched wild-type or heterozygote clutches as indicated. The intensity of expression was qualitatively categorized by high (abbreviated hi), medium (med), or low (lo), relative to the median intensity of the control group for a given clutch. For runx1/cmyb expression at 36hpf, a score of “hi” typically reflects continuous aortic signal with robust thickness, indicating numerous cell clusters, “medium” denotes modest, yet continuous aortic signal, and low signal indicates spotty/discontinuous aortic signal. At least 25 embryos from a minimum of two independent clutches were analyzed per treatment condition and/or genotype, with phenotype scoring independently confirmed. WISH images were captured on a Zeiss Axio Imager A1 equipped with Axio Vision software; images were selected to represent the median expression level for each treatment/condition. Area of stained kidney marrows were quantified in images using Fiji software (Schindelin et al., 2012).
EdU labeling and fluorescence microscopy
Embryos were exposed to 500μM EdU in E3 water (10% DMSO final) for 1 hour at 4°C. Embryos were fixed in 4% paraformaldehyde, permeabilized with 1% Triton for 1 hour, and labeled with Alexa Fluor 647 using the Click-iT reaction (Thermo Fisher Scientific) for 1 hour at room temperature according to the manufacturer’s instructions. Embryos were mounted in 3% methylcellulose and captured on a Zeiss LSM880 Inverted Confocal Microscope. Live Tg(pycard:pycard-EGFP) and Tg(il1b:EGFP) embryos were mounted in 1.5% low melting agarose and imaged on a Zeiss LSM880 Inverted Confocal Microscope or a Nikon Ti2 inverted microscope equipped with a CSU-W1 spinning disk confocal unit and iXon888 life camera. Tg(CD41:EGFP) and Tg(EF1a:mAG-zGem(1/100))rw0410h live embryos were imaged on a Zeiss Discovery V8 (Axio Vision software). Cell counts were performed manually in Fiji software. Image brightness and contrast were adjusted in Fiji or Powerpoint.
Glucose measurement assay
Embryos from two independent clutches were reared at 28.2°C and treated at 24hpf with phenylthiourea (0.003%) to prevent pigmentation. Groups of 24 embryos were then dechorionated at specified timepoints and flash frozen in 1xPBS on dry ice. Samples were homogenized and spun down to remove excess debris; supernatants were stored at −80°C. Supernatants were incubated with substrate from an Amplex Red Glucose measurement kit and glucose measurements were obtained from absorbance readings according to the manufacturer’s instructions (Fisher Scientific).
Flow cytometry
Pools of zebrafish embryos/larvae were dechorionated and resuspended in 500μL Liberase TM (Millipore Sigma) solution (75μg/mL in 1xPBS/1mM EDTA), incubated at 34°C and dissociated with a 1000μL pipette every 20–30 minutes until embryo trunks are no longer visible (Frame et al., 2017); each data point represents 4 or 5 zebrafish. Adult male kidney marrow was manually triturated in 1xPBS/1mM EDTA with a 1000μL pipette. Zebrafish samples were passed through a 30μm filter, washed in PBS, and labeled with Sytox Red (Invitrogen) to 5nM prior to analysis. Human cells were blocked with TruStain FcX and labeled with antibodies (listed in the Key Resources Table) at room temperature for 20 minutes, followed by DAPI addition (to 0.5μg/mL final) to exclude dead cells. Flow cytometric data were collected on a FACSCanto, LSRII, or LSRFortessa (BD) and cell sorting was performed on a FACSAria II (BD). Data were analyzed using FlowJo X (BD). CD41+ zebrafish HSPCs were gated separately from the CD41hi thrombocyte population (Lin et al., 2005; Ma et al., 2011).
