Abstract
Chronic respiratory diseases account for high morbidity and mortality, with asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (CF) being the most prevalent globally. Even though the diseases increase in prevalence, the exact underlying mechanisms have still not been fully understood. Despite their differences in nature, pathophysiologies, and clinical phenotypes, a growing body of evidence indicates that the presence of lung microbiota can shape the pathogenic processes underlying chronic inflammation, typically observed in the course of the diseases. Therefore, the characterization of the lung microbiota may shed new light on the pathogenesis of these diseases. Specifically, in chronic respiratory tract diseases, the human microbiota may contribute to the disease’s development and severity. The present review explores the role of the microbiota in the area of chronic pulmonary diseases, especially COPD, asthma, and CF.
Keywords: microbiota, asthma, COPD, cystic fibrosis, lungs
Introduction
The Human Microbiome Project launched in 2007 approximated the human microbiome’s complexity and led to significant growth in understanding its role in health and disease (Moffatt and Cookson 2017). In 2020, an international panel of experts proposed an updated definition of the microbiota, describing it as the assemblage of living microorganisms belonging to different kingdoms (Prokaryotes [Bacteria, or Archea], Eucaryotes [e.g., Protozoa, Fungi, and Algae]) (Berg et al. 2020). The human microbiome represents not only the entire community of commensal, symbiotic, and pathogenic microorganisms but also their “theatre of activity”, involving the whole spectrum of molecules produced by the microorganisms (their structural elements and metabolites), and molecules produced by coexisting hosts embedded in the environmental habitat (Berg et al. 2020). This complex ecosystem plays a fundamental role in controlling most aspects of physiology. Inhabiting various anatomical body sites, such as skin, mucosa, the gastrointestinal tract, and the respiratory tract, the microbiota is crucial in regulating the homeostasis and metabolism of hematopoiesis, inflammation, and immunity of its host (Moffatt and Cookson 2017). The microbiome’s size and composition evolve in response to environmental and host factors, and in any imbalance, a negative impact can be seen upon human health (Ogunrinola et al. 2020). Shifts in the microbiota composition may lead to dysbiosis by decreasing the number of symbionts and increasing potential dangerous pathogens. Microbial dysbiosis has been found to be involved in a growing list of human diseases, including chronic respiratory diseases (Paudel et al. 2020).
Chronic respiratory diseases account for high morbidity and mortality, with asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (CF) being the most prevalent globally (Paudel et al. 2020). Even though the diseases increase in prevalence, the exact underlying mechanisms have still not been fully understood. Despite their differences in nature, pathophysiologies, and clinical phenotypes, a growing body of evidence indicate that the presence of lung microbiota has the potential to shape the pathogenic processes underlying chronic inflammation typically observed in the course of the diseases (Dima et al. 2019; Loverdos et al. 2019). The characterization of the lung microbiota may therefore shed new light on the pathogenesis of these diseases. Specifically, in chronic respiratory tract diseases, the human microbiota may contribute to the disease’s development and severity. The present review aims at exploring the role of the microbiota in chronic pulmonary diseases, especially COPD, asthma, and CF, and, in particular, the possible mechanisms of airway microbiome contributing to the pathogenesis of these diseases. The main hypothesis is that interactions between the bacterial microbiome and host inflammation are related to chronic lung diseases and their exacerbations. For this purpose, a comprehensive literature search was conducted using PubMed to collect current studies concerning lung microbiome in COPD, asthma, and CF. The following keywords were used 1. microbiome and chronic pulmonary disease; 2. asthma; 3. COPD; 4. CF; 5. microbiota and immunity; 6. airway microbiome; 7. airway microbiota and combinations thereof.
Healthy lung microbiota
A microbiome analysis based on 16S rRNA sequencing has identified unique microbiota in organs previously considered sterile, such as the lower respiratory tract. Currently, there is no doubt that the mucous membrane of the lungs has its own resident microbiome (Huffnagle et al. 2017). Moreover, increasing evidence indicates that pulmonary microbiota, acting on resident immune cells, plays a key role in maintaining homeostasis in the organs (Dickson et al. 2013).
