Abstract
Severe cases of coronavirus disease 2019 (COVID-19), caused by infection with SARS-CoV-2, are characterized by a hyperinflammatory immune response that leads to numerous complications. Production of proinflammatory neutrophil extracellular traps (NETs) has been suggested to be a key factor in inducing a hyperinflammatory signaling cascade, allegedly causing both pulmonary tissue damage and peripheral inflammation. Accordingly, therapeutic blockage of neutrophil activation and NETosis, the cell death pathway accompanying NET formation, could limit respiratory damage and death from severe COVID-19. Here, we demonstrate that synthetic glycopolymers that activate signaling of the neutrophil checkpoint receptor Siglec-9 suppress NETosis induced by agonists of viral toll-like receptors (TLRs) and plasma from patients with severe COVID-19. Thus, Siglec-9 agonism is a promising therapeutic strategy to curb neutrophilic hyperinflammation in COVID-19.
Short abstract
In COVID-19, viral pathogen associated molecular patterns and viral-induced cytokines can induce NETosis of neutrophils at the site of infection and in the periphery. Siglec-9 agonists inhibit COVID-19 plasma-induced NETosis, potentially preventing deleterious hyperinflammatory responses.
Introduction
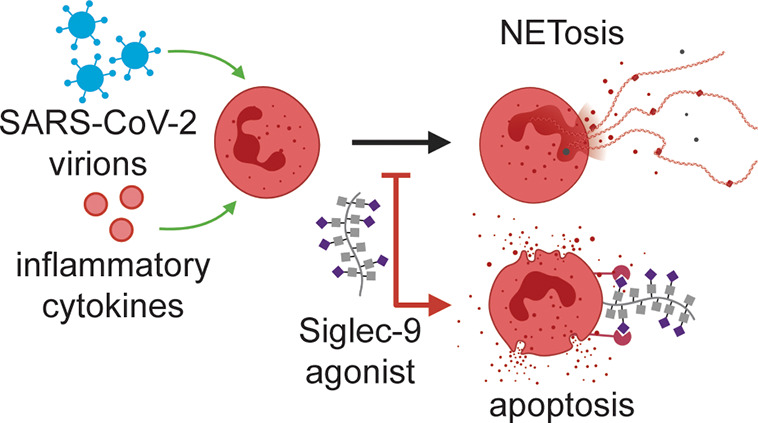
Runaway inflammation in coronavirus disease 2019 (COVID-19) is thought to lead to numerous complications, including potentially fatal pneumonia and acute respiratory distress syndrome (ARDS).1−3 While the specific causal factors of inflammation in COVID-19-related ARDS are unknown and likely multifarious, an emerging hypothesis posits that hyperactivation of neutrophils initiates and drives this response (Figure 1).4−12 Neutrophils are immune cells of the myeloid lineage that are involved in numerous innate immune functions. It has been suggested that neutrophils drive a hyperinflammatory response in COVID-19 through a death process called NETosis, in which neutrophils rapidly decondense chromatin and spew out a neutrophil extracellular trap (NET), an amalgam of genomic DNA, intracellular proteins (e.g., histones), and tissue-damaging enzymes (e.g., neutrophil elastase, myeloperoxidase).13,14 Extracellular DNA and tissue damage from NET-associated enzymes act as proinflammatory signals to other immune cells15−17 and are proposed to initiate the hyperinflammatory cascade in COVID-19, leading to ARDS and potentially death. Consistent with this hypothesis, NETs have been extensively observed both at the site of infection (i.e., pulmonary tissue)18−21 and in the periphery (i.e., sera and plasma).19,21
Figure 1.
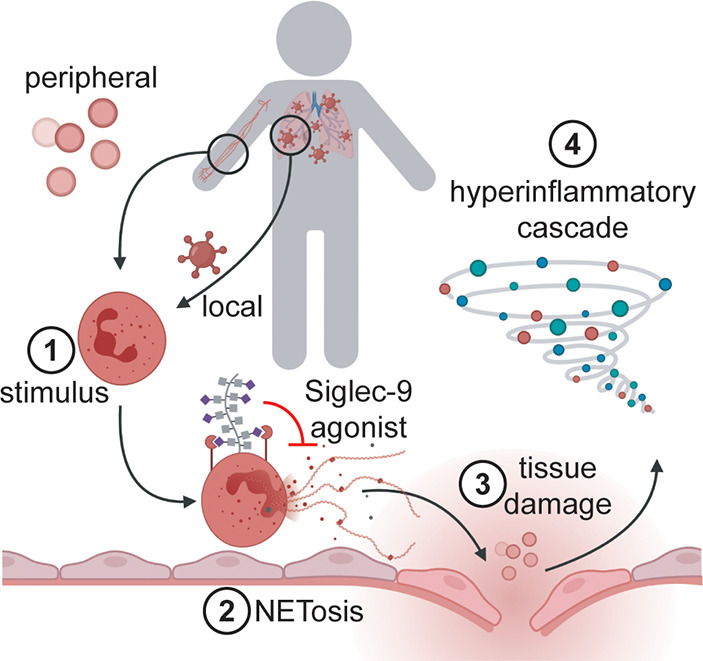
Local and peripheral inflammatory stimuli induce NETosis and a subsequent hyperinflammatory cascade in COVID-19. Both local inflammatory stimuli at the site of SARS-CoV-2 infection (e.g., virions) and peripheral inflammatory stimuli (e.g., the proinflammatory cytokines IL-8 and G-CSF) associated with COVID-19 have been shown to induce NETosis in vitro. These factors are suspected to be causative agents of NETosis in vivo as well, initiating a deleterious hyperinflammatory cascade leading to the symptoms of moderate and severe COVID-19. Agonists of the neutrophil-associated checkpoint receptor Siglec-9 could inhibit NETosis in COVID-19.
Both SARS-CoV-2 virions and serum/plasma from COVID-19 patients have been shown to induce NETosis of neutrophils isolated from healthy donors in vitro, consistent with the local and peripheral inflammatory responses observed in COVID-19.19,21,22 However, the specific signals that induce NETosis in viral disease remain an open question; viral ligands for toll-like receptors (TLRs), host damage-associated molecular patterns, antiviral cytokines (e.g., IL-8 and IFNγ), and activated platelets have all been implicated, but which if any of these is sufficient to induce NETosis is still debated.21,23 Beyond viral disease, NETosis has been linked to numerous inflammatory pathologies, including thrombosis and sepsis, both of which are observed in patients with COVID-19.4 During NETosis, inflammatory stimuli signal neutrophils to import calcium ions, which activate protein arginine deiminase 4 (PADI4).24,25 PADI4 mediates the conversion of arginine to the deiminated citrulline on histones.25 The loss of positive charges induces rapid unwinding of genomic DNA, which eventually ruptures the nucleus and the cell.25 When this happens, intracellular contents including genomic DNA, active PADI4, tissue-damaging NET-associated enzymes, and citrullinated histones are emitted into the extracellular space, all of which provoke an inflammatory response.24,25 Thus, strategies to curb neutrophil-mediated inflammation could treat both COVID-19 as well as other neutrophilic inflammatory pathologies.