RNA isolation and quantitative PCR
Total zebrafish embryo RNA was isolated from pools of >20 embryos using the RNAqueous kit according to the manufacturer’s protocol (Invitrogen). Briefly, embryos were mechanically homogenized in the lysis buffer and treated with DNAse I (Invitrogen) after elution from the column. For sorted zebrafish myeloid cell and CD41+ populations, RNA was isolated using an RNEasy Micro kit (Qiagen) and DNAse was applied directly on the column per the manufacturer’s protocol. cDNA was synthesized using Superscript III Supermix or Superscript VILO cDNA Synthesis kit (Invitrogen). Flk1+/cMyb+ sorted cell cDNAs were prepared using the NuGEN Ovation RNA Amplification System V2 according to the manufacturer’s protocol (Cortes et al., 2016; Lim et al., 2017). RNA was isolated from iPSC-derived populations using the RNEasy Micro kit Plus (Qiagen) and cDNA was transcribed using the Maxima Reverse Transcriptase kit (Thermo Fisher Scientific). Quantitative PCR was performed on a CFX384 (BioRad) or ABI7900 (Applied Biosystems) using either SYBR green Master mix or Taqman universal master mix (Applied Biosystems). Absolute expression level in each sample was calculated relative to reference genes (eef1a1l1 for Taqman assays; and b-actin, 18s, or GAPDH for SYBR green) (Esain et al., 2015) using the Delta/delta Ct Method or the PCR Miner interface (Zhao and Fernald, 2005), fold change was calculated relative to the negative control or untreated sample population within each biological clutch or group of clutches. For sorted cell populations, fold change was calculated relative to the bulk negative control fraction. Primer sequences are listed in Table S1.
Hematopoietic colony assay
Hematopoietic colony potential of the live floating cells (4.25k-15k) on day 7 of EHT culture was assessed in methylcellulose cultures (MethoCult™ SF H4636; StemCell Technologies). Dishes were kept in a humidified chamber in the incubator for 14 days, then scored manually and blindly.
Monocyte co-cultures
Human THP-1 monocytes (Invivogen) were pre-stimulated with LPS (1 μg/ml) overnight according to the manufacturer’s instructions. The following day, 25K THP-1 cells were seeded in the upper well of transwell inserts (0.4μm pore size, Corning); while 50K iPSC-derived CD34+ MACS-purified hemogenic endothelial cells were seeded on the bottom 2 days prior in EHT medium. Nigericin (10 nM) was subsequently added to the EHT co-culture.
T cell differentiation
Human iPSCs were differentiated to form embryonic bodies, and CD34+ hemogenic endothelium was enriched from dissociated day 8 EBs as above. CD34+ cells were plated under T lymphoid differentiation conditions using the StemSpan T cell generation kit (#09940, Stem Cell Technologies) according to the manufacturer’s instructions. Nigericin (10 nM) or EtOH (for controls) was added on day 2, and CD45+CD5+CD7+ pro-T cells were detected after 2 weeks.
Transplantation
Human iPSC-derived hematopoietic progenitors were transplanted intrafemorally into 8–12 week old female immunodeficient NOD.Cg-KitW−41J Tyr + Prkdcscid Il2rgtm1Wjl/ThomJ (NBSGW) mice in the absence of irradiation. After 8 weeks, femoral bone marrow was harvested using a mortar and pestle and analyzed by flow cytometry.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data analyses were performed using GraphPad Prism. Error bars indicate SD unless otherwise indicated; statistical significance was determined by two-tailed Student’s T-test unless otherwise indicated. In some cases, differences in cell number, frequency, or expression were normalized to the average control value within clutches, or to the average control well in iPSC differentiation assays. Significance of WISH phenotype distributions was determined by Chi-square analysis.
Supplementary Material
Movie S1 (related to Figure 6). Budding of a Pycard+ cell from the aortic endothelium of a Tg(pycard:pycard-EGFP;flk1:mCherry) embryo from 34–36hpf.
