Abstract
The soil bacterial communities have been widely investigated. However, there has been little study of the bacteria in Qinghai-Tibet Plateau, especially about the culturable bacteria in highland barley cultivation soil. Here, a total of 830 individual strains were obtained at 4°C and 25°C from a highland barley cultivation soil in Qamdo, Tibet Autonomous Region, using fifteen kinds of media. Seventy-seven species were obtained, which belonged to 42 genera and four phyla; the predominant phylum was Actinobacteria (68.82%), followed by Proteobacteria (15.59%), Firmicutes (14.29%), and Bacteroidetes (1.30%). The predominant genus was Streptomyces (22.08%, 17 species), followed by Bacillus (6.49%, five species), Micromonospora (5.19%, four species), Microbacterium (5.19%, four species), and Kribbella (3.90%, three species). The most diverse isolates belonged to a high G+C Gram-positive group; in particular, the Streptomyces genus is a dominant genus in the high G+C Gram-positive group. There were 62 species and 33 genera bacteria isolated at 25°C (80.52%), 23 species, and 18 genera bacteria isolated at 4°C (29.87%). Meanwhile, only eight species and six genera bacteria could be isolated at 25°C and 4°C. Of the 77 species, six isolates related to six genera might be novel taxa. The results showed abundant bacterial species diversity in the soil sample from the Qamdo, Tibet Autonomous Region.
Keywords: Qinghai-Tibet Plateau; Streptomyces; 16S rRNA, novel taxa; high-altitude area
Introduction
Bacteria constitute a major proportion of biodiversity in soil ecosystems; they are the main driving force for the conversion and circulation of carbon, nitrogen, and phosphorus, and also the prominent participants in biochemical processes of soil organic matter decomposition and humus formation (Fulthorpe et al. 2008; Řeháková et al. 2015; Malard et al. 2019). Bacterial assemblages are essential components of soils in arid ecosystems, especially in remote high-elevation mountains (Margesin et al. 2009; Yuan et al. 2014). While global surveys of microbial diversity and functional activity have already been conducted (Bodelier 2011; Delgado-Baquerizo et al. 2018), the number of Qinghai-Tibet Plateau samples is restricted, and, therefore bacterial data is still lacking in this area, especially in the most high-altitude area (Zhang et al. 2016).
Highland barley (Hordeum vulgare L.) is the fourth most consumed grain worldwide, only ranked after rice, wheat, and maize (Shen et al. 2016; Deng et al. 2020). Highland barley is a hulless barley cultivar and used as the main staple food for the Tibetan people widely grown in Qinghai-Tibet Plateau in China (He et al. 2019; Zhang et al. 2019). Extreme environments such as cold and hypoxia in Tibet have promoted the unique ecological environment and soil bacterial composition (Zhang et al. 2007; 2010a). However, the extreme environments also have led to the decline of soil bacterial activity and the impoverishment of soil for growing highland barley (Yu et al. 2009; Zhao et al. 2014). The research of soil bacteria in the highland barley planting field has important significance for highland barley yield increase, pest control, and soil quality improvement (Bailly and Weisskopf 2012). At present, there were few studies on bacteria in the soil of the highland barley-planting field (Liu et al. 2019). Significantly, the culturable bacteria isolated from highland barley cultivation soil have not been reported systematically.
The Qamdo region’s temperature is between 20°C and 28°C from June to September, a significant growth period for highland barley. While the temperature is below 10°C from November to March, no crops were planted on the land during this period. So the culturable bacteria were isolated from a high-altitude highland barley cultivation soil collected in Qamdo using 15 media at 4°C and 25°C to simulate the temperature conditions over these two periods in this study. The composition of bacterial communities was characterized based on the 16S rRNA gene (Furlong et al. 2002; Li et al. 2019). Our aims were: (1) to reveal the diversity of culturable bacteria isolated from highland barley cultivation soil in the high-altitude area; and (2) to study the effect of different culture temperatures on the species of culturable bacteria in highland barley cultivation soil.
Experimental
Materials and Methods
Study site and samples collection. The sampling site was located in the Zhu Village, Banbar County, Qamdo, Tibet Autonomous Region (30°55’48.9’N, 94°58’13.4’E, Altitude: 4,011 m); the sampling site is the typical high-altitude patches farmland in Qamdo, which is about one-third of Qamdo’s farmland. The sample site belongs to the plateau temperate subhumid climate type, the air temperature range is –40–29°C, the annual average air temperature is –1°C, and the yearly frozen period is from September to April. The soil type was sandy loam, and the pH value is 7.6. The previous crop was highland barley, and the yield is about 1,000–1,800 kg/hm2 in this area. A highland barley cultivation soil sample was collected from a depth of 5–15 cm using the five-point method and kept in sterilized paper bags in April 2018. Once retrieved, the soil sample was immediately stored at 4°C, and bacteria were isolated in the laboratory in Lhasa in May and June 2018.
Isolation and maintenance of bacteria. The bacteria in highland barley cultivation soil sample were isolated using X1, R, L1, ISP2, GW1, DSM372, F1, F2, M1, M5, M6, M7, M8, HV, and GS media, as shown in Table I. Gram-negative bacteria and Actinobacteria were isolated by using the dilution plating technique as described by Kuklinsky-Sobral et al. (2004) and Zhang et al. (2016), respectively, with some modifications. 0.2 ml of 10–2, 10–3, and 10–4 soil suspensions were spread onto F1, F2, M1, M5, M6, M7, M8, HV, and GS media to isolate Actinobacteria. While, 0.2 ml of 10–4, 10–5, and 10–6 soil suspension was spread onto X1, R, L1, ISP2, GW1, and DSM372 media to isolate Gram-negative bacteria. Two sets of plates were incubated at 4°C and 25°C, respectively; the bacterial strains were obtained across 3–60 days. The pure culture isolates were preserved in glycerol suspensions (20%, v/v) at –80°C for further research.
Table I.
