Abstract
Photoactivation and photodissociation have long proven useful tools in tandem mass spectrometry, but implementation often involves cumbersome and potentially dangerous configurations. Here we redress this problem by using a fiber optic cable to couple an infrared (IR) laser to a mass spectrometer for robust, efficient, and safe photoactivation experiments. Transmitting 10.6 μm IR photons through a hollow-core fiber, we show that such fiber-assisted activated ion-electron transfer dissociation (AI-ETD) and infrared multiphoton dissociation (IRMPD) experiments can be carried out as effectively as traditional mirror-based implementations. We report on transmission efficiency of the hollow-core fiber for conducting photoactivation experiments and perform various intact protein and peptide analyses to illustrate the benefits of fiber-assisted AI-ETD: namely a simplified system for irradiating the two-dimensional linear ion trap volume concurrent with ETD reactions to limit uninformative non-dissociative events and thereby amplify sequence coverage. We also describe a calibration scheme for the routine analysis of IR laser alignment and power through the fiber and into the dual cell quadrupolar linear ion trap. In all, these advances allow for a more robust, straightforward, and safe instrumentation platform, permitting implementation of AI-ETD and IRMPD on commercial mass spectrometers and broadening the accessibility of these techniques.
Graphical Abstract
INTRODUCTION
A centerpiece of mass spectrometry (MS) technology, tandem MS (MS/MS) requires efficient and effective peptide fragmentation to enable accurate proteome characterization.1-8 Dissociation is commonly achieved by collisions with inert gases, as in collisionally activated dissociation (CAD), also referred to as collision-induced dissociation (CID). By contrast, electron-transfer dissociation (ETD) reacts multiply protonated polypeptides with small molecule radical anions, resulting in peptide backbone cleavage.9 Relative to CAD, this method preserves labile post-translational modifications (PTMs) during backbone fragmentation, often yielding site-specific PTM localization.10 Now widely available on a variety of commercial mass spectrometers, ETD has become a routine activation method for the dissociation of biomolecules for protein sequence analysis.9,11-13 Its advent has advanced both shotgun proteomic investigations, where mixtures are protease digested before MS analysis,11,12 and top-down techniques for intact proteins and proteoforms, which often rely on activation by electron-based methods.14-16
Electron-based dissociation methods – ETD as well as electron-capture dissociation (ECD) – have been limited by their inefficiency for precursors of low charge density.13 For these precursors, secondary gas-phase structure can prevent dissociation, even after a free electron has been introduced into the system. These non-dissociative electron transfer events (termed ETnoD and ECnoD17-19) result in non-informative, seemingly intact products that simply appear as charge-reduced versions of the precursor ion. Many previous studies have alleviated the challenge of ETnoD20-24 and ECnoD25-33 via secondary activation. In the solution we designed – activated ion-electron-transfer dissociation (AI-ETD) – the ion trapping region is concurrently irradiated with IR photons during the ETD reaction. These photons are intended to disrupt noncovalent interactions and gas-phase secondary structure and thereby increase the generation of sequence-informative product ions following electron transfer.22,34,35 The photon flux is kept low enough so that labile PTMs and peptide backbone bonds are not cleaved.36-39 Previous reports have shown promise of this concurrent photoactivation regime for the analysis of molecules ranging from tryptic peptides and RNA molecules (~2–7 kDa)24,40-42 to intact proteins (> 30 kDa)16,43,44 and monoclonal antibodies (~ 150 kDa).45 Moreover, the concurrent nature of AI-ETD enables efficient analysis without increasing scan time.
While AI-ETD provides an exciting platform for proteome and proteoform characterization, its commercial utility has yet to be realized because the nuances of previous versions – namely multiple mirrors and exposed photons – limit robustness and introduce safety hazards. IR photons are invisible to the human eye and at the powers used here are extremely dangerous.22,34,40 Indeed, nearly all photoactivation MS/MS methods to date have been inhibited by such challenges.46,47 Several FT-ICR systems and UVPD-enabled systems present safe designs, but contain multiple mirrors. The ideal solution is a simple and self-contained configuration. Here we present a novel platform for IR photoactivation on a quadrupole Orbitrap linear ion-trap hybrid system (Orbitrap Fusion Lumos), utilizing a hollow-core fiber optic cable to transmit the 10.6 μm IR light rather than traditional mirrors and lenses. Compared to previous renditions, this straightforward, fully contained system enables higher analytic replicability for improved robustness and addresses health requirements necessary for widespread use in both expert and non-expert labs. Moreover, this setup allows for on-demand infrared photons that can be used for AI-ETD or infrared multiphoton dissociation (IRMPD), another approach not yet widely available on commercial mass spectrometers.
To demonstrate the effectiveness of this design, we show that AI-ETD via fiber-optic transmission increases sequence coverage for peptides and intact proteins over ETD alone, predominantly through increased c- and z•-type ion formation. We also establish a laser calibration platform that allows for the routine determination of IR laser power and alignment to promote facile adoption of AI-ETD and IRMPD across all user and laboratory settings. With this technological innovation, lasers can now be routinely implemented with fiber-optic transmission for AI-ETD and IRMPD analyses of peptides, intact proteoforms, or other biomolecules across a variety of MS instruments.
EXPERIMENTAL SECTION
Materials
Protein standards ubiquitin from bovine erythrocytes (UniProt Accession P63048) and myoglobin from equine skeletal muscle (UniProt Accession P68082) were obtained from Sigma-Aldrich (St. Louis, MO, USA) for intact protein analysis. The proteins were desalted using a Sep-Pak C2 column (Waters, Milford, MA). For electrospray MS analysis, protein solutions were diluted to ~5 pmol/μL in 50% acetonitrile, 49.8% water, and 0.2% acetic acid. The 34-amino acid-long non-tryptic peptide with sequence ELVNDAYDISKQTEDIGNAISEREAVVTNELEGR was obtained from New England Peptides (Gardner, MA, USA), and Angiotensin I with sequence DRVYIHPFHL was obtained from Sigma-Aldrich (St. Louis, MO, USA). Flexmix Calibration Solution (Thermo Fisher Scientific, San Jose, CA), which contains the small peptide MRFA, was obtained and used without modification. For electrospray MS analysis, peptide solutions were diluted to ~2 pmol/μL in 50% methanol, 49.8% water, and 0.2% formic acid and directly infused into the mass spectrometer for experiments with ETD, AI-ETD, and IRMPD.