Lungs, being constantly in contact with environmental air, full of microorganisms and inhaled particles, are at the frontline of immunity (Wang et al. 2017; Sommariva et al. 2020). The lower respiratory tract is a rather hostile environment for microbes. Ciliary epithelium and numerous mucin-releasing secretory cells of bacteriostatic properties mainly line it, creating an immune barrier. Moreover, the immunity of healthy lungs is conditioned by the presence of lung-resident lymphocytes and alveolar macrophages, which additionally protects them against external threats (Malinowska et al. 2017; Wang et al. 2017). In addition to its essential role in immunity against pathogens, perhaps the pulmonary immune system’s most important function is to maintain a state of immunotolerance to non-dangerous environmental particles and self-antigens (Ramírez-Labrada et al. 2020; Sommariva et al. 2020). Cross-talk between alveolar macrophages, dendritic cells, and regulatory T cells (Treg) is the most critical to maintaining an immunological tone of the airways (Lloyd and Marsland 2017; Ramírez-Labrada et al. 2020; Sommariva et al. 2020). Of note, depletion of naturally occurring regulatory cells is characteristic of many chronic inflammatory diseases, including allergies, CF, and COPD, and their downregulation are strongly connected with disease progression (Ramírez-Labrada et al. 2020; Sommariva et al. 2020). Accumulating evidence shows that resident pulmonary Treg cells’ functionality can be significantly shaped by local lung microbiota (Ramírez-Labrada et al. 2020; Sommariva et al. 2020).
Lung microbiota is a relatively small bacterial community containing 103–105 cells/g of bronchial tissue. Comparatively, colon microbiota, which represents the most abundant ecosystem, comprises 1011–1012 cells/g of luminal content (Loverdos et al. 2019). Similar to other body sites, lungs present a complex bacterial community (Sommariva et al. 2020). According to numerous studies, the most common bacterial phyla that constitute the lung microbial ecosystem belong to Bacteroides and Firmicutes genera. It was also found that oral commensals, such as Prevotella, Veilonella, and Streptococcus, are the most prominent in healthy individuals’ lungs (Ramírez-Labrada et al. 2020; Sommariva et al. 2020; Xu et al. 2020). This neutral community is acquired mostly during microaspiration of pharyngeal particles or direct migration along airway mucosa, and all their members equally participate in creating the regional growth conditions, i.e., the lung microenvironment (Ramírez-Labrada et al. 2020). Thus, it appears that the microbes’ composition in the lungs is dynamic, mainly determined by the balance between microbial migration from the upper respiratory tract during microaspiration, elimination by coughing, mucociliary clearance, and immune system activity (Ramírez-Labrada et al. 2020; Sommariva et al. 2020; Xu et al. 2020). Different external and internal factors can influence the composition of the lung microbiome. Age, nutrition, lifestyle, pollution or tobacco smoke, inherited genes, and underlying diseases all can shape the type and number of lung microbiota, eventually leading to dysbiosis (Ogunrinola et al. 2019).
The respiratory microbiota both impacts and is impacted by immunity and disease. To avoid chronic inflammation under healthy condition, the lung microbiota has a fundamental role in shaping pulmonary immune tolerance. Microbes boost and calibrate innate and adaptive immunity, contribute to metabolic activities, and provide resistance to invasion by respiratory pathogens. Commensals can promote the establishment of a hostile environment and protect its host from pathogenic colonization (Ramírez-Labrada et al. 2020). This growth restriction may be caused by several mechanisms, such as altering of nutrient availability and production of antimicrobial metabolites (Ramírez-Labrada et al. 2020). Aside from the fact that microbiota can act on invasive microbes preventing their growth and spread, it plays a pivotal role in regulating immune tolerance in the lung environment. For homeostasis to be effectively managed, a balance must be maintained between several factors, including Th cell activation and suppression by regulatory Treg cells (Belkaid and Hand 2014; Belkaid and Harrison 2017). As mentioned before, a local lung commensal can significantly modulate the activity of resident pulmonary immune cells. Indeed, microbiota participates in shaping an immune tolerant environment influencing the recruitment and activation of Treg cells as well as macrophages and tolerogenic dendritic cells (Ramírez-Labrada et al. 2020; Sommariva et al. 2020).
An optimal microbiota-host interaction impacts the formation of a symbiotic relationship between commensal bacteria and their host. Beneficial as microbiota may be in maintaining health, its perturbation disrupts homeostatic processes promoting dysfunction and disease (Sommariva et al. 2020).
Lung microbiota in chronic pulmonary diseases
Inflammation represents a host response to many harmful stimuli, including pro-inflammatory mediators, environmental toxins, and chronic infection (Huffnagle et al. 2017). In contrast to acute inflammatory response, chronic inflammation represents a long-term reaction, characteristic of predominant recruitment of mononuclear leucocytes. A growing body of evidence suggests that the dysregulation of the human lung microbiota is involved in shaping the pathogenetic processes underlying nearly all kinds of chronic inflammatory respiratory diseases (Wang et al. 2017; Loverdos et al. 2019). This has been achieved by culture-independent sequencing techniques that in patients with chronic pulmonary diseases microbiota can be found that profoundly differs from that of healthy lungs, which indeed, determines a chronic inflammatory status. The reason for the problem – inflammation – suggests that microbiota can modulate the environment, in which it lives. Admittedly, disturbances in the microbial community can drastically change the local growth conditions by changing the resident immune cells’ activation state. During chronic inflammation, the lung habitat becomes unstable, and its species composition changes from “healthy” to a “pathogenic” state (Kovaleva et al. 2019).