Isolation media.
| Media | Composition |
|---|---|
| X1 | peptone 2.0 g, yeast extract 0.5 g, FePO4 · 4H2O 0.1 g, MgSO4 · 7H2O 0.5 g, CaCO3 0.2 g, NaCl 0.5 g, agar 18.0 g, ddwater 1,000 ml, pH 7.0 |
| R | peptone 10.0 g, yeast extract 5.0 g, maltose extract 5.0 g, casein amino acid 5.0 g, beef extract 2.0 g, glycerol 2.0 g, Tween-80 50.0 mg, MgSO4 · 7H2O 1.0 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.6 |
| L1 | NaCl 100.0 g, K2HPO4 5.0 g, MgSO4 · 7H2O 7.5 g, hydrolyzed casein 1.0 g, yeast extract 5.0 g, Na3C6H5O7 · 2H2O 3.0 g, FeSO4 · 7H2O 0.1 g, MnCl2 · 4H2O 0.1 g, ZnSO4 · 7H2O 0.1 g, agar 18.0 g, ddwater 1,000 ml, pH 7.0-8.0 |
| ISP2 | NaCl 100.0 g, dextrose 4.0 g, yeast extract 4.0 g, maltose extract 10.0 g, MgSO4 · 7H2O 0.5 g, CaCO3 2.0 g, FeSO4 10 mg, agar 18.0 g, ddwater 1,000 ml, pH 7.0-8.0 |
| GW1 | NaCl 100.0 g, casein 0.3 g, mannitol 1.0 g, NaHCO3 2.0 g, CaCO3 0.2 g, (NH4)2SO4 2.0 g, KNO3 2.0 g, K2HPO41.0 g, MgSO4 · 7H2O 2.0 g, FeSO4 10.0 mg, Trace-salt 10.0 mg/l, Agar 18.0 g, ddwater 1,000 ml, pH natural |
| DSM372 | NaCl 100.0 g, hydrolyzed casein 5.0 g, yeast extract 5.0 g, Na3C6H5O7 · 2H2O 3.0 g, Na2CO3 · 10H2O 8.0 g, NaC5H8NO4 1.0 g, KCl 2.0 g, MgSO4 · 7H2O 2.0 g, agar 18.0 g, ddwater 1,000 ml, pH natural |
| F1 | glycerol 5.0 g, alanine 3.0 g, arginine 1.0 g, (NH4)2SO4 2.64 g, KH2PO4 2.38 g, K2HPO4 5.65 g, MgSO4 · 7H2O 1.0 g, CuSO4 · 5H20 0.0064 g, FeSO4 · 7H2O 0.0011 g, MnCl2 · 4H2O 0.0079 g, ZnSO4 · 7H2O 0.0015 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| F2 | MgSO4 · 7H2O 0.5 g, CaCO3 0.2 g, FeSO4 10.0 mg, NaCl 0.5 g, MnCl2 · 4H2O 1.4 g, Na2MoO4 · 2H2O 0.39 g, Co(NO3)2 · 6H2O 0.025 g, ZnSO3 · 7H2O 0.222 g, NaHCO3 2.0 g, NaH2PO4 · 2H2O 0.05 g, agar 18.0 g, ddwater 1,000 ml, pH natural (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| M1 | soluble starch 10.0 g, casein 0.3 g, KNO3 2.0 g, K2HPO4 2.0 g, MgSO4 · 7H2O 0.05 g, FeSO4 · 7H2O 0.01 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| M5 | yeast extract 4.0 g, soluble starch 15.0 g, K2HPO4 1.0g, FeSO4 · 7H2O 0.01 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.6 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| M6 | raffinose 10.0 g, L-histidine 1.0 g, MgSO4 · 7H2O 0.5 g, FeSO4 · 7H2O 0.01 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| M7 | L-aspartic acid 0.1 g, peptone 2.0 g, sodium propionate 4.0 g, FeSO4 · 7H2O 0.01 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| M8 | glycerine 6.0 ml, arginine 1.0 g, MgSO4 · 7H2O 0.5 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| HV | humic acid 1.0g, Na2HPO4 0.5 g, KCl 1.7 g, MgSO4 0.5 g, FeSO4 0.01 g, CaCO3 0.02 g, agar 18.0 g, ddwater 1,000 ml, pH 7.2-7.4 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
| GS | soluble starch 20.0 g, NaCl 0.5 g, KNO3 1.0 g, K2HPO4 · 3H2O 0.5 g, MgSO4 · 7H2O 0.5 g, FeSO4 · 7H2O 0.01 g, agar 18.0 g, ddwater 1,000 ml, pH 7.4-7.6 (add 25 μg/ml nalidixic acid and 100 μg/ml nystatin) |
PCR amplification and sequencing of the 16S rRNA gene. According to the manufacturer’s protocol, the genomic DNA of bacteria was extracted using a bacterial genomic DNA FastPrep Extraction Kit (TIAN-GEN DP302). Polymerase chain reaction (PCR) amplification of the partial 16S rRNA gene was performed using the universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGAC TT-3’), PCR was performed using the extracted highly purified genomic DNA as a template under the following conditions: 95°C for 10 min, followed by 94°C for 45 s, 55°C for 45 s, and 72°C for 90 s for 30 cycles with a final 10 min extension at 72°C. The PCR products were detected by agarose gel electrophoresis and then sent to GENEWIZ.lnc for the 16S rRNA gene sequencing. The phylogenetic status of the species was determined by a reaction of 700–750 bp (V1-V4) using the universal primers 27F, if the similarity was less than 98.65% (Kim et al. 2014), then the phylogenetic status of the species was further analyzed by nearly full-length 16S rRNA gene (1,300–1,400 bp).
Phylogenetic analysis. Similarity searches of the 16S rRNA gene sequences were performed in the NCBI and EzBiocloud database for BLAST, then the 16S rRNA gene sequences with the highest homology were obtained for phylogenetic analysis. The sequence alignments were performed using Clustal X, the phylogenetic trees were constructed from evolutionary distances using the neighbor-joining method with a bootstrap of 1,000 repetitions, and the phylogenetic analysis was conducted using the MEGA 7 software (Kumar et al. 2016b).
Nucleotide sequence accession numbers. The full and partial 16S rRNA gene sequences of the strains were submitted to the NCBI GenBank database under the accession numbers (MT611248-MT611324).
