Abstract
Lung cancer is the most common fatal malignancy in adults worldwide, and non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer diagnoses. Computed tomography (CT) is routinely used in clinical practice to determine lung cancer treatment and assess prognosis. Here, we developed LungNet, a shallow convolutional neural network for predicting outcomes of NSCLC patients. We trained and evaluated LungNet on four independent cohorts of NSCLC patients from four medical centers: Stanford Hospital (n = 129), H. Lee Moffitt Cancer Center and Research Institute (n = 185), MAASTRO Clinic (n = 311) and Charité – Universitätsmedizin (n=84). We show that outcomes from LungNet are predictive of overall survival in all four independent survival cohorts as measured by concordance indices of 0.62, 0.62, 0.62 and 0.58 on cohorts 1, 2, 3, and 4, respectively. Further, the survival model can be used, via transfer learning, for classifying benign vs malignant nodules on the Lung Image Database Consortium (n = 1010), with improved performance (AUC=0.85) versus training from scratch (AUC=0.82). LungNet can be used as a noninvasive predictor for prognosis in NSCLC patients and can facilitate interpretation of CT images for lung cancer stratification and prognostication.
Introduction
Lung cancer is the most common fatal malignancy in adults worldwide, and non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer diagnoses1. Over 1.6 million people die per year as a result of lung cancer and the 5-year survival rates remain low2. Computed tomography (CT) has been a major diagnostic tool to enable risk assessment of lung cancer in both clinical trials and clinical practice3–5. CT imaging provides in vivo capabilities to measure the extent and location of lung lesions and information on morphological manifestation guiding therapeutic decisions for lung cancer patients6. Despite these advances, qualitative analysis of CT images is limited to what is visible by the human eye causing intra- and inter-reader variability influencing care across clinical centers. There remains an unmet need for robust, fast interpretation of CT images to improve patient stratification, accurate clinical prognostication and treatment selection.
Quantitative image analysis has demonstrated that radiological images, such as CT scans of lung cancer patients and beyond, contain more minable information than what is observed by radiologists7,8,17–19,9–16 Examples include lung nodule segmentation20, lesion detection21, and clinical outcome classification11,22,23. Recent advances in machine learning, especially convolutional neural networks (CNN)24–26, have led to a class of powerful models that show promise to achieve accurate diagnosis and improve medical decision-making27. The use of CNN-based models on imaging data can identify predictive features with clinical importance previously not appreciated or not visible by the human eye. However, to date, the lack of large publicly available clinical imaging cohorts with follow-up data has been an impediment the development and validation of CNN-based models. Recently, however, Ardila et al.28 has developed an end-to-end deep learning model for prediction of cancer risk using low-dose screening lung CTs using the very large National Lung Screening Trial (NLST)29,30 cohort and has shown performance at par or better than trained radiologists.
In this paper, we focus on a different problem – predicting overall survival for patients with confirmed NSCLC. To that end we develop a shallow convolutional neural network, LungNet, for analyzing CT images across multi-institutional cohorts. Our results show that LungNet can predict clinical outcome better than clinical models in multi-institutional cohorts enabling accurate stratification of patients. Through a transfer-learning framework (Fig. 1), we also show that a model pretrained on a survival prediction task can be useful on the nodule malignancy prediction task.
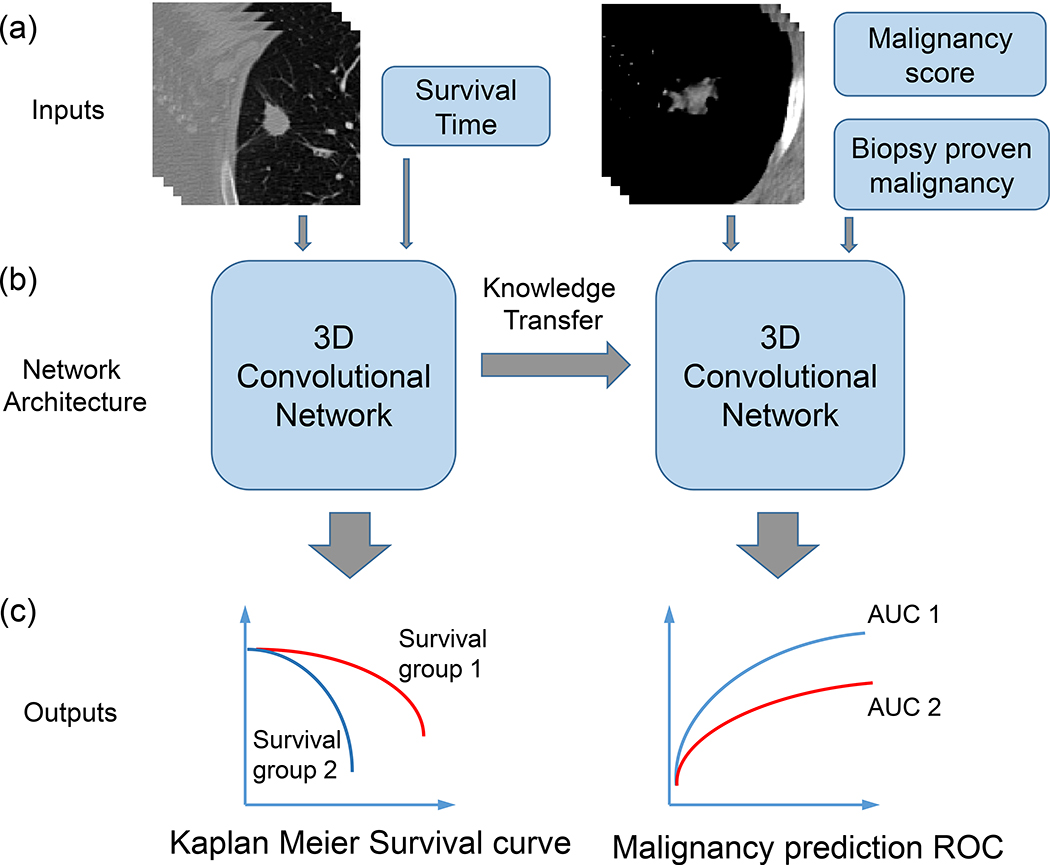
Fig. 1.
Illustration of the proposed computational framework: (a) Input data for the two transfer learning tasks: CT images with survival time and CT images with malignancy scores and for a subset of patients the biopsy proven malignancy. (b) Training and validation of LungNet, a convolutional neural network model including transfer learning between the two tasks. (c) Evaluation of the two tasks using Kaplan Meier survival curves, and ROC curves.
