Abstract
Background
Mouse mammary tumor virus (MMTV) is thought to have a role in human breast cancer (BC) pathogenesis. BRCA1 and 2 genes mutations are well-established risk factors for BC. The purpose of this study was to evaluate the presence of MMTV in familial and non-familial Egyptian breast cancer patients. We also aimed to establish a correlation between BRCAs genes mutations and MMTV infection in those patients.
Patients and Methods
The study was included 80 BC patients and 10 healthy women were included as a control group. We used PCR to amplify a 250-bp MMTV-like env sequence. We also used PCR followed by direct sequencing to identify the genetic variation of exons 2, 13, 19 of BRCA1 gene and exon 9 and region f of exon 11 of BRCA2 gene. High resolution melting (HRM) analysis was used to screen the selected exons of BRCA1/2 genes in order to detect different variants.
Results
MMTV DNA-like env sequences were detected in 70%, 76% of familial and non-familial BC patients, respectively, and it was not detected in any of the control subjects. The presence of viral sequences was associated with larger tumor size in the sporadic patients. Seventy BC patients showed variations in BRCA1/2 genes according to HRM analysis and sequencing analysis showed two different sequences of polymorphism among 22 familial and non-familial BC patients.
Conclusion
MMTV DNA was present among BC patients and it was associated with increased tumor growth. This indicates a potential role for MMTV in BC patients with and without deleterious mutation in BRCA1/2 genes.
Keywords: breast cancer, MMTV, BRCA1/2, HRM, Egypt
Introduction
Breast cancer (BC) is the deadliest cancer among women in several countries representing a quarter of cancer cases diagnosed in women.1,2 In Egypt, BC represents 32% of women’s cancers.3
Studying of BC has improved its detection and treatments leading to a significant decrease in the BC-associated death.4 However, little is known about the main causes, etiology, or carcinogenesis of the BC due to its great complexity. BC is highly heterogeneous comprising several types and subtypes with distinct genetic background, pathogenesis, response to treatment, and prognosis indicating the presence of other causative agents that may be implicated in BC carcinogenesis. Genetic mutations play a role in the multi-step process of BC malignancy.5–7 In addition, somatic mutations are also involved in the familial BC cases.6 BC cases are mostly sporadic, also are associated with a low frequency of familial predisposition. Therefore, relying on identifying genetic causes will only explain a limited number of BC cases.8 In 2017, it was reported that 154,794 women living with metastatic breast cancer (MBA) in the United States indicating the increase of MBA cases in USA.9 Besides, environmental factors such as alcohol consumption, smoking, lifestyle, obesity, and body mass index affect the BC incidence and prognosis.10−13 Moreover, infection is another risk factor for BC development. Infection plays a role in 16% of human cancers.14 These cancers are caused by viral oncogenic genes such as human papilloma virus (HPV) which cause cervical cancer, or by viruses that diminish the host immune response such as human immunodeficiency virus (HIV), or by viruses that cause chronic inflammation and damage such as HCV and HBV. Interestingly, mouse mammary tumors were caused by Mouse Mammary Tumor Virus (MMTV) which is transmitted in breast milk.15,16 Viral infections play a role in the etiology and pathogenesis of BC such as HPV, polyomavirus, and MMTV.17,18 The MMTV-like env gene sequences of the MMTV-like virus, previously known as mouse milk factor, were identified at a significantly high level in breast cancers compared to the non-cancer tissues.19–21 MMTV-like virus was identified in 30–74% of BC tissues in several countries.16,22–24 MMTV initiates BC via the insertional mutagenesis process which causes uncontrolled cellular proliferation leading to cancer initiation and progression.16
Genetic mutations increase the BC susceptibility among certain families, especially the tumor suppressor genes such as breast cancer anti-estrogen resistance-1 and 2 (BRCA1 and BRCA2), which are found to be frequently mutated in hereditary cancers.5–7 BRCA1 and BRCA2 mutations account for 30% of breast and ovarian cancers.25 The estimated cumulative breast cancer risk at age 70 years was 0.46 in BRCA1 women and 0.43 in BRCA2 women.26 However, the role of MMTV in BC initiation and progression in patients with BRCA mutations is not completely understood.
Up to our knowledge, there is no report linking the role of MMTV and BRCA1 and 2 mutations in the BC development among Egyptian women. In this study, we tested the presence of MMTV-like gene among Egyptian BC patients and correlated between MMTV-like gene, BRCA1 and two genetic mutations, and clinicopathological parameters of BC.
Patients and Methods
Patients
The study is done on BC patients (n=80) admitted to surgery, and pathology departments, National Cancer Institute, Cairo University, Egypt between March 2011 to July 2013, it is a retrospective analysis. BC patients, enrolled in this study, were divided into two groups; group 1: BC patients with family history (n=30) and group 2: sporadic BC patients (n=50). The inclusion criteria were women suffering from BC and did not receive any chemotherapy treatment before surgery. BC women received chemotherapy prior to surgery were not included in this study. Ten (n=10) healthy women served as controls. The control subjects were age-matched with the BC groups. The sample size was selected based on Al Dossary et al,20 MMTV env proviral sequences were detected in 5.9% (6/101) of breast cancer tissues in a group of Saudi women. Based on these findings, a sample size of 80 patients with breast cancer is required for estimating the expected proportion with 5% absolute precision and 95% confidence. The study was approved by the Institution Review Board (IRB) of the National Cancer Institute, Cairo University (IRB No. 200801002.2). A written informed consent was obtained from each participant. All of the experiments were performed in compliance with relevant laws and institutional guidelines and in accordance with the ethical standards of the Declaration of Helsinki.
Methods
Sample Collection and Storage
Breast tumor tissues and blood were collected from each BC patient and healthy controls and were stored at −80°C until use. Total genomic DNA was extracted from blood samples using QIAamp DNA Mini kit (Qiagen, Hilden, and Germany), according to the manufacturer’s instructions. Tumor tissues were processed as described previously.27 The concentration and quality of the extracted DNA were determined by Nanodrop ND2000 spectrophotometer (Nanodrop Technologies, USA).
Molecular Detection
Nucleic Acid Extraction
DNA was extracted from breast cancer tissues using the phenol–chloroform method as previously described.27 MTV DNA extraction from MCF-7 cell line was done using QIA amp DNA extraction kit (Qiagen, Valencia, USA) according to the manufacturer’s instructions. MCF-7 was obtained from VACSERA (Egypt). The concentration and quality of MMTV DNA were assessed by spectrophotometry using a Nano-Drop 2000 spectrophotometer. About100 ng of DNA template was used as a positive control in each PCR run.
