Abstract
Male infertility is a prevalent condition, affecting 5–10% of men. So far, few genetic factors have been described as contributors to spermatogenic failure. Here, we report the first re-sequencing study of the Y-chromosomal Azoospermia Factor c (AZFc) region, combined with gene dosage analysis of the multicopy DAZ, BPY2, and CDYgenes and Y-haplogroup determination. In analysing 2324 Estonian men, we uncovered a novel structural variant as a high-penetrance risk factor for male infertility. The Y lineage R1a1-M458, reported at >20% frequency in several European populations, carries a fixed ~1.6 Mb r2/r3 inversion, destabilizing the AZFc region and predisposing to large recurrent microdeletions. Such complex rearrangements were significantly enriched among severe oligozoospermia cases. The carrier vs non-carrier risk for spermatogenic failure was increased 8.6-fold (p=6.0×10−4). This finding contributes to improved molecular diagnostics and clinical management of infertility. Carrier identification at young age will facilitate timely counselling and reproductive decision-making.
Research organism: Human
Introduction
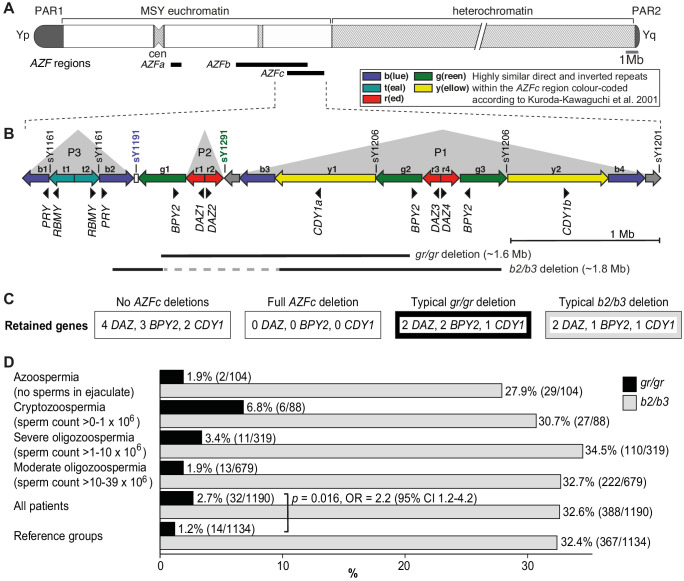
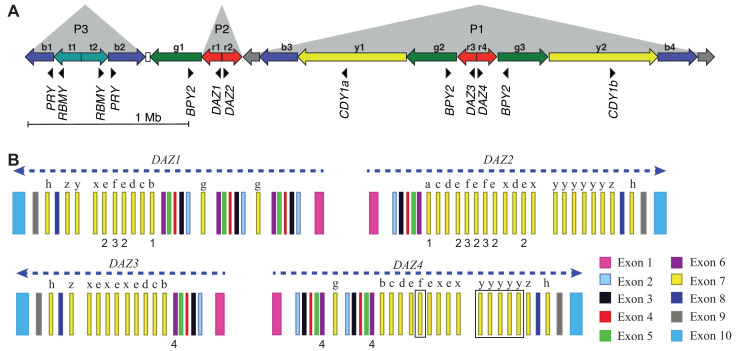
The diagnosis of male factor infertility due to abnormal semen parameters concerns ~10% of men (Jungwirth et al., 2012; Datta et al., 2016). In today’s andrology workup, ~60% of patients with spermatogenic failure remain idiopathic (Punab et al., 2017). Among the known causes, the most widely considered genetic factors are karyotype abnormalities (up to 17% of patients) and recurrent de novo microdeletions of the Y-chromosomal Azoospermia Factor a (AZFa) (~0.8 Mb), AZFb (~6.2 Mb), and AZFc (~3.5 Mb) regions (2–10%) (Punab et al., 2017; Olesen et al., 2017; Tüttelmann et al., 2011). For more than 15 years, testing for AZF deletions has been strongly recommended in the diagnostic workup for infertility patients with sperm concentration of <5 × 106/ml (ASRM, 2015; Krausz et al., 2014). Most deletion carriers represent patients with either azoospermia (no sperm) or cryptozoospermia (>0–1 million sperm/ejaculate) (Punab et al., 2017; Kohn et al., 2019; Stahl et al., 2010). The most prevalent deletion type is AZFc (~80%), followed by the loss of AZFa (0.5–4%), AZFb (1–5%), and AZFbc (1–3%) regions (Figure 1A). Excess of recurrent AZFc deletions is promoted by the region’s complex genomic structure comprised of long direct and inverted amplicons of nearly identical DNA segments that lead to aberrant meiotic rearrangements in gametogenesis (Kuroda-Kawaguchi et al., 2001; Skaletsky et al., 2003; Figure 1B). The AZFc full deletions remove all the multicopy DAZ (deleted in azoospermia 1), BPY2 (basic charge Y-linked 2), and CDY1 (chromodomain Y-linked 1) genes that are expressed in a testis-enriched manner and considered important in spermatogenesis (Figure 1C).