Results
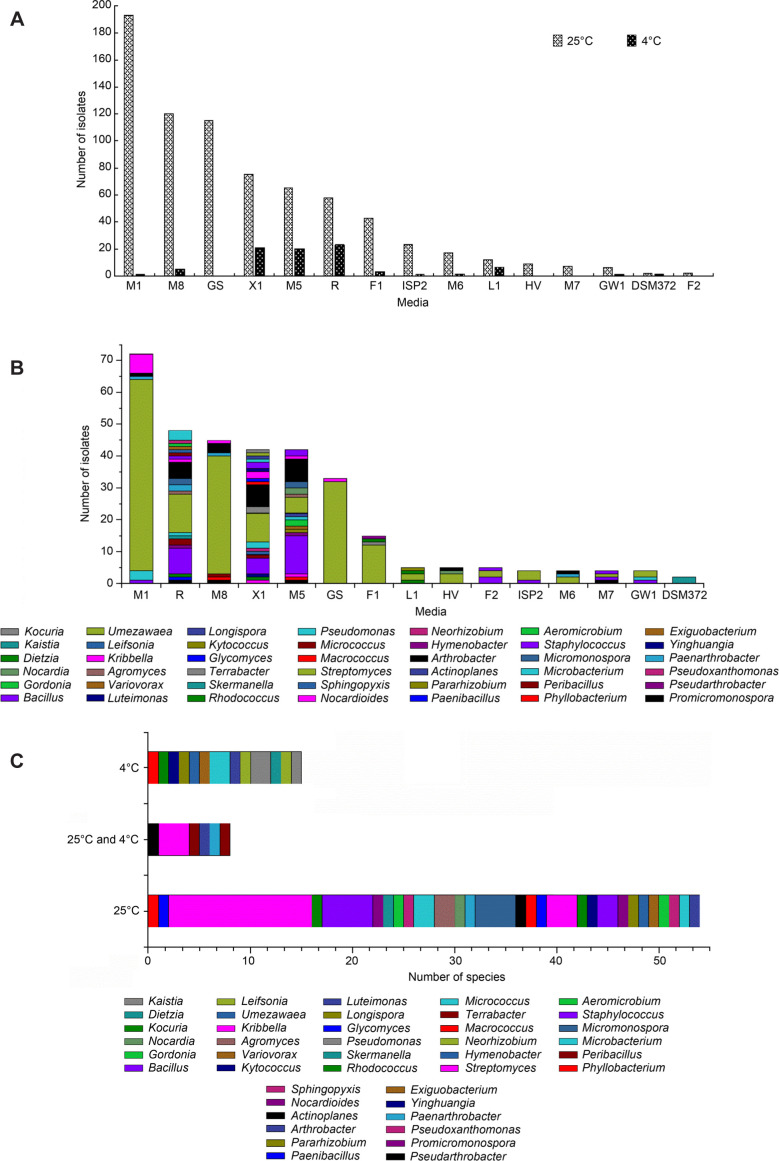
The isolated strains. Bacterial populations were successfully isolated from the highland barley cultivation soil sample using fifteen kinds of media, a total of 830 individual strains were obtained at different culture temperatures (4°C and 25°C) (Fig. 1A). Eighty-three and 747 strains of bacteria were isolated from these media at 4°C and 25°C, respectively. The results showed that X1, R, F1, M1, M5, M8, and GS culture media had a better effect on isolating bacteria at 25°C; however, X1, R, and M5 culture media had a better effect on isolating bacteria at 4°C, none of the bacteria was isolated from the F2, M7, HV, and GS media at 4°C.
Fig. 1.
The number and diversity of bacteria.
A) The numbers of bacteria isolated from different media at 4° and 25°. B) Diversity of bacteria isolated from different culture media. C) Diversity of bacteria isolated from different temperature. D) The numbers of dominant species isolated from 4° and 25°.
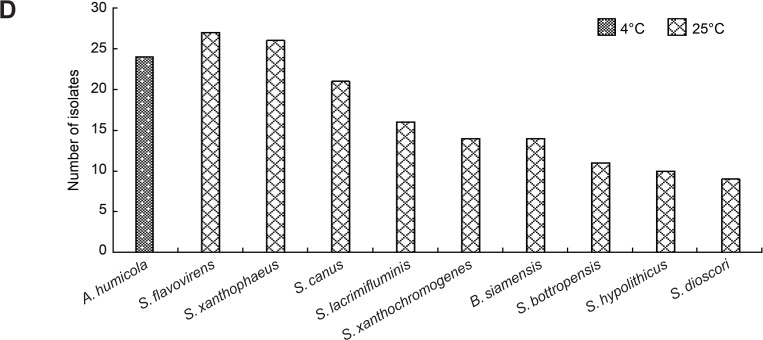
Phylogenetic analysis of culturable strains by the 16S rRNA gene sequence. According to the morphological characteristics of bacteria, 330 strains were screened for the 16S rRNA gene sequence analysis using the universal primers 27F/1492R, and 98.65% of the 16S rRNA gene sequences were used as the species boundary of prokaryotes. After combining more than 98.65% of the 16S rRNA gene sequences with the same species, the sequences of 77 species were obtained, which belonged to 42 genera and four phyla (Actinobacteria, Proteobacteria, Firmicutes, and Bacteroidetes), as shown in Table II. Phylogenetic tree based on the 16S rRNA gene sequences of representative bacteria strains were shown (Fig. 2).
Table II.
Genera distributed in each of the four phyla.
| Actinobacteria | Proteobacteria | Firmicutes | Bacteroidetes | |
|---|---|---|---|---|
| Actinoplanes | Micrococcus | Kaistia | Bacillus | Hymenobacter |
| Aeromicrobium | Micromonospora | Luteimonas | Exiguobacterium | |
| Agromyces | Nocardia | Neorhizobium | Macrococcus | |
| Arthrobacter | Nocardioides | Pararhizobium | Paenibacillus | |
| Dietzia | Paenarthrobacter | Phyllobacterium | Peribacillus | |
| Glycomyces | Promicromonospora | Pseudomonas | Staphylococcus | |
| Gordonia | Pseudarthrobacter | Pseudoxanthomonas | ||
| Kocuria | Rhodococcus | Skermanella | ||
| Kribbella | Streptomyces | Sphingopyxis | ||
| Kytococcus | Terrabacter | Variovorax | ||
| Leifsonia | Umezawaea | |||
| Longispora | Yinghuangia | |||
| Microbacterium | ||||
Fig. 2.
Phylogenetic tree based on 16S rRNA gene sequences of soil isolates and related species.
There were 53 species and 25 genera in Actinobacteria, accounting for 68.82% of the species’ total number. The predominant genus was Streptomyces (22.08%, 17 species), followed by Micromonospora (5.19%, four species), Microbacterium (5.19%, four species), and Kribbella (3.90%, three species). Some rare Actinobacteria were also isolated, for example, Leifsonia, Longispora, Nocardia, Nocardioides, Terrabacter, Umezawaea, and Kribbella. There were 12 species and ten genera in Proteobacteria, accounting for 15.59% of the total number of species, but no dominant genus was found in Proteobacteria. There were 11 species and six genera in Firmicutes, accounting for 14.29% of the total number of species; the predominant genus was Bacillus (6.49%, five species). Only one species was found in Bacteroidetes, classified as Hymenobacter (1.30%, one species) (Table III).