Detection of MMTV-DNA
Detection of MMTV-DNA was done using primers targeting env genomic region targeting the nucleotide position from 1386 to 1640 bp of MMTV genome (GenBank ID AF346816). The PCR reaction was done as described previously28 at the following conditions: 94°C for 5 minutes, followed by 35 cycles of: 95°C for 1–5 min, 55°C for 1 min, and 72°C for 5 min, with a final step at 72°C for 10 minutes. The PCR product was run on a 2.5% agarose gel. The expected PCR product size appeared at 250 bp.
Sensitivity and Specificity of MMTV PCR Assay
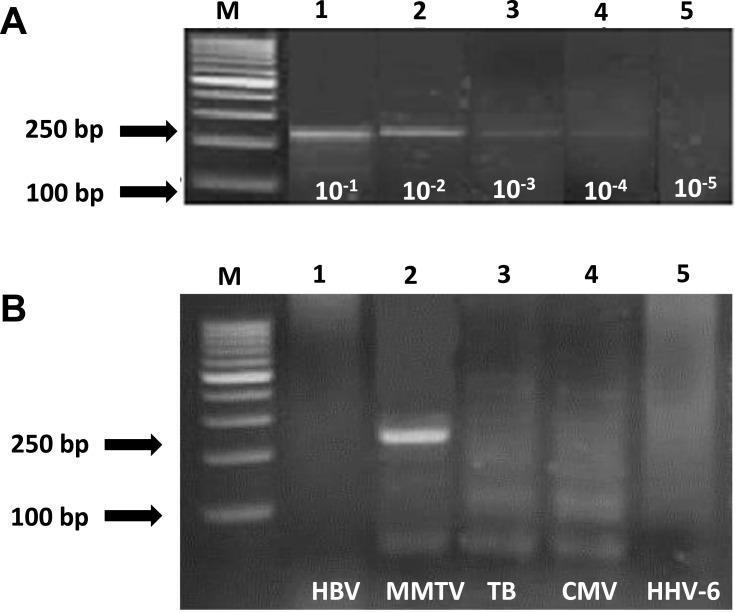
The sensitivity of MMTV PCR assay was evaluated by making serial dilutions of positive control (MMTV extracted from MCF-7 cell line) from 10−1 up to 10–5 (Figure 1). The specificity of the PCR assay was assessed by testing the PCR reaction against other pathogens such as HBV, TB, CMV, and HHV-6 (Figure 1).
Figure 1.
Sensitivity and specificity of PCR assay for detection of MMTV DNA-like env sequences extracted from MCF-7 cell line. (A) Ethidium bromide–stained gel electrophoresis shows sensitivity assay of primers used to amplify MMTV DNA-like env sequences. Positive signals are 250 bp. Lane M = 100-bp ladder; lanes 1–5 = positive lanes of serial dilutions from 10−1-10−5. (B) Ethidium bromide–stained gel electrophoresis of specificity assay of primers used to amplify MMTV DNA-like env sequences. Positive signal at 250 bp. For MMTV DNA product (lane 2), lanes 1, 3, 4, 5 are negatives, Lane M = 100bp ladder.
HRM Assay of BRCA1/2 Variations and Sequencing for the Variants
PCR reactions were done to amplify the exons (2, 13, and 19) of BRCA1 and exons (9 and 11f) of BRCA2 using sets of primers previously described29 and mentioned in Table 1, using Applied Biosystems 7500 Fast Real-Time PCR system, USA (SN.275014218, Singapore). The PCR and HRM were performed in a single run on a 7500 Fast Real-Time PCR System in a reaction mix containing MeltDoctor HRM Master Mix (Thermofisher, applied biosystem, Vilnius, Lithuania), 200 nM of each PCR primers and 20 ng of genomic DNA. The PCR reaction was run as the followings: an activation step at 95°C for 10 minutes followed by 55 cycles of 95°C for 10 seconds, a touch down of 65°C to 55°C for 10 seconds (1◦C/cycle) and 72°C for 30 seconds. The products were heated to 95°C for 1 minute prior to the high-resolution melting step and the HRM was carried out over the range from 72°C to 95°C rising at 1°C per second with 30 acquisitions per degree. The melting curves obtained from control samples (healthy women, n=10) were used for normalization and comparison with BC samples. Analysis of the obtained curves was performed using Applied Biosystems HRM v2.0.2 software. The HRM software plots fluorescence signal over temperature. BRCA1 and BRCA2 PCR products which show variations by HRM analysis were sequenced (Macrogen, Korea). The PCR products were sequenced using the same primers used for amplification reactions.
Table 1.
List of Primers Used to Amplify the Selected Exons from BRCA1 and 2 Genes
| Gene Accession No. |
Primers | Nucleotide Sequence | Nucleotide Positions |
Amplicon Size |
|---|---|---|---|---|
|
BRCA1 MIM# 113705 |
Exon 2 | F: 5ʹ- CTT TTA AAA AGA TAT ATA TAT ATG TTT TTC TAA TGT GT-3ʹ R: 5ʹ- TCC CAA ATT AAT ACA CTC TTG TGC TGA-3’ |
93 826–93 863 93 999–93 981 |
173bp |
| Exon 13 | F: 5ʹ- GAT GTC TAC AAT TTC ACC TTT CT-3ʹ R: 5ʹ- TTG CCA AAA TGA CGA ACA CA-3’ |
138 557–138 579 138 690–138 669 |
133bp | |
| Exon 19 | F: 5ʹ- CTT TCT CTT ATC CTG ATG GGT TGT G-3ʹ R: 5ʹ- GAG TGG TGG GGT GAG ATT TTT GTC-3ʹ |
160 808–160 833 160 978–160 955 |
170bp | |
|
BRCA2 MIM# 600185 |
Exon 9 | F: 5ʹ- ATA AGG GGG GAC TAC TAC TAT ATG TG-3ʹ R: 5ʹ- CAA AAA AAC CTG TAG TTC AAC TAA ACA GAG-3ʹ |
20 368–20 393 20 588–20 558 |
220bp |
| Exon 11F | F: 5ʹ- GCA GGA TTT TAA TTC AAA CCA TAA TTT AAC AC3ʹ R: 5ʹ- CCC TAA ACC CCA CTT CAT TTT CAT C3’ |
27 158–27 189 27 528-27 504 |
371bp |
Statistical Analysis
Data were analyzed using SPSS advanced statistics version 20 (SPSS Inc., IBM, Chicago, IL) and GraphPad Prism 6 (La Jolla, California). Numerical data were expressed as mean and standard deviation or median and range as appropriate. Qualitative data were expressed as frequency and percentage. Chi-square test (Fisher’s exact test) was used to examine the relation between qualitative variables. For paired qualitative data, comparison was done using McNemar test. For quantitative data, comparison between two groups was done using either Student’s t-test or Mann–Whitney test. P-value < 0.05 was considered significant.