Although the role of specific bacteria in the development of specific lung pathologies is still being established, a shift often occurs towards Proteobacteria the bacterial class of common lung-associated Gram-negative pathogens (mostly Neisseria, Moraxella, and Haemophilus) (Huffnagle et al. 2017; Sommariva et al. 2020). Particular metabolites and toxins with immunoregulatory properties are released, and other permissive niches for other species to reside there are created (O’Dwyer et al. 2016; Toraldo end Conte 2019). There is growing evidence that their metabolites, toxins, and cell lysis products by enriching the local microenvironment activate the host’s inflammatory cells and, therefore, contribute to lung diseases’ pathogenesis (Huffnagle et al. 2017; Sommariva et al. 2020).
Key environmental factors that influence commensal bacteria growth and survival, such as temperature, pH, oxygen tension, and host immune cells, change dramatically. Nutrients, limited during the steady-state, are made available in abundance with the introduction of mucus and vascular permeability (Huang et al. 2017; Sommariva et al. 2020). These profound changes in the community composition of the respiratory microbiota, directly influencing the immune system regulation, are now believed to be involved in a rapid increase of chronic inflammatory diseases (Belkaid and Hand 2014; O’Dwyer et al. 2016). It must also be considered that numerous byproducts of inflammation: catecholamines, and inflammatory cytokines, positively influence the growth and survival of some potentially pathogenic groups of bacteria, especially members of Proteobacteria. During chronic inflammation, bacteria that can benefit from inflammation outcompete bacteria that cannot thrive in such an unfavorable environment, resulting in a reduction in microbiome diversity (Huffnagle et al. 2017; Loverdos et al. 2019).
The basis of chronic lung diseases is a variety of biological and genetic mechanisms and dysbiosis, which significantly contributes to injury to the host. The microbiota’s metabolites and toxins are responsible for the changes in maintaining the steady-state of the lung microenvironment. However, it is not a single species or bacterial product responsible for this imbalance, but the entire cascade of events is an underlying cause of lung diseases. Thus, pathological changes in the composition and abundance of the lung microbiota might contribute to and result from chronic inflammation (Huang et al. 2017; Huffnagle et al. 2017; Sommariva et al. 2020) (Fig. 1).
Fig. 1.
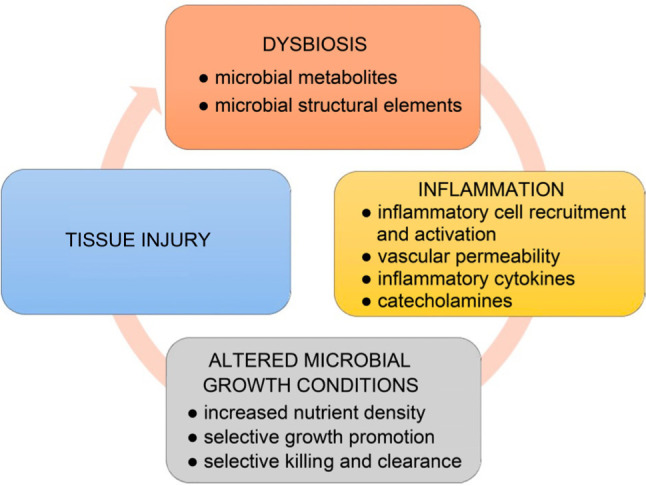
A circle of dysbiosis and inflammation in chronic pulmonary diseases.
Airway microbiome in asthma
Asthma is a complex disease of the respiratory airways characteristic of chronic inflammation in the lungs, reversible airflow obstruction, mucus overproduction, and tissue neutrophilia or eosinophilia (Paudel et al. 2020). There is not a single cause of asthma (Kozik and Huang 2019). Existing data point to genetic and exogenous risks, including cigarette smoke, viral and bacterial infections, and obesity, associated with severe asthma and exacerbations of the disease (Kozik and Huang 2019; Paudel et al. 2020).