Results
Patient demographics in all cohorts
Examination of the clinical characteristics of our four cohorts shows heterogeneity (Table 1): The four cohorts have different histology with Cohort 1 containing more adenocarcinoma (76.7%) than Cohort 2 (57.8%), Cohort 3 (10.3%) and Cohort 4 (42.9%), one-way chi-square test with cohort 1 as reference p-values: 2.4e-6, 2.9e-58, 1.2e-16, and cohorts 1, 2 and 4 have longer average survival follow-up times (889 days 1021 days, and 944 days, respectively) compared to Cohort 3 (609 days), Welch t-test p-values: 3.0e-4, 9.1e-19 and 5.0e-5. Next, the coefficient of variation of survival times in cohort 2 (0.49) is quite different from that of the remaining cohorts (0.75 in each case). For the benign vs malignant classification task, we used the LIDC-IDRI cohort of lung lesions, which contains malignancy scores annotated by radiologists, for n=1010 patients. For a subset of these patients (n=131), the diagnosis was confirmed by biopsy; therefore the ground truth labels for malignancy are available in these cases. In this subset of LIDC-IDRI cohort with biopsy-proven diagnosis, there were 37 benign cases and 94 malignant cases. Malignant cases included NSCLC patients (n = 43) and malignant metastases (n = 51) from 11 cancer types, including head and neck cancer, colon cancer and metastatic melanoma. The LIDC-IDRI contains lung nodules with diameters ranging from 3 mm to 30 mm. The inclusion of survival cohorts and malignancy labels of LIDC-IDRI allows us to comprehensively evaluate the predictive performance for diagnosis and prognosis of lung cancer patients (Fig. 1).
Table 1.
Patient demographics: the number in parentheses represents percentage.
| Characteristic | cohort 1 | cohort 2 | cohort 3 | cohort 4 |
|---|---|---|---|---|
| Number of patients | 129 | 185 | 311 | 84 |
| Age (yrs., Mean ± SD) | 69.4 ± 8.47 | 67.65 ± 10.13 | 67.05 ±9.07 | |
| Sex (n Male, %) | 101 (78.3) | 83 (44.3) | 220 (70.7) | 64(76.2) |
| Smoking history | 20(15.5) | 38(20.5) | ||
| Histology | ||||
| Adenocarcinoma | 100(77.5) | 107 (57.8) | 32(10.3) | 36 (42.9) |
| Squamous Carcinoma | 29(22.5) | 50 (27) | 84 (27) | 44 (52.4) |
| Other histology type(s) | 28(15.2) | 195(62.7) | 4 (4.8) | |
| Survival time (days, Mean) | 889 | 1021 | 609 | 944 |
| Survival time (days, SD) | 671 | 504 | 457 | 710 |
| Staging Status | ||||
| Stage 1 | 67 (51.9) | 97(52.4) | 81 (26.0) | 5 (6.0) |
| Stage 2 | 42 (32.6) | 32(17.3) | 26(8.4) | 10(11.9) |
| Stage 3 | 15(11.6) | 38(20.5) | 73 (23.5) | 69 (82.1) |
| Stage 4 | 5(3.9) | 17(9.7) | 131 (42.1) |
LungNet predicts survival outcome across institutions
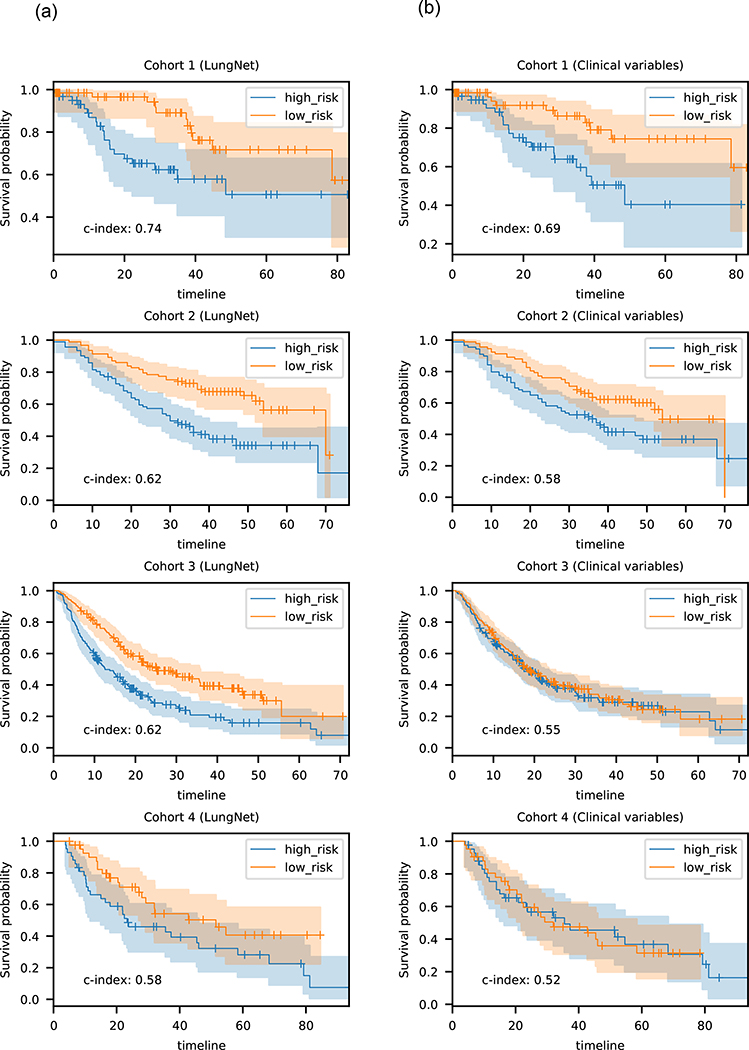
First, we evaluated whether CNNs can be built to predict overall survival across multiple cohorts. We built two versions of LungNet, one using images only as the input, and the other incorporating clinical variables of age, sex, histology and cancer stage along with the CT images. Both versions of LungNet were evaluated in two stages. First, the model was trained on two cohorts and tested on the third in a round robin fashion. The smaller Cohort 4 was kept aside in this phase. Next, the model was trained on cohorts 1, 2 and 3 together, and tested on Cohort 4. In the first phase, with round robin training and evaluation, LungNet achieved validation CI of 0.62 on cohorts 1, 2 and 3. In comparison, a Cox proportional hazards model trained using clinical features of age, sex, histology and cancer stage with one-hot encoding for sex, histology and stage as covariates, achieved CIs of 0.69, 0.58 and 0.55, respectively. The risk scores predicted by LungNet also stratified patients into high risk and low risk groups. The groups showed significant separation in terms of Kaplan-Meier curves for all three cohorts, with two-sided log-rank P values: 2.59e-03, 7.82e-05, 1.10e-05 on cohorts 1, 2 and 3, respectively. The clinical features only model, by comparison, achieved much poorer stratification, with log-rank P values: 7.92e-03, 9.98e-03, and 6.90e-01 on cohorts 1, 2 and 3, respectively (Fig. 2b). While the clinical only model outperformed the images-only LungNet on cohort 1 in terms of CI, the LungNet model incorporating clinical features performed better, achieving CI of 0.73 with P=5.97e-03 on cohort 1. In cohorts 2, and 3, however, incorporating clinical features did not improve the performance of the images-only LungNet model. In the second stage in Cohort 4, we obtained similar results: the images-only LungNet model achieved CI of 0.58 with significant stratification into high and low risk categories (log-rank P value: 5.15e-02), outperforming a clinical only model (CI = 0.52, log-rank P value: 9.33e-01) and thus adding clinical data did also not improve the prognostic performance in Cohort 4.