Results
The mean age of the familial and non-familial BC groups was 47.0±9.4 and 50.9±11.4, respectively (p=0.113). The mean tumor size was significantly larger in patients with non-familial BC than in familial BC patients (5.7±3.9 cm vs 4.3±2.6 cm, respectively, p=0.031). Clinicopathological characteristics including age, estrogen receptor status (ER), progesterone receptor status (PR), lymph node involvement, safety margin among familial and non-familial BC are presented in Table 2. Positive ER was significantly higher among non-familial BC compared to familial BC (84% vs 63.3%, respectively, p=0.036). Most of the familial BC patients were premenopausal, while the majority of non-familial BC patients were postmenopausal (p=0.033).
Table 2.
Clinicopathological Features in Breast Cancer Patients
| Parameters | Non-Familial Breast Cancer N= 50 (%) |
Familial Breast Cancer N=30 (%) |
p-value |
|---|---|---|---|
| Age (years) | 0.16 | ||
| <45 | 16 (32.0) | 14 (46.7) | |
| ≥45 | 34 (68.0) | 16 (53.3) | |
| Tumor Size (Mean±SD) | 5.7±3.9 | 4.3±2.6 | 0.031 |
| ER-status | 0.036 | ||
| Positive | 42 (84) | 19 (63.3) | |
| Negative | 8 (16) | 11 (36.7) | |
| PR-Status | |||
| Positive | 37 (74) | 20 (66.7) | 0.48 |
| Negative | 13 (26) | 10 (33.3) | |
| Lymph Node Involvement | |||
| Positive | 38 (76) | 24 (80) | 0.67 |
| Negative | 12 (24) | 6 (20) | |
| Safety Margin | |||
| Positive | 4 (8) | 3 (10) | 0.75 |
| Negative | 46 (92) | 27 (90) | |
| Tumor Grade | |||
| G I, II | 45 (90) | 27 (90) | 1.00 |
| G III | 5 (10) | 3 (10) | |
| Pathological Type | |||
| Invasive duct carcinoma | 46 (92) | 26 (86.7) | 0.44 |
| Invasive lobular carcinoma | 4 (8) | 4 (13.3) | |
| Menopausal Status | |||
| Premenopausal | 21 (42) | 20 (66.7) | 0.033 |
| Postmenopausal | 29 (58) | 10 (33.3) | |
| Laterality | |||
| Right | 31 (63.3) | 15 (50) | 0.25 |
| Left | 18 (36.7) | 15 (50) | |
| Stage | |||
| II | 25 (50) | 19 (63.3) | 0.54 |
| III | 21 (42) | 10 (33.3) | |
| IV | 4 (8) | 1 (3.3) | |
| Hormonal Therapy | 0.119 | ||
| Yes | 41 (82) | 20 (66.7) | |
| No | 9 (18) | 10 (33.3) |
Presence of MMTV DNA in All Studied Groups
Sensitivity and specificity of qualitative PCR assay for detection of MMTV extracted from MCF-7:
The sensitivity level of the assay was found to be approximately 100 copies/μL (Figure 1A). The PCR was found to be highly specific for MMTV DNA as none of the other tested viruses were amplified (HBV, CMV, TB, and HHV6) (Figure 1B). MMTV-DNA-like env sequences were detected in 38 of 50 (76%) of non-familial BC tissues and in 21 of 30 (70%) of familial BC tissues (p=0.555) (Table 3). It was not detected in the 10 healthy control women.
Table 3.
MMTV-Like env Sequence and Its Association with Clinicopathological Features in Breast Cancer Patients
| Non-Familial | Familial | |||
|---|---|---|---|---|
| N= 50 (%) | N=30 (%) | |||
| MMTV | MMTV | |||
| Positive | Negative | Positive | Negative | |
| N=38(76) | N=12(24) | N=21(70) | N=9(30) | |
| Age (years) | ||||
| <45 | 12 (75) | 4(25) | 11(78.6) | 3(21.4) |
| 45 | 26(76.5) | 8(23.5) | 10(62.5) | 6(37.5) |
| p-value | 0.91 | 0.44 | ||
| ER Status | ||||
| Positive | 31 (73.8) | 11(26.2) | 14(73.7) | 5(26.3) |
| Negative | 7 (87.5) | 1 (12.5) | 7(63.6) | 4(36.4) |
| p-value | 0.406 | 0.563 | ||
| PR Status | ||||
| Positive | 29 (78.4) | 8(21.6) | 13(65) | 7(35) |
| Negative | 9 (69.2) | 4 (30.8) | 8(80) | 2(20) |
| p-value | 0.506 | 0.398 | ||
| Tumor Size | ||||
| <5cm | 14 (60.9) | 9 (39.1) | 12 (66.7) | 6 (33.3) |
| ≥5cm | 24 (88.9) | 3 (11.1) | 9(75) | 3 (25) |
| p-value | 0.012 | 0.626 | ||
| Lymph Node | ||||
| Positive | 30 (78.9) | 8(21.1) | 17 (70.8) | 7(29.2) |
| Negative | 8 (66.7) | 4(33.3) | 4(66.7) | 2(33.3) |
| p-value | 0.385 | 1.000 | ||
| Tumor Grade | ||||
| G I, II | 33(73.3) | 12(26.7) | 19(70.4) | 8(29.6) |
| G III | 5(100) | 0.0(0) | 2(66.7) | 1(33.3) |
| p-value | 0.319 | 1.000 | ||
| Safety Margin | ||||
| Positive | 4(100) | 0 (0.0) | 2(66.7) | 1(33.3) |
| Negative | 34(73.9) | 21(26.1) | 19(70.4) | 8(29.6) |
| p-value | 0.56 | 1.000 | ||
| Pathological Type | ||||
| Invasive duct carcinoma | 35 (76.1) | 11(23.9) | 17(65.4) | 9(34.6) |
| Invasive lobular carcinoma | 3(75) | 1 (25) | 4 (100) | 0(0.0) |
| p-value | 1.000 | 0.287 | ||
| Menopausal Status | ||||
| Premenopausal | 15(71.4) | 6(28.6) | 14(70) | 6(30) |
| Postmenopausal | 23(79.3) | 6(20.7) | 7(70) | 3(30) |
| p-value | 0.52 | 1.000 | ||
| Laterality | ||||
| Right | 24(77.4) | 7(22.8) | 12(80) | 3(20) |
| Left | 13(72.2) | 5(27.8) | 9(60) | 6(40) |
| p-value | 0.683 | 0.427 | ||
| Stage | ||||
| II | 18(72) | 7(28) | 13(68.4) | 6(31.6) |
| III+IV | 20(80) | 5(20) | 8(72.7) | 3(27.3) |
| p-value | 0.508 | 0.804 | ||
| Hormonal Therapy | ||||
| Yes | 8(88.9) | 1(11.1) | 5(50) | 5(50) |
| No | 30 (73.2) | 11(26.8) | 16(80) | 4(20) |
| p-value | 0.317 | 0.091 | ||
MMTV DNA-Like env Sequences and Clinicopathological Factors
The presence of MMTV-DNA like env sequences was not associated with different clinicopathological factors in both BC groups except larger tumor size in the non-familial group (p=0.012) (Table 3).
