Abstract
Leptospirosis is a ubiquitous zoonotic disease and a major clinical challenge owing to the multitude of clinical presentations and manifestations that are possibly attributable to the diversity of Leptospira, the understanding of which is key to study the epidemiology of this emerging global disease threat. Sri Lanka is a hotspot for leptospirosis with high levels of endemicity as well as annual epidemics. We carried out a prospective study of Leptospira diversity in Sri Lanka, covering the full range of climatic zones, geography, and clinical severity. Samples were collected for leptospiral culture from 1,192 patients from 15 of 25 districts in Sri Lanka over two and half years. Twenty-five isolates belonging to four pathogenic Leptospira species were identified: L. interrogans, L. borgpetersenii, L. weilii, and L. kirschneri. At least six serogroups were identified among the isolates: Autumnalis (6), Pyrogenes (4), Icterohaemorrhagiae (2), Celledoni (1), Grippotyphosa (2) and Bataviae (1). Seven isolates did not agglutinate using available antisera panels, suggesting new serogroups. Isolates were sequenced using an Illumina platform. These data add 25 new core genome sequence types and were clustered in 15 clonal groups, including 12 new clonal groups. L. borgpetersenii was found only in the dry zone and L. weilii only in the wet zone. Acute kidney injury and cardiovascular involvement were seen only with L. interrogans infections. Thrombocytopenia and liver impairment were seen in both L. interrogans and L. borgpetersenii infections. The inadequate sensitivity of culture isolation to identify infecting Leptospira species underscores the need for culture-independent typing methods for Leptospira.
Author summary
There is a huge diversity in pathogenic Leptospira species worldwide, and our knowledge of the currently circulating species is deficient owing to limited isolation and identification of Leptospira species from endemic countries. This prospective study reveals the wide pathogen diversity that causes human leptospirosis in Sri Lanka, representing four species, more than six serogroups, and fifteen clonal groups. Further, the different geographic and climatic zone distributions and clinical manifestations observed underscores the need for prospective studies to expand the molecular epidemiological approaches to combat leptospirosis.
Introduction
Leptospirosis is caused by a group of pathogenic Leptospira species of the phylum Spirochetes and is considered one of the commonest zoonotic diseases worldwide [1,2]. Leptospira spp. colonize proximal convoluted tubules of kidney tissue of various mammals particularly rodents, dogs and various domesticated livestock, and such reservoir hosts excrete the bacteria to the environment via urine [3,4]. Humans are incidental hosts who acquire the disease by direct contact with urine or tissues of reservoir animals or, more frequently by indirect contact with contaminated water sources [5,6]. The number of cases due to leptospirosis has been estimated to be 1.03 million annually worldwide, with 58,900 deaths [1]. The majority of tropical countries in Oceania, southeast Asia, the Caribbean region, South America, central and eastern sub-Saharan Africa, and south Asia are estimated to have substantial morbidity and mortality that is attributable to leptospirosis.[7]
Understanding the diversity of infecting Leptospira has been a major global focus, especially in recent years. Substantial changes in the classification of Leptospira based on whole-genome sequencing have led to the identification of 43 new Leptospira species during the period 2018 to 2020 [8–11]. In addition, the more robust classification of Leptospira strains beyond the species level using core-genome multi-locus sequence typing (cgMLST) [9,12] and single nucleotide polymorphism typing methods have rapidly expanded our knowledge of the molecular epidemiology of Leptospira. However, the goal of reducing the global burden of this deadly disease will require enhanced understanding of pathogen types and applications and linkage to disease distribution, transmission, virulence, clinical presentations, and outcome.
The global leptospirosis disease burden study [1] highlighted Sri Lanka as leptospirosis hyperendemic with an estimated morbidity of 300.6 and mortality of 17.98 per 100,000 population per year. Disease incidence tends to be higher during the rainy seasons, i.e., the southwest and northeast monsoons. Cases, however, are not confined exclusively to the wet zone and have been reported in the dry zone as well where most residents are engaged in farming activities. Outbreaks have also occurred in the dry zone following extreme weather events like flooding [13].
As in many other endemic countries, understanding Leptospira diversity in Sri Lanka is limited because of a lack of knowledge of the circulating pathogenic species and serovars. Studies that utilized culture-based isolation of Leptospira species were carried out in Sri Lanka during the period from 1950 to 1970 in the wet zone only. Several pathogenic strains of the species L. interrogans [14–16], L. borgpetersenii [17], L. kirschneri [18], and L. santarosai were detected during that time [19,20]. Since the 1970s, no culture based isolation studies were reported until 2018, when two human isolates belonging to L. interrogans were recovered from the wet zone [21]. Despite the availability of next-generation sequencing methods for many years, whole-genome sequencing data for Sri Lankan isolates were not available until recently [22].
A systematic review published in 2016 revealed the large diversity of Leptospira strains in Sri Lanka based on historical data [23]. Being an island with a high leptospirosis disease burden makes Sri Lanka an ideal location to study pathogen diversity linked with epidemiological and clinical patterns of the disease. Low-passage isolates from human sources with high-resolution genetic typing in a place with high pathogen diversity would enhance our global knowledge of leptospirosis. This study was designed to provide a comprehensive understanding of the circulating pathogenic Leptospira species and serotypes responsible for human leptospirosis in Sri Lanka, covering different clinical presentations and geographical locations as well as epidemic and endemic disease over a period of two and half years.
Methods
Ethics statement
Written informed consent was obtained from all patients prior to sample collection. This study is approved by the Ethics Review Committee of the Faculty of Medicine and Allied Sciences, Rajarata University of Sri Lanka. Protocol No. ERC/2015/18
The present study was embedded in a larger clinical-epidemiological study on leptospirosis, in Sri Lanka and the study protocol has been published [24]. Specific details related to Leptospira diversity and methods in brief are given here.
