Abstract
The majority of CpG dinucleotides in the human genome are methylated at cytosine bases. However, active gene regulatory elements are generally hypomethylated relative to their flanking regions, and the binding of some transcription factors (TFs) is diminished by methylation of their target sequences. By analysis of 542 human TFs with methylation-sensitive SELEX (systematic evolution of ligands by exponential enrichment), we found that there are also many TFs that prefer CpG-methylated sequences. Most of these are in the extended homeodomain family. Structural analysis showed that homeodomain specificity for methylcytosine depends on direct hydrophobic interactions with the methylcytosine 5-methyl group. This study provides a systematic examination of the effect of an epigenetic DNA modification on human TF binding specificity and reveals that many developmentally important proteins display preference for mCpG-containing sequences.
The methylation of cytosine at CpG dinucleotides (mCpG) plays an important role in the regulation of human genome architecture and activity. Most CpG dinucleotides in mammalian genomes are methylated, but the methylation pattern is not uniform. Nucleosome-associated DNA has a lower rate of methylation than the more accessible linker sequences located between nucleosomes (1, 2). In addition, methylation patterns vary between cell types (3), and the changes correlate with gene expression. Gene bodies of highly expressed genes are heavily methylated (4, 5), whereas active gene regulatory elements have a low degree of methylation (6–8).
Methylation of DNA is thought to regulate transcription both directly and indirectly. CpG methylation can directly repress transcription by preventing binding of some transcription factors (TFs) to their recognition motifs [for example, (9–12)]. In addition, mCpG dinucleotides can be recognized by a specific class of proteins, the methyl-CpG domain–binding proteins, some of which can recruit histone deacetylases and are thought to promote local chromatin condensation (7, 13). It is generally thought that methylation serves as a barrier to reprogramming (14, 15) and that binding of TFs to previously methylated sites (16) or removal of CpG methylation (7) is involved in cellular differentiation or reprogramming.
Methylation patterns are inherited across cell divisions. This is accomplished by the methyltransferase DNMT1, which associates with the DNA replication fork, methylating the newly synthesized DNA strand at positions where its template strand is methylated (7). This process, together with the inheritance of cytoplasmic determinants such as TFs, forms the basis of the epigenetic memory that allows cellular inheritance of acquired characteristics, such as the state of differentiation [for example, (14)]. The role of TFs and DNA methylation in epigenetic inheritance is thus well established. Several studies have also characterized the interplay between these key determinants by analyzing binding of individual TFs to methylated sites (9, 10, 12, 17, 18) and/or analyzing binding of multiple TFs to a limited number of sequences (19, 20). However, systematic analysis of binding of a large collection of TFs to all possible DNA sequences has so far not been conducted.
HT-SELEX in the presence and absence of CpG methylation reveals TF binding specificities
To globally characterize the effect of cytosine methylation on TF binding, we performed a HT-SELEX [high-throughput systematic evolution of ligands by exponential enrichment (21, 22)] analysis of ~1000 human TF extended DNA binding domains (eDBDs; details are given in table S1 and the methods) and ~550 full-length TFs. This collection included 84% of the high-confidence TFs described by Vaquerizas et al. (23) (classes a and b; table S1). The assay was performed with unmethylated DNA ligands and DNA ligands that were methylated (24) using the CpG-specific cytosine 5-methylase M.SssI before each selection cycle (see the methods for details; Fig. 1A). The ligands contained a 40–base pair (bp) random sequence and were sequenced before the assay and after each selection cycle. The resulting data were analyzed using the previously described Autoseed pipeline, a de novo binding motif discovery method based on identification of distinct seed sequences that are subsequently used to generate position weight matrix (PWM) models [see (25) and the methods]. We (21, 26, 27) and others have previously established that motifs generated using HT-SELEX are similar to those obtained using other state-of-the-art methods, such as protein-binding microarrays or bacterial one-hybrid assays (28, 29), and are biologically relevant on the basis of their ability to predict TF binding in vivo (30–32).
Fig. 1. Methyl-SELEX.
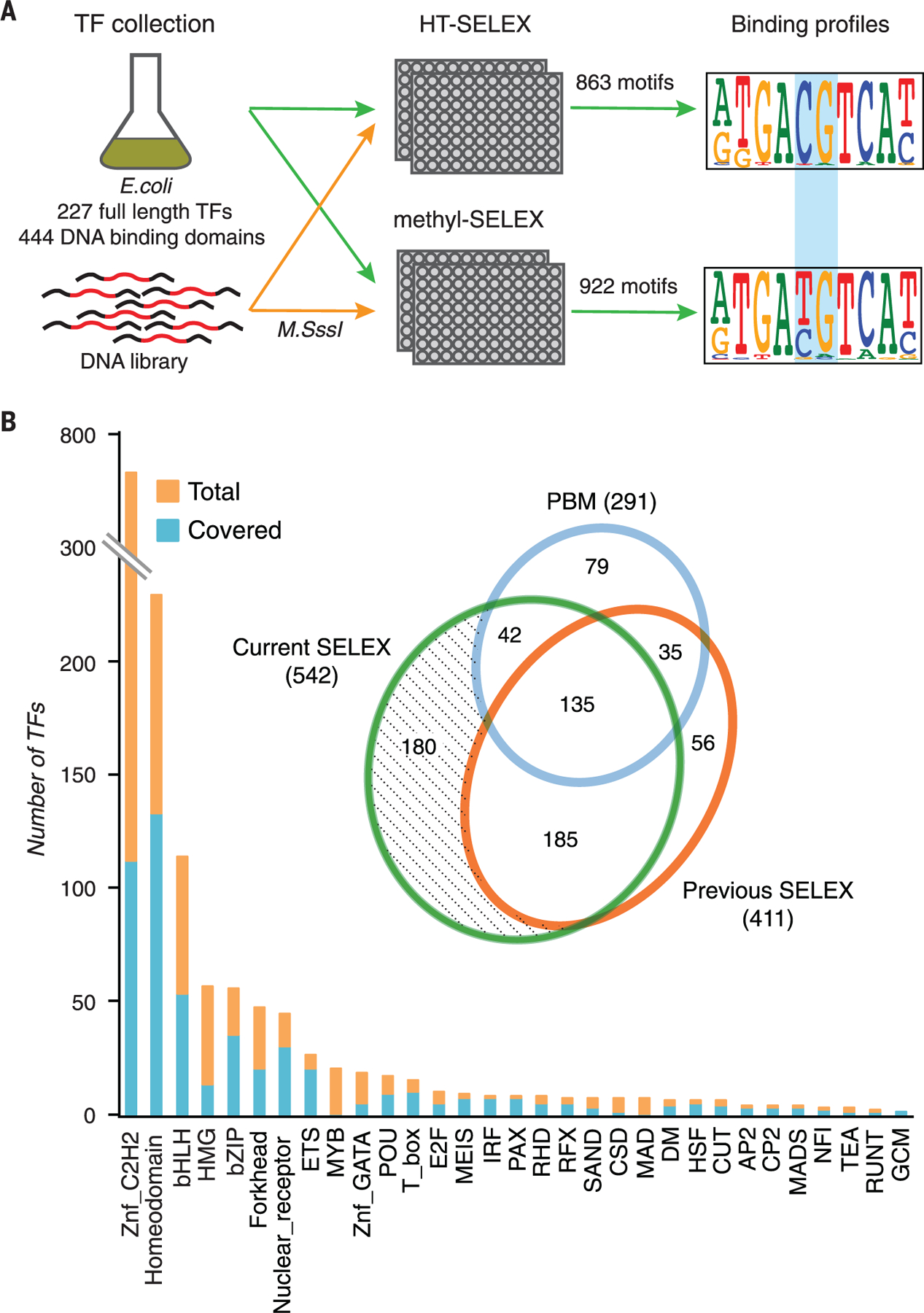
(A) Schematic representation of the SELEX process that allows identification of the binding specificity of TFs for all DNA sequences, including sequences containing methylated and unmethylated CpG dinucleotides. The process uses two parallel reactions with either unmethylated DNA (top, HT-SELEX) or DNA that is methylated at each selection cycle (bottom, methyl-SELEX). Numbers of full-length TFs and extended DBDs for which motifs were obtained are indicated. The blue rectangle indicates the position of a CpG dinucleotide that is affected by methylation. (B) Coverage of TFs by family. The inset is a Venn diagram comparing coverage of mammalian TFs in this work versus in previous large studies using protein-binding microarrays (PBMs) (35, 36) and HT-SELEX (21, 25, 26). Znf, zinc finger.
Compared with the other methods, HT-SELEX is able to detect longer binding motifs because of the high complexity of the input library (26). Although SELEX measures enrichment of sequences and not affinity of binding per se, the order in which sequences are enriched is the same as the order of their affinities. In addition, we have previously shown that the motifs obtained from early HT-SELEX cycles are similar to those obtained by methods that more directly measure affinity, such as oligonucleotide competition assays (33) or assays that compare enrichment across a single SELEX cycle (26). Thus, the motifs presented here can be directly used for motif matching using a threshold, where only the rank of the affinities affects the outcome. However, obtained scores should be considered as estimates rather than true affinity measures.
The median success rate of motif discovery per TF was 47%, and in total, data were obtained for 444 eDBDs and 227 full-length TFs (table S2). As described previously (26), a relatively low rate of success was observed for C2H2 zinc finger proteins, SMAD proteins, and SANT/Myb proteins, likely because of the very long recognition motifs of some C2H2 proteins, the fact that SMAD proteins act as obligate heterotrimers, and the misclassification of many SANT/Myb proteins as TFs despite their lack of key amino acids required for DNA binding (25, 34).
The median coverage for the TF families was 60% (Fig. 1B).The coverage of individual TFs was considerably higher than that reported in previous systematic studies (26, 35–37). For example, this study, using both unmethylated and methylated ligands, and our previously published HT-SELEX data (26) respectively recovered models for 542 and 411 TFs, representing 343 and 239 distinctly different specificities (Fig. 2 and table S2). The motifs obtained in this study were highly consistent with earlier data for TFs that have been studied previously (figs. S1 and S2A). Most motifs that were different from any of the previously determined HT-SELEX motifs were for C2H2 zinc finger proteins whose specificity had not been determined earlier or represented known TFs whose preference for mCpG was not previously recognized, such as the motifs newly identified for a collection of homeodomain proteins in this study (fig. S2). In total, HT-SELEX models now exist for 632 of the ~1400 (23) human TFs.
Fig. 2. Similarity of motifs.
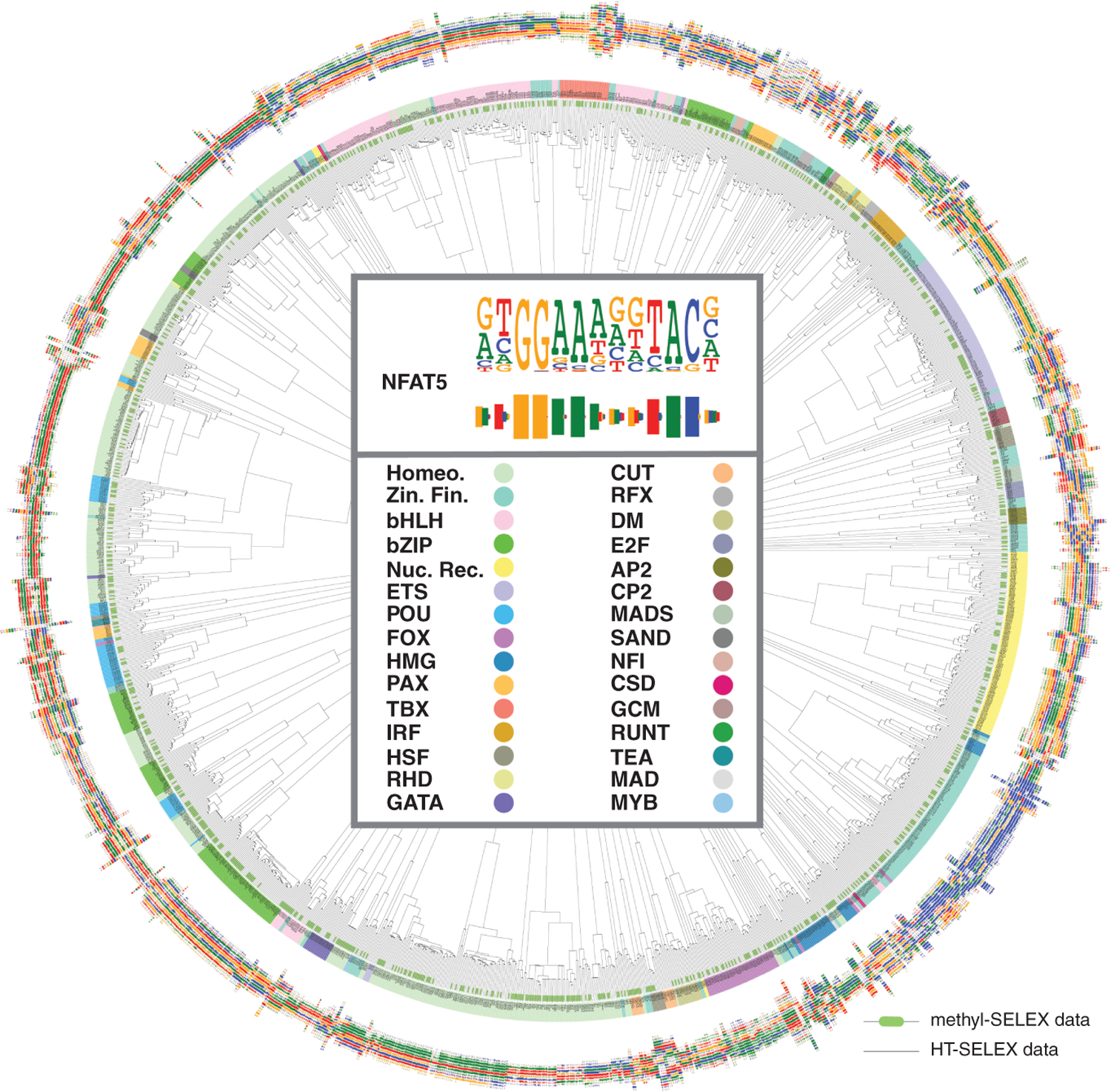
The dendrogram indicates similarities between the motifs from HT-SELEX (thin dendrogram lines) and methyl-SELEX (thick green bars at the end of dendrogram lines). Barcode logos (25) for each factor are also shown. The center of the dendrogram shows an example of the conversion of a sequence logo into a barcode logo (top) and the color key for the TF families (bottom). Motifs for TFs in the same structural families are generally similar to each other, and motifs from methyl-SELEX and HT-SELEX are also closely related in most cases (green and black ends are found in the same branches). This is because many TFs do not have CpGs in their motifs, and the changes induced by methylation generally only affect one dinucleotide in a motif. Homeo., homeodomain; zin. fin., zinc finger; nuc. rec., nuclear receptor.