Table III.
BLAST results based on 16S rRNA gene sequences of 77 bacterial species.
| Strain number | Name of strain having the highest 16S rRNA gene similarity | The highest similarity (%) |
|---|---|---|
| T74* | Actinoplanes digitatis IFO 12512 | 98.82 |
| T203 | Aeromicrobium ginsengisoli Gsoil 098 | 99.82 |
| T96* | Agromyces binzhouensis OAct353 | 98.62 |
| T229* | Agromyces humatus CD5 | 98.74 |
| T805 | Arthrobacter crystallopoietes DSM 20117 | 99.85 |
| T763 | Arthrobacter humicola KV-653 | 100 |
| T65 | Bacillus siamensis KCTC 13613 | 100 |
| T94 | Bacillus cereus ATCC 14579 | 100 |
| T228* | Bacillus drentensis LMG 21831 | 99.34 |
| T59 | Bacillus pumilus ATCC 7061 | 100 |
| T115 | Bacillus selenatarsenatis SF-1 | 99.6 |
| T822 | Dietzia kunjamensis subsp DSM 44907 | 99.86 |
| T230 | Exiguobacterium mexicanum 8NT | 100 |
| T183* | Glycomyces algeriensis NRRL B-16327 | 98.9 |
| T64 | Gordonia otitidis NBRC 100426 | 100 |
| T830* | Hymenobacter humi DG31A | 98.60 |
| T769* | Kaistia defluvii B6-12 | 99.72 |
| T144 | Kocuria sediminis FCS-11 | 99.43 |
| T145 | Kribbella albertanoniae BC640 | 100 |
| T214* | Kribbella catacumbae DSM 19601 | 99.6 |
| T422 | Kribbella karoonensis Q41 | 99.87 |
| T823 | Kytococcus schroeteri DSM 13884 | 99.73 |
| T781 | Leifsonia flava SYP-B2174 | 99.73 |
| T146 | Longispora urticae NEAU-PCY-3 | 99.88 |
| T181* | Luteimonas composti CC-YY255 | 98.9 |
| T156 | Macrococcus canis KM 45013 | 99.86 |
| T489 | Microbacterium maritypicum DSM 12512 | 99.55 |
| T773 | Microbacterium natoriense TNJL143-2 | 99.87 |
| T804 | Microbacterium phyllosphaerae DSM 13468 | 99.73 |
| T133 | Microbacterium thalassium IFO 16060 | 98.93 |
| T226 | Micrococcus luteus NCTC 2665 | 99.63 |
| T47 | Micromonospora cremea DSM 45599 | 99.87 |
| T206 | Micromonospora luteifusca GUI2 | 99.87 |
| T197* | Micromonospora palomenae NEAU-CX1 | 98.74 |
| T92 | Micromonospora saelicesensis Lupac 09 | 100 |
| T786* | Neorhizobium vignae CCBAU 05176 | 98.70 |
| T62 | Nocardia salmonicida subsp R89 | 99.47 |
| T105* | Nocardioides caeni MN8 | 98.01 |
| T218 | Paenibacillus odorifer DSM 15391 | 99.63 |
| T608 | Paenarthrobacter aurescens NBRC 12136 | 99.07 |
| T236 | Paenarthrobacter nitroguajacolicus G2-1 | 100 |
| T808* | Pararhizobium herbae CCBAU 83011 | 98.79 |
| T209 | Peribacillus simplex NBRC 15720 | 100 |
| T811 | Phyllobacterium ifriqiyense STM 370 | 100 |
| T274* | Phyllobacterium zundukense Tri-48 | 98.57 |
| T63 | Promicromonospora alba 1C-HV12 | 100 |
| T193* | Pseudarthrobacter siccitolerans 4J27 | 99.34 |
| T755 | Pseudomonas laurylsulfativorans AP3_22 | 99.73 |
| T776 | Pseudomonas lini CFBP 5737 | 100 |
| T174* | Pseudoxanthomonas sacheonensis BD-c54 | 99.34 |
| T127* | Rhodococcus jostii DSM 44719 | 99.32 |
| T788 | Rhodococcus qingshengii JCM 15477 | 100 |
| T185* | Skermanella aerolata 5416T-32 | 98.86 |
| T93 | Sphingopyxis fribergensis Kp5.2 | 99.87 |
| T45 | Staphylococcus caprae ATCC 35538 | 100 |
| T61 | Staphylococcus cohnii subsp ATCC 49330 | 100 |
| T666 | Streptomyces albogriseolus NRRL B-1305 | 100 |
| T313 | Streptomyces atroolivaceus NRRL ISP-5137 | 100 |
| T234 | Streptomyces bottropensis ATCC 25435 | 99.87 |
| T130 | Streptomyces caniferus NBRC 15389 | 99.87 |
| T235 | Streptomyces canus DSM 40017 | 99.73 |
| T690 | Streptomyces dioscori A217 | 99.47 |
| T532 | Streptomyces flavovirens NBRC 3716 | 99.85 |
| T674* | Streptomyces humidus NBRC 12877 | 98.8 |
| T296 | Streptomyces hydrogenans NBRC 13475 | 99.46 |
| T219 | Streptomyces hypolithicus HSM10 | 99.46 |
| T426 | Streptomyces kurssanovii NBRC 13192 | 99.6 |
| T569 | Streptomyces lunaelactis MM109 | 99.2 |
| T348 | Streptomyces niveus NRRL 2466 | 99.46 |
| T84 | Streptomyces phaeoluteigriseus DSM 41896 | 99.6 |
| T581 | Streptomyces turgidiscabies ATCC 700248 | 100 |
| T110* | Streptomyces xanthochromogenes NRRL B-5410 | 98.97 |
| T100 | Streptomyces xanthophaeus NRRL B-5414 | 99.71 |
| T111* | Terrabacter ginsengisoli Gsoil 653 | 99.19 |
| T160 | Umezawaea tangerina NRRL B-24463 | 99.18 |
| T812 | Variovorax boronicumulans BAM-48 | 99.47 |
| T134* | Yinghuangia seranimata YIM 45720 | 98.73 |
– shown that the full length 16S rRNA gene of this bacterium was sequenced
Diversity of culturable strains recovered from different culture media. Among the 330 identified bacteria strains, the number of bacterial isolates recovered from M1 was the largest (21.82%, 72 strains), followed by R (14.55%, 48 strains), M8 (13.64%, 45 strains), X1 (12.73%, 42 strains), M5 (12.73%, 42 strains), GS (10.00%, 33 strains), F1 (4.55%, 15 strains), L1 (1.52%, five strains), F2 (1.52%, five strains), HV (1.52%, five strains), ISPT2 (1.21%, four strains), M6 (1.21%, four strains), M7 (1.21%, four strains), GW1 (1.21%, four strains), and DSM372 (0.61%, two strains). The number of bacteria isolated from M1, M8, and GS was larger, while the main genus was only Streptomyces. The R, X1, and M5 yielded higher genera diversity (21 genera, 20 genera, and 17 genera, respectively). Meanwhile, media R, X1, and M5 were more useful than other media for isolation of rare genera of bacteria, such as Nocardioides, Leifsonia, Terrabacte, Umezawaea, Variovorax, Neorhizobium, and Pararhizobium (Fig. 1B). Here, we presumed that single-nutrition was the main reason, especially when non-monosaccharide was used as the carbon source (Zhang et al. 2010b; Kurm et al. 2019). This study demonstrated that it is necessary to use various isolation media types to increase the number and diversity of bacteria from highland barley cultivation soil samples.