Mass Spectrometry
All MS and MS/MS experiments were performed on a quadrupole-Orbitrap-quadrupole linear ion trap (q-OT-QLT) hybrid mass spectrometry system (Orbitrap Fusion Lumos, Thermo Fisher Scientific, San Jose, CA) that was modified to include a Firestar Ti-60 Synrad 60 Watt (W) CO2 continuous wave laser (Mukilteo, WA) to allow for the excitation of precursor ions within the quadrupolar linear ion trap during ion-ion reactions.48,49 A multi-mode hollow-core fiber capable of transmitting mid-IR wavelength photons was obtained (Opto-Knowledge Systems, Torrance, CA) and coupled between the laser and hybrid mass spectrometer.
Proteins and peptides were infused at 3-5 μL/min using a Hamilton GasTight Valco syringe (Reno, NV) and a Chemyx Fusion 101 syringe pump (Stafford, TX), and precursors were ionized using electrospray ionization at 3-4 kV with respect to ground. The inlet capillary was held at 275 °C and the ion funnel RF was held at 30%. For ETD experiments, precursor cations were selected in the quadrupole and accumulated in the middle section of the high-pressure trap of the quadrupole linear ion trap, followed by accumulation of fluoranthene reagent anion (202 m/z) within the front section of the trap, and subsequent charge sign independent trapping for the ETD reaction.50 Laser powers for AI-ETD ranged from 10–50 Watts (W). All MS/MS scans were conducted within the Orbitrap at a resolving power of 120,000 at 200 m/z.
A calibration routine was written into the instrument code to allow for the direct calculation of laser-based precursor dissociation within the 2-dimensional quadrupolar linear ion trap. The calibration was centered on the dissociation of the singly protonated peptide MRFA (m/z = 524.26) via IR laser irradiation within the high-pressure and low-pressure traps of the 2D-QLT. Monitoring precursor dissociation within the high-pressure trap and low-pressure trap allowed for the routine examination of beam alignment and laser power due to the design of the 2D-QLT (Figure S1).
Data Analysis
For increased signal-to-noise (S/N) ratio in MS/MS spectra, 10 or 25 individual scans were summed, with averaging performed in the vendor’s post-acquisition software (XCalibur Qual Browser, version 2.2). No microscans were performed. Note electrospray MS spectra for all analytes are shown in Figure S2. MS/MS spectra were deconvoluted with XTRACT (Thermo Fisher Scientific) using default parameters and a signal-to-noise ratio threshold of three. Peak lists from XTRACT and protein sequences from UniProt were searched via ProSight Lite which reported number of bonds cleaved and P-score for each method.51 For combination of spectra, peak lists from each spectrum were concatenated. All fragment matches were made within a 10-ppm tolerance. Note, a C# script using the C# Mass Spectrometry Library (CSMSL, https://github.com/dbaileychess/CSMSL) was written to extract fragment ions and their intensities.
RESULTS AND DISCUSSION
Instrumental modifications for fiber-enabled photoactivation
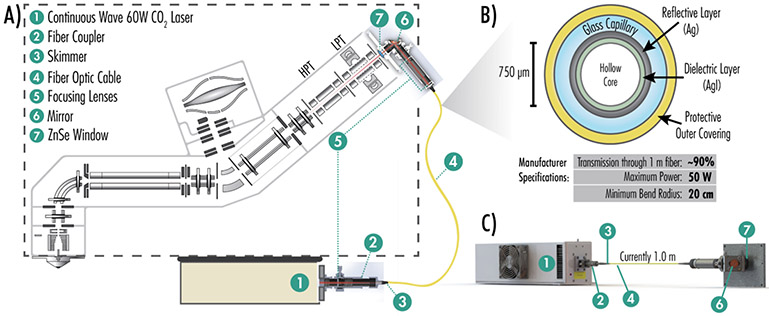
To implement IR photoactivation in a robust, safe manner we developed a contained hollow-core fiber waveguide setup. Key to this arrangement is a suitable fiber – only recently made available – capable of transmitting mid-IR photons. Figure 1A provides an overview of the implementation of optical fiber-enabled photoactivation on a quadrupole Orbitrap linear ion-trap hybrid system (Orbitrap Fusion Lumos) and the components required for this platform. The dashed line indicates the instrument chassis to show that the laser and fiber optic are attached to the exterior of the mass spectrometer. Still, all IR light is contained within the fiber optic cable. Figure 1B details the cross-section of the fiber integrated here, which includes a 750 μm hollow core through which the mid-IR light travels efficiently (~90% for a one-meter fiber). The fiber itself comprises a thin silver iodide dielectric layer, a reflective silver layer, a glass capillary, and a thicker protective coating, allowing for transmission of photons up to 50 W. A continuous wave 60 W CO2 laser was attached to a fiber coupler for transmission into the fiber. A three-dimensional model of the scheme is shown in Figure 1C. During initial testing, IR light quickly decomposed the end of the fiber, so an in-house skimmer was created to funnel light into the optical fiber. Prior to photons passing through the skimmer and entering the fiber, a focusing lens narrows the photon beam to a waist of approximately 1.25 mm. After photons exit the fiber, an internal mirror directs them through a zinc selenide window and into the trapping volume of the linear ion trap where they interact with the ion cloud with radius ~ 1.0 mm and length ~ 30 cm.52,53 In all these results emphasize two primary advantages to the optical fiber compared to mirror-based delivery for instrument configuration: first, the laser can be positioned in a non-constrained manner, i.e., anywhere in or near the instrument, and second, the adapter cover with focusing lens can be simply swapped in place of the standard vacuum cover, requiring no further alignment.54-57
Figure 1. Instrument modifications for photodissociation via a fiber-optic cable capable of transmitting mid-IR photons.
A) Instrumental schematic summarizes the retrofitting of a Fusion Lumos mass spectrometer with a fiber optic cable capable of transmitting IR light. A detailed schematic illustrates instrumental modifications necessary for IR transmission into the dual cell linear ion trap to implement AI-ETD and IRMPD. Modifications include the CO2 laser, fiber coupler, IR light skimmer, fiber optic cable, focusing lenses, guiding mirrors, and an entrance window added to the vacuum manifold. B) The cross-section and manufacturer specifications of the hollow-core fiber used in this study are shown. C) A three-dimensional depiction of the hollow-core fiber and components transmitting light from an IR laser and through a custom machined ion trap mount. Note, the illustrations in panels A-C are not to scale.