Analysis of the new HT-SELEX data also revealed a mechanism of evolution of TF binding specificity—utilized by the BARX homeodomain, ELF3 ETS-family TFs, and bHLH-family TF NEUROD1—whereby the addition of an AT-hook domain next to the main DBD results in preference for a flanking AT-rich sequence (Fig. 3).
Fig. 3. Diversification of specificity of paralogs by AT-hook addition.
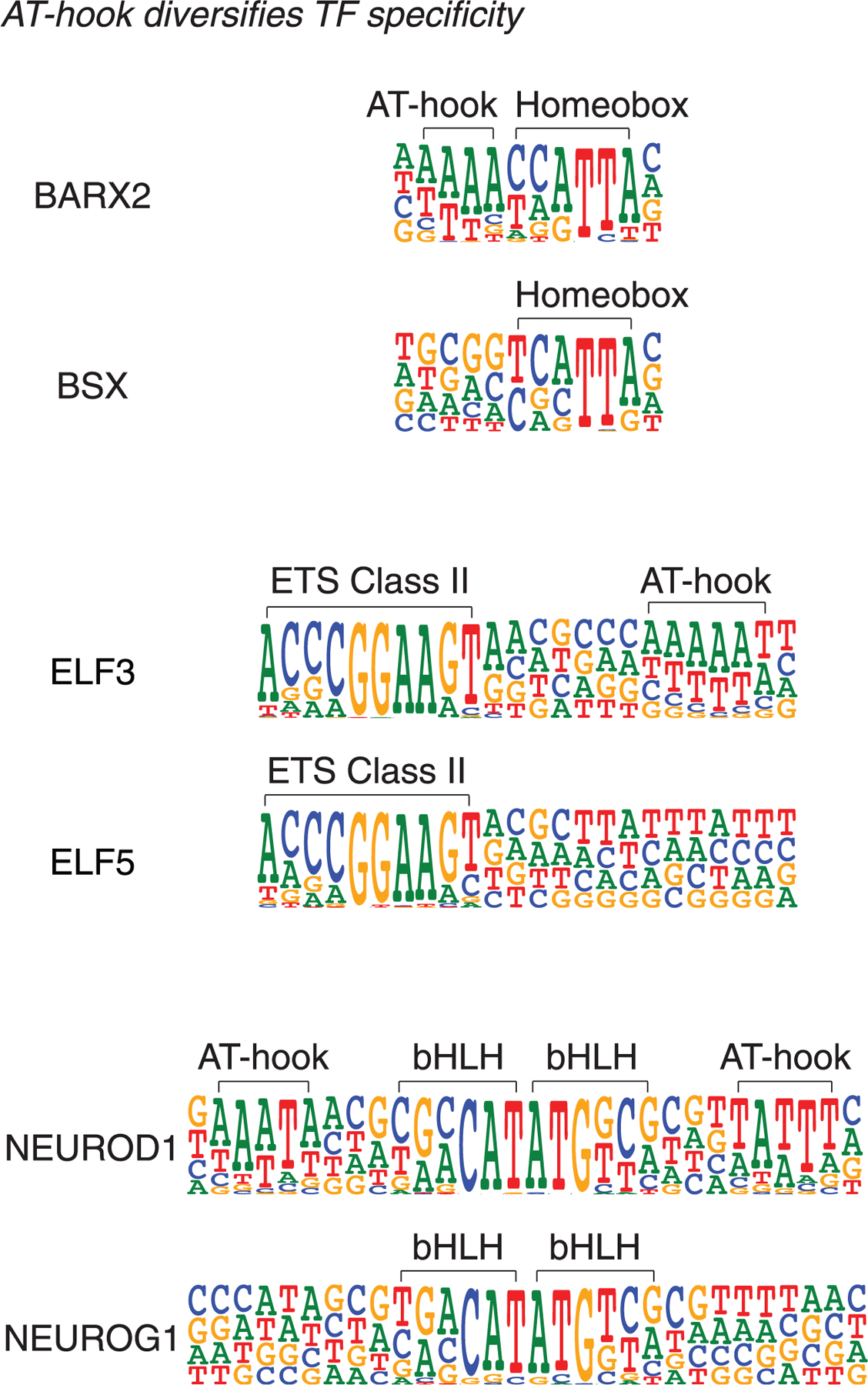
The evolution of TF binding specificity by addition of an AT-hook peptide motif is illustrated. The specificities of the homeodomain TF BARX2, the ETS factor ELF3, and the bHLH protein NEUROD1 have diverged from the related TFs because of the addition of AT-hook–like amino acid sequences, which recognize a short AT-rich sequence.
CpG methylation has a major effect on TF-DNA binding
Using the methyl-SELEX process (see the methods), it is possible to determine the effect of CpG methylation on TF binding, if the TF enriches both sequences that do and do not contain CpG dinucleotide(s). It is, however, difficult to determine the effect of methylation on TFs that have a very strong or an absolute requirement for a CpG in their motif, because they can yield the same CpG-containing motif or fail to yield any motif when DNA is methylated. To allow measurement of the effect of CpG methylation in such cases and to validate the results of methyl-SELEX, we subjected most of the TFs whose recognition motifs contained CpG sequences to one cycle of bisulfite-SELEX (Fig. 4A; see the methods). Using this method, it is possible to determine in one reaction the preference of TFs for unmethylated or methylated forms of their CpG-containing recognition sequences. The results from bisulfite-SELEX confirmed most of the results of methyl-SELEX, and because of ts higher sensitivity, bisulfite-SELEX also revealed many additional TFs with preferences for unmethylated or methylated CpG (for a comparison, see table S3).
Fig. 4. Examples of effects of mCpG on TF binding.
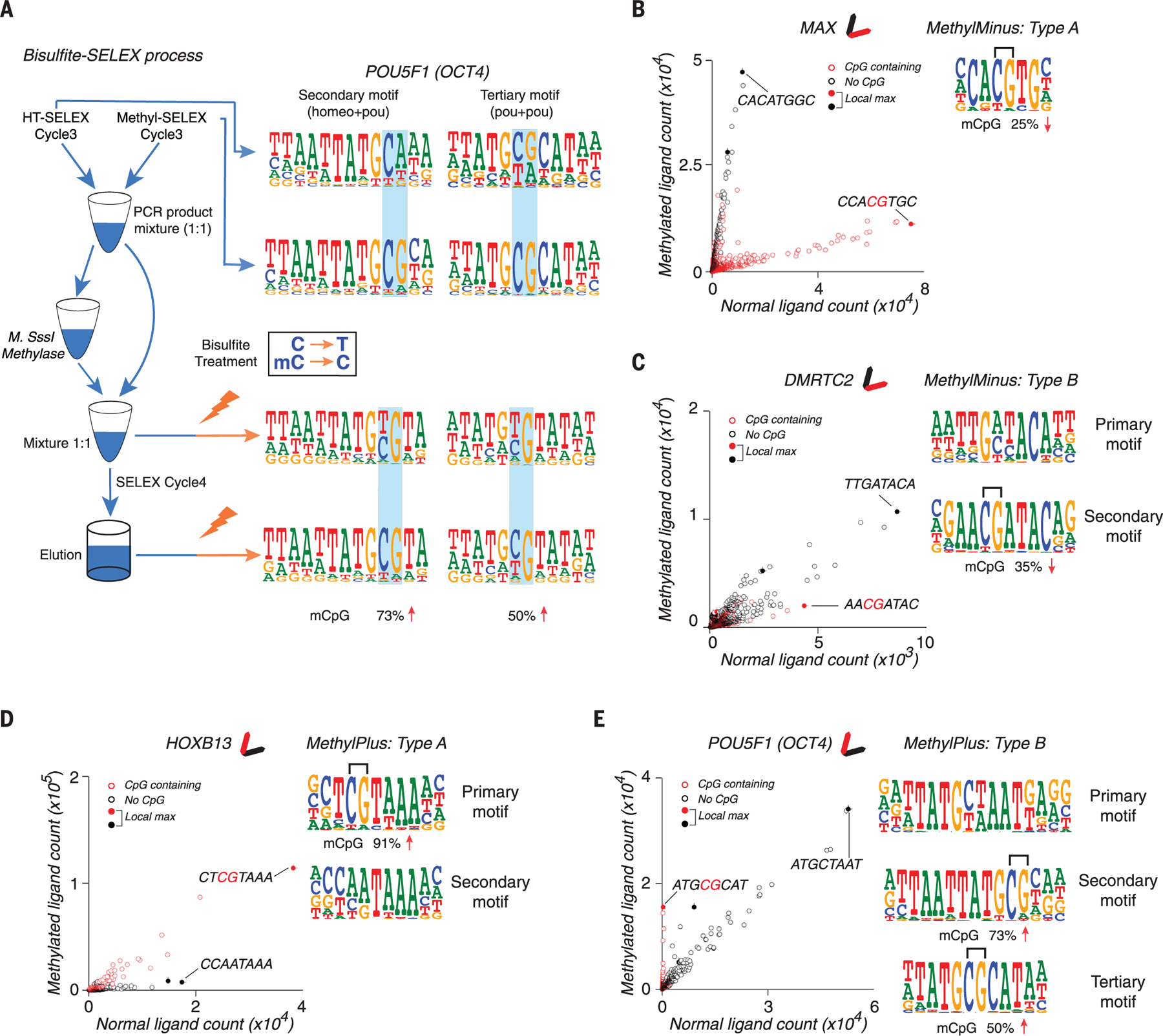
(A) Bisulfite-SELEX. Two models for POU5F1 (OCT4) are recovered from different stages of the bisulfite-SELEX process. OCT4 can bind to both unmethylated and methylated sequences corresponding to the indicated motifs, but it prefers to bind the sequences when the indicated CpG is methylated (remains CpG after bisulfite treatment, indicated in the box). Lightning bolts represent bisulfite treatment, and blue shading highlights dinucleotides affected by methylation. Numbers at the bottom show the increased percentage of mCpG from cycle 3 to cycle 4. (B) Example of a type A methyl-minus TF, MAX (Myc-associated factor X). The scatterplot (left) shows the counts of all 8-mer subsequences from methyl-SELEX (y axis) and HT-SELEX (x axis) at cycle 4. Filled circles indicate subsequences that are more enriched than any other subsequence within a Huddinge distance (25) of 1.The most enriched sequence (CCACGTGC) is also indicated. Because methylation of CpG inhibits MAX binding, the population of the red circles (sequences with CpG) forms an elongated pattern that is located below the population of the black circles (sequences without CpG); this is also shown by the simplified glyph (top). When binding to the optimal site is blocked, other sequences (CACATGGC) that bind more weakly enrich more strongly. The logo of the MAX motif is also shown (right), with the effect of methylation of the CpG in bisulfite-SELEX shown below it. MAX is classified as type A because the consensus of its motif contains a CpG (bracket). (C) A type B methyl-minus TF, DMRTC2, for which the primary motif (right, top) is not affected by methylation, but a CpG in the secondary motif (right, bottom) is. Sequences matching the consensus of its two motifs are indicated on the scatterplot. (D and E) As in (B) and (C), but for the type A methyl-plus TF HOXB13 (D) and the type B methyl-plus TF POU5F1 (OCT4) (E). The subsequence ATGCGCAT is much more enriched by POU5F1 (OCT4) in the presence of CpG methylation. OCT4 also enriches the subsequence ATGCTAAT, which does not contain a CpG and is not affected by methylation.
Combining the results of methyl-SELEX and bisulfite-SELEX showed that some TFs did not recognize sites with CpG sequences, and their binding was thus not influenced by CpG methylation (fig. S3A; “no CpG” class). A second class of TFs recognized CpG-containing sequences, but methylation of the CpG had little effect on binding (fig. S3B; “little effect” class) (38, 39). A third class of TFs did not bind to, or bound more weakly to, methylated versions of their recognition sequences (Fig. 4, B and C, and fig. S3C; “methyl-minus” class). In most cases (82%), the methylation affected the sequence with the highest score (consensus sequence) of the motif with the highest enrichment (primary motif). Such TFs were classified as methyl-minus type A (Fig. 4B and fig. S4; see the methods for details). In the remaining cases, the consensus sequence of the primary motif did not contain a CpG, but the binding of the TF to other enriched sites that did contain a CpG could be affected by methylation. These TFs were classified as methyl-minus type B (Fig. 4C and fig. S5). The methyl-minus group included several proteins for which previous evidence exists for inhibition of binding by mCpG (9, 10, 12, 17, 18) (fig. S6A), indicating that the methyl-SELEX method has high sensitivity and specificity.
In addition to the previously known neutral and negative effects of CpG methylation on TF binding, we also found a fourth class of TFs that preferred to bind to some methylated sequences over the corresponding unmethylated sequence (Fig. 4, D and E, and fig. S3D; “methyl-plus” class). This class included TFs that have previously been reported to weakly prefer mCpG, such as CEBPB (40), KLFs (19, 20, 41), and RFX5 (20) (fig. S6, B to D). In addition, we identified many TFs that, in our assay, displayed stronger preference for mCpG than these proteins (fig. S6D and table S3). In about half of the TFs in this class, the methylation affected the consensus sequence of the primary motif (Fig. 4D and fig. S7; methyl-plus type A; see the methods for details), and in the remaining cases, a weaker site with mCpG was preferentially bound over the corresponding unmethylated site (Fig. 4E and fig. S8; methyl-plus type B). To validate the results by a different experimental method, we enzymatically methylated (11) protein-binding microarrays (35); seven of the eight assignments based on methyl-SELEX were confirmed by this analysis (fig. S9).