Fig. 2.
Kaplan-Meier analysis of LungNet. (a) Kaplan-Meier survival performance of LungNet on four lung cancer survival cohorts. For Cohort 1, the LungNet model incorporates clinical features; for the other cohorts, the images-only version of LungNet was used. LungNet demonstrates stratification of low- and high-risk survival subgroups on four independent cohorts. (b) Kaplan-Meier survival performance of clinical-only models on four lung cancer survival cohorts. The median of the predicted risk scores was used to stratify patients into high and low risk groups.
Prediction of survival for early stage cancers
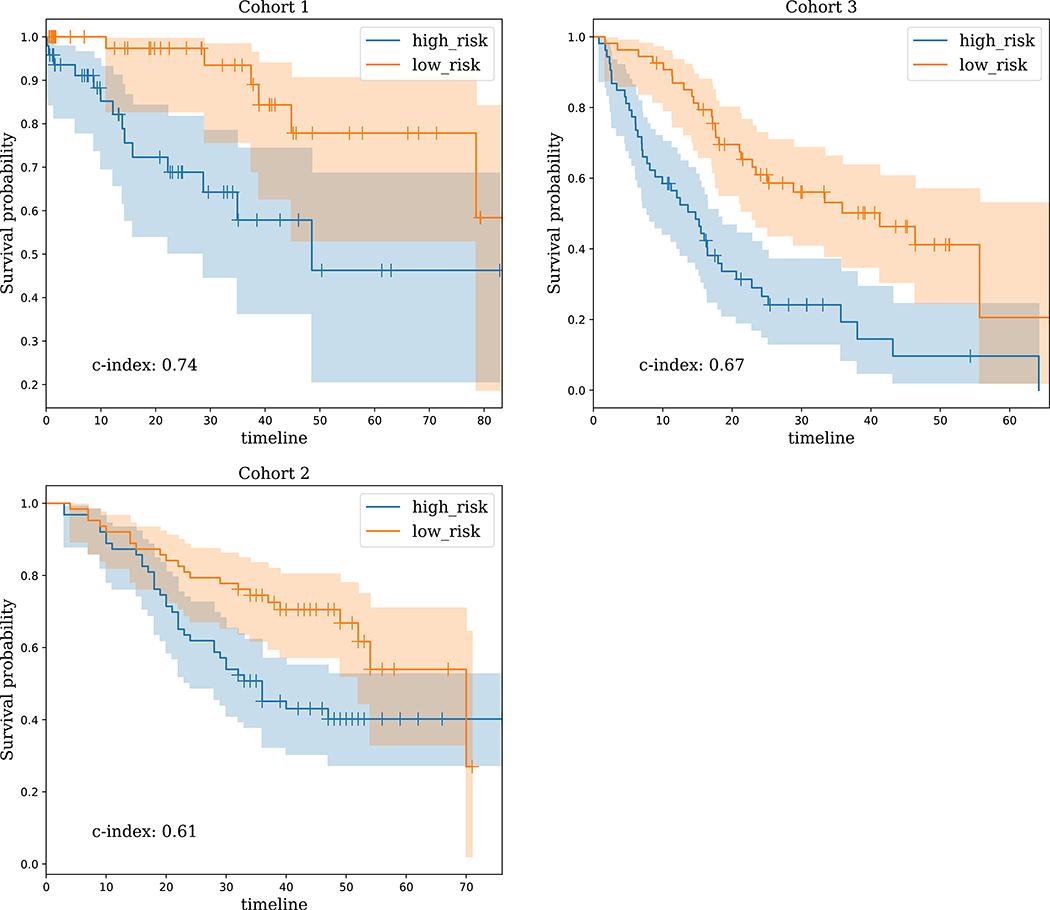
LungNet was effective in predicting survival for early stage cancers, defined as stage 1 and stage 2 NSCLC. Using the images-only LungNet model on Cohort 1, LungNet achieved CI of 0.59 (log-rank P value= 2.81e-02 in stratifying high risk vs low risk patients), and incorporating the clinical features improved the performance to CI = 0.74 (log-rank P = 3.92e-03). On cohorts 2 and 3, the images-only LungNet model achieved CIs of 0.61 (P=9.24e-03) and 0.67 (P=1.70e-05) on cohorts 2 and 3, respectively (Fig. 3), while adding clinical features did not improve performance of LungNet on these cohorts. Since the number of early stage cancers was only 15 in Cohort 4, we did not assess its performance on early stage cancers for Cohort 4.
Fig. 3.
Kaplan-Meier survival performance of LungNet on early stage cancers. It shows that LungNet can stratify low- and high-risk survival subgroups on three independent cohorts for early stage cancers. For cohort 1, the LungNet model incorporates clinical features; for the other cohorts, the images-only version of LungNet is used.
Comparison of LungNet with radiomics
Next, we compared LungNet with prediction of overall survival using radiomics features. Under the same experimental setting, our models based on radiomics features performed worse than LungNet for each of the three cohorts, achieving CI = 0.52, 0.53 and 0.55 for the cohorts 1, 2 and 3 respectively.
