Abstract
Several studies have shown that an increased risk of metabolic and immune disorders associated with cesarean section mode of delivery may exist. However, such studies have not been conducted in the Chinese population. Stool sample sequencing of the gene encoding the 16S rRNA of 82 prospectively enrolled 3- and 30–42-day-old vaginal and cesarean section delivered newborns was performed to study the composition and predicted function of the intestinal microbiota. In the samples from the 3-day-old neonates, the levels of Escherichia-Shigella in the two groups were similar. The genera Bifidobacterium, Lactobacillus, and Bacteroides were more prominent in the vaginal delivery than in the cesarean section group, which showed a predominance of Staphylococcus, Streptococcus, and Corynebacterium. The differences between the two groups were statistically significant (p < 0.05). In the samples from 30- to 42-day-old infants, Bifidobacterium, Lactobacillus, Escherichia-Shigella, and Bacteroides were the main genera present in the vaginal delivery group, while in the cesarean section delivery group; the predominant genera were Escherichia-Shigella, Bifidobacterium, Bacteroides, and Staphylococcus. Predicted functions of the vaginal delivery group revealed higher metabolic and biodegradation rates of carbohydrates, vitamins, and xenobiotics than those in the cesarean section group, which contributed to the stability of the microbiota in the former. The abundance of probiotic bacteria such as Bifidobacterium and Lactobacillus, and the negative correlation between obesity and Bacteroides presence were higher in vaginally delivered infants than in cesarean-delivered infants at both studied time points.
Keywords: vaginal delivery, cesarean section delivery, early infants, intestinal microbiota, predicted function
Introduction
Previous studies have found that the intestinal microbiota composed of colonizing bacteria plays an important regulatory role in human metabolism, cell differentiation, and immune function (Blaser and Falkow 2009). The interaction between the intestinal microbiota and their host is mechanistically involved in health and disease pathogenesis (Hegazy et al. 2017; Smith and Ravel 2017; Meisel et al. 2018). The colonization and maturity of the intestinal microbiota are affected by various factors, such as the modes of delivery, gestational age, feeding methods, and the use of antibiotics, especially in infants and young children (Madan et al. 2016; Chu et al. 2017; Uberos 2020).
In recent years, owing to an increase in cesarean deliveries worldwide, several studies on the effects of delivery modes on the structure and predicted function of the intestinal microbiota in infants had been conducted. These studies have shown that an increased risk of metabolic disorders, such as respiratory illness, and immune disorders, such as allergies and autoimmune diseases, may be associated with the cesarean section mode of delivery (Baumfeld et al. 2018; Reyman et al. 2019). However, to the best of our knowledge, such studies have not been conducted in the Chinese population.
We sought to determine the effects of the delivery modes and their potential confounders or modifiers on the structure and predicted function of intestinal microbiota in early infants in China.
Experimental
Materials and Methods
Volunteers and samples. We enrolled 82 healthy newborns (39 boys and 43 girls), of which 51 were delivered vaginally and 31 by cesarean section. The 82 recruited infants were born between the 15th of January and the 30th of November of 2019. All infants belonged to the Han Chinese ethnicity, were born between the gestational ages of 37 and 42 weeks, presented a weight at birth of 2.5–4 kg, and did not receive perinatal antibiotics. In all cases, the time between premature rupture of membranes and delivery was less than 18 hours. There were no significant differences between the two groups in gender, gestational age, birth weight, time of premature rupture of membranes, and Hb. Cesarean section was indicated according to the expert consensus on the procedure by the National Health Commission. The detailed information is presented in Table I.
Table I.
Description of 82 participants of this study.