Figure 1. Y-chromosomal AZFc region and its partial deletions in the study group.
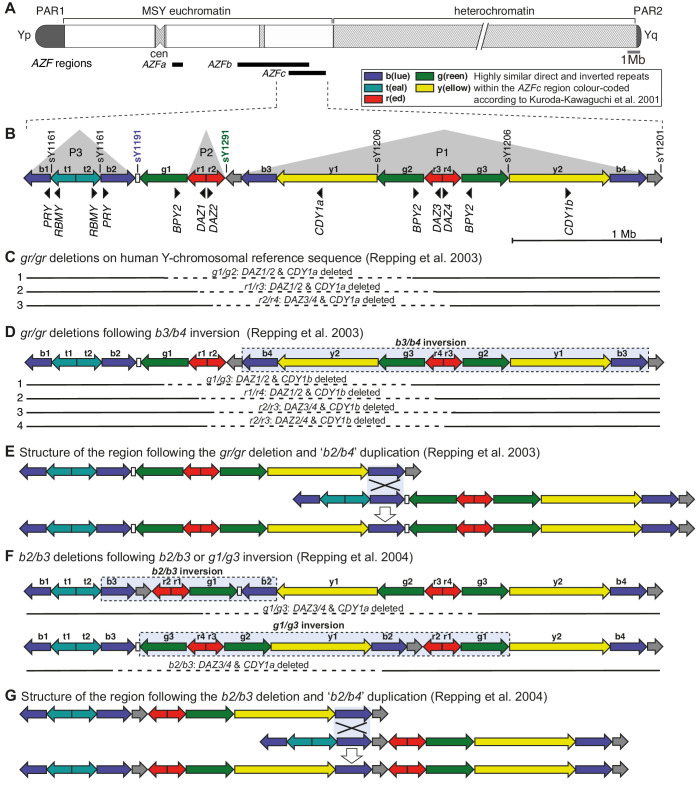
(A) Schematic representation of the human Y chromosome with the AZFa, AZFb, and AZFc regions shown as black bars. (B) Magnified structure of the AZFc region with approximate locations of multicopy protein-coding genes, STS (sY) markers for the detection of AZFc partial deletions and the span of typical gr/gr and b2/b3 deletions (Kuroda-Kawaguchi et al., 2001). P1–P3 (gray triangles) denote palindromic genomic segments consisting of two ‘arms’ representing highly similar inverted DNA repeats (>99.7% sequence identity) that flank a relatively short distinct ‘spacer’ sequence. Of note, the occurrence of the b2/b3 deletion requires a preceding inversion in the AZFc region and therefore its presentation on the reference sequence includes also the retained segment (gray dashed line). Full details about alternative gr/gr and b2/b3 deletion types are presented in Figure 1—figure supplement 1. (C) Dosage of multicopy genes on human Y chromosomes with or without AZFc deletions. (D) Prevalence of the gr/gr and b2/b3 deletions detected in the subgroups of this study. Fisher’s exact test was used to test the statistical significance in the deletion frequencies between the groups. PAR, pseudoautosomal region; MSY, male-specific region of the Y chromosome; cen, centromere; AZF, azoospermia factor region.
Figure 1—figure supplement 1. The human Y chromosome and the Azoospermia Factor (AZF) regions.
