Abstract
Ticks exist across diverse environments and transmit numerous pathogens. Due to their long and unique life cycles, these arthropods likely evolved robust epigenetic mechanisms that provide sustainable responses and buffers against extreme environmental conditions. Herein, we highlight how the study of the epigenetic basis of tick biology and vectorial capacity will enrich our knowledge of tick-borne infections.
Origin and Geographic Expansion of Ticks and Tick-Borne Diseases
Molecular systematics studies on the evolution of ticks suggest that these ancient ectoparasites appeared as early as the Carboniferous period of the Paleozoic era, which began 354 million years ago [1], with later dispersal during the Tertiary period [2]. Ticks parasitize a wide range of vertebrates, thrive in variable settings from the Arctic to the tropical regions, and transmit a greater variety of pathogens than any other group of arthropod vectors. In past decades, both the incidence of tick-borne diseases and the number of recognized tick-borne pathogens have substantially increased in the US [3] and in the entire world.
Sequenced Tick Genomes
A group of scientists deciphered the ~2.1 Gbp nuclear genome of the Lyme disease-transmitting tick, Ixodes scapularis [4], which is a prolific tick vector. Notably, this study found that about 60% of tick genes have arthropod orthologs, while roughly 22% are paralogs, resulting from two gene duplication events that occurred within the last 40 million years. However, due to its large size and high repeat content, only about 57% of the I. scapularis genome was mapped. Later, another I. scapularis genome assembly from the Ixodes ISE6 cell line was published, although the estimated genome size differs in these studies [5]. A recent report described the high-quality genomes of six ixodid tick species that were collected from different parts of China, including details of their sequencing, assembly, and comparative analyses [6]. The ecological and geographic factors, as shown in the study, are the prime determinants for the genetic diversity and pathogen composition in the different tick species; presumably, epigenetic factors also play a similar influential role. Therefore, these studies further underscore the unmet need for high-quality genome sequencing data in additional tick species, such as I. scapularis or Ixodes ricinus, which feature wide distribution across North America and Europe, respectively, and transmit the most diverse human and animal pathogens. Cutting-edge technologies, including long-read DNA/RNA sequencing, single cell RNA sequencing, chromosome conformation capture (3C) sequencing, and single molecule imaging, can be used to construct an accurate and complete tick genome atlas.
Epigenetic Regulation of Gene Expression
The precise regulation of gene expression drives cell fate decision, lineage commitment, body plan establishment, and organ formation, which are fundamental to the proper growth and development of an organism. Spatiotemporal transcription regulation is controlled by the cell’s ‘epigenetic landscape’, a term coined by Conrad H. Waddington in 1942. Epigenetics is now broadly defined as the study of the molecules and mechanisms that can generate gene activity states that differ from a given DNA sequence. Some of the widely studied epigenetic mechanisms include histone modifications, DNA methylation, and noncoding RNAs. In Drosophila, genetic screens for mutations that alter cell fate first identified the Polycomb (PcG) and Trithorax (TrxG) epigenetic regulators. Many PcG and TrxG genes encode components of histone-modifying enzyme complexes [e.g., histone acetyl transferases (HATs), histone methyl transferases (HMTs), and histone demethylases (HDMs)] and chromatin remodeling complexes (Table 1). In addition to their roles in development, these important genes regulate many aspects of Drosophila biology, including immune responses. Another major epigenetic modification is the methylation of cytosine residues, as catalyzed by DNA methyltransferases (DNMTs), typically at CpG repeats. Although there is a lack of substantial evidence of CpG methylation in Drosophila, the machinery is present in specific arthropod lineages, all invertebrate chordates, and vertebrates. Finally, the roles of various noncoding RNAs (ncRNAs) in the epigenetic regulation of gene expression, organism development, and immunity are now well-established (Table 1). For example, a particular ncRNA is induced in Drosophila upon bacterial infection, playing a role in fly immunity.
Table 1.
