Abstract
The role of food nutrients in mediating the positive effect of dietary restriction (DR) on longevity has been extensively characterized, but how non-nutrient food components regulate lifespan is not well understood. Here, we show that food-associated odors shorten the lifespan of C. elegans under DR but not those fed ad libitum, revealing a specific effect of food odors on DR-mediated longevity. Food odors act on a neural circuit comprising the sensory neurons ADF and CEP, and the interneuron RIC. This olfactory circuit signals the gut to suppress DR-mediated longevity via octopamine, the invertebrate homolog of norepinephrine, by regulating the energy sensor AMPK through a Gq-PLCβ-CaMKK-dependent mechanism. In mouse primary cells, we find that norepinephrine signaling regulates AMPK through a similar mechanism. Our results identify a brain-gut axis that regulates DR-mediated longevity by relaying olfactory information about food abundance from the brain to the gut.
Introduction
Food is a primary environmental factor that affects aging1,2. Dietary restriction (DR) represents one of the most effective interventions to extend lifespan in all tested organisms, ranging from yeast to mammals1,2. Previous efforts have mostly focused on investigating how nutrients in the food affect aging. This has led to the identification of several nutrient signaling pathways, particularly AMPK and mTOR signaling, that regulate longevity2–4.
However, food is not solely composed of nutrients, as other components are also associated with food, such as volatile odors and various types of non-volatile, non-nutrient chemicals. Like nutrients, these food components may also affect aging5. For example, in C. elegans, worms defective in chemosensation show altered lifespan, suggesting a role of chemical cues in lifespan regulation6,7; in Drosophila, odorants from food can inhibit DR longevity8. However, it is unclear how non-nutrient chemicals in the food regulate longevity. In particular, it is not known whether and how these non-nutrient food chemicals interact with the brain, and if so, how the brain then engages the rest of the body to alter lifespan. Here, we sought to address these questions in C. elegans, a genetic model organism widely used for studying the biology of aging.
Results
Food odors suppress the lifespan of worms under dietary restriction (DR) but not those fed ad libitum (AL)
Lifespan assays in C. elegans are usually conducted on NGM agar plates. Though not as common, lifespan may also be performed in liquid culture. As such, two major types of DR regimens have been developed: solid-phase DR (sDR) assayed on agar plates, and liquid-phase DR (bDR) carried out in liquid culture, with each supplied with a reduced amount of bacteria food9. Different DR regimens may engage different signaling mechanisms4,9. sDR has been widely used in the community due to its easy handing. Because of this and the fact that it is rather difficult to expose food odors to worms cultured in liquid, we chose to focus on sDR. In this protocol, bacteria are killed to prevent their further growth10. Previous effort found that application of the supernatant from bacteria culture to the assaying plates shortened the lifespan of worms deprived of food (bacteria deprivation or DD)11,12. However, as this protocol did not separate volatile odors from various water-soluble chemicals, it remains unclear whether food odors have an effect on worm longevity.
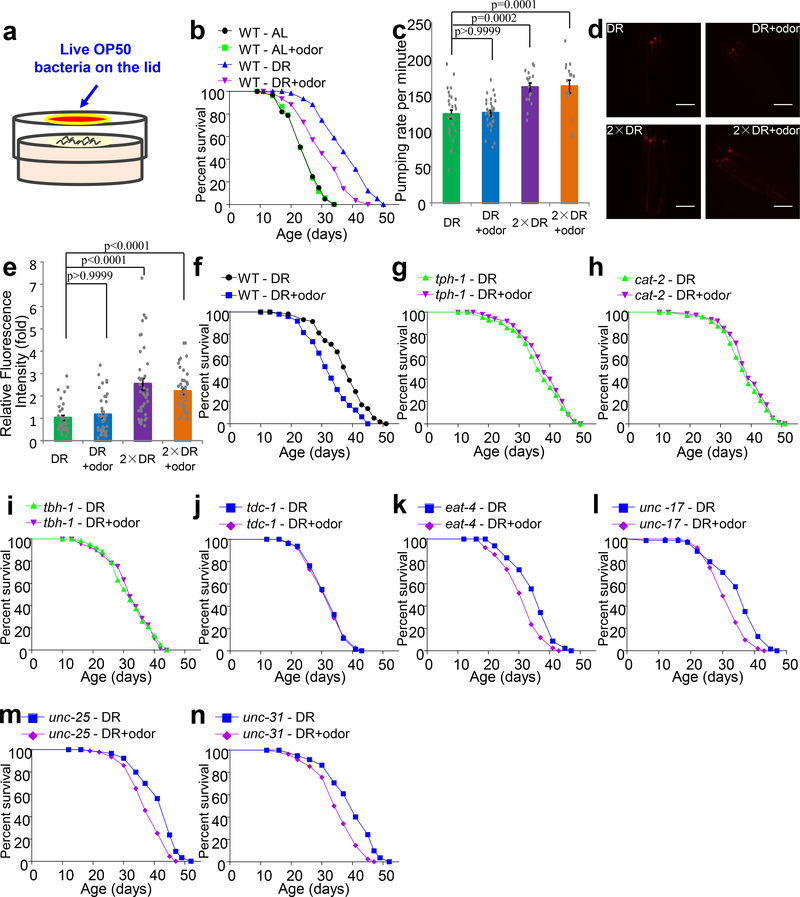
To directly test the effect of food odors, we developed a protocol to deliver food odors to worms in lifespan assays. To do so, we placed a small piece of NGM agar with a thin lawn of live OP50 bacteria on the lid of the NGM plate (Fig.1a). As a control, the same size of NGM agar piece was placed on the lid but without OP50 bacteria. Worms on the plate did not make direct contact with bacteria on the lid, but were exposed to the odors emitted from these bacteria (Fig.1a). Exposing such food-derived odors to worms under DR-mediated longevity by ~50%, while exposing food odors to worms fed ad libitum (AL) did not have a notable effect on lifespan (Fig.1b), revealing a specific effect of food odors on DR longevity.
Fig.1 |. Food odors suppress the lifespan of worms under DR but not those fed AL, and this requires neurotransmission.
(a-b) Food-derived odors shorten the lifespan of worms under DR, but not those fed AL. (a) Schematic describing the assay. (b) Lifespan curves.
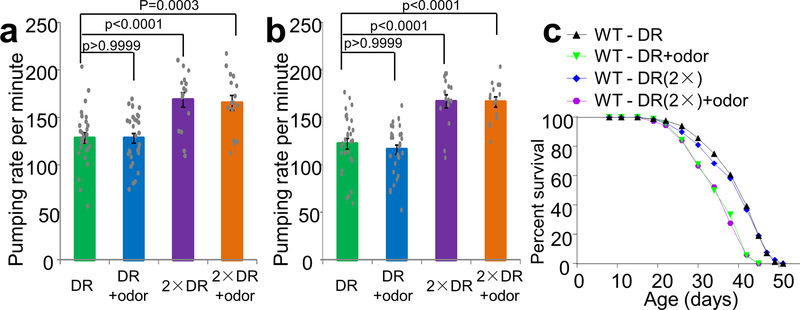
(c) Food odors do not change the pumping rate of worms under DR. Pumping rate was counted at 1 hr after worms were transferred to the DR assaying plates. 2xDR: twice amount of bacterial food. n = 30 (DR), 30 (DR + odor), 16 (2xDR) and 16 (2xDR + odor) biologically independent animals.
(d-e) Food odors do not change the amount of bacteria ingested by worms under DR. tdTomato-expressing OP50 bacteria were used as food source. Scale bar, 200 μm. (d) Sample images. (e) Bar graph. n = 34 (DR), 40 (DR + odor), 40 (2xDR) and 40 (2xDR + odor) biologically independent animals.
(f-n) Food odors shorten the lifespan of WT (f), eat-4 mutant (k), unc-17 mutant (l), unc-25 mutant (m), and unc-31 mutant (n) worms under DR condition. However, loss of tph-1 (g), cat-2 (h), tbh-1 (i), or tdc-1 (j) blocks the ability of food odors to suppress DR longevity.
Data are presented as mean ± s.e.m. p values were calculated with one-way ANOVA with Bonfferronìs test.
See Supplementary Table 1 for lifespan statistics.
One potential concern is that food odors might have stimulated feeding, thereby shortening lifespan due to increased food ingestion. This, however, was not the case, as the pharyngeal pumping rate of DR worms was not affected by food odors compared to the control (Fig.1c and Extended Data Fig.1a–b). In addition, the amount of ingested bacteria was similar under the two conditions, which was determined by quantifying the fluorescence of worm gut bacteria expressing tdTomato (Fig.1d–e). Slightly increasing the amount of bacteria food (2x) fed to DR worms stimulated their pharyngeal pumping rate and increased the amount of ingested bacteria in the gut (Fig.1c–e), yet had no notable effect on lifespan (Extended Data Fig.1c). By contrast, the inhibitory effect of food odors on the lifespan of DR worms persisted under this condition (Extended Data Fig.1c). This set of control experiments indicates that the two assays used have the sensitivity to detect small increases in feeding and food intake that would have been induced by food odors. We thus conclude that food odors inhibit longevity in a DR-dependent manner in C. elegans.
Food odor-induced suppression of DR longevity requires neurotransmission
How do food odors suppress DR longevity? As odors are usually detected by the nervous system, which involves neurotransmission, we first asked whether and which types of neurotransmission mediate the odor effect. By screening different neurotransmission mutants, we found that mutations abolishing the synthesis of serotonin (tph-1), dopamine (cat-2), octopamine (tbh-1), or tyramine (tdc-1) all prevented food odors from inhibiting DR longevity (Fig.1f–j), indicating a requirement of these neurotransmitters. Other neurotransmitter mutants such as eat-4 (glutamate), unc-17 (ACh), unc-25 (GABA) and unc-31 (neuropeptides) did not exhibit a notable defect (Fig.1k–n). These results raise the possibility that serotonin, dopamine and octopamine/tyramine neurons may form a chemosensory circuit to sense and process odor signals to inhibit DR longevity.