Transcriptomic analyses of immune cells from severe COVID-19 patients show that neutrophils upregulate the myeloid checkpoint receptor Siglec-9, a member of the sialic acid-binding immunoglobulin-like lectin (Siglec) family that is also found on macrophages and activated T cells.8,9,26−29 This sialoglycan-binding immunosuppressive receptor has an intracellular signaling domain similar to the prominent lymphoid checkpoint receptor PD-1 and the myeloid suppressive receptor SIRPα.30−32 Clustering of Siglec-9 by ligand engagement leads to inhibitory signaling that quenches activation of the immune cells.28 Both erythrocytes and host-mimicking pathogens have been shown to engage Siglec-9 to suppress neutrophil-mediated immunity, including inhibiting NETosis.28,33−35 Furthermore, engagement of Siglec-9 on primary neutrophils has been shown to induce apoptotic pathways,26 in a manner similar to the engagement of Siglec-8 on eosinophils as occurs with an FDA-approved Siglec-8 agonist for eosinophilic inflammatory conditions.36 Given that Siglec-9 is both an anti-inflammatory and pro-apoptotic checkpoint molecule, we hypothesized that engagement of Siglec-9 could simultaneously inhibit proinflammatory NETotic cell death and induce quiet apoptotic cell death in COVID-19-related inflammation. Notably, an agonist of the related myeloid checkpoint receptor Siglec-10 (CD24Fc, trade name SACCOVID) has recently shown great promise in suppressing viral hyperinflammation and is in a Phase III clinical trial.37,38 However, unlike the case for Siglec-10 for which CD24 is a high-affinity and specific ligand, there is no comparable glycoprotein ligand known for Siglec-9 and thus no biological starting point for the design of therapeutic agonists.39
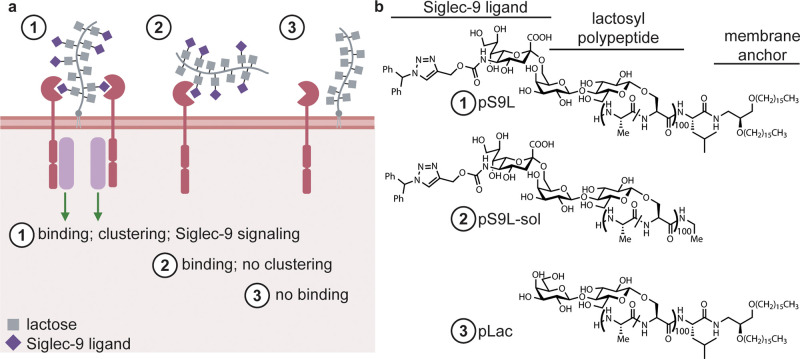
We recently reported40 the design and synthesis of a potent Siglec-9 agonist comprising a lipid-conjugated glycopolypeptide bearing modified sialic acid residues that Paulson and co-workers had previously found to bind Siglec-9 with high affinity and specificity41 (pS9L, Figure 2). The lipid group enabled passive insertion into cell membranes, leading to engagement of Siglec-9 in cis on macrophage cell surfaces. This cell-surface clustering, in turn, induced Siglec-9 signaling and suppressed macrophage activation.40 We hypothesized that this was due to induced distribution of Siglec-9 into actively signaling clusters that we and others have observed.28,40 In this former study, we also designed control glycopolypeptides lacking either Siglec-9 binding glycans (i.e., the lactose-functionalized glycopolypeptide pLac) or a membrane anchoring lipid moiety (i.e., the soluble glycopolypeptide pS9L-sol) (Figure 2). Notably, potent Siglec-9 agonism required membrane anchoring and cis-engagement; the soluble analogue pS9L-sol was unable to stimulate Siglec-9 signaling and suppress macrophage activity, which we proposed was due to clustering efficiency of membrane-associated ligands binding cis versus soluble ligands binding in trans. We hypothesized that pS9L might also suppress neutrophil activation and NETosis by clustering Siglec-9 on neutrophils.
Figure 2.
Synthetic glycopolypeptides bearing high-affinity Siglec-9 ligands engage Siglec-9 and induce clustering and signaling. (a) Membrane-anchored, cis binding glycopolypeptide 1 (pS9L) induces Siglec-9 signaling, while a soluble control polypeptide 2 (pS9L-sol) or a nonbinding but membrane-anchored control polypeptide 3 (pLac) does not. (b) Structures of the polypeptides pS9L, pS9L-sol, and pLac. Polypeptides are all based on an O-lactosyl poly serine-co-alanine scaffold and in some cases bear terminal Siglec-9-binding sialic acid analogues and/or C-terminal membrane-anchoring lipids.
Here, we demonstrate that a synthetic cis-binding Siglec-9 agonist (pS9L, Figure 2b)40 inhibits NETosis in primary neutrophils in models of local (TLR-7/8 agonist) and peripheral (COVID-19 plasma) COVID-19-associated inflammation. Using live cell microscopy, we showed that TLR-7/8 activation by the nucleoside analogue resimiquod (R848) induces NETosis in primary human neutrophils. R848 treatment induces rapid citrullination of histone substrates, consistent with PADI4-mediated NETosis, and this process was blocked by Siglec-9 signaling induced by pS9L. Significantly, pS9L inhibited neutrophil NETosis induced by treatment with plasma from severe COVID-19 patients. In light of these data, we propose that Siglec-9 agonists could be therapeutic agents that inhibit COVID-19-associated inflammation.
Results and Discussion
TLR-7/8 Agonist R848 Induces NETosis of Primary Neutrophils in Vitro
In COVID-19, evidence of extensive NETosis can be observed in infected lungs,18−21 and SARS-CoV-2 virions have been shown to infect and induce NETosis of healthy neutrophils in vitro.20 These reports implicate TLR-7 and/or TLR-8 in inducing NETosis of neutrophils at the site of infection.20,42 Notably, TLR-7 and TLR-8 are single-stranded RNA receptors with numerous substrates identified in the SARS-CoV-2 genome.43 Consistent with the hypothesis that SARS-CoV-2 induces TLR-7/8-mediated immunity, human genetic variations in TLR7 are associated with severe COVID-19.44 Thus, agonists of TLR-7/8 may provide a convenient means of modeling local inflammation induced by viral infection in vitro without using live virus.