Study setting
This study was carried out from June 2016 through January 2019 at several locations in Sri Lanka that differed with respect to mean temperature, rainfall, elevation, ecology, human activities, and leptospirosis endemicity. The main data collection sites were the Teaching Hospital Anuradhapura (THA) and Teaching Hospital Peradeniya (THP). THA is in the dry zone located at low elevation with low humidity, high temperature, large rice paddy fields, water reservoir–based irrigation systems, and low endemicity for the disease. THP is in the wet zone located at high elevation, low temperature, rainfall-based farming activities, and high endemicity. Samples for Leptospira diversity assessment were collected through two approaches. First, as a part of the main study described in Agampodi et al. 2019, prospective data and sample collection was done in THA and THP. In addition, during an outbreak of leptospirosis in 2017, we set up the same procedure at Base Hospital Avissawella and Provincial General Hospital Rathnapura from June to September. These two wet-zone areas have high endemicity, representing low and intermediate elevations. As a part of the service component of this study, we offered diagnostic services to all requesting physicians and collected additional culture samples. This resulted in sample collection from District General Hospital Kegalle, Base Hospital Karawanella, Sri Jayewardenepura General Hospital, and General Hospital Polonnaruwa (GHP) again representing different geographical locations. These study sites (Fig 1) represent seven districts belonging to four provinces of the country, and the patients who visited these hospitals came from all nine provinces.
Fig 1. Geographical distribution of the seven hospitals from which the culture collection was done to identify 25 new strains of pathogenic Leptospira from Sri Lanka.

The base map used in the figure is freely available from The United Nations Office for the Coordination of Humanitarian Affairs https://data.humdata.org/dataset/sri-lanka-administrative-levels-0-4-boundaries.
Study samples
Culture collection was done from three types of patients. Acute undifferentiated febrile (temperature >38°C) patients who presented to adult wards (age >13 years) of THA, THP, Provincial General Hospital Rathnapura (PGHR) and Base Hospital Avissawella (BHA) (both outpatient department and hospitalized patients) were included as possible cases of leptospirosis. A possible case was defined as any acute undifferentiated febrile patient with headache, myalgia and prostration. Probable cases of clinical leptospirosis which were defined as those who were having the classical clinical features of leptospirosis with an exposure history, were included from GHP. Culture samples from Sri Jayewardenepura General Hospital and Base Hospital Karawanella were included only if they came from clinically confirmed cases of leptospirosis. These cases were defined according to the surveillance case definition for leptospirosis set by epidemiology unit of Sri Lanka[25]. Physician-diagnosed probable or definite acute bacterial meningitis or lower respiratory tract infections (e.g., consolidated lobar pneumonia), traumatic or post-operative fever per physician discretion, fever owing to nosocomial infections, and any patient with confirmed diagnosis as a cause for the fever were excluded. Epidemiological data were collected from each patient using a fully structured, interviewer administered questionnaire which was described in detail in the study protocol paper published elsewhere[24]
Once the isolation is done, all culture-positive patients were later contacted and/or visited to collect additional data. The additional data were collected mainly to identify exact type of exposure if possible and residing places of patients preceding the illness. In addition, diagnosis cards of these patients were also traced to extract any missing data during hospital stay. Each patient’s clinical records were also retrieved from the corresponding hospital.
Sample collection and isolation of Leptospira
Blood (7 ml) was collected into EDTA tubes from all eligible patients. Bedside inoculation of 2 and 4 drops (100–400 μl) was done into two tubes with 9 ml Ellinghausen-McCullough-Johnson-Harris (EMJH) semisolid medium with added antibiotics (5-fluorouracil and neomycin). These cultures were kept at room temperature (usually 28–32°C) until transfer to the Leptospirosis Research Laboratory of the Faculty of Medicine and Allied Sciences, Rajarata University, Sri Lanka, and then incubated at 30°C until the cultures become positive or for 6 months. Samples from THA were transferred on the same day to the research laboratory whereas other samples collected from distant places were transferred within 2 days of collection.
In brief, EMJH semisolid media were prepared by adding 2.3 g of EMJH base (Difco), 1.5 g bacteriological agar, and 100 mg sodium pyruvate into 785 ml distilled water and adjusting the pH to 7.4. The media were autoclaved, and once cooled to ~50°C, 100 ml Leptospira enrichment media and 100 ml fetal bovine serum were added. To suppress the growth of possible contaminating bacteria, 5-fluorouracil (100 μg/ml, final) and neomycin (25 μg/ml) were added. Each inoculated medium was inspected by taking approximately 50μl of volume into a clean glass slide after mixing the culture tubes well by inverting several times. Prepared slides were examined under 40X objective of a dark-field microscope to check for the presence of motile spirochetes; this was done initially after 3 weeks and then on a monthly basis. However, samples were inspected before 3 weeks if quantitative PCR of the corresponding whole-blood sample indicated a positive reaction. The procedure for qPCR on clinical samples is described elsewhere in the published protocol paper[24]. Culture tubes were inspected for consecutive six months before reporting as negative. When growth was detected, subcultures were made into liquid and semisolid media, and an aliquot was fixed with 5% dimethyl sulfoxide and stored at –80°C. Isolates were sub-cultured every two weeks on liquid media and on semisolid media every three months. Certain isolates required weekly subculture into liquid media to maintain viability. None of the isolates became contaminated during the subculture process, although two positive original clinical samples were contaminated with bacilli. For those two samples, subcultures were made into liquid media and subsequently filtered through a 0.2 μm pore-size microfilter to overcome the problem of contamination.
Next-generation sequencing, cgMLST and phylogenetic tree
DNA was extracted from culture using the PureLink Genomic DNA Mini kit (Invitrogen, Dublin, Ireland) and Wizard Genomic DNA Purification Kit (Promega, Southampton, UK) according to manufacturer instructions. NGS was performed using Nextera XT DNA Library Preparation kit and the NextSeq 500 sequencing systems (Illumina, San Diego, CA, USA) at the Mutualized Platform for Microbiology (P2M) at Institut Pasteur. The data were analyzed using CLC Genomics Workbench 9 software (Qiagen, Hilden, Germany). cgMLST typing was performed for strain taxonomy using a scheme based on 545 highly conserved genes with BIGSdb (http://bigsdb.pasteur.fr/leptospira), and a phylogenetic tree was generated using cgMLST with Interactive Tree of Life v3, and GrapeTree [26] was used to visualize the core genomic relationships among the isolates and the previously reported Sri Lankan isolates [9,27,28]. A Clonal Group (CG) is defined as a group of cgMLST allelic profiles differing by no more than 40 allelic mismatches, out of 545 gene loci, from at least one other member of the group.
Serotyping of new isolates
Serotyping of newly isolated Leptospira strains was done at the Pasteur Institute, France. Microscopic agglutination test using a standard battery of rabbit antisera raised against 24 reference serovars representing the main serogroups was used for this study [29,30].