ADF, CEP and RIC neurons form an olfactory circuit mediating odor-induced suppression of DR longevity
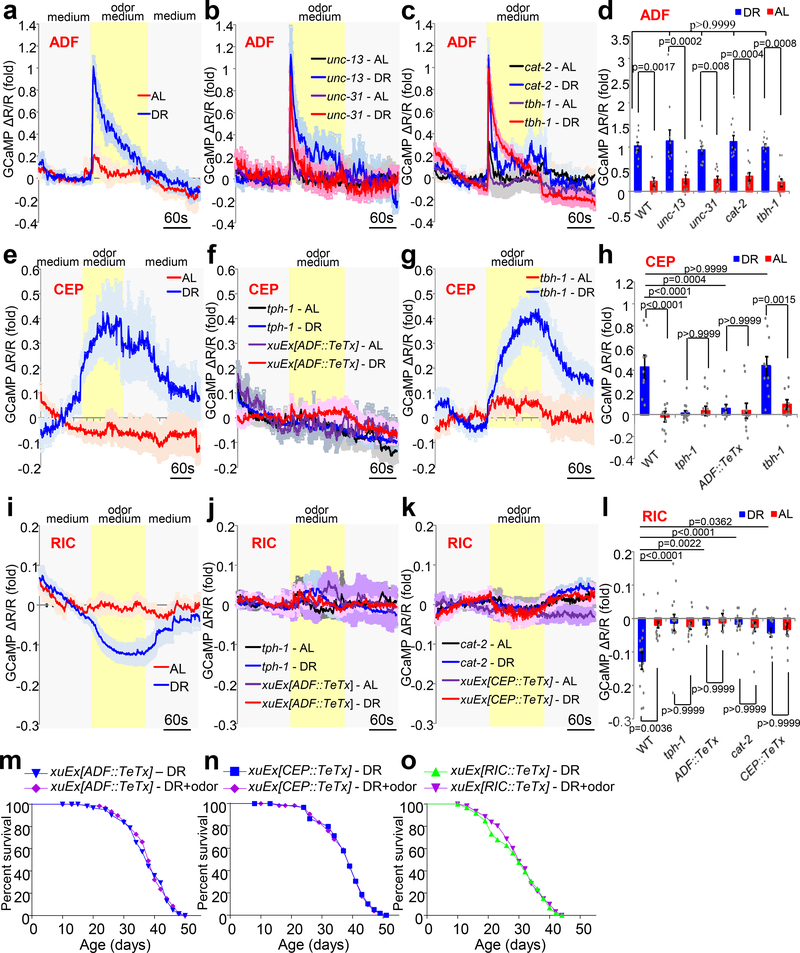
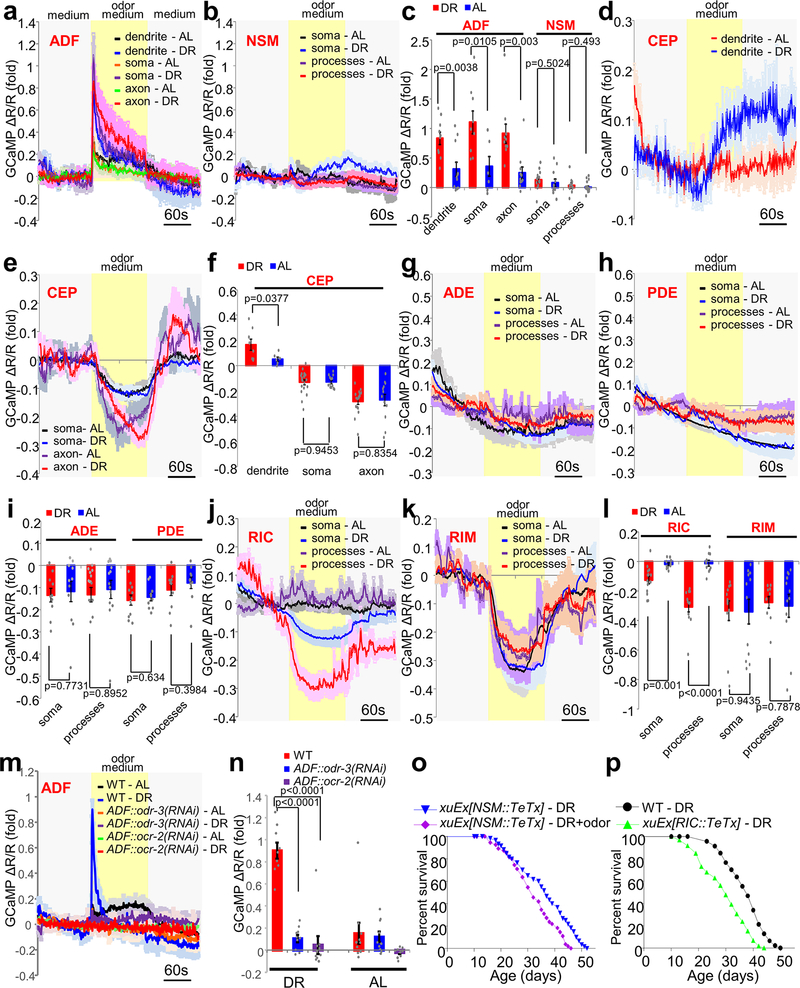
Among serotonin, dopamine and octopamine/tyramine neurons, only ADF serotonin neurons are chemosensory neurons13; the other major type of serotonin neurons NSM is classified as motor neurons14, while dopamine neurons (CEP/ADE/PDE) are mechanosensory neurons15–17, and octopamine/tyramine neurons (RIC and RIM) are interneurons18. As ADF neurons are known to sense volatile odors19, ADF neurons might function as the olfactory neurons in the circuit to sense food odors. To test this, we recorded the activity of both ADF and NSM serotonin neurons in response to food odors by calcium imaging. ADF neurons from DR worms responded robustly to medium containing odors from OP50 bacteria food (Fig.2a, 2d). Strikingly, ADF neurons from AL worms responded very weakly to food odors (Fig.2a, 2d), revealing a DR-specific effect. By contrast, food odors evoked no notable response in NSM serotonin neurons (Extended Data Fig.2b, 2c), suggesting that NSM neurons are not part of the circuit. To test whether ADF neurons sense food odors cell-autonomously, we repeated the imaging experiment in unc-13 and unc-31 mutants, and obtained a similar result (Fig.2b, 2d). Mutations in unc-13 and unc-31 block neurotransmitter release from synaptic vesicles and dense core vesicles, respectively20,21. Thus, ADF neurons likely responded to food odors cell-autonomously, suggesting that these chemosensory neurons act as primary olfactory sensory neurons to sense food odors in DR worms.
Fig.2 |. Food odors act on an olfactory circuit, which is composed of ADF, CEP and RIC neurons, to suppress DR longevity.
(a) ADF neurons from DR worms respond robustly to medium containing odors from OP50 bacteria food, while very weak, if any, calcium response is evoked by food odors in ADF neurons from AL worms. G-CaMP6f was expressed as a transgene in ADF neurons under tph-1(L) promoter. DsRed was co-expressed to enable ratiometric imaging.
(b) Food odor-evoked calcium response in AFD neurons remains normal in unc-13 and unc-31 mutant worms.
(c) Blocking dopamine signaling (using cat-2 mutation) or octopamine signaling (using tbh-1 mutation) does not notably affect food odor-evoked calcium response in ADF neurons.
(d) Bar graph summarizing the data in (a), (b), and (c). n=8 (WT- AL), 10 (WT - DR), 10 (unc-13 - AL), 10 (unc-13 - DR), 10 (unc-31 - AL), 11 (unc-31 - DR), 11 (cat-2 - AL), 11 (cat-2 - DR), 10 (tbh-1 - AL) and 10 (tbh-1 - DR) biological independent animals.
(e) CEP neurons from DR worms but not AL worms respond to food odors. To facilitate dendrite imaging, we expressed myr-GCaMP6f (membrane-targeted) in CEP using the dat-1 promoter. Note: the dat-1 promoter also drives expression in ADE and PDE dopamine neurons.
(f) Blocking serotonin signaling (using either tph-1 mutation or ADF::TeTx transgene) eliminates CEP calcium response evoked by food odors.
(g) Blocking octopamine signaling (using tbh-1 mutation) has no effect on CEP calcium response evoked by food odors.
(h) Bar graph summarizing data in (e), (f), and (g). n=11 (WT - AL), 9 (WT - DR), 13 (tph-1 - AL), 13 (tph-1 - DR), 10 (ADF::TeTx - AL), 11 (ADF::TeTx - DR), 10 (tbh-1 - AL) and 10 (tbh-1 - DR) biological independent animals.
(i) RIC neurons from DR worms but not AL worms respond to food odors. tbh-1 promoter was used to drive transgene expression in RIC.
(j-k) Blocking serotonin signaling (using tph-1 mutation or ADF::TeTx transgene (j)) or inhibiting dopamine signaling (using cat-2 mutation or CEP::TeTx transgene) (k)) abolishes RIC calcium response evoked by food odors.
(l) Bar graph summarizing the data in (i), (j), and (k). n=11 (WT - AL), 14 (WT - DR), 12 (tph-1 - AL), 14 (tph-1 - DR), 8 (ADF::TeTx - AL), 8 (ADF::TeTx - DR), 13 (cat-2 - AL), 15 (cat-2 - DR), 8 (CEP::TeTx - AL) and 9 (CEP::TeTx - DR) biological independent animals.
(m-o) Blocking the output from ADF (m), CEP (n), and RIC (o) neurons using TeTx transgene abrogates the ability of food odors to suppress DR longevity. A fragment of the bas-1 promoter (bas-1(prom7)), dat-1 promoter and tbh-1 promoter was used to drive TeTx expression in ADF, CEP and RIC neurons, respectively.
Data are presented as mean ± s.e.m. Shades along the calcium imaging traces represent error bars ( ± s.e.m). p values were calculated with one-way ANOVA with Bonfferronìs test.
See Supplementary Table 1 for lifespan statistics.
We then asked whether ADF neurons are important for mediating the inhibitory effort of food odors on DR longevity. We ablated the output of ADF neurons by expressing TeTx (tetanus toxin) as a transgene specifically in these neurons. TeTx cleaves synaptobrevin, an essential SNARE subunit, to block exocytosis22. Ablating the output of ADF neurons but not NSM neurons abolished the effect of food odors (Fig.2m and Extended Data Fig. 2o), indicating that ADF neurons are required for food odors to suppress DR longevity. This data, together with our calcium imaging results, suggests that ADF chemosensory neurons act as primary olfactory sensory neurons to sense food odors to suppress longevity in DR worms.
Using the same strategy, we interrogated the potential roles of dopamine neurons (CEP/ADE/PDE), octopamine neurons (RIC), and tyramine neurons (RIM) in the circuit. CEP dopamine neurons responded robustly to food odors in a DR-dependent manner (Fig.2e, 2h), while ADE and PDE dopamine neurons did not (Extended Data Fig.2g–i). Interestingly, RIC neurons also displayed DR-dependent calcium response to food odors, except that food odors inhibited rather than stimulated their activity (Fig.2i–l). These results suggest that CEP and RIC neurons are part of the circuit. One notable observation is that while different sub-compartments of ADF and RIC neurons (dendrites/soma/axon for ADF and soma/processes for RIC) all responded similarly to food odors (Extended Data Fig.2a, 2c, 2j, and 2l), in CEP neurons only the dendrites responded to food odors in a DR-dependent manner (Extended Data Fig.2d–f). This is not surprising, as many worm neurons show compartmentalized calcium responses23,24. By contrast, RIM neurons exhibited no specificity towards DR, as these neurons from DR and AL worms responded similarly to food odors (Extended Data Fig.2k–l), suggesting that RIM neurons are not part of the circuit. These calcium imaging results suggest that in addition to ADF neurons, CEP and RIC neurons are also part of the circuit. Consistent with this model, blocking the output of CEP and RIC neurons using a TeTx transgene eliminated the inhibitory effect of food odors on DR longevity (Fig.2n–o), indicating that CEP and RIC are required for food odors to suppress lifespan in DR worms. We thus conclude that ADF, CEP and RIC are essential components of an olfactory circuit that senses and processes odor signals from food to suppress longevity in DR worms.
We then sought to map the position of ADF, CEP and RIC neurons in the circuit. Given that our data showed that ADF neurons are the primary olfactory neurons sensing food odors, CEP and RIC would be expected to act downstream of ADF in the circuit. If so, blocking the output of the upstream ADF sensory neurons shall abolish the sensitivity of the downstream CEP and RIC neurons to food odors. This appears to be the case: blunting the output of ADF with a TeTx transgene rendered both CEP and RIC neurons insensitive to food odors (Fig.2f, 2h, 2j, and 2l); so did mutations in tph-1 that abolished serotonin release from ADF neurons (Fig.2f, 2h, 2j, and 2l). On the other hand, eliminating the output of the downstream neurons CEP and RIC using cat-2 mutation (blocking dopamine release from CEP) or tbh-1 mutation (blocking octopamine release from RIC), respectively, had no effect on the response of ADF to food odors (Fig.2c–d). This set of experiment places ADF neurons upstream of CEP and RIC neurons.
Using a similar strategy, we tested CEP and RIC neurons. While blocking the output of CEP neurons (using a TeTx transgene and cat-2 mutation) abrogated RIC’s odor sensitivity (Fig. 2k–l), inhibiting RIC’s output (using tbh-1 mutation) had no effect on CEP’s sensitivity to food odors (Fig.2g–h). This places CEP upstream of RIC neurons. As food odors stimulated ADF and CEP neurons but inhibited RIC neurons, this suggests a circuit mechanism by which food odors suppress DR longevity by stimulating ADF and CEP neurons to inhibit RIC neurons (Fig.3l). Though CEP neurons are mechanosensory neurons, as they act downstream of ADF, these CEP neurons may in fact function as interneurons in the circuit.
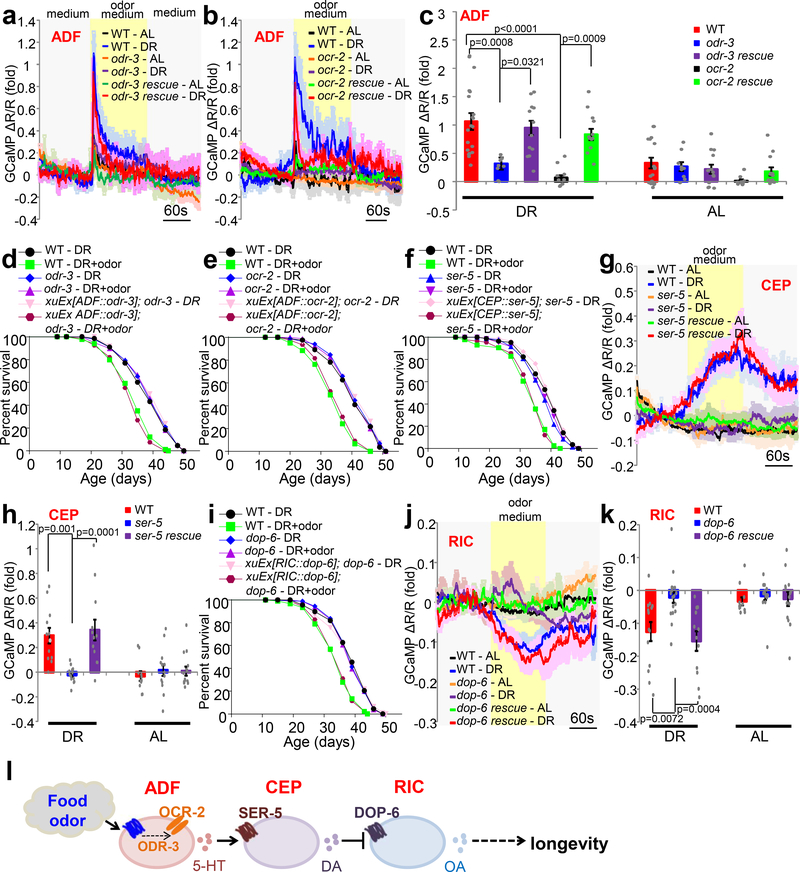
Fig.3 |. The molecular basis by which the olfactory circuit senses and processes odor signals from food.