We assayed TLR agonists using the live-cell imaging techniques described by Gupta and co-workers.45 In this assay, freshly isolated neutrophils are cultured in low-serum media in the presence of a fluorogenic and membrane impermeable DNA-intercalating dye (Cytotox Green). Upon genomic DNA-externalization by NETosis, dye intercalates and fluorescence increases. As previously demonstrated,45 because NETs are much larger than the nuclei of apoptotic cells, NETotic cells yield much larger areas of fluorescence than apoptotic cells, as observed by microscopy. Thus, apoptotic cells can be filtered out by only counting large (i.e., ≫100 μm2) fluorescent objects.
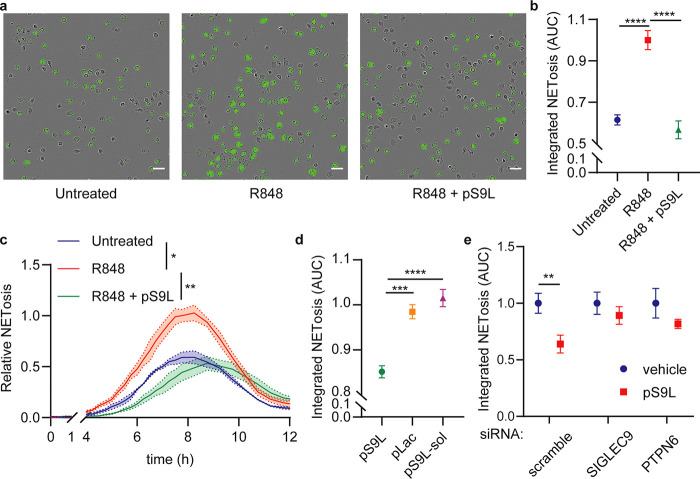
We found that a TLR-7/8 agonist, R848, was sufficient to induce NETosis of healthy neutrophils in vitro (Figure 3a–c, Figure S1). We also assayed the citrullination status of the PADI4 substrate H3 by Western blot and observed that R848 rapidly induced citrullination at R2, R8, and R17 (Figure S2). While citrullination is an important aspect of NETosis,24 the extent of citrullination is not necessarily indicative of the extent of NETosis as, for example, PMA-induced NETosis only yields moderate citrullination (Figure S2).46 Additionally, we performed quantitative phosphoproteomics47 with lysates of neutrophils treated with media, phorbol-12-myristate-13-acetate (PMA), or R848 (Figure S3, Table S1). We observed results similar to previously published data sets using neutrophils stimulated with either R84848 or PMA.49 Furthermore, several phosphosites were found to be differentially regulated in both data sets, including those involved in neutrophil degranulation and calcium flux, consistent with the described mechanism of NETotic cell death.24,49 These results indicate that the TLR-7/8 agonist R848 induces NETosis in primary neutrophils. Thus, this compound can be used to model local inflammation associated with viral infection, including in COVID-19.
Figure 3.
A cis-binding Siglec-9 agonist (pS9L) inhibits R848-induced NETosis via Siglec-9 and SHP-1. (a–c) Primary neutrophils were cotreated with R848 (10 μM) and glycopolypeptide (500 nM) in IMDM supplemented 0.5% hiFBS containing the membrane impermeable DNA intercalators Cytotox Green or Red (250 nM). Images were acquired by fluorescence microscopy every 15 min for 12 h. The area of all green fluorescent objects >300 μm2 was quantified, and the total area was averaged across three images per well. Relative NETosis was determined by normalizing to the maximal NET area from R848 treatment alone (t = 8 h). (a) Representative phase contrast and fluorescence images from t = 8 h. Scale bars indicate 40 μm. (b) Quantitation of NETosis over time as area under the curve in (c). Error bars represent SD. (c) NET formation and degradation as a function of time. Error bands represent SEM. (d) Treatment of R848-stimulated neutrophils with various glycopolypeptides. Error bars represent SD. (e) HL-60 cells were transfected with siRNAs against SIGLEC9 (encoding Siglec-9), PTPN6 (encoding SHP-1), or a scrambled control and then grown for 2 days. Cells were then cotreated with R848 (10 μM) and vehicle or pS9L (500 nM). Relative NETosis is determined as in (b), except all objects >200 μm2 were quantified and the R848 maximum in dHL-60s was observed at 2.5 h post induction. Error bars represent SD. Statistics were determined by two-way ANOVA (c) or one-way ANOVA (b,d,e). * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
A Siglec-9 Agonist Inhibits TLR-7/8-Induced NETosis via SHP-1
Previous work by von Gunten, Varki, and their respective co-workers has shown that engagement of Siglec-9 leads to apoptotic and nonapoptotic death pathways as well as immunosuppression in neutrophils.26,50 Thus, we hypothesized that Siglec-9-mediated immunosuppression and cell death could override the NETotic effect of antiviral TLR signaling. To test this notion, we used our previously described Siglec-9 agonist, pS9L(40) as well as the two control glycopolypeptides pLac and pS9L-sol (Figures 2 and S4). We assayed anti-NETotic activity by cotreatment of glycopolypeptide (500 nM) with R848 (10 μM) in primary neutrophils in the live-cell assay described above (Figure 3). We observed that pS9L was sufficient to inhibit NETosis induced by R848 treatment (Figure 3a–c). Moreover, neither control polymer inhibited R848-induced NETosis (Figure 3d). We also confirmed that pS9L inhibits NETosis comparably to high concentrations of cross-linked anti-Siglec-9 antibody (clone 191240) (Figure S5).51,52 Previously, von Gunten and co-workers described the generation of mitochondrial-derived reactive oxygen species (ROS) as an important signaling step of Siglec-9-induced apoptotic signaling.26 We found that treatment with pS9L, in the absence of any TLR ligand so as to avoid NADPH-derived ROS in inflammatory signaling, induced an oxidative burst, as did treatment with a cross-linked anti-Siglec-9 antibody (Figure S6). Furthermore, the oxidative burst was inhibited by the addition of the SHP-1/2 inhibitor NSC-87877, suggesting that SHP-1 and/or SHP-2 mediate pS9L-induced oxidative burst in neutrophils, consistent with Siglec-9 engagement (Figure S6b).