| Group | Vaginal delivery (n = 51) | Cesarean delivery (n = 31) | p-value |
|---|---|---|---|
| Sex (%) | 0.907 | ||
| Boys | 24 (47.1%) | 15 (48.4%) | |
| Girls | 27 (52.9%) | 16 (51.6%) | |
| Gestational age (w) | 39.0 ± 0.9 | 39.2 ± 1.0 | 0.532 |
| Birth weight (g) | 3,091.0 ± 299.7 | 3,117.4 ± 260.4 | 0.685 |
| Hb (g/l) | 163.9 ± 12.0 | 161.2 ± 10.2 | 0.296 |
| Premature rupture of membranes (h) | 3.2 ± 2.1 | 3.3 ± 2.5 | 0.836 |
| Feeding (%) 3-day-old | 0.687 | ||
| Breast-fed | 24 (47.1%) | 12 (38.7%) | |
| Mixed-fed | 16 (31.4%) | 10 (32.3%) | |
| Fomula-fed | 11 (21.6%) | 9 (29.0%) | |
| Feeding (%) 30-42-day-old | 0.853 | ||
| Breast-fed | 21 (41.2%) | 11 (35.5%) | |
| Mixed-fed | 16 (31.3%) | 10 (32.3%) | |
| Fomula-fed | 14 (27.5%) | 10 (32.3%) | |
Mean ± SD for continuous variables: p-value was calculated by linear regression model.
% for categorical variables: p-value was calculated by chi-square test.
Sequencing analysis of the gene encoding the 16S rRNA was performed in stool samples on days 3 and 30–42 after delivery. The structure and predicted functions of the stool microbiota were analyzed. The rate of cesarean delivery was similar to the Chinese incidence (38.1% versus 36.7%, respectively). The Ethics Board of The First People’s Hospital of Xiaoshan (2019-XS-04) approved this project, and written informed consents were obtained from all participants.
Sequencing experiment flow. Herein, 200 mg of fresh fecal samples collected by the parents were transferred to 2 ml centrifuge tubes. Next, 1 ml RNA was added to each centrifuge tube, and the samples were mixed thoroughly and incubated at 4°C for 8–12 hours before freezing at –80°C. The samples were then transported over dry ice to Bio-science (Hangzhou, Zhejiang) for 16S rRNA sequencing. The DNA extracted from the stool samples was pooled and sequenced using 1% agarose gel electrophoresis, PCR amplification, fluorescence quantification, construction, and MiSeq library sequencing.