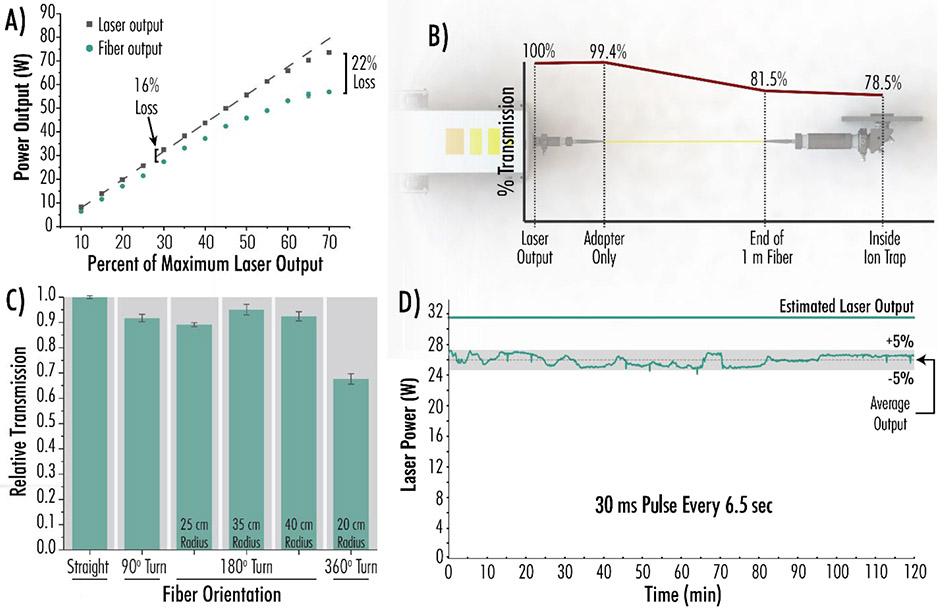
Having implemented this IR delivery mechanism, we next sought to characterize its performance across the four parameters depicted in Figure 2: laser power, transmission efficiency, bending, and temporal stability. Our initial question was whether the fiber optic could deliver high photon flux at powers up to 100 W. Figure 2A illustrates the expected output from a 100 W laser (dashed line), the measured output from the laser (grey squares), and the measured output from a 1.0 m optical fiber (green circles). At laser powers in excess of 55 W, the laser output began to drop below expected values. At 30% of the maximal laser power of 100 W, a 16% loss was observed when comparing power output from the laser to power output from the fiber. (Typical AI-ETD and IRMPD experiments require 30–50 W). This fiber-induced power loss increased to 22% when the laser was operated at 70 W, or 70% of the maximum laser output. Operating the laser at lower power levels minimized this phenomenon. Figure 2B illustrates transmission losses due to various components of the set up. The laser-fiber adapter was quite efficient, reducing the relative percent transmission to 99.4%. After transmission through a 1.0 m fiber at 30 W, the transmission was reduced to 81.5% of the power produced by the laser. Finally, after navigating the focusing lenses, mirror, and ion trap mount, 78.5% of the original laser power was retained. This compares favorably to the traditional mirror-based approach, which typically gave ~60 – 70% transmission. Notably, the photon beam no longer has a Gaussian distribution after passing through the fiber and isn’t concentrated in a single mode.
Figure 2. Laser Power, Transmission Efficiency, Bending, and Temporal Stability of Fiber Optic IR Delivery.
A) Power output before and after fiber optic transmission. Measurements were made at the end of the 1.0 m fiber. B) Visual depiction of percent light transmission through the fiber coupler, straight fiber optic cable, lenses, and mirror for a 30 W laser output. C) Relative percent transmission through the fiber as a function of varying fiber orientations. Experiments are normalized to transmission through a straight fiber and were performed in triplicate. Error bars represent one standard deviation. Measurements were made at the end of the 1.0 m fiber after a 6 W laser output. D) Stability test, monitoring power output from the laser (operating at 30% maximal laser power) through the 1.0 m fiber with a 30 ms pulse every 6.5 seconds over 120 minutes.
We next wondered how the fiber optic affected laser power, particularly for indirect photon transmission routes (i.e. fiber bending). While laser and all fiber optic components will ideally be mounted inside the mass spectrometer itself, such an implementation would require the fiber optic to take an indirect path. Figure 2C demonstrates a set of experiments testing the fiber’s ability to transmit 10.6 μm IR light at 6 W when bent to varying degrees, with transmission measurements presented as the average of triplicate experiments. The fiber has a minimum bend radius of 20 cm, preventing hairpin turns. After normalizing light transmission to the transmission through a straight fiber, we observed the following losses. A 90° turn accounted for ~ 8% transmission loss relative to a straight fiber, whereas three 180° turns accounted for 5–12% transmission loss. Specifically, even a very tight 25 cm radius turn (the tightest of three tested) only reduced transmission by 12%. A 360° bend at the manufacturer-specified minimum radius, 20 cm, transmitted ~ 65% of the IR light that a straight fiber transmitted. We conclude that likely losses due to bending are minimal and would permit incorporation of the light source on the mass spectrometer’s interior. The laser beam travelling out of the fiber optic can enter anywhere in the vacuum chamber if an appropriate combination of lenses and mirrors can direct the beam to the target region, making this approach translatable to other platforms like time-of-flight systems.
Finally, we assessed the extent to which the system can consistently deliver high power over extended periods of time by performing a power stability experiment lasting 120 minutes (Figure 2D). The laser was fired at ~ 32 W for 30 ms every 6.5 seconds. Laser power output was measured at the ion trap after transmission through a 1.0 m fiber and gave an average value of ~ 26 W across the 120-minute duration. The 26 W mean output represents a 17% transmission loss, similar to the power loss calculated at 30% laser power in Figure 2A. According to the manufacturer and our experience, the fiber is very robust and gives consistent photon transmission month-to-month. We have used the fiber without issue for several months steady. Overall, these fiber transmission and laser efficiency data illustrate the utility of an optical fiber in the transmission of long-wave IR light and show that IR light is successfully transported into the linear ion trap for subsequent AI-ETD or IRMPD.