The role of the lung microbiome in the pathogenesis of asthma has not yet been fully elucidated. However, the most constant finding among lung microbiome studies is an observed increase in bacterial load and dominance of Proteobacteria, particularly Haemophilus spp. and Moraxella catarrhalis (Castro-Nallar et al. 2015; Noval Rivas et al. 2016; Loverdos et al. 2019). Dysbiotic communities may substantially contribute to the course or severity of the disease. On the other hand, a reduction in bacterial diversity could influence the inflammatory asthma phenotypes (Chung 2017; Sverrild et al. 2017). Different studies have shown that in patients with neutrophilic asthma, the microbiota composition is altered, and organisms, such as M. catarrhalis, Haemophilus spp. and Streptococcus spp. predominate. Their presence is associated with severe airflow obstruction, longer asthma duration, and neutrophilic infiltration, possibly via Th-17 – driven mechanism (Chung et al. 2017; Sverrild et al. 2017; Kozik and Huang 2019; Goto 2020; Paudel et al. 2020). In patients with worse asthma symptoms, airway enrichment with potentially pathogenic Proteobacteria members correlates with the level of IL-8, the neutrophilic pro-inflammatory marker. Interestingly, neutrophilic asthma is also associated with inadequate response to the first-line corticosteroid treatment. In corticosteroid-resistant patients, an overgrowth of Haemophilus parainfluenzae has been noted. Moreover, Haemophilus influenzae and Tropheryma have been reported in sputum samples of patients with poorly controlled severe cases (Simpson et al. 2016; Chung 2017). As inhaled steroids are a mainstay of the therapy, the involvement of particular bacterial genera in resistance mechanisms may explain the cause of steroidunresponsive asthma.
Eosinophilic asthma results from the activation of Th2 response that leads to airway infiltration by eosinophils. Here, in contrast to neutrophilic phenotype, microbiota’s role is less clear and more heterogeneous (Taylor et al. 2018; Barcik et al. 2020). As observed, eosinophil infiltration of bronchial tissue is associated with low bacterial burden and diversity (Denner et al. 2016; Cait et al. 2018). The same studies found a negative correlation between Proteobacteria and Firmicutes and lung eosinophilia (Huang et al. 2015; Denner et al. 2016; Cait et al. 2018). It is now suggested that fungi, such as Aspergillus spp. rather than bacteria play a more significant role in eosinophilic asthma. This finding is supported by the fact that fungi, being significant allergens, can induce a strong eosinophilic allergic reaction in asthmatic patients. Nevertheless, the role of fungal microbiota in asthma remains poorly understood (Durack et al. 2017; Kozik and Huang 2019; Loverdos et al. 2019).
Respiratory viruses, mainly rhinoviruses, are detected in more than 75% of patients with acute exacerbation of asthma. For a long time, the bacterial role in asthma exacerbations has not been considered, since traditionally, no microbes were grown from patients’ clinical specimens. Also, no clinical benefits of early antibiotic therapy were noticed in patients with acute exacerbation of the disease. However, culture-independent approaches, surprisingly, demonstrated the presence of atypical bacteria, such as Mycoplasma pneumoniae and Chlamydia pneumoniae (Dickson et al. 2014). However, their role in exacerbations of asthma is less clear than that of viral infections. It is believed that these bacterial pathogens may be somewhat involved in asthma persistence than exacerbations (Papadopoulos et al. 2011).
Airway microbiome in cystic fibrosis
Although cystic fibrosis (CF) is a multisystem disease, the disease’s most tremendous burden falls upon the respiratory tract, characteristic of chronic airway infection and inflammation (Cribbs and Beck 2016). Recent studies have demonstrated that the lung microbiome is associated with the pathogenesis of this disease.
Cystic fibrosis arises from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene that is required for the homeostatic control of chloride ions in the lung. The CFTR gene’s role is crucial, and its mutations lead to mucin overproduction along airways and disruption of the regular mucociliary activity and transport (Cribbs and Beck 2016; Françoise and Héry-Arnaud 2020). This defect also promotes polymicrobial proliferation and microbial imbalance along the respiratory tract, as mucin is a rich nutritional resource, which can support bacterial growth (Françoise and Héry-Arnaud 2020). In CF, the subsequent activation of neutrophilic inflammatory response leads to uncontrolled inflammation and progressive lung disease that severely limits pulmonary function (LiPuma 2010; Belkaid and Hand 2014). Polymicrobial chronic airway infections, which are characteristic of CF, lead to persistent inflammation and periodic episodes of acute pulmonary exacerbation.
Traditionally, only a few bacteria have been associated with CF. Pseudomonas aeruginosa, Staphylococcus aureus, H. influenzae, and Burkholderia cepacia are the most often CF airway constituents (Belkaid and Hand 2014; Surette 2014; Cuthbertson et al. 2020). However, next-generation sequencing significantly advanced our knowledge about bacterial communities within a CF lung. Lines of evidence now show that CF lung microbiome is more complex, polymicrobial, highly diverse with overexpression of Actinobacteria and Proteobacteria, and potentially pathogenic anaerobes (Gillanders et al. 2011; Cuthbertson et al. 2020; Françoise and Héry-Arnaud 2020). The changes in the bacterial community might play an essential role in CF airway microenvironment condition. Bacteria within the CF lung contribute to horizontal gene transfer, especially in the presence of biofilm. They may affect the host by shedding different bioactive molecules that interact with host cells in shaping immune response and triggering inflammatory cytokines and growth factors (Cribbs and Beck 2017).