Results
From June 2016 through January 2019, we acquired blood cultures from 1192 patients. Patients were from 14 districts of Sri Lanka, representing all 9 provinces (Fig 2).
Fig 2. Distribution of probable exposure sites/residence of the 1192 febrile patients from whom the cultures were collected for the isolation of 25 new strains of pathogenic Leptospira in Sri Lanka.
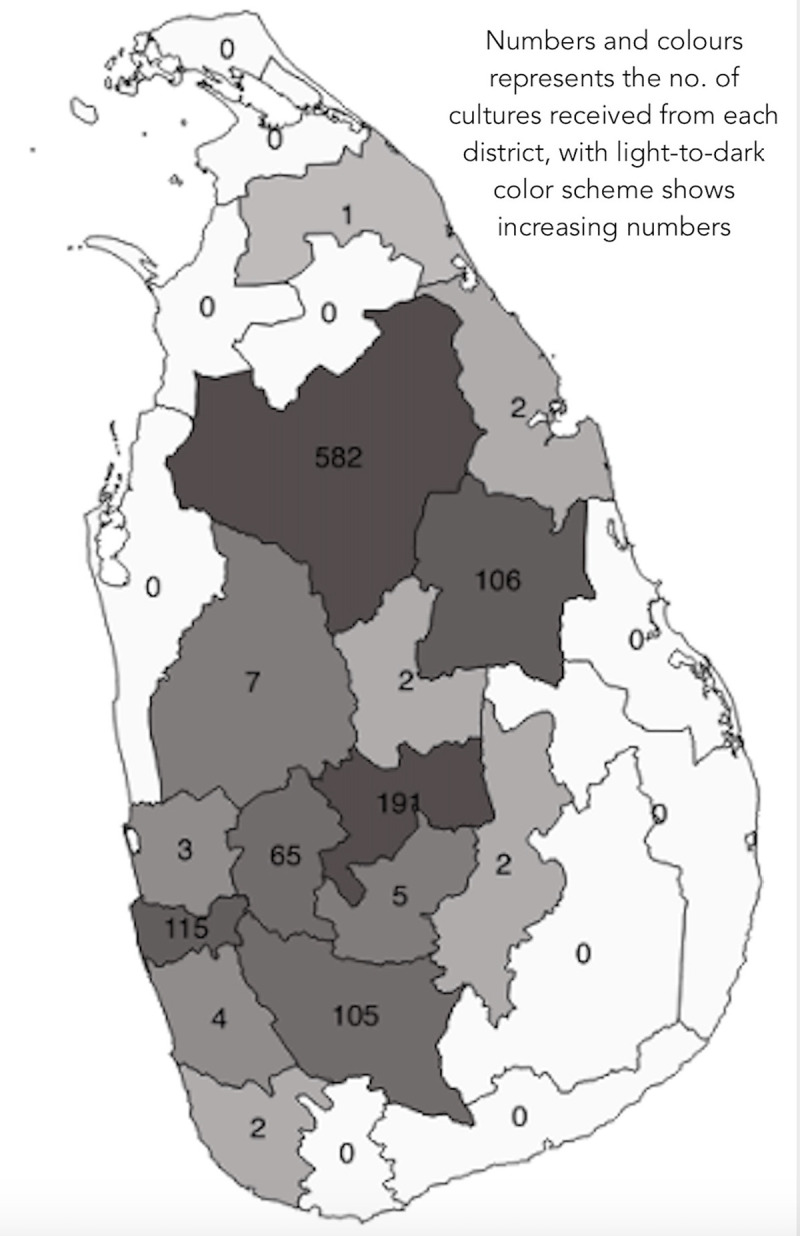
The base map used in the figure is freely available from The United Nations Office for the Coordination of Humanitarian Affairs https://data.humdata.org/dataset/sri-lanka-administrative-levels-0-4-boundaries.
The majority of patients were male (n = 985, 82.6%). The mean age of the sample was 43.4 years (SD 14.7). Most of the cultures (96%) were obtained from hospital inpatients, and the remaining 4% (48 cultures) were from outpatients. Of the 1192 cultures, 80 (6.7%) were received from hospitals where only typical clinical cases of leptospirosis were sampled (from Sri Jayewardenepura General Hospital, Base Hospital Karawanella and Provincial General Hospital Rathnapura). Another 107 (9.0%) were from GHP, where patients represented only probable cases of leptospirosis. The remaining 1005 patients (84.3%) were from THA, THP, Base Hospital Avissawella and Provincial General Hospital Rathnapura, representing patients who presented with acute undifferentiated fever.
Of the 1192 patients, 25 isolates had been identified by January 2019. Among the acute undifferentiated febrile patients, 1.5% (16/1047) had culture-positive leptospirosis; among the probable and clinically confirmed cases of leptospirosis, culture positivity was 4.7% (5/107) and 5.0% (4/80), respectively. The incubation period required to detect growth (assessed with dark-field microscopy) varied between 1 and 17 weeks. For each patient, both 2 drops and 4 drops of blood-inoculated media gave positive results. The median incubation period was 15 weeks for the first 9 isolates and was 6 weeks for the remaining 16 isolates (S1 Fig).
Leptospira was isolated from one female and 24 male patients who presented with fever. Only three patients were from outpatient departments, and the rest were inpatients. Of the three outpatients, two were later admitted to a hospital owing to increased disease severity.
cgMLST analysis revealed that the 25 isolates represented four species: L. interrogans (15 isolates, 60%), L. borgpetersenii (7 isolates, 28%), L. weilii (2 isolates, 8%), and L. kirschneri (1 isolate, 4%). The predominant clonal groups (CG) were L. borgpetersenii CG267 (7/25), followed by L. interrogans CG266 (3/25), CG10 (2/25), and CG263 (2/25).
The two L. weilii isolates clustered in different clonal groups (CG262, CG264). Interpretable data for serogroup assay was available for only 19 isolates. Seven samples resulted in no agglutination, probably owing to new, previously unreported serogroups/serovars. The assay revealed that the 19 seropositive isolates represented at least six serogroups, namely Autumnalis, Pyrogenes, Icterohaemorrhagiae, Grippotyphosa, Celledoni, and Bataviae. Table 1 shows the species identity, serogroup status and distribution of clonal groups among the 25 isolates.