(a-c) Mutations in odr-3 (a) and ocr-2 (b) abolish food odor-evoked calcium response in ADF neurons, a phenotype rescued by transgenic expression of wild-type odr-3 and ocr-2 genes in ADF neurons. A fragment of the bas-1 promoter (bas-1(prom7)) was used to drive expression of odr-3 and ocr-2 cDNA specifically in ADF neurons. (c) Bar graph summarizing the data in (a) and (b). n=17 (WT - AL), 20 (WT - DR), 9 (odr-3 - AL), 8 (odr-3 - DR), 13 (odr-3 rescue - AL), 12 (odr-3 rescue - DR), 9 (ocr-2 - AL), 9 (ocr-2 - DR), 13 (ocr-2 rescue - AL) and 13 (ocr-2 rescue - DR) biological independent animals.
(d-e) Mutations in odr-3 and ocr-2 prevent food odors from suppressing DR longevity, a phenotype rescued transgenic expression of wild-type odr-3 and ocr-2 cDNA in ADF neurons.
(f) Loss of ser-5 eliminates the ability of food odors to suppress DR longevity, a phenotype rescued by transgenic expression ser-5 cDNA in CEP neurons using the dat-1 promoter.
(g-h) Loss of ser-5 abolishes CEP calcium response evoked by food odors, a defect rescued by transgenic expression ser-5 cDNA in CEP neurons. (h) Bar graph. n=11 (WT - AL), 12 (WT - DR), 14 (ser-5 - AL), 13 (ser-5 - DR), 15 (ser-5 rescue - AL) and 12 (ser-5 rescue - DR) biological independent animals.
(i) Loss of dop-6 prevents food odors from suppressing DR longevity, a phenotype rescued by transgenic expression dop-6 cDNA in RIC neurons.
(j-k) Loss of dop-6 abolishes RIC calcium response evoked by food odors, a defect rescued by transgenic expression dop-6 cDNA in RIC neurons. (k) Bar graph with individual datapoints. n=12 (WT - AL), 14 (WT - DR), 15 (dop-6 - AL), 17 (dop-6 - DR), 14 (dop-6 rescue - AL) and 12 (dop-6 rescue - DR) biological independent animals.
(l) A schematic model of the olfactory circuit that senses and processes odor signals from food to suppress DR longevity.
Data are presented as mean ± s.e.m. Shades along the calcium imaging traces represent error bars ( ± s.e.m). p values were calculated with one-way ANOVA with Bonfferronìs test.
See Supplementary Table 1 for lifespan statistics.
The molecular mechanisms by which the olfactory circuit senses and processes odor signals
To gain a molecular understanding of how the olfactory circuit senses and processes odor signals from food, we went on to identify the receptors that act in each neuron to sense and process odor signals. Olfactory transduction in C. elegans sensory neurons is a G protein-mediated process that culminates in the opening of downstream transduction channels, leading to sensory neuron activation13. We thus examined the G protein ODR-3 and the transduction channel subunit OCR-2 known to function in ADF sensory neurons to mediate chemosensation13,25. Mutations in both odr-3 and ocr-2 not only abolished ADF neuron’s calcium response to food odors (Fig.3a–c), but also the ability of food odors to suppress DR longevity (Fig.3d–e). Both phenotypes were rescued by transgenic expression of wild-type odr-3 and ocr-2 genes in ADF neurons (Fig.3a–e), demonstrating that ODR-3 and OCR-2 act in ADF neurons to sense food odors. Additional evidence came from cell-specific knockdown experiments, where we expressed dsRNA of odr-3 or ocr-2 as a transgene specifically in ADF neurons. RNAi of odr-3 and ocr-2 in ADF neurons abolished their sensitivity to food odors (Extended Data Fig.2m–n), providing further evidence that the G protein ODR-3 and the transduction channel OCR-2 act in ADF neurons to sense food odors. These experiments are also consistent with the notion that ADF neurons are primary olfactory sensory neurons19.
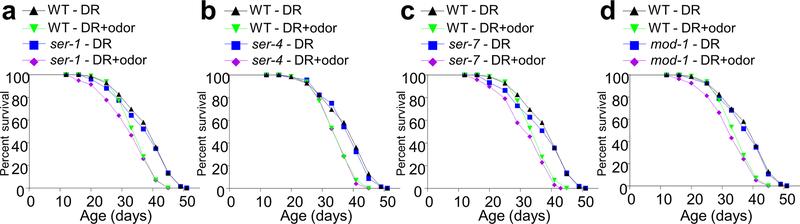
ADF neurons are serotoninergic. Given that CEP neurons act downstream of ADF neurons, we reasoned that a serotonin receptor might act in CEP neurons to transmit odor signals by responding to serotonin released from ADF neurons. We thus examined all the five serotonin receptor genes encoded by the worm genome: ser-1, ser-4, ser-5, ser-7 and mod-126. Mutations in ser-5, but not the other four serotonin receptor genes, blocked the ability of food odors to suppress DR longevity, a phenotype that was rescued by transgenic expression of wild-type ser-5 gene in CEP neurons (Fig.3f and Extended Data Fig.3a–d). Thus, SER-5 may function as the serotonin receptor in CEP neurons to transmit odor signals. In support of this idea, no food odor-evoked calcium response in CEP neurons was detected in ser-5 mutant worms, and this phenotype was rescued by a ser-5 transgene expressed in CEP neurons (Fig.3g–h). These results identify SER-5 as the serotonin receptor that acts in CEP neurons to transmit odor signals in the circuit.
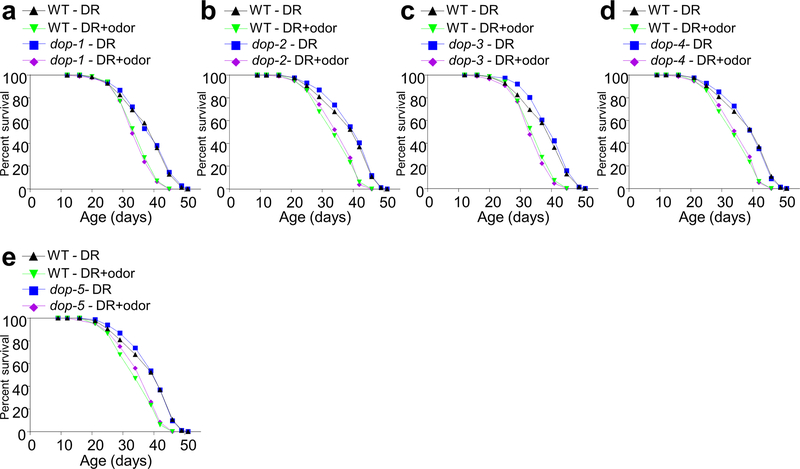
CEP neurons are dopaminergic. We thus hypothesized that a dopamine receptor may act in the downstream RIC neurons to transmit odor signals by responding to dopamine released from CEP neurons. By screening mutants of all the six dopamine receptor genes encoded by the worm genome: dop-1, dop-2, dop-3, dop-4, dop-5 and dop-626, we found that food odors lost the ability to suppress DR longevity in dop-6 but not the other five dop mutant worms (Fig.3i and Extended Data Fig.4a–e). Similarly, RIC neurons in dop-6 mutant worms lost the ability to respond to food odors in calcium imaging assay (Fig.3j–k). Both phenotypes were rescued by transgenic expression of wild-type dop-6 gene in RIC neurons (Fig.3i–k). Thus, DOP-6 may function as the dopamine receptor in RIC neurons to transmit odor signals in the circuit.
Together, our data suggest a model that food odors suppress DR longevity via an olfactory circuit comprising three pairs of neurons: ADF, CEP and RIC (Fig.3l). In this circuit, we suggest that ADF neurons function as primary sensory neurons to sense food odors through a G protein-mediated transduction mechanism; ADF neurons then stimulate CEP neurons, which in turn inhibit RIC neurons; SER-5 and DOP-6 function in CEP and RIC neurons to transmit odor signals by responding to the neurotransmitter serotonin and dopamine released from upstream neurons, respectively (Fig.3l). As food odors inhibit RIC neurons to suppress DR longevity, this suggests that the normal output of RIC neurons is to promote longevity. If so, then inhibiting the output of RIC neurons should mimic the inhibitory effect of food odors on DR longevity. Indeed, inhibiting the output of RIC neurons with a TeTx transgene shortened DR longevity (Extended Data Fig.2p), providing further evidence that food odors suppress DR longevity by inhibiting the output of RIC octopamine neurons.
The olfactory circuit signals the intestine to regulate the energy sensor AMPK
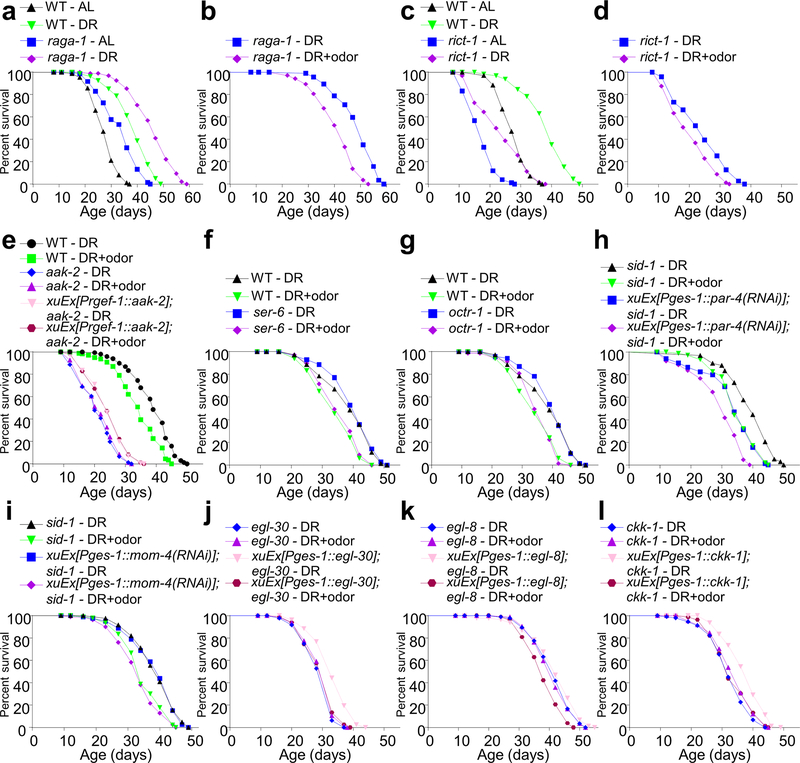
We then wondered how the olfactory circuit engages the rest of the animal body to regulate DR longevity. As DR longevity pathways all converge on nutrient signaling2–4, we first asked which nutrient signaling underlies DR longevity in our lifespan assay. AMPK and mTOR signaling are the two primary nutrient signaling that regulate DR longevity in worms2,3. Consistent with previous work10, we found that loss of the C. elegans AMPKα ortholog AAK-2 abolished the ability of DR to extend lifespan (Fig.4a), demonstrating that AAK-2 is required for DR longevity. This supports the notion that AMPK mediates the longevity under the sDR regimen10. Notably, aak-2 mutant worms were also insensitive to food odors (Fig.4a), indicating a requirement of AMPK for food odors to suppress DR longevity. By contrast, DR longevity persisted in raga-1 and rict-1 mutant worms (Extended Data Fig.5a, 5c), which were deficient in mTORC1 and mTORC2 signaling, respectively 27, and these two mutants also remained sensitive to food odors (Extended Data Fig.5b, 5d). Thus, both DR longevity and its sensitivity to food odors require AMPK but not mTOR signaling in our DR regimen. Notably, transgenic expression of wild-type aak-2 gene in the intestine rescued both the longevity and odor sensitivity defects in aak-2 mutant worms (Fig.4b). By contrast, expression of aak-2 as a transgene in neurons only had a slight rescue effect on the longevity defect (Extended Data Fig.5e). Neuronal expression of aak-2 also failed to rescue the odor sensitivity defect in aak-2 mutant worms. Thus, AAK-2 primarily acts in the intestine. Additional evidence came from an RNAi experiment, in which we knocked down aak-2 expression in the intestine by expressing dsRNA of aak-2 as a transgene specifically in the intestine, and found that it blocked the ability of food odors to suppress DR longevity (Fig.4c). This experiment was conducted in sid-1 background to restrict RNAi to the tissue of interest28. These results together suggest that the olfactory circuit signals the intestine to regulate the energy sensor AMPK to suppress DR longevity, revealing a brain-gut signaling axis.
