Abstract
Background:
Per- and polyfluoroalkyl substances (PFASs) exposure is ubiquitous among the US population and has been linked to adverse health outcomes including cardiometabolic diseases, immune dysregulation and endocrine disruption. However, the metabolic mechanism underlying the adverse health effect of PFASs exposure is unknown.
Objective:
The aim of this project is to investigate the association between PFASs exposure and altered metabolic pathways linked to increased cardiometabolic risk in young adults.
Methods:
A total of 102 young adults with 82% overweight or obese participants were enrolled from Southern California between 2014 and 2017. Cardiometabolic outcomes were assessed including oral glucose tolerance test (OGTT) measures, body fat and lipid profiles. High-resolution metabolomics was used to quantify plasma exposure levels of three PFAS congeners and intensity profiles of the untargeted metabolome. Fasting concentrations of 45 targeted metabolites involved in fatty acid and lipid metabolism were used to verify untargeted metabolomics findings. Bayesian Kernel Machine Regression (BKMR) was used to examine the associations between PFAS exposure mixture and cardiometabolic outcomes adjusting for covariates. Mummichog pathway enrichment analysis was used to explore PFAS-associated metabolic pathways. Moreover, the effect of PFAS exposure on the metabolic network, including metabolomic profiles and cardiometabolic outcomes, was investigated.
Results:
Higher exposure to perfluorooctanoic acid (PFOA) was associated with higher 30-minute glucose levels and glucose area under the curve (AUC) during the OGTT (p < 0.001). PFAS exposure was also associated with altered lipid pathways, which contributed to the metabolic network connecting PFOA and higher glucose levels following the OGTT. Targeted metabolomics analysis indicated that higher PFOA exposure was associated with higher levels of glycerol (p = 0.006), which itself was associated with higher 30-minute glucose (p = 0.006).
Conclusions:
Increased lipolysis and fatty acid oxidation could contribute to the biological mechanisms linking PFAS exposure and impaired glucose metabolism among young adults. Findings of this study warrants future experimental studies and epidemiological studies with larger sample size to replicate.
Keywords: Perfluoroalkyl substances, Cardiometabolic dysfunction, Metabolomics, β-oxidation, Lipolysis, Young adults
1. Introduction
Metabolic dysfunction including dysregulated glucose and lipid metabolism in young adults has far-reaching impact on morbidity and all-cause mortality in later adulthood (Juonala et al., 2016; Morrison et al., 2007). Beyond well-known risk factors such as unhealthy diet and physical inactivity, there is growing concern over environmental exposures to endocrine disrupting chemicals including per- and polyfluoroalkyl substances (PFASs) for their influence on cardiometabolic health (Sunderland et al., 2019; Lee, 2018).
PFASs have been used for decades as industrial surfactants in textile coatings, fire-fighting foams and many consumer products (Grandjean and Clapp, 2014). Ingestion of contaminated water and food are thought to be the main contributors to non-occupational PFAS exposures in humans (Domingo, 2012; Jain, 2014). A recent study has shown ubiquitous exposures to major PFASs, such as perfluorooctanoic acid (PFOA), perfluorooctane sulfonate (PFOS), and perfluorohexane sulfonic acid (PFHxS) among the US population (Kato et al., 2011). Perinatal and/or early postnatal exposures to PFASs induce insulin resistance and impaired glucose tolerance in rodents (Hines et al., 2009; Lv et al., 2013). Epidemiological studies have also shown that higher peripheral concentrations of PFASs are associated with increased type 2 diabetes risk in children (Domazet et al., 2016; Alderete et al., 2019) and adults (He et al., 2018; Cardenas et al., 2019).
Animal studies suggest that PFASs may act on various nuclear receptors such as peroxisome proliferator-activated receptors (PPARs) and alter glucose and lipid metabolism (Le Magueresse-Battistoni et al., 2017; Li et al., 2019). However, the mechanism of linking PFAS exposure and metabolic dysfunction in human is unclear. Recent metabolomics studies in adults and children have found that PFAS exposure is associated with dysregulated metabolism in glycerophospholipids, fatty acids and amino acids (Alderete et al., 2019; Lin et al., 2019). However, there are knowledge gaps about the contribution of altered metabolic pathways to the association between PFAS exposure and metabolic dysfunction in young adults. Therefore, the overall aim of this project was to identify key metabolic pathways that link PFAS exposure and adverse effects on obesity and cardiometabolic outcomes in young adults by using advanced metabolomics approaches.
2. Methods
2.1. Study design and recruitment
Participants of this study comprised of 103 young adults (age 17–22 years), who were enrolled in the original Meta-AIR study18 between 2014 and 2017. Details of the study design and methods of the Meta-AIR study are described elsewhere (Kim et al., 2019). All Meta-AIR participants were part of the larger Southern California Children’s Health Study (CHS) (Chen et al., 2015), which recruited children from schools across Southern California communities and followed them from kindergarten or first grade (starting in year 2002) through high school graduation. All subjects of the Meta-AIR study had a history of being overweight or obese (age- and sex-specific BMI percentiles ≥85th) during their high school years in 2011–2012. Participants who had diabetes or any major illness since birth were excluded from the study. By October 2017, there were 103 participants enrolled in the Meta-AIR study, which were selected for high-resolution untargeted metabolomics analysis, supported by the Children’s Health Exposure Analysis Resource (CHEAR) Program. Written informed assents and consents were obtained from study participants. The Institutional Review Board at the University of Southern California (USC) approved this study.
The Meta-AIR study visit included extensive phenotyping of obesity and cardiometabolic outcomes as well as health and lifestyle questionnaires conducted at the USC Diabetes and Obesity Research Institute and the Clinical Trials Unit. Details of clinical measures and health questionnaire data collected during the study visit are described in the Supplementary Material.
2.2. Glucose and insulin traits
All participants completed a 2-hour OGTT, and blood samples were collected during the OGTT for glucose and insulin measures. Following a minimum 10-hour fast, a 2-hour oral glucose tolerance test (OGTT) was administered using a load of anhydrous glucose dissolved in water for 1.75 g per kilogram of body weight with a max dose of 75 g. All participants received the maximum glucose load. Blood glucose and insulin samples were collected at fasting (pre-glucose load) and then post glucose challenge at 30-, 60-, 90-, and 120-minutes. OGTT-derived outcomes analyzed in this study included fasting and post glucose challenge at 30- and 120-minute glucose and insulin levels, glucose and insulin area under the curve (AUC), homeostatic model assessment for insulin resistance (HOMA-IR) (Matthews et al., 1985), and the Matsuda Index for insulin sensitivity (Matsuda and DeFronzo, 1999). More details about OGTT-derived measures are described in Supplementary Material.
2.3. Laboratory analysis
Plasma samples were assayed for glucose concentration by hexokinase-mediated reaction assay run on Roche Covas C501. Insulin concentration in plasma samples were measured by Human Insulin ELISA Kit (EZHI-14BK). Four quality control (QC) plasma samples were added to each analytical plate across a total of 6 batches. The coefficient of variation (CV) of insulin concentrations across QC samples of all batches had a median of 0.08 (range: 0.03, 0.16). Lipids profiles (triglycerides, total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), and very-low-density lipoprotein (VLDL) cholesterols) were assessed from fasting serum samples using Fujifilm Wako Diagnostics enzymatic assay. More details are described in the Supplementary Material.
2.4. Adiposity measures
Several anthropometric and body composition measures were taken to estimate adiposity: 1) body mass index (BMI) = weight/height2 (kg/m2); 2) dual-energy X-ray absorptiometry (DEXA) scan to determine total body fat percent; and 3) 3 T magnetic resonance imaging (MRI) abdominal scan to determine subcutaneous abdominal adipose tissue (SAAT), visceral adipose tissue (VAT), and hepatic fat fraction (HFF). The ratio of VAT to SAAT (VAT-SAAT ratio) was further calculated (Kaess et al., 2012). Obesity was defined as BMI ≥ 30 kg/m2, overweight was defined as BMI ≥ 25 kg/m2 and normal weight was defined as BMI < 25.0 kg/m2.
2.5. Untargeted metabolomics profiles
Untargeted metabolomics was used to characterize plasma samples collected at fasting and post glucose challenge at 30 min using established methods (Liu et al., 2016; Valvi et al., 2020). Plasma samples were treated with acetonitrile containing the following 14 internal standards that were selected to include a range of chemical properties: [13C6)-D-glucose, [15 N]-indole, [2-15N]-L-lysine dihydrochloride, [13C5]-L-glutamic acid, [13C7]-benzoic acid, [3,4-13C2]- cholesterol, [15N]-L-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13 C2]-palmitic acid, [15N,13C5]-L-methionine, [15N]-choline chloride, and 2′-deoxyguanosine-15N2,13C10-5′-monophosphate (Soltow et al., 2013). Sample extracts were analyzed using liquid chromatography and Orbitrap high-resolution mass spectrometry (LC-HRMS; Dionex Ultimate 3000, Q-Exactive HF, Thermo Scientific) (Soltow et al., 2013). Six CHEAR pooled human plasma samples were added to each of the five analytical batches for quality control purposes and reference standardization. Sample preparation and process protocol is described in the Supplementary Material.