Table 1. Putative species, serogroups, cgMLST and clonal groups for the 25 newly isolated pathogenic Leptospira from Sri Lanka.
| Strain ID | Species | Serogroup | cgMLST* | CG* |
|---|---|---|---|---|
| FMAS_KW1 | L. interrogans | Pyrogenes | 784 | 10 |
| FMAS_KW2 | L. interrogans | Autumnalis | 631 | 291 |
| FMAS_AW1 | L. interrogans | Autumnalis | 555 | 74 |
| FMAS_RT1 | L. weilii | No agglutination | 556 | 262 |
| FMAS_AW2 | L. interrogans | Autumnalis | 567 | 269 |
| FMAS_AW3 | L. interrogans | Pyrogenes | 557 | 9 |
| FMAS_RT2 | L. interrogans | Autumnalis | 569 | 271 |
| FMAS_PD1 | L. interrogans | Pyrogenes | 558 | 263 |
| FMAS_PD2 | L. weilii | Celledoni | 559 | 264 |
| FMAS_KG1 | L. interrogans | Bataviae | 560 | 265 |
| FMAS_KG2 | L. interrogans | Pyrogenes | 561 | 263 |
| FMAS_AP1 | L. interrogans | Autumnalis | 562 | 266 |
| FMAS_AP2 | L. borgpetersenii | No agglutination | 563 | 267 |
| FMAS_AP3 | L. borgpetersenii | No agglutination | 575 | 267 |
| FMAS_AP4 | L. borgpetersenii | No agglutination | 564 | 267 |
| FMAS_AP5 | L. interrogans | Pyrogenes | 565 | 10 |
| FMAS_AP6 | L. interrogans | Pyrogenes | 785 | 321 |
| FMAS_AP7 | L. interrogans | Autumnalis | 786 | 266 |
| FMAS_PN1 | L. borgpetersenii | No agglutination | 787 | 267 |
| FMAS_PN2 | L. interrogans | Icterohaemorrhagiae | 788 | 322 |
| FMAS_PN3 | L. interrogans | Autumnalis | 789 | 266 |
| FMAS_PN4 | L. borgpetersenii | No agglutination | 790 | 267 |
| FMAS_AP8 | L. borgpetersenii | No agglutination | 791 | 267 |
| FMAS_AP9 | L. borgpetersenii | No agglutination | 792 | 267 |
| FMAS_PN5 | L. kirschneri | Grippotyphosa | 793 | 323 |
cgMLST- core genome Multi Locus Sequence Typing, CG: clonal group defined as a group of cgMLST allelic profiles differing by no more than 40 allelic mismatches, out of 545 gene loci, from at least one other member of the group.
* cgMLST and CG strain types reported in the table were identified through the database BIGSdb at http://bigsdb.pasteur.fr/leptospira
A phylogenetic tree was constructed from the cgMLST data for our 25 isolates (labeled as FMAS) together with previously reported local isolates (n = 9) and worldwide pathogenic Leptospira isolates. Fig 3 shows the phylogenetic tree constructed using the cgMLST data.
Fig 3. Phylogenetic tree showing the distribution of species and serogroups of the 25 new pathogenic Leptospira isolates from the present study (labeled as FMAS) along with the previously reported Sri Lankan isolates and other species.
The phylogenetic tree was generated using cgMLST with Interactive Tree of Life v3, and GrapeTree.
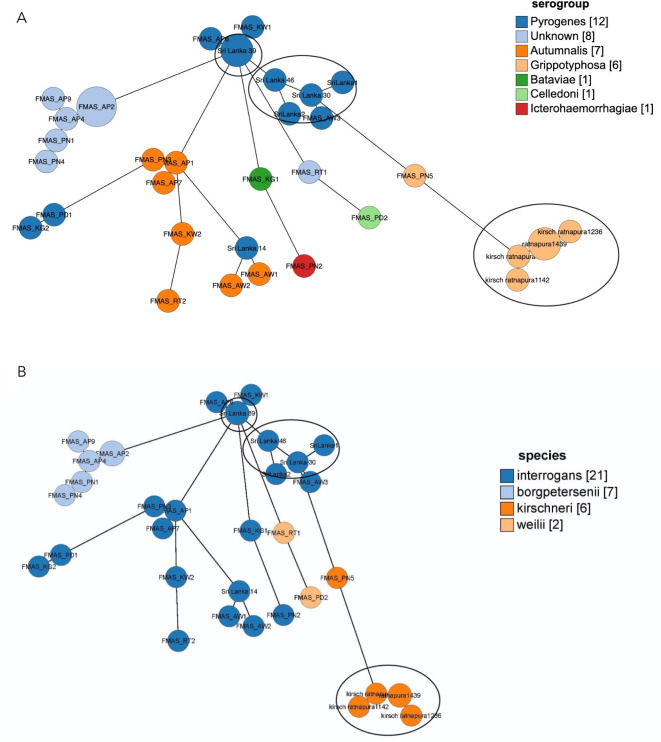
Most of the clonal groups found in the local isolates were unique and not found in other countries. The three isolates of L. borgpetersenii (FMAS_AP2, FMAS_AP3 and FMAS_AP8) were from patients in the same district of dry zone. Autumnalis from dry zone shows significant core genome relatedness and clustered together (Fig 4). Genomic relatedness between FMAS_AP5 of dry zone and Sri Lanka 39 was observed; the latter isolate is an old isolate recovered from wet zone. (metadata for the GrapeTree is included in S1 Table)
Fig 4. Genome GrapeTree showing the core-genome relationship among 25 new and 11 previously isolated Leptospira strains from Sri Lanka.
4A: species, 4B:serogroups. New isolates are having the prefix Faculty of Medicine and Allied Sciences. Old isolates are flagged with a circle.
Culture-positive leptospirosis patients were distributed widely in the study areas (Fig 5). L. borgpetersenii was exclusively isolated from patients in areas of the dry zone at low elevation, with hot and dry conditions. The single isolate of L. kirschneri was from the same setting. In contrast, L. weilii was isolated from patients in the wet zone, whereas L. interrogans was isolated from patients in all geographical areas (Fig 5). The serogroup distribution also revealed a specific pattern for all nonagglutinating isolates, which mainly were from the dry zone at low elevation. The most frequent serogroup, Autumnalis, was observed in all geographical settings. All L. borgpetersenii isolates were in CG267, the majority of which failed to agglutinate with reference antisera (slight agglutination was seen with antisera against serovars Grippotyphosa and Louisiana).
Fig 5. Geographic distribution of 25 newly isolated pathogenic Leptospira species, serogroups and clonal groups from Sri Lanka.