Diversity of culturable strains at different temperature. There were 62 species and 33 genera bacteria isolated at 25°C, accounting for 80.52% of the species’ total number. The predominant genus was Streptomyces (22.08%, 17 species), followed by Bacillus (6.49%, five species), Micromonospora (5.19%, four species), Kribbella (3.90%, three species), and Paenarthrobacter (3.90%, three species). There were 23 species and 18 genera bacteria isolated at 4°C, accounting for 29.87% of the total species, but no dominant genus was found. Meanwhile, only eight species and six genera of bacteria could be isolated at 25°C and 4°C (Fig. 1C). Most common bacteria could be isolated at 25°C, but some rare bacteria could be isolated at 4°C without the inhibitory effect of dominant species, promoting the diversity of bacteria (Margesin 2012; Collins and Margesin 2019). The numbers of dominant species mainly isolated at 4°C were Arthrobacter humicola (7.25%, 24 strains), while the numbers of dominant species mainly isolated at 25°C were Streptomyces flavovirens (8.19%, 27 strains), Streptomyces xanthophaeus (7.58%, 25 strains), Streptomyces canus (6.36%, 21 strains), and Bacillus siamensis (4.24%, 14 strains) (Fig. 1D). The species of culturable bacteria and the numbers of dominant species were significantly different at 4°C and 25°C in this study.
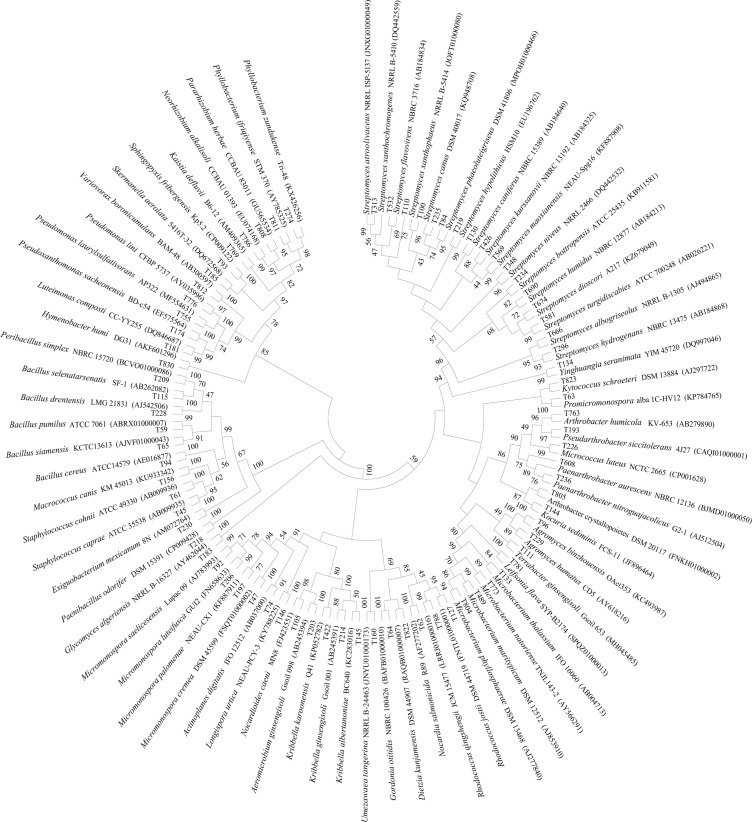
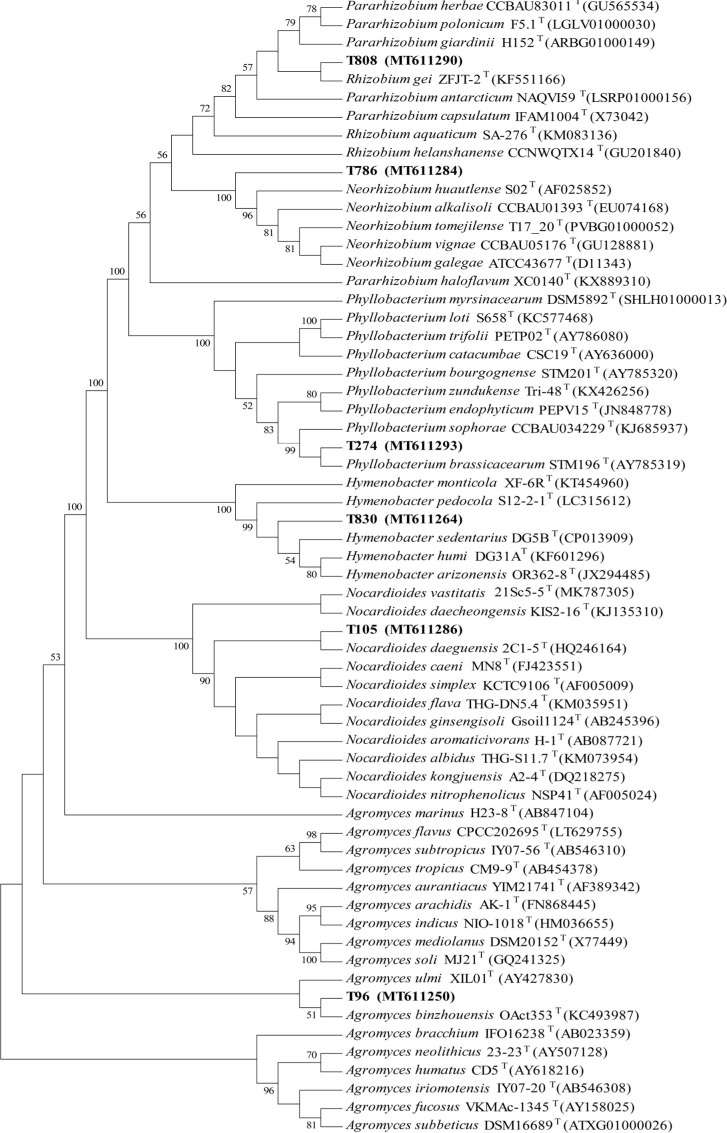
Potential new species information. Among the 77 species, four bacterial strains exhibited low 16S rRNA gene sequence similarities (< 98.65 %) with validly described species based on the results of the BLAST search in EzBiocloud (Table IV), which indicated that these isolates could represent novel taxa. Neorhizobium gen. nov. was a new genus of rhizobia established by Mousavi et al. (2014); so far, only five species had been published. The T786 strain had 98.70%, 98.47%, 98.24%, 98.16%, 97.79%, and 96.55% sequence similarity with Neorhizobium vignae CCBAU 05176T (GU128881), Neorhizobium alkalisoli CCBAU01393T (EU074168), Neorhizobium tomejilense T17_20T (PVBG01000052), Neorhizobium huautlense S02T (AF025852), Neorhizobium galegae ATCC43677T(D11343), and Neorhizobium lilium 24NRT (MK386721), respectively (Fig. 3). Further data analysis suggested that the dDDH and ANI values between strain T786 and N. vignae CCBAU 05176T, N. alkalisoli CCBAU 01393T, N. tomejilense T17_20T, N. huautlense S02T, and N. galegae ATCC 43677T were 20.20–20.50% and 76.64–80.01%, respectively, which were lower than the threshold values of 70% and 95–96% for species discrimination (unpublished). Pararhizobium gen. nov. was a new genus of rhizobia also established by Mousavi et al. (2015); so far, only seven species had been published. The T808 strain had high similarity with Pararhizobium herbae CCBAU83011T (GU565534) (98.79%), Pararhizobium polonicum F5.1T (LGLV01000030) (98.65%), and Pararhizobium giardinii H152T (ARBG01000149) (98.50%). The 16S rRNA sequence of strain T808 had about 40 more bases in the V1-V2 region than the seven validly published species of Pararhizobium, while the NCBI database showed that T808 had 98.54–99.15% similarity with Uncultured bacterium clone barrow_ FF_26 (JX668750.1), Rhizobium sp.Ia8 (KF444807), and Rhizobiaceae bacterium strain FW305-C-27(MN067584), all of which were uncultured bacteria without lacking the 40 bases in V1-V2 region (Fig. 4). Based on the above analysis, T808 might be a potentially new species of Pararhizobium. Neorhizobium and Pararhizobium were important non-symbiotic species of rhizobia with poor nodulation or nitrogen fixation genes, which have the important microbial niche value (Shen et al. 2018; Soenens et al. 2019).
Table IV.
The sequence analyses based on almost full-length of the 16S rRNA gene of six potential new species.
| Strain number | Name of strain having the highest 16S rRNA gene similarity | Separation medium | The highest similarity (%) | Separation temperature (°C) |