Microbial diversity is a marker of lung function (Françoise and Héry-Arnaud 2020). In CF, bacterial diversity correlates with the patient’s age, the disease stage, and recurrent antibiotic therapies. A frequent broad-spectrum antibiotic therapy to treat pulmonary infections together with airway inflammation appears to be a significant driver of a fall in bacterial diversity that increases with age. This reduction is additionally associated with the establishment of species that are regarded as pathogens, usually, H. influenzae, S. aureus, and P. aeruginosa (Cox et al. 2010; Rogers et al. 2017; Françoise and Héry-Arnaud 2020). Consequently, the collapse in community diversity is associated with reduced lung function and disease progression (Cox et al. 2010). Data on strain changes in patients with pulmonary exacerbations remain unclear and contradictory (Carmody et al. 2013; Surette et al. 2014; Acosta et al. 2017). Many authors indicate that a total load of bacteria does not generally change at exacerbation onset. They also noticed that exacerbations are not related to significant in microbial diversity changes, but the microbiota composition that was more diverse at baseline tended to change over time (Hoffman and Surette 2013). Some studies report that P. aeruginosa and anaerobes are the critical components of pulmonary exacerbations. According to the observations of Carmody et al. (2013) facultatively anaerobic bacteria of the genus Gemella increased by 83% during exacerbation. Although the pathogenic potential of Gemella is not known, its abundance in patients during exacerbation makes it a potential biomarker candidate of the acute stage of the disease (Hoffman and Surette 2013).
Airway microbiome in chronic obstructive pulmonary disease (COPD)
Chronic obstructive pulmonary disease (COPD), one of the most prevalent respiratory diseases, is characteristic of persistent symptoms and airway obstruction due to inflammation. Traditionally, culture-based methods of analysis of COPD samples frequently revealed H. influenzae, S. pneumoniae, M. catarrhalis and P. aeruginosa as the most prominent pathogenic bacteria involved in the pathogenesis of the disease. Recent use of high throughput 16S rRNA gene-based sequencing has provided additional evidence of the presence of dysbiosis in COPD. Culture-independent methods confirmed that in COPD, aerobic and anaerobic bacteria colonize the airways, with Proteobacteria (Haemophilus spp.) being more common than Bacteroides (Prevotella spp.) being significantly reduced (Einarsson et al. 2016; Haldar et al. 2020). In addition, in stable COPD, a higher level of colonization with potentially pathogenic bacteria, such as H. influenzae, S. pneumonia, and M. catarrhalis is associated with a greater chance of exacerbation and a more severe airflow obstruction (Martinez et al. 2013; Ditz et al. 2020). These bacteria are found in more than 75% of cases and are regarded as a “core” pulmonary microbiome in COPD (Martinez et al. 2013; Einarsson et al. 2016).
The disease is characteristic of a chronic inflammatory state, and bacteria colonizing COPD patients’ airways are believed to participate in this dysregulation and provoke the host inflammatory response (Dima 2019; Haldar et al. 2020). Even at the state of bacterial colonization in stable COPD, many authors have reported excessive inflammation. In patients with COPD, persistent neutrophilic inflammation likely represents a local host immune response to chronic microbial colonization (Burgel et al. 2017). In addition, it has been confirmed that the profile of colonization-induced inflammation is similar to that observed during exacerbation. Therefore, it is likely that chronic microbial infection with potential bacterial pathogens serves as an activator of an immune response (Martinez et al. 2013; Wang et al. 2016; Burgel et al. 2017).
In patients with acute exacerbation, augmenting neutrophilic or eosinophilic inflammation is typically observed. As expected, bacteria-associated COPD exacerbations are associated with airway neutrophilia, accompanied by elevated levels of the primary mediators of inflammation, including IL-1β, IL-8, and TNF. Subsequently, an increased number of neutrophils and inflammatory markers in the airways can cause significant damage to the respiratory tissue (Burgel et al. 2017; Ditz et al. 2020).
Conclusions and perspectives
While the study of the exact influence of the microbiome on lung health is still a relatively new field, it is clear that respiratory microbiome changes accompany chronic lung diseases of a non-infectious etiology, as previously believed. The composition of the lung microbiome is increasingly well characterized, and a specific spectrum of pathogenic microorganisms observed in association with several chronic respiratory diseases such as asthma, cystic fibrosis, and chronic obstructive pulmonary disease is already described. Indeed, the lung microbiome in chronic pulmonary diseases has a dynamic nature and is influenced by many factors, such as age, environmental exposure, treatment, etc.