Laser alignment and power calibration
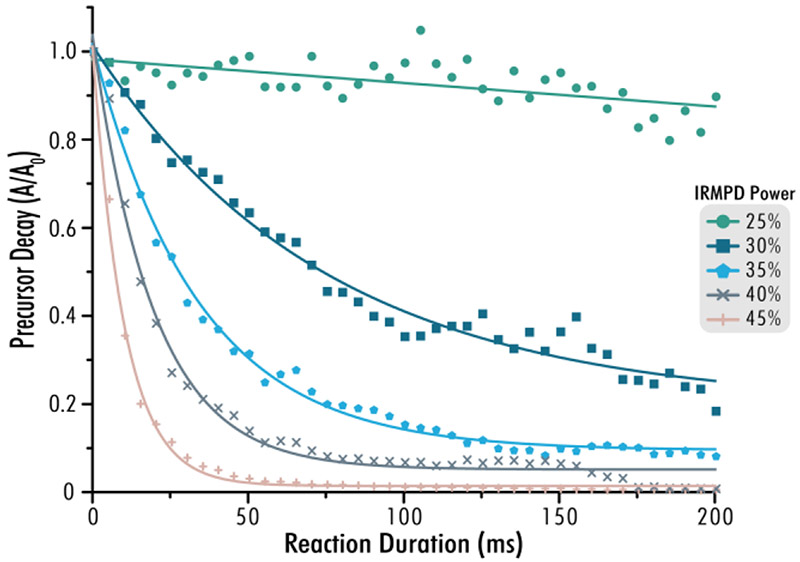
Having shown that IR light could be successfully transmitted into the 2D-QLT by means of an optical fiber, we next established a calibration platform based on precursor decay rates. This platform allows for facile testing of laser alignment and power on a routine basis, similar to ultraviolet photodissociation (UVPD) calibrations.58 The laser settings were tuned to allow for proper analyses by monitoring the dissociation of a small peptide precursor MRFA. Specifically, z = 1 precursor ions (m/z = 524.26, AGC = 10,000) were isolated with the quadrupole using an isolation width of 2 Th and shuttled to either the high-pressure or low-pressure cell of the 2D-QLT where they were irradiated for increasing duration and laser power. Precursor decay is monitored as a function of precursor ion signal, A, following photoactivation, as directly compared to precursor signal prior to irradiation, A0.59 The instrument proceeds with 41 individual experiments varying in reaction duration from 0.5 to 200.5 ms and each consisting of ten microscans, at each laser power. Overall, twelve laser powers are tested within the high-pressure and low-pressure traps for a total of 492 measurements, unique in laser power and reaction duration (Figure S1). Calibration proceeds by adjusting for imbalances in laser power and fine-tuning of laser alignment can be performed until expected precursor decay values are met.60 Values lower than expected in the high-pressure cell, low-pressure cell, or both, are indicative of the nature of beam mis-alignment. Typically, adjustment of the pre-optic lenses helps maximize power output. Manipulation of the single internal mirror or a xy-translational mount at the fiber’s end enable facile alignment of the IR beam path before iteration through the MRFA photodissociation routine. This calibration platform now allows for robust AI-ETD and IRMPD with replicable experimental methods across different instruments. Figure 3 reveals how increasing IR laser power greatly increases the rate of precursor dissociation for this fiber-enabled configuration, thus enabling IRMPD analyses on a chromatographic timescale. Our prior work suggests that slightly lower laser powers are helpful for phosphoproteomics, while glycopeptide fragmentation is optimal at the same laser power used for unmodified peptides. Future work will need to address these PTM-centric tests across multiple systems to understand if laser power can be “calibrated” for different PTM enrichments, much like chromatographic gradients are optimized for different PTM enrichments.
Figure 3. IRMPD of the small peptide MRFA across laser powers.
Dissociation of the singly protonated peptide MRFA (m/z = 524.26) tracked as a function of precursor decay, where A0 is the precursor intensity before irradiation and A is the precursor intensity after a given reaction duration. Each point represents an average of ten transients after photoirradiation via the designated power level (corresponding to ~ 26, 32, 38, 44, and 50 W laser output, from top-to-bottom, respectively). Curves represent an exponential decay asymptotic fit.
Dissociation of peptide and protein cations with AI-ETD
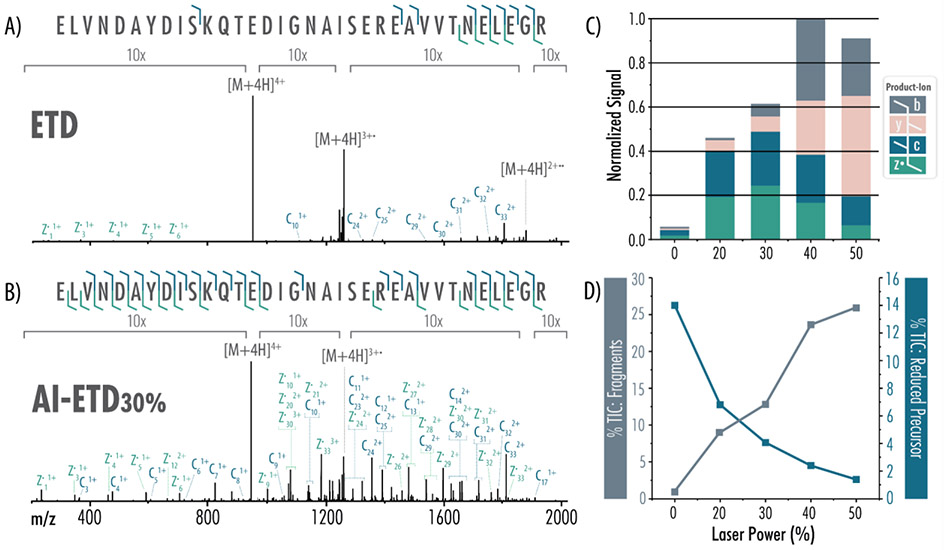
We tested the efficacy of fiber-enabled protein fragmentation with AI-ETD to ensure that results were comparable to traditional mirror-based implementations. Figure 4 provides evidence that this new set-up can conduct high-quality AI-ETD experiments (as compared to ETD) for the analysis of a 34-amino acid-long non-tryptic peptide, ELVNDAYDISKQTEDIGNAISEREAVVTNELEGR. For a z = 4 precursor (945.21 m/z), the ETD spectrum in Figure 4A is dominated by charge-reduced peaks representing 26.5% of the total ion current (TIC). Relatively few sequence-informative product ions are detected; the charge-reduced species confer no sequence information as they simply appear as intact precursors. By contrast, AI-ETD significantly diminishes ETnoD events with increasing laser power. The charge-reduced precursor peaks are reduced more than two-fold following AI-ETD at 20% laser power (12.5% TIC) and are reduced more than three-fold following AI-ETD at 30% laser power (8% TIC) (Figure 4B). These spectra have an abundance of c- and z•-type ion products: when signal intensities of all fragment types (b, c, y, z•) are summed for each method and normalized to the highest value across all five conditions, AI-ETD clearly generates more c- and z•-type ion signal than ETD alone, while also increasing the generation of y-type ions (Figure 4C). Additionally, at higher laser powers (above ~ 32 W), spectra demonstrate a prominent increase in b- and y-type product ions, indicating that dissociation proceeds through vibrational channels, as is typical of IRMPD and other slow-heating methods. While c- and z•-type ion signal for this 34-amino acid peptide peaks at 30% laser power, the generation of b- and y-type ions at ~ 44 W and ~ 56 W laser powers causes the percent of TIC explained by fragment ions to continue increasing (Figure 4D). Noticeably, the percent TIC explained by charge-reduced intact precursors decreases across laser power, reaching 1.7% of the TIC during AI-ETD at ~ 56 W laser power – a 15-fold reduction relative to ETD.