|---|---|---|---|---|
| T96 | Agromyces binzhouensis OAct353T | 98.62 | M5 | 25 |
| T105 | Nocardioides caeni MN8T | 98.01 | M5 | 25 |
| T274 | Phyllobacterium zundukense Tri-48T | 98.57 | M8 | 25 |
| T786 | Neorhizobium vignae CCBAU 05176T | 98.70 | R | 4 |
| T808 | Pararhizobium herbae CCBAU 83011T | 98.79 | M5 | 4 |
| T830 | Hymenobacter humi DG31AT | 98.60 | F1 | 4 |
Fig. 3.
Phylogenetic tree based on the16S rRNA gene sequences of new candidates and related species.
Fig. 4.
The Clustal X analysis of strain T808.
Three potential new species were isolated from media M5, and one species was isolated from media M8, R, and F1, respectively. Half of six potential new species were cultured at 4°C, while others were cultured at 25°C. The culture medium and temperature have a significant influence on the separation of new species. All six potential new species will be further identified with a polyphasic approach (including chemotaxonomic properties, DNA-DNA hybridization analysis) to determine their taxonomic positions.
Discussion
Together with the incubation of the highland barley cultivation soil sample using fifteen kinds of media at 25°C and 4°C, a total of 830 individual strains were purified. The 16S rRNA gene sequence analysis results are consistent with a previous report, in which Actinobacteria, Proteobacteria, Firmicutes were found to be dominant phyla in the arctic-alpine area, especially in the Qinghai-Tibet plateau (Jiang et al. 2006; Kumar et al. 2016a; Tang et al. 2016). The predominant genus was Streptomyces, followed by Bacillus, Micromonospora, and Microbacterium. The most diverse isolates belonged to high the G+C Gram-positive group; in particular, the Streptomyces genus is a dominant genus in the high G+C Gram-positive group. The bacteria in arctic-alpine areas are mainly the spore producing, stress-resistant, and thick cell walls microorganisms (Zhang et al. 2010b; Rao et al. 2016).
The Actinobacteria are widely dispersed throughout the highland barley cultivation soil, while few studies are on it. The bacteria in highland barley cultivation soil in Lhasa analyzed by high-throughput sequencing technology showed that the main actinomycetes were Gaiiella, Arthrobacter, and Nocardioides (Liu et al. 2020), which was quite different from our study using the culturable technique. In other previous reports, the main genus in the highland barley cultivation soil was Streptomyces, Arthrobacter,and Nocardioides.Most Actinomycetes had a wide spectrum of inhibitory activity against pathogenic bacteria, highly IAA production, and phosphate solubilization, which were in similarity with our study (Qi et al. 2017; Yin et al. 2017; Gao et al. 2019). As the most well-known genus in Actinobacteria, Streptomyces contains 960 species (http://www.bacterio.net/streptomyces) and 4227 genome assemblies available (https://www.ncbi.nlm.nih.gov/genome/streptomyces) at the time of writing. Members of the genus Streptomyces are well known as the primary sources of antibiotics with diverse biological activities and chemical structures (Jones and Elliot 2017; Li et al. 2018). In this study, 17 species of Streptomyces were found in the highland barley cultivation soil, the larger numbers of dominant species of Streptomyces were Streptomyces flavovirens, Streptomyces xanthophaeus and Streptomyces canus, which were mainly isolated at 25°C. The Qamdo region’s temperature is between 20°C and 28°C from June to September, which is also a critical growth period for highland barley. We believe that these Streptomyces that can produce many biological activities have an essential role in the growth of highland barley in this period. The other dominant isolates in highland barley cultivation soil were Arthrobacter humicola and Bacillus siamensis, which are important plant growth-promoting rhizobacteria (PGPR) (Bai et al. 2015).