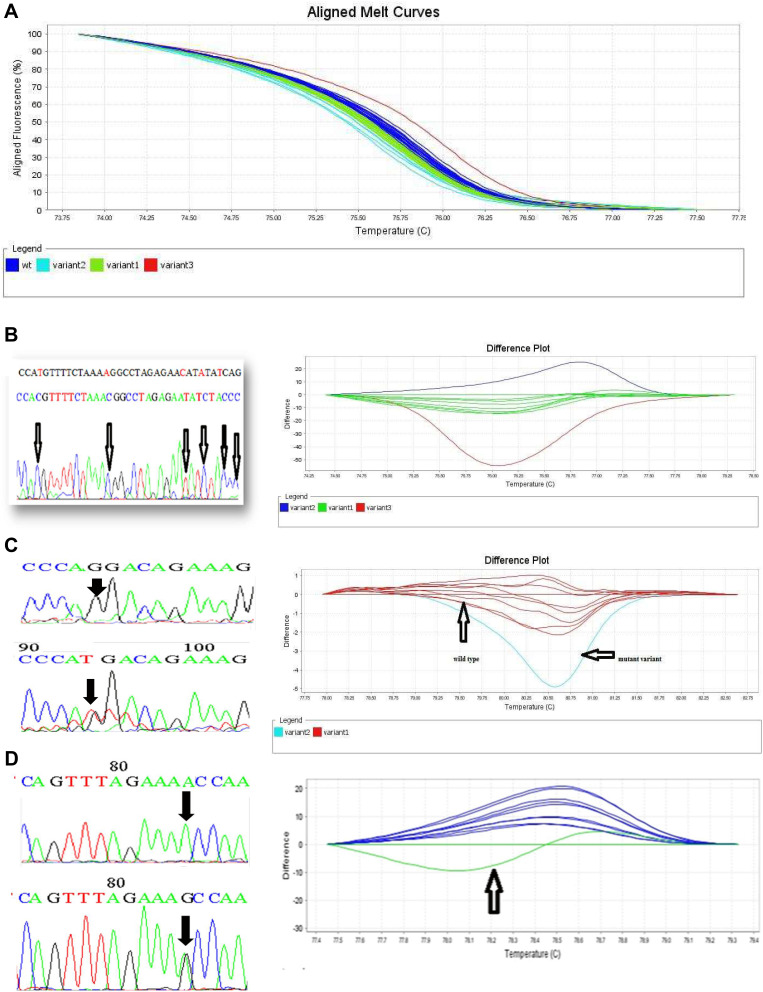
Detection of BRCA1 and 2 Gene Variations Using HRM
The aligned melt curve graph and difference plots for the selected exons of BRCA1/2 genes are shown in (Figure 2). Variations in BRCA1/2 genes in the selected exons are summarized in (Table 4). Patients with familial BC showed more frequent variations (2–4) in exon 13 of BRCA1 gene compared to non-familial BC (50% vs 26%, p=0.029). In contrast, significantly more frequent variations (2–4) (same as the previous comment) in exon 9 of BRCA2 gene were noticed in the non-familial BC compared to familial BC (62% vs 33%, p=0.013) Table 4. Forty-two samples that showed HRM variations were subjected to sequencing analysis to identify the type of mutations.
Figure 2.
HRM results and its sequencing analysis. (A) Aligned melt curves of BRCA1 gene exon 2 by HRM. 2a. Dark blue: wt (wild type), light blue: variant 2, green: variant1, and red: variant 3 (B) HRM results and its sequencing analysis amplifying 133bps of exon 13, BRCA1 gene. Dark blue: variant 2, green: variant 1, red: variant 3 (C) HRM results and its sequencing analysis amplifying 170bps of exon 19, BRCA1 gene. Red: wild type and light blue: mutant variant (D) HRM results and its Sequencing analysis amplifying 371bps of BRCA2 gene exon 11f of BRCA2 gene. Dark blue: wild type and green: mutant variant.
Table 4.