Figure 4. ETD and AI-ETD tandem mass spectra and fragmentation profiles.
Each tandem mass spectrum, representing an average of 25 transients, was generated following 10 ms of A) ETD or B) AI-ETD of the z = 4 peptide precursor. Spectra are on the same intensity scale with labeled regions given a 10x zoom. C) Increasing laser power increases generation of sequence-informative c-, z•-, and y-type product ions over ETD alone. D) AI-ETD drastically improves the percent of the total ion chromatogram (%TIC) explained by product ion fragments (c-, z•-, b-, and y-type) as laser power increases (grey bars, left axis). Additionally, AI-ETD greatly reduces the charge-reduced intact precursor signal, modeled as a percentage of the TIC (blue line, right axis). Laser powers denote the percentage of maximal output and correspond to 0, 20, 32, 44, and 56 W, from left-to-right, respectively. Each graph is normalized to the maximum intensity across all conditions.
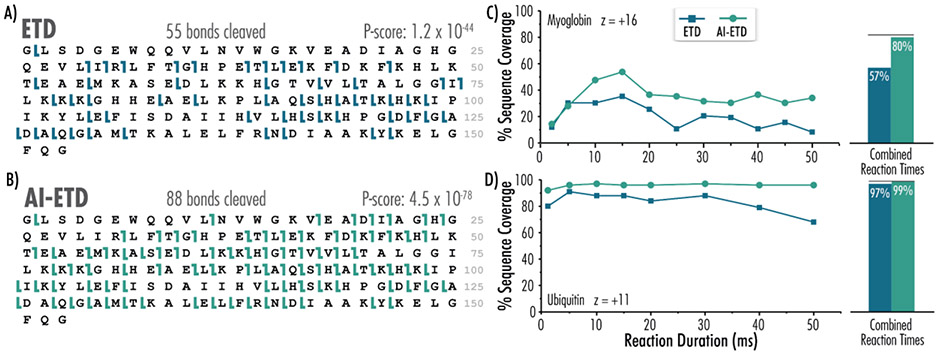
Having shown that this setup works effectively for mid-sized precursors, we next sought to assess its performance for small proteins. Panels A and B of Figure 5 demonstrate the sequence coverage attained for an intact protein investigation of myoglobin with ETD and AI-ETD, respectively. The z = 16 precursor was selected for the ~ 16.9 kDa protein which corresponds to a low-to-intermediate charge density precursor ion (Figure S2). ETD at 15 ms succesfully cleaved 55 bonds within the 153-residue protein, and AI-ETD increased the number of bonds cleaved by 160% to 88. This boost compares well with what was achieved with a comparable number of averaged scans (10 scans) with the previous traditional mirror-based AI-ETD setup (Figure S3).44 Figure 5C demonstrates the ability to perform many ETD and AI-ETD experiments across varying reaction durations and shows their respective sequence coverage. For the z = 16 myoglobin precursor, sequence coverage peaked at about 15 ms for both ETD and AI-ETD, corresponding to sequence coverages of 32% and 58%, respectively. When all reaction timepoints were summed, we observed increases in overall sequence coverage (57% for ETD and 80% for AI-ETD), in keeping with previous reports on the benefits of combining results from multiple reaction times.16,61 These data were then further concatenated to attain a miminal increase in sequence coverage to 82%, denoted by the black line in Figure 5C – an unsurprising outcome considering AI-ETD already generates the majority of fragments achieved by ETD.
Figure 5. Fiber-enabled AI-ETD outperforms ETD for top-down analyses of myoglobin and ubiquitin.
Myoglobin sequence coverage maps following A) ETD and B) AI-ETD (with ~ 32 W laser output) of the z = 16 precursor ion are shown. AI-ETD (again with ~ 32 W laser output) elicits greater coverage compared to ETD across reactions durations for both C) myoglobin and D) ubiquitin. Black lines above bars indicate sequence coverage when both ETD and AI-ETD data points are included.
Ubiquitin ETD and AI-ETD data were similarly compared across reaction durations, with peak sequence coverage coming at 5 ms for ETD and 10 ms for AI-ETD (Figure 5D). When experimental data across reaction durations were summed, AI-ETD attained slightly higher coverage than ETD alone (99% to 97% coverage, respectively). Representative ubiquitin ETD and AI-ETD spectra for the z = 11 charge state precursor with laser power at ~ 32 W are shown in Figure S4. For this small protein with a molecular weight of ~8.6 kDa, charge-reduced intact species are significant across the 15 ms ETD spectrum. In the AI-ETD spectrum, charge-reduced species are noticeably diminished and, in turn, more c- and z•-ion products were generated. Specifically, whereas ETD generated 91 unique product ions yielding 84% sequence coverage, AI-ETD generated 164 unique products, a 180% boost in turn generating 96% sequence coverage (Figure S4C). These results are comparable to our previous AI-ETD characterization on this molecule using the traditional mirror-based IR design.43,44 Collectively these data highlight the effectiveness of fiber-optic enabled AI-ETD to produce spectra for all peptides and proteins tested here. Further, the calibration routine and improved safety of this implementation position this technology for broad dissemination.
CONCLUSION
Here we report the coupling of an IR laser to a mass spectrometer using an optical fiber to enable flexible photoactivation options, including AI-ETD and IRPMD. Few fiber waveguide implementations have been reported since Hogan and co-workers (now nearly thirty years ago) coupled a laser to a mass spectrometer with fiber optics to affect laser desorption ionization of peptides, polymers and porphyrins within an FT-ICR instrument.62-68 Our report is the first description of fiber waveguide technology on a commercially available instrument for ETD-related studies, providing a direct path for a safe commercial implementation of AI-ETD. We show that fiber-optic-enabled AI-ETD maintains the benefits in product ion generation and sequence coverage previously reported for AI-ETD, while now providing a direct route to robust implementation across different instruments within and across laboratories. We also introduce a new calibration scheme to test laser power settings and alignment to aid in this effort. While not fully explored here, the fiber-optic IR delivery now makes IRMPD highly accessible and worthy of further investigation. As the field of proteomics moves forward, more diverse audiences of MS researchers will benefit from access to the advantages AI-ETD provides for investigations of post-translationally modified peptides and complex proteoform mixtures. With this fiber optics approach, photoactivation methodologies should be substantially more accessible and will allow entirely new applications of the method as it reaches new users.