It is also now becoming clear that chronic respiratory diseases’ pathogenesis is a cascade of events in a self-amplifying cycle of altered microbial growth conditions, respiratory dysbiosis, and airway inflammation. In this cycle of events, changes in lung microbial community composition and abundance provoke airway inflammation, injuring the delicate lung tissue. And then, inflammation of the airways alters microbial environmental growth conditions promoting dysbiosis.
Lung microbiota does not seem to be the only microbial factor contributing to chronic pulmonary diseases. The gut microbiota’s influence on lung immunity has been well documented and linked to disease development in the lungs with immune response changes when the gut microbiota community is altered (Pulvirenti et al. 2019). Mounting evidence indicates that the immune cells and cytokines induced by the gut microbiota components or metabolites, especially short-chain fatty acids (SCFAs), can modulate lung immunity and inflammatory responses, with specific taxa able to influence the pathogenesis of chronic pulmonary diseases (Samuelson et al. 2015; Cait et al. 2018). Although there is a growing body of evidence for the role of the gut/lung axis in lung diseases, several unanswered questions still exist for its clinical relevance.
Environmental factors such as pollution, cigarette smoke, antibiotics, and diet can also modulate microbiota composition and affect disease susceptibility. Indeed, it is broadly accepted that cigarette smoking and air pollution are the most substantial risk factors for developing COPD (Allais et al. 2016). At the same time, an increased risk of asthma has been associated with microbial dysbiosis in the gut in early life. Simultaneously, the use of antibiotics in children has also been implicated in the development of asthma (Yang et al. 2019).
Although many of the underlying mechanisms are likely disease-specific, the microbiome’s exploration is promising and provides a solid background for developing innovative strategies for diagnostic, preventative, and therapeutic approaches.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Acosta N, Whelan FJ, Somayaji R, Poonja A, Surette MG, Rabin HR, Parkins MD. The evolving cystic fibrosis microbiome: a comparative cohort study spanning 16 years. Ann Am Thorac Soc. 2017. Aug;14(8):1288–1297. 10.1513/AnnalsATS.201609-668OC [DOI] [PubMed] [Google Scholar]
- Allais L, Kerckhof FM, Verschuere S, Bracke KR, De Smet R, Laukens D, Van den Abbeele P, De Vos M, Boon N, Brusselle GG, et al. Chronic cigarette smoke exposure induces microbial and inflammatory shifts and mucin changes in the murine gut. Environ Microbiol. 2016. May;18(5):1352–1363. 10.1111/1462-2920.12934 [DOI] [PubMed] [Google Scholar]
- Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020. Feb 18;52(2):241–255. 10.1016/j.immuni.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014. Mar;157(1):121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017. Apr;46(4):562–576. 10.1016/j.immuni.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Rybakova D, Fischer D, Cernava T, Vergès MCC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020. Dec;8(1):103. 10.1186/s40168-020-00875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgel P-R, Contoli M, López-Campos JL. Acute exacerbations of the pulmonary diseases. Sheffield (UK): European Respiratory Society; 2017. 10.1183/2312508X.erm7717 [DOI] [Google Scholar]
- Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, Reynolds LA, Hacker L, Mohr J, Finlay BB, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018. May;11(3):785–795. 10.1038/mi.2017.75 [DOI] [PubMed] [Google Scholar]
- Carmody LA, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB, Li JZ, LiPuma JJ. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc. 2013. Jun; 10(3):179–187. 10.1513/AnnalsATS.201211-107OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Nallar E, Shen Y, Freishtat RJ, Pérez-Losada M, Manimaran S, Liu G, Johnson WE, Crandall KA. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med Genomics. 2015. Dec;8(1):50. 10.1186/s12920-015-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KF. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J Allergy Clin Immunol. 2017. Apr; 139(4):1071–1081. 10.1016/j.jaci.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010. Jun 23;5(6):e11044. 10.1371/journal.pone.0011044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs SK, Beck JM. Microbiome in the pathogenesis of cystic fibrosis and lung transplant-related disease. Transl Res. 2017. Jan; 179:84–96. 10.1016/j.trsl.2016.07.022 [DOI] [PubMed] [Google Scholar]