Frequency of Variations in the Selected Exons of BRCA1/2 Genes in Breast Cancer Patients as Detected by HRM Assay
| Non-Familial | Familial | p-value | |||
|---|---|---|---|---|---|
| N=50 (%) | N=30 (%) | ||||
| Variant | Variants | Variant | Variants | ||
| (1) | (2–4) | (1) | (2–4) | ||
| BRCA1 | |||||
| Exon 2 | 31(62) | 19(38) | 23(76.7) | 7(23.3) | 0.175 |
| Exon 19 | 30(60) | 20(40) | 18(60) | 12(40) | 1.000 |
| Exon 13 | 37(74) | 13 (26) | 15(50) | 15(50) | 0.029 |
| BRCA2 | |||||
| Exon 9 | 19(38) | 31(62) | 20(66.7) | 10(33.3) | 0.013 |
| Exon 11 F | 39(78) | 11(22) | 25(83.3) | 5(16.7) | 0.564 |
Mutational Analysis of BRCA Genes Using Sequencing Assay
Only 22/42 (52%) familial and non-familial BC patients had changes in exons 13, 19 of BRCA1 gene and exon 11f of BRCA2, whereas exon 2 of BRCA1 and exon 9 of BRCA2 did not demonstrate any changes in their sequences. Sequencing analysis of exon 11f of BRCA2 gene showed nine different sequences, submitted to gene bank for annotation under the following accession numbers and entry ID: LC331907, LC331908, LC331909, LC331910, LC331911, LC331912, LC331708, LC331709, LC331710. Regarding, LC331909 and LC331911, SNP rs1801206 was detected at anchored position 27,272 of NG_012772.3 on chromosome 13 (A/C/G) (Table 5, Figure 3A). LC331912 demonstrated SNP rs1799952 at anchored position 27,392 of NG_012772.3 on chromosome 13 (A/G/T).
Table 5.
Polymorphism in BRCA1/2 Genes
| Program Database |
Accession Number | Refseq Genome | Anchored Position | BRCA/Exon | Allele | Variant | No. of Observation |
|---|---|---|---|---|---|---|---|
| BLASTN Human G+T |
LC331909 LC331911 |
NG_012772.3 | 27,272 | BRCA2/11 | A/C/G | rs1801406 | 2 |
| BLASTN Human G+T |
LC331912 | NG_012772.3 | 27,392 | BRCA2/11 | A/G/T | rs1799952 | 2 |
| BLASTN Human G+T |
LC368290 | NC_000017.11 | 43,057,061 | BRCA1/19 | C/T/A | rs571834423 | 1 |
Figure 3.
Polymorphism of BRCA1 and BRCA2. (A) Blast result of LC331909 (exon 11f of BRCA2) using NG_012772.3 demonstrated SNP. (B) Blast result of LC368290 (exon 13 of BRCA1) using NC_000017 demonstrated SNP.
As for sequence analysis of exon 13, 11 variations were detected in the same sequences, but we could not process them for annotation due to low quality of data; further studies will be performed on this exon. Our results showed one sequence in exon 19, has annotated with accession number and entry ID of LC368290. This sequence demonstrated prominent SNP rs571834423, detected at anchored position 43,057,061 of NC_000017.11 on chromosome 17 (Table 5, Figure 3B).
Only one sporadic BC patient showed mutation in exon 19 of BRCA1 and was associated with negative ER.
Relation Between MMTV-Like env Sequence, BRCA1/2 Genes Polymorphism and Clinicopathological Factors
MMTV DNA was present in all BC patients who demonstrated polymorphism in exons 19 of BRCA1 and exon 11f of BRCA2 genes. Most BC patients were >45 years and most of them were positive ER (>75%). Distribution of data is presented in Table 6.
Table 6.
Presence of MMTV, BRCA Variations by HRM and Some Clinical Parameters Among 80 Women with Breast Cancer Disease. (A) In Familial Breast Cancer. (B) In Non-Familial Breast Cancer
| (A) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | Familial Breast Cancer Patients N=30 |
|||||||||
| HRM Variant (1) | HRM Variants (2–4) | |||||||||
| BRCA1 Exons |
BRCA2 Exons |
BRCA1 Exons |
BRCA2 Exons |
|||||||
| 2 N=23 |
13 N=15 |
19 N=18 |
9 N=20 |
11f N=25 |
2 N=7 |
13 N=15 |
19 N=12 |
9 N=10 |
11f N=5 |
|
|
Age (years) <45 (N=14) ≥45 (N=16) |
12 11 |
6 9 |
10 8 |
9 11 |
13 12 |
2 5 |
8 7 |
4 8 |
5 5 |
1 4 |
|
ER-status Pos. (N=19) Neg. (N=11) |
14 9 |
9 6 |
12 6 |
12 8 |
16 9 |
5 2 |
10 5 |
7 5 |
7 3 |
3 2 |
|
MMTV Pos. (N=21) Neg. (N=9) |
15 8 |
10 5 |
11 7 |
13 7 |
16 9 |
6 1 |
11 4 |
10 2 |
8 2 |
5 0 |
| (B) | ||||||||||
| Parameters |
Non-Familial Breast Cancer Patients N=50 |
|||||||||
| HRM Variant (1) | HRM Variants (2–4) | |||||||||
|
BRCA1 Exons |
BRCA2 Exons |
BRCA1 Exons |
BRCA2 Exons |
|||||||
|
2 N=31 |
13 N=37 |
19 N=30 |
9 N=19 |
11f N=39 |
2 N=19 |
13 N=13 |
19 N=20 |
9 N=31 |
11f N=11 |
|
|
Age <45 (N=14) ≥45 (N=16) |
20 11 |
25 12 |
21 9 |
12 7 |
27 12 |
13 6 |
9 4 |
12 8 |
21 10 |
7 4 |
|
ER-status Pos. (N=19) Neg. (N=11) |
26 5 |
31 6 |
24 6 |
14 5 |
33 6 |
16 3 |
8 5 |
14 6 |
28 3 |
9 2 |
|
MMTV Pos. (N=21) Neg. (N=9) |
22 9 |
31 6 |
23 7 |
14 5 |
33 6 |
16 3 |
8 5 |
15 5 |
24 7 |
4 7 |
Discussion
BC is classified into several types and subtypes which differ significantly in its pathogenesis, response to treatment, and prognosis. Genetic mutations, especially in BRCA1 and 2 genes significantly increase the risk of BC development. Viral infections play a role in BC pathogenesis in several populations. However, there have not been any studies that linked the viral infection and genetic mutations in the development of BC in Egyptian patients. So, we thought to be the first study to investigate this critical matter.