Well-Studied Examples of Epigenetic Regulators
| Types of epigenetic regulators | General functions | Refs |
|---|---|---|
| Histone methyltransferases (HMTs, e.g., IsDot1L, I. scapularis) | Transfer methyl groups from S-adenosyl methionine (SAM) to conserved lysine and arginine residues of histone proteins | [11] |
| Histone demethylases (HDMs, e.g., IsLSD1A, I. scapularis) | Catalyze the removal of methyl groups from histones | [11] |
| Histone acetyltransferases (HATs, e.g., Isp300/CBP, I. scapularis) | Catalyze the transfer of acetyl groups from acetyl-CoA to conserved lysine residues of histone proteins | [11] |
| Histone deacetylases (HDACs, e.g., IsHDAC1, I. scapularis) | Hydrolyze an acetyl-lysine residue of a histone to yield a deacetylated histone | [11] |
| DNA methyltransferases (e.g., DNMT1, I. ricinus) | Add methyl groups to DNA molecules; use SAM as the methyl donor | [7] |
| DNA demethylases (e.g., TET, human) | Remove or modify the methyl group from DNA | |
| Noncoding RNAs (e.g., microRNAs, Haemaphysalis longicornis) | Regulate gene expression at the transcriptional and post-transcriptional levels | [6] |
Impact on Tick Biology
Ticks thrive in a complex enzootic life cycle spanning several years, consisting of short on-host phases of blood meal engorgement and long off-host periods during the intermolt or molting stages, possibly assisted by epigenetic reprogramming events (Figure 1). These arthropods have unparalleled abilities to ingest and store large blood meals (often 100-fold greater than their own body volume) during their limited yet voracious feeding cycles, aided by rapid bursts of tissue development. Additionally, ticks survive through prolonged periods of environmental extremities and metabolic quiescence and maintain complex reproductive and postembryonic developmental processes. Furthermore, while subadult ticks parasitize a diverse set of hosts, adults display a specific host preference. These distinctive characteristics suggest the evolution of an underlying genetic and epigenetic plasticity that enables them to thrive in new and disparate environments. A recent study has reported the existence of DNMTs in I. ricinus ticks [7]. In addition, the HMT Dot1L is shown to be involved in the larval molting and nymphal feeding of the soft tick, Ornithodoros moubata [8]. Some organs, particularly the salivary gland and gut, play crucial roles in the tick life cycle and in the trans-stadial maintenance and transmission of tick-borne pathogens. Like Drosophila and other organisms, ticks maintain a layer of intestinal stem cells (ISC) that are important for organ homeostasis and function, which are likely to be regulated epigenetically. Some of the known signaling pathways (such as JAK/STAT, IMD-like, Wnt, Hippo, Notch, and EGFR) and genetic/epigenetic components (e.g., escargot, nubbin/pdm1, cohesin, and PcGs/TrxGs) that regulate ISC function are also present in ticks. Further investigation into the epigenetic determinants of tick organs and important