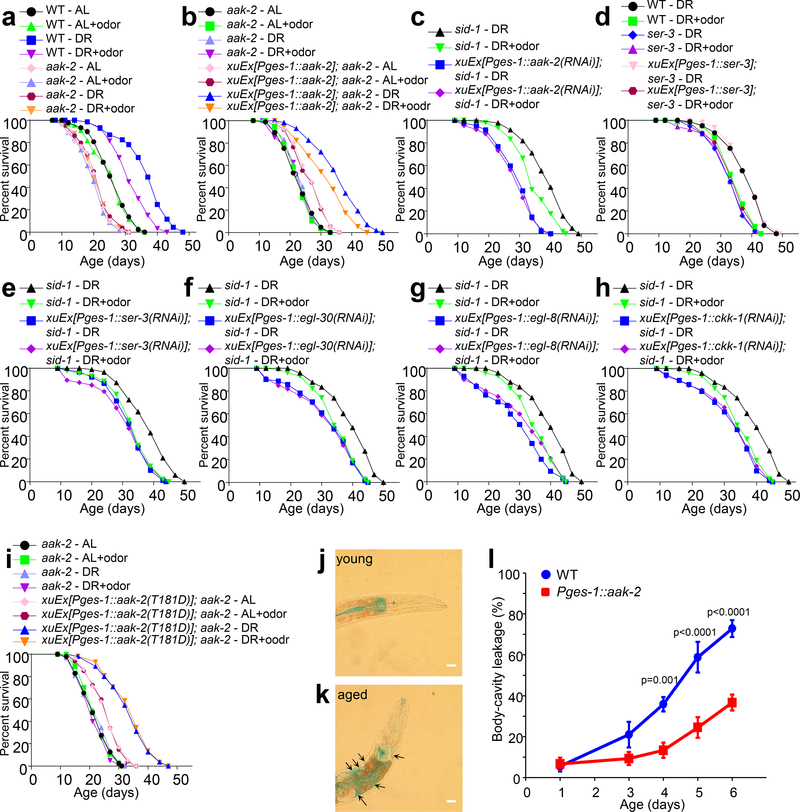
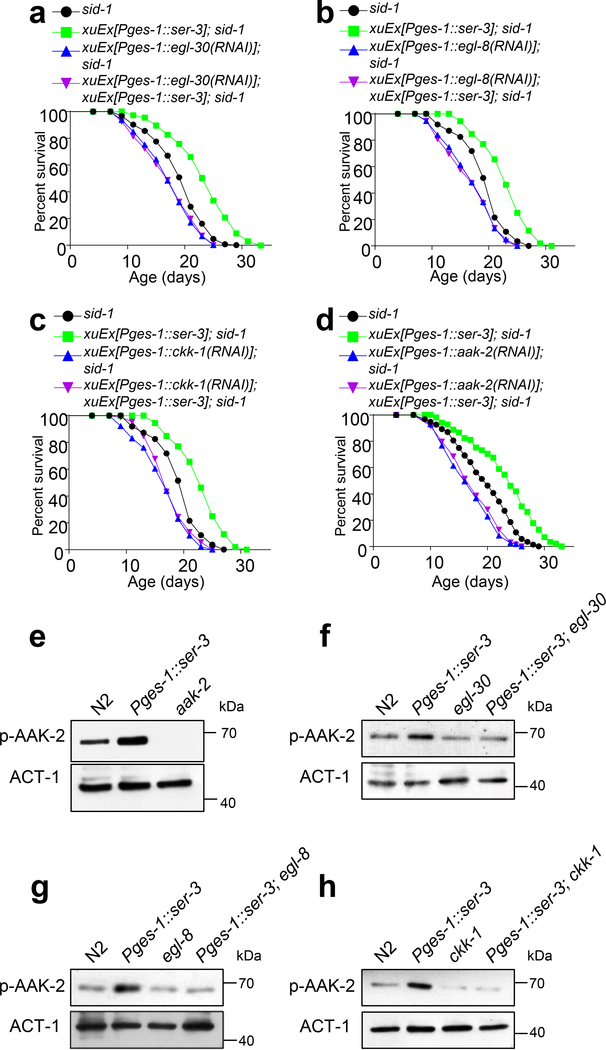
Fig.4 |. The olfactory circuit signals the intestine to regulate the energy sensory AAK-2/AMPK via Gq-PLCβ-CaMKK-dependent norepinephrine signaling.
(a) aak-2 mutant worms are completely insensitive to DR and food odors.
(b) Transgenic expression of wild-type aak-2 gene in the intestine rescues both the longevity and odor sensitivity phenotypes in aak-2 mutant worms.
(c) RNAi of aak-2 in the intestine abolishes the ability of food odors to suppress DR longevity. dsRNA against aak-2 was expressed as a transgene specifically in the intestine using the ges-1 promoter. All experiments were carried out in sid-1 mutant background where the systemic effect of RNAi is absent.
(d) Loss of ser-3 abolishes the ability of food odors to suppress DR longevity, a phenotype rescued by transgenic expression of wild-type ser-3 gene in the intestine using the ges-1 promoter.
(e) Intestine-specific RNAi of ser-3 by a dsRNA transgene prevented food odors from suppressing DR longevity.
(f-h) Intestine-specific RNAi of egl-30/Gq (f), egl-8/PLCβ (g) or ckk-1/CaMKK (h) by dsRNA transgenes abolishes the ability of food odors to suppress DR longevity. (f), (g) and (h) share the same sid-1 control curves, as these experiments were performed at the same time.
(i) aak-2(T181D) transgene rescues the DR longevity defect but not the odor sensitivity defect of aak-2 mutant worms.
(j-l) Intestinal expression of AAK-2/AMPK slows down the age-dependent decline in intestinal barrier function. The blue dye (FD&C blue), which is impermeable to the intestine epithelium, was taken up into the intestine by the worm. The dye was confined inside the intestinal lumen in young worms; but as worms age, the dye leaked into the body cavity. Worms were cultured at 25oC. (j) Young wild-type worm (day 1). (k) Aged wild-type worm (day 6), showing that the dye leaked into the body cavity. Arrows point to the dye outside of the intestine. Scale Bar: 25 μm. (l) Bar graph showing that the intestinal aak-2 transgene Pges-1::aak-2 slowed down the age-dependent dye leakage. The percentage of worms showing body-cavity dye leakage was quantified. Each data point was derived from 10 worms and repeated five times. Data are presented as mean ± s.e.m. p values were calculated with two-way ANOVA with Bonfferronìs test.
See Supplementary Table 1 for lifespan statistics.
Given that AAK-2/AMPK expression in the intestine extended lifespan (Extended Data Fig.6a), we wondered if it could also promote the health of the intestine. A primary function of the intestine epithelium is to forms a selective barrier that allows it to absorb nutrients, ions and water but remain impermeable to toxic substances and microorganisms29,30. Like other animals29,31, the barrier function of C. elegans intestine declines with age progressively, which renders the intestine of aged worms permeable to otherwise impermeable chemicals, resulting in their leak into the body cavity30 (Fig. 4j–k). Intestinal expression of AAK-2 greatly slowed down the aged-dependent decline in the intestinal barrier function (Fig.4l), indicating that AMPK can promote the health of the intestine.
Gq-PLCβ-CaMKK-dependent octopamine signaling regulates AMPK in the intestine
How does the olfactory circuit signal the intestine to regulate AMPK? As RIC neurons are octopamine neurons acting downstream in the circuit (Fig.3l), we reasoned that RIC neurons might signal the intestine via octopamine. Consistent with this model, food odors failed to suppress DR longevity in tbh-1 mutant worms deficient in octopamine production (Fig.1i). We then attempted to identify the octopamine receptor that functions in the intestine to transmit odor signals to regulate AMPK. Among all the three worm octopamine receptor genes (i.e. ser-3, ser-6 and octr-1)26, loss of ser-3 but not the other two receptor genes abrogated the ability of food odors to suppress DR longevity (Fig.4d and Extended Data Fig.5f–g). Transgenic expression of wild-type ser-3 gene in the intestine rescued the odor sensitivity defect in ser-3 mutant worms (Fig.4d). In addition, RNAi of ser-3 specifically in the intestine of wild-type worms recapitulated the ser-3 mutant phenotype (Fig.4e). These data identify SER-3 as the octopamine receptor that acts in the intestine to transmit signals from the olfactory circuit to regulate AMPK.
The question arises as to how the octopamine receptor SER-3 regulates AMPK in the intestine. SER-3 is coupled to Gq/PLCβ-mediated calcium signaling32. We thus examined the worm Gq ortholog EGL-30 and PLCβ ortholog EGL-8. RNAi of egl-30 and egl-8 specifically in the intestine prevented food odors from suppressing DR longevity (Fig.4f–g), indicating a requirement for Gq and PLCβ in the pathway. Then how does Gq-PLCβ couple SER-3 to AMPK? AMPK activation requires phosphorylation of its catalytic α subunit by an AMPK kinase33. Among the three AMPK kinases CaMKK, LKB1 and TAK1, CaMKK is the only one that is activated by calcium signaling33. As Gq-PLCβ activation triggers calcium signaling34, CaMKK emerges as a candidate AMPK kinase that couples SER-3-Gq-PLCβ to AMPK. Indeed, RNAi of the worm CaMKK gene ckk-1 specifically in the intestine prevented food odors from suppressing DR longevity (Fig.4h), while RNAi of the other two putative AMPK kinase genes par-4/LKB1 and mom-4/TAK1 did not (Extended Data Fig.5h–i). This result suggests that CKK-1/CaMKK is an AMPK kinase that acts downstream of Gq-PLCβ to activate AMPK. As a complementary approach, we examined egl-30, egl-8, and ckk-1 mutant worms, and found that they all lost the sensitivity to food odors in lifespan assay, a phenotype that was rescued by transgenic expression of wild-type egl-30, egl-8, and ckk-1 genes in the intestine, respectively (Extended Data Fig.5j–l). These observations together suggest that the octopamine receptor SER-3 regulates AMPK in the intestine via a Gq-PLCβ-CaMKK-dependent mechanism.
To provide additional evidence, we overexpressed the octopamine receptor SER-3 as a transgene in the intestine, and found that it extended lifespan under normal condition (Fig.5a), suggesting that activation of octopamine signaling in the intestine promotes longevity. This is also consistent with our observation that the output of RIC octopamine neurons in the olfactory circuit was to promote longevity. Importantly, the SER-3-dependent longevity was fully suppressed by intestine-specific RNAi of all the downstream components, i.e. egl-30/Gq, egl-8/PLCβ, ckk-1/CaMKK, and aak-2/AMPK (Fig.5a–d). This provides additional evidence that the octopamine receptor SER-3 regulates AMPK in the intestine via a Gq-PLCβ-CaMKK-dependent mechanism.
Fig. 5 |. Octopamine signaling in the intestine promotes lifespan and stimulates AMPK phosphorylation via a Gq-PLCβ-CaMKK-dependent mechanism.
(a-d) Transgenic expression of the octopamine receptor SER-3 in the intestine (Pges-1::ser-3) extends lifespan under normal condition (a), and this lifespan-extending effect is blocked by intestine-specific RNAi of egl-30/Gq (a), egl-8/PLCb (b), ckk-1/CaMKK (c), and aak-2 (d) with corresponding dsRNA transgenes. (b) and (c) share the same sid-1 control curves, as these experiments were performed at the same time.
(e) Transgenic expression of SER-3 in the intestine (Pges-1::ser-3) stimulates AAK-2/AMPK phosphorylation. Samples collected from aak-2 mutant worms was used to demonstrate the specificity of the antibody. Actin (ACT-1) was used as a loading control.
(f-h) Loss of egl-30/Gq (f), egl-8/PLCb (g) or ckk-1/CaMKK (h) inhibits SER-3-depedent stimulation of AAK-2/AMPK phosphorylation.
See Supplementary Table 1 for lifespan statistics.