We performed quantitative phosphoproteomics using lysates of R848-stimulated primary neutrophils cotreated with vehicle, pS9L, or pLac (Figure S3, Table S2). Notably, we found increased phosphorylation of hyccin (HYCCI/FAM126A), a key component in phosphorylation of phosphoinositides,53 a class of signaling molecules implicated in mediating NETosis.54 Additionally, we observed increased phosphorylation of RASAL3 (RASL3), a negative regulator of the MAPK signaling pathway.55 These data suggest that pS9L inhibits the calcium flux and NADPH activity necessary for NETosis, as well as the MAPK-suppressive effects that have been previously described for pS9L in macrophages.40
To determine whether the anti-NETotic effect of pS9L is specifically mediated by Siglec-9 signaling, we recapitulated our results in the promyelocytic leukemia cell line HL-60. These cells can be differentiated into neutrophil-like cells (dHL-60) using all-trans retinoic acid (ATRA, 100 nM) and dimethyl sulfoxide (DMSO, 1.25% v/v). Notably, dHL-60 cells have previously been used to study NETosis in vitro.31,56 Consistent with those prior reports, R848 induced NETosis in dHL-60 cells (Figure S7). Furthermore, we observed that pS9L inhibited NETosis and that siRNA knockdown of Siglec-9 (encoded by SIGLEC9) or SHP-1 (encoded by PTPN6) abrogated the effect of pS9L (Figures 3e, S8 and S9). Therefore, the Siglec-9 agonist pS9L inhibits TLR7/8-induced NETosis via Siglec-9 and SHP-1.
Siglec-9 is Upregulated in Severe COVID-19 and Can Suppress NETosis Induced by COVID-19 Plasma
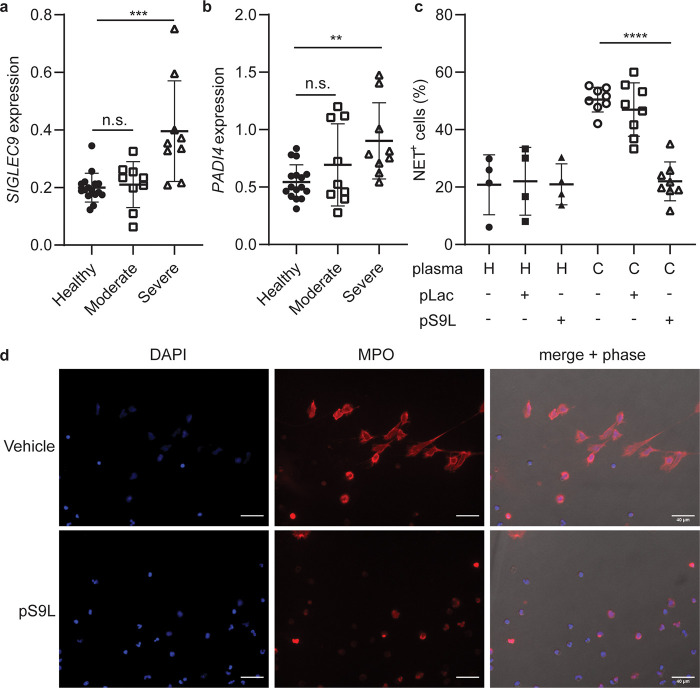
Sera and plasma from COVID-19 patients have been shown to induce NETosis of neutrophils isolated from healthy donors in vitro.19,21 The causative components are unclear; however, potential factors include viral TLR ligands, damage-associated molecular patterns that bind TLRs, activated platelets, and (pro)inflammatory cytokines. Recent reports have described increased levels of neutrophil-activating cytokines in COVID-19 plasma, predominantly IL-8 and G-CSF.57 We also observed that the combination of IL-8 and G-CSF was sufficient to induce NETosis in vitro (Figure S10). Additionally, transcriptomic analyses of peripheral myeloid cells8 and neutrophils9 in COVID-19 patients have revealed increased SIGLEC9 expression (Figure 4a, S11) and PADI4 expression (Figures 4b and S11). We hypothesize that this is an exhaustion-like phenotype in which Siglec-9 expression is induced on hyper-NETotic neutrophils, similar to what has been observed with Siglec-9 on exhausted tumor-infiltrating T cells.51 These observations further support Siglec-9 an attractive target for therapeutic blockade of hyperinflammatory NETosis in COVID-19.
Figure 4.
Siglec-9 agonist pS9L inhibits NETosis of neutrophils induced by COVID-19 plasma. (a, b) Analysis of publicly available single-cell transcriptomics data8 for SIGLEC9 expression (a) and PADI4 expression (b) on neutrophils in peripheral blood from healthy donors or COVID-19 patients. Error bars represent SD. Statistics were determined using mixed effects model. ** = p < 0.01; *** = p < 0.001 (c, d) Primary neutrophils were cultured in undiluted and citrate anticoagulated plasma from healthy donors or COVID-19 patients for 4 h. Cells were fixed, stained for extracellular myeloperoxidase, and imaged in DAPI imaging media by fluorescence microscopy. Cells were treated in technical triplicate and imaged across multiple fields of view. (c) Proportion of NET-positive cells (%) across all fields of view. Each dot represents and individual plasma sample. (d) Representative images from a COVID-19 patient plasma sample with or without pS9L. Error bars represent SD. Statistics were determined using mixed effects models to account for samples using repeat neutrophil donors. **** = p < 0.0001.
To test the hypothesis that pS9L can inhibit NETosis induced by COVID-19 plasma, we treated neutrophils isolated from whole blood of healthy donors with citrate-anticoagulated heterologous plasma from healthy donors or COVID-19 patients. Neutrophils in undiluted plasma were cotreated with pS9L (500 nM), the nonbinding analogue pLac (500 nM), or vehicle. To satisfy biosafety restrictions, cells were incubated in the presence of COVID-19 plasma for 4 h and then fixed before assaying for extracellular complexes of myeloperoxidase (MPO) and DNA (DAPI) (Figure 4c,d). The combination of these stains, which when observed extracellularly is indicative of NETosis, has been previously used to identified NET+ cells in the context of COVID-19.20 We observed that COVID-19 plasma induced NETosis of neutrophils from healthy donors, as indicated by the formation of web-like NET structures (Figure 4d). As in previous experiments with R848, COVID-19 plasma-stimulated NETosis was inhibited by pS9L treatment (Figure 4c,d). Furthermore, pLac did not inhibit NETosis induced by COVID-19 plasma, and neither pS9L nor pLac affected basal NETosis of in vitro cultured neutrophils (Figure 4c). We performed similar experiments staining neutrophils treated with 10% plasma in IMDM (Figure S12) or undiluted plasma (Figure S13) for extracellular H1/DNA complexes, another marker of NETs,58−60 and observed comparable results.