- Cuthbertson L, Walker AW, Oliver AE, Rogers GB, Rivett DW, Hampton TH, Ashare A, Elborn JS, De Soyza A, Carroll MP, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome. 2020. Dec;8(1):45. 10.1186/s40168-020-00810-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, Sperling AI, Solway J, Naureckas ET, Gilbert JA, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. 2016. May; 137(5):1398–1405.e3. 10.1016/j.jaci.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013. Jun;7(3):245–257. 10.1586/ers.13.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014. Aug; 384(9944): 691–702. 10.1016/S0140-6736(14)61136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima E, Kyriakoudi A, Kaponi M, Vasileiadis I, Stamou P, Koutsoukou A, Koulouris NG, Rovina N. The lung microbiome dynamics between stability and exacerbation in chronic obstructive pulmonary disease (COPD): Current perspectives. Respir Med. 2019. Oct;157:1–6. 10.1016/j.rmed.2019.08.012 [DOI] [PubMed] [Google Scholar]
- Ditz B, Christenson S, Rossen J, Brightling C, Kerstjens HAM, van den Berge M, Faiz A. Sputum microbiome profiling in COPD: beyond singular pathogen detection. Thorax. 2020. Apr;75(4):338–344. 10.1136/thoraxjnl-2019-214168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, Dyer AM, Israel E, Kraft M, Martin RJ, et al. ; National Heart, Lung and Blood Institute’s “AsthmaNet” . Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017. Jul;140(1):63–75. 10.1016/j.jaci.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsson GG, Comer DM, McIlreavey L, Parkhill J, Ennis M, Tunney MM, Elborn JS. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax. 2016. Sep;71(9):795–803. 10.1136/thoraxjnl-2015-207235 [DOI] [PubMed] [Google Scholar]
- Françoise A, Héry-Arnaud G. The microbiome in cystic fibrosis pulmonary disease. Genes (Basel). 2020. May 11;11(5):536. 10.3390/genes11050536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillanders LJ, Elborn JS, Gilpin DF, Schneiders T, Tunney MM. The airway microbiome in cystic fibrosis: challenges for therapy. Therapy. 2011. Nov;8(6):645–660. 10.2217/thy.11.81 [DOI] [Google Scholar]
- Goto T. Airway microbiota as a modulator of lung cancer. Int J Mol Sci. 2020. Apr 26;21(9):3044. 10.3390/ijms21093044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar K, George L, Wang Z, Mistry V, Ramsheh MY, Free RC, John C, Reeve NF, Miller BE, Tal-Singer R, et al. The sputum microbiome is distinct between COPD and health, independent of smoking history. Respir Res. 2020. Dec;21(1):183. 10.1186/s12931-020-01448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L, Surette M. What to expect when you’re expectorating: cystic fibrosis exacerbations and microbiota. Ann Am Thorac Soc. 2013. Jun;10(3):249–250. 10.1513/AnnalsATS.201304-086ED [DOI] [PubMed] [Google Scholar]
- Huang YJ, Erb-Downward JR, Dickson RP, Curtis JL, Huffnagle GB, Han MK. Understanding the role of the microbiome in chronic obstructive pulmonary disease: principles, challenges, and future directions. Transl Res. 2017. Jan;179:71–83. 10.1016/j.trsl.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, Boushey H. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015. Oct;136(4):874–884. 10.1016/j.jaci.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017. Mar;10(2):299–306. 10.1038/mi.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva OV, Romashin D, Zborovskaya IB, Davydov MM, Shogenov MS, Gratchev A. Human lung microbiome on the way to cancer. J Immunol Res. 2019. Jul 29;2019:1394191. 10.1155/2019/1394191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik AJ, Huang YJ. The microbiome in asthma: Role in pathogenesis, phenotype, and response to treatment. Ann Allergy Asthma Immunol. 2019. Mar;122(3):270–275. 10.1016/j.anai.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010. Apr;23(2):299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM, Marsland BJ. Lung homeostasis: influence of age, microbes and the immune system. Immunity. 2017. Apr;46(4):549–561. 10.1016/j.immuni.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Loverdos K, Bellos G, Kokolatou L, Vasileiadis I, Giamarellos E, Pecchiari M, Koulouris N, Koutsoukou A, Rovina N. Lung microbiome in asthma: Current perspective. J Clin Med. 2019. Nov 14; 8(11):1967. 10.3390/jcm8111967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowska M, Tokarz-Deptuła B, Deptuła W. [Mikrobiom człowieka] (in Polish). Postepy Mikrobiol. 2017;56(1):33–42. [Google Scholar]