All individual samples were analyzed in triplicate using hydrophilic interaction liquid chromatography (HILIC) with positive electrospray ionization (ESI) and C18 hydrophobic reversed-phase chromatography with negative ESI for metabolomic profiling. All data was acquired in MS1 mode only. Raw data files were then extracted using apLCMS (Yu et al., 2009) with modifications by xMSanalyzer. (Uppal et al., 2013) Uniquely detected ions consisted of m/z, retention time and ion abundance, referred to as metabolomic features. Metabolomic feature data is available at CHEAR Data Center (https://cheardatacenter.mssm.edu/). The coefficients of variation (CVs) of pooled samples across five analytical batches suggested that batch effect of untargeted metabolomics data was very small in our analysis. The medians (inter-quartile ranges) of CVs across 8945 detected HILIC positive features and 9136 C18 negative features were 0.40 (0.27, 0.62) and 0.51 (0.27, 0.67), respectively (Supplementary Fig. 1). Details of the quality control and data normalization and transformation of metabolomic intensity data are described in the Supplementary Material. In the main statistical analyses, we focused on metabolomic features with relatively low cross-batch variability (CVs ≤ 0.3). We also performed the sensitivity analyses including all features without the restriction on CV threshold. After removing metabolomic features detected in less than 50% of the study samples and samples with < 80% detected metabolomic features, our final sample size of untargeted metabolomics was 101 for fasting plasma samples and 102 for plasma samples collected at 30 min after the OGTT.
2.6. Assessment of plasma PFASs exposure levels
Concentrations of PFOA, PFOS and PFHxS were quantified from plasma samples by reference standardization using the LC-HRMS method described previously with reverse phase chromatography for analyte separation and negative mode ESI (details are presented in the Supplementary Material) (Alderete et al., 2019; Go et al., 2015). Calculated limit of detection (LOD) for PFOA, PFOS and PFHxS was 0.02, 0.1 and 0.03 ng/mL, respectively. There were 3 participants with PFHxS exposure below the LOD, which were treated as zero exposure to PFHxS in the final analysis. In order to reduce measurement errors, we measured PFASs concentrations in both fasting and 30-minutes post glucose challenge plasma samples, assuming plasma PFASs concentrations do not vary after oral glucose challenge (correlations of repeated measures for each PFAS chemical are presented in Supplementary Fig. 2). Based on the repeated measurements, medians (interquartile ranges) of CVs of three PFAS chemical exposure (PFOA, PFOS and PFHxS) across all analytical batches were 0.09 (0.04, 0.17), 0.06 (0.03, 0.14) and 0.07 (0.03, 0.16), respectively (Supplementary Fig. 3). Individual PFAS exposure levels were estimated by the average of plasma PFAS concentrations analyzed from both fasting and 30-minutes post glucose challenge samples. PFOA, PFOS, and PFHxS were detected in 100%, 100%, and 97.2% of participants.
2.7. Targeted metabolomics analysis of lipid and fatty acid metabolism
Serum samples were stored at −80 °C and were sent to Duke Molecular Physiology Institute Metabolomics/Biomarker Core Laboratory on dry ice for targeted metabolomics analysis. Serum concentrations of 45 targeted metabolites including 43 acylcarnitines, non-esterified free fatty acids (NEFA) and glycerol were assayed. (Newgard et al., 2009) These metabolites were selected to complement the untargeted metabolomics for investigating lipolysis and fatty acid oxidation, which could be dysregulated by PFAS exposures. (Le Magueresse-Battistoni et al., 2017; Li et al., 2019) Absolute serum concentrations of targeted metabolites were quantified by inclusion of stable isotope-labeled internal standards for metabolites. A Beckman Unicel DxC 600 autoanalyzer was used for analysis of NEFA and glycerol. The analysis of acylcarnitine was conducted by flow injection-tandem mass spectrometry (MS/MS). Concentrations of acylcarnitines with various lengths of carbon chains and with or without hydroxyl-/dicarboxyl (OH/DC) groups were measured. These acylcarnitine signatures were used to investigate mitochondrial and peroxisomal β-oxidation (non-OH/DC acylcarnitines), as well as microsomal ω-oxidation (OH/DC acylcarnitines) of various sources of fatty acids. Moreover, total concentrations of non-OH/DC and OH/DC acylcarnitines were calculated for each participant by various lengths of carbon chains (short-chain: < 6 carbons; medium-chain: 6 to13 carbons; and long chain: ≥ 14 carbons).
2.8. Covariates
We administered questionnaires detailing sociodemographic characteristics, education and smoking history. Self-reported physical activity status was assessed by the questions “Have you taken any exercise classes, lessons, or special programs (e.g., dance, martial arts, aerobics, gymnastics or tumbling and swimming) during the past 12 months?” and “Please place yourself on the scale (0–100) to rate your usual physical activity”. The self-evaluation of physical activity scale was further categorized into three categories of low- (0–40), moderate-(50–60) and high- (70–100) activity levels. Furthermore, two non-consecutive 24-hour diet recalls including serving sizes of 168 food items were collected at the study visit and a phone call on a separate day after the study visit. (Hoffmann et al., 2002) These diet data were processed using the Nutrition Data System for Research (version 2014, University of Minnesota).
2.9. Statistical analysis
1). PFAS association with cardiometabolic outcomes:
Triglycerides, VLDL-cholesterol, insulin traits, HOMA-IR and Matsuda Index were log transformed to approximate normal distributions. Linear regression was used to assess the associations between each PFAS chemical exposure (PFOA, PFOS and PFHxS) and each adiposity outcome adjusting for age, sex, parental education, race/ethnicity, cigarette and e-cigarette smoking status in the past week, physical activity levels and dietary covariates (total calorie intake, percent calorie intake from fat and protein, as well as glycemic index). Body fat percent was additionally adjusted for in analyses of OGTT-derived measures and lipid profiles. The Benjamini-Hochberg procedure was used to control the false discovery rate (FDR) (Benjamini and Hochberg, 1995) across multiple testing over cardiometabolic outcomes. Statistical tests were considered significant with an FDR value < 0.05.
For cardiometabolic outcomes that were found to be associated with PFAS exposures in the linear regression analysis, Bayesian Kernel Machine Regression (BKMR) (Bobb et al., 2015) (R package “bkmr”) was further used to examine the joint effects of the three PFAS chemicals (PFOA, PFAS and PFHxS) accounting for correlations among chemical exposures (Pearson correlation r from 0.4 to 0.6, Supplementary Fig. 1) and potential nonlinear associations between PFAS exposures and cardiometabolic outcomes.
2). Dysregulated metabolic pathways associated with PFAS exposures:
Metabolome-wide association analysis (MWAS) was performed to assess linear associations between each of the three PFAS chemicals and intensities of each metabolomic feature adjusting for covariates as previously described. To include all possible metabolomic features that were involved in PFAS-associated metabolic pathways, metabolomic features having marginally significant associations with PFAS exposures (raw p < 0.05) were selected for further metabolic pathway analyses. Mummichog pathway enrichment analysis (Li et al., 2013) with 5000 permutations was used to predict chemical annotations of metabolomic features and identify key metabolic pathways associated with PFAS exposures.
3). Connections between PFAS exposures and the metabolic network of metabolomic signatures and cardiometabolic outcomes:
Among metabolic pathways that were significantly associated with PFAS exposures (p for pathway enrichment test < 0.05), we further extracted the intensity data of metabolites with previously confirmed chemical identities using MS2 spectra compared with authentic compounds analyzed under the identical experimental condition according to the Metabolomics Standards Initiative (MSI) level 1 criteria. (Sumner et al., 2007) Among all detected metabolomic features, there were 467 metabolites identified from features assayed by HILIC positive and C18 negative platforms. (Liu et al., 2020) Among all confirmed metabolites, there were 54 lipids, 131 amino acids, 99 organic acids and derivatives which include key tricarboxylic acid (TCA) metabolites such as pyruvate, citric acid and succinic acid, as well as 30 carbohydrates. (Liu et al., 2020).
The xMWAS integrated network analysis (R package “xMWAS”) (Uppal et al., 2017) was used to investigate the associations among three PFAS chemical exposure levels, intensities of annotated metabolites and a spectrum of cardiometabolic outcomes. In the network analysis, each PFAS exposure variable, metabolites and outcome variables were treated as nodes. The eigenvector centrality measure (ECM: ranging from 0 to 1) was used to evaluate the importance of nodes in the network. If a node is pointed to by many nodes (which also have high eigenvector centrality), then that node will have high eigenvector centrality and larger influence on the network.
4). Targeted metabolite analysis to explore contributions of lipolysis and fatty acid oxidation to the PFAS associations with cardiometabolic outcomes:
Linear regression was performed to examine associations between individual PFAS exposure and targeted metabolites (acylcarnitines, NEFA and glycerol) adjusting for covariates as previously described. If any metabolites were found to be associated with PFAS exposures and cardiometabolic outcomes, the Sobel test (Sobel, 1982) for mediation analysis was further conducted to examine the mediation effect of metabolites in the associations between PFAS exposure and cardiometabolic outcomes.
3. Results
The mean age of 102 participants was 19.2 ± 0.8 years. There were 50 (49.0%) overweight and 34 (33.3%) obese participants at the study visit. A total of 61 (60%) participants were Hispanic. Sociodemographic characteristic and metabolic measures of our study participants are presented in Table 1 and Supplementary Table 1. The mean plasma concentrations of the three PFAS compounds in our cohort were close to those reported by National Health and Nutrition Examination Survey (NHANES) among US adults in 2013–2014 (Table 2) (CDC, 2019). In our study, Hispanic participants had a lower average PFAS levels compared to non-Hispanic participants.
Table 1.
Sociodemographic characteristics of 102 meta-AIR young adults enrolled from 2014 to 2017.
| Sample size N (%) | |
|---|---|
| Age (years) | 19.2 (0.8)* |
| Sex | |
| Male | 58 (56.9) |
| Female | 44 (43.1) |
| Parental Education | |
| Less than high school | 34 (33.3) |
| Completed high school | 35 (34.3) |
| Some college or higher | 26 (25.5) |
| Unknown | 7 (6.9) |
| Race/Ethnicity | |
| Non-Hispanic White | 29 (28.4) |
| Hispanic White | 61 (59.8) |
| Other † | 12 (11.8) |
| Ever used e-cigarette | |
| Ever | 33 (32.4) |
| Never | 69 (67.6) |
| Current cigarette smoker ‡ | |
| Yes | 7 (6.9) |
| No | 95 (93.1) |
| Participate in exercise class in the last year § | |
| Yes | 29 (28.4) |
| No | 73 (71.6) |
| Self-reported physical activity levels | |
| Less active | 25 (24.5) |
| Moderately active | 38 (37.3) |
| More active | 39 (38.2) |
| Dietary variables | |
| Total calorie intake (KJ/day) | 1988.8 (627.5)* |
| Percent calorie from fat | 34.3 (8)* |
| Percent calorie from protein | 16.5 (4.9)* |
| Glycemic index | (43.5)* |
These variables are presented as mean (standard deviation) rather than N (%).