The base map used in the figure is freely available from The United Nations Office for the Coordination of Humanitarian Affairs https://data.humdata.org/dataset/sri-lanka-administrative-levels-0-4-boundaries.
Clinical profile of culture-positive patients
Complete demographic and clinical profiles for the culture-positive patients are included as additional data (S1 Table). No fatalities were reported for patients with culture-positive leptospirosis. Among hospitalized patients, the median duration of hospital stay was 4 days (interquartile range, 3–5 days), and the longest stay was 12 days followed by 9 days (both patients had acute kidney injury requiring hemodialysis). Another two had renal involvement with elevated serum creatinine but not severe acute kidney failure. Twelve patients had elevated serum glutamic-oxaloacetic transaminase and serum glutamic-pyruvic transaminase, and three other patients had elevated serum bilirubin. Thrombocytopenia was common in those 12 patients, with a platelet count <100,000 per microliter. Two patients who underwent hemodialysis also had cardiac involvement with hypotension. Notably, all patients who had one or more severe complications were infected with L. interrogans. Infection with L. interrogans or L. borgpetersenii was associated with thrombocytopenia and liver involvement.
Discussion
This study is the first to report information on the diversity of pathogenic Leptospira species in Sri Lanka and probably one of the few prospective studies on Leptospira disease diversity in the literature representing all geographical regions of an entire country. The study also reports the first isolation of Leptospira from dry zones in Sri Lanka. In addition, we report here the first isolation of L. weilii from humans in Sri Lanka, the existence of which was suggested based on molecular studies of clinical samples [31]. This study also provides the first evidence of serogroups Celledoni and Bataviae circulating in Sri Lanka. Moreover, with the exception of L. santarosai, our study identified all pathogenic Leptospira species that were reported to have existed in Sri Lanka during the 1960s and 70s (as reviewed by Naotunna et al. in 2015), confirming the breadth of Leptospira diversity in Sri Lanka [23]. Recent reports indicated that L. interrogans, L. borgpetersenii and L. kirschneri are widespread species in other tropical regions of the world [32–37]. However, the geographical distribution of certain Leptospira species is limited, such as L. santarosai, which has been mainly reported in South America [38–41], and L. weilii in Asia [37,42,43]. Only limited reports have described the existence of L. weilii outside Asia, for which cattle and rodents are the dominant reservoir hosts [37,44–46].
We identified 15 distinct clonal groups of Leptospira, underscoring the diversity of pathogenic Leptospira circulating in Sri Lanka. Genomic relatedness was not observed between most of old Sri Lankan isolates with these new isolates probably due to emergence or introduction of new strains rather than genetic drift. The distribution of certain clonal groups suggests geographical demarcation of Leptospira spp. found in humans between dry and wet zones. Interestingly, all three L. borgpetersenii isolates belonged to a same cluster and originated from patients from the same district in the dry zone. These observations might reflect similarities in climate, reservoir hosts and environmental conditions in a particular geographical area. Clustering of L. interrogans into several clonal groups and shared genomic relatedness across different geographical areas could possibly be due to diversity of reservoir hosts and adaptability of the species to different environmental conditions. These observations suggest that different environmental drivers of leptospirosis operate in distinct ways for different pathogenic species and their serovars in these climatic zones. Notably, areas where L. borgpetersenii was found in our study were all in the dry zone where cattle and buffalo are commonly used in rice paddy farming activities.
Majority of the total study population and culture positive patients were males and this is likely to be due to exposure to contaminated sources as males engage in outdoor activities more frequently than women and this pattern of involvement is seen across most of the published studies worldwide[1,7,47,48]. According to available clinical data, renal, hepatic, haematological and cardiovascular complications are observed in infections with L.interrogans, which is consistent with worldwide-published data [49–51]. Though the severe pulmonary infections are reported as common in some parts of Sri Lanka[52], pulmonary complications were not observed among the 25 culture positive patients.
The diversity of circulating serogroups in Sri Lanka that has been known since the 1960s has been replicated in this study, and 19 isolates belonging to 6 different serogroups, and the non-agglutinated cultures might reflect other unidentified serovars. Serological and molecular assays done on veterinary field has already identified the role of rodents, cattle and dogs as reservoir hosts in Sri Lanka[53–55]. Nityananda and Harvey carried out extensive investigations of reservoir hosts and isolated Leptospira from dog (serogroups Icterohaemorrhagiae, Hebdomadis, Javanica, Pomona), brown rat (Icterohaemorrhagiae), rice field rat (serogroups Icterohaemorrhagiae, Grippotyphosa, Javanica), bandicoot rat (serogroups Icterohaemorrhagiae, Javanica) and shrew (serogroups Icterohaemorrhagiae, Javanica) [53]. The observed diversity in serogroups suggests a wider range of reservoir hosts even though Sri Lanka is a small island.
Although culture isolation remains essential for in-depth molecular epidemiological studies, the present study highlights the constraints faced for culture isolation of Leptospira, for which both a high level of skill and procedural optimization are required. Leptospira spp. are fastidious organisms, and their growth requirements differ from those of many other bacterial genera. Leptospira tend to have a relatively long incubation period, as the lag period during in vitro culture range from days to many weeks [56]. Other culture-isolation studies have reported an incubation period of ~3 weeks, but with a wide range [57–59]. We attribute the relatively lengthy incubation period of 15 weeks required for the first nine cultures in our study partly to lack of experience to detect scanty growth of Leptospira during the first phase of the study. However, it is also possible that those isolates were fastidious and required lengthy incubation periods. Isolation is essential for genomic and vaccine studies pertaining to Leptospira, and for that purpose specific skills are needed. Although culture isolation has 100% positive predictive value for diagnosing leptospirosis, its sensitivity is low, consistently <10% in most studies [34,60,61]. Similarly, our study yielded low sensitivity, which can be attributed to a few possible causes: Because the study population consisted of patients with acute febrile illness, their fever may have been caused by another unrelated illness; Use of antibiotics prior to culture; also, performing blood cultures during the late phase of illness and infection with fastidious Leptospira spp. may account for the low sensitivity during culture isolation.
One limitation of this study is the lack of isolates from Southern part of the country where high level of mortality is reported[62]. Extremely low sensitivity of culture procedures resulted in 25 isolates, a number inadequate for statistically significant analysis of patient outcomes. Further, while L. santarosi has been previously reported in Sri Lanka, isolates of this species were not obtained in this study.