Meanwhile, Arthrobacter humicola was mainly isolated at 4°C, producing cold lipase and biopolymeric flocculant (Agunbiade et al. 2017). The low-temperature adaptation and ecological function of A. humicola in highland barley cultivation soil need to be studied in-depth. Some rare Actinobacteria were also isolated from the soil sample, for example, Leifsonia, Longispora, Nocardia, Nocardioides, Terrabacter, Umezawaea, and Kribbella. Rare Actinobacteria are also important sources in discovering novel antibiotics and have been seldom studied (Cai et al. 2018; Bundale et al. 2019).
In summary, this study has demonstrated a rich diversity of bacteria (especially Actinobacteria) and some undiscovered bacteria species in the highland barley cultivation soil of Qinghai-Tibet plateau it suggests that these strains might represent a valuable source of new taxa for further microbial development and utilization. Additionally, this study indicates that cultivating Actinobacteria in highland barley cultivation soil of Qinghai-Tibet plateau could be interesting for further study.
Acknowledgments
National Natural Science Foundation of China (32060025), the Special Financial Item of Tibet Autonomous Region (XZNKY-2018-C-026) and the “Double First-Class” construction project of Hunan Agricultural University (SYL201802002) funded this work.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication
Literature
- Agunbiade MO, Van Heerden E, Pohl CH, Ashafa AT. Flocculating performance of a bioflocculant produced by Arthrobacter humicola in sewage waste water treatment. BMC Biotechnol. 2017. Dec;17(1):51. 10.1186/s12896-017-0375-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015. Dec;528(7582):364–369. 10.1038/nature16192 [DOI] [PubMed] [Google Scholar]
- Bailly A, Weisskopf L. The modulating effect of bacterial volatiles on plant growth: current knowledge and future challenges. Plant Signal Behav. 2012. Jan;7(1):79–85. 10.4161/psb.7.1.18418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier PLE. Toward understanding, managing, and protecting microbial ecosystems. Front Microbiol. 2011;2:80. 10.3389/fmicb.2011.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundale S, Singh J, Begde D, Nashikkar N, Upadhyay A. Rare actinobacteria: a potential source of bioactive polyketides and peptides. World J Microbiol Biotechnol. 2019. Jun;35(6):92. 10.1007/s11274-019-2668-z [DOI] [PubMed] [Google Scholar]
- Cai Y, Tao WZ, Ma YJ, Cheng J, Zhang MY, Zhang YX. Leifsonia flava sp. nov., a novel actinobacterium isolated from the rhizosphere of Aquilegia viridiflora. J Microbiol. 2018. Aug;56(8):549–555. 10.1007/s12275-018-8061-z [DOI] [PubMed] [Google Scholar]
- Collins T, Margesin R. Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl Microbiol Biotechnol. 2019. Apr;103(7):2857–2871. 10.1007/s00253-019-09659-5 [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. A global atlas of the dominant bacteria found in soil. Science. 2018. Jan 19;359(6373):320–325. 10.1126/science.aap9516 [DOI] [PubMed] [Google Scholar]
- Deng N, Zheng B, Li T, Liu RH. Assessment of the phenolic profiles, hypoglycemic activity, and molecular mechanism of different highland barley (Hordeum vulgare L.) varieties. Int J Mol Sci. 2020. Feb 11;21(4):1175. 10.3390/ijms21041175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe RR, Roesch LFW, Riva A, Triplett EW. Distantly sampled soils carry few species in common. ISME J. 2008. Sep;2(9):901–910. 10.1038/ismej.2008.55 [DOI] [PubMed] [Google Scholar]
- Furlong MA, Singleton DR, Coleman DC, Whitman WB. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl Environ Microbiol. 2002. Mar;68(3):1265–1279. 10.1128/AEM.68.3.1265-1279.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Gu YF, Nyima T, Liu GY, Liu T, Liu Y, Pubu G. Analysis of the genetic diversity and promoting functions of the culturable Actinomycetes in the rhizosphere of highland barley in Tibet. Sichuan Nongye Daxue Xuebao. 2019;37(6):777–784. [Google Scholar]
- He Q, Wang X, He L, Yang L, Wang S, Bi Y. Alternative respiration pathway is involved in the response of highland barley to salt stress. Plant Cell Rep. 2019. Mar;38(3):295–309. 10.1007/s00299-018-2366-6 [DOI] [PubMed] [Google Scholar]
- Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol. 2006. Jun;72(6):3832–3845. 10.1128/AEM.02869-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Elliot MA. Streptomyces exploration: competition, volatile communication and new bacterial behaviours. Trends Microbiol. 2017. Jul;25(7):522–531. 10.1016/j.tim.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014. Feb 01;64(Pt_2):346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Kuklinsky-Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol. 2004. Dec;6(12):1244–1251. 10.1111/j.1462-2920.2004.00658.x [DOI] [PubMed] [Google Scholar]
- Kumar M, Männistö MK, van Elsas JD, Nissinen RM. Plants impact structure and function of bacterial communities in Arctic soils. Plant Soil. 2016a. Feb;399(1–2):319–332. 10.1007/s11104-015-2702-3 [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016b. Jul;33(7):1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurm V, van der Putten WH, Hol WHG. Cultivation-success of rare soil bacteria is not influenced by incubation time and growth medium. PLoS One. 2019. Jan 10;14(1):e0210073. 10.1371/journal.pone.0210073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Liu S, Lu Q, Zheng H, Osterman IA, Lukyanov DA, Sergiev PV, Dontsova OA, Liu S, Ye J, et al. Studies on antibacterial activity and diversity of cultivable actinobacteria isolated from mangrove soil in Futian and Maoweihai of China. Evid Based Complement Alternat Med. 2019. Jun 09;2019:1–11. 10.1155/2019/3476567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wei K, Zheng G, Liu X, Chen S, Jiang W, Lu Y. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol. 2018. Jul 06;84(18):e00827-18. 10.1128/AEM.00827-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Narsing Rao MP, Dong ZY, Wang JP, Chen Z, Liu B, Li WJ. Two novel alkaliphiles, Bacillus alkalisoli sp. nov., and Bacillus solitudinis sp. nov., isolated from saline-alkali soil. Extremophiles. 2019. Nov;23(6):759–764. 10.1007/s00792-019-01127-2 [DOI] [PubMed] [Google Scholar]