Other races = Asian, African American, Other/Mixed Races.
Current cigarette smoker = smoked in the past 7 days.
Exercise Class = took any exercise classes, lessons, or special programs during the past 12 months (outside of school only).
Table 2.
Plasma concentrations of PFAS exposure presented as geometric means (95% confidence intervals) among 102 Meta-AIR young adults and stratified by racial/ethnic groups.
| PFAS levels (μg/L) | All Samples (N = 102) | Hispanic White (N = 61) | Non-Hispanic White (N = 31) | Other Race/Ethnicity (N = 12) |
|---|---|---|---|---|
| PFOA* | 2.26 (1.61, 3.18) | 2.24 (1.58, 3.17) | 2.35 (1.69, 3.25) | 2.18 (1.57, 3.02) |
| PFOS† | 4.29 (1.61, 11.47) | 4.23 (1.48, 12.12) | 4.27 (1.75, 10.44) | 4.67 (1.94, 11.24) |
| PFHxS§ | 1.37 (0.32, 5.79) | 1.39 (0.36, 5.32) | 1.48 (0.34, 6.37) | 1.07 (0.17, 6.74) |
PFOA = perfluorooctanoic acid.
PFOS = perfluorooctane sulfonate.
PFHxS = perfluorohexane sulfonic acid.
3.1. PFAS exposure associations with cardiometabolic outcomes
We assessed linear associations between exposures to each of the three PFAS compounds and cardiometabolic outcomes adjusting for covariates (Table 3, Supplementary Tables 2 and 3). Results suggested that higher exposure to PFOA was significantly associated with higher 30-minute glucose levels and glucose AUC during the OGTT (FDR values < 0.05). Stratified analysis by race/ethnicity further suggested that the association effect sizes between PFOA exposure and 30-minute glucose levels and glucose AUC were larger among Hispanic White participants (p for interactions between PFOA and race/ethnicity = 0.033 and 0.006, respectively). No significant associations were found between PFOA and adiposity, insulin resistance and lipid measures (FDR values > 0.05). Also, no significant associations were found between PFOS and PFHxS exposures with cardiometabolic outcomes (FDR values > 0.05).
Table 3.
Adjusted* associations between perfluorooctanoic acid (PFOA) exposure and cardiometabolic outcomes among all 102 young adults and stratified by race/ethnicity and obesity.
| Outcomes | All Samples (N = 102) | Hispanic White (N = 61) | Other Races‖ (N = 41) | Non-obese (N = 68) | Obese (N = 34) |
|---|---|---|---|---|---|
| Adiposity | |||||
| Body mass index, BMI (kg/m2) | −0.46 (−1.55, 0.63) | −0.29 (−1.85, 1.27) | −1.05 (−2.58, 0.48) | NA | NA |
| Body fat percent (%) | −1.46 (−3.01, 0.10) | −0.36 (−2.18, 1.46) | −2.93 (−6.73, 0.87) | NA | NA |
| Visceral adipose tissue, VAT (L)‡ | −0.04 (−0.2, 0.11) | 0.03 (−0.18, 0.23) | −0.16 (−0.52, 0.20) | 0.02 (−0.18, 0.21) | 0.03 (−0.25, 0.31) |
| SAAT (L)§ | −0.42 (−1.01, 0.18) | −0.19 (−0.96, 0.58) | −0.95 (−2.09, 0.19) | −0.23 (−0.76, 0.30) | 0.72 (−0.87, 2.30) |
| Hepatic fat fraction (%)‡ | 0.06 (−0.05, 0.17) | 0.21 (0.05, 0.36) | −0.20 (−0.38, −0.03) | 0.10 (−0.03, 0.23) | 0.05 (−0.31, 0.41) |
| VAT-to-SAAT Ratio | 0.06 (−0.03, 0.16) | 0.05 (−0.08, 0.19) | 0.11 (−0.08, 0.30) | 0.11 (−0.01, 0.23) | −0.08 (−0.29, 0.12) |
| Glucose Metabolism Traits | |||||
| Fasting glucose (mg/dL) | 0.78 (−0.85, 2.41) | 1.93 (−0.10, 3.97) | 1.94 (−1.79, 5.66) | 0.84 (−0.97, 2.65) | 0.95 (−4.56, 6.47) |
| 30-min glucose after OGTT (mg/dL)§ | 9.20 (4.42, 13.98) † | 12.32 (6.13, 18.51) † | 7.94 (−1.46, 17.35) | 7.76 (2.20, 13.33) | 10.58 (0.10, 21.07) |
| 2-hour glucose after OGTT (mg/dL)§ | 5.57 (−0.43, 11.56) | 8.31 (−0.02, 16.64) | 5.96 (−7.16, 19.08) | 7.81 (0.32, 15.31) | −4.69 (−24.28, 14.91) |
| OGTT glucose area under the curve§ | 1096.83 (554.50, 1639.17) † | 1383.30 (726.82, 2039.77) † | 1214.45 (−38.87, 2467.77) | 1116.15 (422.10, 1810.19) | 959.07 (−313.92, 2232.06) |
| Fasting insulin (μU/mL)‡ | 0.25 (−0.01, 0.52) | 0.31 (0.01, 0.61) | −0.08 (−0.78, 0.63) | 0.17 (−0.15, 0.49) | 0.32 (−0.39, 1.03) |
| 30-min insulin after OGTT (μU/mL)‡§ | 0.22 (0.004, 0.43) | 0.30 (0.08, 0.53) | 0.14 (−0.52, 0.81) | 0.06 (−0.28, 0.40) | 0.02 (−0.31, 0.36) |
| 2-hour insulin after OGTT (μU/mL)‡§ | 0.25 (0.02, 0.49) | 0.38 (0.05, 0.71) | 0.24 (−0.29, 0.78) | 0.07 (−0.25, 0.40) | −0.16 (−0.74, 0.43) |
| OGTT insulin area under the curve‡§ | 0.21 (0.08, 0.35) | 0.28 (0.10, 0.46) | 0.20 (−0.10, 0.50) | 0.06 (−0.14, 0.26) | 0.03 (−0.30, 0.36) |
| HOMA-⇧ | 0.23 (−0.03, 0.49) | 0.25 (−0.02, 0.53) | −0.16 (−0.83, 0.52) | 0.14 (−0.16, 0.45) | 0.32 (−0.37, 1.00) |
| HOMA-IR‡§ | 0.26 (−0.01, 0.53) | 0.33 (0.02, 0.64) | −0.05 (−0.77, 0.67) | 0.18 (−0.15, 0.50) | 0.33 (−0.39, 1.06) |
| Matsuda index‡ | −0.27 (−0.45, −0.09) | −0.35 (−0.58, −0.12) | −0.11 (−0.57, 0.34) | −0.15 (−0.40, 0.09) | −0.21 (−0.75, 0.32) |
| Hemoglobin A1c, HbA1c (mmol/mol) | −0.02 (−0.09, 0.05) | 0.01 (−0.09, 0.11) | −0.11 (−0.26, 0.05) | −0.003 (−0.09, 0.09) | 0.05 (−0.13, 0.23) |
| Lipids | |||||
| LDL (mg/dL)§ | −1.08 (−8.59, 6.42) | 7.63 (−2.44, 17.70) | −1.52 (−8.83, 5.78) | −2.28 (−12.32, 7.77) | 2.64 (−19.53, 24.8) |
| VLDL (mg/dL)‡§ | 0.11 (−0.02, 0.25) | 0.22 (0.04, 0.40) | −0.11 (−0.44, 0.23) | 0.10 (−0.07, 0.28) | −0.20 (−0.51, 0.11) |
| HDL (mg/dL)§ | −2.55 (−4.59, −0.52) | −2.86 (−5.22, −0.51) | −1.77 (−6.99, 3.46) | −2.87 (−5.55, −0.19) | 0.11 (−5.03, 5.26) |
| Triglycerides (mg/dL)‡ | 0.11 (−0.02, 0.25) | 0.22 (0.04, 0.40) | −0.11 (−0.44, 0.23) | 0.10 (−0.07, 0.28) | −0.20 (−0.51, 0.11) |
| Total cholesterol (mg/dL) | −0.92 (−9.6, 7.76) | 9.83 (−1.80, 21.46) | −4.87 (−14.70, 4.97) | −2.58 (−14.56, 9.39) | −1.23 (−26.16, 23.69) |
Linear regression was used to investigate the associations between PFOA exposure and individual cardiometabolic outcomes adjusting for age, sex, education, race/ethnicity, cigarette and e-cigarette smoking status in the past week, physical activity levels and dietary covariates including total calorie intake, percent calorie intake from fat and protein and glycemic index. For the outcomes of glucose metabolism and lipids, percent body fat was additionally adjusted for in the model. PFOA exposure was scaled by one standard deviation = 0.41. Association estimates β (95% confidence intervals) are presented in the table. False discovery method (FDR) was used to adjust for multiple testing.
Significant associations with FDR values < 0.05 are bolded.
Log transformation was applied to specific outcome variables to approximate normal distributions.
SAAT = Subcutaneous adipose tissue; OGTT = Oral glucose tolerance test; HOMA-β = Homeostatic model assessment-β-cell function; HOMA-IR = Homeostatic model assessment-insulin resistance; LDL = Low-density lipoprotein; VLDL = Very low-density lipoprotein; HDL = High-density lipoprotein.