Biological information analysis process. We sequenced the hypervariable regions of the 16S ribosomal gene from the extracted DNA. Then, an interactive cloud analysis of the diversity in the microbial community was conducted. The diversity within samples (α-diversity) and between samples (β-diversity) was evaluated using the operational taxonomic unit (OTU) table. For α-diversity measurements, the indices were calculated using the sobs index, and the significance was determined using Student’s t-test. Phylo-)genetic (UniFrac) distance matrices were determined for β-diversity measurements. Community heatmaps were generated using the R package vegan. Cumulative distribution plots of β-diversity distances were generated using Qiime 1.9.1. Heatmap-based KEGG pathways were generated using PICRUSt 1.0.0 for the predicted function.
Statistical analysis. SPSS 22.0 software was used for statistical analysis. The Student’s t-test was used for normal distribution data, and the Wilcoxon signed-rank test was used for non-normal distribution data to compare the two groups. P-values < 0.05 were considered statistically significant.
Results
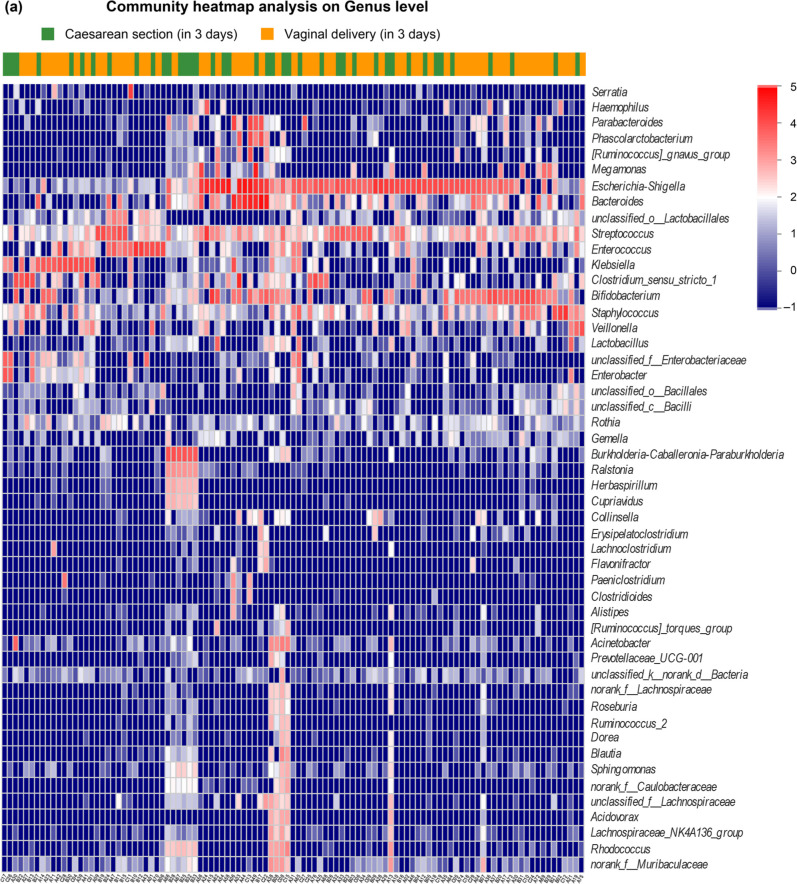
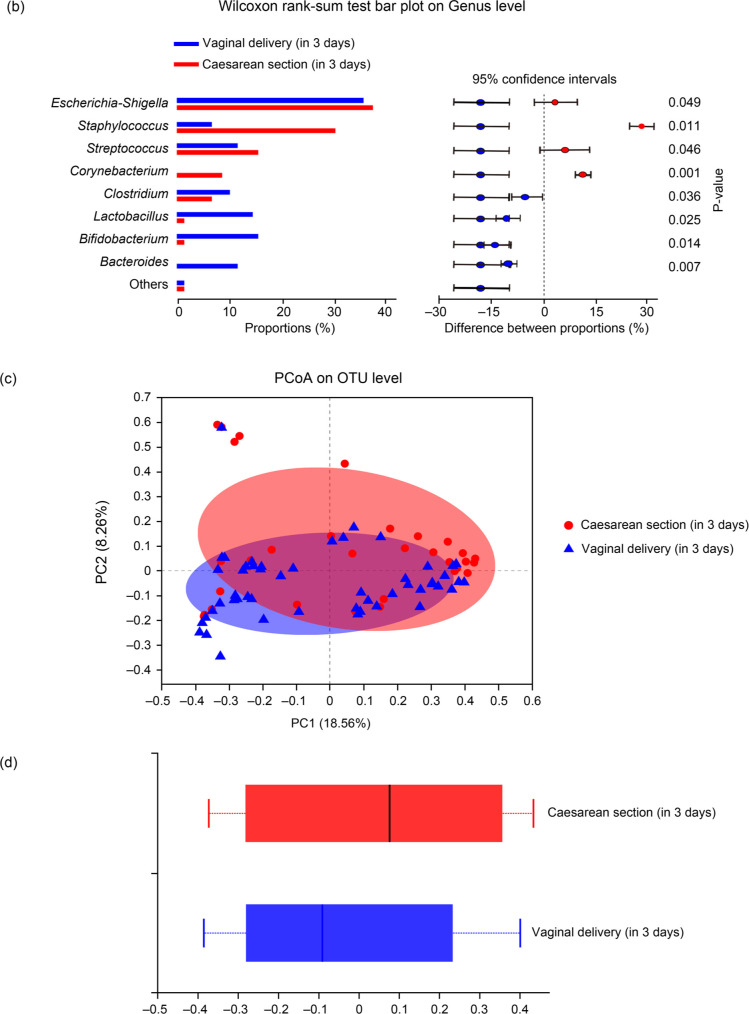
Effects of the delivery mode on the structure of intestinal microbiota. The intestinal microbiota structure of the two groups (vaginal delivery and cesarean section) in 3-day-old neonates and 30–42-day-old infants is shown in Fig. 1 and 2, respectively. As shown in Fig. 1, in the 3-day-old neonates, there were no significant differences in the levels of Escherichia-Shigella in the two groups (p > 0.05). The genera Bifidobacterium, Lactobacillus, and Bacteroides were more prominent in the vaginal delivery group than the cesarean section group, which had higher levels of Staphylococcus, Streptococcus, and Corynebacterium. The difference between the two groups was statistically significant (p < 0.05). Furthermore, as shown in Fig. 2, in the 30–42-day-old early infants, Bifidobacterium, Lactobacillus, Escherichia-Shigella, and Bacteroides were the prominent genera in the vaginal delivery group, whereas Escherichia-Shigella, Bifidobacterium, Bacteroides, and Staphylococcus were the main observed genera in the cesarean section group. The difference between the two groups was statistically significant (p < 0.05).