physiological processes, such as gut development and blood meal digestion, is important to the understanding of tick biology. Additional studies are required to explore diverse biotic factors (e.g., host-specific factors or microbiota), abiotic components (such as light, temperature, and moisture), and their impacts on epialleles and epigenetic homeostasis, which together facilitate the tick’s overall development and fitness [9,10]. It also remains unknown as to how the resident microbiota and tick-borne pathogens exert their epigenetic influences on tick biology. Intracellular tick-borne pathogens, like Anaplasma phagocytophilum, are known to manipulate the transcriptional programs of their host cells via epigenetic mechanisms, such as the preferential modulation of certain histone-modifying enzymes, possibly impacting tick biology [11].
Figure 1. Epigenetic Reprogramming during Ixodes scapularis Enzootic Life Cycle.
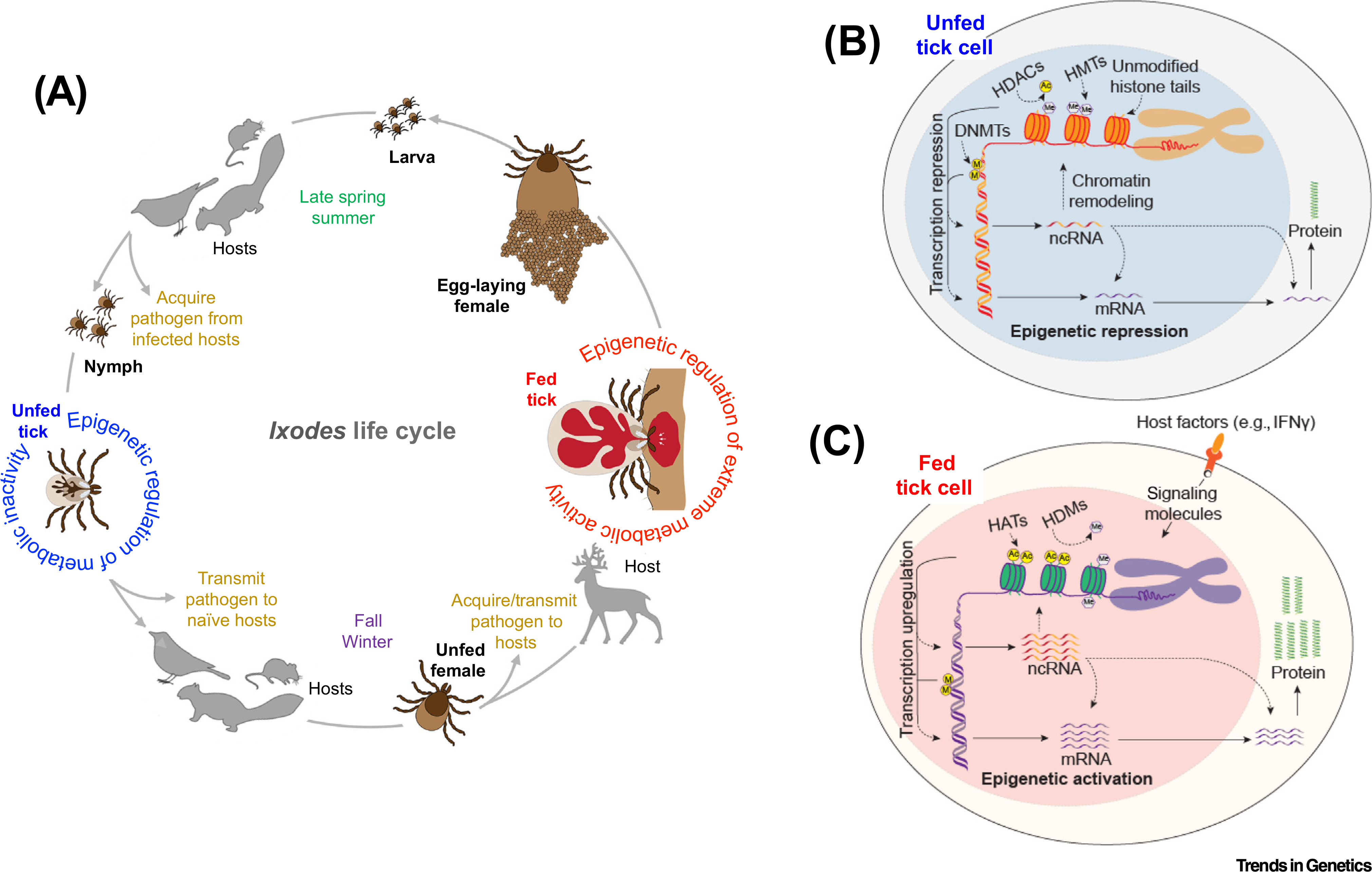
(A) Enzootic life cycle of Ixodes ticks. After hatching in late spring and summer, naïve six-legged larvae seek and feed on a host, typically a bird, small rodent, or other mammal, and molt to eight-legged nymphs. Questing nymphs ingest blood from the same set of hosts that is parasitized by larvae and then molt to the adult stage. The newly emerged adult ticks search for a preferred host, typically deer, on which they mate and engorge with a final blood meal. After detachment from the host, fully engorged and fertilized females oviposit an egg mass (containing hundreds to thousands of eggs) and die. While taking a blood meal from an infected host, ticks can acquire pathogens, like the agents of Lyme disease or anaplasmosis, maintain them trans-stadially, and subsequently transmit them to a new host during the next blood meal. (B) Possible epigenetic regulation in unfed and metabolically inactive ticks. The largely quiescent chromatin in unfed tick cells is epigenetically transcriptionally repressed and is refractory to transcriptional activation. (C) Hypothetical epigenetic regulation in metabolically active and engorged ticks. During feeding, signaling molecules from the host propel a massive epigenetic reprogramming within the tick cells, via chromatin regulators, to accommodate and store an enormous blood meal. The expression of these chromatin regulators/epigenetic factors is likely to be dynamically regulated during the feeding process. Moreover, transcriptional changes at specific gene loci are generally accompanied by local and global alterations in chromatin structure. Ticks inherit epigenetic memory throughout their life cycles through mitotic divisions (from larvae to adults) and possibly through meiotic divisions (from adults to fertilized eggs). This epigenetic memory can be further reinforced by tick-borne pathogens. Abbreviations: DNMT, DNA methyl transferase; HAT, histone acetyl transferase; HDAC, histone deacetylase; HDM, histone demethylase; HMT, histone methyl transferase; IFNγ, interferon γ; ncRNA, noncoding RNA.