The origin of MMTV has not been well identified but it is strongly thought to be transmitted from mice to humans.30 The incidence of MMTV among BC patients is variable.30–33 The incidence of BC is higher in areas where certain types of mice with rich MMTV in its genome, Mus domesticus, are common.22,30 MMTV is capable of infecting human cells, suggesting MMTV transmission to humans.34
Our results show that MMTV DNA-like env sequences were detected in 70% and 76% in familial and non-familiar BC tissues, respectively. This prevalence is considerably higher than the prevalence rates reported elsewhere.22,28,33,35,36 MMTV-like sequences have been found in human breast cancer tissues in wide geographical variations (0.8–74%).24,35 Several studies have demonstrated the presence of MMTV-like env sequences in 30–40% of BC cases in several Western countries including the United States, Italy, Mexico, Brazil, Argentina, and Australia.22 In Southeast Asia, where BC incidences are known to be of low rate, the presence of MMTV-like env sequences in BC was 12% in Japan and 10–17% in China.37 However, in Vietnamese BC has been reported to be only 0.8% MMTV-like positive.24,33 In Tunisia, BC was reported to have a high proportion of MMTV-like env sequences, almost 74%,24 but later MMTV-like env sequences were reported at a lower rate.23
PEV positive tumors were reported to be more aggressive compared to PEV negative tumors and most tumors containing MMTV-like sequences are considered aggressive tumors, which propose a link between MMTV-like and inflammatory BC.24 Therefore, the difference in MMTV prevalence between our study and that reported elsewhere could be explained by a higher frequency of inflammatory BC in our patients. Especially that a higher rate of inflammatory BC was reported in Egypt compared to other countries.38 The MMTV-like prevalence reported here concords with other studies performed in countries with a high incidence of BC such as the United States which reports MMTV-like sequences in 30–40% of BC patients.39–41 Other studies from Argentina, Italy, and Australia report MMTV-like prevalence in 31.7%, 37.7%, and 42.2% of BC tumors, respectively.28,33,36 However, studies from other countries could not identify MMTV-like sequences in any cases.42–44 This discrepancy could be largely attributed to variation in detection procedures or tissue heterogeneity.22 When comparing PEV-positive tumors with PEV-negative tumors in the latter series, a higher proportion of aggressive tumors contained the MMTV-like sequences, suggesting a correlation between MMTV-like and inflammatory BC. Importantly, MMTV-like env sequences could not be identified in any of the tissues obtained from apparently healthy women included in this study.
We could not establish a correlation between MMTV DNA-like env sequences and any clinicopathological parameters of BC such as age, hormonal receptors, histologic subtype, or tumor grade, similar findings were reported elsewhere.45 However, our results show a significant association between the presence of MMTV DNA-like env sequences and larger tumor size (>5cm) in non-familial BC patients. This indicates the detection of MMTV DNA as a risk factor for BC alongside age, menopausal status, hormonal receptor.46 Nonetheless, it cannot be considered as major risk factors that significantly affect BC pathogenesis, since we could not find any significant difference in presence of MMTV DNA-like env sequences between familial and non-familial BC. It has been reported before that MMTV is correlated with tumor grade in BC, where MMTV-like env sequences were found in 26.3% of ductal carcinoma in situ and 53.8% of infiltrating ductal carcinoma.33 In addition, MMTV identified in BC tumors was associated with inflammatory tumors compared to tumors without MMTV-like env sequences.23 To this end, large-scale studies are required to rule out whether MMTV is truly correlated with specific clinicopathological parameters that complicate BC carcinogenesis.
Genetic factors are risk factors for BC susceptibility, specifically BRCA1 and BRCA2 gene mutations. Identification of founder and recurrent mutations in those genes could be a major step towards the improvement of genetic counseling. In BC, such improvements largely rely on identifying relevant mutations present in certain ethnic groups, this will eventually lead to developing rapid and less expensive tests for the determined human population.47 In this regard, high resolution melting (HRM) assay has proven to be an attractive choice of mutation screening; it is rapid, sensitive, and relatively inexpensive.48,49 The current study investigated the sequence variation of exons 2,13 and 19 of BRCA1 and exons 9 and certain regions of exon 11 (11f) of the BRCA2 gene in Egyptian familial and non-familial BC patients.
Sequencing analysis was performed on a total of 70 familial and non-familial BC patients who showed sequence variations as detected by HRM assay. HRM results showed a frequency of variants in exon 13 of the BRCA1 gene to be more prominent among familial patients, whereas the frequency of variants in exon 9 of the BRCA2 gene was more evident among the sporadic cases. This establishes HRM as a sensitive, cost-effective and rapid tool for the detection of genetic mutations in BC patients. Results showed that 42 of 70 (60%) BC patients showed different types of polymorphisms. The BRCA1 gene did not show the classical mutation 185delAG instead it showed a variation at rs571834423 at the position of 43,057,061 of NC_000017.11. Similarly, exon 9 of the BRCA2 gene did not show the previously reported mutation (3492InsT).50 Instead, polymorphism in BRCA2 exon 11 (rs1801406 and rs1799952) was observed in 12% of BC patients. Eleven of 42 (26%) BC patients showed the same changes in exon 13 but could not be analyzed.
Our results showed that MMTV-like env sequences are present in BC, independent of BRCA1 and 2 variations; however, the fact that it is present in patients with polymorphisms highlights it as a potential risk factor, but this observation remains to be further verified. In light of our findings, the sequences of BRCA genes have not been completely studied in Egyptian patients and further analysis of these genes is urgently required as they harbor different genetic backgrounds than those reported in other populations. Genetic variation in BRCA genes could be playing a role in determining response to treatment and BC prognosis.
Until now, the biological implication of MMTV-like env sequences in BC remains largely unknown, and whether these sequences are of no importance to BC etiology and maybe just an epiphenomenon needs to be further verified. On the other hand, the fact that MMTV-like env sequences are only detected in cancerous and not normal breast tissues suggests MMTV-like as a risk factor for BC. In addition, results from this study show a link between MMTV-like sequences and larger tumor size which supports a role for MMTV-like in BC. In this regard, viral infections, along with hormones and some genetic variations, were suggested as contributing factors in a multifactorial model for BC; our findings support this model.51
Conclusion
In conclusion, our findings show a high prevalence of MMTV-like env sequences among Egyptian BC patients and not in healthy women. In addition, MMTV-like was correlated with increased tumor size, which suggests MMTV as a risk factor for BC. Also, our results confirm the importance of reducing viral sequences from Egyptian BC patients compared to other populations. This increase in MMTV-like env sequences suggests higher exposure to the virus. Finally, the genetic sequence of BRCA genes in our patients showed a different genetic variation than that reported in other BC patients, which suggest further analysis of whole functional sequences of these genes. Finally, we report the presence of MMTV-like env sequences in BC patients with genetic polymorphisms which indicate a role of these sequences in BC carcinogenesis.