- Liu QH, Pan H, Da WZM, Tian Y, Liu HH, Wang C, Lu XY, Bai JP. [High-throughput analysis of bacterial diversity in highland barley cultivation soil in Lhasa] (in Chinese). J Biol. 2020;37(2):46–51. [Google Scholar]
- Malard LA, Anwar MZ, Jacobsen CS, Pearce DA. Biogeographical patterns in soil bacterial communities across the Arctic region. FEMS Microbiol Ecol. 2019. Sep 01;95(9):fiz128. 10.1093/femsec/fiz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R, Jud M, Tscherko D, Schinner F. Microbial communities and activities in alpine and subalpine soils. FEMS Microbiol Ecol. 2009. Feb;67(2):208–218. 10.1111/j.1574-6941.2008.00620.x [DOI] [PubMed] [Google Scholar]
- Margesin R. Psychrophilic microorganisms in alpine soils. In: Lütz C, editor. Plants in Alpine regions. Vienna (Austria): Springer; 2012. p. 187–198. [Google Scholar]
- Mousavi SA, Österman J, Wahlberg N, Nesme X, Lavire C, Vial L, Paulin L, de Lajudie P, Lindström K. Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol. 2014. May;37(3):208–215. 10.1016/j.syapm.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindström K. Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol. 2015. Mar;38(2):84–90. 10.1016/j.syapm.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Qi SS, Zhou LH, Hu JP, Liu M, Zhao H, Xiong Y. [Isolation, identification and diversity of soil bacteria in multiple regions from Tibetan Plateau] (in Chinese). Xi Nan Nong Ye Xue Bao. 2017; 30(7):1629–1635. [Google Scholar]
- Rao S, Chan OW, Lacap-Bugler DC, Pointing SB. Radiation-tolerant bacteria isolated from high altitude soil in Tibet. Indian J Microbiol. 2016. Dec;56(4):508–512. 10.1007/s12088-016-0604-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Řeháková K, Chroňáková A, Krištůfek V, Kuchtová B, Čapková K, Scharfen J, Čapek P, Doležal J. Bacterial community of cushion plant Thylacospermum ceaspitosum on elevational gradient in the Himalayan cold desert. Front Microbiol. 2015. Apr 16;6:304. 10.3389/fmicb.2015.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Li Y, Zhao Z, Han YF, Zhang WW, Yu XY, Zhang CY, Sun C, Wu M. Polyphasic taxonomic characterisation of a novel strain as Pararhizobium haloflavum sp. nov., isolated from soil samples near a sewage treatment tank. Antonie van Leeuwenhoek. 2018. Apr;111(4):485–491. 10.1007/s10482-017-0969-5 [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhang H, Cheng L, Wang L, Qian H, Qi X. In vitro and in vivo antioxidant activity of polyphenols extracted from black highland barley. Food Chem. 2016. Mar;194:1003–1012. 10.1016/j.foodchem.2015.08.083 [DOI] [PubMed] [Google Scholar]
- Soenens A, Gomila M, Imperial J. Neorhizobium tomejilense sp. nov., first non-symbiotic Neorhizobium species isolated from a dry-land agricultural soil in southern Spain. Syst Appl Microbiol. 2019. Mar;42(2):128–134. 10.1016/j.syapm.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Tang JY, Ma J, Li XD, Li YH. Illumina sequencing-based community analysis of bacteria associated with different bryophytes collected from Tibet, China. BMC Microbiol. 2016. Dec;16(1):276. 10.1186/s12866-016-0892-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin MY, He JQ, Zhang GJ. [Biological activity and diversity of psychrophilic Actinomycetes in Tibet] (in Chinese). J Northwest Agric Forest Univer (Nat Sci Ed). 2017;45(6):221–234. [Google Scholar]
- Yu Y, Guo Z, Wu H, Kahmann JA, Oldfield F. Spatial changes in soil organic carbon density and storage of cultivated soils in China from 1980 to 2000. Global Biogeochem Cy. 2009;23: GB2021. 10.1029/2008GB003428 [DOI] [Google Scholar]
- Yuan Y, Si G, Wang J, Luo T, Zhang G. Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol Ecol. 2014. Jan;87(1):121–132. 10.1111/1574-6941.12197 [DOI] [PubMed] [Google Scholar]
- Zhang G, Niu F, Ma X, Liu W, Dong M, Feng H, An L, Cheng G. Phylogenetic diversity of bacteria isolates from the Qinghai-Tibet Plateau permafrost region. Can J Microbiol. 2007. Aug;53(8): 1000–1010. 10.1139/W07-031 [DOI] [PubMed] [Google Scholar]
- Zhang K, Yang J, Qiao Z, Cao X, Luo Q, Zhao J, Wang F, Zhang W. Assessment of β-glucans, phenols, flavor and volatile profiles of hulless barley wine originating from highland areas of China. Food Chem. 2019. Sep;293:32–40. 10.1016/j.foodchem.2019.04.053 [DOI] [PubMed] [Google Scholar]
- Zhang S, Yang G, Wang Y, Hou S. Abundance and community of snow bacteria from three glaciers in the Tibetan Plateau. J Environ Sci (China). 2010a. Sep;22(9):1418–1424. 10.1016/S1001-0742(09)60269-2 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dong S, Gao Q, Liu S, Zhou H, Ganjurjav H, Wang X. Climate change and human activities altered the diversity and composition of soil microbial community in alpine grasslands of the Qinghai-Tibetan Plateau. Sci Total Environ. 2016. Aug;562:353–363. 10.1016/j.scitotenv.2016.03.221 [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Liu HY, Chen J, Yuan LJ, Sun W, Zhang LX, Zhang YQ, Yu LY, Li WJ. Diversity of culturable actinobacteria from Qinghai-Tibet plateau, China. Antonie van Leeuwenhoek. 2010b. Aug;98(2): 213–223. 10.1007/s10482-010-9434-4 [DOI] [PubMed] [Google Scholar]
- Zhao NN, Guggenberger G, Shibistova O, Thao DT, Shi WJ, Li XG. Aspect-vegetation complex effects on biochemical characteristics and decomposability of soil organic carbon on the eastern Qinghai-Tibetan Plateau. Plant Soil. 2014. Nov;384(1–2):289–301. 10.1007/s11104-014-2210-x [DOI] [Google Scholar]