To obtain further evidence, we assessed whether the octopamine receptor SER-3 can promote AMPK activity, and if so, whether it acts in a Gq-PLCβ-CaMKK-dependent manner. Transgenic expression of SER-3 in the intestine stimulated AAK-2 phosphorylation (Fig.5e), demonstrating that intestinal SER-3 can promote the activity of AMPK. Importantly, this SER-3-dependent AAK-2 activation was abrogated in egl-30/Gq, egl-8/PLCβ and ckk-1/CaMKK mutant worms (Fig.5f–h). This experiment provides biochemical evidence that the octopamine receptor SER-3 promotes AMPK activity in a Gq-PLCβ-CaMKK-dependent manner.
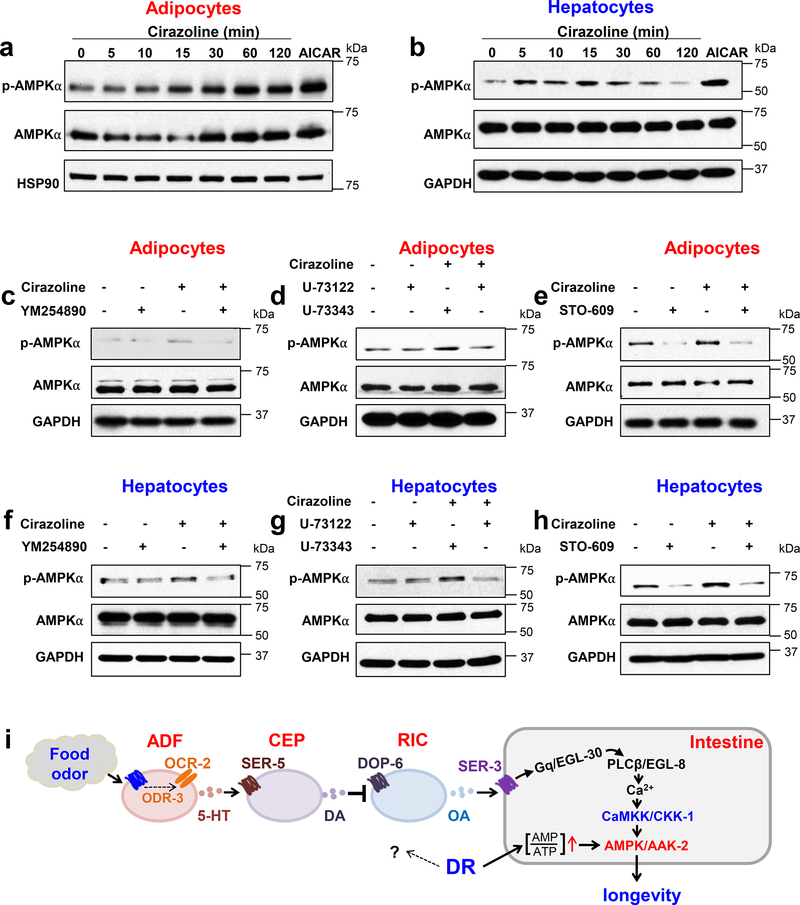
Activation of norepinephrine signaling in mouse primary cells regulate AMPK via a similar Gq-PLCβ-CaMKK-dependent mechanism
Octopamine is the invertebrate homolog of norepinephrine. We then wondered if norepinephrine signaling could regulate AMPK in mammalian cells via a similar Gq-PLCβ-CaMKK-dependent mechanism. In C. elegans, the intestine also fulfills many of the complex functions of mammalian fat and liver tissues35. We thus characterized mouse primary subcutaneous adipocytes and primary hepatocytes, both of which express the Gq-coupled α1A adrenergic receptor36,37. Selective activation of α1A adrenergic receptor with cirazoline stimulated AMPK phosphorylation in both mouse primary subcutaneous adipocytes and primary hepatocytes (Fig.6a–b), indicating that activation of norepinephrine signaling can promote AMPK activity in these primary cells. Importantly, this AMPK activation was blocked by inhibitors of Gq (YM254890), PLC (U73122 but not its inactive analog U73343), and CaMKK (STO-609) (Fig.6c–h). This set of data suggests that activation of norepinephrine signaling in mouse primary cells can stimulate AMPK via a Gq-PLCβ-CaMKK-dependent mechanism.
Fig.6 |. Activation of norepinephrine signaling stimulates AMPK via a Gq-PLCβ-CaMKK-dependent mechanism in mouse primary cells.
(a) Selective activation of α1A-adrenergic receptor activates AMPK in mouse primary subcutaneous adipocytes. Cirazoline (100 μM), a selective agonist for α1A-adrenergic receptor, can stimulate the phosphorylation of T172 site in AMPKα. AICAR (500 μM, 15 min), an AMPK activator, was used as a positive control.
(b) Selective activation of α1A-adrenergic receptor activates AMPK in mouse primary hepatocytes. Cirazoline (100 μM), a selective agonist for α1A-adrenergic receptor, can stimulate the phosphorylation of T172 site in AMPKα. AICAR (500 μM, 15 min), an AMPK activator, was used as a positive control.
(c-e) Inhibition of Gq (c), PLCβ (d) or CaMKK (e) abolishes cirazoline-induced (100 mM, 60 min) phosphorylation of T172 site in AMPKα in mouse primary subcutaneous adipocytes. Cells were pre-treated with the following inhibitors for 30 min: the Gq inhibitor YM254890 (25 μM), the PLC inhibitor U73122 (5 μM), U73343 (5 μM) that is an inactive analog of U73122, and the CaMKK inhibitor STO-609 (5 μM).
(f-h) Inhibition of Gq (f), PLCβ (g) or CaMKK (h) abolishes cirazoline-induced (100 μM, 15 min) phosphorylation of T172 site in AMPKα in mouse primary hepatocytes. Cells were pre-treated with the following inhibitors for 30 min: the Gq inhibitor YM254890 (25 μM), the PLC inhibitor U73122 (5 μM), U73343 (5 μM) that is an inactive analog of U73122, and the CaMKK inhibitor STO-609 (5 μM).
(i) Schematic model. 5-HT: serotonin. DA: dopamine. OA: octopamine.
Food odors and DR converge on AMPK to regulate longevity
One notable observation is that among all the genes characterized in this study, AMPK is unique in that it is the only one that, when mutated, blocked the effects from both DR and food odors. Namely, aak-2/AMPK mutants were completely insensitive to both DR and food odors (Fig.4a). By contrast, though mutants of other genes in the olfactory circuit and the downstream octopamine signaling failed to respond to food odors, they remained sensitive to DR at least partially in lifespan assay (Supplementary Table 1). This unique feature of AMPK suggests that food odors and DR may converge on AMPK to regulate lifespan. AMPK is an energy sensor sensitive to DR3,33. Specifically, DR leads to an increase in AMP::ATP ratio, resulting in activation of AMPK3,33. Notably, AMPK is under dual-regulation, as its activation also requires phosphorylation by AMPK kinases such as CaMKK3,33, which we showed is regulated by octopamine signaling residing downstream of the olfactory circuit. Thus, AMPK may sit at a unique position to integrate signals from DR and food odors (Fig.6i). To test this, we mutated the CKK-1/CaMKK phosphorylation site in AAK-2/AMPK from Thr to Asp (i.e. AAK-2(T181D)) to render it insensitive to CKK-1/CaMKK, thereby uncoupling it from food odors. This AAK-2(T181D) is functional, as it rescued the DR longevity defect in aak-2 mutant worms (Fig.4i). Remarkably, worms expressing AAK-2(T181D) were insensitive to food odors (Fig.4i), indicating that this mutant form of AAK-2/AMPK lost the ability to integrate signals from DR and food odors. This provides further evidence that food odors and DR converge on AAK-2/AMPK to regulate longevity.
Discussion
In the current study, we investigated how food-associated odors regulate DR longevity in C. elegans. The fact that food odors suppress the lifespan of worms under DR but not those fed AL highlights the notion that in addition to the actual food abundance (nutrient level), the perception of food abundance (from food odors) is also important for longevity5. Our results suggest a model that food odors suppress DR longevity by acting on an olfactory circuit, which signals the gut intestine via octopamine, the invertebrate homology of norepinephrine, to regulate the energy sensor AMPK through a Gq-PLCβ-CaMKK-dependent mechanism (Fig.6i). Interestingly, norepinephrine signaling can also regulate AMPK through a similar mechanism in mouse primary cells. These results identify a brain-gut axis through which food odors suppress DR-mediated longevity, illustrating how non-nutrient food components may regulate lifespan by altering animals’ perception of food abundance.
The olfactory circuit comprises three pairs of neurons: ADF, CEP and RIC. In this circuit, food odors stimulate ADF to inhibit RIC via CEP (Fig.6i). We also characterized the molecular mechanisms by which the circuit senses and process odor signals by identifying some of the key components mediating sensory transduction and sensory processing (Fig.6i). While CEP forms direct connections with RIC, no such connections are found between ADF and CEP14, suggesting that part of this circuit is neuroendocrine by nature. We thus do not exclude the possibility that other neurons or cells may also be part of this circuit. For example, some other chemosensory neurons have also been suggested to modulate longevity6,12,38; however, whether these neurons can directly sense food odors to regulate lifespan remains to be determined. One interesting observation is that food odors elicited robust calcium responses in ADF/CEP/RIC neurons from DR worms, but not those fed AL, revealing a DR-specific effect. It is possible that DR sensitized the neurons in the circuit. Alternatively but not mutually exclusively, AL may suppress the circuit responsiveness. In humans, fasting potentiates olfactory sensitivity, while satiety suppresses it39, unveiling an interesting similarity between worms and mammals. Future studies will determine how DR and AL differentially regulate the sensitivity of the olfactory system to food odors.
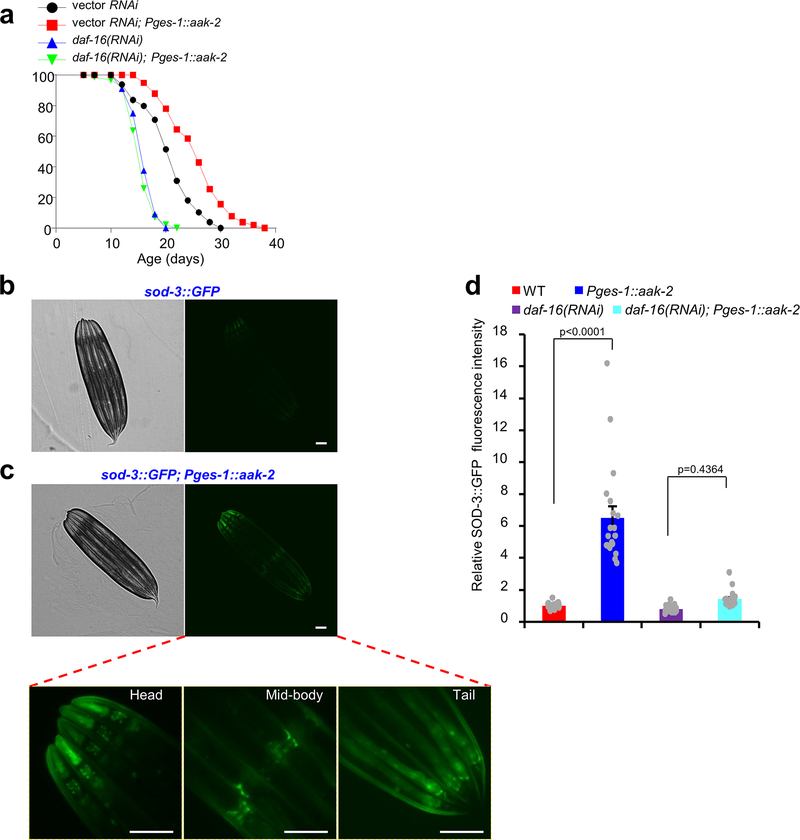
AMPK sits at a unique position to integrate signals from both food odors and DR due to its dual-regulation mode by AMP::ATP ratio (DR) and CaMKK (food odors). AMPK primarily acts in the intestine to mediate this effect. Nevertheless, we do not exclude a role for AMPK in other tissues such as neurons, given that AMPK has the capacity to regulate lifespan cell-non-autonomously40. AMPK is probably not the only substrate regulated by both DR and food odors. For example, DR also regulates the sensitivity of the olfactory circuit to food odors. Nonetheless, our results suggest that AMPK is a primary site where food odors and DR converge to regulate longevity. Then what act downstream of AMPK? One downstream effector of AMPK is the transcription factor DAF-16/FOXO10, which is a master regulator of longevity. AMPK regulates DAF-16/FOXO via direct phosphorylation10,41. We found that AAK-2/AMPK-dependent lifespan extension requires DAF-16 (Extended Data Fig.6a), and that AAK-2/AMPK expression in the intestine promoted the expression of the DAF-16/FOXO target gene sod-3 in a DAF-16-dependent manner (Extended Data Fig.6b–d). SOD-3 expression was up-regulated cell-non-autonomously in many other tissues, indicating that the longevity signal was disseminated throughout the body (Extended Data Fig.6c), consistent with the notion that the intestine is a signaling hub for longevity regulation1. Another effector of AMPK could be mitochondria, as AMPK regulates mitochondria function and dynamics to modulate longevity42. Thus, it is likely that multiple AMPK effectors may act downstream to regulate DR longevity.