Transfer learning between lung cancer overall survival and lung lesion malignancy
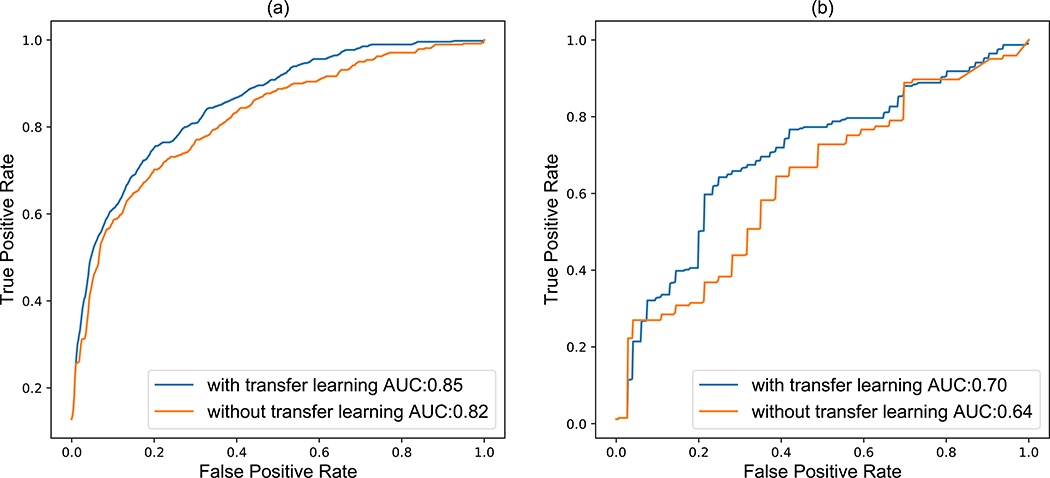
We assessed if transfer learning – fine-tuning the model pretrained for the survival prediction task for predicting nodule malignancy – improves the performance of the model in the malignancy prediction task. Our experimental results showed that applying LungNet via transfer learning led to a significant improvement (P = 0.05 31) for predicting malignancy scores (AUC = 0.85) over the result without applying transfer-learning (AUC = 0.82) (Fig. 4). Choosing a threshold to ensure sensitivity of 0.8, we obtain a specificities of 0.3 and 0.36 with and without transfer learning, respectively. Next, using transfer learning we achieved an AUC of 0.70 for predicting biopsy proven malignancy significantly outperforming (P = 0.0326) the model without transfer learning (AUC = 0.64). Again, choosing a threshold to ensure sensitivity of 0.8, we obtain a specificities of 0.54 and 0.64 with and without transfer learning, respectively. Overall, we found that transfer learning is effective and helpful in improving prediction performance. Note that our tasks of survival prediction and malignancy prediction are likely strongly related to each other; however the outputs (hazard ratio for survival prediction, class probability for malignancy prediction) and loss metrics used for training are very different (Cox regression loss for survival, cross-entropy loss for malignancy prediction).
Fig. 4.
Receiver operating characteristic (ROC) curves for maligancy outcome prediction comparison with and without transfer learning for (a) magliancy by radiologist assessment and (b) biopsy-proven for the LIDC-IDRI cohort.
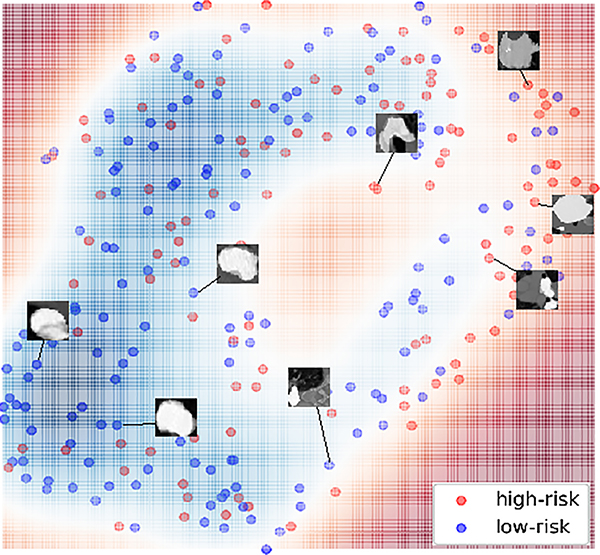
Visualization of LungNet in 2D space
To better understand the predictions of LungNet, we used t-SNE32 to visualize the decision map of LungNet (Fig. 5). We observed that high-risk patients are clustered away from the low-risk patients. Visual inspection of representative cases showed that low-risk lung cancer patients appeared to contain lesions with larger regularity and uniformity around nodule edges compared to high-risk patients who exhibited lesions with sharp and irregular margins. Overall, note that the decision boundary between high risk and low-risk samples in the 2D t-SNE embedding is highly nonlinear, and there is significant overlap among them, pointing to significant heterogeneity in lung cancer appearance between patients10,33,34.
Fig. 5.
Visualization of lung nodules and their survival outcomes in 2D space using t-SNE. The output features of LungNet are embedded into a two-dimensional manifold via a t-distributed stochastic neighbor embedding (t-SNE). The color-coded map is created based on the median survival time. High-risk patients (below the median survival threshold) are highlighted in red and clustered to the far right while low-risk patients (above the median survival threshold) are blue and clustered in the bottom left.
Discussion
We developed and validated a shallow CNN, LungNet, to automatically predict the overall survival of individuals using pre-treatment CT images. Despite being trained on cohorts from various clinical centers with demographical differences, the outcome of LungNet successfully stratified the overall survival of patients in each survival cohort. Additionally, we evaluated LungNet using transfer learning and we demonstrated that pre-training a model for prognostication improves the performance of the malignancy prediction task.
We propose LungNet as a shallow CNN with only seven layers, in contrast to many previous approaches developed on non-biomedical images from the ImageNet database24,27,35. Previous CNN architectures such as Inception36, ResNet and Inception-ResNet37 and DenseNet38 have shown good performance for image classification; however, these deep models also require large databases for training millions of parameters (e.g. Inception has 22 layers and ResNet can have up to 152 layers) from scratch. 3D variants of these networks require even more data, and even large datasets such as UCF-101 (>13,000 action instances), HMDB-51 (>7,500 videos) and ActivityNet (>28,000 action instances) may not be sufficient to train them from scratch 39, These are typically not available for most biomedical applications.
Transfer learning can be an effective tool for addressing the dearth of data. However, it has been demonstrated in the literature that transfer learning from natural images to medical images may not provide much benefit over training a smaller network from scratch 40. Transfer learning within the ambit of medical images can, however, be beneficial. For example, the task of predicting biopsy-proven malignancy using deep learning can be difficult due to the scarcity of ground-truth labels. In this paper, we demonstrated the use of transfer learning, leveraging the survival data in one cohort for improving the prediction of malignancy in another cohort (Fig 4). Specifically, we found that training LungNet on overall survival data of 625 patients resulted in improved accuracy for malignancy classification on a different lung lesion cohort of 1010 patients using 10-fold cross validation. Although previous work with dedicated models for malignancy classification have higher predictive performance41, this use of transfer learning shows that a model can be trained on the prognosis task and subsequently improve the performance when tested on the malignancy prediction task.
Recently, Ardila et al.28 has developed an end-to-end deep learning model for prediction of cancer risk using low-dose screening lung CTs using the very large National Lung Screening Trial (NLST)29,30 cohort and has shown performance at par or better than trained radiologists. In our work, we did not consider the problem of nodule detection and segmentation, which has been extensively studied in the literature in recent years 42–46, instead relying instead on segmentations done by radiologists. We focus on a different problem – predicting overall survival for patients with confirmed NSCLC. In addition, we show that the model pretrained on the survival prediction task can also be useful for the malignancy prediction task as well.