Highlights:
Developmental metabolic shifts prime inflammasome-associated il1b expression in vivo
Loss of inflammasome function inhibits HSPC production in the zebrafish embryo
IL1β+ macrophages promote production of Il1rl1+ HSPCs via inflammasome action in vivo
Inflammasome activation enhances HSPC production in human hemogenic cultures
Acknowledgments
We thank the Boston Children’s Hospital flow cytometry core, Yi Zhou for genomic expertise, Mohamad Najia for flow cytometry help, Wade Sugden for cloning expertise, Maria Leptin for pycard:EGFP line and caspa mutants, David Tobin for pycard mutants, David Traver for the UAS:dnikbaa line, and Leonard Zon for mpeg:NTR-mCherry and flk1:mCherry lines. Work was funded by the National Institutes of Health: T32 HL007893 (JMF), R01 DK098241 (TEN), R01 HL152636 (TEN), R24 DK092760 (GQD/TEN), U01 HL134812 (GQD/TEN), and the German Research Foundation (CK). Graphical abstract created from Biorender.com.
Declaration of Interests
GQD holds equity in 28/7 Therapeutics and Epizyme, has received sponsored research support from Megakaryon, and holds patents pertaining to HSPC production from iPSCs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abderrazak A, Syrovets T, Couchie D, El Hadri K, Friguet B, Simmet T, and Rouis M (2015). NLRP3 inflammasome: From a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 4, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge MT, King KY, Boles NC, Weksberg DC, and Goodell MA (2010). Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 465, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, et al. (2009). Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. Baltim. Md 1950 183, 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, and Traver D (2007). Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134, 4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, and Traver D (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böiers C, Carrelha J, Lutteropp M, Luc S, Green JCA, Azzoni E, Woll PS, Mead AJ, Hultquist A, Swiers G, et al. (2013). Lymphomyeloid Contribution of an Immune-Restricted Progenitor Emerging Prior to Definitive Hematopoietic Stem Cells. Cell Stem Cell 13, 535–548. [DOI] [PubMed] [Google Scholar]
- Bukrinsky A, Griffin KJP, Zhao Y, Lin S, and Banerjee U (2009). Essential role of spi-1-like (spi-1l) in zebrafish myeloid cell differentiation. Blood 113, 2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camell C, Goldberg E, and Dixit VD (2015). Regulation of Nlrp3 inflammasome by dietary metabolites. Semin. Immunol 27, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, and Goodell MA (2010). Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp. Hematol 38, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanput W, Mes JJ, and Wichers HJ (2014). THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol 23, 37–45. [DOI] [PubMed] [Google Scholar]
- Clements WK, and Traver D (2013). Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat Rev Immunol 13, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Chen MJ, Stachura DL, Liu SY, Kwan W, Wright F, Vo LT, Theodore LN, Esain V, Frost IM, et al. (2016). Developmental Vitamin D Availability Impacts Hematopoietic Stem Cell Production. Cell Rep. 17, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel MG, Pereira C-F, Lemischka IR, and Moore KA (2016). Making a Hematopoietic Stem Cell. Trends Cell Biol. 26, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon SS, Torell F, Donten M, Lundstedt-Enkel K, Bennett K, Rännar S, Trygg J, and Lundstedt T (2019). Metabolic profiling of zebrafish embryo development from blastula period to early larval stages. PLOS ONE 14, e0213661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ignazio L, Bandarra D, and Rocha S (2016). NF–κB and HIF crosstalk in immune responses. Febs J. 283, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA (1999). IL-18: A TH1 -inducing, proinflammatory cytokine and new member of the IL-1 family. J. Allergy Clin. Immunol 103, 11–24. [DOI] [PubMed] [Google Scholar]
- Dinarello CA (2009). Immunological and Inflammatory Functions of the Interleukin-1 Family. Annu. Rev. Immunol 27, 519–550. [DOI] [PubMed] [Google Scholar]
- Dinarello CA (2011). A clinical perspective of IL-1β as the gatekeeper of inflammation. Eur. J. Immunol 41, 1203–1217. [DOI] [PubMed] [Google Scholar]