Supplementary Material
Figure S1 Calibration routine for facile laser power and alignment modifications
Figure S2 Annotated electrospray mass spectra of peptide and protein standards
Figure S3 Direct comparison of fiber- and mirror-based implementations
Figure S4 Ubiquitin MS/MS spectra and resulting sequence coverages
Figure S5 Laser power output data as a function of maximum percentage
Figure S6 ETD and AI-ETD spectra across increasing laser power.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health [P41GM108538 to J.J.C.] and the National Human Genome Research Institution through a training grant to the Genomic Sciences Training Program [5T32HG002760 to T.PC.]. We thank the University of Wisconsin–Madison, Thermo Fisher Scientific, and the National Institutes of Health for support. We acknowledge invaluable support from John E. P. Syka, Christopher Mullen, Graeme McAlister, Jae Schwartz, and Alicia Williams.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
J.J.C. is a co-inventor of ETD and consultant for Thermo Fisher Scientific. Raw data are available online on Chorus (Project ID 1686).
REFERENCES
- (1).Hunt DF; Iii JRY; Shabanowitz J; Winston S; Hauer CR Protein Sequencing by Tandem Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A 1986, 83, 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Aebersold R; Mann M Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [DOI] [PubMed] [Google Scholar]
- (3).Coon JJ; Syka JEP; Shabanowitz J; Hunt DF Tandem Mass Spectrometry for Peptide and Protein Sequence Analysis. Biotechniques 2005, 38 (4), 519–523. [DOI] [PubMed] [Google Scholar]
- (4).Washburn MP; Wolters D Large-Scale Analysis of the Yeast Proteome by Multidimensional Protein Identification Technology. Nat. Biotechnol 2001, 19, 242–247. [DOI] [PubMed] [Google Scholar]
- (5).Sleno L; Volmer DA Ion Activation Methods for Tandem Mass Spectrometry. J. Mass Spectrom 2004, 39, 1091–1112. 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- (6).Nielsen ML; Hubner NC; Fro F; De Godoy LMF; Olsen JV; Walther TC; Mann M Comprehensive Mass-Spectrometry-Based Proteome Quantification of Haploid versus Diploid Yeast. Nat. Lett 2008, 455, 1251–1255. 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- (7).Horn DM; Zubarev RA; Mclafferty FW Automated de Novo Sequencing of Proteins by Tandem High-Resolution Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A 2000, 97 (19), 10313–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Palzs B; Suhal S Fragmentation Pathways of Protonated Peptides. Mass Spectrom. Rev 2005, 24 (4), 508–548. 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- (9).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (26), 9528–9533. 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Coon JJ Collisions or Electrons? Protein Sequence Analysis in the 21st Century. Anal. Chem 2009, 81 (9), 3208–3215. 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhurov KO; Fornelli L; Wodrich MD; Tsybin YO Principles of Electron Capture and Transfer Dissociation Mass Spectrometry Applied to Peptide and Protein Structure Analysis. Chem. Soc. Rev 2013, 42, 5014–5030. 10.1039/c3cs35477f. [DOI] [PubMed] [Google Scholar]
- (12).Lermyte F; Valkenborg D; Loo JA; Sobott F Radical Solutions: Principles and Application of Electron-Based Dissociation in Mass Spectrometry-Based Analysis of Protein Structure. Mass Spectrom. Rev 2018, 37 (6), 750–771. 10.1002/mas.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Riley NM; Coon JJ The Role of Electron Transfer Dissociation in Modern Proteomics. Anal. Chem 2018, 90, 40–64. 10.1021/acs.analchem.7b04810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Smith LM; Kelleher NL Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10 (3), 186–187. 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kelleher NL Peer Reviewed: Top-Down Proteomics. Anal. Chem 2004, 76 (11), 196 A–203 A. 10.1021/ac0415657. [DOI] [PubMed] [Google Scholar]
- (16).Riley NM; Westphall MS; Coon JJ Sequencing Larger Intact Proteins (30-70 KDa) with Activated Ion Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom 2018, 29 (1), 140–149. 10.1007/s13361-017-1808-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Pitteri SJ; Chrisman PA; Mcluckey SA Electron-Transfer Ion/Ion Reactions of Doubly Protonated Peptides: Effect of Elevated Bath Gas Temperature. Anal. Chem 2005, 77 (17), 5662–5669. 10.1021/ac050666h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu J; Mcluckey SA Electron Transfer Dissociation: Effects of Cation Charge State on Product Partitioning in Ion/Ion Electron Transfer to Multiply Protonated Polypeptides. Int. J. Mass Spectrom 2012, 332, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Xia Y; Han H; McLuckey SA Activation of Intact Electron-Transfer Products of Polypeptides and Proteins in Cation Transmission Mode Ion/Ion Reactions. Anal. Chem 2008, 80 (4), 1111–1117. 10.1021/ac702188q. [DOI] [PubMed] [Google Scholar]
- (20).Swaney DL; Mcalister GC; Wirtala M; Schwartz JC; Syka JEP; Coon JJ Supplemental Activation Method for High-Efficiency Electron-Transfer Dissociation of Doubly Protonated Peptide Precursors. Anal. Chem 2007, 79 (2), 477–485. 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Frese CK; Altelaar AFM; Van Den Toorn H; Nolting D; Griep-Raming J; Heck AJR; Mohammed S Toward Full Peptide Sequence Coverage by Dual Fragmentation Combining Electron-Transfer and Higher-Energy Collision Dissociation Tandem Mass Spectrometry. Anal. Chem 2012, 84 (22), 9668–9673. 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- (22).Ledvina AR; Mcalister GC; Gardner MW; Smith SI; Madsen JA; Schwartz JC; Stafford GC; Syka JEP; Brodbelt JS; Coon JJ Infrared Photoactivation Reduces Peptide Folding and Hydrogen- Atom Migration Following ETD Tandem Mass Spectrometry. Angew. Chemie Int. Ed 2009, 48, 8526–8528. 