Hosny et al.47 explored the use of deep learning for a coarse prognostication task: predicting dead or alive status for lung cancer patients 2 years after start of treatment, showing AUC around 0.70, with a retrospective multi-center cohort (n=1194). In contrast, our paper focuses on the more challenging task of predicting hazard ratios for each patient. Of course, the predicted hazard ratios can be used to stratify patients into high and low risk groups and the corresponding KM curves showed significant separation for every cohort. Overall, the images-only LungNet model shows consistent performance across the different cohorts, outperforming a clinical only model; however, in cohorts where the clinical variables are strongly predictive of survival, they can be effectively incorporated into LungNet to achieve better performance for prognostication than a clinical only model. Overall, we found that the results of LungNet were predictive of overall survival on all four survival cohorts.
Currently, molecular profiles are used for lung cancer prognostication48–51. LungNet however may be used as a noninvasive, fast and cost-effective complement to the use of molecular biomarkers in predicting prognosis, especially since lung CT scans are typically available and they may be able to capture intratumor heterogeneity better than single biopsies. Thus, quantitative imaging can complement molecular phenotypes obtained from biopsies, and in situations where biopsies are not possible, quantitative imaging can be an alternative.
In this paper, we have focused our efforts on predicting overall survival. Other oncological endpoints such as progression free survival, local control and metastases free survival are also of interest in treating cancer patients. LungNet can be extended to predict other endpoints using transfer learning. If patients can be accurately stratified into high and low risk categories with respect to overall survival or other oncological endpoints, their treatment can be tailored to their predicted risks. For example, treatment may be intensified for high risk patients with higher radiation doses or additional cycles of chemotherapy, while de-intensifying treatment for low risk patients. This can lead to improved quality of life and better clinical outcomes for both high and low risk patients.
Our study has the following limitations. First, one of the inputs to our models is a binary nodule mask created by manual nodule segmentation. The tumor mask delineations for patients in different cohorts were done by different radiologists. We did not assess the impact of inter-reader variability of segmentations on our performance. However, we use random crops as a data augmentation step during training, which introduces variation in segmentations, and therefore, we expect our models to be robust to minor variability of segmentations. Secondly, we did not assess the effect of variability in CT acquisition parameters across patients and institutions on the performance of our models. However, the data augmentation step of random brightness shifts during training is expected to make our models robust to variability in CT acquisition parameters. Indeed, the results we obtain with our rigorous validation strategy with round-robin training and validation on cohorts 1, 2 and 3, followed by validation on a fourth external cohort, suggests that our models are robust to variation in segmentations and CT acquisition parameters.
In conclusion, we developed LungNet, a CNN that uses pretreatment CT imaging for prediction of lung cancer overall survival and nodule malignancy. The performance of LungNet highlights the potential of using CNNs to stratify patients into high and low risk groups based on their CT images. While we have focused on the stratification of patients into only two risk groups, the output of LungNet is a risk score which can be used to stratify patients into three or more risk groups (e.g., high, medium, low) as well, depending on clinical need and setting. Moreover, our transfer learning strategy offers an efficient means to model multiple patient cohorts to address two different tasks: the diagnostic task of malignancy prediction and the prognostic task of survival prediction. Based on its performance on multiple cohorts, we expect LungNet to generalize to other institutional cohorts as well.
Materials and Methods
Study cohorts
This multi-institutional study was approved by the institutional review board of each participating institution, and conducted in compliance with the Health Insurance Portability and Accountability Act (HIPAA), with all patient records deidentified before analysis. For model development and validation, we obtained pretreatment CT images from three unrelated institutions: Cohort 1 from Stanford Hospital (n = 129 patients), Cohort 2 from H. Lee Moffitt Cancer Center and Research Institute (n = 185 patients) and Cohort 3 from MAASTRO Clinic, The Netherlands (n = 311 patients).. The subjects in Cohort 1 were selected from a pool of NSCLC patients referred for surgical treatment between 2008 and 2012 with diagnostic CT performed prior to surgical procedures. Cohort 2 included patients with diagnosed primary tumors who underwent surgical resection and collected contrast-enhanced CT scans obtained within 60 days of the diagnosis between years 2006 and 2009. Cohort 3 comprised patients with confirmed primary tumors who received surgery. We also obtained CT images from a fourth independent institution: Cohort 4 from Charité – Universitätsmedizin, Berlin (n = 84 patients) for final external validation of a fully developed model. For all these cohorts, patients with synchronous malignancies or receiving palliative treatment were excluded. Patients’ overall survival, age, histological subtypes, cancer staging (I, II, III, and IV), and gender information were collected from respective institutions. The CT acquisition parameters in the different cohorts are summarized in Table 2.
Table 2.
CT acquisition parameters: the number in parentheses represents percentage.
| CT parameters | cohort 1 | cohort 2 | cohort 3 | cohort 4 |
|---|---|---|---|---|
| Convolution Kernel | ||||
| STANDARD | 43 (37.2) | 6(3-2) | 4(4.8) | |
| BONEPLUS | 21(16.3) | |||
| LUNG | 26 (20.2) | 1 (0.5) | 4 (4.8) | |
| B45f | 12(9.3) | |||
| BONE | 5(3.9) | |||
| B40f | 1(0.8) | 83 (44.9) | ||
| B41f | 59 (31.9) | 12 (3.9) | ||
| B30f | 13 (7.0) | 67 (21.5) | 13(15.5) | |
| B19f | 66 (21.2) | |||
| BBlf | 1 (0.8) | 25 (8.0) | 2 (2.4) | |
| B31s | 2(1.1) | 29 (9.3) | 26(31.0) | |
| B18f | 17(5.5) | |||
| Other/NA | 15(11.6) | 21(11.3) | 95 (30.5) | 35 (41.7) |
| Peak Kilovoltage | ||||
| 120 | 119 (92.2) | 154(83.2) | 155(49.8) | 81 (96.4) |
| 130 | 4(2.2) | 3(3.6) | ||
| 140 | 27 (14.6) | 67 (21.5) | ||
| Other/NA | 10 (7.8) | 89 (28.6) | ||
| Manufacturer | ||||
| GE | 97(75.2) | 8(4.3) | 6(7.1) | |
| Siemens | 13(10.1) | 169 (91.4) | 222 (71.4) | 50(59.5) |
| Philips | 2 (1.6) | 4(2.2) | 20(23.8) | |
| Toshiba | 1(0.8) | 4(2.2) | 8(9.5) | |
| Other/NA | 89 (28.6) | |||
| Slice Thickness | ||||
| ≤1mn | 21 (16.3) | 12 (14.3) | ||
| ϵ (1, 2] mm | 75 (58.1) | 47 (56.0) | ||
| ϵ (2, 3] mm | 15 (11.6) | 18 (9.7) | 311 (100) | 11 (13.1) |
| ϵ (3, 4] mm | 7 (5.4) | 19 (10.3) | 4 (3.1) | |
| ϵ (4, 5] mm | 7 (5.4) | 122 (65.9) | 10 (11.9) | |
| > 5 mm | 4 (3.1) | 26 (14.1) |
Next, the public Lung Image Database Consortium image collection (LIDC-IDRI)52 cohort was included to measure the performance of malignancy prediction using transfer learning. The LIDC-IDRI cohort provides lung nodule CT images with malignancy scores (i.e., 0 to 4, indicating an increasing degree of malignancy status) assessed by four radiologists (n = 1010). For a subset of patients (n=131), the diagnosis was confirmed with biopsy, and therefore, the ground truth labels for malignancy are available for these cases. We dichotomized the radiologists’ malignancy scores into malignant (score 3 or 4) and benign (score 0 and 1) as previously described22, resulting in 880 benign nodules and 495 malignant nodules. These binary labels (benign vs malignant) were used for training the LungNet model for malignancy prediction.