- Ditadi A, Sturgeon CM, Tober J, Awong G, Kennedy M, Yzaguirre AD, Azzola L, Ng ES, Stanley EG, French DL, et al. (2015). Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol 17, 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror E, Dalmas E, Meier DT, Wueest S, Thévenet J, Thienel C, Timper K, Nordmann TM, Traub S, Schulze F, et al. (2017). Postprandial macrophage-derived IL-1β stimulates insulin, and both synergistically promote glucose disposal and inflammation. Nat. Immunol 18, 283–292. [DOI] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nuñez G, Schnurr M, et al. (2010). NLRP3 inflamasomes are required for atherogenesis and activated by cholesterol crystals that form early in disease. Nature 464, 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E, and Speck NA (2008). Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 9, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, and Lieschke GJ (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esain V, Kwan W, Carroll KJ, Cortes M, Liu SY, Frechette GM, Sheward LMV, Nissim S, Goessling W, and North TE (2015). Cannabinoid Receptor-2 Regulates Embryonic Hematopoietic Stem Cell Development via Prostaglandin E2 and P-Selectin Activity: CNR2 Impacts Embryonic HSC Development. STEM CELLS 33, 2596–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Palazón R, Stachura DL, Campbell CA, García-Moreno D, Del Cid N, Kim AD, Candel S, Meseguer J, Mulero V, and Traver D (2014). Proinflammatory Signaling Regulates Hematopoietic Stem Cell Emergence. Cell 159, 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplin BL, Shimazu T, Welner RS, Garrett KP, Nie L, Zhang Q, Humphrey MB, Yang Q, Borghesi LA, and Kincade PW (2011). Chronic Exposure to a TLR Ligand Injures Hematopoietic Stem Cells. J. Immunol 186, 5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MAG, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, and Trumpp A (2009). IFNα activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908. [DOI] [PubMed] [Google Scholar]
- Frame JM, Lim S-E, and North TE (2017). Hematopoietic stem cell development: Using the zebrafish to identify extrinsic and intrinsic mechanisms regulating hematopoiesis. Methods Cell Biol. 138, 165–192. [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Castaño J, Bueno C, Díaz de la Guardia R, Delgado M, Bigas A, Espinosa L, and Menendez P (2017). Proinflammatory signals are insufficient to drive definitive hematopoietic specification of human HSCs in vitro. Exp. Hematol 45, 85–93.e2. [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB, and Ting JP-Y (2015). Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med 21, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G, Santana AKM, Gomes CM, Turatti A, Milanezi CM, Bueno Filho R, Fuzo C, Almeida RP, Carregaro V, Roselino AM, et al. (2019). Inflammasome gene expression is associated with immunopathology in human localized cutaneous leishmaniasis. Cell. Immunol 341, 103920. [DOI] [PubMed] [Google Scholar]
- Hall CJ, Sanderson LE, Lawrence LM, Pool B, Kroef M.van der, Ashimbayeva E, Britto D, Harper JL, Lieschke GJ, Astin JW, et al. (2018). Blocking fatty acid–fueled mROS production within macrophages alleviates acute gouty inflammation. J. Clin. Invest 128, 1752–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneklaus M, and O’Neill LAJ (2015). NLRP3 at the interface of metabolism and inflammation. Immunol. Rev 265, 53–62. [DOI] [PubMed] [Google Scholar]
- Harris JM, Esain V, Frechette GM, Harris LJ, Cox AG, Cortes M, Garnaas MK, Carroll KJ, Cutting CC, Khan T, et al. (2013). Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood 121, 2483–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T, Hall CJ, Crosier PS, Abe G, Kawakami K, Kudo A, and Kawakami A (2017). Transient inflammatory response mediated by interleukin-1β is required for proper regeneration in zebrafish fin fold. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Zhang C, Wang L, Zhang P, Ma D, Lv J, and Liu F (2015). Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 125, 1098–1106. [DOI] [PubMed] [Google Scholar]
- He Y, Hara H, and Núñez G (2016). Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci 41, 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BM, Bos FL, Bussmann J, Witte M, Chi NC, Duckers HJ, and Schulte-Merker S (2009). Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat. Genet 41, 396–398. [DOI] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, and Latz E (2008). Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol 9, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MM, and O’Neill LAJ (2018). Metabolic regulation of NLRP3. Immunol. Rev 281, 88–98. [DOI] [PubMed] [Google Scholar]