To determine the effect of methylation of individual CpGs on TF binding, we calculated the percentage of increase of mCpG at specific positions within motifs during the bisulfite-SELEX experiment (see the methods). Methylation of individual CpG sites commonly affected TF binding, with effects ranging from −100 to +380% (Fig. 5A). The effects were observed both for high-affinity sites and for more moderate-affinity sites (figs. S4, S5, S7, and S8 and table S3). Thus, depending on the presence or absence of a CpG dinucleotide, many TFs can bind in a biologically relevant affinity range to both methylation-sensitive and -insensitive sites.
Fig. 5. Classification of TFs based on methyl-SELEX and bisulfite-SELEX.
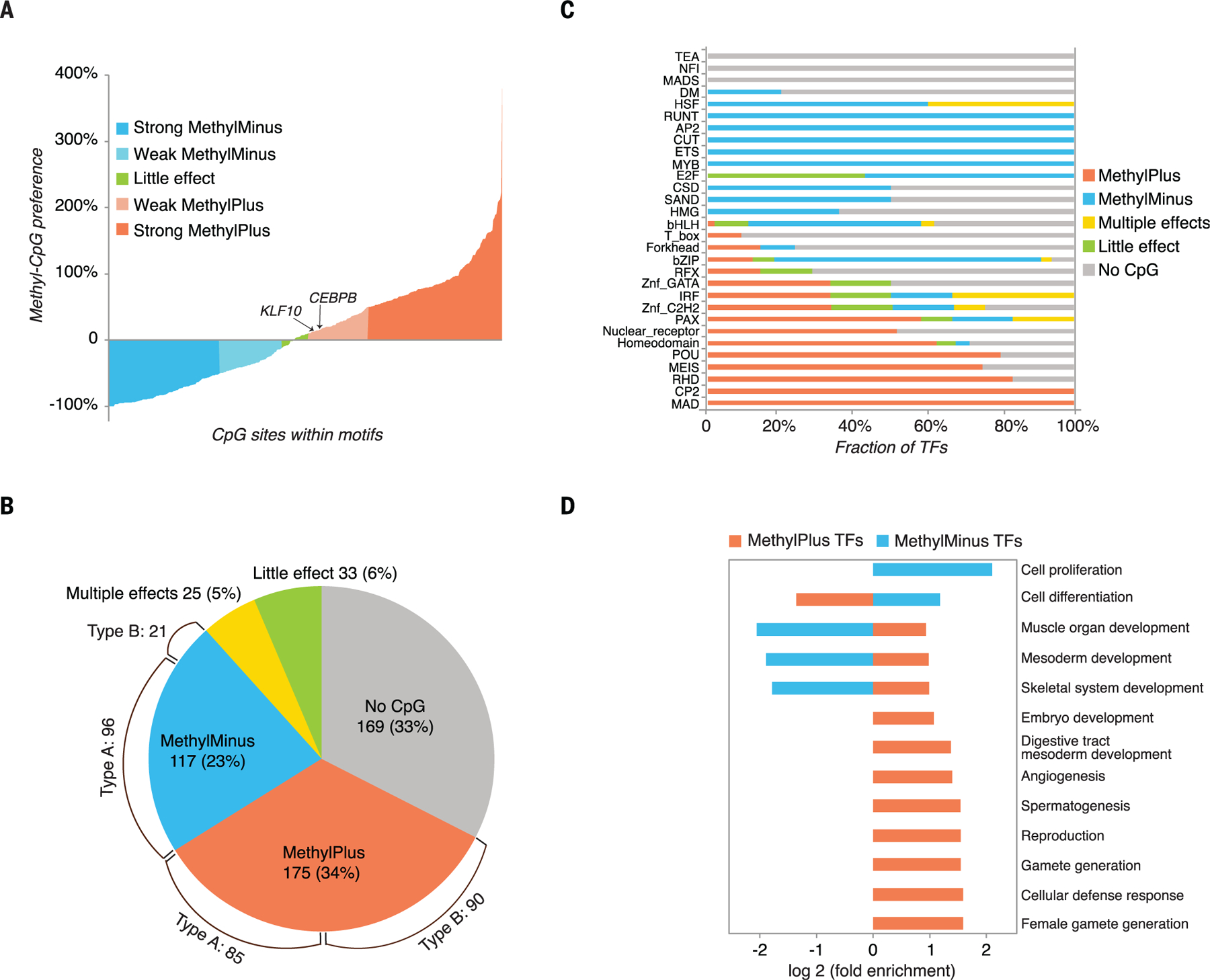
(A) Effect of methylation of individual CpG dinucleotides on binding of human TFs. Percent increases of all mCpG dinucleotides in TF binding motifs during one round of bisulfite-SELEX are shown. Methylation of most CpGs has either a negative (blue) or positive (orange) impact on TF binding. (B) Classification of TFs based on combined analysis of methyl-SELEX and bisulfite-SELEX data (see table S3 for details for each factor). The pie chart shows the fraction of TFs that are not affected by cytosine methylation (no CpG or little effect), that prefer unmethylated CpG (methyl-minus), or that prefer methylated CpG (methyl-plus). In addition, 25 TFs exhibit differential preferences to mCpG dinucleotides at the different positions of their binding sequences or at the different motifs (multiple effects). TFs that can bind to multiple motifs were classified on the basis of the motif that contained CpG dinucleotide(s), if such motif existed. Brackets indicate the numbers of type A and type B TFs of the methyl-minus and methyl-plus groups. (C) Fraction of TFs in each group for each structural TF family. (D) Gene ontology enrichment analysis of methyl-plus and methyl-minus TFs. Biological process classes that are significantly (corrected P < 0.005) enriched or depleted (more than twofold relative to random expectation, based on all the TFs for which motifs were obtained) are included.
Binding of most TFs can be affected by CpG methylation
We next classified all TFs by using a combination of the bisulfite-SELEX and methyl-SELEX data. In cases where a TF bound to two or more motifs, the classification was based on the motif containing a CpG dinucleotide (see the methods, table S3, and data S1 and S2 for details for each TF). This analysis revealed that of the 519 TFs that could be classified, 60% could bind to one or more highly or moderately (>10% of maximum) enriched sequences whose enrichment was influenced by CpG methylation (Fig. 5B); of these TFs, binding of 117 TFs (23%) was inhibited, binding of 175 (34%) was enhanced, and 25 (5%) displayed distinct effects for different motifs or at different CpG positions in a single motif (“multiple effects” class; methods and table S3). The remaining 40% of TFs were not affected by CpG methylation (Fig. 5B). Of these, 169 TFs (33%) did not have CpG dinucleotides in their recognition sequences, and 33 TFs (6%) could bind to a CpG-containing motif but did not display a marked preference for methylated or unmethylated CpG. For this analysis, a threshold of an effect of ±10% was used, based on previous data that reported the preference of CEBPB (40) and multiple KLF proteins (41) for mCpG. The classification of TFs into the five classes (methyl-minus, methyl-plus, multiple effects, little effect, and no CpG) was robust; of 129 cases for which data were obtained for both full-length and eDBD constructs of the same TFs, 125 were classified similarly (data S2). Replicate experiments using independent expression constructs also confirmed the robustness of the analysis (fig. S10A).
TF families differ in CpG methylation sensitivity
We next compared the obtained motifs to determine whether specific TF structural families had common characteristics. This analysis showed that bHLH-, bZIP-, and ETS-family TFs were generally inhibited by mCpG, whereas NFAT (RHD) factors and many members of the extended homeodomain family (e.g., homeodomain, POU, and NKX) preferred to bind to mCpG-containing sequences (Fig. 5C and data S2). However, differences existed within families, with several bZIP proteins binding to mCpG-containing sites with unchanged or slightly higher affinity (Fig. 5C and data S2). Similarly, many, but not all, homeodomain proteins bound to the methylated sequence TmCGTTA in addition to their canonical TAATTA consensus motif (data S2 and table S2). In general, methyl-plus TFs often preferred to bind to unmethylated and methylated sites that were different from each other (fig. S10B and data S2). The differences observed were, in almost all cases, due to changes in the frequency of one or more CpG dinucleotide(s) (data S2). In contrast, the HT-SELEX and methyl-SELEX sites derived for the other groups were very similar to each other (fig. S10B and data S2).
To facilitate the global analysis of distinct binding specificities, we generated a minimal collection of representative motifs that, given a similarity cut-off, can represent all motifs in the whole collection [see (26, 42) and the methods for details]. This analysis revealed that binding to 42 representative binding motifs can be inhibited by CpG methylation (fig. S11A); in all of these, the affected CpG was included in the consensus sequence. In addition, a total of 44 representative binding motifs were preferentially bound when the CpG was methylated (fig. S11B); of these, all but four had the affected CpG in the motif consensus. Analysis of mammalian conservation of the motif matches showed that many of the newly discovered motifs for both groups were conserved, suggesting that they are biologically relevant (figs. S11 and S12). The observed conservation is particularly striking given the high mutational load on CpG sequences (fig. S12C) [for example, (43)].
Panther GO-slim gene ontology enrichment analysis of the TFs whose binding was inhibited by mCpG revealed that cell proliferation and cell differentiation were the most enriched biological processes (Fig. 5D). In contrast, TFs that preferred mCpG are commonly involved in embryonic and organismal developmental processes (Fig. 5D). Such enrichment analysis can determine if a particular class of TFs is enriched relative to all the TFs for which we obtained a motif. However, the reason why the TFs are enriched cannot be determined in this way. The enrichment could be caused by the fact that many paralogous TFs could have inherited their biological roles and methylation specificity from one original TF. Alternatively, the enrichment could be biologically driven—for instance, to ensure that TFs involved in cell proliferation cannot bind and activate the methylated and silenced regulatory elements located at genes that control development, and conversely, that TFs involved in embryonic development are able to bind to methylated loci and induce major changes in cellular chromatin states. In this regard, it is important to note that most methyl-plus TFs belong to a key family of developmental TFs, the homeodomain proteins. Examples include the homeodomain factors that specify the embryonic anterior-posterior axis (e.g., HOXC11 and HOXB13), the NKX proteins that define cell lineages during development, and the pluripotency regulator POU5F1 (OCT4) (44–48).
Effect of CpG methylation on TF binding in vivo
To determine whether TFs also display the expected preferences in vivo, we used existing chromatin immunoprecipitation sequencing (ChIP-seq) data and new ChIP-exonuclease (ChIP-exo) experiments to locate key TFs and full-genome bisulfite sequencing to identify mCpG sequences in two human colorectal cancer cell lines, LoVo and GP5d. The results were broadly consistent with the in vitro analyses (data S3 and table S4). However, in every case, TF-occupied sites displayed lower levels of methylation than their corresponding flanking regions. In addition, sites bound by some methyl-plus TFs were devoid of methylation. These results are consistent with the earlier finding that TF binding induces loss of local DNA methylation, potentially using the TET/TDG-dependent demethylation pathway (7, 49, 50).
Because the TF binding–induced changes in methylation state can confound the ChIP-seq analyses, we next tested TF binding in vivo under three different conditions in which methylation is perturbed. First, we introduced HOXC11 sites to a luciferase reporter construct that is otherwise completely devoid of CpG dinucleotides. Cotransfection of HOXC11 with the unmethylated and methylated reporter construct showed that, as expected from methyl-SELEX, the methylation of the recognition sequences led to an increase in transcriptional activity (fig. S13A). In a second set of experiments, we analyzed TF binding in vivo in mouse embryonic stem (ES) cells displaying increased or reduced levels of CpG methylation. To generate an ES cell line with a high CpG methylation level, we used CRISPR-Cas9 to delete all three TET enzymes. Whole-genome bisulfite sequencing revealed an increased level of methylation of deoxyribonuclease (DNase) I hypersensitive sites in this cell line (fig. S13C), compared with control cells or a previously described ES cell line that lacks CpG methylation [Dnmt triple-knockout (Dnmt-TKO) cell line (51)]. We then performed ChIP-seq analysis of the methyl-plus TF OCT4 and the methyl-minus TF n-Myc in all three ES cell lines. This analysis showed that the alterations in the methylation state resulted in the expected changes in TF binding (Fig. 6A; figs. S13, B to D, and S14; and data S3, C and D). In addition to increased methylation, the Tet-TKO cell line also lacks 5-hydroxymethylcytosine, which has been shown to specifically affect TF binding (52). However, it is unlikely that the effects observed in this study were due to loss of this modified base, because it is present at low per-allele frequency (53), and the differences in OCT4 binding to CpG-containing motifs were also observed in comparisons between the Dnmt-TKO and wild-type cell lines (fig. S14C).
Fig. 6. ChIP-seq analysis.
