Abstract
In this study, the complete mitochondrial (mt) genome of the New Zealand parasitic blowfly Lucilia sericata (green bottle blowfly) field strain NZ_LucSer_NP was generated using next-generation sequencing technology. The length of complete the mt genome is 15,938 bp, with 39.4% A, 13.0% C, 9.3% G, and 38.2% T nucleotide distribution. The complete mt genome consists of 13 protein-coding genes (PCGs), two ribosomal RNAs, 22 transfer RNAs, and a 1124 bp non-coding region, similar to most metazoan mt genomes. Phylogenetic analysis showed that L. sericata NZ_LucSer_NP forms a monophyletic cluster with the remaining six Lucilia species and the Calliphoridae are polyphyletic. This study provides the first complete mt genome sequence for a L. sericata blowfly species derived from New Zealand to facilitate species identification and phylogenetic analysis.
Keywords: Diptera, Calliphoridae, Luciliinae, complete mitochondrial genome, Lucilia sericata
Members of the Calliphoridae (blowflies) are important for medical and veterinary management. The large and highly variable mitochondrial (mt) genomes of blowflies are ideal sources of molecular markers suitable for studying population genetic structures and evolution. Lucilia sericata (Meigen) NZ_LucSer_NP was selected for genome sequencing as a representative of an NZ field strain of L. sericata. The L. sericata NZ_LucSer_NP mitogenome resource will facilitate investigation of phylogenomic relationships of both species- and strain-level diversity among Calliphoridae. In addition, future comparative studies exploring the features of other available representative mitogenomes derived from Australia may shed light on the evolutionary provenance of L. sericata.
The L. sericata specimen was collected from the Palmerston North area (40°21.3′S, 175°36.7′E), and is stored and available upon request from AgResearch Ltd., Grasslands Research Center (accession number: NPY120886). High molecular weight genomic DNA was isolated from entire L. sericata adult males using a modified phenol:chloroform protocol explained in our previous articles (Palevich, Kelly, Ganesh, et al. 2019; Palevich, Kelly, Leahy, et al. 2019; Palevich et al. 2019b). Genomic DNA was prepared for whole-genome sequencing using an Illumina TruSeq Nano library preparation kit (Illumina, Inc., San Diego, CA) according to the manufacturer’s instructions. The Illumina NovaSeq™ 6000 (PE150, Novogene, Beijing, China) platform was used to amplify the entire mt genome sequence (BioProject ID: PRJNA667961, GenBank accession number: MW266393). The mt genome was assembled and annotated using the NOVOPlasty pipeline version 3.1 with default parameters (Dierckxsens et al. 2016), as previously described (Palevich et al. 2019a; Palevich, Maclean, Mitreva, et al. 2019; Palevich et al. 2020, Palevich and Maclean 2021).
The mitogenome (15,938 bp) is standard in size and comparable to other L. sericata strains and isolates (Nelson et al. 2012). For example: genes are transcribed in both directions, it contains 13 protein-coding genes (PCGs), two rRNAs, 22 tRNAs, and an AT-rich region (1124 bp). Among these 23 are located on the heavy strand with the remaining 14 genes located on the light strand, consistent with other Lucilia species AJ422212, JX913758, and JX913744 (Stevens J et al. 2008; Nelson et al. 2012) and isolates. The studied genome has a high T content (38.2%) and a low G content (9.3%), resulting in a very strong A + T bias (77.7%) and in particular the AT-rich region (90.1%). Gene sizes and all common organization features are relatively conserved among the blowfly and fly mitogenomes (usually 14.5–19.6 kb) (Stevens J and Wall 2001; Stevens JR 2003; Stevens JR and Wallman 2006).
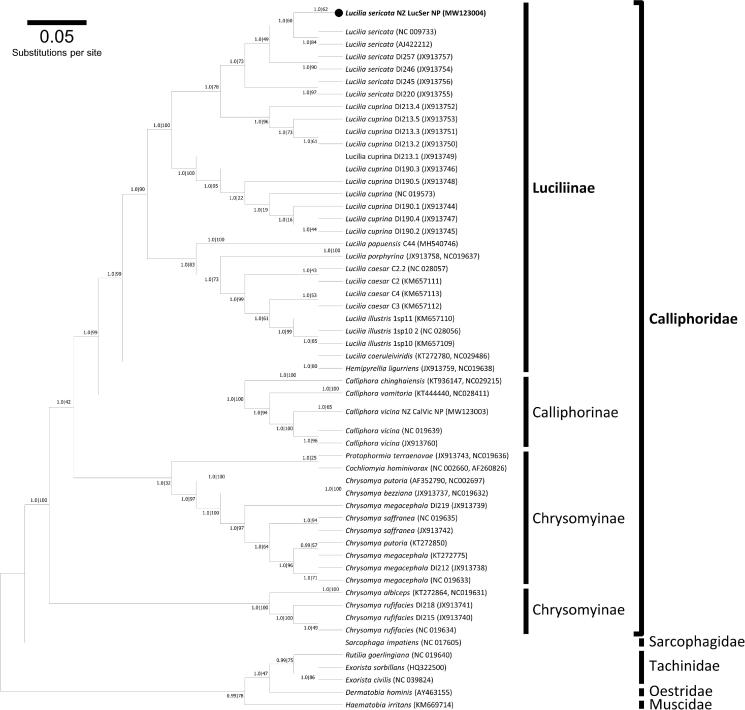
The phylogenetic position of L. sericata NZ_LucSer_NP was estimated using maximum-likelihood, implemented in RAxML version 8.2.11 (1000 bootstrap replications) (Stamatakis 2014), and the Bayesian inference (BI), implemented in MrBayes version 3.2.6 (default settings, four MCMC chains) (Huelsenbeck and Ronquist 2001) approaches. Complete mitogenome sequences of 27 available blowfly species were retrieved from GenBank and phylogenetic analyses performed on the concatenated mt PCGs (Figure 1). Within the Calliphoridae, L. sericata NZ_LucSer_NP formed a monophyletic cluster with the remaining Luciliinae species, there was also strong support for the sister-grouping of Calliphorinae with Luciliinae. Overall, the dendrogram topology is highly congruent with the previous results of Nelson et al. (2012). In the pursuit of improving the phylogenetic resolution within the phylum Calliphoridae, future efforts should focus on the availability of more complete mitogenomes across all blowfly species, and especially for different strains/isolates.