- Martinez FJ, Erb-Downward JR, Huffnagle GB. Significance of the microbiome in chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2013. Dec;10 Supplement:S170–S179. 10.1513/AnnalsATS.201306-204AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Cookson WO. The lung microbiome in health and disease. Clin Med (Lond). 2017. Dec;17(6):525–529. 10.7861/clinmedicine.17-6-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noval Rivas M, Crother TR, Arditi M. The microbiome in asthma. Curr Opin Pediatr. 2016. Dec;28(6):764–771. 10.1097/MOP.0000000000000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer DN, Dickson RP, Moore BB. The lung microbiome, immunity and the pathogenesis of chronic lung disease. J Immunol. 2016. Jun 15;196(12):4839–4847. 10.4049/jimmunol.1600279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunrinola GA, Oyewale JO, Oshamika OO, Olasehinde GI. The human microbiome and its impacts on health. Int J Microbiol. 2020. Jun 12;2020:1–7. 10.1155/2020/8045646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos NG, Christodoulou I, Rohde G, Agache I, Almqvist C, Bruno A, Bonini S, Bont L, Bossios A, Bousquet J, et al. Viruses and bacteria in acute asthma exacerbations – A GA2LEN--DARE* systematic review. Allergy. 2011. Apr;66(4): 458–468. 10.1111/j.1398-9995.2010.02505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel KR, Dharwal V, Patel VK, Galvao I, Wadhwa R, Malyla V, Shen SS, Budden KF, Hansbro NG, Vaughan A, et al. Role of lung microbiome in innate immune response associated with chronic lung diseases. Frontiers in Medicine. 2020. Sep 18;7:554. 10.3389/fmed.2020.00554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvirenti G, Parisi GF, Giallongo A, Papale M, Manti S, Savasta S, Licari A, Marseglia GL, Leonardi S. Lower airway microbiota. Front Pediatr. 2019. Sep 27;7:393. 10.3389/fped.2019.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Labrada AG, Isla D, Artal A, Arias M, Rezusta A, Pardo J, Gálvez EM. The influence of lung microbiota on lung carcinogenesis, immunity and immunotherapy. Trends Cancer. 2020. Feb;6(2):86–97. 10.1016/j.trecan.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Rogers GB, Bruce KD, Hoffman LR. How can the cystic fibrosis respiratory microbiome influence our clinical decision-making? Curr Opin Pulm Med. 2017. Nov;23(6):536–543. 10.1097/MCP.0000000000000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. 2015. Oct 7;6:1085. 10.3389/fmicb.2015.01085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Hugenholtz P, Willner D, et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J. 2016. Mar;47(3):792–800. 10.1183/13993003.00405-2015 [DOI] [PubMed] [Google Scholar]
- Sommariva M, Le Noci V, Bianchi F, Camelliti S, Balsari A, Tagliabue E, Sfondrini L. The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol Life Sci. 2020. Jul;77(14):2739–2749. 10.1007/s00018-020-03452-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG. The cystic fibrosis lung microbiome. Ann Am Thorac Soc. 2014. Jan;11 Suppl 1:S61–S65. 10.1513/AnnalsATS.201306-159MG [DOI] [PubMed] [Google Scholar]
- Sverrild A, Kiilerich P, Brejnrod A, Pedersen R, Porsbjerg C, Bergqvist A, Erjefält JS, Kristiansen K, Backer V. Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J Allergy Clin Immunol. 2017. Aug;140(2): 407–417.e11. 10.1016/j.jaci.2016.10.046 [DOI] [PubMed] [Google Scholar]
- Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Jenkins C, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018. Jan;141(1):94–103.e15. 10.1016/j.jaci.2017.03.044 [DOI] [PubMed] [Google Scholar]
- Toraldo DM, Conte L. Influence of the lung microbiota dysbiosis in chronic obstructive pulmonary disease exacerbations: the controversial use of corticosteroid and antibiotic treatments and the role of eosinophils as a disease marker. J Clin Med Res. 2019;11(10):667–675. 10.14740/jocmr3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li F, Tian Z. Role of microbiota on lung homeostasis and diseases. Sci China Life Sci. 2017. Dec;60(12):1407–1415. 10.1007/s11427-017-9151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Bafadhel M, Haldar K, Spivak A, Mayhew D, Miller BE, Tal-Singer R, Johnston SL, Ramsheh MY, Barer MR, et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J. 2016. Apr;47(4):1082–1092. 10.1183/13993003.01406-2015 [DOI] [PubMed] [Google Scholar]
- Xu N, Wang L, Li C, Ding C, Li C, Fan W, Cheng C, Gu B. Microbiota dysbiosis in lung cancer: evidence of association and potential mechanisms. Transl Lung Cancer Res. 2020. Aug;9(4):1554–1568. 10.21037/tlcr-20-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Feng H, Zhan X, Zhang C, Cui R, Zhong L, Ying S, Chen Z. Early-life vancomycin treatment promotes airway inflammation and impairs microbiome homeostasis. Aging (Albany NY). 2019. Apr 13; 11(7):2071–2081. 10.18632/aging.101901 [DOI] [PMC free article] [PubMed] [Google Scholar]