LungNet: a convolutional neural network for analyzing lung CT imaging
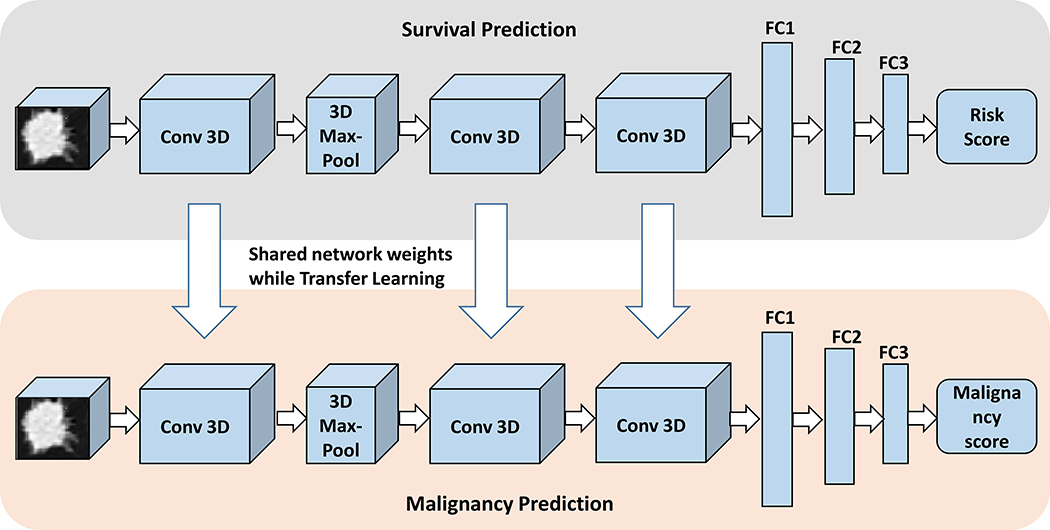
LungNet is a shallow convolutional neural network (CNN) that extracts discriminative CT-based features (Fig. 6). The CNN architecture incorporates three 3D convolutional layers with size 16×3×3 along with a 3D max-pooling layer with kernel size = 2, stride = 2. Three fully connected layers with decreasing sizes of feature vectors (i.e. 128, 64, and 64) were concatenated to reduce feature dimensions towards convergence of model training. The network architecture of LungNet, including the defined 3D convolutional filters and fully-connected layers, were empirically tested on all three survival cohorts towards the shallowest model without overfitting. The output of LungNet is a “risk score”, which represents the logarithm of the ratio of the individual hazard to the baseline hazard. Recall that in the usual Cox Proportional-hazard model, this is assumed to be a linear function of the input covariates; here, it can be a highly non-linear function represented by the CNN. Cox Proportional-hazard loss53 and cross-entropy loss54 functions were used for the survival prediction and the malignancy prediction tasks, respectively.
Fig. 6.
Illustration of LungNet’s convolutional neural network (CNN) architecture. LungNet is a CNN architecture that is designed to address the survival prediction task. In addition, we show that LungNet can be used for the malignancy prediction task using transfer learning. It consists of three 3D convolutional layers with along with a 3D max-pooling layer. Three fully-connected layers were concatenated to reduce feature dimensions. Cox Proportional-hazard loss and Cross-entropy loss functions were used for the survival prediction and malignancy prediction tasks, respectively.
Training, evaluation, and comparison of LungNet
We used a rigorous two-stage evaluation strategy for validating our models, first, using cohorts 1, 2 and 3, we trained our model on two of the cohorts and tested on the third cohort, measuring prediction performance in a round robin fashion. Next, we trained our model on a combination of cohorts 1, 2 and 3 and tested it on cohort 4 for external validation of the developed model.
The input to LungNet is based on 3D volume-of-interest (VOI) regions centered on lung lesions and lesion masks. For patients with multiple lesions, only the largest lesion (by volume) was included. The VOIs were extracted from the original chest CT scans by centering at the nodule location and cropping and resizing to a fixed size (i.e., 64×64×64) using interpolation. Nodule mask delineations for cohorts 1, 2, 3 and 4 were provided by radiologists with manual annotation. We used VOI cropping and random flipping to augment the training data as follows: the 64×64×64 input images were randomly cropped to size 60×60×60 during model training. We also used random left-right and up-down flips and random brightness shifts (between 0.5 and 1) for further data augmentation during training. While training, 20% of the training samples were used as a validation set for monitoring validation loss. We used a cyclic learning rate policy55 (triangular2) for training, and continued the training for 100 epochs with no early stopping. Along the way, we saved the model weights with the lowest validation loss. The whole training process was repeated 20 times, and the model with the lowest validastion loss was chosen as the final trained model, which was then evaluated on the testing cohort. Weight decay was used as L2 regularization. For the survival prediction task, Cox regression loss was used for training.
Regarding the task of malignancy prediction, we kept the same network architecture, but used cross-entropy loss for training. We trained LungNet by applying 10-fold cross-validation on the LIDC-IDRI cohort. We report the average area-under-the-curve (AUC) after repeating the experiment 20 times. The training of network layers was performed by stochastic gradient descent in batch with a learning rate of 0.001. LungNet was implemented in TensorFlow (v 1.4)56 and we used an NVIDIA Titan X GPU for training and testing.
We compared our survival findings with a clinical-only model. We used clinical data including age, histology type (squamous cell carcinoma, small cell carcinoma, or adenocarcinoma), cancer staging (I, II, III, and IV), and sex information to train a Cox Proportional Hazard (C-PH) regression model for survival prediction.
Besides the clinical-only model, we additionally compared with conventional radiomics7,57 analysis, which defined and extracted hand-crafted quantitative features for clinical outcome prediction. We followed the usual radiomics workflow as in prior studies58. We used the same resized VOIs (64×64×64) and the corresponding segmentation masks that were used as input for the LungNet model for feature extraction. We extracted 2131 radiomic features, including intensity-based features, shape and size features, texture features as well as filter-based features, using an in-house radiomics features pipeline 16,59,60 (available at: https://github.com/gevaertlab/radiomics_pipeline). Description of all features and their feature classes are provided in the Supplement S1. These features were used to build a multivariate Cox proportional hazards regression model. Due to the high dimensionality of the feature space, we imposed an l1 penalty on the feature weights for feature selection and regularization, following Goeman61. The l1 penalty tends to assign non-zero weights to a small number of features and set the weights of remaining features to zero.