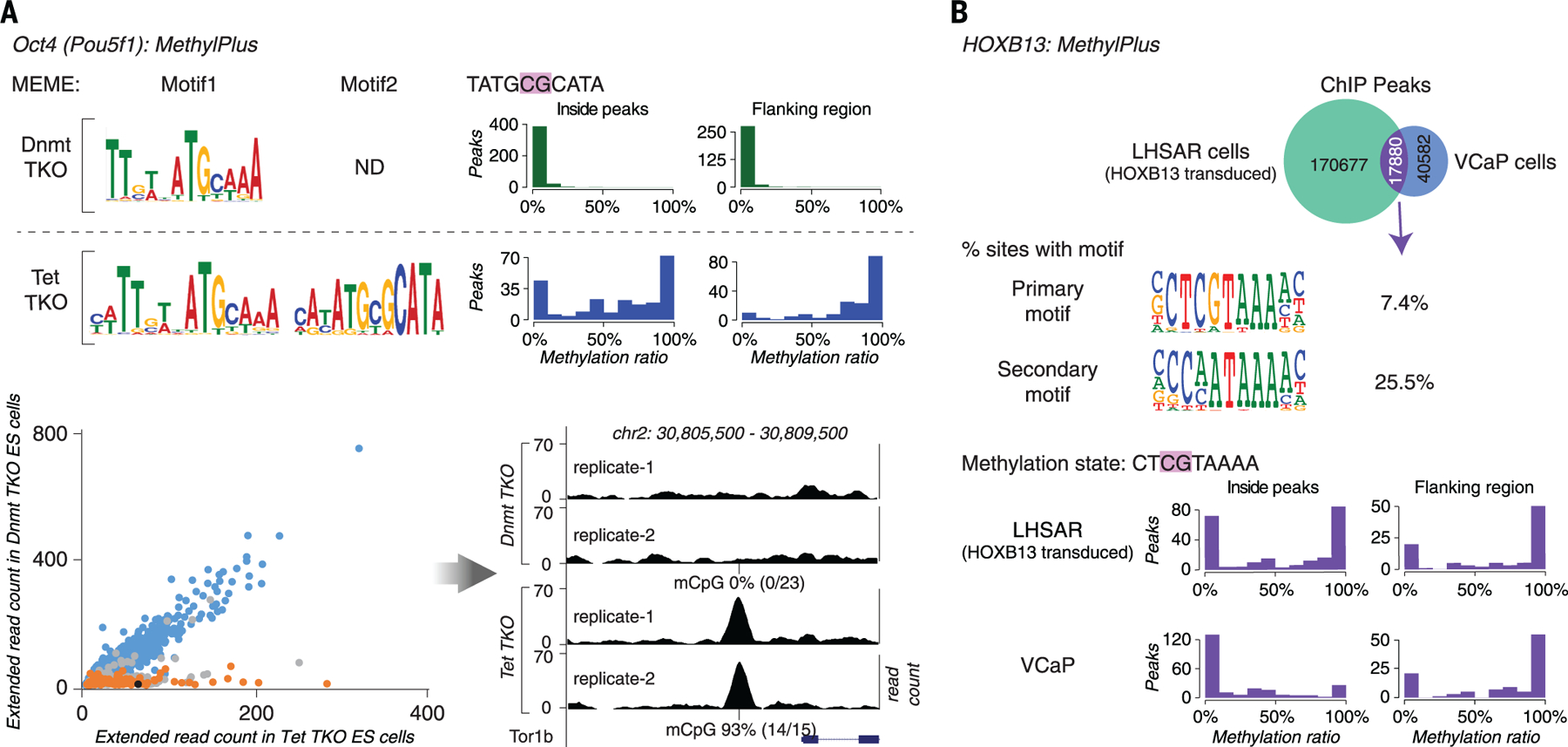
(A) OCT4 prefers a methylated motif in vivo. ChIP-seq analysis of OCT4 was performed in ES cells lacking methylcytosine (Dnmt-TKO) or displaying increased methylation of gene regulatory regions (Tet-TKO). Motif enrichment analysis with MEME (top left) recovered the methyl-plus motif (motif 2) of OCT4 only from peaks from the Tet-TKO cells. Most of the OCT4 occupied sites containing motif 2 were fully methylated in Tet-TKO cells (blue histograms) but not in Dnmt-TKO cells (green histograms) (top right). The scatterplot (bottom left) shows ChIP extended read coverage at motif match positions from peaks in the Tet- (x axis) and Dnmt-TKO (y axis) cells. The ChIP-seq peak heights at the motif 1 match positions (blue) are similar among the cell types, whereas peaks at motif 2 match positions whose methylation state changes (orange) are taller in Tet-TKO cells. Only sites that overlapped with two or more bisulfite-sequencing reads were analyzed. Peaks containing motif 2 matches whose methylation does not change or changes less than the cut-off (from ≤ 20% in Dnmt-TKO to ≥80% in Tet-TKO) are in gray. The black dot indicates the example peak site shown in the bottom right panel. (B) Exogenously introduced HOXB13 binds to methylated sites in the primary prostate epithelial cell line LHSAR. ChIP-seq analysis for HOXB13 was performed in VCaP prostate cancer cells and LHSAR cells transduced with HOXB13-expressing lentivirus. Analysis of peaks that were common to both cell lines (top) showed that HOXB13 can bind to two different motifs, one of which (SELEX primary motif) commonly contains a CpG dinucleotide. The positions of most of the common peaks containing CpG were methylated in LHSAR cells, indicating that HOXB13 can bind to methylated sites. The methylation level of the occupied sites is generally either very low or very high, consistent with the fact that methylation is either present or absent at a given allele. Methylation is lower in the VCaP prostate cancer cells, potentially because of binding-induced demethylation (7).
Last, to rule out effects due to alteration of the methylation state by a constitutively bound TF, we analyzed the in vivo binding specificity of the methyl-plus TF HOXB13, which was exogenously introduced into a prostate epithelial cell line that does not express this protein. It has been previously shown that the chromatin state of normal prostate epithelium cells becomes more similar to that of prostate cancer cells upon expression of exogenous HOXB13 and FOXA1 (54). Consistent with the evidence from methyl-SELEX, ChIP-seq analysis from HOXB13-transduced cells revealed strong binding of HOXB13 to the mCpG-containing site in vivo (Fig. 6B, fig. S15, and data S3C). Within the 48-hour incubation period, the methylation state of the bound sites was not strongly affected (fig. S15). However, in the prostate cancer cell line VCaP, the corresponding peaks displayed lower levels of methylation, suggesting that decreased methylation of these regions may contribute to the reprogramming of chromatin during tumorigenesis (Fig. 6B, fig. S15, and data S3C). Consistent with the ability of HOXB13 to also bind to unmethylated CpG, albeit with lower affinity, it appears that it is able to remain bound to many unmethylated sites in vivo. It remains to be determined whether there exists a subset of mCpG sites that are transiently bound before they are unmethylated. It is tempting to speculate, however, that such sites could have specific biological roles in transitional cellular states, such as in short-lived developmental progenitor cells or in adult transit-amplifying cell populations. The three types of TF methylation preference (plus, minus, and little effect), two methylation states (mC and C), and three possible consequences of TF binding for the methylation state (methylation, demethylation, or no change) suggest that other mechanisms (table S5) for reinforcement of epigenetic states and negative and positive feedbacks could contribute to biological processes.
Structural basis of mCpG preference
To validate our findings and to determine the molecular basis of the observed preference of homeodomain TFs for methylated cytosine, we solved the structure of HOXB13 bound to dually methylated versions of its preferred site CTCGTAAA in the presence or absence of its heterodimeric partner MEIS1. The proteins were expressed in Escherichia coli, purified, and crystallized bound to synthetic double-stranded DNA fragments containing the monomeric CTmCGTAAA and the heterodimeric CTmCGTAAAAcTGTCA motifs. Solving the structures and comparing them with previously solved HOX protein structures (55–60) revealed a very similar architecture of the DBD, with an expected core consisting of three α-helices (Fig. 7A and fig. S16). As in all known homeodomain structures, two parts of the HOXB13 DBD interact with DNA: the recognition helix α3, which tightly packs into the major groove, and the N-terminal tail that interacts with the minor groove (Fig. 7A; 3.0-Å resolution). Analysis of the DNA contacts showed that HOXB13 recognizes mCpG by direct hydrophobic interactions between amino acids and the 5-methyl groups of both methylcytosines of the CpG dinucleotide. HOXB13 Ile262 forms a hydrophobic contact with the first methylcytosine, whereas Val269 recognizes the second methylcytosine opposite to the guanine of the TCG sequence (Fig. 7A). In addition, the aliphatic chain of Arg258 interacts with Ile262 and contributes to the hydrophobic environment of this region. The hydrophobic interactions were also present in the HOXB13:MEIS1-DNA structure, indicating that the methyl groups of both cytosines are robustly recognized by HOXB13 in multiple physiologically relevant contexts (Fig. 7B; 2.54-Å resolution).
Fig. 7. Molecular basis of recognition of mCpG by homeodomain proteins.
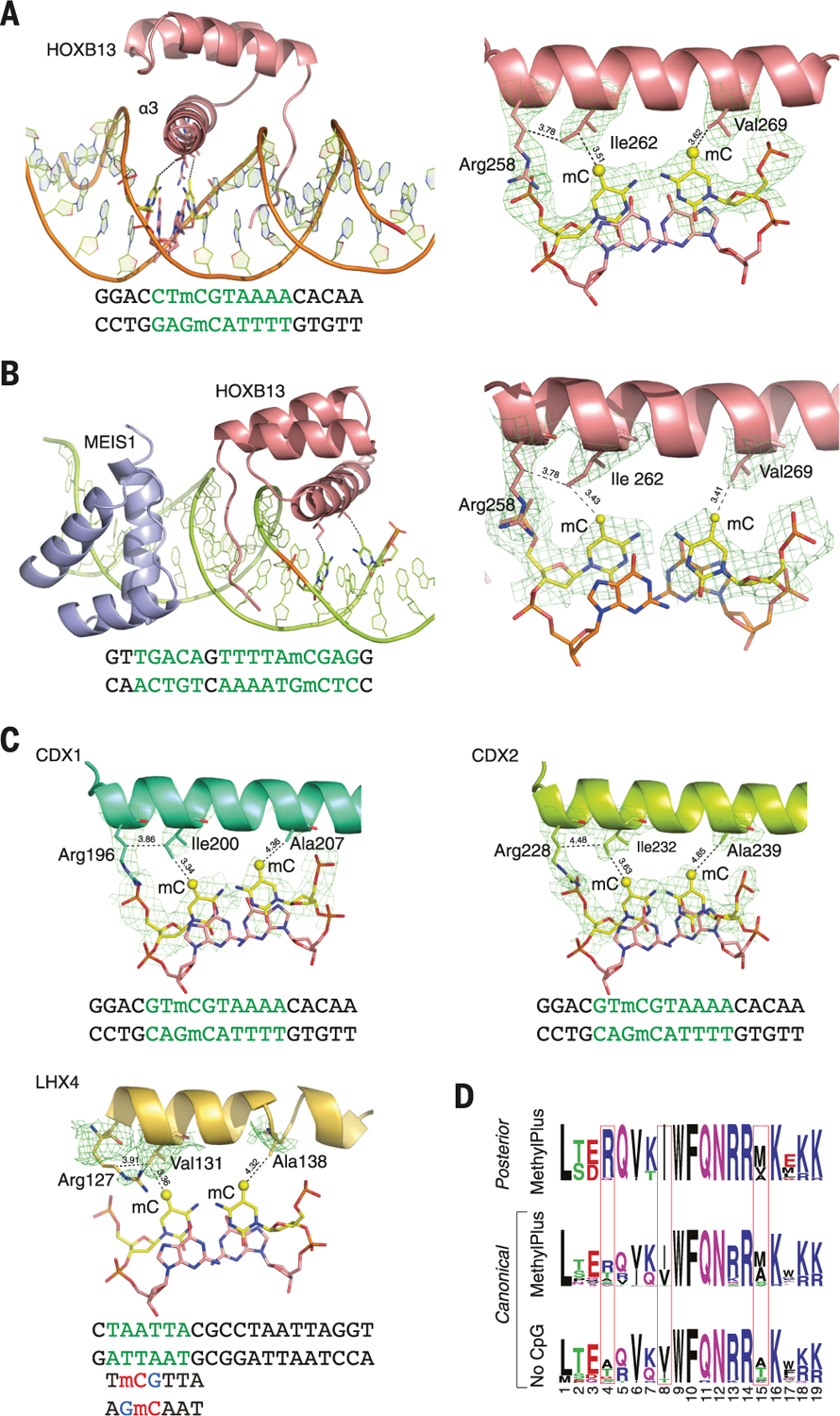
(A) The structure of HOXB13 bound to methylated DNA reveals a mechanism by which posterior homeodomain proteins recognize methylated cytosine. Shown on the left is the overall structure of HOXB13 bound to methylated DNA. Residues that recognize the methylated CpG are shown as ball-and-stick models, and the DNA sequence used in crystallization is presented under the structure. Shown on the right is the composite omit electron density map for the residues of the HOXB13 DBD recognition helix that form hydrophobic interactions with both of the methylated cytosines. The contacts of the model are shown with dashed lines; numbers are distances in angstroms. Ile262 interacts with the mC of the TmCG sequence, whereas Val269 interacts with the mC from the complementary strand. The aliphatic chain of Arg258 also contributes to the local hydrophobic environment. Green letters highlight the bases specifically bound by the TFs. (B) Overview of the HOXB13:MEIS1 heterodimer bound to a methylated DNA. HOXB13 is colored pink, MEIS1 is colored blue, the methylated base pairs are shown as ball-and-stick models, the contacts are presented as dashed lines, and the residues and methylated bases are labeled. Similar to the HOXB13 monomer, the two methylated cytosines are respectively recognized by Ile262 and Val269. (C) Composite omit electron density maps indicate residues of CDX1, CDX2, and LHX4 that recognize mCpG. (D) Sequence logo showing similarity between the strongly methyl-plus posterior homeodomain proteins and canonical homeodomains that prefer or do not bind to mCpG. The identities of the residues at positions identified by the structural analysis (boxes) explain the different preferences of these proteins with respect to mCpG. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C,Cys; D, Asp; E,Glu; F, Phe; G,Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
To determine the thermodynamic parameters of HOXB13 binding to its unmethylated and methylated sites, we performed isothermal titration calorimetry (ITC) experiments. HOXB13 bound to the unmethylated site with a similar change in free energy (ΔG = −10015 cal/mol; fig. S17A) to what has previously been reported for this class of proteins (61). Consistent with the bisulfite-SELEX and methyl-SELEX data, the binding to the methylated site was stronger (ΔG = −10824 cal/mol).