Other races = Non-Hispanic White, Asian, African American, Other/Mixed Races.
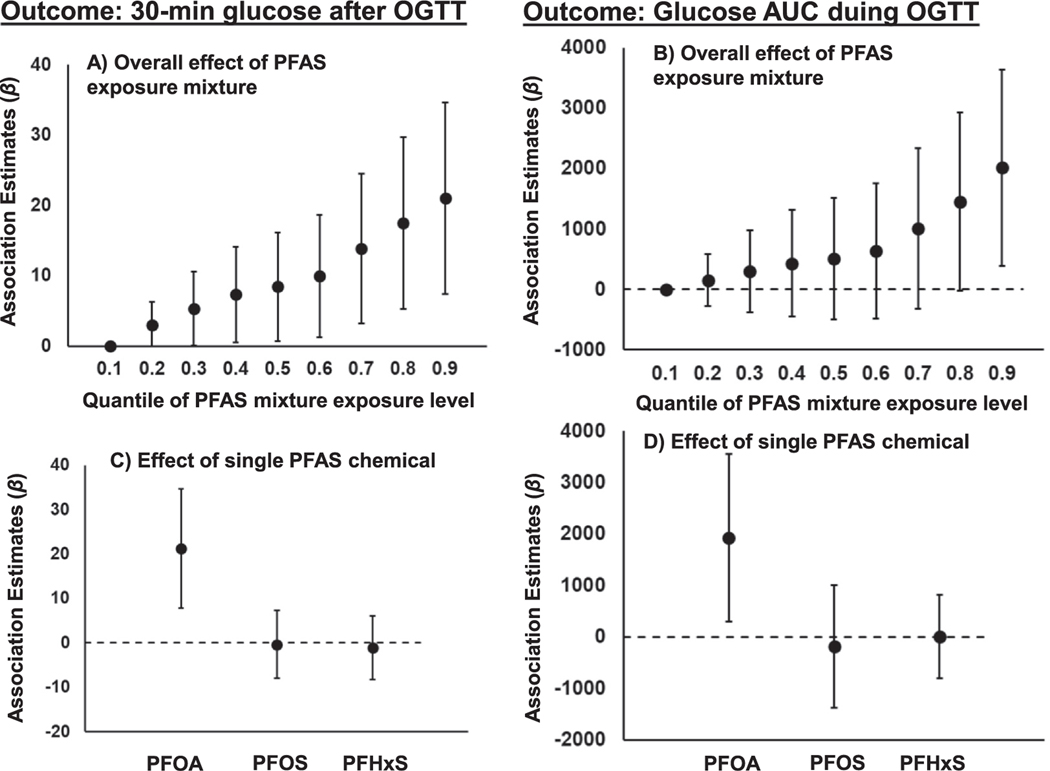
For significant PFAS-metabolite relationships indicated by the linear association analysis, BKMR analysis of the joint associations between mixtures of three PFAS congeners and cardiometabolic outcomes indicated that higher exposure to PFAS mixture was associated with higher 30-minute glucose level and glucose AUC during the OGTT (Fig. 1). For example, participants with exposure levels of all three PFAS chemicals in the top 90th percentile had 21 (95% confidence interval, CI = 7, 35) mg/dL higher 30-min glucose and 2015 (95% CI = 389, 3641) mg/dL × min higher glucose AUC, compared to participants who had all three PFAS chemical exposure levels below the 10th percentile.
Fig. 1.
Associations of perfluoroalkyl substances (PFASs) exposure mixture with 30-minute glucose levels and glucose area under the curve (AUC) measured during the oral glucose tolerance test (OGTT). Panels A) and B) present total effects of PFAS exposure mixture on glucose outcomes by quantiles of exposure levels. Panels C) and D) present differences of glucose outcomes in participants with individual PFAS chemical exposure [perfluorooctanoic acid (PFOA), perfluorooctane sulfonate (PFOS) and perfluorohexane sulfonic acid (PFHxS)] level at 90th percentile to participants with the exposure level at 10th percentile, while conditioned on the other two PFAS chemical exposures both at 50th percentiles across all participants.
Furthermore, the adverse effect of PFAS exposure mixture was largely driven by PFOA exposure (Fig. 1). Conditional on PFOS and PFHxS exposure levels at the 50th percentile, participants with PFOA exposure level at the top 90th percentile had 21 (95% CI = 9,35) mg/dL higher 30-min glucose and 1925 (95% CI = 253, 3557) mg/dL × min higher glucose AUC, compared to participants whose PFOA exposure levels were lower than 10th percentile. No significant association was found for PFOS and PFHxS exposures with 30-minute glucose levels and glucose AUC. Additionally, BKMR analysis indicated that there was no significant non-linear relationship between PFAS exposure and glucose outcomes (data not shown).
3.2. Altered metabolic pathways associated with PFAS exposures
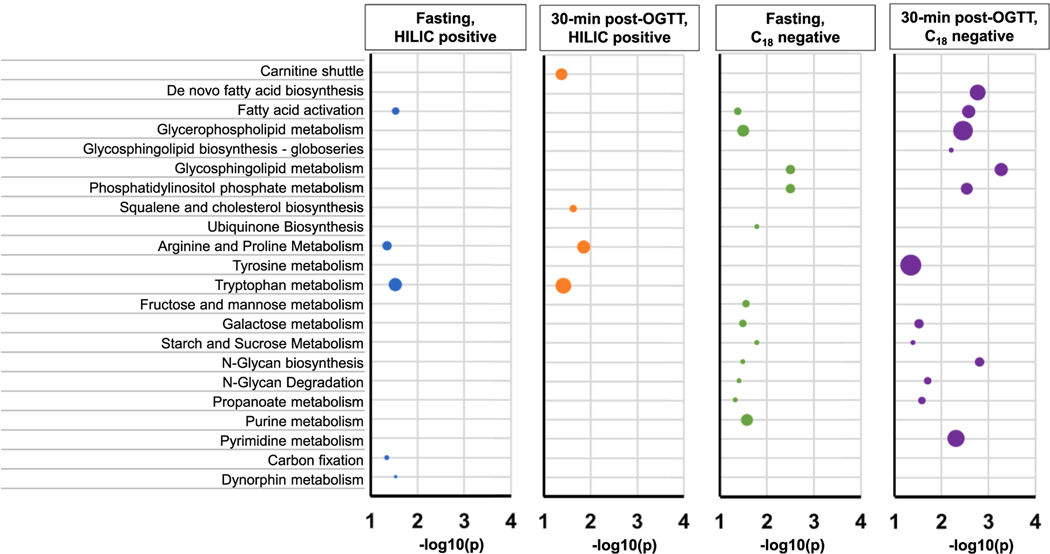
MWAS of fasting plasma found that 231 metabolomic features from 2164 HILIC positive features (all CVs ≤ 0.3) and 239 metabolomic features from 2004 C18 negative features (all CVs ≤ 0.3) had marginally significant associations with at least one of the three PFAS congeners (p < 0.05) (Supplementary Figures 4–5). Using 30-minute post glucose challenge plasma samples, there were 372 metabolomic features from 2154 HILIC positive features (all CVs ≤ 0.3) and 518 metabolomic features from 2001 C18 negative features (all CVs ≤ 0.3) had marginally significant associations with at least one of the three PFAS exposures (p < 0.05) (Supplementary Figures 6–7).
Significant metabolomic features revealed by the MWASs were further included in the Mummichog pathway analysis. In general, PFAS exposure was significantly associated with dysregulated metabolism of lipids, fatty acids, amino acids such as arginine, proline, and tryptophan, as well as hexoses (Fig. 2). These altered pathways were consistently found from the analyses of untargeted metabolomic profiles of fasting and 30-minute post glucose challenge samples. The sensitivity analysis of all metabolomic features without restriction on CV threshold (CVs ranged from 0.001 to 3.73) found similar results that PFAS exposure was associated with dysregulated lipid metabolism such as glycerophospholipid and glycosphingolipid metabolism (Supplementary Figures 8–12).
Fig. 2.
Metabolic pathways that are associated with PFAS exposures revealed by the analysis of high-resolution metabolomics data (metabolomic features having coefficients of variation ≤ 0.3 across 5 analytical batches) from fasting and 30-minute post glucose challenge plasma samples using both HILIC positive and C18 negative modes. The horizontal scale indicates the significance levels of pathway enrichment tests conducted by Mummichog software.
3.3. Metabolomic signatures linking PFAS exposures and cardiometabolic risk
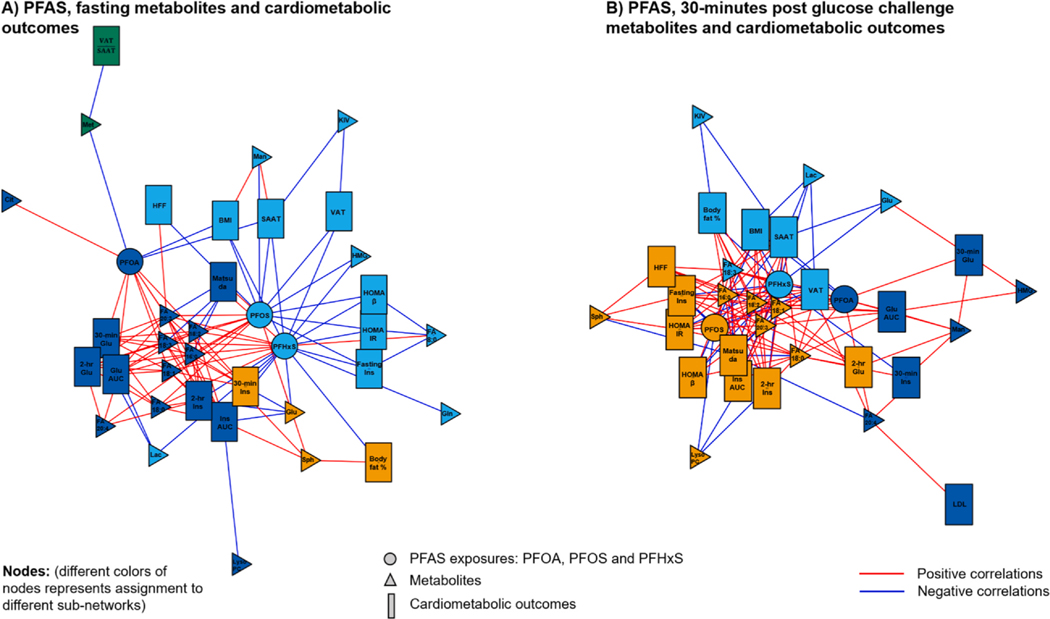
Among extracted metabolomic features (CVs ≤ 0.3) from Mummichog-identified PFAS-related metabolic pathways, we confirmed the chemical identities of 7 features from HILIC positive mode and 12 features from C18 negative mode according to the MSI level 1 criteria by comparing MS2 spectra with authentic chemical compounds (Supplementary Table 4). (Sumner et al., 2007) We then performed an integrated network analysis using xMWAS (Uppal et al., 2017) among three PFAS chemical exposures, 19 identified metabolites in 16 metabolic pathways, and cardiometabolic traits including adiposity, OGTT and lipid measures to investigate the influence of PFAS exposures on a metabolic network involving metabolites and cardiometabolic outcomes.