Collectively, these data demonstrate that Siglec-9 agonism inhibits NETosis induced by COVID-19 patient plasma and thus could inhibit peripheral inflammation in patients with COVID-19. Additionally, Siglec-9 agonists could resolve NET-associated pathologies observed in COVID-19 and elsewhere such as immunothrombosis21 and sepsis.4,5
Safety Statement
For experiments using plasma from patients with COVID-19, all experiments were performed in a certified BSL-2+ biosafety cabinet with appropriate institutional approval for working with blood products derived from patients with COVID-19. All items that came in contact with plasma were disinfected with 10% bleach for 30 min or fixed in 4% formaldehyde solution for 15 min before being removed from the biosafety cabinet. Otherwise, no unexpected or unusually high safety hazards were encountered.
Conclusion
We have demonstrated that Siglec-9 agonists can inhibit NETosis induced by COVID-19-associated proinflammatory signals. Thus, Siglec-9 is a therapeutic target to inhibit potentially fatal hyperinflammation associated with COVID-19 in an analogous fashion to the highly effective therapeutics currently aimed at the Siglec-10/CD24 interaction. A CD24-Fc fusion has been shown to engage Siglec-10 as an immune checkpoint on macrophages and sequester the nuclear protein HMGB1, which can act as a damage associated molecular pattern by engaging TLR4.61 The Siglec-9 agonists described here have previously been shown to inhibit macrophage TLR4 signaling and engage macrophage Siglec-9.40 Thus, Siglec-9 agonists may be multipurpose therapeutics, able to inhibit both the clinically unaddressed problem of proinflammatory NETosis and also subsequent inflammatory signaling from tissue damage that is currently being clinically investigated.
The glycopolypeptides described here may have direct translational potential. As lipid conjugates, they may have sufficient reversible albumin binding activity as to achieve long plasma residence times approaching those of antibodies or Fc fusion proteins. As they are products of chemical synthesis, modifications to enhance preferred drug properties would be quite straightforward. Indeed, glycopolymers with lipid variants that enhance membrane association or plasma membrane residence time have been used for other purposes in our lab.62,63 More broadly, however, the work herein provides motivation to develop Siglec-9 agonists of any molecular classes, including monoclonal antibodies or Fc fusion proteins. Finally, Siglec-9 agonists have the potential to expand beyond ARDS to other NET-related pathologies such as thrombosis,64,65 atherosclerosis,66 and cystic fibrosis.67
Acknowledgments
The authors thank the following individuals at the Stanford COVID-19 Biobank for enabling this work: Nancy Zhao, Rosemary Vergara, Julia McKechnie, Lauren de la Parte, Kathleen Whittle Dantzler, Maureen Ty, Nimish Kathale, Arjun Rustagi, Giovanny Martinez-Colon, Geoff Ivison, Ruoxi Pi, Maddie Lee, Rachel Brewer, Taylor Hollis, Andrea Baird, Michele Ugur, Drina Bogusch, Georgie Nahass, Kazim Haider, Kim Quyen Thi Tran, Laura Simpson, Michal Tal, Iris Chang, Evan Do, Andrea Fernandes, Shu-Chen Lyu, Wenming Zhang, Monali Manohar, James Krempski, Jonasel Roque, Hena Naz DinRosen Mann, Anita Visweswaran, Elizabeth J. Zudock, Kathryn Jee, Komal Kumar, Jennifer A. Newberry, James V. Quinn, Donald Schreiber, and Andra L. Blomkalns. The authors also thank Rishi Kulkarni and Susan Holmes for their advice on statistics; and Gabby Tender for her assistance with fluorescence microscopy. This work was supported by a grant from the National Institutes of Health to C.R.B. (CA227942-18). C.S.D was supported by a National Science Foundation Graduate Research Fellowship (DGE-114747) and a ChEM-H affiliated Stanford Interdisciplinary Gradate Fellowship. N.M.R. was funded through an NIH Predoctoral to Postdoctoral Transition Award (Grant K00 CA212454-05). J.C.S. was supported by an NIH/NCI F32 postdoctoral fellowship (Grant 1F32CA250324-01). A.J.W. was supported by the Stanford Medical Scientist Training Program (T32 GM007365-44) and the Stanford Bio-X Interdisciplinary Graduate Fellowship. C.A.B. was supported by the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases #1016687 and the Bill and Melinda Gates Foundation (OPP1113682). C.A.B. is the Tashia and John Morgridge Faculty Scholar in Pediatric Translational Medicine from the Stanford Maternal Child Health Research Institute and an Investigator of the Chan Zuckerberg Biohub. Figure illustrations were created using BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01669.
The authors declare the following competing financial interest(s): C.S.D. and C.R.B. are coinventors on a patent application for cis-binding Siglec agonist glycopolymers as immune suppressants (USPTO63046140). C.R.B. is a co-founder and Scientific Advisory Board member of Lycia Therapeutics, Palleon Pharmaceuticals, Enable Bioscience, Redwood Biosciences (a subsidiary of Catalent), OliLux Inc., and InterVenn Bio, and a member of the Board of Directors of Eli Lily & Company. C.A.B. is a Scientific Advisory Board member of Catamaran Bio.