- Imtiyaz HZ, and Simon MC (2010). Hypoxia-inducible factors as essential regulators of inflammation. Curr. Top. Microbiol. Immunol 345, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, and Medvinsky A (2017). Human haematopoietic stem cell development: from the embryo to the dish. Development 144, 2323–2337. [DOI] [PubMed] [Google Scholar]
- Jeong J-Y, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, and Guo S (2007). Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development 134, 127–136. [DOI] [PubMed] [Google Scholar]
- Jo E-K, Kim JK, Shin D-M, and Sasakawa C (2016). Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol 13, 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Roy N, Bajwa R, Gut P, Lipson K, Yang C, Covassin L, Racki WJ, Rossini AA, Phillips N, et al. (2011). Dynamic glucoregulation and mammalian-like responses to metabolic and developmental disruption in zebrafish. Gen. Comp. Endocrinol 170, 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, and Poss KD (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KY, and Goodell MA (2011). Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol 11, 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, and Herbomel P (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. [DOI] [PubMed] [Google Scholar]
- Kuri P, Schieber NL, Thumberger T, Wittbrodt J, Schwab Y, and Leptin M (2017). Dynamics of in vivo ASC speck formation. J. Cell Biol 216, 2891–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, and Kanki JP (2007). The Tol2kit: A multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Dev. Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Purcell MK, Winton JR, and Hansen JD (2008). A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol. Biol 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam EYN, Hall CJ, Crosier PS, Crosier KE, and Flores MV (2010). Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116, 909–914. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, et al. (2003). Myc-Induced T Cell Leukemia in Transgenic Zebrafish. Science 299, 887–890. [DOI] [PubMed] [Google Scholar]
- Lee H-M, Kim J-J, Kim HJ, Shong M, Ku BJ, and Jo E-K (2013). Upregulated NLRP3 Inflammasome Activation in Patients With Type 2 Diabetes. Diabetes 62, 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao K, Shao T, Fan D, Hu C, Sun C, Dong W, Lin A, Xiang L, and Shao J (2018). Characterization of an NLRP1 Inflammasome from Zebrafish Reveals a Unique Sequential Activation Mechanism Underlying Inflammatory Caspases in Ancient Vertebrates. J. Immunol 201, 1946–1966. [DOI] [PubMed] [Google Scholar]
- Li J-Y, Wang Y-Y, Shao T, Fan D-D, Lin A-F, Xiang L-X, and Shao J-Z (2020). The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J. Biol. Chem 295, 1120–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lahvic JL, Binder V, Pugach EK, Riley EB, Tamplin OJ, Panigrahy D, Bowman TV, Barrett FG, Heffner GC, et al. (2015). Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature 523, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Esain V, Teng L, Xu J, Kwan W, Frost IM, Yzaguirre AD, Cai X, Cortes M, Maijenburg MW, et al. (2014). Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S-E, Esain V, Kwan W, Theodore LN, Cortes M, Frost IM, Liu SY, and North TE (2017). HIF1α-induced PDGFRβ signaling promotes developmental HSC production via IL-6 activation. Exp. Hematol 46, 83–95.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-F, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, and Handin RI (2005). Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 106, 3803–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Muñoz A, Sepulcre MP, Roca FJ, Figueras A, Meseguer J, and Mulero V (2011). Evolutionary conserved pro-inflammatory and antigen presentation functions of zebrafish IFNγ revealed by transcriptomic and functional analysis. Mol. Immunol 48, 1073–1083. [DOI] [PubMed] [Google Scholar]