Based on metabolites from fasting samples, xMWAS results suggested that three PFAS congeners were assigned to three sub-networks (Fig. 3). PFOA, PFOS and PFHxS were all highly influential in the entire exposure-metabolites-cardiometabolic traits network (ECM = 0.6, 1 and 0.5, respectively). PFOA exposure was assigned to the sub-network, linking to cardiometabolic traits, including 30-minute glucose, 2-hr glucose and insulin levels, as well as glucose and insulin AUC after the OGTT (ECM > 0.4, Supplementary Table 5). In addition, metabolites including palmitate, linolenic acid, linoleic acid, oleic acid, stearic acid, arachidonic acid, and homolinoleic acid were also connected to the PFOA subnetwork (ECM > 0.3). In contrast, metabolites and cardiometabolic outcomes connected to PFOS and PFHxS exposures were not important to the whole network (all ECM < 0.1).
Fig. 3.
Integrated network analysis among PFAS exposures, metabolites with confirmed identity and multiple cardiometabolic outcomes including adiposity measures, oral-glucose tolerance test-derived glucose and insulin measures, insulin resistance index and lipid profiles. Each PFAS exposure, metabolite and outcome variable is treated as a node in the entire network and are plotted using different shapes. Sub-networks are classified by different colors and represent more connections between specific PFAS congener with a group of metabolomic signatures and cardiometabolic outcomes. Panel A presents the network among PFAS exposure, metabolite intensity in fasting plasma samples and cardiometabolic outcomes. Panel B presents the network among PFAS exposure, metabolite intensity in 30-minute post glucose challenge plasma samples and cardiometabolic outcomes. PFAS exposures: PFOA = Perfluorooctanoic acid; PFOS = Perfluorooctane sulfonate; PFHxS = Perfluorohexane sulfonic acid. Metabolites: LysoPC = LysoPC (18:0); FA8:0 = FA 8:0 (Octanoate); FA16:0 = FA 16:0 (Palmitate); FA18:3 = FA 18:3n-3 or n-6 (Linolenic acid); FA18:2 = FA 18:2 (Linoleic acid); FA18:1 = FA 18:1 (Oleic acid); FA18:0 = FA 18:0 (Stearic acid); FA20:4 = FA 20:4 (Arachidonic acid); FA20:3 = FA 20:3 (Homolinoleic acid); Sph = Sphingosine; Cit = Citrulline; Met = Methionine; Glu = Glucose; Man = Mannose/Galactose; Lac = Lactate; Gln = Glutamine; HMG = Hydroxymethylglutarate; KIV = Oxovalerate/Ketoisovalerate. Cardiometabolic outcomes: 30-min GLU = 30-min glucose after OGTT; 2-hr GLU = 2-hour glucose after OGTT; Glu AUC = OGTT glucose area under the curve; Fasting Ins = Fasting insulin; 30-min Ins = 30-min insulin after OGTT; 2-hr Ins = 2-hour insulin after OGTT; Ins AUC = OGTT insulin area under the curve; HFF = Hepatic fat fraction; HOMA-β = Homeostatic model assessment β-cell function; Matsuda = Matsuda index; HOMA-IR = Homeostatic model assessment insulin resistance; SAAT = Subcutaneous adipose tissue; BMI = Body mass index; Body fat % = Body fat percent; VAT = Visceral adipose tissue; = VAT-to-SAAT ratio; LDL = Low-density lipoprotein.
Next, by analyzing metabolites from 30-minute post glucose challenge samples, we found that PFOA exposure had the largest influence to the entire network compared to PFOS and PFHxS exposures (ECM for PFOA, PFOS and PFHxS = 0.7, 0.03 and 0, respectively) (Fig. 3). PFOA exposure was assigned to a sub-network that correlated with 30-minute glucose and insulin levels, glucose AUC (ECM = 0.1, 0.2 and 0.5, respectively). The metabolites connected to the PFOA sub-network were arachidonic acid and mannose/galactose (both ECM = 0.2, Supplementary Table 5). Although PFOS and PFHxS exposure was not important to the entire network, they were connected to the subnetworks including long-chain saturated and unsaturated fatty acids and sphingosine, as well as cardiometabolic outcomes of fasting and 2-hr insulin levels, HOMA-IR, percent body fat, visceral adipose tissue, subcutaneous adipose tissue and hepatic fat fraction (ECM > 0.3, Supplementary Table 5).
3.4. Analysis of targeted metabolites involved in lipid metabolism
Based on untargeted metabolomics findings of dysregulated metabolism of lipids and fatty acids that link PFAS exposure and 30-minute glucose levels and glucose AUC, we further verified our findings using absolute concentrations of targeted metabolites including 43 acylcarnitines, NEFA and glycerol (Table 4, Supplementary Tables 6–8). A consistent finding across three PFAS exposures was their positive associations with glycerol (p < 0.05). Moreover, higher PFOA exposure was associated with higher fasting levels of short-chain non-OH/DC acylcarnitines (p = 0.046, Table 4). Stratified analyses by obesity status suggested that higher PFOA exposure was associated with higher levels of acylcarnitines related to β-oxidation such as acetylcarnitine (C2) and 3-hydroxybutyrylcarnitine (C4-OH) (p = 0.028 and 0.045, respectively, Supplementary Table 6) among non-obese participants. Borderline associations were also observed among obese participants (p = 0.09 and 0.06, respectively). Stratified analyses further suggested that PFHxS exposure was associated with glycerol, short-chain non-OH/DC acylcarnitines, and a spectrum of acylcarnitines including C2, C4-OH, 9,12-hexadecadienoylcarnitine (C16:2), 9-hexadecenoylcarnitne (C16:1) and stearoylcarnitine (C18) among obese participants (p < 0.05, Table 4 and Supplementary Table 8). The interaction between PFHxS and obesity status was statistically significant for the association with C4-OH (p = 0.0001).
Table 4.
Adjusted* associations between PFAS exposure and targeted metabolites involved in the fatty acid and lipid metabolism in the entire Meta-AIR cohort and stratified by race/ethnicity and obesity.