We identify octopamine as a signaling molecule that mediates brain-gut communications. Specifically, our data suggest that food odors suppress octopamine signaling in the intestine by inhibiting octopamine release from the olfactory circuit. We also show that octopamine signaling regulates AMPK in the intestine via a Gq-PLCβ-CaMKK-dependent mechanism. Octopamine is the invertebrate homolog of norepinephrine. Remarkably, activation of norepinephrine signaling in mouse primary cells can stimulate AMPK in a similar manner, suggesting a conserved mechanism. Interestingly, blocking olfactory sensation in mice stimulates norepinephrine release from sympathetic nerves and promotes norepinephrine signaling in fat tissues, leading to improved energy metabolism and protection against obesity43. Conversely, enhancing olfaction in mice causes insulin resistance and obesity43. This raises the intriguing possibility that olfaction might inhibit longevity in mice through norepinephrine signaling, pointing to another potential similarity between worms and mice. It would be interesting to test whether those olfaction deficient mice are long-lived.
Materials and methods
Strains
C. elegans strains were maintained at 20°C on nematode growth medium (NGM) plates seeded with OP50 bacteria unless otherwise specified. Transgenic lines were generated by injecting plasmid DNA directly into hermaphrodite gonad following standard protocol. Mutant strains were outcrossed at least six times before use. For genetic crosses, all genotypes were confirmed using PCR and, if necessary, followed by Sanger sequencing to verify single nucleotide mutations. The strains used are:
| TQ3030 | N2 (Wild-type) |
| TQ4931 | tph-1(mg280) II |
| TQ4935 | cat-2(e1112) II |
| TQ4927 | tbh-1(n3247) X |
| TQ4929 | tdc-1(n3419) II |
| TQ4933 | eat-4(ky5) III |
| TQ6452 | unc-17(e245) IV |
| TQ4168 | unc-25(e156) III |
| TQ1280 | unc-31(e169) IV |
| TQ9677 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2] |
| TQ9984 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat-1::mcherry2] |
| TQ10132 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2] |
| TQ10049 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherrry2]; unc-13(e51) I |
| TQ10046 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; unc-31(e169) IV |
| TQ10047 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; cat-2(e1112) II |
| TQ10042 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; tbh-1(n3247) X |
| TQ10003 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat-1::mcherry2]; tph-1(mg280) II |
| TQ10011 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat-1::mcherry2]; xuEx54a[Pbas-1(prom7)::TeTx::sl2::mcherry2+Punc-122delta::gfp] |
| TQ10045 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat-1::mcherry2]; tbh-1(n3247) X |
| TQ9923 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]; tph-1(mg280) II |
| TQ10012 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]; xuEx54a[Pbas-1(prom7)::TeTx::sl2::mcherry2+Punc-122delta::gfp] |
| TQ9858 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]; cat-2(e1112) II |
| TQ9939 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]; xuEx201a[Pdat-1::TeTx::sl2::mcherry2] |
| TQ9573 | xuEx54a[Pbas-1(prom7)::TeTx::sl2::mcherry2+Punc-122delta::gfp] |
| TQ9571 | xuEx201a[Pdat-1::TeTx::sl2::mcherry2] |
| TQ9570 | xuEx197a[Ptbh-1::TeTx::sl2::mcherry2] |
| TQ600 | odr-3(n2150) V |
| TQ5663 | ocr-2(ak47) IV |
| TQ9883 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; odr-3(n2150) V |
| TQ9860 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; ocr-2(ak47) IV |
| TQ10107 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; xuEx3402[Pbas-1 (prom7)::odr-3(cDNA)::sl2::cfp]; odr-3(n2150)V |
| TQ10044 | xuEx255a[Ptph-1(L)::GCaMP6f+Ptph-1(L)::mcherry2]; xuEx3399[Pbas-1 (prom7)::ocr-2(cDNA)::sl2::cfp]; ocr-2(ak47)IV |
| TQ9996 | xuEx3364[Pbas-1(prom7)::odr-3(cDNA)::sl2::yfp1]; odr-3(n2150) V |
| TQ9931 | xuEx3357[Pbas-1(prom7)::ocr-2(cDNA)::sl2::yfp1]; ocr-2(ak47) IV |
| TQ1615a | ser-5(ok3087) I |
| TQ9688 | xuEx191a[Pdat-1::ser-5(cDNA)::sl2::mcherry2]; ser-5(ok3087) I |
| TQ10005 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat-1::mcherry2]; ser-5(ok3087) I |
| TQ10025 | xuEx3388[Pdat-1::myr-GCaMP6f+Pdat- 1::mcherry2]; xuEx3401[Pdat-1::ser-5(cDNA)::sl2::cfp]; ser-5(ok3087)I |
| TQ9820 | dop-6(n2090) X |
| TQ9932 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]; dop-6(ok2090) X |
| TQ10024 | xuEx3312[Ptbh-1::GCaMP6f+Ptbh-1::mcherry2]); xuEx3400[Ptbh-1::dop-6(cDNA)::sl2::cfp]; dop-6(ok2090) X |
| TQ9449 | xuEx3222[Ptbh-1::dop-6(cDNA)::sl2::yfp1]; dop-6(ok2090) X |
| TQ6682 | aak-2(ok524) X |
| TQ6511 | xuEx2334[Pges-1::aak-2(cDNA)::sl2::mcherry2]; aak-2(ok524) X |
| TQ1809a | sid-1(qt9) V |
| TQ9550 | xuEx3239[Pges-1::DsRed+Pges-1::aak-2(RNAi, s+as)]; sid-1(qt9) V |
| TQ9779 | ser-3(ok1995) I |
| TQ9503 | xuEx235a[Pges-1::ser-3(cDNA)::sl2::yfp1]; ser-3(ok1995) I |
| TQ9770 | xuEX3307[Pges-1::DsRed+Pges-1::ser-3(RNAi,s+as)]; sid-1(qt9) V |
| TQ9557 | xuEx3229[Pges-1::DsRed+Pges-1::egl-30(RNAi,s+as)]; sid-1(qt9) V |
| TQ9555 | xuEx3227[Pges-1::DsRed+Pges-1::egl-8(RNAi,s+as)]; sid-1(qt9) V |
| TQ9551 | xuEx3223[Pges-1::DsRed+Pges-1::ckk-1(RNAi,s+as)]; sid-1(qt9) V |
| TQ9548 | xuEx235a[Pges-1::ser-3(cDNA)::sl2::yfp1]; sid-1(qt9) V |
| TQ9458 | xuEx235a[Pges-1::ser-3(cDNA)::SL2::yfp1]; xuEx3229[Pges 1::DsRed+Pges-1::egl-30(RNAi,s+as)]; sid-1(qt9) V |
| TQ9456 | xuEx235a[Pges-1::ser-3(cDNA)::SL2::yfp1]; xuEx3227[Pges-1::DsRed+Pges-1::egl-8(RNAi,s+as)]; sid-1(qt9) V |
| TQ9452 | xuEx235a[Pges-1::ser-3(cDNA)::SL2::yfp1]; xuEx3223[Pges-1::DsRed+Pges-1::ckk-1(RNAi,s+as)]; sid-1(qt9) V |
| TQ9581 | xuEx235a[Pges-1::ser-3(cDNA)::sl2::yfp1]; xuEx3239[Pges-1::DsRed+Pges-1::aak-2(RNAi, s+as]; sid-1(qt9) V |
| TQ9505 | xuEx3232[Pges-1::aak-2(T181D)::sl2::mcherry2]; aak-2(ok524) X |
| TQ3114a | xuEx235a[Pges-1::ser-3::sl2::yfp1]; egl-30(ep271) I |
| TQ3115a | xuEx235a[Pges-1::ser-3::sl2::yfp1]; egl-8(n488) V |
| TQ3116a | xuEx235a[Pges-1::ser-3::sl2::yfp1]; ckk-1(ok1033) III |
Chemical reagents
Collagenase D (#11088882001) and dispase II (#04942078001) were purchased from Roche. All the cell culture media, including DMEM (#11995–073), DMEM-low glucose (#11885–084) and DMEM/F12 GlutaMAX (#10565–042), were obtained from Life Technologies. Erioglaucine disodium salt (#861146), 5-Fluoro-2’-deoxyuridine (FUDR, #F0503), 3-Isobutyl-1-methylxanthine (IBMX, #I7018), dexamethasone (#D4902), insulin (#I5500) and fetal bovine serum (FBS) (#F2442) were acquired from Sigma Aldrich. Cirazoline hydrochloride (#0888) and AICAR (#2840) were purchased from Tocris. STO-609 (#15325) and rosiglitazone (#71740) were purchased from Cayman Chemical. YM2548909 (#257–00631) was purchased from FUJIFILM Wako Chemicals. Collagenase Type IV (#LS004188) was obtained from Worthington Biochemical.
Mice
Animal care and experimental protocol were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Michigan. Wild-type C57BL/6J mice (Stock no. 00664) were obtained from the Jackson Laboratory and housed under 12 h light/12 h dark cycle with a standard rodent chow diet (5L0D, PicoLab).
Molecular biology
The following promoters were used to drive gene expression in specific neurons and tissues: tph-1(L) promoter (2.1 kb 5’UTR and 0.5 kb coding region) in serotonin neurons, including both ADF and NSM; tph-1(s) promoter (1.7 kb) in NSM neurons; bas-1(prom7) promoter (166 bp) in ADF neurons; cex-1 promoter (1.1 kb) in RIM neurons44; dat-1 promoter (0.8 kb) in CEP, ADE and PDE neurons; tbh-1 promoter (4.4 kb) in RIC neurons; rgef-1 promoter (3.4 kb) in all neurons; ges-1 promoter (3.2 kb) in the intestine. odr-3, ocr-2, ser-5, dop-6, ser-3, egl-30, egl-8, ckk-1, aak-2 cDNA were cloned by RT-PCR from total RNA isolated from wild-type (N2) worms. To generate dsRNA plasmids, ser-3, egl-30, egl-8, ckk-1, aak-2, par-4, and mom-4 fragments were amplified from genomic DNA extracted from wild-type (N2) worms. The first 12 amino acids from NCS-2 protein were fused with GCaMP6f to make myr-GCaMP6f, a membrane-targeting form of GCaMP6f. The dat-1 promoter was used to drive myr-G-CaMP6f and DsRed expression in CEP neurons to facilitate imaging of the dendritic compartments of these neurons. To make tdTomato-expressing OP50 bacteria, tdTomato coding sequence was cloned into pGEX-5x-3 vector and transformed into OP50 component cells.
Lifespan assays
Lifespan experiments were performed on NGM plates seeded with OP50 at 20°C. In all experiments, the first day of adulthood was scored as day 1. Worms that crawled off the plate, exploded or bagged were censored. DR lifespan assays (sDR) were performed as described in previous literature45. All assays were conducted on OP50. To harvest bacteria food, fresh OP50 colonies were inoculated in LB medium overnight (14–16 hr), and bacteria pellets were collected by high-speed centrifugation (4000 rpm, 4°C, 20 min). Bacteria pellets were re-suspended in 10 ml fresh LB medium and pelleted again by centrifugation, and this step was repeated one more time. Lastly, bacteria pellets were re-suspended in fresh LB medium containing 5 mg/mL carbenicillin to a final concentration of 1011 cfu/mL. Ad libitum (AL) plates were prepared with a bacteria concentration of 1011 cfu/mL (200 μl/per plate), and DR plates with a bacteria concentration of 109 cfu/mL (200 μl/per plate). 5-Fluoro-2’-deoxyuridine (FUDR) was added to NGM plates at a final concentration of 2.5 μg/ml to prevent egg-laying and bagging. Unless indicated otherwise, about 80–100 worms were transferred onto AL and DR plates at Day 4 and then transferred every 2–4 days to fresh AL or DR NGM plates at a density of 15 worms per plate until day 19–20 for DR and 15–16 for AL conditions. For lifespan assays under normal condition, about 80–100 worms of each strain were included and transferred every 2–4 days until day 11–12 as described previously46. All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, Inc.) and IBM SPSS Statistics 21 (IBM, Inc.). p values were calculated using the log-rank (Kaplan-Meier) method.