Figure 1.
A summary of the molecular phylogeny of the Calliphoridae complete mitochondrial genomes. The evolutionary relationship of Lucilia sericata field strain NZ_LucSer_NP (black circle) was compared to the complete mitochondrial genomes of 67 blowfly species or isolates retrieved from GenBank (accession numbers in parentheses) and nucleotide sequences of all protein-coding genes were used for analysis. Phylogenetic analysis was conducted using the Bayesian approach implemented in MrBayes version 3.2.6 (Huelsenbeck and Ronquist 2001) and maximum likelihood (ML) using RAxML version 8.2.11 (Stamatakis 2014). The mtREV with Freqs. (+F) model was used for amino acid substitution and four independent runs were performed for 10 million generations and sampled every 1000 generations. For reconstruction, the first 25% of the sample was discarded as burnin and visualized using Geneious Prime (Kearse et al. 2012). Nodal support is given: Bayes posterior probabilities RAxML bootstrap percentage. The phylogram provided is presented to scale (scale bar = 0.05 estimated number of substitutions per site) with the species Haematobia irritans from the family Muscidae used as the outgroup.
Funding Statement
This research was supported by the Agricultural and Marketing Research and Development Trust (AGMARDT) Postdoctoral Fellowships Programme [No. P17001] and the AgResearch Ltd Strategic Science Investment Fund (SSIF) [No. PRJ0098715] of New Zealand.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW266393. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA667961, SRR13038366, and SAMN16745530, respectively.
References
- Dierckxsens N, Mardulyn P, Smits G.. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL.. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142. [DOI] [PubMed] [Google Scholar]
- Palevich N, Kelly WJ, Ganesh S, Rakonjac J, Attwood GT.. 2019. Butyrivibrio hungatei MB2003 competes effectively for soluble sugars released by Butyrivibrio proteoclasticus B316T during growth on xylan or pectin. Appl Environ Microbiol. 85(3):e02056–02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N, Kelly WJ, Leahy SC, Denman S, Altermann E, Rakonjac J, Attwood GT.. 2019. Comparative genomics of rumen Butyrivibrio spp. uncovers a continuum of polysaccharide-degrading capabilities. Appl Environ Microbiol. 86(1):e01993–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N, Maclean PH. 2021. Sequencing and reconstructing helminth mitochondrial genomes directly from genomic next-generation sequencing data. In Parasite Genomics Protocols. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Palevich N, Maclean P, Baten A, Scott R, Leathwick DM.. 2019a. The complete mitochondrial genome of the New Zealand parasitic roundworm Haemonchus contortus (Trichostrongyloidea: Haemonchidae) field strain NZ_Hco_NP. Mitochondrial DNA Part B. 4(2):2208–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N, Maclean PH, Baten A, Scott RW, Leathwick DM.. 2019b. The genome sequence of the anthelmintic-susceptible New Zealand Haemonchus contortus. Genome Biol Evol. 11(7):1965–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N, Maclean PH, Choi Y-J, Mitreva M.. 2020. Characterization of the complete mitochondrial genomes of two sibling species of parasitic roundworms, Haemonchus contortus and Teladorsagia circumcincta. Front Genet. 11(1066):573395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N, Maclean PH, Mitreva M, Scott R, Leathwick D.. 2019. The complete mitochondrial genome of the New Zealand parasitic roundworm Teladorsagia circumcincta (Trichostrongyloidea: Haemonchidae) field strain NZ_Teci_NP. Mitochondrial DNA Part B. 4(2):2869–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Wall R.. 2001. Genetic relationships between blowflies (Calliphoridae) of forensic importance. Forensic Sci Int. 120(1–2):116–123. [DOI] [PubMed] [Google Scholar]
- Stevens J, West H, Wall R.. 2008. Mitochondrial genomes of the sheep blowfly, Lucilia sericata, and the secondary blowfly, Chrysomya megacephala. Med Vet Entomol. 22(1):89–91. [DOI] [PubMed] [Google Scholar]
- Stevens JR, Wallman JF.. 2006. The evolution of myiasis in humans and other animals in the old and new worlds (part I): phylogenetic analyses. Trends Parasitol. 22(3):129–136. [DOI] [PubMed] [Google Scholar]
- Stevens JR. 2003. The evolution of myiasis in blowflies (Calliphoridae). Int J Parasitol. 33(10):1105–1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW266393. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA667961, SRR13038366, and SAMN16745530, respectively.