To determine whether the mechanism of recognition of mCpG by homeodomains is general, we solved the structures of three additional homeodomain proteins: CDX1, CDX2, and LHX4 (Fig. 7C; 3.2-, 2.7-, and 2.7-Å resolutions, respectively). The structures of CDX1 and CDX2 bound to their preferred GTmCGTAAA site indicated that they also directly recognize the 5-methyl group of methylcytosine by using amino acids in the same relative positions (Fig. 7C). Whereas the posterior-type paraHOX proteins CDX1 and CDX2 bind strongly to mCpG in their TmCGTAAA motif, LHX4 binds to the canonical TAATTA site and displays somewhat weaker binding to the mCpG-containing sequence TmCGTTA, with no detectable binding to the unmethylated TCGTTA site in methyl-SELEX or ITC experiments (fig. S17C and data S2). The structure of LHX4 bound to the canonical TAATTA site showed hydrophobic residues Val131 and Ala138 in positions suitable for formation of hydrophobic contacts with methylcytosines in both strands of a TCGTTA sequence. The aliphatic chain of Arg127 also supports the hydrophobic interaction (Fig. 7C). These three residues are conserved in all LHX proteins, explaining their strong preference for mC. In contrast, in DLX3, a homeodomain that shows weaker preference for mC, the key residues corresponding to Arg127 and Ala138 were replaced by a threonine and a serine, respectively, leading to a decrease in hydrophobicity (fig. S18A and data S2). Furthermore, in TLX2, which does not bind to TmCGTTA at all, hydrophobicity of the entire binding site was completely lost (fig. S18B and data S2).
Analysis of the amino acid sequences of different homeodomains that either do or do not bind to mCpG-containing sites confirmed a critical role in mCpG recognition for the three residues located at the beginning and end of the recognition helix (Fig. 7D). Analysis of structures of the methyl-plus TFs, including NKX-, IRX-, and NFAT-family proteins, further confirmed that the preference for methylcytosine is based on hydrophobic interactions with its 5-methyl group (fig. S18C). Most TFs that preferred mCpG also bound to sites containing the dinucleotides TG or AA because of the similarity of the shapes of thymine and methylcytosine. Structural analyses also confirmed the mechanism by which methylation of cytosine inhibits TF-DNA binding; in all cases examined, the negative impact of methylation was due to steric hindrance (fig. S18D).
Conclusion
Biochemical studies and experiments using model organisms, cell lines, and in vitro reconstituted systems have resulted in the discovery of the principal mechanisms that control gene expression. The language by which the genome imparts when and where genes are expressed is thus understood at a conceptual level. However, reading the genomic instructions also requires knowledge of binding specificities of all TFs, which constitute the words of the gene regulatory language. In this work, we performed systematic analysis of DNA binding specificities of full-length TFs and eDBDs using unmethylated and CpG-methylated DNA ligands. Compared with our earlier work (26), this analysis used an extended clone collection, and it resulted in identification of binding motifs for 222 TFs for which a HT-SELEX motif was not available. Moreover, an improved computational pipeline (25) allowed us to identify secondary binding profiles for 57 TFs. Taken together, the full data set containing 596 previously unknown motifs (table S2) substantially expands the known lexicon of TF binding specificities.
The results also shed light on the mechanisms of diversification of TF binding specificity during evolution. We found that specific members of three different structural TF families used an AT-hook domain to recognize an AT-rich sequence that flanked the motifs recognized by the TF and its paralogs. This mechanism of diversification of TF specificity is similar to one reported previously (26), whereby an addition of an arginine residue close to the DBD results in preference for an AAAA or TTTT sequence adjacent to the core binding motif. As a whole, these results suggest that TF specificity commonly evolves through addition of simple amino acid features that diversify the sequences recognized by paralogous proteins.
Our work shows that the effect of the most prevalent human epigenetic DNA modification, CpG methylation, on TF binding specificity is more widespread than previously appreciated. In contrast to most previously reported cases [for example, (9, 10, 12, 17, 18)], we found that methylation can affect TF binding positively in many cases [see also (16, 62)]. For example, earlier work has found that only 4% of plant TFs prefer cytosine-methylated sites. This difference from our results is explained by four mechanisms. First, some variance is caused by the fact that the sets of TFs for which data have been obtained in this and previous studies are not complete and do not represent a random sample of the entire TF compendium. Second, all the major families of TFs that prefer mCpG in humans are absent in plants, including canonical homeodomain, POU, and NFAT proteins. Third, plants methylate repeat elements but rarely regulatory sequences. Fourth, non-CpG methylation is very common in plants, and many TFs that O’Malley et al. (63) studied are affected by this modification. We find here that most TFs can bind to a site whose methylation alters the binding affinity of the TF. However, most of these TFs can also bind to sites that do not contain a CpG dinucleotide. Thus, two regulatory elements responding to the same factor can differ in their sensitivity to DNA methylation. In addition, some DNA sequences, such as some motifs containing a CGGAA subsequence, can be bound by TFs that are either negatively (ETS) or positively (NFAT) influenced by methylation. Thus, methylation can act to select the factor that binds to a target sequence. Such a selective effect can have a major impact on transcription, particularly in cases where one factor acts as an activator and the other as a repressor.
Our structural analysis of homeodomains reveals that mCpG is recognized through direct hydrophobic interactions between amino acids and the 5-methyl group of the methylcytosine. Given the structural similarity of thymine and methylcytosine and the identical position of their 5-methyl groups, it is difficult to determine why the preference of TFs for mCpG has evolved. It could represent a simple consequence of high-affinity recognition of thymine. However, several findings suggest that this may not be the whole explanation. For example, all canonical homeodomains prefer similar core sequences containing an AATT sequence. However, their preference for mCpG varies as a function of the hydrophobicity of amino acid residues at three positions. In addition, the strongest preference for mCpG is observed in the posterior homeodomain proteins, a family that has expanded in the vertebrate lineage, where genome-wide DNA methylation is obligatory. Regardless of how the specificity for mCpG has evolved, our results show that it influences binding of TFs to DNA both in vitro and in vivo, leading to changes in the occupancy and activity of TFs based on methylation of their recognition motifs.
In this work, we identified many developmentally important TFs from several TF families that prefer to bind to mCpG, and we also determined the molecular mechanism behind the differential preference of homeodomain proteins for mCpG. TFs that can bind to methylated sequences may be particularly important for reprogramming of cells because CpG methylation functions as a barrier for cellular reprogramming (14, 15). In this regard, it is of particular interest that several factors that regulate ES cell self-renewal—including PRDM4, Nanog, and POU homeodomain factor POU5F1 (OCT4)—are capable of binding to mCpG sites. This may explain in part the ability of POU5F1 to reprogram differentiated cells toward a pluripotent fate (48). Our finding that many TFs prefer methylated CpGs, together with the genome-scale resource on TF binding specificities that we have generated, will be important for future analyses of epigenetic inheritance and transcriptional regulation.
Materials and methods
Clones, protein expression, and purification
Bacterial protein expression Gateway recipient vectors that incorporated a N-terminal Thioredoxin-6×His tag, with either a C-terminal streptavidin binding peptide (SBP) or 3×FLAG tag were constructed using pETG20A-plasmid as a backbone. Inserts for protein expression were derived either by polymerase chain reaction (PCR) from Mammalian gene collection (MGC), ORFeome, Megaman cDNA library, or by gene synthesis (Genscript; see table S1 for protein sequences and domains), or from previously published Gateway donor clones (21).
Protein expression and purification from E. coli cells was performed as described in (64), with the following modification: 30 µM ZnSO4 was included in the culture medium to facilitate expression and folding of zinc finger proteins. The expression of the purified proteins were checked by SDS-PAGE electrophoresis (E-PAGE protein gels, Invitrogen) and Coomassie brilliant blue staining. 50% glycerol was added to the proteins before storage at −20°C. Comparison of results revealed that motifs recovered using such recombinant bacterial proteins were highly similar (fig. S1 and table S2) to those from experiments where TFs were expressed in cultured human cells (21, 26).
HT-SELEX assays
HT-SELEX was performed essentially as described in Nitta et al. (25). Briefly, selection ligands consisting of a 40 bp random sequence flanked by barcodes and Illumina sequencing adapters were generated by primer extension from single-stranded templates. The ligands (~1.5 µg at cycle 0 and ~200 ng thereafter) were then incubated with hexahistidine-thioredoxin tagged purified E. coli recombinant proteins (100 to 200 ng) in microwell plates in the presence of poly dI:dC competitor (75 ng), and the proteins and bound DNA were then recovered by nickel affinity beads. The process was repeated up to four times, and the ligands were amplified by PCR, and sequenced after each cycle. The reactions were performed in a buffer containing somewhat lower salt concentration (4% glycerol, 1 mM DTT, 500 µM EDTA, 50 mM NaCl in 10 mM Tris-Cl, pH 7.5) than that found inside the nucleus in order to prevent excessive dissociation of TFs from DNA during the SELEX wash steps (lower salt strengthens the non-sequence specific ionic interactions between TFs and the DNA backbone). The motifs obtained were similar to those we obtained using physiological level of KCl (140 mM; fig. S19B).
The initial amount of DNA was sufficient to contain the majority of all gapped and ungapped 20 bp sequences. Although not all 40 bp sequences were interrogated due to the limiting amount of DNA, TFs do not generally bind to exact matches of 40 bp (80 bits of information); using the amounts of DNA used, HT-SELEX allows for identification of motifs whose information content is in the order of ~40 bits, exceeding information content of most human TFs [~15 bits (26)]. Additional details of the HT-SELEX method and analysis are available in (21, 26).
In SELEX, the first cycle underestimates affinity due to saturation of high affinity sites by TFs, and the following cycles yield exponential enrichment of the high affinity sites. In late cycles (>4), most sequences will contain a sequence that binds the TF, and high affinity sequences will start to compete out even moderate affinity sites, finally yielding very few individual sequences. We have previously compared PBMs and HT-SELEX with methods that more directly measure (relative) affinity, and the obtained motifs are very similar for PBMs, and for early cycle (2 or 3) SELEX data (21, 26, 33). For this reason, the motifs are generated here using relatively early SELEX cycles (cycle indicated for each motif on table S2). In addition, as the ranks of affinities and enrichment are the same, the precise affinity does not matter for methods, which use a threshold for motif matching. The relative values obtained from SELEX PWMs are, however, only rough estimates of affinity, and calibration of the motifs using standards, and/or methods such as Spec-seq (65) should be used if precise relative affinity values are desired.
Methyl-SELEX
The methyl-SELEX process is based on HT-SELEX (21, 26) with the addition of a DNA methylation step prior to each selection cycle. The CpG methylation is performed by the CpG specific methylase M.SssI. The protocol for methylation was adapted from (11) as follows: 2.5 µl (10U, for initial library) or 1.25 µl (5 U, for cycle 1 to 3) of CpG methyltransferase enzyme M.SssI (NEB; 2-fold excess units in cycle 0 and 10-fold excess thereafter), was added to DNA ligands together with 0.4 µl (for initial library) or 0.2 µl (for cycle 1 to 3) of S-adenosylmethionine, 3.4 µl of 50 mM MgCl2, and 0.2 µl of 100 mM DTT in a total volume of 20 µl. The mixture was incubated at 37°C for 3 hours to methylate the CpG dinucleotides in the double stranded DNA and then at 65°C for 20 min to inactivate the M.SssI enzyme prior to each selection cycle. The methylation reaction was optimized by testing the methylation state of the ligands by the methylation-specific restriction enzyme BstBI (fig. S19A). In addition, in the screen, control methyl-specific TFs (HOXB13 and/or ATFs) were included in each plate.
The CpG methylated DNA ligands and unmethylated DNA ligands were subjected to SELEX assay in parallel, in different 96-well plates for each protein and the selection process was repeated up to 4 cycles to enrich the binding sequences as described in (21, 26). The enriched oligos from all four cycles were subjected to sequencing.
To address the potential effect of salt concentration on hydrophobic interactions, which could affect preference of TFs to mCpG (66), we also performed a control experiment in the presence of 140 mM KCl. The experiment with higher salt concentration had lower yield of successful experiments, but the motifs obtained were not materially different from those obtained using the 50 mM NaCl buffer (fig. S19B).
Bisulfite-SELEX
The methyl-SELEX can be used to classify TFs that bind with some affinity to both sites that contain and do not contain a CpG. In cases where methylation blocks binding to the highest-affinity sites, lower affinity sequences that do not contain CpG will enrich more than in the absence of methylation. The relative depletion of CpG-containing motifs will indicate that the TF is methyl-minus. However, TFs that can only bind to CpG-containing sequences are difficult to classify, as weak binding will still yield a motif, and even TFs that are completely blocked from binding by mCpG may still yield more weakly enriched CpG-containing motif in methyl-SELEX due to incomplete methylation of DNA by M.SssI. For this reason, we developed bisulfite-SELEX, which allows analysis of quantitative effect of CpG methylation in a single SELEX round. In this analysis, the mCpG status is directly measured, and thus partial methylation of DNA by M.SssI cannot affect the results. However, some hemimethylated DNA is likely to exist in the assay. Using bisulfite-SELEX it is possible to specifically analyze hemimethylated DNA as the motif is generated only from one strand of DNA; however, hemimethylation analysis requires generation of hemimethylated DNA by for example performing one cycle of PCR after methylation. This analysis was not performed here, as based on the structural analyses, the effect of hemimethylation is expected to be intermediate between the fully methylated and unmethylated states.
In bisulfite-SELEX, mixed HT-SELEX and methyl-SELEX enriched selection ligands are partially methylated, and subjected to one more round of SELEX. Analysis of the input mixtures and ligands selected in the additional SELEX round using both standard and bisulfite sequencing is then used to determine the preference of each TF toward methylated CpG-containing subsequences.