10.1002/anie.200903557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cannon JR; Holden DD; Brodbelt JS Hybridizing Ultraviolet Photodissociation with Electron Transfer Dissociation for Intact Protein Characterization. Anal. Chem 2014, 86 (21), 10970–10977. 10.1021/ac5036082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Peters-Clarke T; Quan Q; Brademan D; Hebert AS; Westphall M; Coon JJ Ribonucleic Acid Sequence Characterization by Negative Electron Transfer Dissociation Mass Spectrometry. Anal. Chem 2020, 92, 4436–4444. 10.1021/acs.analchem.9b05388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Horn DM; Ge Y; Mclafferty FW Activated Ion Electron Capture Dissociation for Mass Spectral Sequencing of Larger (42 KDa) Proteins. Anal. Chem 2000, 72 (20), 4778–4784. 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- (26).Breuker K; Oh H; Horn DM; Cerda BA; Mclafferty FW Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J. Am. Chem. Soc 2002, 124, 6407–6420. 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- (27).Horn DM; Breuker K; Frank AJ; Mclafferty FW; V CU; York N; August RV Kinetic Intermediates in the Folding of Gaseous Protein Ions Characterized by Electron Capture Dissociation Mass Spectrometry. J. Am. Chem. Soc 2001, 123, 9792–9799. 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- (28).Tsybin YO; Witt M Combined Infrared Multiphoton Dissociation and Electron Capture Dissociation with a Hollow Electron Beam in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Rapid Commun. Mass Spectrom 2003, 17, 1759–1768. 10.1002/rcm.1118. [DOI] [PubMed] [Google Scholar]
- (29).Mihalca R; Van Der Burgt YEM; Mcdonnell LA; Duursma M; Cerjak I; Heck AJR; Heeren RMA Combined Infrared Multiphoton Dissociation and Electron-Capture Dissociation Using Co-Linear and Overlapping Beams in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Rapid Commun. Mass Spectrom 2006, 20, 1838–1844. 10.1002/rcm. [DOI] [PubMed] [Google Scholar]
- (30).Ge Y; Lawhorn BG; ElNaggar M; Strauss E; Park JH; Begley TP; McLafferty FW Top down Characterization of Larger Proteins (45 KDa) by Electron Capture Dissociation Mass Spectrometry. J. Am. Chem. Soc 2002, 124 (4), 672–678. 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- (31).Håkansson K; Cooper HJ; Emmett MR; Costello CE; Marshall AG; Nilsson CL Electron Capture Dissociation and Infrared Multiphoton Dissociation MS/MS of an N-Glycosylated Tryptic Peptide to Yield Complementary Sequence Information. Anal. Chem 2001, 73 (18), 4530–4536. 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- (32).Håkansson K; Chalmers MJ; Quinn JP; McFarland MA; Hendrickson CL; Marshall AG Combined Electron Capture and Infrared Multiphoton Dissociation for Multistage MS/MS in a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem 2003, 75 (13), 3256–3262. 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- (33).Mikhailov VA; Cooper HJ Activated Ion Electron Capture Dissociation (AI ECD) of Proteins: Synchronization of Infrared and Electron Irradiation with Ion Magnetron Motion. J. Am. Soc. Mass Spectrom 2009, 20, 763–771. 10.1016/j.jasms.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ledvina AR; Rose CM; Mcalister GC; Syka JEP; Westphall MS; Griep-Raming J; Schwartz JC; Coon JJ Activated Ion ETD Performed in a Modified Collision Cell on a Hybrid QLT-Oribtrap Mass Spectrometer. J. Am. Soc. Mass Spectrom 2013, 24, 1623–1633. 10.1007/s13361-013-0621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ledvina AR; Beauchene NA; McAlister GC; Syka JEP; Schwartz JC; Griep-Raming J; Westphall MS; Coon JJ Activated-Ion Electron Transfer Dissociation Improves the Ability of Electron Transfer Dissociation to Identify Peptides in a Complex Mixture. Anal. Chem 2010, 82 (24), 10068–10074. 10.1021/ac1020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Riley NM; Hebert AS; Westphall MS; Coon JJ Capturing Site-Specific Heterogeneity with Large-Scale N-Glycoproteome Analysis. Nat. Commun 2019, 10 (1), 1311. 10.1038/S41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Riley NM; Hebert AS; Dü G; Stanek F; Mechtler K; Westphall MS; Coon JJ Phosphoproteomics with Activated Ion Electron Transfer Dissociation. Anal. Chem 2017, 89, 6367–6376. 10.1021/acs.anal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Buch-Larsen SC; Hendriks IA; Lodge JM; Rykaer M; Furtwängler B; Shishkova E; Westphall MS; Coon JJ; Nielsen ML Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation (AI-ETD). bioRxiv 2020. 10.1101/2020.01.27.921650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Leung KK; Wilson GM; Kirkemo LL; Riley NM; Coon JJ; Wells JA Broad and Thematic Remodeling of the Surfaceome and Glycoproteome on Isogenic Cells Transformed with Driving Proliferative Oncogenes. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (14), 7764–7775. 10.1073/pnas.1917947117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Riley NM; Westphall MS; Hebert AS; Coon JJ Implementation of Activated Ion Electron Transfer Dissociation on a Quadrupole-Orbitrap-Linear Ion Trap Hybrid Mass Spectrometer. Anal. Chem 2017, 89, 6358–6366. 10.1021/acs.analchem.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Riley NM; Rush MJP; Rose CM; Richards AL; Kwiecien NW; Bailey DJ; Hebert AS; Westphall MS; Coon JJ The Negative Mode Proteome with Activated Ion Negative Electron Transfer Dissociation (AI-NETD). Mol. Cell. Proteomics 2015, 14 (10), 2644–2660. 10.1074/mcp.M115.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Shaw JB; Kaplan DA; Brodbelt JS Activated Ion Negative Electron Transfer Dissociation of Multiply Charged Peptide Anions. Anal. Chem 2013, 85 (9), 4721–4728. 10.1021/ac4005315. [DOI] [PubMed] [Google Scholar]