This large collection of pathogenic Leptospira isolates from clinical samples will be a great addition to the global knowledgebase for leptospirosis. Whole-genome sequencing and genomic analysis of this set of isolates will reveal the pathogenic diversity and evolution of Leptospira species, in comparison to archived Leptospira isolated from Sri Lanka more than 50 years ago. The first three isolates from this study are already published[22] and available in NCBI genome database (accession numbers: GCA_005222565.1, GCA_005222585.1, GCA_005222625.1.
Whole genome sequencing and comparative genomic analysis of this collection will facilitate ongoing studies on identifying the putative virulence genes, pathogenic mechanisms with specific host adaptations, horizontal gene transfer mechanisms, and microbial resistance as shown in studies on the diversity and epidemiology of other microorganisms [63–68].
Supporting information
(JPG)
(XLSX)
Acknowledgments
We would like to thank Ms. Thilakanjali Gamage, Mr. K.M.R. Premathilaka, Mr. S.K. Senevirathna, and Mr. Milinda Perera for technical assistance, Mr. Shalka Srimantha and Ms. Chamila Kappagoda for culture maintenance and laboratory support, and Dr. Muditha Abeykoon, Dr. Chamida Wickramasinghe, and Dr. Shanika Gamage for additional culture collections. We also thank all the physicians and healthcare staff in the various participating hospitals, team of core facility P2M (Institut Pasteur, Mutualized Platform for Microbiology) for genomic sequencing and members of the National Reference Center for Leptospirosis (Institut Pasteur) for technical assistance with the cultures of Leptospira.
Data Availability
All genomic data are available at BIGSdb-Pasteur: the genomic-based strain taxonomy and nomenclature platform, under the Leptospira sequence typing at http://bigsdb.pasteur.fr/leptospira Hyperlink to each cgMLST is given in the Table 1. All sequences are available through the direct link.
Funding Statement
This work was supported by the United States Public Health Service by grant U19AI115658 (JV, MM, SA) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis. 2015;9. 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12: 351–357. 10.1016/j.ijid.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 3.Adler B, editor. Leptospira and Leptospirosis | Springer. Berlin, Heidelberg: Springer Berlin Heidelberg; 2015. 10.1007/978-3-662-45059-8 [DOI] [Google Scholar]
- 4.Human leptospirosis: guidance for diagnosis, surveillance and control. Rev Inst Med Trop Sao Paulo. 2005;45: 292–292. 10.1590/s0036-46652003000500015 [DOI] [Google Scholar]
- 5.Levett PN. Systematics of leptospiraceae. Curr Top Microbiol Immunol. 2015;387: 11–20. 10.1007/978-3-662-45059-8_2 [DOI] [PubMed] [Google Scholar]
- 6.Haake DA, Levett PN. Leptospirosis in humans. Curr Top Microbiol Immunol. 2015;387: 65–97. 10.1007/978-3-662-45059-8_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torgerson PR, Hagan JE, Costa F, Calcagno J, Kane M, Martinez-Silveira MS, et al. Global Burden of Leptospirosis: Estimated in Terms of Disability Adjusted Life Years. PLoS Negl Trop Dis. 2015;9: e0004122. 10.1371/journal.pntd.0004122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibeaux R, Iraola G, Ferrés I, Bierque E, Girault D, Soupé-Gilbert ME, et al. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb genomics. 2018. 10.1099/mgen.0.000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guglielmini J, Bourhy P, Schiettekatte O, Zinini F, Brisse S, Picardeau M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl Trop Dis. 2019. 10.1371/journal.pntd.0007374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019. 10.1371/journal.pntd.0007270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casanovas-Massana A, Hamond C, Santos LA, de Oliveira D, Hacker KP, Balassiano I, et al. Leptospira yasudae sp. Nov. and Leptospira stimsonii sp. nov., two new species of the pathogenic group isolated from environmental sources. Int J Syst Evol Microbiol. 2020. 10.1099/ijsem.0.003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, et al. What Makes a Bacterial Species Pathogenic?:Comparative Genomic Analysis of the Genus Leptospira. Small PLC, editor. PLoS Negl Trop Dis. 2016;10: e0004403. 10.1371/journal.pntd.0004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agampodi SB, Dahanayaka NJ, Bandaranayaka AK, Perera M, Priyankara S, Weerawansa P, et al. Regional Differences of Leptospirosis in Sri Lanka: Observations from a Flood-Associated Outbreak in 2011. Lukehart S, editor. PLoS Negl Trop Dis. 2014;8: e2626. 10.1371/journal.pntd.0002626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nityananda K. Isolation of Leptospira in Ceylon. Ceylon Med J. 1962;7: 95–6. [Google Scholar]
- 15.Nityananda K SC. A New Leptospiral Serotype in the Icterohaemorrhagiae Serogroup from Ceylon. Ceylon J Med Sci. 1972;21: 9–13. [PubMed] [Google Scholar]
- 16.Nityananda K, Sulzer CR. A new serotype of Leptospira belonging to the autumnalis serogroup. J Trop Med Hyg. 1971;74: 184–186. [PubMed] [Google Scholar]
- 17.Nityananda K, Sulzer CR. A new leptospiral serotype in the Javanica serogroup from Ceylon. Trop Geogr Med. 1969/06/01. 1969;21: 207–209. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=5799577 [PubMed] [Google Scholar]
- 18.Kokovin IL, Chernukha IG. [Serological classification of leptospirae of the grippotyphosa serogroup—new serological type ratnapura]. Zh Mikrobiol Epidemiol Immunobiol. 1970;47: 102–5. [PubMed] [Google Scholar]
- 19.Nityananda K, Harvey T. Leptospirosis_in_Ceylon-Epidemiological and Laboratory Studies. Ceylon J Med Sci. 1971;20: 5–14. [Google Scholar]
- 20.Kmety E, Dikken H. Classification of the species Leptospira interrogans and history of its serovars. University Press; Groningen; 1993. p. 104. [Google Scholar]
- 21.Nisansala GGT, Muthusinghe D, Gunasekara TDCP, Weerasekera MM, Fernando SSN, Ranasinghe KNP, et al. Isolation and characterization of Leptospira interrogans from two patients with leptospirosis in Western Province, Sri Lanka. J Med Microbiol. 2018. 10.1099/jmm.0.000800 [DOI] [PubMed] [Google Scholar]