Feeding and food ingestion measurements
To quantify feeding rate, day 4 adult worms were transferred to NGM plates seeded with DR or 2xDR amount of bacteria food and randomly divided into two groups, with one group exposed to food odors and the other group not. Pharyngeal pumping rate was then counted under a stereoscope at 1 hr, 24 hr, and 96 hr after worms were transferred to DR plates at 20oC.
To quantify food ingestion, OP50-tdTomato bacteria were used as food. Worms were transferred to NGM plates seeded with DR or 2×DR amount of bacteria food and randomly divided into two groups, with one group exposed to food-derived odors and the other group not. At 1 hr post-transfer, worms were immobilized in 20 mM sodium-azide/M13 solution, mounted on 2% agarose pads, and imaged on Olympus IX73 inverted microscope under a 10x objective using ORCA-Flash4.0 LT+ Digital CMOS camera (Hammatsu Inc.) with MetaMorph software (Molecular Devices Inc.). ImageJ software was used to quantify the images.
Calcium imaging
A microfluidic system was used to perform calcium imaging as previously described47. 1–2 days after transferred onto AL or DR plates, worms were loaded onto a microfluidic device mounted on Olympus IX73 inverted microscope and incubated in M13 solution (30 mM Tris-Cl, 100 mM NaCl, 10 mM KCl, pH7.0). Images were acquired under a 40x objective using an ORCA-Flash4.0 LT+ Digital CMOS camera (Hammatsu Inc.) with MetaFluor software (Molecular Devices Inc.) at 1 Hz. GCaMP6f and DsRed were co-expressed as a transgene in specific neurons using corresponding promoters to enable ratiometric imaging under blue (484 nm) and yellow (565 nm) light. As food odor-evoked calcium responses are similar in different sub-compartments in ADF and RIC neurons, we focused on recording soma signals in these two neurons. For CEP neurons, we focused on recording dendritic signals. This is because only dendritic responses in CEP neurons are DR-dependent, while soma and axon responses do not depend on DR or the input from the upstream ADF neurons, indicating that CEP soma and axon responses are irrelevant to the DR pathway. We expressed myr-GCaMP6f in CEP to facilitate dendritic imaging. Worms were first exposed to blue-yellow light cycles for 2 min in LB medium to establish a basal line before challenged with LB medium containing food odors. To prepare LB medium containing food odors, OP50 bacteria were cultured in LB medium overnight (14–16 hr), and the medium was collected by two rounds of high-speed centrifugation (each at 4000 rpm, 4°C, 20 min) to remove all bacteria, followed by filtering through a 0.45 μm filter. Such medium was freshly prepared each time for the imaging experiment. Background fluorescence was subtracted when calculating the ratio of GCaMP/DsRed. The peak fold change in the ratio of GCaMP/DsRed fluorescence was analyzed.
Mouse primary cell isolation and culture
To prepare mouse primary subcutaneous adipocytes, the stromal vascular fraction (SVF) was first isolated from inguinal adipose tissues of C57BL/6J mice (age: 6–8 weeks; gender: male and female) cultured and differentiated as described previously48. Specifically, inguinal adipose tissues were dissected, washed in PBS, minced and digested in PBS containing collagenase D (1.5 U/mL), dispase II (2.4 U/mL) and 10 mM CaCl2 for 20 min in a 37 °C water bath with agitation. Digestion reaction was terminated by addition of culture medium consisting of DMEM/F12 GlutaMAX supplemented with 10 % FBS and 1% penicillin/streptomycin. Tissue suspension was passed through a 100 μm cell strainer and centrifuged at 300–500 × g for 5 min to pellet SVF cells. The cell pellet was resuspended in culture medium, filtered through a 40 μm cell strainer and centrifuged as above. Isolated SVF cells were plated onto a collagen-coated 10 cm cell culture dish and grown in culture medium. Adipogenesis was induced with culture medium supplemented with 0.5 μg/mL insulin, 5 μM dexamethasone, 1 μM rosiglitazone and 0.5 mM IBMX in confluent cells. After 2 days of induction, cells were maintained in culture medium containing 0.5 μg/mL insulin for 3 days. Differentiated adipocytes were stimulated with 100 μM Cirazoline for varying time durations as indicated in the figure or with 500 μM AICAR (15 min). To test inhibitors, cells were pre-treated with 25 μM YM254890, 5 μM U73122, 5 μM U73343 or 5 μM STO-609 for 30 min, followed by 100 μM Cirazoline treatment for 60 min before harvested for immunoblotting.
Primary hepatocytes were isolated from C57BL/6J mice as previously reported 49. Mouse was anesthetized, and the liver was perfused via an inferior vena cava with washing buffer (HBSS buffer containing 0.5 mM EGTA and 25 mM HEPES, pH 7.4), followed by digestion medium (DMEM-low glucose supplemented with 200 mg/L CaCl2, 1% penicillin/streptomycin, 15 mM HEPES and 100 U/mL collagenase IV). The digested liver was excised, diced in digestion medium and filtered through a 70 μm cell strainer. Isolated hepatocytes were washed twice and plated onto collagen-coated culture plates with isolation medium (DMEM/F12 GlutaMAX supplemented with 10% FBS, 1% penicillin/streptomycin, 1 μM dexamethasone and 0.1 μM insulin) for one hour. The medium was replaced with culture medium (DMEM-low glucose containing 10% FBS, 1% penicillin/streptomycin, 0.1 μM dexamethasone and 1 nM insulin). After 3 hours, hepatocytes were maintained in culture medium without FBS overnight until treatment with 100 μM Cirazoline (varying time durations as indicated in the figure) or 500 μM AICAR (15 min). To test inhibitors, hepatocytes were pre-treated with 25 μM YM254890, 5 μM U73122, 5 μM U73343 or 5 μM STO-609 for 30 min, followed by 100 μM Cirazoline treatment for 15 min before harvested for immunoblotting.
Immunoblotting.
Mouse primary cells were lysed in ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Roche) and phosphatase inhibitors (10 mM NaF, 60 mM β-glycerolphosphate, pH 7.5, 2 mM sodium orthovanadate and 10 mM sodium pyrophosphate). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with the following primary antibodies obtained from Cell Signaling Technology: phospho-AMPKαT172 (#2531), AMPKα (#2532), GAPDH (#5174) and HSP90 (#4874).
Total proteins from ~120 day-1 adult worms grown on OP50 bacteria at 20°C were extracted with 120 μl RIPA lysis buffer supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitors (Sigma-Aldrich) by ultrasound sonication. The samples were then heated to 100°C for 10 min, followed by high-speed centrifugation (13000 rpm, 4°C, 5 min). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes, which were probed with the following primary antibodies obtained from Cell Signaling Technology: phospho-AMPKαT172 (#2531), β-actin (#4970). The phospho-AMPKαT172 antibody recognizes the phosphorylated form of C. elegans AAK-2/AMPK 50. As AMPKα (#2532) antibody does not recognize C. elegans AAK-2/AMPK, we used β-actin as a control for protein loading.
Intestinal barrier function assay
Worms were cultured at 25°C and were removed at different days from NGM plates and incubated for 3 hours in M9 medium containing fresh E.coli OP50 (OD = 0.5–0.6) mixed with 5% FD&C blue No.1 dye (Erioglaucine disodium salt, Sigma-Aldrich). Subsequently, worms were collected and washed with fresh M9 buffer for 3 times and transferred to fresh NGM plates with fresh OP50 bacteria. Next, worms were immobilized on an agarose pad with 5 mM sodium azide, and images were acquired on an Olympus upright microscope with a digital camera (Canon). We quantified the percentage of worms with body-cavity leakage, characterized by the presence of the dye outside the intestine.
Statistics and reproducibility
Samples were randomized and treated under the same conditions. The sample sizes were not pre-determined with a statistical method, but they are similar to those reported previously46,51,52. The number of independent replicates was indicated in Supplemental Table 1 for lifespan assay. For other assays, experiments were repeated independently at least twice with similar results. Data collection and analysis were not performed blindly. We assumed data distribution to be normal, but did not test it formally. No data were excluded from the analysis. Quantification and statistical parameters were indicated in the legends of each figure or directly marked in the figures, including the statistical method, error bars, n numbers, and p values. We applied one-way ANOVA, two-way ANOVA, student`s t-test and Log-Rank test to determine statistical significance. Specifically, for those analyses involving multiple group comparisons, we applied one-way ANOVA followed by a post hoc test (Bonferroni test). In the case of factor analysis (Fig. 4l), we applied two-way ANOVA followed by a post hoc test (Bonferroni test). For those only involving two groups, we applied two-tailed student`s t-test. Life span comparisons were calculated by Log-Rank test. P values less than 0.05 are considered statistically significant. One-way or two-way ANOVA tests were performed using Prism 8 (GraphPad Software). Two-tailed student`s t tests were performed using Excel (Microsoft Office). All Lifespan analyses were performed using SPSS Statistics software (IBM, Inc).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Additional data related to food odor suppression of DR longevity without increasing food ingestion.
(a-b) Food odors do not change the pumping rate of worms under DR. Pumping rate was counted at 24 hr (a), and 96 hr (b), after worms were transferred to the DR assaying plates. 2xDR: twice amount of bacterial food, which stimulated the pumping rate. n = 30 (DR), 30 (DR + odor), 16 (2xDR) and 16 (2xDR + odor) biologically independent animals in (a). n = 30 (DR), 30 (DR + odor), 15 (2xDR) and 14 (2xDR + odor) biologically independent animals in (b). Data are presented as mean ± s.e.m. p values were calculated with one-way ANOVA with Bonfferronìs test. See Fig.1c for data of the pumping rate counted at 1 hr post-transfer. (c) Feeding worms with twice amount of bacteria food (2x DR) does not affect DR longevity, but food odors can still suppress the lifespan of these worms.
Extended Data Fig. 2. Additional data related to the olfactory circuit.
a) ADF dendrite, soma and axon from DR, but not AL worms, all respond robustly to medium containing food odors. GCaMP6f was expressed as a transgene in ADF neurons under tph-1 long promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM).
(b) NSM neurons (dendrite/soma/axon) from DR and AL worms do not respond to medium containing food odors. GCaMP6f was expressed as a transgene in the NSM neurons using tph-1(s) promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM).
(c) Bar graph summarizing data in (a) and (b). n=11 (ADF dendrite - AL), 10 (ADF dendrite - DR), 8 (ADF soma - AL), 9 (ADF soma - DR), 11 (ADF axon - AL), 10 (ADF axon - DR), 11 (NSM soma - AL), 14 (NSM soma - DR), 13 (NSM processes - AL), and 10 (NSM processes - DR) biologically independent animals.
(d) CEP dendrite from DR worms, but not AL worms, responds robustly to medium containing food odors. GCaMP6f was expressed as a transgene in CEP neurons using dat-1 promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM). (e) CEP soma and axon from both DR and AL worms respond to medium containing food odors, showing no specificity towards DR. Shades along the traces represent error bars (SEM). (f) Bar graph summarizing data in (d) and (e). n=6(CEP dendrite - AL), 8(CEP dendrite - DR), 13(CEP soma - AL), 20(CEP soma - DR), 12(CEP axon - AL) and 17(CEP axon - DR) biologically independent animals. (g-h) ADE and PDE neurons (soma and processes) from DR and AL worms do not respond to medium containing food odors. GCaMP6f was expressed as a transgene in ADE and PDE neurons using dat-1 promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM). (i) Bar graph summarizing data in (g) and (h). n=11 (ADE soma - AL), 10 (ADE soma - DR), 13 (ADE processes - AL), 15 (ADE processes - DR), 10 (PDE soma - AL), 11 (PDE soma - DR), 10 (PDE processes - AL) and 11 (PDE processes - DR) biologically independent animals. (j) RIC soma and processes from DR worms, but not AL worms, respond robustly to medium containing food odors. GCaMP6f was expressed as a transgene in RIC neurons under tbh-1 promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM). The soma curves are duplicates of those presented in Fig.2i, as these experiments were performed at the same time. (k) RIM soma and axon from both DR and AL worms respond to medium containing food odors, showing no specificity towards DR. GCaMP6f was expressed as a transgene in RIM neurons using cex-1 promoter. DsRed was co-expressed to enable ratiometric imaging. Shades along the traces represent error bars (SEM). (l) Bar graph summarizing data in (j) and (k). n=11 (RIC soma - AL), 14 (RIC soma - DR), 11 (RIC processes - AL), 12 (RIC processes - DR), 12 (RIM soma - AL), 12 (RIM soma - DR), 12 (RIM processes - AL) and 10 (RIM processes - DR) biologically independent animals. (m-n) RNAi knockdown of odr-3 and ocr-2 specifically in ADF neurons eliminates food odor-evoked calcium responses in these neurons. dsRNA against odr-3 and ocr-2 gene was expressed as a transgene specifically in ADF neurons using the bas-1(prom7) promoter. (m) Calcium imaging traces. Shades along the traces represent error bars (SEM). (n) Bar graph summarizing the data in (m). n=10 (WT - AL), 9 (WT - DR), 10 (ADF odr-3(RNAi) - AL), 10 (ADF odr-3(RNAi) - DR), 10 (ADF ocr-2(RNAi) - AL) and 10 (ADF ocr-2(RNAi) - DR) biologically independent animals. (o) NSM neurons are not required for food odors to suppress DR longevity. tph-1(s) promoter was used to drive the expression of TeTx transgene specifically in NSM neurons.