Transfer learning
In this study, transfer learning was employed to improve the performance on the prediction of nodule malignancy. For this, we first pretrained LungNet on all three survival cohorts combined, with CT images and survival labels (Fig. 6), until convergence. The weights of convolutional layers were frozen and used as fixed feature extractors. Next, this initialized CNN model was further trained on the LIDC-IDRI cohort to predict malignancy using a 10-fold cross-validation evaluation strategy (Fig. 6). The retraining process fine-tunes the weights of the convolutional layers by unfreezing and updating network weights for the malignancy prediction task. We did not try to use the malignancy classification model for survival prediction via transfer learning.
Visualization
To illustrate LungNet, we visualized the decision map of LungNet in 2D. We used the output of the penultimate layer of LungNet as the extracted output features, which were then projected into a two-dimensional manifold via a t-distributed stochastic neighbor embedding (t-SNE)32. Next, we used a two-color scheme to refer to high risk (i.e. red) and low risk (i.e. blue) based on the median survival time. Selected 2D CT image patches were sampled from the patient’s 3D VOI.
Statistical analysis
Statistical analysis was conducted using Python 3.6. Statistical significance levels were all two-sided, with statistical significance set at P < 0.05. Evaluation metrics include the Concordance Index (CI) and the Log-rank p-value in combination with Kaplan-Meier survival analysis using the Lifelines62 package (v 0.8.0.1). Receiver operating characteristics (ROC) and the area under the curve (AUC) in ROC were used to measure classification and prediction performance. ROC curves display the true positive (sensitivity) versus the false positive rate (1 – specificity) over the 10-fold cross-validation on LDIC-IDRI cohort. ROC were generated using classification probabilities of malignant labels versus benign labels of total number of testing images with Python scikit-learn library (v 0.19.1). ROC curves were compared statistically according to Hanley and McNeil method31.
Data Availability
Cohort 1 (Stanford Hospital, n=129): The data is publicly available on The Cancer Imaging Archive (TCIA) at: http://doi.org/10.7937/K9/TCIA.2017.7hs46erv 63
- Cohort 2 (H. Lee Moffitt Cancer Center and Research Institute, n=185): A portion of the data (54/185) is available from TCIA at:
Cohort 3 (MAASTRO Clinic, The Netherlands, n=311): The data is publicly available on TCIA1,5 at http://doi.org/10.7937/K9/TCIA.2015.PF0M9REI 66
Cohort 4 (Charité – Universitätsmedizin, Berlin, n=84): This data is not publicly available yet.
LIDC-IDRI (n=1010): The data is available on TCIA at http://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX 67
Code Availability
Code for the LungNet is available at https://doi.org/10.24433/CO.0612256.v1 68.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB020527 and R56EB020527. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A Titan X Pascal used for this research was donated by the NVIDIA Corporation.
Footnotes
Competing interests: All authors report no conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.
Supplementary Materials
Data supplement S1: Description of radiomic features used for building the radiomics model
References
- 1.Ferlay J et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer (2015). doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Hirsch FR et al. Lung cancer: current therapies and new targeted treatments. The Lancet (2017). doi: 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 3.Swensen SJ et al. CT Screening for Lung Cancer: Five-year Prospective Experience. Radiology (2005). [DOI] [PubMed] [Google Scholar]
- 4.Swensen SJ et al. Lung cancer screening with CT: Mayo Clinic experience. Radiology (2003). doi: 10.1148/radiol.2263020036 [DOI] [PubMed] [Google Scholar]
- 5.Martel S et al. Probability of Cancer in Pulmonary Nodules Detected on First Screening CT. N. Engl. J. Med. (2013). doi: 10.1056/nejmoa1214726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henschke CI et al. Early Lung Cancer Action Project: Overall design and findings from baseline screening. Lancet (1999). doi: 10.1016/S0140-6736(99)06093-6 [DOI] [PubMed] [Google Scholar]
- 7.Gillies RJ, Kinahan PE & Hricak H Radiomics: Images Are More than Pictures, They Are Data. Radiology (2015). doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambin P et al. Radiomics: The bridge between medical imaging and personalized medicine. Nature Reviews Clinical Oncology (2017). doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 9.Thawani R et al. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer (2018). doi: 10.1016/j.lungcan.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 10.Zhou M et al. Non–Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology (2017). doi: 10.1148/radiol.2017161845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts HJWL et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl. Oncol. (2017). doi: 10.1016/j.tranon.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattonen SA et al. [18F] FDG Positron Emission Tomography (PET) Tumor and Penumbra Imaging Features Predict Recurrence in Non-Small Cell Lung Cancer. Tomogr. (Ann Arbor, Mich.) 5, 145–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL & Gillies RJ Quantitative imaging of cancer in the postgenomic era: Radio(geno)mics, deep learning, and habitats. Cancer 124, 4633–4649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamimoto R et al. Prediction of EGFR and KRAS mutation in non-small cell lung cancer using quantitative 18F FDG-PET/CT metrics. Oncotarget 8, 52792–52801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevaert O et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. (2017). doi: 10.1038/srep41674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Griethuysen JJM et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aerts HJWL Data Science in Radiology: A Path Forward. Clin. Cancer Res. 24, 532–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosny A, Parmar C, Quackenbush J, Schwartz LH & Aerts HJWL Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehmeshki J, Amin H, Valdivieso M & Ye X Segmentation of pulmonary nodules in thoracic CT scans: A region growing approach. IEEE Trans. Med. Imaging (2008). doi: 10.1109/TMI.2007.907555 [DOI] [PubMed] [Google Scholar]
- 21.Lee Y, Hara T, Fujita H, Itoh S & Ishigaki T Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. IEEE Trans. Med. Imaging (2001). doi: 10.1109/42.932744 [DOI] [PubMed] [Google Scholar]
- 22.Shen W, Zhou M, Yang F, Yang C & Tian J Multi-scale convolutional neural networks for lung nodule classification. in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (2015). doi: 10.1007/978-3-319-19992-4_46 [DOI] [PubMed] [Google Scholar]
- 23.Xu Y et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res. (2019). doi: 10.1158/1078-0432.CCR-18-2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krizhevsky A, Sutskever I & Hinton GE ImageNet Classification with Deep Convolutional Neural Networks. in ImageNet Classification with Deep Convolutional Neural Networks (2012). doi: 10.1061/(ASCE)GT.1943-5606.0001284 [DOI] [Google Scholar]
- 25.Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature (2017). doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi WL et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA. Cancer J. Clin. 69, caac.21552 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin HC et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging (2016). doi: 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ardila D et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature Medicine (2019). doi: 10.1038/s41591-019-0447-x [DOI] [PubMed] [Google Scholar]