Supplementary Material
References
- Cao X. COVID-19: Immunopathology and Its Implications for Therapy. Nat. Rev. Immunol. 2020, 20, 269–270. 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R. J. Pathogenesis of COVID-19 from a Cell Biology Perspective. Eur. Respir. J. 2020, 55 (4), 9–11. 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M. Z.; Poh C. M.; Rénia L.; MacAry P. A.; Ng L. F. P. The Trinity of COVID-19: Immunity, Inflammation and Intervention. Nat. Rev. Immunol. 2020, 20 (6), 363–374. 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A. R.; Roch B.. NETs By-Products and Extracellular DNA May Play a Key Role in COVID-19 Pathogenesis: Incidence on Patient Monitoring and Therapy. Preprints 2020, 10.20944/preprints202004.0238.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B. J.; Adrover J. M.; Baxter-Stoltzfus A.; Borczuk A.; Cools-Lartigue J.; Crawford J. M.; Daßler-Plenker J.; Guerci P.; Huynh C.; Knight J. S.; et al. Targeting Potential Drivers of COVID-19: Neutrophil Extracellular Traps. J. Exp. Med. 2020, 217 (6), e20200652. 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Liu Y.; Xiang P.; Pu L.; Xiong H.; Li C.; Zhang M.; Tan J.; Xu Y.; Song R.; et al. Neutrophil-to-Lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. medRxiv 2020, 10.1101/2020.02.10.20021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendib I.; De Chaisemartin L.; Granger V.; Schlemmer F.; Maitre B.; Hüe S.; Surenaud M.; Beldi-Ferchiou A.; Carteaux G.; Razazi K.; et al. Neutrophil Extracellular Traps Are Elevated in Patients with Pneumonia-Related Acute Respiratory Distress Syndrome. Anesthesiology 2019, 130 (4), 581–591. 10.1097/ALN.0000000000002619. [DOI] [PubMed] [Google Scholar]
- Schulte-Schrepping J.; Reusch N.; Paclik D.; Baßler K.; Schlickeiser S.; Zhang B.; Krämer B.; Krammer T.; Brumhard S.; Bonaguro L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419. 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner A. C.; Mouktaroudi M.; Krämer B.; Antonakos N.; et al. Disease Severity-Specific Neutrophil Transcriptomes Stratify COVID-19 Patients Signatures in Blood. medRxiv 2020, 10.1101/2020.07.07.20148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A. J.; Rustagi A.; Zhao N. Q.; Roque J.; Martínez-Colón G. J.; McKechnie J. L.; Ivison G. T.; Ranganath T.; Vergara R.; Hollis T.; et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients with Severe COVID-19. Nat. Med. 2020, 26 (7), 1070–1076. 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P.; De Sanctis F.; Canè S.; Ugel S.; Donadello K.; Castellucci M.-N.; Eyal D.; Fiore A.; Anselmi C.; Barouni R. M.; et al. Deciphering the State of Immune Silence in Fatal COVID-19 Patients. medRxiv 2020, 10.1101/2020.08.10.20170894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K. A.; Shishkova E.; Miller I. J.; Balnis J.; Bernstein M. N.; Peters-Clarke T. M.; Meyer J. G.; Quan Q.; Muehlbauer L. K.; Trujillo E. A.; et al. Large-Scale Multi-Omic Analysis of COVID-19 Severity. Cell Syst. 2020, 1–18. 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q.; Kuijpers T. W.; Wirawan E.; Lippens S.; Vandenabeele P.; Vanden Berghe T. Dying for a Cause: NETosis, Mechanisms behind an Antimicrobial Cell Death Modality. Cell Death Differ. 2011, 18 (4), 581–588. 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V.; Reichard U.; Goosmann C.; Fauler B.; Uhlemann Y.; Weiss D. S.; Weinrauch Y.; Zychlinsky A. Neutrophil Extracellular Traps Kill Bacteria. Science (Washington, DC, U. S.) 2004, 303 (5663), 1532–1535. 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Daniel C.; Leppkes M.; Muñoz L. E.; Schley G.; Schett G.; Herrmann M. Extracellular DNA Traps in Inflammation, Injury and Healing. Nat. Rev. Nephrol. 2019, 15 (9), 559–575. 10.1038/s41581-019-0163-2. [DOI] [PubMed] [Google Scholar]
- Xu J.; Zhang X.; Monestier M.; Esmon N. L.; Esmon C. T. Extracellular Histones Are Mediators of Death through TLR2 and TLR4 in Mouse Fatal Liver Injury. J. Immunol. 2011, 187 (5), 2626–2631. 10.4049/jimmunol.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood C.; Blanco L. P.; Purmalek M. M.; Carmona-Rivera C.; De Ravin S. S.; Smith C. K.; Malech H. L.; Ledbetter J. A.; Elkon K. B.; Kaplan M. J. Neutrophil Extracellular Traps Enriched in Oxidized Mitochondrial DNA Are Interferogenic and Contribute to Lupus-like Disease. Nat. Med. 2016, 22 (2), 146–153. 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermecker C.; Detrembleur N.; Guiot J.; Cavalier E.; Henket M.; Cataldo D.; Delvenne P.; Marichal T.; Chemistry M.; Biology D. Neutrophil Extracellular Traps Infiltrate the Lung Vascular, Interstitial and Airway Compartments in Severe Covid-19. J. Exp. Med. 2020, 217 (12), e20201012. 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y.; Yalavarthi S.; Shi H.; Gockman K.; Zuo M.; Madison J. A.; Blair C.; Weber A.; Barnes B. J.; Egeblad M. Neutrophil Extracellular Traps in COVID19. J. Clin. Invest. 2020, 5 (11), e138999. 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F. P.; Pontelli M.; Silva C.; Toller-Kawahisa J.; de Lima M.; Nascimento D.; Schneider A.; Caetite D.; Rosales R.; Colon D.; Tavares L. A.; Paiva I. M. SARS-CoV-2 Triggered Neutrophil Extracellular Traps (NETs) Mediate COVID-19 Pathology. J. Exp. Med. 2020, 217 (12), e20201129. 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E. A.; He X.-Y.; Denorme F.; Campbell R. A.; Ng D.; Salvatore S. P.; Mostyka M.; Baxter-Stoltzfus A.; Borczuk A. C.; Loda M. Neutrophil Extracellular Traps (NETs) Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood 2020, 136 (10), 1169–1179. 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras F.; et al. SARS-CoV-2 Triggered Neutrophil Extracellular Traps (NETs) Mediate COVID-19 Pathology. MedRxiv 2020, 10.1101/2020.06.08.20125823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrich G.; Raftery M. J. Neutrophil Extracellular Traps Go Viral. Front. Immunol. 2016, 7, 11–14. 10.3389/fimmu.2016.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam H. R.; Wong S. L. Cellular Mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218. 10.1146/annurev-cellbio-020520-111016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshner M.; Wang S.; Lewis C.; Zheng H.; Chen X. A.; Santy L.; Wang Y. PAD4Mediated Histone Hypercitrullination Induces Heterochromatin Decondensation and Chromatin Unfolding to Form Neutrophil Extracellular Trap-like Structures. Front. Immunol. 2012, 3 (Oct 4), 307. 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gunten S.; Yousefi S.; Seitz M.; Jakob S. M.; Schaffner T.; Seger R.; Takala J.; Villiger P. M.; Simon H. U. Siglec-9 Transduces Apoptotic and Nonapoptotic Death Signals into Neutrophils Depending on the Proinflammatory Cytokine Environment. Blood 2005, 106 (4), 1423–1431. 10.1182/blood-2004-10-4112. [DOI] [PubMed] [Google Scholar]