| PFAS Exposure | Metabolite Outcomes | All samples (N = 102) | Hispanic White (N = 61) | Other Races** (N = 41) | Non-obese (N = 68) | Obese (N = 34) |
|---|---|---|---|---|---|---|
| PFOA# (μg/L) | NEFA¶ (mmol/L) | 2.58E−02 (−3.80E−03, 5.54E−02) | 3.07E−02 (−1.03E−02, 7.17E−02) | −1.41E−02 (−7.39E−02, 4.57E−02) | 3.19E−02 (−3.22E−03, 6.70E−02) | 2.99E−02 (−4.70E−02, 1.07E−01) |
| Glycerol (mg/dL) | 1.06E−01 (3.69E−02, 1.75E−01) | 1.55E−01 (6.88E−02, 2.42E−01) | −1.39E−02 (−1.72E−01, 1.44E−01) | 7.79E−02 (−1.03E−02, 1.66E−01) | 2.19E−01 (9.03E−02, 3.48E−01) | |
| Total non-OH/DC† (μmol/L) | 6.16E−02 (3.40E−04, 1.23E−01) | 6.38E−02 (−1.79E−02, 1.45E−01) | −7.74E−02 (−1.99E−01, 4.44E−02) | 8.73E−02 (9.76E−03, 1.65E−01) | 1.59E−01 (−2.05E−02, 3.39E−01) | |
| Total OH/DC†(μmol/L) | 3.04E−02 (−3.07E−02, 9.15E−02) | −3.08E−02 (−1.07E−01, 4.58E−02) | −2.51E−03 (−1.18E−01, 1.13E−01) | 9.33E−02 (2.26E−02, 1.64E−01) | 7.33E−02 (−1.89E−01, 3.35E−01) | |
| Short-chain non-OH/DC‡(μmol/L) | 6.55E−02 (2.14E−03, 1.29E−01) | 7.35E−02 (−1.12E−02, 1.58E−01) | −7.41E−02 (−2.02E−01, 5.40E−02) | 9.31E−02 (1.22E−02, 1.74E−01) | 1.72E−01 (−9.51E−03, 3.53E−01) | |
| Short-chain OH/DC‡(μmol/L) | 2.59E−02 (−4.56E−02, 9.73E−02) | −8.29E−03 (−1.13E−01, 9.68E−02) | 5.02E−02 (−8.36E−02, 1.84E−01) | 6.18E−02 (−1.82E−03, 1.25E−01) | 1.19E−01 (−1.81E−01, 4.19E−01) | |
| Medium-chain non-OH/DC§(μmol/L) | 5.77E−02 (−2.54E−02, 1.41E−01) | 1.56E−02 (−9.41E−02, 1.25E−01) | −7.80E−02 (−2.64E−01, 1.08E−01) | 7.76E−02 (−2.43E−02, 1.79E−01) | 8.64E−02 (−1.61E−01, 3.33E−01) | |
| Medium-chain OH/DC§(μmol/L) | 4.42E−02 (−3.90E−02, 1.27E−01) | −5.32E−02 (−1.45E−01, 3.84E−02) | −3.26E−02 (−2.28E−01, 1.62E−01) | 1.41E−01 (2.39E−02, 2.57E−01) | 4.81E−02 (−2.08E−01, 3.04E−01) | |
| Long-chain non-OH/DC‖(μmol/L) | 3.14E−02 (−2.28E−02, 8.56E−02) | 2.27E−02 (−4.28E−02, 8.82E−02) | −6.04E−02 (−1.93E−01, 7.22E−02) | 3.71E−02 (−2.89E−02, 1.03E−01) | 1.06E−01 (−5.95E−02, 2.71E−01) | |
| Long-chain OH/DC‖(μmol/L) | 3.87E−02 (−1.64E−02, 9.39E−02) | 1.86E−03 (−7.11E−02, 7.48E−02) | −3.72E−03 (−1.03E−01, 9.53E−02) | 6.63E−02 (−1.11E−03, 1.34E−01) | 1.27E−01 (−6.07E−02, 3.14E−01) | |
| PFOS# (μg/L) | NEFA¶ (mmol/L) | 1.03E−02 (−2.15E−02, 4.20E−02) | 5.23E−03 (−3.81E−02, 4.86E−02) | 3.46E−02 (−2.38E−02, 9.30E−02) | −1.75E−03 (−4.71E−02, 4.36E−02) | 1.63E−02 (−6.57E−02, 9.83E−02) |
| Glycerol (mg/dL) | 9.90E−02 (2.53E−02, 1.73E−01) | 7.82E−02 (−2.17E−02, 1.78E−01) | 1.71E−01 (3.48E−02, 3.07E−01) | 1.11E−01 (2.43E−03, 2.20E−01) | 1.57E−01 (−1.60E−02, 3.30E−01) | |
| Total non-OH/DC† (μmol/L) | 2.34E−02 (−4.27E−02, 8.96E−02) | 4.05E−03 (−8.26E−02, 9.07E−02) | 7.63E−02 (−4.71E−02, 2.00E−01) | 1.00E−02 (−9.20E−02, 1.12E−01) | 1.60E−01 (−2.92E−02, 3.50E−01) | |
| Total OH/DC†(μmol/L) | −1.27E-02 (−7.75E−02, 5.21E−02) | −4.98E−02 (−1.28E−01, 2.80E−02) | 5.12E−02 (−6.29E−02, 1.65E−01) | 2.26E−02 (−7.20E−02, 1.17E−01) | 4.90E−02 (−2.27E−01, 3.25E−01) | |
| Short-chain non-OH/DC‡(μmol/L) | 2.38E−02 (−4.47E−02, 9.22E−02) | 7.52E−03 (−8.29E−02, 9.79E−02) | 7.56E−02 (−5.37E−02, 2.05E−01) | 8.55E−03 (−9.81E−02, 1.15E−01) | 1.77E−01 (−1.25E−02, 3.67E−01) | |
| Short-chain OH/DC‡(μmol/L) | 7.49E−03 (−6.81E−02, 8.31E−02) | −4.41E−02 (−1.51E−01, 6.31E−02) | 1.25E−01 (2.30E−03, 2.48E−01) | 4.72E−02 (−3.42E−02, 1.29E−01) | 7.29E−02 (−2.46E−01, 3.92E−01) | |
| Medium-chain non-OH/DC§(μmol/L) | 3.48E−02 (−5.36E−02, 1.23E−01) | −4.42E−03 (−1.17E−01, 1.09E−01) | 9.63E−02 (−9.00E−02, 2.83E−01) | 4.63E−02 (−8.32E−02, 1.76E−01) | 6.96E−02 (−1.90E−01, 3.30E−01) | |
| Medium-chain OH/DC§(μmol/L) | −2.64E−02 (−1.15E−01, 6.18E−02) | −6.68E−02 (−1.60E−01, 2.66E−02) | −1.01E−02 (−2.08E−01, 1.88E−01) | 1.50E−02 (−1.40E−01, 1.70E−01) | 3.61E−02 (−2.31E−01, 3.04E−01) | |
| Long-chain non-OH/DC‖(μmol/L) | −4.01E−03 (−6.17E−02, 5.36E−02) | −2.94E−02 (−9.66E−02, 3.78E−02) | 6.00E−02 (−7.41E−02, 1.94E−01) | −9.60E−03 (−9.29E−02, 7.37E−02) | 7.64E−02 (−1.04E−01, 2.57E−01) | |
| Long-chain OH/DC‖(μmol/L) | −4.16E−04 (−5.93E−02, 5.85E−02) | −1.48E−02 (−8.97E−02, 6.02E−02) | 5.49E−02 (−4.14E−02, 1.51E−01) | −1.14E−02 (−9.89E−02, 7.61E−02) | 8.65E−02 (−1.20E−01, 2.93E−01) | |
| PFHxS# (μg/L) | NEFA¶ (mmol/L) | 9.55E−03 (−2.07E−02, 3.98E−02) | 5.51E−03 (−3.70E−02, 4.80E−02) | 1.70E−02 (−3.30E−02, 6.71E−02) | 2.03E−03 (−3.56E−02, 3.97E−02) | 3.76E−02 (−5.06E−02, 1.26E−01) |
| Glycerol (mg/dL) | 7.33E−02 (1.80E−03, 1.45E−01) | 3.06E−02 (−6.98E−02, 1.31E−01) | 9.66E−02 (−2.79E−02, 2.21E−01) | 7.54E−02 (−1.61E−02, 1.67E−01) | 3.06E−01 (2.09E−01, 4.03E−01) | |
| Total non-OH/DC† (μmol/L) | 1.91E−02 (−4.40E−02, 8.22E−02) | −2.07E−02 (−1.05E−01, 6.40E−02) | 4.77E−02 (−5.74E−02, 1.53E−01) | −1.35E−02 (−9.81E−02, 7.11E−02) | 2.56E−01 (8.42E−02, 4.28E−01) | |
| Total OH/DC†(μmol/L) | −1.03E−02 (−7.20E−02, 5.15E−02) | −2.04E−02 (−9.80E−02, 5.72E−02) | −1.31E−02 (−1.10E−01, 8.42E−02) | −3.46E−02 (−1.13E−01, 4.34E−02) | 2.04E−01 (−7.27E−02, 4.81E−01) | |
| Short-chain non-OH/DC‡(μmol/L) | 2.5È−02 (−3.94E−02, 9.10E−02) | −1.93E−02 (−1.08E−01, 6.92E−02) | 6.10E−02 (−4.72E−02, 1.69E−01) | −3.66E−03 (−9.22E−02, 8.48E−02) | 2.69E−01 (9.75E−02, 4.41E−01) | |
| Short-chain OH/DC‡(μmol/L) | 2.69E−02 (−4.49E−02, 9.87E−02) | −8.00E−03 (−1.14E−01, 9.80E−02) | 7.83E−02 (−2.98E−02, 1.86E−01) | 1.27E−02 (−5.58E−02, 8.11E−02) | 2.72E−01 (−3.85E−02, 5.82E−01) | |
| Medium-chain non-OH/DC§(μmol/L) | −9.07E−03 (−9.36E−02, 7.55E−02) | −2.14E−02 (−1.32E−01, 8.92E−02) | −1.84E−02 (−1.79E−01, 1.42E−01) | −7.05E−02 (−1.76E−01, 3.54E−02) | 2.09E−01 (−4.97E−02, 4.67E−01) | |
| Medium-chain OH/DC§(μmol/L) | − 5.01E −02 ( −1.34E −01, 3.33E −02) | − 4.27E −02 ( −1.36E −01, 5.03E −02) | − 1.01E −01 ( − 2.58E −01, 5.70E −02) | − 8.91E −02 (− 2.15E −01, 3.65E −02) | 1.87E −01 ( −8.38E −02, 4.58E −01) | |
| Long-chain non-OH/DC‖(μmol/L) | − 1.74E −02 ( −7.22E −02, 3.74E −02) | − 3.46E −02 ( −1.00E −01, 3.09E −02) | − 1.46E −03 (− 1.16E −01, 1.13E −01) | − 3.56E −02 (− 1.04E −01, 3.27E −02) | 1.50E −01 ( −3.28E −02, 3.32E −01) | |
| Long-chain OH/DC‖(μmol/L) | 9.07E −03 ( −4.70E −02, 6.52E −02) | − 1.90E −03 ( −7.55E −02, 7.17E −02) | − 8.78E −03 ( −9.22E −02, 7.46E −02) | − 1.63E −02 ( −8.88E −02, 5.62E −02) | 1.85E −01 ( −1.80E −02, 3.89E −01) |
Linear regression was performed to examine the associations between individual PFAS exposure and targeted metabolites adjusting for covariates including age, sex, education, race/ethnicity, cigarette and e-cigarette moking status in the past week, percent body fat, physical activity levels and dietary covariates including total calorie intake, percent calorie intake from fat and protein and glycemic index. Metabolites as the outcome variables were log transformed in the model. Association estimates β (95% confidence intervals) are presented in the table. Significant associations with p-values < 0.05 are bolded.
non-OH/DC = non-hydroxyl‐/dicarboxyl‐ acylcarnitine. OH/DC = hydroxyl‐/dicarboxyl‐acylcarnitine. Units for acylcarnitines are μmol/L
Short-chain acylcarnitines are sum of concentrations of acylcarnitines with chains of<6 carbons (Supplementary Tables 13–15).