- 29.National Lung Screening Trial Research Team et al. The National Lung Screening Trial: Overview and Study Design. Radiology 258, 243–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Team, N.L.S.T.R. et al. Results of Initial Low-Dose Computed Tomographic Screening for Lung Cancer. N. Engl. J. Med. (2013). doi: 10.1056/nejmoa1209120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanley JA & McNeil BJ A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology (2014). doi: 10.1148/radiology.148.3.6878708 [DOI] [PubMed] [Google Scholar]
- 32.Maaten, L. van der & Hinton G Visualizing Data using t-SNE. J. Mach. Learn. Res. (2008). doi: 10.1007/s10479-011-0841-3 [DOI] [Google Scholar]
- 33.Jamal-Hanjani M et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. (2017). doi: 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 34.Parmar C, Grossmann P, Bussink J, Lambin P & Aerts HJWL Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. (2015). doi: 10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Fei-Fei et al. ImageNet: A large-scale hierarchical image database. in (2009). doi: 10.1109/cvprw.2009.5206848 [DOI] [Google Scholar]
- 36.Szegedy C et al. Going deeper with convolutions. in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (2015). doi: 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- 37.Szegedy C, Ioffe S, Vanhoucke V & Alemi AA the Impact of Residual Connections on Learning. in AAAI Conference on Artificial Intelligence (2017). [Google Scholar]
- 38.Huang G, Liu Z, Van Der Maaten L & Weinberger KQ Densely connected convolutional networks. in Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 (2017). doi: 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 39.Hara K, Kataoka H & Satoh Y Can Spatiotemporal 3D CNNs Retrace the History of 2D CNNs and ImageNet? in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (2018). doi: 10.1109/CVPR.2018.00685 [DOI] [Google Scholar]
- 40.Raghu M, Zhang C, Kleinberg J & Bengio S Transfusion: Understanding Transfer Learning for Medical Imaging. (2019). [Google Scholar]
- 41.Causey JL et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci. Rep. 8, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S et al. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 40, 172–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu W, Liu C, Fan W & Xie X DeepLung: Deep 3D dual path nets for automated pulmonary nodule detection and classification. in Proceedings - 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018 (2018). doi: 10.1109/WACV.2018.00079 [DOI] [Google Scholar]
- 44.Shen W et al. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. (2017). doi: 10.1016/j.patcog.2016.05.029 [DOI] [Google Scholar]
- 45.Cao H et al. Dual-branch residual network for lung nodule segmentation. (2019). doi: 10.1016/j.asoc.2019.105934 [DOI] [PubMed] [Google Scholar]
- 46.Liu H et al. A cascaded dual-pathway residual network for lung nodule segmentation in CT images. Phys. Medica (2019). doi: 10.1016/j.ejmp.2019.06.003 [DOI] [PubMed] [Google Scholar]
- 47.Hosny A et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLOS Med. 15, e1002711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gentles AJ et al. Integrating Tumor and Stromal Gene Expression Signatures with Clinical Indices for Survival Stratification of Early-Stage Non-Small Cell Lung Cancer. J. Natl. Cancer Inst. (2015). doi: 10.1093/jnci/djv211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang C et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non—Small Cell Lung Cancer. Radiology (2016). doi: 10.1148/radiol.2016152234 [DOI] [PubMed] [Google Scholar]
- 50.Shedden K et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat. Med. (2008). doi: 10.1038/nm.1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo NL et al. Confirmation of gene expression - based prediction of survival in non-small cell lung cancer. Clin. Cancer Res. (2008). doi: 10.1158/1078-0432.CCR-08-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armato SG et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys (2011). doi: 10.1118/1.3528204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox DR Regression Models and Life-Tables. J. R. Stat. Soc. Ser. B (2018). doi: 10.1111/j.2517-6161.1972.tb00899.x [DOI] [Google Scholar]
- 54.De Boer PT, Kroese DP, Mannor S & Rubinstein RY A tutorial on the cross-entropy method. Ann. Oper. Res. (2005). doi: 10.1007/s10479-005-5724-z [DOI] [Google Scholar]
- 55.Smith LN Cyclical learning rates for training neural networks. in Proceedings - 2017 IEEE Winter Conference on Applications of Computer Vision, WACV 2017 (2017). doi: 10.1109/WACV.2017.58 [DOI] [Google Scholar]
- 56.Abadi M et al. TensorFlow : A System for Large-Scale Machine Learning This paper is included in the Proceedings of the TensorFlow : A system for large-scale machine learning. Proc 12th USENIX Conf. Oper. Syst. Des. Implement. (2016). doi: 10.1126/science.aab4113.4 [DOI] [Google Scholar]
- 57.Lambin P et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer (2012). doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gevaert O et al. Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology (2012). doi: 10.1148/radiol.12111607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gevaert O et al. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology (2015). doi: 10.1148/radiol.2015154019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang C et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine 45, 70–80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goeman JJ L1 penalized estimation in the Cox proportional hazards model. Biometrical J (2010). doi: 10.1002/bimj.200900028 [DOI] [PubMed] [Google Scholar]
- 62.Davidson-Pilon C et al. CamDavidsonPilon/lifelines: v0.21.1. (2019). doi: 10.5281/ZENODO.2652543 [DOI] [Google Scholar]
- 63.Bakr S et al. Data descriptor: A radiogenomic dataset of non-small cell lung cancer. Sci. Data (2018). doi: 10.1038/sdata.2018.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalpathy-Cramer J et al. A Comparison of Lung Nodule Segmentation Algorithms: Methods and Results from a Multi-institutional Study. J. Digit. Imaging (2016). doi: 10.1007/s10278-016-9859-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grove O et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One (2015). doi: 10.1371/journal.pone.0118261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aerts HJWL et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. (2014). doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Armato SG et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Med. Phys. 38, 915–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukherjee P, Zhou M, Lee E & Gevaert O LungNet: A Shallow Convolutional Neural Network Predicts Prognosis of Lung Cancer Patients in Multi-Institutional CT-Image Data. CodeOcean https://codeocean.com/capsule/5978670/tree/v1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cohort 1 (Stanford Hospital, n=129): The data is publicly available on The Cancer Imaging Archive (TCIA) at: http://doi.org/10.7937/K9/TCIA.2017.7hs46erv 63
- Cohort 2 (H. Lee Moffitt Cancer Center and Research Institute, n=185): A portion of the data (54/185) is available from TCIA at:
Cohort 3 (MAASTRO Clinic, The Netherlands, n=311): The data is publicly available on TCIA1,5 at http://doi.org/10.7937/K9/TCIA.2015.PF0M9REI 66
Cohort 4 (Charité – Universitätsmedizin, Berlin, n=84): This data is not publicly available yet.
LIDC-IDRI (n=1010): The data is available on TCIA at http://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX 67