Fig. 1.
Intestinal microbiota community structure of 2 groups (vaginal delivery and cesarean section) in 3 days neonates.
(A) Intestinal microbiota community heatmap analysis on the genus level of the two groups.
(B) Wilcoxon rank-sum test bar plot on the genus level between the two groups.
(C) Principal coordinate analysis (PCoA) on unweighted UniFrac distances between the neonatal microbiota is shown along the first two principal coordinates (PC) axes. Each point represents a single sample and is colored by a delivery mode: vaginal delivery, blue; cesarean section, red. The closer the two sample points are, the more similar the species composition is.
(D) PCoA box diagram. Represents the discrete distribution of different groups of samples on the PC1 axis: vaginal delivery, blue; cesarean section, red.
Fig. 2.
Intestinal microbiota community structures of two groups (vaginal delivery and cesarean section) in 30–42 days infants.
(A) Intestinal microbiota community heatmap analysis on the genus level of the two groups.
(B) Wilcoxon rank-sum test bar plot on the genus level between the two groups.
(C) Principal coordinate analysis (PCoA) on unweighted UniFrac distances between the infants’ intestinal microbiota is shown along the first two principal coordinates (PC) axes. Each point represents a single sample and is colored by delivery mode: vaginal delivery, blue; cesarean section, red. The closer the two sample points are, the more similar the species composition is.
(D) PCoA box diagram. Represents the discrete distribution of different groups of samples on the PC1 axis: vaginal delivery, red; cesarean section, blue.