- Ma D, Zhang J, Lin H, Italiano J, and Handin RI (2011). The identification and characterization of zebrafish hematopoietic stem cells. Blood 118, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani SA, Li Z, Rice S, Krieg C, Fragkogianni S, Robinson M, Vink CS, Pollard JW, and Dzierzak E (2019). Pro-inflammatory Aorta-Associated Macrophages Are Involved in Embryonic Development of Hematopoietic Stem Cells. Immunity 50, 1439–1452.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SL, Dunne A, Subramanian SL, Hull RL, Tannahill GM, Sharp FA, Becker C, Franchi L, Yoshihara E, Chen Z, et al. (2010). Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol 11, 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SL, Gerlic M, Metcalf D, Preston S, Pellegrini M, O’Donnell JA, McArthur K, Baldwin TM, Chevrier S, Nowell CJ, et al. (2012). NLRP1 Inflammasome Activation Induces Pyroptosis of Hematopoietic Progenitor Cells. Immunity 37, 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto J, Zhou W, Chen FF, Su F, Kuwada JY, Hidaka E, Katsuyama T, Sagara J, Taniguchi S, Ngo-Hazelett P, et al. (2003). Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J. Biol. Chem 278, 4268–4276. [DOI] [PubMed] [Google Scholar]
- Matty MA, Knudsen DR, Walton EM, Beerman RW, Cronan MR, Pyle CJ, Hernandez RE, and Tobin DM (2019). Potentiation of P2RX7 as a host-directed strategy for control of mycobacterial infection. ELife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, and Palis J (2015). Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 11, 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, and Taoudi S (2011). Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031. [DOI] [PubMed] [Google Scholar]
- Mirantes C, Passegué E, and Pietras EM (2014). Pro-inflammatory cytokines: Emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res 329, 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J-S, Hisata S, Park M-A, DeNicola GM, Ryter SW, Nakahira K, and Choi AMK (2015). mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep. 12, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Navarro-Montero O, Ayllon V, Lamolda M, López-Onieva L, Montes R, Bueno C, Ng E, Guerrero-Carreno X, Romero T, Romero-Moya D, et al. (2017). RUNX1c Regulates Hematopoietic Differentiation of Human Pluripotent Stem Cells Possibly in Cooperation with Proinflammatory Signaling: RUNX1c Promotes Human Embryonic Hematopoiesis. STEM CELLS 35, 2253–2266. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, and Ganz T (2004). IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J. Clin. Invest 113, 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Chi M, Phan QT, Gonzalez C, Dubremetz J-F, Levraud J-P, and Lutfalla G (2014). Transient infection of the zebrafish notochord with E. coli induces chronic inflammation. Dis. Model. Mech 7, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang I-H, Grosser T, et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, et al. (2009). Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell 137, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko NV, Lewis A, Wilson HL, Meijer AH, Renshaw SA, and Elks PM (2019). Hif-1α–Induced Expression of Il-1β Protects against Mycobacterial Infection in Zebrafish. J. Immunol 202, 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelio C, Haak E, Peeters M, and Dzierzak E (2008). Interleukin-1-mediated hematopoietic cell regulation in the aorta-gonad-mesonephros region of the mouse embryo. Blood 112, 4895–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelio C, Peeters M, Haak E, van der Horn K, and Dzierzak E (2009). Interleukin-1 regulates hematopoietic progenitor and stem cells in the midgestation mouse fetal liver. Haematologica 94, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, Wang C-H, Lieschke GJ, Herbomel P, Schwartz O, et al. (2013). Real-Time Whole-Body Visualization of Chikungunya Virus Infection and Host Interferon Response in Zebrafish. PLOS Pathog. 9, e1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MN, Carroll RG, Galván-Peña S, Mills EL, Olden R, Triantafilou M, Wolf AI, Bryant CE, Triantafilou K, and Masters SL (2017). Inflammasome Priming in Sterile Inflammatory Disease. Trends Mol. Med 23, 165–180. [DOI] [PubMed] [Google Scholar]
- Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, Lakshminarasimhan R, Chin CP, Techner J-M, Will B, et al. (2016). Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol 18, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progatzky F, Sangha NJ, Yoshida N, McBrien M, Cheung J, Shia A, Scott J, Marchesi JR, Lamb JR, Bugeon L, et al. (2014). Dietary cholesterol directly induces acute inflammasome-dependent intestinal inflammation. Nat. Commun 5, 5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, and Whyte MKB (2006). A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978. [DOI] [PubMed] [Google Scholar]