Tick Vectorial Capacity
Arthropod vectorial capacity is a complex process, as it is governed by a multifaceted suite of genetic mechanisms. First, interaction with a new pathogen generally induces antimicrobial molecular activity, thereby establishing a strong vectorial resistance. Second, the novel metabolite pool resulting from the initial (and potentially recurrent) pathogenic infection may impose permanent and unique changes in the vector’s existing epigenetic landscape, thus initiating vectorial tolerance. Finally, during future infections, the newfound vectorial tolerance could usher a mutually beneficial relationship between the vector and pathogen. Unlike other insect disease vectors, practically nothing is known about the tick’s genetic/epigenetic mechanisms of vector competence. We identified more than 234 immunity-related genes in I. scapularis [4,12], some of which may influence vectorial capacity. Studies on the interactions of I. scapularis with pathogens have identified a handful of genes and signaling pathways, which is likely the tip of the iceberg for a vastly unidentified network of ligand–receptor interactions, which may impact epigenetic reprogramming and cell fate decision [10,13]. Furthermore, the discovery of a cross-species interferon signaling event shows how the epigenetic landscape of a host blood meal, after being altered by infection with Borrelia burgdorferi, can initiate a gene expression cascade, yielding microbicidal immune responses in ticks [14]. Most pathogens are acquired by ticks during blood meal engorgement on infected hosts; nonetheless, cyclical communication with pathogens, in addition to transovarial transmission of the ‘epigenetic memory’ that is acquired during different pathogenic conditions throughout the life cycle, can provide better fitness to tick progeny in a challenging environment [15]. This process must be understood in order to better control ticks and the diseases that they transmit.
Concluding Remarks
Tick-borne pathogens likely modulate the epigenetic landscape of ticks at multiple levels by affecting certain processes, including the signaling pathways leading to epigenetic changes, such as histone modifications or DNA methylation patterns. The biological significance of such epigenetic components and mechanisms warrants further exploration, as they may contribute greatly to the remarkable plasticity of tick vectors. These ancient ectoparasites’ unparalleled tolerance to a wide range of microbial pathogens and environmental extremities makes them increasingly more dangerous to human and animal health. This article is intended to act as a primer for novel ideas and future research on the epigenetic regulation of tick biology and vectorial capacity, which can help us to combat tick-borne infections.
Acknowledgments
Support for studies involving tick-borne pathogens was received from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (Award Numbers R01AI080615, R01AI116620, and P01AI138949 to U.P.).
References
- 1.Beati L and Klompen H (2019) Phylogeography of ticks (Acari: Ixodida). Annu. Rev. Entomol 64, 379–397 [DOI] [PubMed] [Google Scholar]
- 2.de la Fuente J (2003) The fossil record and the origin of ticks (Acari: Parasitiformes: Ixodida). Exp. Appl. Acarol 29, 331–344 [DOI] [PubMed] [Google Scholar]
- 3.Sonenshine DE (2018) Range expansion of tick disease vectors in North America: implications for spread of tick-borne disease. Int. J. Environ. Res. Public Health 15, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulia-Nuss M et al. (2016) Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun 7, 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JR et al. (2018) A draft genome sequence for the Ixodes scapularis cell line, ISE6. F1000Res. 7, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia N et al. (2020) Large-scale comparative analyses of tick genomes elucidate their genetic diversity and vector capacities. Cell 182, 1328–1340 [DOI] [PubMed] [Google Scholar]
- 7.Kotsarenko K et al. (2020) Newly identified DNA methyltransferases of Ixodes ricinus ticks. Ticks Tick Borne Dis 11, 101348. [DOI] [PubMed] [Google Scholar]
- 8.Gobl J et al. (2020) Histone methyltransferase DOT1L is involved in larval molting and second stage nymphal feeding in Ornithodoros Moubata. Vaccines (Basel) 8, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L et al. (2015) Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58, 216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Fuente J et al. (2017) Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front. Cell Infect. Microbiol 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabezas-Cruz A et al. (2016) Anaplasma phagocytophilum increases the levels of histone modifying enzymes to inhibit cell apoptosis and facilitate pathogen infection in the tick vector Ixodes scapularis. Epigenetics 11, 303–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AA and Pal U (2014) Immunity-related genes in Ixodes scapularis–perspectives from genome information. Front. Cell. Infect. Microbiol 4, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurokawa C et al. (2020) Interactions between Borrelia burgdorferi and ticks. Nat. Rev. Microbiol 18, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith AA et al. (2016) Cross-species interferon signaling boosts microbicidal activity within the tick vector. Cell Host Microbe 20, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De S and Kassis JA (2017) Passing epigenetic silence to the next generation. Science 356, 28–29 [DOI] [PubMed] [Google Scholar]