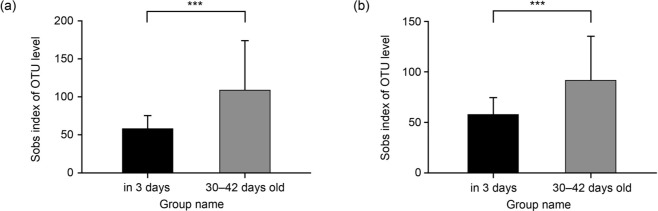
Sobs index analysis for α-diversity measurement. The Student’s t-test was used to calculate the significance of the intestinal microbiota’s sobs index in the vaginal delivery and cesarean section groups in 3- and 30–42-day-old infants shown in Fig. 3. The microbiota was detected in all the stool samples from the 3-day-old neonates, but their sobs index (α-diversity) was at significantly lower levels than that of the 30–42-day-old group, irrespective of vaginal or cesarean section modes of delivery (p < 0.001).
Fig. 3.
Student’s t-test for sobs index of vaginal delivery and cesarean section groups’ intestinal microbiota at two points (in 3 days and 30–42 days old). (A) Sobs index at the two points of vaginal delivery infants. (B) Sobs index at the two points of cesarean section infants.
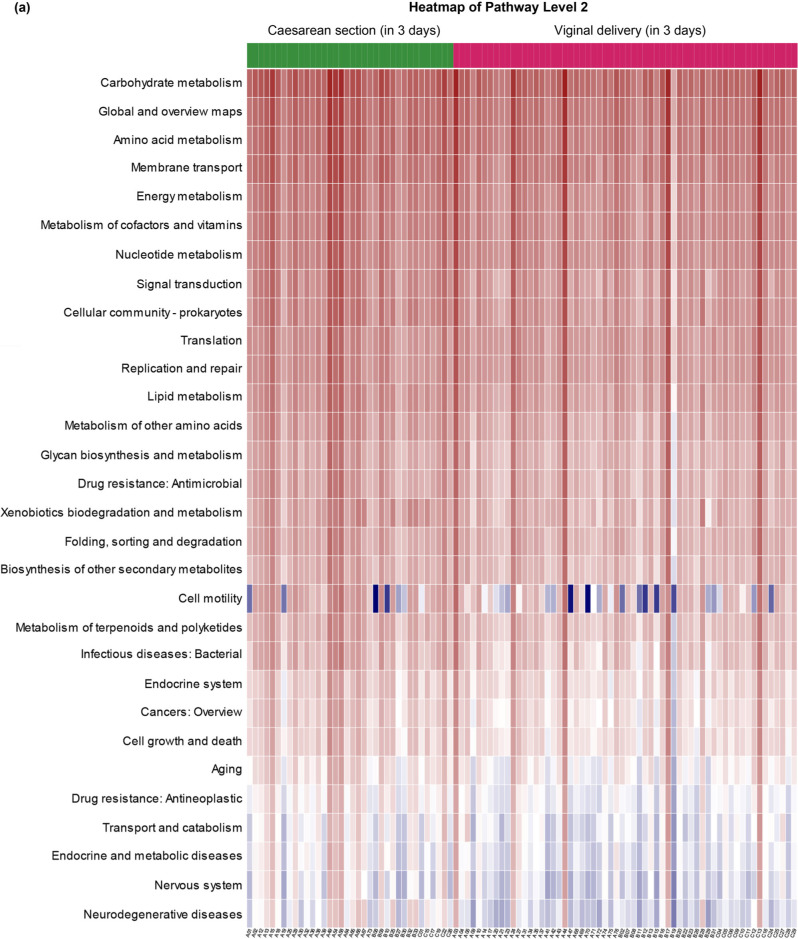
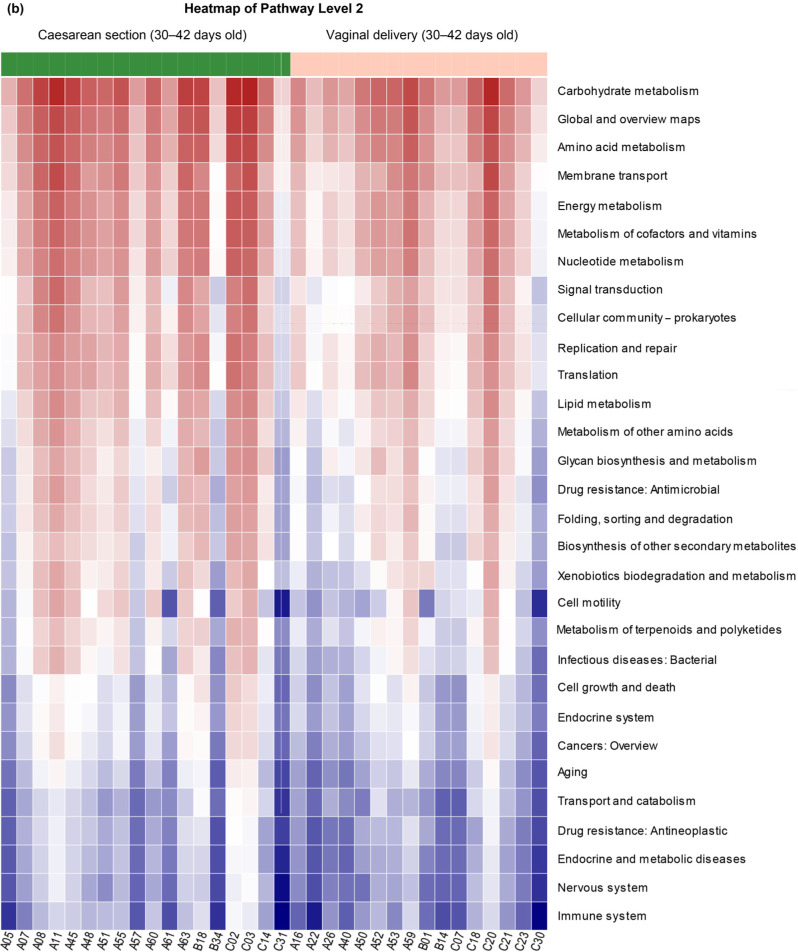
Predicted function of the intestinal microbiota. The predicted function of the intestinal microbiota of the vaginal delivery and cesarean section groups of both 3- and 30–42-day-old infants is shown in Fig. 4. Heatmap of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway level 2 at the two points belonged to the categories that showed statistical differences between vaginal delivery and cesarean section groups. Compared to the cesarean delivery group, the vaginal delivery group presented a higher carbohydrate and vitamin metabolism level. A higher rate of biodegradation and metabolism of xenobiotics in both neonates and infants favors their microbiota community’s stability.
Fig. 4.