- Rock KL, Latz E, Ontiveros F, and Kono H (2010). The Sterile Inflammatory Response. Annu. Rev. Immunol 28, 321–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamiphak S, Kontarakis Z, and Stainier DYR (2014). Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev. Cell 31, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif O, Bolshakov VN, Raines S, Newham P, and Perkins ND (2007). Transcriptional profiling of the LPS induced NF-κB response in macrophages. BMC Immunol. 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieger D, Stein C, Neifer D, van der Sar AM, and Leptin M (2009). The role of gamma interferon in innate immunity in the zebrafish embryo. Dis. Model. Mech 2, 571–581. [DOI] [PubMed] [Google Scholar]
- Son M-Y, Kwak JE, Seol B, Lee DY, Jeon H, and Cho YS (2015). A novel human model of the neurodegenerative disease GM1 gangliosidosis using induced pluripotent stem cells demonstrates inflammasome activation. J. Pathol 237, 98–110. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, and Seed B (2010). PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Awong G, Kennedy M, and Keller G (2014). Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol 32, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu Y-F, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S.-i, and Miyawaki A (2009). Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci 106, 20812–20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Ganchi P, Ballard D, and Greene W (1993). NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science 259, 1912–1915. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Regoes RR, Boddupalli CS, Bonhoeffer S, and Manz MG (2011). Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med 208, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. (2013). Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Xu J, Feng S, He S, Zhao S, Zhu L, Jin W, Dai Y, Luo L, Qu JY, et al. (2017). The first wave of T lymphopoiesis in zebrafish arises from aorta endothelium independent of hematopoietic stem cells. J. Exp. Med 214, 3347–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirziu D, Jaba IM, Yu P, Larrivée B, Coon BG, Cristofaro B, Zhuang ZW, Lanahan AA, Schwartz MA, Eichmann A, et al. (2012). Endothelial NFκB - Dependent Regulation of Arteriogenesis and Branching. Circulation 126, 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, and Zon LI (2003). Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol 4, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Tseng HHL, Vong CT, Kwan YW, Lee SM-Y, and Hoi MPM (2016). TRPM2 regulates TXNIP-mediated NLRP3 inflammasome activation via interaction with p47 phox under high glucose in human monocytic cells. Sci. Rep 6, 35016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, and Tada K (1980). Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176. [DOI] [PubMed] [Google Scholar]
- Tyrkalska SD, Pérez-Oliva AB, Rodríguez-Ruiz L, Martínez-Morcillo FJ, Alcaraz-Pérez F, Martínez-Navarro FJ, Lachaud C, Ahmed N, Schroeder T, Pardo-Sánchez I, et al. (2019). Inflammasome Regulates Hematopoiesis through Cleavage of the Master Erythroid Transcription Factor GATA1. Immunity 51, 50–63.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtech LN, Scharping N, Woodson JC, and Hansen JD (2012). Roles of Inflammatory Caspases during Processing of Zebrafish Interleukin-1β in Francisella noatunensis Infection. Infect. Immun 80, 2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EM, Cronan MR, Beerman RW, and Tobin DM (2015). The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PLOS ONE 10, e0138949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, and Fernald RD (2005). Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. J. Comput. Biol. J. Comput. Mol. Cell Biol 12, 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Ma C, O’Connell RM, Mehta A, DiLoreto R, Heath JR, and Baltimore D (2014). Conversion of Danger Signals into Cytokine Signals by Hematopoietic Stem and Progenitor Cells for Regulation of Stress-Induced Hematopoiesis. Cell Stem Cell 14, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 (related to Figure 6). Budding of a Pycard+ cell from the aortic endothelium of a Tg(pycard:pycard-EGFP;flk1:mCherry) embryo from 34–36hpf.
Data Availability Statement
This study did not generate datasets or code requiring public database deposition.