(p) Blocking the output of RIC neuron shortens DR longevity. tbh-1 promoter was used to drive the expression of TeTx transgene in RIC neurons. Data are presented as mean ± s.e.m. p values in c, f, i and l: two-tailed student`s t test. P values in n: one-way ANOVA with Bonfferronìs test.
Extended Data Fig. 3. Other serotonin receptors are not required for food odors to suppress DR longevity.
Food odors can still suppress DR longevity in ser-1 (a), ser-4 (b), ser-7 (c) and mod-1 (d) mutant worms. (a-d) share the same control group, as these experiments were performed at the same time
Extended Data Fig. 4. Other dopamine receptors are not required for food odors to suppress DR longevity.
Food odors can still suppress DR longevity in dop-1 (a), dop-2 (b), dop-3 (c) , dop-4 (d) and dop-5 (e) mutant worms. (a) and (c) share the same control group, as these experiments were performed at the same time. (b), (d) and (e) share the same control group, as these experiments were performed at the same time.
Extended Data Fig. 5. Additional data related to regulation of DR longevity by AMPK and octopamine signaling.
a) DR can extend the lifespan of raga-1 mutant worms. (b) Food odors can suppress DR longevity in raga-1 mutant worms. (c) DR can extend the lifespan of rict-1 mutant worms. (d) Food odors can suppress DR longevity in rict-1 mutant worms. (e) Pan-neuronal expression of aak-2 gene only has a slight rescue effect on the longevity defect of aak-2 mutant worms. This aak-2 neuronal transgene also does not rescue the odor sensitivity defect of aak-2 mutant worms. rgef-1 promoter was used to drive the expression of aak-2 cDNA in neurons. (f-g) Food odors can still suppress DR longevity in ser-6 (f) and octr-1 (g) mutant worms. (f) and (g) share the same control group, as these experiments were performed at the same time. (h) Intestine-specific knock-down of par-4/LKB1 by dsRNA transgene (Pges-1::par-4(RNAi)) does not prevent food odors from suppressing DR longevity, though it partially inhibits DR longevity. (i) Intestine-specific knock-down of mom-4/TAK1 by dsRNA transgene (Pges-1::mom-4(RNAi)) does not prevent food odors from suppressing DR longevity; nor does it affect DR longevity.
(j-l) Mutations in egl-30 (j), egl-8 (k), and ckk-1 (l) abolish the ability of food odors to suppress DR longevity, a defect that is rescued by transgenic expression of corresponding wild-type genes in the intestine using ges-1 promoter.
Extended Data Fig. 6. AAK-2/AMPK-dependent lifespan extension requires DAF-16/FOXO.
(a) Lifespan extension mediated by intestinal expression of aak-2 requires daf-16. daf-16 RNAi blocked the lifespan-extension effect of the intestinal aak-2 transgene. (b-d) Intestinal expression of aak-2 promotes sod-3 gene expression in multiple tissues in a daf-16-dependent manner. sod-3::gfp is a transgene reporting the expression level of sod-3 gene. (b) Sample images showing a low level of sod-3::gfp expression. Left: bright field image. Right: fluorescent image. (c) Sample images showing that the Pges-1::aak-2 transgene increased the expression of sod-3::gfp. Top left: bright field image. Top right: fluorescent image. Bottom: zoomed-in images showing sod-3::gfp expression in multiple tissues, including pharynx (head), neurons (head), body-wall muscles, vulval muscles (mid-body), intestine, etc. Scale Bar: 100 μm. (d) Bar graph summarizing the data in (b) and (c). n=24 (WT), 20 (Pges-1::aak-2), 43 (daf-16(RNAi)) and 22 (daf-16(RNAi); Pges-1::aak-2) biologically independent animals. Data are presented as mean ± s.e.m. p values were calculated with one-way ANOVA with Bonfferronìs test.
Supplementary Material
Acknowledgments
We thank Rui Xiao for comments on the manuscript, and Jianke Gong, Si Zhang and Hao Chen for assistance. Some strains were obtained from the CGC. J.L. received funding support from the NSFC (81872945 and 81720108031 to J.L.). J.W. received funding support from the NIDDK (R01DK107583 to J.W.). X.Z.S.X received funding support from the NIGMS (R35GM126917 to X.Z.S.X).
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
The datasets generated and analyzed during this study are either included within the manuscript or are available from the corresponding author on reasonable request.
References
- 1.Kenyon CJ The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Fontana L, Partridge L & Longo VD Extending healthy life span--from yeast to humans. Science 328, 321–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greer EL, Banko MR & Brunet A AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann N Y Acad Sci 1170, 688–692, doi: 10.1111/j.1749-6632.2009.04019.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapahi P, Kaeberlein M & Hansen M Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res Rev 39, 3–14, doi: 10.1016/j.arr.2016.12.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gendron CM et al. Neuronal Mechanisms that Drive Organismal Aging Through the Lens of Perception. Annu Rev Physiol 82, 227–249, doi: 10.1146/annurev-physiol-021119-034440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcedo J & Kenyon C Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41, 45–55 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Apfeld J & Kenyon C Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804–809, doi: 10.1038/45544 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Libert S et al. Regulation of Drosophila life span by olfaction and food-derived odors. Science 315, 1133–1137 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Greer EL & Brunet A Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127, doi: 10.1111/j.1474-9726.2009.00459.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer EL et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 17, 1646–1656, doi: 10.1016/j.cub.2007.08.047 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith ED et al. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol 8, 49, doi: 10.1186/1471-213X-8-49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artan M et al. Food-derived sensory cues modulate longevity via distinct neuroendocrine insulin-like peptides. Genes Dev 30, 1047–1057, doi: 10.1101/gad.279448.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bargmann CI Chemosensation in C. elegans. WormBook, 1–29 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White JG, Southgate E, Thomson JN & Brenner S The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 15.Kang L, Gao J, Schafer WR, Xie Z & Xu XZSC elegans TRP family protein TRP-4 is a pore-forming subunit of a native mechanotransduction channel. Neuron 67, 381–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawin ER, Ranganathan R & Horvitz HRC elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Li W, Feng Z, Sternberg PW & Xu XZS A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature 440, 684–687 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkema MJ, Hunter-Ensor M, Ringstad N & Horvitz HR Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Shao J et al. Serotonergic neuron ADF modulates avoidance behaviors by inhibiting sensory neurons in C. elegans. Pflugers Arch 471, 357–363, doi: 10.1007/s00424-018-2202-4 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Richmond JE, Davis WS & Jorgensen EM UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speese S et al. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27, 6150–6162, doi: 10.1523/JNEUROSCI.1466-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Link E et al. Tetanus toxin action: inhibition of neurotransmitter release linked to synaptobrevin proteolysis. Biochem Biophys Res Commun 189, 1017–1023 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Hendricks M, Ha H, Maffey N & Zhang Y Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature 487, 99–103, doi: 10.1038/nature11081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Liu J, Zheng M & Xu XZ Encoding of both analog- and digital-like behavioral outputs by one C. elegans interneuron. Cell 159, 751–765, doi: 10.1016/j.cell.2014.09.056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokolchik I, Tanabe T, Baldi PF & Sze JY Polymodal sensory function of the Caenorhabditis elegans OCR-2 channel arises from distinct intrinsic determinants within the protein and is selectively conserved in mammalian TRPV proteins. J Neurosci 25, 1015–1023, doi: 10.1523/JNEUROSCI.3107-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobert O The neuronal genome of Caenorhabditis elegans. WormBook, 1–106, doi: 10.1895/wormbook.1.161.1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackwell TK, Sewell AK, Wu Z & Han M TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics 213, 329–360, doi: 10.1534/genetics.119.302504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winston WM, Molodowitch C & Hunter CP Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459, doi: 10.1126/science.1068836 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Rera M, Clark RI & Walker DW Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A 109, 21528–21533, doi: 10.1073/pnas.1215849110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelino S et al. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS Genet 12, e1006135, doi: 10.1371/journal.pgen.1006135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dambroise E et al. Two phases of aging separated by the Smurf transition as a public path to death. Sci Rep 6, 23523, doi: 10.1038/srep23523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida M, Oami E, Wang M, Ishiura S & Suo S Nonredundant function of two highly homologous octopamine receptors in food-deprivation-mediated signaling in Caenorhabditis elegans. J Neurosci Res 92, 671–678, doi: 10.1002/jnr.23345 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Burkewitz K, Zhang Y & Mair WB AMPK at the nexus of energetics and aging. Cell Metab 20, 10–25, doi: 10.1016/j.cmet.2014.03.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neves SR, Ram PT & Iyengar R G protein pathways. Science 296, 1636–1639 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Dimov I & Maduro MF The C. elegans intestine: organogenesis, digestion, and physiology. Cell Tissue Res 377, 383–396, doi: 10.1007/s00441-019-03036-4 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Lafontan M & Berlan M Fat cell adrenergic receptors and the control of white and brown fat cell function. J Lipid Res 34, 1057–1091 (1993). [PubMed] [Google Scholar]
- 37.Graham RM, Perez DM, Hwa J & Piascik MT alpha 1-adrenergic receptor subtypes. Molecular structure, function, and signaling. Circ Res 78, 737–749, doi: 10.1161/01.res.78.5.737 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Finger F et al. Olfaction regulates organismal proteostasis and longevity via microRNA-dependent signaling. Nat Metab 1, 350–359, doi: 10.1038/s42255-019-0033-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albrecht J et al. Olfactory detection thresholds and pleasantness of a food-related and a non-food odour in hunger and satiety. Rhinology 47, 160–165 (2009). [PubMed] [Google Scholar]
- 40.Burkewitz K et al. Neuronal CRTC-1 Governs Systemic Mitochondrial Metabolism and Lifespan via a Catecholamine Signal. Cell 160, 842–855, doi: 10.1016/j.cell.2015.02.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer EL et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282, 30107–30119, doi: 10.1074/jbc.M705325200 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Weir HJ et al. Dietary Restriction and AMPK Increase Lifespan via Mitochondrial Network and Peroxisome Remodeling. Cell Metab 26, 884–896 e885, doi: 10.1016/j.cmet.2017.09.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riera CE et al. The Sense of Smell Impacts Metabolic Health and Obesity. Cell Metab 26, 198–211 e195, doi: 10.1016/j.cmet.2017.06.015 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Piggott BJ, Liu J, Feng Z, Wescott SA & Xu XZS The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell 147, 922–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ching TT & Hsu AL Solid plate-based dietary restriction in Caenorhabditis elegans. J Vis Exp, doi: 10.3791/2701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B et al. Brain-gut communications via distinct neuroendocrine signals bidirectionally regulate longevity in C. elegans. Genes Dev 32, 258–270, doi: 10.1101/gad.309625.117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Li G, Liu J, Liu J & Xu XZ TMC-1 Mediates Alkaline Sensation in C. elegans through Nociceptive Neurons. Neuron 91, 146–154, doi: 10.1016/j.neuron.2016.05.023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jun H et al. An immune-beige adipocyte communication via nicotinic acetylcholine receptor signaling. Nat Med 24, 814–822, doi: 10.1038/s41591-018-0032-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiao X et al. Protein Arginine Methyltransferase 1 Interacts With PGC1alpha and Modulates Thermogenic Fat Activation. Endocrinology 160, 2773–2786, doi: 10.1210/en.2019-00504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang AB et al. Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 111, E4458–4467, doi: 10.1073/pnas.1411199111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao R et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 152, 806–817, doi:j.cell.2013.01.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B et al. Environmental Temperature Differentially Modulates C. elegans Longevity through a Thermosensitive TRP Channel. Cell Rep 11, 1414–1424, doi: 10.1016/j.celrep.2015.04.066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during this study are either included within the manuscript or are available from the corresponding author on reasonable request.