Predicted function: Heatmap of KEGG pathway level 2.
(A) Heatmap showing distinct microbial gene (KEGG pathway level 2) profiles of the two groups’ stool in 3 days after delivery: vaginal delivery, r)ed; cesarean section, green.
(B) Heatmap showing distinct microbial gene (KEGG pathway level 2) profiles of 2 groups in 30–42 days old: vaginal delivery, green; cesarean section, red.
Discussion
In this study, we found that Escherichia-Shigella levels were significantly high in all stool samples of the 3-day-old infants. These data confirmed previous observations on the typical microbial constituents of early infant stools at this age (Backhed et al. 2015; Nagpal et al. 2017). Previously reported OTUs were considered to be derived from the maternal stool (Chu et al. 2017). Additionally, the levels of Escherichia-Shigella did not vary significantly between the modes of delivery. Furthermore, we found that the delivery mode has a much more significant influence on the presence and abundance of several other notable taxa in early infants’ intestinal microbiota. Accordingly, the vaginally delivered infants’ intestinal microbiota has high levels of Bifidobacterium, Lactobacillus, and Bacteroides, whereas the cesarean section-delivered infants harbor Staphylococcus, Streptococcus, and Corynebacterium. In infants delivered by cesarean section, the amniotic membrane does not rupture at birth; this prevents the mother’s birth-tract flora from entering the baby. Thus, vaginally delivered infants are enriched in bacterial communities resembling those found in the maternal vagina, whereas the cesarean-section delivered infants harbor skin microbiota. In the stool samples of the vaginally delivered 30–42-day-old early infants, Bifidobacterium, Lactobacillus, Escherichia-Shigella, and Bacteroides were the prominent genera, whereas those in the cesarean section group showed a predominance of Escherichia-Shigella, Bifidobacterium, Bacteroides, and Staphylococcus. The differences in each genus’s levels between the two groups were statistically significant (p < 0.05). Although microbiota was detected in all the first-pass stool samples, their α-diversity was relatively lower in the 3-day-old neonates than in the 30–42-day-old early infants, irrespective of the mode of delivery (p < 0.001). It has been previously shown that the abundance and growth of the intestinal microbiota progress with age (Chu et al. 2017). An endpoint of approximately 30–42 days postpartum was chosen because the infants at this age have limited person-to-person contact and are not yet exposed to the wide variety of environmental microbes. At 30–42 days of age, significant levels of Bifidobacterium and Lactobacillus continued to be present in the vaginally delivered infants. However, the infants delivered by cesarean section showed delayed colonization of bifidobacteria and a gradual rise in lactobacilli levels compared to those delivered vaginally (Wampach et al. 2017; Reyman et al. 2019). Bifidobacterium is a probiotic bacterium that promotes gut health and provides defense against pathogens (Tamburini et al. 2016). Acetate and lactate are the primary end products of bifidobacterial fermentation and important energy sources for colonocytes (Fukuda et al. 2011). Moreover, intestinal bifidobacteria produce essential nutrients, including riboflavin and folate (Sugahara et al. 2015). Lactobacillus is also a probiotic bacterium that regulates the gastrointestinal tract’s normal microbiota and maintains the micro-ecological balance to reduce serum cholesterol, improve gastrointestinal function, inhibit the growth of intestinal putrefactive bacteria, and increase immune functions. The abundance of lactobacilli was lower than that of bifidobacteria in the gut of infants delivered vaginally at both time points studied; our findings are similar to those reported previously (Yang et al. 2019). Furthermore, there is evidence that Bifidobacterium and Lactobacillus supplementation has positive effects in protecting the human gut from different intestinal infections (Tamburini et al. 2016) and has also been associated with the production of beneficial metabolites (Arboleya et al. 2015).