For bisulfite-SELEX, HT-SELEX and methyl-SELEX processes were performed up to cycle 3 for the proteins with CpG subsequence in their binding sites. The enriched DNA oligos from CpG methylated and unmethylated DNA ligands were then mixed together, after which half of the mixed oligos was subjected to methylation process as describe above and then mixed back with the unmethylated oligos. The mixture of CpG methylated and unmethylated oligos was subsequently subjected to an additional cycle (cycle 4) of the SELEX process and the enriched oligos were eluted in 70 μl of milli-Q water. 13 μl of the elution was amplified by PCR (Phusion DNA polymerase, Fisher Scientific; 65°C for 10 s, 72°C for 36 s, 97°C for 15 s for annealing, elongation and denaturation, respectively, for 20 cycles) and 3 μl aliquot was analyzed by qPCR (Roche LightCycler 480) to monitor progress of the experiment. Subsequently, 40 µl of the elution, and 5 µl of the mixture of CpG methylated and unmethylated oligos from cycle 3 were subjected to bisulfite treatment (EZ-96 DNA Methylation-Gold kit, ZYMO RESEARCH) and amplified by PCR (PfuTurbo Cx Hotstart DNA Polymerase, Agilent Technologies; 60°C for 30 s, 72°C for 60 s, 95°C for 30 s for annealing, elongation and denaturation, respectively, for the first 2 cycles and then 65°C for 30 s, 72°C for 60 s, 95°C for 30 s for the subsequent 13 or 25 cycles). The PCR products of cycle 4 amplified from normal elution and bisulfite-treated elution, the mixture of CpG methylated and unmethylated oligos and the PCR product from bisulfite-treated mixture of CpG methylated and unmethylated oligos from cycle 3 were all subjected to sequencing.
To calculate the percentage of increase of mCpG within motifs, the frequency of dinucleotides at specific positions was determined from subsequences that perfectly matched the bisulfite-SELEX seed (table S3) at all other positions except the dinucleotide position that was interrogated. For positions (indicated in bold in table S3) where either methyl-SELEX or HT-SELEX cycle 3 CpG count was above 10%, the increase of mCpG frequency from cycle 3 to cycle 4 was calculated as follows: fimCG = (fnmCG_cycle4/fnmCG_cycle3 − 1) ×100%. For both cycles 3 and 4, the normalized frequency of methylated fnmCG was calculated from the following equation: fnmCG = sf × fmCG, where size factor sf = 1/(fumCG + fmCG + pseudocount), and frequency of unmethylated CG fumCG = [(fYG − fTG) + (fCG − fmCG)]/2; YG is the TG count after bisulfite treatment (umCGand TG) and pseudocount of 10−9 was included to avoid division by zero.
Generation of PWM models
Binding models for each individual TF from unmethylated and methylated ligands were analyzed using the Autoseed pipeline as described previously (25, 64). Briefly, 8 and 10 bp ungapped subsequences, and subsequences containing a gap in the middle were counted, and similarity between them was analyzed by employing the “Huddinge distance” measure (25). Huddinge distance is defined as d − a, where d is the maximum number of defined bases in either of the two compared subsequences, and a is the maximum number of bases that can be perfectly aligned between them without introduction of new gaps. Initial seeds for each TF were generated using locally maximal subsequences (subsequences with higher count than any of their neighbors at Huddinge distance of 1). Initial PWM models were generated using these seeds using the multinomial method (21), and the seed was subsequently refined by expert analysis [see (26, 64)]. Exact seeds, SELEX cycles, and multinomial models used are indicated in table S2.
TF classification
The classification of each TF to the methyl-plus, methyl-minus, little effect, or multiple effects class was based on bisulfite-SELEX (Fig. 5A and table S3), except for cases where bisulfite experiment had low enrichment, low complexity seed, or no data; in these cases, bisulfite-SELEX data was not considered and the classification is based on methyl-SELEX. The 23 cases where data of bisulfite-SELEX and methyl-SELEX or replicates between two bisulfite-SELEX experiments were inconsistent were not classified and are labeled “inconclusive” in table S3. First, each motif was classified to the multiple effects, methyl-plus, methyl-minus, little effect, or no CpG class. Then, TFs with a single motif were assigned their motif’s class, and TFs with multiple motifs were classified as follows: If a TF had any motif in the multiple effects class, or two or more motifs that displayed different effects toward CpG methylation (different motifs belonged to two or more of the following classes: methyl-plus, methyl-minus, or little effect), it was classified to multiple effects. If a TF had a motif with no CpG, and a motif with CpG(s), it was classified according to the motif with a CpG (see data S2 for details).
In addition, methyl-plus and methyl-minus TFs were subclassified to type A or type B to indicate whether the most enriched sites or the moderately enriched sites (lower than maximum but >10% of max) are affected, respectively. For each TF, this classification was based on the consensus sequence derived from their primary motif (motif with highest count of occurrence). An approach based on a consensus sequence was used instead of kmers of specific length, because motifs of different TFs have different length, and many individual TFs also enrich two or more motifs of different length. If the consensus sequence contained a CpG, the TF was classified to type A, and if not, to type B. Methyl-plus and methyl-minus TFs were classified based on the motifs enriched using the methylated and unmethylated ligands, respectively.
Protein-binding microarrays
For protein-binding microarray analyses, theDBD clones for DLX3, POU5F1, MAX, NFATC2, CUX1, and CUX2 were transferred to pDEST15 with an N-terminal GST tag using gateway LR reaction. DBD of Nkx2.5 was amplified from pDONR clone and cloned into NcoI-SacI restriction sites of pETGEXCT (67). DBD of LHX9 with N-terminal GST tagwas obtained fromDr. Timothy R.Hughes lab (University of Toronto). All clones were sequence verified.
The 16× HK array design with 40,000 unique DNA features [as described in (11, 35)] were double stranded as described in (11). Methylation of the CpG dinucleotides on the double stranded arrays was performed with 10 μl of CpG methyltransferase enzyme M.SssI (20 units/µl) (NEB), 1 μl of S-adenosylmethionine, and 15 μl of 10× NEB Buffer 2. Reaction volume was adjusted to 150 µl with 0.005% Triton X-100 and incubated at 37°C for 3 hours. Addition of Triton X-100 was critical for complete methylation of the array.
The protein binding reactions were carried out as described in (35). Briefly, the double-stranded microarrays were blocked with 4% nonfat dried milk in 1×PBS (Sigma) for 1 hour. Microarrays were then washed once with PBS with 0.1% (vol/vol) Tween-20 for 5 min and once with PBS with 0.01% Triton X-100 for 2 min. DBDs with GST tag were expressed using PURExpress In Vitro Protein Synthesis Kit (NEB) as per manufacturers instructions. 25 μl of IVT reactions were added to make a total volume of 150 μl protein binding reaction containing PBS with 2% (wt/vol) milk, 51.3 ng/μl salmon testes DNA (Sigma), and 0.2 μg/μl bovine serum albumin (NEB), and incubated for 1 hour at 20°C. Preincubated protein binding mixtures were applied to individual chambers of 40K arrays and incubated for 1 hour at 20°C. Microarrays were washed in a Coplin jar once with 0.5% (vol/vol) Tween-20 in PBS for 3 min, once with 0.01% Triton X-100 in PBS for 2 min, and then finally washed with PBS. Alexa Fluor 647-conjugated GST antibody (Invitrogen) was applied to each chamber and incubated for 1 hour at 20°C. Finally,microarrays were washed twice with PBS with 0.05% (vol/vol) Tween-20 for 3 min each, and once in PBS for 2 min. Every protein in this study was assayed in duplicate. Protein-bound microarrays were scanned to detect Alexa Fluor 647-conjugated anti-GST at 640 nm. Microarray images were analyzed using ImaGene (BioDiscovery), and the extracted data were used for further analysis (GEO: GSE94634). To estimate the relative preference for each 8-mer, the Z-score was calculated from the average signal intensity across the 16 or 32 spots containing each 8-mer (11).
Cell culture, cell transduction, ChIP-seq, and ChIP-exo
Tet-TKO (deficient in Tet1, Tet2, and Tet3) was generated in the same manner and genetic background as the Dnmt-TKO line (51) with the following modification: After puromycin selection, clones were originally genotyped based on amplification and RFLP digestion as previously described (68) using the following primer and restriction enzyme combinations: Tet1 (TTGTTCTCTCCTCTGACTGC, TGATTGATCAAATAGGCCTGC, SacI), Tet2 (CAGATGCTTAGGCCAATCAAG, AGAAGCAACACACATGAAGATG, EcoRV), Tet3 (CCACCTCTGAGCGCAGAGTG, GATGAACACAGTTCCTGACAG, XhoI). The positive clone used in these studies had a 9 bp homozygous deletion in Tet1, 8 bp and 10 bp deletions in Tet2, and homozygous 8 bp deletion in Tet3.
Wild-type, Dnmt-TKO (51) (deficient in Dnmt1, Dnmt3a, and Dnmt3b) and Tet-TKO cells were cultured without feeder cells on 0.2% gelatin-coated dishes in DMEM, supplemented with 15% fetal calf serum, 1× non-essential amino acids, 2mM L-glutamine, LIF and 0.001% β-mercaptoethanol (37°C, 7% CO2) (51).
Immortalized human prostate epithelial cells expressing wild-type androgen receptor (LSH-AR) were a kind gift from Prof. William Hahn (Dana-Farber Cancer Institute, Boston). The cells were cultured in PrEBM prostate epithelium basal medium (Lonza) with growth factor supplements supplied as PrEGM SingleQuots (Lonza) and passaged as described previously (69).
The full length HOXB13 ORF was cloned into pLenti6/V5 lentiviral expression vector using gateway recombination system. Viruses were generated by co-transfection of expression vectors with packaging vectors psPAX2 and pMD2.G (Addgene) into 293FT cells with Lipofectamine 2000 (Thermo Fisher Scientific). The following day the cells were replenished with fresh culture media and virus containing media was collected after 48 hours. The virus was concentrated using Lenti-X concentrator (Clontech). The transduction was performed in the presence of 8 µg/ml polybrene. The medium for LSH-AR cells was replaced 16 hours after transduction with fresh medium and was further incubated for 48 hours.
The chromatin immunoprecipitation (ChIP) was performed as previously described with minor modifications (70) by using antibodies for OCT4, KLF4, n-Myc (Abcam cat. no. ab19857 and R&D Systems cat. no. AF1759, AF3158, and AF3640, and Abcam cat. no. ab16898,) in wild-type and defective ES cells or by using antibodies for HOXB13 in transfected LHSAR cells. Briefly, the cells were fixed in 1% formaldehyde for 10 min at room temperature followed by addition of 0.125 M glycine. The cells were washed with ice-cold PBS twice and collected in lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, and 0.5% NP-40). The cell suspension was centrifuged and the pellet resuspended for lysis in 300 µl RIPA buffer (1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS in 1×PBS) containing protease inhibitors (Roche). The chromatin was sonicated to an average fragment size of 100–300 bp using Bioruptor (Diagenode), after which the samples were centrifuged at 13,000 rpm for 15 min at +4 C to collect the supernatant. Dynal protein-G magnetic beads (Invitrogen) were pre-washed with 5mg/ml BSA in PBS for a total of 5 times and resuspended in 100 ul of wash buffer. Antibodies specific for the protein of interest were coupled to magnetic beads overnight with rotation at +4 C. For each immunoprecipitation (IP), 100 µl of sonicated chromatin was diluted 1:10 with 900 µl of RIPA buffer and 10% was stored as input fraction. To the remaining, 100 µl of antibody-coupled magnetic beads were added and incubated on a rotator overnight at 4°C. After incubation, beads were washed 5 times with LiCl wash buffer (100 mM Tris-Cl, pH 7.5, 500 mM LiCl, 1% NP-40, and 1% sodium deoxycholate) and followed by two washes with 10 mM Tris-Cl (pH 8.0) containing 1 mM EDTA. Chromatin-antibody samples were eluted from beads by incubating for 1 hour at 65°C in IP elution buffer (1% SDS, 0.1 M NaHCO3, in 10 mM Tris-Cl, pH 7.5), followed by overnight incubation at 65°C to reverse cross-linking. The eluted DNA was purified using phenol-chloroform followed by library preparation for Illumina sequencing.
LoVo (ATCC, cat. no. CCL229TM) cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics. When the confluence reaches 60 to 70%, ChIP-exo experiments were performed essentially as described by Rhee and Pugh (71) with modifications from Katainen et al. (43) by using antibodies for CEBPB and MAX (and Santa Cruz Biotechnology cat. nos. sc-150 X, sc-197, and Cell Signaling Technology cat. 4732S). ChIP-exo data of KLF5 from LoVo cells were from Katainen et al. (43).
Raw sequencing reads from ChIP libraries were mapped to the human reference genome (hg19) or mouse reference genome (mm9), using bwa (72) with default parameters. For ChIP-exo peak-calling, we used Peakzilla with default parameters (73). For ChIP-seq peak calling, we used MACS (v1.4) (74) with the following parameters: unadjusted P < 10−5; fold change over IgG control ≥2; false discovery rate ≤5%. ChIP-seq data of LoVo, GP5D and VCaP cells used were from Yan et al. (27) and Huang et al. (75). The peak calls were transformed from hg18 to hg19 coordinates by using UCSC liftOver. See table S1 for the sequences of the Illumina sequencing adapters, and table S4 for the number of aligned reads and E-values for the enriched motifs in the peak regions detected by MEME (76) for each experiment. Overlap of peaks was calculated using BEDtools (v2.24.0) requiring minimum of 20% overlap. Overlapping peaks were used for the downstream analyses whenever available (table S4).