Acknowledgment
The authors acknowledge Virology & Immunology Unit, Cancer Biology Department, National Cancer Institute, Cairo University for funding the current research.
Funding Statement
This study was part of PhD grant funded by National Cancer Institute, Cairo University. A.A.A was supported by an NIH-funded Cancer Therapeutics Training Program (CT2, T32 CA121938) and P.G by the NIH (CA238042, CA100768 and CA160911).
Ethical Approval
The research has been approved by National Cancer Institute, Cairo University, IRB No. 200801002.2.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583 [DOI] [PubMed] [Google Scholar]
- 2.Azamjah N, Soltan-Zadeh Y, Zayeri F. Global trend of breast cancer mortality rate: a 25-year study. Asian Pac J Cancer Prev. 2019;20(7):2015–2020. doi: 10.31557/apjcp.2019.20.7.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;2014:437971. doi: 10.1155/2014/437971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke M, Collins R, Darby S, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;366(9503):2087–2106. doi: 10.1016/S0140-6736(05)67887-7 [DOI] [PubMed] [Google Scholar]
- 5.Kwong A, Shin VY, Au CH, et al. Detection of germline mutation in hereditary breast and/or ovarian cancers by next-generation sequencing on a four-gene panel. J Mol Diagn. 2016;18(4):580–594. doi: 10.1016/j.jmoldx.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 6.Chen B, Zhang G, Li X, et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY). 2020;12(4):3140–3155. doi: 10.18632/aging.102783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Klemp JR, Kimler BF, et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat. 2014;145(3):707–714. doi: 10.1007/s10549-014-2980-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariotto AB, Etzioni R, Hurlbert M, Penberthy L, Mayer M. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomark Prev. 2017;26(6):809–815. doi: 10.1158/1055-9965.Epi-16-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwase M, Matsuo K, Koyanagi YN, et al. Alcohol consumption and breast cancer risk in Japan: a pooled analysis of eight population-based cohort studies. Int J Cancer. 2021. doi: 10.1002/ijc.33478 [DOI] [PubMed] [Google Scholar]
- 11.Hauner D, Rack B, Friedl T, Hepp P, Janni W, Hauner H. Rationale and description of a lifestyle intervention programme to achieve moderate weight loss in women with non-metastatic breast cancer: the lifestyle intervention part of the SUCCESS C Study. BMJ Nutr Prev Health. 2020;3(2):213–219. doi: 10.1136/bmjnph-2020-000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li F, Zhang X, Li Z, Li H. Smoking increases risks of all-cause and breast cancer specific mortality in breast cancer individuals: a dose-response meta-analysis of prospective cohort studies involving 39725 breast cancer cases. Oncotarget. 2016;7(50):83134–83147. doi: 10.18632/oncotarget.13366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendall BJ, Wilson LF, Olsen CM, et al. Cancers in Australia in 2010 attributable to overweight and obesity. Aust N Z J Public Health. 2015;39(5):452–457. doi: 10.1111/1753-6405.12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–615. doi: 10.1016/S1470-2045(12)70137-7 [DOI] [PubMed] [Google Scholar]
- 15.Johal H, Ford C, Glenn W, Heads J, Lawson J, Rawlinson W. Mouse mammary tumor like virus sequences in breast milk from healthy lactating women. Breast Cancer Res Treat. 2011;129(1):149–155. doi: 10.1007/s10549-011-1421-6 [DOI] [PubMed] [Google Scholar]
- 16.Lawson JS, Glenn WK, Salmons B, et al. Mouse mammary tumor virus-like sequences in human breast cancer. Research Support, Non-U.S. Gov’t. Cancer Res. 2010;70(9):3576–3585. doi: 10.1158/0008-5472.CAN-09-4160 [DOI] [PubMed] [Google Scholar]
- 17.Joshi D, Buehring GC. Are viruses associated with human breast cancer? Scrutinizing the molecular evidence. Breast Cancer Res Treat. 2012;135(1):1–15. doi: 10.1007/s10549-011-1921-4 [DOI] [PubMed] [Google Scholar]
- 18.Lehrer S, Rheinstein PH. The virology of breast cancer: viruses as the potential causative agents of breast tumorigenesis. Discov Med. 2019;27(148):163–166. [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkar NH. Mouse mammary tumor virus derived from wild mice does not target Notch-4 protooncogene for the development of mammary tumors in inbred mice. Research Support, Non-U.S. Gov’t. Virology. 2009;388(1):121–127. doi: 10.1016/j.virol.2009.02.035 [DOI] [PubMed] [Google Scholar]
- 20.Al Dossary R, Alkharsah KR, Kussaibi H. Prevalence of Mouse Mammary Tumor Virus (MMTV)-like sequences in human breast cancer tissues and adjacent normal breast tissues in Saudi Arabia. Research Support, Non-U.S. Gov’t. BMC Cancer. 2018;18(1):170. doi: 10.1186/s12885-018-4074-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson JS, Salmons B, Glenn WK. Oncogenic Viruses and Breast Cancer: mouse Mammary Tumor Virus (MMTV), Bovine Leukemia Virus (BLV), Human Papilloma Virus (HPV), and Epstein-Barr Virus (EBV). Front Oncol. 2018;8:1. doi: 10.3389/fonc.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland JF, Pogo BG. Mouse mammary tumor virus-like viral infection and human breast cancer. Clin Cancer Res. 2004;10(17):5647–5649. doi: 10.1158/1078-0432.CCR-04-1234 [DOI] [PubMed] [Google Scholar]
- 23.Hachana M, Trimeche M, Ziadi S, et al. Prevalence and characteristics of the MMTV-like associated breast carcinomas in Tunisia. Cancer Lett. 2008;271(2):222–230. doi: 10.1016/j.canlet.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 24.Levine PH, Pogo BG, Klouj A, et al. Increasing evidence for a human breast carcinoma virus with geographic differences. Cancer. 2004;101(4):721–726. doi: 10.1002/cncr.20436 [DOI] [PubMed] [Google Scholar]