- Angata T.; Varki A. Cloning, Characterization, and Phylogenetic Analysis of Siglec-9, a New Member of the CD33-Related Group of Siglecs: Evidence for Co-Evolution with Sialic Acid Synthesis Pathways. J. Biol. Chem. 2000, 275 (29), 22127–22135. 10.1074/jbc.M002775200. [DOI] [PubMed] [Google Scholar]
- Lizcano A.; Secundino I.; Dohrmann S.; Corriden R.; Rohena C.; Diaz S.; Ghosh P.; Deng L.; Nizet V.; Varki A. Erythrocyte Sialoglycoproteins Engage Siglec-9 on Neutrophils to Suppress Activation. Blood 2017, 129 (23), 3100–3110. 10.1182/blood-2016-11-751636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams O. J.; Stanczak M. A.; von Gunten S.; Läubli H. Targeting Sialic Acid-Siglec Interactions to Reverse Immune Suppression in Cancer. Glycobiology 2017, 28 (9), 640–647. 10.1093/glycob/cwx108. [DOI] [PubMed] [Google Scholar]
- Duan S.; Paulson J. C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020, 38 (1), 365–395. 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- Zhang J. Q.; Nicoll G.; Jones C.; Crocker P. R. Siglec-9, a Novel Sialic Acid Binding Member of the Immunoglobulin Superfamily Expressed Broadly on Human Blood Leukocytes. J. Biol. Chem. 2000, 275 (29), 22121–22126. 10.1074/jbc.M002788200. [DOI] [PubMed] [Google Scholar]
- Morrissey M. A.; Vale R. D. CD47 Suppresses Phagocytosis by Repositioning SIRPA and Preventing Integrin Activation. bioRxiv 2019, 752311. 10.1101/752311. [DOI] [Google Scholar]
- Bornhöfft K. F.; Galuska S. P. Glycans as Modulators for the Formation and Functional Properties of Neutrophil Extracellular Traps : Used by the Forces of Good and Evil. Front. Immunol. 2019, 10 (May), 959. 10.3389/fimmu.2019.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A. F.; Uchiyama S.; Chang Y. C.; Lewis A. L.; Nizet V.; Varki A. Molecular Mimicry of Host Sialylated Glycans Allows a Bacterial Pathogen to Engage Neutrophil Siglec-9 and Dampen the Innate Immune Response. Blood 2009, 113 (14), 3333–3336. 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatua B.; Bhattacharya K.; Mandal C. Sialoglycoproteins Adsorbed by Pseudomonas Aeruginosa Facilitate Their Survival by Impeding Neutrophil Extracellular Trap through Siglec-9. J. Leukocyte Biol. 2012, 91 (April), 641–655. 10.1189/jlb.0511260. [DOI] [PubMed] [Google Scholar]
- Youngblood B. A.; Brock E. C.; Leung J.; Falahati R.; Bochner B. S.; Rasmussen H. S.; Peterson K.; Bebbington C.; Tomasevic N. Siglec-8 Antibody Reduces Eosinophils and Mast Cells in a Transgenic Mouse Model of Eosinophilic Gastroenteritis. JCI Insight 2019, 4 (19), e126219. 10.1172/jci.insight.126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R. R.; Zhang M. X.; Liu M.; Fang X.; Li D.; Zhang L.; Zheng P.; Zheng Y. T.; Liu Y. CD24Fc Protects against Viral Pneumonia in Simian Immunodeficiency Virus-Infected Chinese Rhesus Monkeys. Cell. Mol. Immunol. 2020, 17 (8), 887–888. 10.1038/s41423-020-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R. R.; Zhang M. X.; Zhang L. T.; Zhang P.; Ma J. P.; Liu M.; Devenport M.; Zheng P.; Zhang X. L.; Lian X. D.; et al. CD24 and Fc Fusion Protein Protects SIVmac239-Infected Chinese Rhesus Macaque against Progression to AIDS. Antiviral Res. 2018, 157 (June), 9–17. 10.1016/j.antiviral.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Läubli H.; Varki A. Sialic Acid – Binding Immunoglobulin - like Lectins (Siglecs) Detect Self - Associated Molecular Patterns to Regulate Immune Responses. Cell. Mol. Life Sci. 2020, 77 (4), 593–605. 10.1007/s00018-019-03288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaveris C. S.; Chiu S. H.; Riley N. M.; Bertozzi C. R. Modulation of Immune Cell Reactivity with Cis-Binding Siglec Agonists. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (3), e2012408118. 10.1073/pnas.2012408118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan C. D.; Schwartz E.; McBride R.; Fokin V. V.; Paulson J. C. Click and Pick: Identification of Sialoside Analogues for Siglec-Based Cell Targeting. Angew. Chem., Int. Ed. 2012, 51 (44), 11014–11018. 10.1002/anie.201205831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppenbrouwers T.; Autar A. S. A.; Sultan A. R.; Abraham T. E.; Van Cappellen W. A.; Houtsmuller A. B.; Van Wamel W. J. B.; Van Beusekom H. M. M.; Van Neck J. W.; De Maat M. P. M. In Vitro Induction of NETosis: Comprehensive Live Imaging Comparison and Systematic Review. PLoS One 2017, 12 (5), 1–29. 10.1371/journal.pone.0176472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Eutimio M. A.; López-Macías C.; Pastelin-Palacios R. Bioinformatic Analysis and Identification of Single-Stranded RNA Sequences Recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV Genomes. Microbes Infect. 2020, 22 (4–5), 226–229. 10.1016/j.micinf.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Made C. I.; Simons A.; Schuurs-Hoeijmakers J.; Van Den Heuvel G.; Mantere T.; Kersten S.; Van Deuren R. C.; Steehouwer M.; Van Reijmersdal S. V.; Jaeger M.; et al. Presence of Genetic Variants among Young Men with Severe COVID-19. J. Am. Med. Assoc. 2020, 324 (7), 663–673. 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Chan D. W.; Zaal K. J.; Kaplan M. J. A High-Throughput Real-Time Imaging Technique To Quantify NETosis and Distinguish Mechanisms of Cell Death in Human Neutrophils. J. Immunol. 2018, 200 (2), 869–879. 