- (43).Riley NM; Westphall MS; Coon JJ Activated Ion Electron Transfer Dissociation for Improved Fragmentation of Intact Proteins. Anal. Chem 2015, 87, 7109–7116. 10.1021/acs.analchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Riley NM; Westphall MS; Coon JJ Activated Ion-Electron Transfer Dissociation Enables Comprehensive Top-down Protein Fragmentation. J. Proteome Res 2017, 16, 2653–2659. 10.1021/acs.jproteome.7b00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lodge JM; Schauer KL; Brademan DR; Riley NM; Shishkova E; Westphall MS; Coon JJ Top-Down Characterization of an Intact Monoclonal Antibody Using Activated Ion-Electron Transfer Dissociation. Anal. Chem 2020. 10.1021/acs.analchem.0c00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Brodbelt JS Ion Activation Methods for Peptides and Proteins. Anal. Chem 2016, 88, 30–51. 10.1021/acs.analchem.5b04563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Stephenson JL; Yost RA Photodissociation in the Ion Trap. In Practical aspects of trapped ion mass spectrometry; March RE, Todd JFJ, Eds.; CRC Press: Boca Raton, 1995; pp 163–204. [Google Scholar]
- (48).Senko MW; Remes PM; Canterbury JD; Mathur R; Song Q; Eliuk SM; Mullen C; Earley L; Hardman M; Blethrow JD; et al. Novel Parallelized Quadrupole/Linear Ion Trap/Orbitrap Tribrid Mass Spectrometer Improving Proteome Coverage and Peptide Identification Rates. Anal. Chem 2013, 85, 11710–11714. 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
- (49).Louris JN; Brodbelt JS; Cooks RG Photodissociation in a Quadrupole Ion Trap Mass Spectrometer Using a Fiber Optic Interface. Int. J. Mass Spectrom. Ion Process 1987, 75, 345–352. [Google Scholar]
- (50).Martens J; Berden G; Oomens J Structures of Fluoranthene Reagent Anions Used in Electron Transfer Dissociation and Proton Transfer Reaction Tandem Mass Spectrometry. Anal. Chem 2016, 88, 6126–6129. 10.1021/acs.analchem.6b01483. [DOI] [PubMed] [Google Scholar]
- (51).Fellers RT; Greer JB; Early BP; Yu X; Leduc RD; Kelleher NL; Thomas PM ProSight Lite: Graphical Software to Analyze Top-down Mass Spectrometry Data. Proteomics 2015, 15 (7), 1235–1238. 10.1002/pmic.201400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Hemberger PH; Nogar NS; Williams JD; Cooks RG; Syka JEP Laser Photodissociation Probe for Ion Tomography Studies in a Quadrupole Ion-Trap Mass Spectrometer. Chem. Phys. Lett 1992, 191 (5), 405–410. [Google Scholar]
- (53).Schwartz JC; Senko MW; Syka JEP A Two-Dimensional Quadrupole Ion Trap Mass Spectrometer. J. Am. Soc. Mass Spectrom 2002, 13, 659–669. [DOI] [PubMed] [Google Scholar]
- (54).Hofstadler SA; Drader JJ Systems and Methods for Inducing Infrared Multiphoton Dissociation with a Hollow Fiber Waveguide. US 6,717,137 B2, 2004.
- (55).Hidaka T Loss Calculations of the Hollow-Core, Oxide- Glass-Cladding, Middle-Infrared Optical Waveguides. J. Appl. Phys 1982, 53 (93), 93–97. 10.1063/1.329961. [DOI] [Google Scholar]
- (56).Hidaka T; Morikawa T; Shimada J Hollow-Core Oxide-Glass Cladding Optical Fibers for Middle-Infrared Region. J. Appl. Phys 1981, 52, 4467–4471. 10.1063/1.329373. [DOI] [Google Scholar]
- (57).Harrington JA; Abel TC; Hirsch J Coherent, Flexible, Coated-Bore Hollow-Fiber Waveguide. US 5,440,664 A, 1995.
- (58).Weisbrod CR; Mullen C; Syka JEP; Schwartz JC Photo-Dissociation Beam Alignment Method. US 9,711,340 B1, 2017.
- (59).Rose CM; Rush MJP; Riley NM; Merrill AE; Kwiecien NW; Holden DD; Mullen C; Westphall MS; Coon JJ A Calibration Routine for Efficient ETD in Large-Scale. J. Am. Soc. Mass Spectrom 2015, 26, 1848–1857. 10.1007/s13361-015-1183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Woodin RL; Bomse DS; Beauchamp JL Multiphoton Dissociation of Molecules with Low Power Continuous Wave Infrared Laser Radiation. J. Am. Chem. Soc 1978, 100 (10), 3248–3250. 10.1021/ja00478a065. [DOI] [Google Scholar]
- (61).Shaw JB; Li W; Holden DD; Zhang Y; Griep-Raming J; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS Complete Protein Characterization Using Top-down Mass Spectrometry and Ultraviolet Photodissociation. J. Am. Chem. Soc 2013, 135, 12646–12651. 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hogan JD; Beu SC; Laude DA; Majidi V Probe-Mounted Fiber Optic Assembly for Laser Desorption/Ionization Fourier Transform Mass Spectrometry. Anal. Chem 1991, 63, 1452–1457. 10.1021/ac00014a020. [DOI] [Google Scholar]
- (63).Mcintosh A; Donovan T; Brodbelt J Axial Introduction of Laser-Desorbed Ions into a Quadrupole Ion Trap Mass Spectrometer. Anal. Chem 1992, 64, 2079–2083. [DOI] [PubMed] [Google Scholar]
- (64).Wu H-F; Brodbelt JS Gas-Phase Complexation of Monopositive Alkaline Earth Metal Ions with Polyethers: Comparison with Alkali Metal Ion and Aluminum Ion Complexations. J. Am. Chem. Soc 1994, 116, 6418–6426. [Google Scholar]
- (65).Alvarez EJ; Wu H-F; Liou C-C; Brodbelt J Collisionally Activated Dissociation of Transition Metal Ion/Polyether Complexes in a Quadrupole Ion Trap. J. Am. Chem. Soc 1996, 118, 9131–9138. [Google Scholar]
- (66).Alvarez EJ; Vartanian VH; Brodbelt JS Metal Complexation Reactions of Quinolone Antibiotics in a Quadrupole Ion Trap. Anal. Chem 1997, 69, 1147–1155. [DOI] [PubMed] [Google Scholar]
- (67).Drader JJ; Hannis JC; Hofstadler SA Infrared Multiphoton Dissociation with a Hollow Fiber Waveguide. Anal. Chem 2003, 75 (15), 3669–3674. 10.1021/ac030157k. [DOI] [PubMed] [Google Scholar]
- (68).Moyer SG; Cotter RJ Atmospheric Pressure MALDI. Anal. Chem 2002, 74, 469–476. 10.1021/ac022091j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Calibration routine for facile laser power and alignment modifications
Figure S2 Annotated electrospray mass spectra of peptide and protein standards
Figure S3 Direct comparison of fiber- and mirror-based implementations
Figure S4 Ubiquitin MS/MS spectra and resulting sequence coverages
Figure S5 Laser power output data as a function of maximum percentage
Figure S6 ETD and AI-ETD spectra across increasing laser power.