To determine whether occupied regions were methylated in vivo, we first identified the best scoring motif-matches within ChIP-exo/seq peak regions and flanking regions that were more than 1 kb but less than 11 kb from the borders of each peak in either direction, and then determined the fraction of methylated cytosine in the motif matches containing a CpG dinucleotide. The motifs enriched from HT-SELEX were used for the TFs in the methyl-minus and little effect categories and from methyl-HT-SELEX for the TFs in the methyl-plus group. Top scoring 300,000 sites recognized by each motif were searched from the human or mouse genome using program MOODS (77) with P value cut-off of 10−4 and score cut-off of 5. The cytosine methylation within CpG subsequence of the best scoring motif-match within peaks and flanking regions was obtained from whole-genome bisulfite sequencing (WGBS) data of the respective cells from which the ChIP-exo/seq data originated. The distributions of cytosine methylation percentages in the peaks and flanking regions were then compared using R histogram plots. All cytosines within CpG subsequence of the best scoring motif-match with ≥2 read coverage were included. However, the results were robust to changes in the cut-off (data S3). OCT4 and n-Myc peaks with motif match from Tet and Dnmt-TKO ChIP-seq data were combined and peak heights were calculated from ChIP-sequencing reads extended by library fragment length using BEDtools genomecov (v2.26.0). Average fragment coverage of the two ChIP-seq experiments for both cells are represented in Fig. 6A and figs. S13D, S14B, and S14C.
Sequencing and bisulfite sequencing
Unselected and selected SELEX libraries were purified by a PCR-purification kit (Qiagen) and sequenced using Illumina HiSeq 2000 [multiplexed 400×, otherwise as in Jolma et al. (21); 55 bp single read length]. Raw sequence reads were demultiplexed, and analyzed using the Autoseed pipeline (25). The range of read counts per cycle and experiment was ~100,000 to 500,000 reads.
The bisulfite-sequencing libraries from GP5d, LoVo, VCaP, LHSAR, wild-type and defective ES cells were prepared as per Illumina’s instructions with minor modifications. The genomic DNA was spiked in with 0.5% unmethylated lambda DNA (Promega) and fragmented in Covaris using the 200-bp target peak size protocol from Covaris. The sonicated DNA samples were end-repaired, dA-tailed and ligated to sequencing adapters. Adapter-ligated DNA fragments were processed for bisulfite conversion using ZYMO EZ DNA Methylation-Gold kit. The bisulfite converted DNA samples were enriched by PCR (4 to 7 cycles). Sequencing was performed with Illumina HiSeq4000 using 100 bp paired-end reads and raw sequencing reads were quality and adapter trimmed with cutadapt version 1.3 in Trim Galore. Low-quality ends trimming was done using Phred score cutoff 30. Adapter trimming was performed using the first 13 bp of the standard Illumina paired-end adapters with default parameters. Read alignment was done against hg19 or mm9 reference genome with Bismark (version v0.10.0) (78) and Bowtie 2 (version 2.2.4) (79). Duplicates were removed using the Bismark deduplicate function. Extraction of methylation calls was done with Bismark methylation extractor discarding first 10 bp of both reads and reading methylation calls of overlapping parts of the paired reads from the first read (–no_overlap parameter). The breadth and depth of WGBS coverage for GP5d, LoVo, VCaP, LHSAR, wild-type and defective ES cells are presented in data S3. Differentially methylated regions were detected with DSS (80). DNAse I hypersensitive sites for mouse ES cells used in fig. S13C were downloaded from ENCODE data at UCSC (wgEncodeUwDnaseEscj7S129ME0PkRep1. narrowPeak), similar to the previous study by Stadler et al. (8) and heatmaps were generated using deepTools (version 2.4.1) (81). All sequence data are deposited in the ENA (European Nucleotide Archive) under accession number PRJEB9797.
ATAC sequencing of ES cells
Open chromatin regions from wild-type and TKO ES cells were captured using assay for transposase-accessible chromatin using sequencing (ATAC-seq). ATAC-seq was essentially performed as described in Buenrostro et al. (82) with the following modifications: Cultured cells (70% confluency)were harvested through trypsinization, resuspended into single cells, washed with ice-cold PBS and centrifuged for 5min at 500 × g. Cell pellet (50,000 cells) was re-suspended in 2× lysis buffer (10mM NaCl, 3mM MgCl2, 0.1% Igepal CA-630 in 10mM Tris-HCl, pH 7.5) and nuclei were pelleted by centrifugation for 30 min at 500 × g at 4°C using a swinging bucket rotor with low acceleration and brake settings. Supernatant was discarded and nuclei were subjected to tagmentation reaction in 25 µl volume containing 2 µl of Tn5 transposase and 12.5 µl of 2× TD buffer (Nextera DNA Library Prep Kit from Illumina, cat. no. FC-121–1030). Tagmentation was performed in shaking incubator (650 rpm) at 37°C for 1 hour. After tagmentation, 5 µl of clean-up buffer (900mMNaCl, 300 mM EDTA), 2 µl of 5% SDS and 2 µl of ProteinaseK (Thermo Fisher Scientific, cat. no.EO0491) were added and reaction further incubated at 40°C for 30minwith vigorous shaking (650 rpm). Tagmented DNA was isolated using 2× Agencourt AMPure XP SPRI beads (Beckman Coulter, cat. no. A63881) and DNA eluted with 22.5 µl of elution buffer (10 mM Tris-Cl, pH 8.0). Library amplification was performed using two sequential PCRs of 6 and 8 cycles, respectively. After the first PCR, library size selection (for fragments less than 800 bp) was performed using reverse phase 0.55X AMPure XP SPRI beads, the supernatant was collected and DNA isolated using MinElute PCR purification kit (QIAGEN, cat. no. 28004). Both sequential PCRs were performed (Nextera DNA Library Prep Reference Guide from Illumina) with 2 µl of indexing primers (Nextera Index Kit from Illumina, cat. no. FC-121–1011) and KAPA HiFi HotStart ReadyMix (Kapa Biosystems, KK2601). Final DNA concentrations were measured with Qubit 3.0 Fluorometer (Thermo Fisher Scientific) and the library size was determined using 2100 Bioanalyzer (Agilent Technologies). Single-end 55 bp sequencing was performed using the Illumina HiSeq 4000 according to the manufacturer’s instructions.
Raw sequencing reads were mapped to the mouse reference genome (mm9) using bwa (72) with default parameters. Duplicates were removed using Picard Tools MarkDuplicates and open chromatin regions were detected using MACS2 (version 2.0.9) (74) broad peak calling with default parameters. ATAC-seq alignment and peak calling statistics are summarized in table S4. Overlapping peaks (minimum of 20% overlap) from two replicates were used for the downstream analyses.
Analysis of motif conservation and similarity
To determine whether matches to the motifs were conserved, the genomic sites matching a methyl-SELEX or HT-SELEX motif were analyzed according to (64) and the procedure detected genomic conservation for 598 out of 900 motifs (66.4%) at family-wise error rate <0.05. Briefly, twenty thousand top affinity sites for each motif was selected from the human constrained elements (not full genome, only sequences conserved in mammals) and checked for conservation in 99 vertebrate species (multiple alignment downloaded from UCSC genome browser, version hg19) according to the motif as explained in (64). To compare the motifs to the substitution patterns at conserved binding sites, the top thousand highest affinity conserved sites, or all conserved sites if there were less than thousand sites, were selected for further analysis. For each position at a conserved binding site the SiPhy program (task 7) was used to estimate the position-specific equilibrium base distribution π based on the multiple alignment at that position. The base distribution π describes the evolutionary constraint acting on the position assuming that positions evolve independently (83). The conservation pattern was constructed by averaging the π frequencies at each position across the conserved sites. The obtained multiple hypothesis testing corrected P values (Holm’s method) and the number of conserved motif sites among tested sites are shown in fig. S12.
This analysis tends to underestimate the conservation of CpG-containing sequences due to the high mutational load on methylated CpGs. We did not, however, correct for the mutation rate, in order to avoid making the measured variable dependent on the correction term. The difference logo between HT-SELEX motif and the conservation pattern was made by subtracting the corresponding base frequencies.
Motif similarity was calculated using SSTAT (42) with a stringent type I error threshold 0.01 to limit the effect of low affinity sites (other parameters 50% GC-content background model, pseudocount regularization). We have previously reported that this approach generally gives similar results as other common methods but performs better when two otherwise dissimilar motifs share a common part (26). Binding models were then connected to each other if their SSTAT similarity score was > 1.5 × 10−5. Minimum dominating set of the resulting network was then used to select the representative PWMs (25, 26). The minimum dominating set is the smallest set of PWMs that are directly connected in the similarity network to all of the PWMs of the original set.
Enrichment analyses
Enrichment analyses were performed by considering the collection of TFs for which we obtained a motif as the reference population to avoid sampling bias. As SELEX enrichment was performed using both methylated and unmethylated ligand, we do not expect that the population would be biased specifically between these two classes. GO enrichment analysis was performed using PANTHER (84) Overrepresentation Test (release 2016/07/15) with annotation version 11.1.
Structural analyses and isothermal titration calorimetry
Expression and purification of the DNA binding domain fragment of human LHX4 (residues 153–220), HOXB13 (residues 209–283), MEIS1 (residues 279–333), CDX1 (residues 154–217), and CDX2 (residues 169–262) was performed as described in (85). The DNA fragments used in crystallization were obtained as single stranded oligonucleotides (Eurofins MWG), and annealed in 20 mM HEPES (pH 7.5) containing 150 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP) and 5% glycerol. The purified and concentrated protein was first mixed with a solution of annealed DNA duplex at a molar ratio of 2:1.2 for LHX and 1:1.2 for HOXB13, CDX1 and CDX2, and 1:1:1.2 for HOXB13:MEIS1 complex, and after 1 hour on ice was subjected to the crystallization trials. The crystallization conditions were optimized using Jena Bioscience JBScreen Nuc-Pro HTS (Jena Bioscience, Jena, Germany). All protein-DNA complexes were crystalized in sitting drops by vapor diffusion technique at room temperature. LHX4-DNA was crystallized from solution containing 50 mM Tris buffer (pH 8.0), 40 mM magnesium formate, 30% (w/v) of polyethylene glycol monomethyl ether [PEGmme (5000)] and 4% of 2-Methyl-2,4-pentanediol (MPD). HOXB13-MethDNA complex was crystallized from the solution containing 50 mM Tris buffer (pH 8.0), 100 mM MgCl2, 150 mM KCl, 24% (w/v) PEG(3350) and 8% of methyl propanol. CDX1-MethDNA and CDX2-MethDNA complexes were crystalized from the solution containing 50 mM Tris buffer (pH 8.0), 100 mM MgCl2, 150 mM KCl, 28.8% (w/v) PEGmme(5000) and 5% of MPD for CDX1_methDNA or 8% of polyethylene glycol (400) for CDX2_methDNA. The crystals of heterodimeric complex of HOXB13, MEIS1 with methylated DNA were grown from solution containing from 50 mM Tris buffer (pH 8.0), 100 mM MgCl2, 150 mM KCl, 24% (w/v) PEG(3350) and 8% of 1-pentanol.
All data sets were collected at European Synchrotron Radiation Facilities (ESRF, Grenoble, France) from a single crystal on beamlines ID29 (LHX4 and HOXB13:MEIS1_methDNA) and ID23–1 (HOXB13_methDNA, CDX1_methDNA and CDX2_methDNA) at 100 K using the reservoir solution as a cryo-protectant. The data collection strategy was optimized with the program BEST (86). Data were integrated with the program XDS (87) and scaled with SCALA (88). Statistics of data collection are presented in table S6.
All structures were solved by molecular replacement technique using program Phaser (89) as implemented in Phenix (90) and CCP4 (88), and Molrep under CCP4 program suite. The structure of HOXB13:MEIS1_methDNA was solved first by molecular replacement using program Phaser with the structure of MEIS1 (PDB entry 4XRM) as a search model. The found solution for MEIS1 was fixed and the coordinates of HOXA9 from HOXA9/PBX1 complex (PDB entry 1PUF) were used to determine the position of HOXB13. After the positioning of both proteins the density of DNA was clear and the molecule was built manually using COOT (91, 92). The rigid body refinement with REFMAC5 was followed by restrain refinement with REFMAC5, as implemented in CCP4 and Phenix.refine (93). The refined model of HOXB13 was later used as a search model to determine the structure of HOXB13 with the methylated DNA as well as for the solving of CDX1_methDNA and CDX2_methDNA structures (sequence identity is 43%). The parts of DNA interacting with a protein were well visible and built manually to the appeared electron density and refined.
The search model for LHX4 was the structure of homeodomain of the rat insulin gene enhancer protein ISL-1 that shows 48% of identity with DBD of LHX4 (PDB entry 1BW5) as a search model. The electron density of the second subunit of LHX4-DNA complex was poorly visible at this stage. The resulted model contained LHX4 and part of DNA was fixed and the position of the second LHX4 molecule was determined by program Molrep with implemented Spherically Averaged Phased Translation Function (SAPTF) as described in (94). After the positioning of both protein molecules the density of DNA was clear and the DNA molecule was built manually using COOT (91, 92).
The resulted models containing one DNA and two protein molecules was refined with Phenix. refine (93) using TLS function. The resulting density of the second subunit is much weaker than the presented one for the first subunit due to the lack of packing contacts. The first 3 and last 4 amino acids from N- and C- termini of the first subunit as well as 7 first amino acids of N-terminal and 4 last of the C-terminal of second one were found disordered and were not included into the final model. The refinement statistics are presented in table S6. The experimental data and atomic coordinates have been submitted to the Protein Data Bank with accession codes 5HOD (LHX4), 5EF6 (HOXB13_methDNA), 5EGO (HOXB13:MEIS1_methDNA), 5LUX (CDX1_methDNA), and 5LTX (CDX2_methDNA).
In order to determine the affinities of ONECUT2, LHX4, HOXA11 and HOXB13 DBDs to their respective methylated and nonmethylated DNA motifs, isothermal titration calorimetry experiments were carried out using an ITC200 microcalorimeter (MicroCal, Northampton, Massachusetts, USA) in PSF (Protein Science Facility at Karolinska Institute, Sweden, and GE Healthcare, Sweden). Binding isotherms of DNAs were measured by direct titration of protein to the cell containing the indicarted double-stranded DNA ligands. The measurements were taken at 20°C. Both protein and DNA were prepared in a buffer containing 20mM HEPES pH7.5, 300mM NaCl, 10% Glycerol and 0.5 mM TCEP. A total of 20 injections were made with 240 s between injections. All data were evaluated using the OriginPro 7.0 software package (Microcal) supplied with the calorimeter. The apparent dissociation constant Kd, binding enthalpy ΔH- and stoichiometry n, together with their corresponding standard deviations, were determined by a nonlinear least-squares fit of the data to standard equations for the binding using a model for one set of independent and identical binding sites as implemented in the package. The entropy and free energy of binding were obtained from the relation ΔG = −RTlnKd = ΔH − TΔS.