Bacteroides counts were low in both groups of 3-day-old neonates. This early reduction in diversity could be due to a decrease in the Bacteroides genus’s diversity within the Bacteroidetes phylum (Fallani et al. 2010). As opposed to a previous study by Wopereis et al. (2014), the stool samples of 30- to 42-day-old early infants showed significantly greater clustering of Bacteroides in the vaginally delivered infants than in the cesarean-delivered infants, irrespective of their ethnic factors. Bacteroides are efficient fermenters of human milk oligosaccharides, which may have been underestimated in early life due to a molecular bias. Furthermore, they may protect against the development of milk allergy (Hoyles and McCartney 2009). Previous studies have also shown that Bacteroides in the gut are negatively correlated to obesity. Here, we showed that the Bacteroides levels were significantly higher in the vaginally-delivered group than in the cesarean-delivered group in the 30–42-day-old infants. These data are consistent with previous findings on the correlation with obesity (Mueller et al. 2019). The abundances of Staphylococcus, Streptococcus, and Corynebacterium were higher in cesarean section delivered infants than in those delivered the vaginally, and these differences between the two groups are consistent with the data on human skin microbiota (Akagawa et al. 2019). Furthermore, these genera are neutral in the intestines of infants in their early life as human symbiotic bacteria or conditional pathogens. Thus, they were not analyzed in this study.
Predicted function revealed that the vaginal delivery group presented higher levels of carbohydrate and vitamin metabolism, biodegradation and metabolism of xenobiotics, and a more stable microbiota when compared to the cesarean delivery group.
In this study, we explored the effects of different delivery modes on the structure and predicted function of intestinal microbiota in neonates and early infants in the Chinese population. Furthermore, we provided scientific data for the construction of an early infant intestinal microbiota bank in China. However, there are certainly worth noting limitations. First, the feeding habits of the infants were variable at the end of the study. The potential impact of feeding habits cannot be overruled as a factor explaining the differences observed between vaginal delivery and cesarean section groups. Second, a study by Savage et al. (2018) reported that maternal diets with a high intake of vegetables and a low intake of processed meats and deep-fried foods present a positive correlation with the Lactobacillus abundance in the intestinal microbiota of infants. Thus, the lack of a food frequency questionnaire about maternal diet constitutes a limitation of our study.
16S rRNA sequencing technology was chosen to study the stool samples. 16S rRNA gene is located on the small subunit of prokaryotic ribosomes, including nine hypervariable regions and ten conserved regions. The conserved regions have few differences among bacteria, and the hypervariable regions have a specificity that varies with the kinship of genus or species. Therefore, 16S rRNA can be used as a characteristic nucleic acid sequence suggesting biological species and is considered to be the most suitable indicator for bacterial phylogeny and taxonomic identification (Caporaso et al. 2011). 16S rRNA amplicon sequencing usually selects one or several mutated regions and universal primers for amplification to obtain PCR products and then performs sequencing, analysis, and strain identification. It is an important means for studying environmental samples for microbial composition and structure (Youssef et al. 2009). At present, 16S rRNA sequencing technology is widely used worldwide and is also being gradually developed in China.
Overall, we have found that the abundance of probiotic bacteria, such as Bifidobacterium and Lactobacillus in vaginally delivered infants was higher than in the infants delivered by cesarean section at both studied time points. The level of Bacteroides in the cesarean section group was lower than that in the vaginal delivery group; hence, the former is associated with the risk of obesity. Further follow-up is needed to monitor the growth of Bacteroides with age dynamically. Screening of infants, especially those delivered by cesarean section, for excess weight gain, may help guide the primordial prevention of obesity. It is suggested that long-term follow-up of growth and development should be undertaken for children delivered by cesarean section to protect against the risk of obesity. Furthermore, this study also concludes that 16S rRNA sequencing technology is effective and reliable in detecting intestinal microbiota in early infants.
Footnotes
Author contributions
KYP, CYZ and JT contributed to the data collection, analysis, and writing of the manuscript. KYP contributed to the study design and editing of the manuscript.
Ethical statement
The Institutional Review Board of The First People’s Hospital of Xiaoshan District approved this study (Protocol Number: 2019-XS-04).
Funding
This research is supported by major scientific and technological plan projects for social development in Xiaoshan District (No. 2018201). Also is supported by Hangzhou Science and Technology Plan Guidance Project in Hangzhou city (No. 20181228Y81).