Medium-chain acylcarnitines are sum of concentrations of acylcarnitines with chains of 6 to13 carbons (Supplementary Tables 13–15).
Long-chain acylcarnitines are sum of concentrations of acylcarnitines with chains of 14 carbons and higher (Supplementary Tables 13–15).
NEFA = non-esterified free fatty acids.
PFOA = perfluorooctanoic acid, PFOS = perfluorooctane sulfonate and PFHxS = perfluorohexane sulfonic acid. PFAS exposures were standardized on standard deviations of 0.41, 2.58 and 1.62, respectively for PFOA, PFOS and PFHxS.
Other races = Asian, African American, Other/Mixed Races.
Meanwhile, higher levels of NEFA, glycerol, short-chain non-OH/DC acylcarnitines and C4-OH were associated with higher 30-minutes glucose levels and glucose AUC (p < 0.05, Supplementary Tables 9–12). Although glycerol was associated with both PFOA exposure and outcomes of 30-minute glucose levels and glucose AUC, no significant mediation effect was found (Sobel test p > 0.14).
4. Discussion
This study investigated the cardiometabolic effect of exposures to three PFAS congeners (PFOA, PFOS and PFHxS) among young adults. Untargeted metabolomics was used to investigate biological mechanisms linking PFAS exposures and cardiometabolic traits. Among the three PFAS chemical exposures, PFOA exposure contributed significantly to the adverse effect of PFAS on glucose metabolism, manifested by higher 30-minute glucose levels and glucose AUC after the OGTT. Dysregulated lipid metabolism may play an important role in the associations between PFAS and impaired glucose metabolism. Furthermore, targeted metabolomics analysis revealed that increased lipolysis and β-oxidation could contribute to the mechanism underlying the relationship between PFAS exposures and glucose intolerance. However, no significant associations were found for PFAS exposure with metabolites in TCA cycle. Also, it should be noted that young adults included in this study had a history of being overweight or obese in their childhood and were mostly (82%) overweight and obese at the Meta-AIR study visit, therefore many of them were potentially insulin resistant. Nonetheless, no significant interaction was found between PFAS exposures and obesity status for the effects on altered lipid and glucose metabolism. It is noted that due to the exploratory nature of untargeted metabolomics research, future studies with larger sample size are needed to validate our findings.
In the last decade, there has been increased concern about the widespread exposure to PFAS chemicals in terms of adverse effects on various health outcomes including cardiometabolic diseases (Sunderland et al., 2019; Lee, 2018). Although long-chain PFAS (PFOA and PFOS) have been phased out in manufactural productions (Agency, 2020), their threats to public health still exists even at low exposure level (Sunderland et al., 2019). Furthermore, health effects of shorter-chain PFAS replacements such as PFHxS have not been well characterized (Sunderland et al., 2019; Lee, 2018). However, few studies have been conducted to examine the PFAS effect in children and young adults. Two previous studies in adolescents suggested that higher exposure to PFOA was associated with hyperglycemia and insulin resistance (Domazet et al., 2016; Alderete et al., 2019, as well as higher total and LDL-cholesterol levels (Geiger et al., 2014). Our findings supported the association between PFOA exposure and decreased glucose tolerance. Although no association was found for PFAS exposure with serum concentrations of cholesterol and triglycerides, the xMWAS network analysis among three PFAS exposures, identity-confirmed metabolites and cardiometabolic outcomes suggested that dysregulated lipid metabolism could play an important role in connecting PFOA and PFHxS exposures with altered glucose metabolism.
In vivo and in vitro studies suggested that the toxicity of PFAS chemicals could be mediated by activations of nuclear receptors such as PPAR-α/γ, which are known to regulate lipid and glucose metabolism in the liver and adipocytes (Le Magueresse-Battistoni et al., 2017; Li et al., 2019). In human metabolomics studies, various metabolic pathways, specifically lipids and urea cycle-related amino acids, have been found to be associated with PFAS exposures (Alderete et al., 2019; Salihovic et al., 2018). However, previous epidemiological studies were largely focused on using untargeted metabolomics approach to explore general metabolic pathways.
To our knowledge, no metabolomics study has been conducted to examine PFAS exposures on metabolomic profiles after the glucose challenge. In this study, both fasting and 30-minute post glucose challenge plasma samples were assayed for HRM. This study design is especially important for our study population of young healthy adults. Early indications of metabolic dysfunction such as insulin resistance and impaired glucose tolerance could only be observed after a glucose challenge in non-diabetic population, while euglycemia is maintained for a long time before the development of diabetes (Xiang et al., 2006). We also found that more PFAS-associated lipid metabolic pathways were revealed by the untargeted metabolomics analysis of 30-minute post-glucose challenge samples then fasting samples.
In order to verify our untargeted metabolomics findings and to delineate biological underpinnings reflected by the identified pathways, we investigated PFAS associations with fasting serum concentrations of metabolites involved in lipid and fatty acid oxidation. Our observations about positive associations of PFOA exposure with acylcarnitines related to fatty acid oxidation including C2 and C4-OH indicated that PFOA exposure might increase mitochondrial β-oxidation.
Moreover, increased glycerol level was observed to be associated with all three PFAS chemical exposures, while NEFA was not associated with PFAS exposures. This finding suggested that increased lipolysis could also be induced by PFAS exposures. Many PFAS congeners such as PFOA and PFOS are xenobiotic agonists of PPAR-α/γ, and PPAR-α has been shown to be more sensitive to PFOA than PFOS exposure (Martin et al., 2007). Activation of PPAR-α, especially at fasting status, is important to stimulate hepatic fatty acid oxidation under the condition of lipid catabolism (Patsouris et al., 2004). Furthermore, activation of PPAR-α can also increase gluconeogenesis by converting glycerol to glucose (Lamichane et al., 2018).
We acknowledge several limitations of this study. First, this project was built upon a cross-sectional study with relatively small sample size. Most of our participants were overweight and obese. But the young adult sample is unique for comprehensive phenotyping of cardiometabolic traits, detailed socio-behavioral covariates, as well as both untargeted and targeted metabolomics data. The cross-sectional study design precludes our opportunities to investigate causal relationships between PFAS exposures and metabolic outcomes. Also, results of this study do not imply causal effects of PFAS exposures on dysregulated lipid pathways and glucose metabolism. Second, the metabolomics assay used in this study could not comprehensively characterize various species of lipids. Findings of this study and other studies have suggested that lipid metabolism could play an important role for PFAS exposure to perturb metabolism (Alderete et al., 2019; Liu et al., 2020; Jin et al., 2020; Salihovic et al., 2019). Therefore, more future studies are warranted to explore lipidomic signatures of PFAS exposure. Also, in the exploratory stage of MWASs, we used a less stringent significance cut-off of p < 0.05 to select features for further Mummichog pathway analysis. Inflated type I error from multiple testing could exist in the MWASs findings. However, the following mummichog pathway analysis, xMWAS network analysis and targeted metabolomics analysis of glycerol, fatty acids and acylcarnitines all consistently suggested that dysregulated lipid metabolism could play a significant role in PFAS-related pathophysiology. Nonetheless, future studies with larger sample size and targeted analysis on metabolites in lipid metabolism are needed to validate our findings. Third, PFAS exposure was measured and quantified by LC-HRMS approach with a single-point collaboration comparing to the reference sample, as described in the methods section. Compared to the traditional targeted metabolomics approach with stable isotope-labeled internal standards added to each sample for specific chemical of interest, the untargeted metabolomics approach could have larger measurement error in quantifying concentrations but increases feasibility and cost efficiency. To ameliorate measurement errors in our PFAS concentration data, we took the average concentrations of two repeated measures from each participant. We also noted that correlations of repeated PFAS measures across all study samples were relatively high, which suggested that the new quantification approach of plasma PFAS exposure is quite robust. Fourth, only three PFAS chemical exposures were quantified for plasma concentrations in this study. Future studies are needed to explore the effects of other widely detected PFAS chemical exposure such as per-fluorononanoic acid (PFNA) (CDC, 2019). Finally, although the BKMR analysis results suggested that PFAS exposure mixture, especially PFOA exposure had adverse effect on higher 30-min glucose and glucose AUC during the OGTT, the toxicity of specific PFAS chemicals and chemical mixture needs to be further investigated by experimental studies.
5. Conclusions
PFAS exposures, especially PFOA exposure, have a significant influence on increasing lipolysis and fatty acid oxidation in young adults, which could further contribute to impaired glucose metabolism. The Hispanic population may be more susceptible to the adverse metabolic effects of the PFOA exposure. Therefore, there is a strong need for increased public attention to reduce PFAS exposures for the sake of improving metabolic health in young adults. Notably, our findings need to be interpreted with caution because participants of this study were mostly overweight or obese young adults. Also, findings of this study largely depend on the exploratory analysis of untargeted metabolomics data, more experimental studies and epidemiological studies with larger sample size and targeted research on lipid metabolism are warranted to validate our findings.
Supplementary Material
Acknowledgement
Z.C. led the study design, statistical analyses and paper writing. T.Y. and C.Q. helped to perform the statistical analysis and paper writing. F.D.G., D.J., D.I.W., D.C.T., E.R.H., L.C. and C.B. all contributed to the study design and development of the study protocol. Z.C., F.D.G., D.I.W., J.K., T.L.A., L.C. and E.R.H. contributed to the data collection. Z.C., E.R.H., D.C.T. D.L., and D.V.C. contributed to the development of statistical methods. All authors reviewed, edited the article and contributed to discussion. Z.C. is the guarantor of this work, and as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors declare that they have no conflict of interest.
Data management of the untargeted metabolomics data was supported by CHEAR Center for Data Science at ICAHN School of Medicine at Mount Sinai, NY (grant U2CES026555). The laboratory analysis of targeted metabolomics data was conducted by Drs. Olga Ilkayeva, Mike Muehlbauer and Christopher Newgard at Duke Molecular Physiology Institute Metabolomics/Biomarker Core Laboratory.