- 22.Seneviratha I, Jayasundara D, Lefler JP, Chaiboonm KL, Warnasekara J, Agampodi S, et al. Complete Genome Sequence of Leptospira interrogans Strains FMAS_KW1, FMAS_KW2 and FMAS_AW1 Isolated from Leptospirosis Patients from Karawanalla and Awissawella, Sri Lanka. J Genomics. 2020. 10.7150/jgen.43953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naotunna C, Agampodi SB, Agampodi TC. Etiological agents causing leptospirosis in Sri Lanka: A review. Asian Pac J Trop Med. 2016;9: 390–394. 10.1016/j.apjtm.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 24.Agampodi S, Warnasekara J, Jayasundara D, Senawirathna I, Gamage C, Kularatne S, et al. Study protocol: characterising the clinical, epidemiological and aetiological aspects of leptospirosis in Sri Lanka: a hospital based clinico- epidemiological study. 2019; 1–8. 10.1136/bmjopen-2018-027850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epidemiologu Unit M of health-2011. G:\Surveillance book.pdf—Surveillance_book.pdf. 2nd Editio. Epidemiology Unit,Ministry of Health, Sri Lanka; 2011. pp. 17–18. [Google Scholar]
- 26.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. Grapetree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28: 1395–1404. 10.1101/gr.232397.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Cj Maiden M. BIGSdb: Scalable analysis of bacterial genome variation at the population level. 2010. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kmety E, Dikken H. Classification of the species Leptospira interrogans and history of its serovars. 1993. p. 104. [Google Scholar]
- 30.Bourhy P, Collet L, Clé Ment S, Huerre M, Ave P, Giry C, et al. Isolation and Characterization of New Leptospira Genotypes from Patients in Mayotte (Indian Ocean). 10.1371/journal.pntd.0000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agampodi SB, Matthias M a., Moreno AC, Vinetz JM. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: Association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis. 2012;54: 1249–1255. 10.1093/cid/cis035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nalam K, Ahmed A, Devi SM, Francalacci P, Baig M, Sechi LA, et al. Genetic affinities within a large global collection of pathogenic Leptospira: implications for strain identification and molecular epidemiology. PLoS One. 2010/09/02. 2010;5: e12637. 10.1371/journal.pone.0012637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourhy P, Collet L, Lernout T, Zinini F, Hartskeerl RA, van der Linden H, et al. Human leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J Clin Microbiol. 2011/12/14. 2012;50: 307–311. JCM.05931-11 [pii] 10.1128/JCM.05931-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A Dominant Clone of Leptospira interrogans Associated with an Outbreak of Human Leptospirosis in Thailand. Picardeau M, editor. PLoS Negl Trop Dis. 2007;1: e56. 10.1371/journal.pntd.0000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarantonelli L, Suanes A, Meny P, Buroni F, Nieves C, Salaberry X, et al. Isolation of pathogenic Leptospira strains from naturally infected cattle in Uruguay reveals high serovar diversity, and uncovers a relevant risk for human leptospirosis. PLoS Negl Trop Dis. 2018;12. 10.1371/journal.pntd.0006694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wuthiekanun V, Sirisukkarn N, Daengsupa P, Sakaraserane P, Sangkakam A, Chierakul W, et al. Clinical diagnosis and geographic distribution of leptospirosis, Thailand. Emerg Infect Dis. 2007;13: 124–126. 10.3201/eid1301.060718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurilung A, Chanchaithong P, Lugsomya K, Niyomtham W, Wuthiekanun V, Prapasarakul N. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res Vet Sci. 2017;115: 146–154. 10.1016/j.rvsc.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 38.Storck CH, Lamaury I. Changes in epidemiology of leptospirosis in 2003–2004, a two El Nino Southern Oscillation period, Guadeloupe archipelago, French West Indies. 2003. 10.1017/S0950268807000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peláez Sanchez RG, Lopez JÁ, Pereira MM, Naranjo MA, Agudelo-F Lórez P. Genetic diversity of leptospira in northwestern Colombia: First report of leptospira santarosai as a recognised leptospirosis agent. Mem Inst Oswaldo Cruz. 2016;111: 737–744. 10.1590/0074-02760160245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno LZ, Miraglia F, Marvulo MFV, Silva JCR, Paula CD, Costa BLP, et al. Characterization of Leptospira santarosai serogroup Grippotyphosa serovar Bananal isolated from Capybara (Hydrochaeris hydrochaeris) in Brazil. J Wildl Dis. 2016;52: 688–693. 10.7589/2015-09-245 [DOI] [PubMed] [Google Scholar]
- 41.Valverde M de los A, Goris MGA, González V, Anchia ME, Díaz P, Ahmed A, et al. New serovars of Leptospira isolated from patients in Costa Rica: Implications for public health. J Med Microbiol. 2013;62: 1263–1271. 10.1099/jmm.0.058545-0 [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Zheng H, Zhang Y, Wang Y, Zhang J, Li Z, et al. Genomic Analysis of a New Serovar of Leptospira weilii Serogroup Manhao. Front Microbiol. 2017;8. 10.3389/fmicb.2017.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haake DA, Dundoo M, Cader R, Kubak BM, Hartskeerl RA, Sejvar JJ, et al. Leptospirosis, Water Sports, and Chemoprophylaxis. Clin Infect Dis. 2002;34: e40–e43. 10.1086/339942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts MW, Smythe L, Dohnt M, Symonds M, Slack A. Serologic-based investigation of leptospirosis in a population of free-ranging eastern grey kangaroos (Macropus giganteus) indicating the presence of Leptospira weilii serovar Topaz. J Wildl Dis. 2010;46: 564–569. 10.7589/0090-3558-46.2.564 [DOI] [PubMed] [Google Scholar]
- 45.Slack AT, Symonds ML, Dohnt MF, Corney BG, Smythe LD. Epidemiology of Leptospira weilii serovar Topaz infections in Australia. Commun Dis Intell. 2007;31: 216–222. [DOI] [PubMed] [Google Scholar]