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Akagawa S, Tsuji S, Onuma C, Akagawa Y, Yamaguchi T, Yamagishi M, Yamanouchi S, Kimata T, Sekiya S, Ohashi A, et al. Effect of delivery mode and nutrition on gut microbiota in neonates. Ann Nutr Metab. 2019;74(2):132–139. 10.1159/000496427 [DOI] [PubMed] [Google Scholar]
- Arboleya S, Sánchez B, Milani C, Duranti S, Solís G, Fernández N, de los Reyes-Gavilán CG, Ventura M, Margolles A, Gueimonde M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr. 2015. Mar;166(3):538–544. 10.1016/j.jpeds.2014.09.041 [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015. Jun;17(6):852. 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Baumfeld Y, Walfisch A, Wainstock T, Segal I, Sergienko R, Landau D, Sheiner E. Elective cesarean delivery at term and the long-term risk for respiratory morbidity of the offspring. Eur J Pediatr. 2018. Nov;177(11):1653–1659. 10.1007/s00431-018-3225-8 [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009. Dec;7(12):887–894. 10.1038/nrmicro2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011. Mar 15;108 Supplement_1:4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017. Mar;23(3):314–326. 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, et al. Other Members of the INFABIO Team . Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010. Jul;51(1):77–84. 10.1097/MPG.0b013e3181d1b11e [DOI] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011. Jan;469(7331):543–547. 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017. Nov;153(5): 1320–1337.e16. 10.1053/j.gastro.2017.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyles L, McCartney AL. What do we mean when we refer to Bacteroidetes populations in the human gastrointestinal microbiota? FEMS Microbiol Lett. 2009. Oct;299(2):175–183. 10.1111/j.1574-6968.2009.01741.x [DOI] [PubMed] [Google Scholar]
- Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, Sogin ML, Li H, Moore JH, Karagas MR. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016. Mar 01; 170(3):212–219. 10.1001/jamapediatrics.2015.3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, Kim B, Brestoff JR, Tyldsley AS, Zheng Q, et al. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018. Dec;6(1):20. 10.1186/s40168-018-0404-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NT, Zhang M, Hoyo C, Østbye T, Benjamin-Neelon SE. Does cesarean delivery impact infant weight gain and adiposity over the first year of life? Int J Obes. 2019. Aug;43(8):1549–1555. 10.1038/s41366-018-0239-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R, Tsuji H, Takahashi T, Nomoto K, Kawashima K, Nagata S, Yamashiro Y. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird’s-eye view. Front Microbiol. 2017. Jul 21;8:1388. 10.3389/fmicb.2017.01388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyman M, van Houten MA, van Baarle D, Bosch AATM, Man WH, Chu MLJN, Arp K, Watson RL, Sanders EAM, Fuentes S, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. 2019. Dec;10(1):4997. 10.1038/s41467-019-13014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage JH, Lee-Sarwar KA, Sordillo JE, Lange NE, Zhou Y, O’Connor GT, Sandel M, Bacharier LB, Zeiger R, Sodergren E, et al. Diet during pregnancy and infancy and the infant intestinal microbiome. J Pediatr. 2018. Dec;203:47-54.e4. 10.1016/j.jpeds.2018.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol. 2017. Jan 15;595(2):451–463. 10.1113/JP271694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara H, Odamaki T, Hashikura N, Abe F, Xiao J. Differences in folate production by bifidobacteria of different origins. Biosci Microbiota Food Health. 2015;34(4):87–93. 10.12938/bmfh.2015-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016. Jul;22(7):713–722. 10.1038/nm.4142 [DOI] [PubMed] [Google Scholar]
- Uberos J. Perinatal microbiota: review of its importance in newborn health. Arch Argent Pediatr. 2020. Jun;118(3):e265–e270. 10.5546/aap.2020.eng.e265 [DOI] [PubMed] [Google Scholar]
- Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, Hugerth LW, Bindl L, Bottu J, Andersson AF, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol. 2017. May 02;8:738. 10.3389/fmicb.2017.00738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. The first thousand days – intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol. 2014. Aug;25(5):428–438. 10.1111/pai.12232 [DOI] [PubMed] [Google Scholar]
- Yang B, Chen Y, Stanton C, Ross RP, Lee YK, Zhao J, Zhang H, Chen W. Bifidobacterium and Lactobacillus composition at species level and gut microbiota diversity in infants before 6 weeks. Int J Mol Sci. 2019. Jul 05;20(13):3306. 10.3390/ijms20133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol. 2009. Aug 15;75(16):5227–5236. 10.1128/AEM.00592-09 [DOI] [PMC free article] [PubMed] [Google Scholar]