- 25.Apostolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int. 2013;2013:747318. doi: 10.1155/2013/747318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Iversen ES, Friebel T, et al. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t. J clin oncol. 2006;24(6):863–871. doi: 10.1200/JCO.2005.03.6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan H, Gulley ML. DNA extraction from fresh or frozen tissues. Methods Mol Med. 2001;49:5–10. doi: 10.1385/1-59259-081-0:5 [DOI] [PubMed] [Google Scholar]
- 28.Pogo BG, Melana SM, Holland JF, et al. Sequences homologous to the mouse mammary tumor virus env gene in human breast carcinoma correlate with overexpression of laminin receptor. Clin Cancer Res. 1999;5(8):2108–2111. [PubMed] [Google Scholar]
- 29.De Leeneer K, Coene I, Poppe B, De Paepe A, Claes K. Rapid and sensitive detection of BRCA1/2 mutations in a diagnostic setting: comparison of two high-resolution melting platforms. Research Support, Non-U.S. Gov’t. Clin Chem. 2008;54(6):982–989. doi: 10.1373/clinchem.2007.098764 [DOI] [PubMed] [Google Scholar]
- 30.Stewart TH, Sage RD, Stewart AF, Cameron DW. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br J Cancer. 2000;82(2):446–451. doi: 10.1054/bjoc.1999.0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imai S. Mouse mammary tumor virus and mammary tumorigenesis in wild mice. Pathol Int. 1996;46(12):919–932. doi: 10.1111/j.1440-1827.1996.tb03570.x [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Pelisson I, Melana SM, Go V, Holland JF, Pogo BG. MMTV-like env gene sequences in human breast cancer. Arch Virol. 2001;146(1):171–180. doi: 10.1007/s007050170201 [DOI] [PubMed] [Google Scholar]
- 33.Ford CE, Tran D, Deng Y, Ta VT, Rawlinson WD, Lawson JS. Mouse mammary tumor virus-like gene sequences in breast tumors of Australian and Vietnamese women. Clin Cancer Res. 2003;9(3):1118–1120. [PubMed] [Google Scholar]
- 34.Indik S, Gunzburg WH, Salmons B, Rouault F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005;65(15):6651–6659. doi: 10.1158/0008-5472.CAN-04-2609 [DOI] [PubMed] [Google Scholar]
- 35.Lawson JS, Mazzanti C, Civita P, et al. Association of mouse mammary tumor virus with human breast cancer: histology, immunohistochemistry and polymerase chain reaction analyses. Front Oncol. 2018;8:141. doi: 10.3389/fonc.2018.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melana SM, Nepomnaschy I, Hasa J, et al. Detection of human mammary tumor virus proteins in human breast cancer cells. J Virol Methods. 2010;163(1):157–161. doi: 10.1016/j.jviromet.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 37.Luo T, Wu XT, Zhang MM, Qian K. [Study of mouse mammary tumor virus-like gene sequences expressing in breast tumors of Chinese women]. Sichuan Da Xue Xue Bao Yi Xue Ban = Journal of Sichuan University Medical Science Edition. 2006;37(6):844–6, 851. Chinese. [PubMed] [Google Scholar]
- 38.Lo AC, Kleer CG, Banerjee M, et al. Molecular epidemiologic features of inflammatory breast cancer: a comparison between Egyptian and US patients. Breast Cancer Res Treat. 2008;112(1):141–147. doi: 10.1007/s10549-007-9833-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Holland JF, Bleiweiss IJ, et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55(22):5173–5179. [PubMed] [Google Scholar]
- 40.Melana SM, Holland JF, Pogo BG. Search for mouse mammary tumor virus-like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res. 2001;7(2):283–284. [PubMed] [Google Scholar]
- 41.Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6(4):1273–1278. [PubMed] [Google Scholar]
- 42.Zangen R, Harden S, Cohen D, Parrella P, Sidransky D. Mouse mammary tumor-like env gene as a molecular marker for breast cancer? Int J Cancer. 2002;102(3):304–307. doi: 10.1002/ijc.10702 [DOI] [PubMed] [Google Scholar]
- 43.Witt A, Hartmann B, Marton E, Zeillinger R, Schreiber M, Kubista E. The mouse mammary tumor virus-like env gene sequence is not detectable in breast cancer tissue of Austrian patients. Oncol Rep. 2003;10(4):1025–1029. [PubMed] [Google Scholar]
- 44.Mant C, Gillett C, D’Arrigo C, Cason J. Human murine mammary tumour virus-like agents are genetically distinct from endogenous retroviruses and are not detectable in breast cancer cell lines or biopsies. Virology. 2004;318(1):393–404. doi: 10.1016/j.virol.2003.09.027 [DOI] [PubMed] [Google Scholar]
- 45.Wang F, Hou J, Shen Q, et al. Mouse mammary tumor virus-like virus infection and the risk of human breast cancer: a meta-analysis. Am J Transl Res. 2014;6(3):248–266. [PMC free article] [PubMed] [Google Scholar]
- 46.Slaoui M, El Mzibri M, Razine R, Qmichou Z, Attaleb M, Amrani M. Detection of MMTV-Like sequences in Moroccan breast cancer cases. Infect Agent Cancer. 2014;9:37. doi: 10.1186/1750-9378-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lugo FI, Diaz NIO, Barbosa LC, Novel GAA. HRM strategy for detection of BRCA mutations on Mexican population. J Carcinogenesis Mutagenesis. 2015;6:4. [Google Scholar]
- 48.Kennerson ML, Warburton T, Nelis E, et al. Mutation scanning the GJB1 gene with high-resolution melting analysis: implications for mutation scanning of genes for Charcot-Marie-Tooth disease. Clin Chem. 2007;53(2):349–352. doi: 10.1373/clinchem.2006.080010 [DOI] [PubMed] [Google Scholar]
- 49.Hung CC, Lee CN, Chang CH, et al. Genotyping of the G1138A mutation of the FGFR3 gene in patients with achondroplasia using high-resolution melting analysis. Research Support, Non-U.S. Gov’t. Clin Biochem. 2008;41(3):162–166. doi: 10.1016/j.clinbiochem.2007.08.014 [DOI] [PubMed] [Google Scholar]
- 50.Lux MP, Fasching PA, Beckmann MW. Hereditary breast and ovarian cancer: review and future perspectives. Review. J Mol Med (Berl). 2006;84(1):16–28. doi: 10.1007/s00109-005-0696-7 [DOI] [PubMed] [Google Scholar]
- 51.Lawson JS, Tran D, Rawlinson WD. From Bittner to Barr: a viral, diet and hormone breast cancer aetiology hypothesis. Breast Cancer Res. 2001;3(2):81–85. doi: 10.1186/bcr275 [DOI] [PMC free article] [PubMed] [Google Scholar]