Sources of Funding Support
This work was supported by [CHEAR project 2016–1448, grants (U2CES026560 and U2CES026555)] from the National Institute of Environmental Health Sciences as part of the Children’s Environmental Health Analysis Resource (CHEAR) and National Exposure Laboratory at Emory University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences (NIEHS). This project was also supported by NIEHS awards R00ES027870 and Southern California Children’s Environmental Health Center grant funded by NIEHS (5P01ES022845–03) and United States Environmental Protection Agency (RD83544101). Additional grants support included NIEHS (5P01ES011627, R00ES027853, and R01ES029944); the Southern California Environmental Health Sciences Center grant (5P30ES007048) funded by NIEHS; and the Hastings Foundation.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2020.106091.
References
- Juonala M, Singh GR, Davison B, et al. , 2016. Childhood metabolic syndrome, inflammation and carotid intima-media thickness. The Aboriginal Birth Cohort Study. Int. J. Cardiol 203, 32–36. [DOI] [PubMed] [Google Scholar]
- Morrison JA, Friedman LA, Gray-McGuire C, 2007. Metabolic syndrome in childhood predicts adult cardiovascular disease 25 years later: the Princeton Lipid Research Clinics Follow-up Study. Pediatrics 120, 340–345. [DOI] [PubMed] [Google Scholar]
- Sunderland EM, Hu XC, Dassuncao C, Tokranov AK, Wagner CC, Allen JG, 2019. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Eposure Sci. Environ. Epidemiol 29, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, 2018. Potential health effects of emerging environmental contaminants perfluoroalkyl compounds. Yeungnam Univ. J. Med 35, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Clapp R, 2014. Changing interpretation of human health risks from perfluorinated compounds. Public Health Rep 129, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo JL, 2012. Health risks of dietary exposure to perfluorinated compounds. Environ. Int 40, 187–195. [DOI] [PubMed] [Google Scholar]
- Jain RB, 2014. Contribution of diet and other factors to the levels of selected poly-fluorinated compounds: data from NHANES 2003–2008. Int. J. Hyg. Environ. Health 217, 52–61. [DOI] [PubMed] [Google Scholar]
- Kato K, Wong LY, Jia LT, Kuklenyik Z, Calafat AM, 2011. Trends in exposure to polyfluoroalkyl chemicals in the U.S. Population: 1999–200. Environ Sci Technol 2011;45:8037–45. [DOI] [PubMed] [Google Scholar]
- Hines EP, White SS, Stanko JP, Gibbs-Flournoy EA, Lau C, Fenton SE, 2009. Phenotypic dichotomy following developmental exposure to perfluorooctanoic acid (PFOA) in female CD-1 mice: low doses induce elevated serum leptin and insulin, and overweight in mid-life. Mol. Cell. Endocrinol 304, 97–105. [DOI] [PubMed] [Google Scholar]
- Lv Z, Li G, Li Y, et al. , 2013. Glucose and lipid homeostasis in adult rat is impaired by early-life exposure to perfluorooctane sulfonate. Environ. Toxicol 28, 532–542. [DOI] [PubMed] [Google Scholar]
- Domazet SL, Grontved A, Timmermann AG, Nielsen F, Jensen TK, 2016. Longitudinal associations of exposure to perfluoroalkylated substances in childhood and adolescence and indicators of adiposity and glucose metabolism 6 and 12 years later: The European Youth Heart Study. Diabetes Care 39, 1745–1751. [DOI] [PubMed] [Google Scholar]
- Alderete TL, Jin R, Walker DI, et al. , 2019. Perfluoroalkyl substances, metabolomic profiling, and alterations in glucose homeostasis among overweight and obese Hispanic children: a proof-of-concept analysis. Environ. Int 126, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Liu Y, Xu B, Gu L, Tang W, 2018. PFOA is associated with diabetes and metabolic alteration in US men: National Health and Nutrition Examination Survey 2003–2012. The Science of the total environment 625, 566–574. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Hivert MF, Gold DR, et al. , 2019. Associations of perfluoroalkyl and polyfluoroalkyl substances with incident diabetes and microvascular disease. Diabetes Care 42, 1824–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Magueresse-Battistoni B, Labaronne E, Vidal H, Naville D, 2017. Endocrine disrupting chemicals in mixture and obesity, diabetes and related metabolic disorders. World J. Biol. Chem 8, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Ren XM, Cao LY, Qin WP, Guo LH, 2019. Investigation of binding and activity of perfluoroalkyl substances to the human peroxisome proliferator-activated receptor beta/delta. Environ. Sci. Process Impacts 21, 1908–1914. [DOI] [PubMed] [Google Scholar]
- Lin PD, Cardenas A, Hauser R, et al. , 2019. Per- and polyfluoroalkyl substances and blood lipid levels in pre-diabetic adults-longitudinal analysis of the diabetes prevention program outcomes study. Environ. Int 129, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Chen Z, Alderete TL, et al. , 2019. Associations of air pollution, obesity and cardiometabolic health in young adults: the Meta-AIR study. Environ. Int 133, 105180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Salam MT, Eckel SP, Breton CV, Gilliland FD, 2015. Chronic effects of air pollution on respiratory health in Southern California children: findings from the Southern California Children’s Health Study. J. Thorac. Dis 7, 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC, 1985. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419. [DOI] [PubMed] [Google Scholar]
- Matsuda M, DeFronzo RA, 1999. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22, 1462–1470. [DOI] [PubMed] [Google Scholar]
- Kaess BM, Pedley A, Massaro JM, Murabito J, Hoffmann U, Fox CS, 2012. The ratio of visceral to subcutaneous fat, a metric of body fat distribution, is a unique correlate of cardiometabolic risk. Diabetologia 55, 2622–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Walker DI, Uppal K, et al. , 2016. High-resolution metabolomics assessment of military personnel: evaluating analytical strategies for chemical detection. J. Occupat. Environ. Med./Am. College Occupat. Environ. Med 58, S53–S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvi D, Walker DI, Inge T, et al. , 2020. Environmental chemical burden in metabolic tissues and systemic biological pathways in adolescent bariatric surgery patients: a pilot untargeted metabolomic approach. Environ. Int 143, 105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP, 2013. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 9, S132–S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP, 2009. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics 25, 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Soltow QA, Strobel FH, et al. , 2013. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinf. 14, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go YM, Walker DI, Liang Y, et al. , 2015. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci.: Off. J. Soc. Toxicol 148, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, et al. , 2009. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Boeing H, Dufour A, et al. , 2002. Estimating the distribution of usual dietary intake by short-term measurements. Eur. J. Clin. Nutr 56 (Suppl 2), S53–S62. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc.: Ser. B (Methodol.) 57, 289–300. [Google Scholar]
- Bobb JF, Valeri L, Claus Henn B, et al. , 2015. Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures. Biostatistics (Oxford, England) 16, 493–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, et al. , 2013. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol 9, e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. , 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Nellis M, Uppal K, et al. , 2020. Reference standardization for quantification and harmonization of large-scale metabolomics. Anal. Chem 92, 8836–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Ma C, Go YM, Jones DP, 2017. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel ME, 1982. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol. Methodol 13, 290–312. [Google Scholar]
- CDC. Fourth national report on human exposure to environmental chemicals. Available from https://www.cdc.gov/exposurereport/index.html. Accessed on November 15th 2019. 2019;1–2.
- Environmental Protection Agency (EPA). Technical Fact Sheet – Perfluorooctane Sulfonate (PFOS) and Perfluorooctanoic Acid (PFOA). Available at https://www.epa.gov/sites/production/files/2017-12/documents/ffrrofactsheet_contaminants_pfos_pfoa_11-20-17_508_0.pdf. Accessed on March 10th, 2020. . 2017.
- Geiger SD, Xiao J, Ducatman A, Frisbee S, Innes K, Shankar A, 2014. The association between PFOA, PFOS and serum lipid levels in adolescents. Chemosphere 98, 78–83. [DOI] [PubMed] [Google Scholar]
- Salihovic S, Fall T, Ganna A, et al. , 2018. Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J. Expo Sci. Environ. Epidemiol [DOI] [PubMed] [Google Scholar]
- Xiang AH, Wang C, Peters RK, Trigo E, Kjos SL, Buchanan TA, 2006. Coordinate changes in plasma glucose and pancreatic β-cell function in latino women at high risk for Type 2 diabetes. Diabetes 55, 1074–1079. [DOI] [PubMed] [Google Scholar]
- Martin PG, Guillou H, Lasserre F, et al. , 2007. Novel aspects of PPARalpha-mediated regulation of lipid and xenobiotic metabolism revealed through a nutrigenomic study. Hepatology 45, 767–777. [DOI] [PubMed] [Google Scholar]
- Patsouris D, Mandard S, Voshol PJ, et al. , 2004. PPARalpha governs glycerol metabolism. J. Clin. Invest 114, 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichane S, Dahal Lamichane B, Kwon SM, 2018. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. International journal of molecular sciences 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang B, Hu Y, et al. , 2020. Associations of perfluoroalkyl substances with blood lipids and apolipoproteins in lipoprotein subspecies: the POUNDS-lost study. Environ. Health: A Glob. Access Sci. Source 19, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, McConnell R, Catherine C, et al. , 2020. Perfluoroalkyl substances and severity of nonalcoholic fatty liver in Children: an untargeted metabolomics approach. Environ. Int 134, 105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salihovic S, Fall T, Ganna A, et al. , 2019. Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J. Eposure Sci. Environ. Epidemiol 29, 196–205. [DOI] [PubMed] [Google Scholar]
- CDC. Fourth National Report on Human Exposure to Environmental Chemicals. Accessed at https://www.cdc.gov/exposurereport/pdf/FourthReport_UpdatedTables_Volume1_Jan2019-508.pdf. November 15, 2019. 2019;1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.