10.4049/jimmunol.1700905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C. L.; Shim D.; Kernien J.; Johnson C. J.; Nett J. E.; Shelef M. A. Insight into Neutrophil Extracellular Traps through Systematic Evaluation of Citrullination and Peptidylarginine Deiminases. J. Immunol. Res. 2019, 2019, 2160192. 10.1155/2019/2160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecha J.; Satpathy S.; Kanashova T.; Avanessian S. C.; Kane M. H.; Clauser K. R.; Mertins P.; Carr S. A.; Kuster B. TMT Labeling for the Masses: A Robust and Cost-Efficient, in-Solution Labeling Approach. Mol. Cell. Proteomics 2019, 18 (7), 1468–1478. 10.1074/mcp.TIR119.001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood C.; Arve S.; Ledbetter J.; Elkon K. B. TLR7/8 Activation in Neutrophils Impairs Immune Complex Phagocytosis through Shedding of FcgRIIA. J. Exp. Med. 2017, 214 (7), 2103–2119. 10.1084/jem.20161512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Chen J. Phosphoproteomic Analyses Provide Insight into Molecular Mechanisms Underlying NETosis. Proteomics 2019, 19, 1900126. 10.1002/pmic.201900126. [DOI] [PubMed] [Google Scholar]
- Lizcano A.; Secundino I.; Dohrmann S.; Corriden R.; Rohena C.; Diaz S.; Ghosh P.; Deng L.; Nizet V.; Varki A. Erythrocyte Sialoglycoproteins Engage Siglec-9 on Neutrophils to Suppress Activation. Blood 2017, 129 (23), 3100–3110. 10.1182/blood-2016-11-751636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczak M. A.; Zippelius A.; Läubli H. J. Clin. Investig. 2018, 128 (11), 4912–4923. 10.1172/JCI120612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läubli H.; Pearce O. M. T.; Schwarz F.; Siddiqui S. S.; Deng L.; Stanczak M. A. Engagement of Myelomonocytic Siglecs by Tumor-Associated Ligands Modulates the Innate Immune Response to Cancer. Proc. Natl. Acad. Sci. U.S.A. 2014, 111 (39), 14211–14216. 10.1073/pnas.1409580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M.; Wu X.; Christiano R.; Oh M. S.; Schauder C. M.; Gazzerro E.; Messa M.; Baldassari S.; Assereto S.; Biancheri R. The Leukodystrophy Protein FAM126A (Hyccin) Regulates PtdIns(4)P Synthesis at the Plasma Membrane. Nat. Cell Biol. 2016, 18 (1), 132–138. 10.1038/ncb3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. K.; Williams S. A.; Bindra G. K.; Lay F. T.; Poon I. K. H.; Hulett M. D. Phosphoinositides : Multipurpose Cellular Lipids with Emerging Roles in Cell Death. Cell Death Differ. 2019, 26, 781–793. 10.1038/s41418-018-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S.; Kawamura T.; Higuchi M.; Kobayashi T.; Yoshita-takahashi M.; Yamazaki M.; Abe M.; Sakimura K.; Kanda Y.; Kawamura H.; et al. RASAL3, a Novel Hematopoietic RasGAP Protein, Regulates the Number and Functions of NKT Cells. Eur. J. Immunol. 2015, 45, 1512–1523. 10.1002/eji.201444977. [DOI] [PubMed] [Google Scholar]
- Moghanloo E.; Ghorbani E.; Beikverdi M. S.; Badameh P.; Rezaei S.; Piroozmand A.; Teimourian S.; Shahidi M.; Khorshidi A. The Netosis Formation of HL-60 Cell Differentiated to Neutrophil-Like Cells by LPS. J. Human, Environ. Heal. Promot. 2018, 4 (3), 138–143. 10.29252/jhehp.4.3.8. [DOI] [Google Scholar]
- Meizlish M. L.; Pine A. B.; Bishai J. D.; Goshua G.; Nadelmann E. R.; Simonov M.; Chang C.-H.; Zhang H.; Shallow M.; Bahel P.; et al. A Neutrophil Activation Signature Predicts Critical Illness and Mortality in COVID-19. medRxiv 2020, 10.1101/2020.09.01.20183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn M. T.; DeLeo F. R.. Neutrophil: Methods and Protocols, 3rd ed.; Springer Science: New York, 2020. [Google Scholar]
- Branitzki-Heinemann K.; Möllerherm H.; Völlger L.; Husein D. M.; de Buhr N.; Blodkamp S.; Reuner F.; Brogden G.; Naim H. Y.; Von Köckritz-Blickwede M. Formation of Neutrophil Extracellular Traps under Low Oxygen Level. Front. Immunol. 2016, 7 (Nov 25), 518. 10.3389/fimmu.2016.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso S.; Neumann A.; Lang I. M.; Etscheid M.; von Köckritz-Blickwede M.; Kanse S. M. Interaction of Factor VII Activating Protease (FSAP) with Neutrophil Extracellular Traps (NETs). Thromb. Res. 2018, 161, 36–42. 10.1016/j.thromres.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Chen G. Y.; Tang J.; Zheng P.; Liu Y. CD24 and Siglec-10 Selectively Repress Tissue Damage - Induced Immune Responses. Science (Washington, DC, U. S.) 2009, 323 (5922), 1722–1725. 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods E. C.; Yee N. A.; Shen J.; Bertozzi C. R. Glycocalyx Engineering with a Recycling Glycopolymer That Increases Cell Survival In Vivo. Angew. Chem., Int. Ed. 2015, 54, 15782–15788. 10.1002/anie.201508783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaveris C. S.; Webster E. R.; Banik S. M.; Boxer S. G.; Bertozzi C. R. Membrane-Tethered Mucin-like Polypeptides Sterically Inhibit Binding and Slow Fusion Kinetics of Influenza A Virus. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (23), 12643–12650. 10.1073/pnas.1921962117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs T. A.; Brill A.; Duerschmied D.; Schatzberg D.; Monestier M.; Myers D. D.; Wrobleski S. K.; Wakefield T. W.; Hartwig J. H.; Wagner D. D. Extracellular DNA Traps Promote Thrombosis. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (36), 15880–15885. 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo J.; Leung H. H. L.; Ahmadi Z.; Yan F.; Chong J. J. H.; Passam F. H.; Chong B. H. Neutrophil Activation and NETosis Are the Major Drivers of Thrombosis in Heparin-Induced Thrombocytopenia. Nat. Commun. 2019, 10 (1), 1322. 10.1038/s41467-019-09160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnatsch A.; Ioannou M.; Wang Q.; Papayannopoulos V. Neutrophil Extracellular Traps License Macrophages for Cytokine Production in Atherosclerosis. Science 2015, 349 (6245), 316–320. 10.1126/science.aaa8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V.; Staab D.; Zychlinsky A. Neutrophil Elastase Enhances Sputum Solubilization in Cystic Fibrosis Patients Receiving DNase Therapy. PLoS One 2011, 6 (12), e28526. 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.