- 46.Corney BG, Slack AT, Symonds ML, Dohnt MF, McClintock CS, McGowan MR, et al. Leptospira weilii serovar Topaz, a new member of the Tarassovi serogroup isolated from a bovine source in Queensland, Australia. Int J Syst Evol Microbiol. 2008/10/10. 2008;58: 2249–2252. 10.1099/ijs.0.64884-0 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Lau CL, Watson CH, Lowry JH, David MC, Craig SB, Wynwood SJ, et al. Human Leptospirosis Infection in Fiji: An Eco-epidemiological Approach to Identifying Risk Factors and Environmental Drivers for Transmission. Picardeau M, editor. PLoS Negl Trop Dis. 2016;10: e0004405. 10.1371/journal.pntd.0004405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanco RM, Romero EC. Fifteen years of human leptospirosis in São Paulo, Brazil. J Epidemiol Res. 2015;2: 56. 10.5430/jer.v2n1p56 [DOI] [Google Scholar]
- 49.Tubiana S, Mikulski M, Becam J, Lacassin F, Lefèvre P, Gourinat AC, et al. Risk Factors and Predictors of Severe Leptospirosis in New Caledonia. PLoS Negl Trop Dis. 2013;7. 10.1371/journal.pntd.0001991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gouveia EL, Metcalfe J, De Carvalho ALF, Aires TSF, Villasboas-Bisneto JC, Queirroz A, et al. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg Infect Dis. 2008;14: 505–508. 10.3201/eid1403.071064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrmann-Storck C, Saint Louis M, Foucand T, Lamaury I, Deloumeaux J, Baranton G, et al. Severe leptospirosis in hospitalized patients, Guadeloupe. Emerg Infect Dis. 2010. 10.3201/eid1602.090139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warnasekara J, Koralegedara I, Agampodi S. Estimating the burden of leptospirosis in Sri Lanka; A systematic review. BMC Infect Dis. 2019;19. 10.1186/s12879-018-3655-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nityananda K, Harvey T. Leptospirosis in Ceylon epidemiological and laboratory studies. Ceylon J Med Sci. 1971;20: 5–14. Available: http://www.dtic.mil/docs/citations/AD0724157 [Google Scholar]
- 54.Gamage CD, Koizumi N, Muto M, Nwafor-Okoli C, Kurukurusuriya S, Rajapakse JRPV, et al. Prevalence and Carrier Status of Leptospirosis in Smallholder Dairy Cattle and Peridomestic Rodents in Kandy, Sri Lanka. Vector Borne Zoonotic Dis. 2011/02/03. 2011;11: 1041–1047. 10.1089/vbz.2010.0153 [DOI] [PubMed] [Google Scholar]
- 55.Gamage CD, Koizumi N, Perera AKC, Muto M, Nwafor-Okoli C, Ranasinghe S, et al. Carrier status of leptospirosis among cattle in Sri Lanka: a zoonotic threat to public health. Transbound Emerg Dis. 2012/09/25. 2014;61: 91–96. 10.1111/tbed.12014 [DOI] [PubMed] [Google Scholar]
- 56.Cameron CE. Leptospiral structure, physiology, and metabolism. Curr Top Microbiol Immunol. 2015;387: 21–41. 10.1007/978-3-662-45059-8_3 [DOI] [PubMed] [Google Scholar]
- 57.Bourhy P, Collet L, Clément S, Huerre M, Ave P, Giry C, et al. Isolation and characterization of new Leptospira genotypes from patients in Mayotte (Indian Ocean). PLoS Negl Trop Dis. 2010;4. 10.1371/journal.pntd.0000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wuthiekanun V, Chierakul W, Limmathurotsakul D, Smythe LD, Symonds ML, Dohnt MF, et al. Optimization of Culture of Leptospira from Humans with Leptospirosis. J Clin Microbiol. 2007;45: 1363–1365. 10.1128/JCM.02430-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girault D, Soupé-Gilbert ME, Geroult S, Colot J, Goarant C. Isolation of Leptospira from blood culture bottles. Diagn Microbiol Infect Dis. 2017;88: 17–19. 10.1016/j.diagmicrobio.2017.01.014 [DOI] [PubMed] [Google Scholar]
- 60.Meny P, Menéndez C, Quintero J, Hernández E, Ríos C, Balassiano IT, et al. Characterization of Leptospira isolates from humans and the environment in Uruguay. Rev Inst Med Trop Sao Paulo. 2017;59. 10.1590/S1678-9946201759079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, Saito M, et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis. 2008;2. 10.1371/journal.pntd.0000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herath N, Uluwattage W, Weliwitiya T, Karunanayake L, Lekamwasam S, Ratnatunga N, et al. Sequel and therapeutic modalities of leptospirosis associated severe pulmonary haemorrhagic syndrome (SPHS); A Sri Lankan experience. BMC Infect Dis. 2019;19. 10.1186/s12879-019-4094-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorge S, Kremer FS, De Oliveira NR, Navarro GDOSV, Guimarães AM, Sanchez CD, et al. Whole-genome sequencing of leptospira interrogans from southern Brazil: Genetic features of a highly virulent strain. Mem Inst Oswaldo Cruz. 2018;113: 80–86. 10.1590/0074-02760170130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beriwal S, Padhiyar N, Bhatt D, Pandit PD, Ansari A, Lata KS, et al. LeptoDB: an integrated database of genomics and proteomics resource of Leptospira. Database. 2018;2018. 10.1093/database/bay057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Llanes A, Restrepo CM, Rajeev S. Whole genome sequencing allows better understanding of the evolutionary history of leptospira interrogans serovar hardjo. PLoS One. 2016;11. 10.1371/journal.pone.0159387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y, Zhu Y, Wang Y, Chang Y-F, Zhang Y, Jiang X, et al. Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira. Sci Rep. 2016;6: 20020. 10.1038/srep20020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurilung A, Keeratipusana C, Suriyaphol P, Hampson DJ, Prapasarakul N. Genomic analysis of Leptospira interrogans serovar Paidjan and Dadas isolates from carrier dogs and comparative genomic analysis to detect genes under positive selection. BMC Genomics. 2019;20: 168. 10.1186/s12864-019-5562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Llanes A, Restrepo CM, Riesgo-Ferreiro P, Rajeev S. Genomic Variability among Field Isolates and Laboratory-Adapted Strains of Leptospira borgpetersenii Serovar Hardjo. Int J Microbiol. 2018;2018. 10.1155/2018/2137036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPG)
(XLSX)
Data Availability Statement
All genomic data are available at BIGSdb-Pasteur: the genomic-based strain taxonomy and nomenclature platform, under the Leptospira sequence typing at http://bigsdb.pasteur.fr/leptospira Hyperlink to each cgMLST is given in the Table 1. All sequences are available through the direct link.