Luciferase assays
Oligonucleotides containing 8 binding sites of HOXC11 were synthesized from EurofinsGenomics Company (table S1) and cloned into pCpGfree-promoter reporter plasmid (InvivoGen), which contains reporter gene (secreted luciferase Lucia) and is completely devoid of CpG dinucleotides, by using Sbf I and Spe I restriction enzymes. The cytosines in the reconstructed pCpGfree_promotor plasmids were methylated by using the CpG specific methylase M.SssI. The plasmids with methylated or unmethylated CpG sites were transfected into HEK293FT cells together with pCDNA3.1–3×FLAG-V5 expression vector containing HOXC11 gene (gift from Mikko Taipale, University of Toronto) and pRL-TK vector containing Renilla Luciferase (Promega) by using FuGENE HD Transfection Reagent (Promega). The Lucia and Renilla Luciferase activities were measured after 36 hours by the Dual-Glo Luciferase Assay System (Promega) and EnVision Multi-label Reader (PerkinElmer).
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Linnarsson, N. Zielke, B. Schmierer, F. Zhu, and J. Zhang for critical review of the manuscript; S. Augsten and the Karolinska Institutet Protein Science Facility for protein production and the opportunity to run ITC experiments; and L. Hu, A. Zetterlund, and M. Hoh for technical assistance. Illumina sequencing reads were deposited in the European Nucleotide Archive under accession number PRJEB9797. The experimental data and atomic coordinates have been submitted to the Protein Data Bank with accession codes 5HOD (LHX4), 5EF6 (HOXB13_methDNA), 5EGO (HOXB13: MEIS1_methDNA), 5LUX (CDX1_methDNA), and 5LTX (CDX2_methDNA). This work was supported by the Academy of Finland Center of Excellence in Cancer Genetics, the European Research Area SynBio project MirrorBio, the Center for Innovative Medicine at Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Göran Gustafsson Foundation, and Vetenskapsrådet. Y.Y., A.J., K.R.N., and J.T. designed the experiments. Y.Y. performed the HT-SELEX, methyl-SELEX, and bisulfite-SELEX experiments. Y.Y. and B.S. performed the ChIP-seq experiment, K.D. performed the ChIP-exo and ATAC-seq experiments, and B.S. performed the whole-genome bisulfite sequencing experiments. S.K.-S. performed the protein-binding microarray experiment. P.K.D. purified proteins for crystallography, E.M. and A.P. performed the x-ray crystallography experiments, and E.M. solved the protein structures. P.A.G. and S.D. generated Tet-TKO ES cells. Y.Y., E.K., K.R.N., A.J., F.Z., T.K., and J.T. contributed to computational analysis. Y.Y., E.K., and K.R.N. prepared the illustrations. Y.Y. and J.T. wrote the manuscript. All authors contributed to data analysis and reviewed the manuscript.
REFERENCES AND NOTES
- 1.Huff JT, Zilberman D, Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156, 1286–1297 (2014). doi: 10.1016/j.cell.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly TK et al. , Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 22, 2497–2506 (2012). doi: 10.1101/gr.143008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird A, DNA methylation patterns and epigenetic memory. Genes Dev 16, 6–21 (2002). doi: 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 4.Ball MP et al. , Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol 27, 361–368 (2009). doi: 10.1038/nbt.1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R et al. , Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). doi: 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hon GC et al. , Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet 45, 1198–1206 (2013). doi: 10.1038/ng.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schübeler D, Function and information content of DNA methylation. Nature 517, 321–326 (2015). doi: 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 8.Stadler MB et al. , DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011). doi: 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]
- 9.Gaston K, Fried M, CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res 23, 901–909 (1995). doi: 10.1093/nar/23.6.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iguchi-Ariga SM, Schaffner W, CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 3, 612–619 (1989). doi: 10.1101/gad.3.5.612 [DOI] [PubMed] [Google Scholar]
- 11.Mann IK et al. , CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res 23, 988–997 (2013). doi: 10.1101/gr.146654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watt F, Molloy PL, Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev 2, 1136–1143 (1988). doi: 10.1101/gad.2.9.1136 [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, Bird AP, Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci 31, 89–97 (2006). doi: 10.1016/j.tibs.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 14.Looney TJ et al. , Systematic mapping of occluded genes by cell fusion reveals prevalence and stability of cis-mediated silencing in somatic cells. Genome Res 24, 267–280 (2014). doi: 10.1101/gr.143891.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polo JM et al. , A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632 (2012). doi: 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rishi V et al. , CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A 107, 20311–20316 (2010). doi: 10.1073/pnas.1008688107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campanero MR, Armstrong MI, Flemington EK, CpG methylation as a mechanism for the regulation of E2F activity. Proc. Natl. Acad. Sci. U.S.A 97, 6481–6486 (2000). doi: 10.1073/pnas.100340697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comb M, Goodman HM, CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res 18, 3975–3982 (1990). doi: 10.1093/nar/18.13.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S et al. , DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726 (2013). doi: 10.7554/eLife.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spruijt CG et al. , Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013). doi: 10.1016/j.cell.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 21.Jolma A et al. , Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res 20, 861–873 (2010). doi: 10.1101/gr.100552.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliphant AR, Struhl K, Defining the consensus sequences of E.coli promoter elements by random selection. Nucleic Acids Res 16, 7673–7683 (1988). doi: 10.1093/nar/16.15.7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM, A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet 10, 252–263 (2009). doi: 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ et al. , DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19, 667–678 (2005). doi: 10.1016/j.molcel.2005.07.021 [DOI] [PubMed] [Google Scholar]
- 25.Nitta KR et al. , Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 4, e04837 (2015). doi: 10.7554/eLife.04837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolma A et al. , DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). doi: 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 27.Yan J et al. , Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813 (2013). doi: 10.1016/j.cell.2013.07.034 [DOI] [PubMed] [Google Scholar]
- 28.Meng X, Brodsky MH, Wolfe SA, A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol 23, 988–994 (2005). doi: 10.1038/nbt1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orenstein Y, Shamir R, A comparative analysis of transcription factor binding models learned from PBM, HT-SELEX and ChIP data. Nucleic Acids Res 42, e63 (2014). doi: 10.1093/nar/gku117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farh KK et al. , Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). doi: 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farley EK et al. , Suboptimization of developmental enhancers. Science 350, 325–328 (2015). doi: 10.1126/science.aac6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vierstra J et al. , Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012 (2014). doi: 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei GH et al. , Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J 29, 2147–2160 (2010). doi: 10.1038/emboj.2010.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer LA, Latek RR, Peterson CL, The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol 5, 158–163 (2004). doi: 10.1038/nrm1314 [DOI] [PubMed] [Google Scholar]
- 35.Badis G et al. , Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009). doi: 10.1126/science.1162327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML, UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res 43, D117–D122 (2015). doi: 10.1093/nar/gku1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weirauch MT et al. , Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014). doi: 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrington MA, Jones PA, Imagawa M, Karin M, Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. U.S.A 85, 2066–2070 (1988). doi: 10.1073/pnas.85.7.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Höller M, Westin G, Jiricny J, Schaffner W, Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev 2, 1127–1135 (1988). doi: 10.1101/gad.2.9.1127 [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee R et al. , High-resolution genome-wide DNA methylation maps of mouse primary female dermal fibroblasts and keratinocytes. Epigenetics Chromatin 7, 35 (2014). doi: 10.1186/1756-8935-7-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y et al. , Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res 42, 4859–4867 (2014). doi: 10.1093/nar/gku134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pape UJ, Rahmann S, Vingron M, Natural similarity measures between position frequency matrices with an application to clustering. Bioinformatics 24, 350–357 (2008). doi: 10.1093/bioinformatics/btm610 [DOI] [PubMed] [Google Scholar]
- 43.Katainen R et al. , CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet 47, 818–821 (2015). doi: 10.1038/ng.3335 [DOI] [PubMed] [Google Scholar]
- 44.Bogani D et al. , The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol. Cell. Biol 33, 3936–3950 (2013). doi: 10.1128/MCB.00498-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kmita M, Duboule D, Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333 (2003). doi: 10.1126/science.1085753 [DOI] [PubMed] [Google Scholar]
- 46.McMahon AP, Neural patterning: The role of Nkx genes in the ventral spinal cord. Genes Dev 14, 2261–2264 (2000). doi: 10.1101/gad.840800 [DOI] [PubMed] [Google Scholar]
- 47.Pearson JC, Lemons D, McGinnis W, Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet 6, 893–904 (2005). doi: 10.1038/nrg1726 [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). doi: 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto H et al. , Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 28, 2304–2313 (2014). doi: 10.1101/gad.250746.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y et al. , WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell 57, 662–673 (2015). doi: 10.1016/j.molcel.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domcke S et al. , Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579 (2015). doi: 10.1038/nature16462 [DOI] [PubMed] [Google Scholar]
- 52.Khund-Sayeed S et al. , 5-Hydroxymethylcytosine in E-box motifs ACAT|GTG and ACAC|GTG increases DNA-binding of the B-HLH transcription factor TCF4. Integr. Biol 8, 936–945 (2016). doi: 10.1039/C6IB00079G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu M et al. , Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012). doi: 10.1016/j.cell.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pomerantz MM et al. , The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet 47, 1346–1351 (2015). doi: 10.1038/ng.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hovde S, Abate-Shen C, Geiger JH, Crystal structure of the Msx-1 homeodomain/DNA complex. Biochemistry 40, 12013–12021 (2001). doi: 10.1021/bi0108148 [DOI] [PubMed] [Google Scholar]
- 56.Joshi R et al. , Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131, 530–543 (2007). doi: 10.1016/j.cell.2007.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaRonde-LeBlanc NA, Wolberger C, Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev 17, 2060–2072 (2003). doi: 10.1101/gad.1103303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK, Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397, 714–719 (1999). doi: 10.1038/17833 [DOI] [PubMed] [Google Scholar]
- 59.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C, Structure of a HoxB1-Pbx1 heterodimer bound to DNA: Role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96, 587–597 (1999). doi: 10.1016/S0092-8674(00)80662-5 [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Larsen CA, Stadler HS, Ames JB, Structural basis for sequence specific DNA binding and protein dimerization of HOXA13. PLOS ONE 6, e23069 (2011). doi: 10.1371/journal.pone.0023069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dragan AI et al. , Forces driving the binding of homeodomains to DNA. Biochemistry 45, 141–151 (2006). doi: 10.1021/bi051705m [DOI] [PubMed] [Google Scholar]
- 62.Quenneville S et al. , In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372 (2011). doi: 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Malley RC et al. , Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). doi: 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jolma A et al. , DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388 (2015). doi: 10.1038/nature15518 [DOI] [PubMed] [Google Scholar]
- 65.Stormo GD, Zuo Z, Chang YK, Spec-seq: Determining protein-DNA-binding specificity by sequencing. Brief. Funct. Genomics 14, 30–38 (2015). doi: 10.1093/bfgp/elu043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khrapunov S et al. , Unusual characteristics of the DNA binding domain of epigenetic regulatory protein MeCP2 determine its binding specificity. Biochemistry 53, 3379–3391 (2014). doi: 10.1021/bi500424z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharrocks AD, A T7 expression vector for producing N- and C-terminal fusion proteins with glutathione S-transferase. Gene 138, 105–108 (1994). doi: 10.1016/0378-1119(94)90789-7 [DOI] [PubMed] [Google Scholar]
- 68.Wang H et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berger R et al. , Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res 64, 8867–8875 (2004). doi: 10.1158/0008-5472.CAN-04-2938 [DOI] [PubMed] [Google Scholar]
- 70.Sahu B et al. , Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J 30, 3962–3976 (2011). doi: 10.1038/emboj.2011.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rhee HS, Pugh BF, Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011). doi: 10.1016/j.cell.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bardet AF, He Q, Zeitlinger J, Stark A, A computational pipeline for comparative ChIP-seq analyses. Nat. Protoc 7, 45–61 (2012). doi: 10.1038/nprot.2011.420 [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Q et al. , A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet 46, 126–135 (2014). doi: 10.1038/ng.2862 [DOI] [PubMed] [Google Scholar]
- 76.Bailey TL, Williams N, Misleh C, Li WW, MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34, W369–W373 (2006). doi: 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korhonen J, Martinmäki P, Pizzi C, Rastas P, Ukkonen E, MOODS: Fast search for position weight matrix matches in DNA sequences. Bioinformatics 25, 3181–3182 (2009). doi: 10.1093/bioinformatics/btp554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krueger F, Andrews SR, Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). doi: 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu H et al. , Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res 43, e141 (2015). doi: 10.1093/nar/gkv715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramírez F et al. , deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165 (2016). doi: 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). doi: 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garber M et al. , Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–i62 (2009). doi: 10.1093/bioinformatics/btp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mi H et al. , PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189 (2017). doi: 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Savitsky P et al. , High-throughput production of human proteins for crystallization: The SGC experience. J. Struct. Biol 172, 3–13 (2010). doi: 10.1016/j.jsb.2010.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bourenkov GP, Popov AN, A quantitative approach to data-collection strategies. Acta Crystallogr. D 62, 58–64 (2006). doi: 10.1107/S0907444905033998 [DOI] [PubMed] [Google Scholar]
- 87.Kabsch W, XDS. Acta Crystallogr. D 66, 125–132 (2010). doi: 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winn MD et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011). doi: 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCoy AJ et al. , Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). doi: 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adams PD et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Emsley P, Cowtan K, Coot: Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 92.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012). doi: 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vagin AA, Isupov MN, Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D 57, 1451–1456 (2001). doi: 10.1107/S0907444901012409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