The palindromic structure of the AZFc region also facilitates partial deletions that are rather frequently detected in the general population (Repping et al., 2003; Repping et al., 2004; Rozen et al., 2012; Fernandes et al., 2004). The most prevalent partial deletion types, named after the involved amplicons as g(reen)-r(ed)/g(reen)-r(ed) (lost segment ~1.6 Mb) and b(lue)2/b(lue)3 (~1.8 Mb) reduce the copy number of DAZ, BPY2, and CDY1 genes by roughly 50% (Figure 1B,C, Figure 1—figure supplement 1). The published data on the contribution of gr/gr and b2/b3 deletions to spermatogenic failure are inconsistent. In European populations, the carrier status of the gr/gr deletion increases a risk to low sperm counts ~1.8-fold (Rozen et al., 2012; Bansal et al., 2016a; Krausz et al., 2009). Its more variable effect on spermatogenesis has been shown in Middle Eastern and Asian populations, where the gr/gr deletion is completely fixed in some Y lineages, for example haplogroups D2 and Q1a that are common in Japan and some parts of China (de Carvalho et al., 2006; Teitz et al., 2018). In contrast, the b2/b3 deletion appears to be a risk factor for spermatogenic impairment in several East Asian and African, but not in European or South Asian populations (Bansal et al., 2016b; Colaco and Modi, 2018). Notably, the b2/b3 deletion is completely fixed in Y haplogroup N3 that has a high frequency (up to 90% in some populations) in Finno-Ugric-, Baltic-, and some Turkic-speaking people living in Northern Eurasia (Rozen et al., 2012; Fernandes et al., 2004; Ilumäe et al., 2016). Thus, it is unlikely that the carriership of a gr/gr or b2/b3 deletion per se has an effect on male fertility potential. It has been proposed that this broad phenotypic variability may be explained by the diversity of gr/gr and b2/b3 deletion subtypes (Machev, 2004). Y chromosomes carrying partial AZFc deletions may differ for the content, dosage, or genetic variability of the retained genes, the overall genetic composition reflected by phylogenetic haplogroups or the presence of additional structural variants. Only limited studies have analyzed the subtypes of gr/gr or b2/b3 deletions, and no straightforward conclusions have been reached for their link to spermatogenic failure (Krausz et al., 2009; Ghorbel et al., 2016; Krausz and Casamonti, 2017).
The current study represents the largest in-depth investigation of AZFc partial deletions in men recruited by a single European clinical center. We analysed 1190 Estonian idiopathic patients with male factor infertility in comparison to 1134 reference men from the same population, including 2000 subjects with sperm parameter data available. Y chromosomes carrying gr/gr or b2/b3 deletions were investigated for additional genomic rearrangements, Y-chromosomal haplogroups, and dosage and sequence variation of the retained DAZ, BPY2, and CDY genes. The study aimed to determine the role and contribution of gr/gr and b2/b3 deletion subtypes in spermatogenic failure and to explore their potential in the clinical perspective.
Results
Enrichment of gr/gr deletions in Estonian idiopathic infertile men with reduced sperm counts
The study analyzed 1190 Estonian men with idiopathic infertility (sperm counts 0–39 × 106/ejaculate) and a reference group comprised of 1134 Estonian men with proven fatherhood (n = 635) or representing healthy young men (n = 499) (Table 1, Supplementary file 1). For all 2324 study subjects, complete AZFa, AZFb, and AZFc deletions were excluded.
Table 1. Characteristics of the patients with male factor infertility and reference groups used for comparison.
| Idiopathic spermatogenic impairment (n = 1190)* | Reference groups (n = 1134) | ||||||
|---|---|---|---|---|---|---|---|
| Parameter | Unit | Azoo-/cryptozoospermia | Severe oligozoospermia | Moderate oligozoospermia | Partners of pregnant women† | Estonian young men cohort‡ | REPROMETA proven fathers§ |
| n | 104/88 | 319 | 679 | 324 | 499 | 311 | |
| Age | Years | 33.2 (23.6–51.8) |
32.2 (23.9–49.5) |
31.7 (23.0–44.6) |
31.0 (22.9–45.0) |
18.6 (17.2–22.9) |
31.0 (21.0–43.0) |
| BMI | kg/m2 | 26.0 (21.2–34.4) |
25.9 (20.2–35.5) |
25.8 (20.1–34.6) |
24.8 (20.0–32.2) |
22.0 (18.7–27.5) |
25.9 (20.2–33.1) |
| Total testis volume | ml | 33.5 (17.0–49.0) |
39.0 (22.0–50.0) |
40.0 (26.0–52.0) |
46.0 (34.0–62.4) |
50.0 (35.0–70.0) |
n.d. |
| Semen volume | ml | 3.3 (0.8–6.6) |
3.3 (1.1–7.0) |
3.6 (1.6–6.9) |
3.7 (1.7–8.0) |
3.2 (1.2–6.4) |
n.d. |
| Sperm concentration | × 106/ml | 0 (0–0.2) |
1.4 (0.4–5.2) |
6.0 (2.2–15.2) |
76.0 (16.7–236.0) |
66.8 (8.2–225.1) |
n.d. |
| Total sperm count | × 106/ ejaculate | 0 (0–0.7) |
4.7 (1.3–9.3) |
23.1 (11.0–37.5) |
295.2 (60.0–980.1) |
221.6 (18.4–788.0) |
n.d. |
| Progressive A+B motility | % | 0 (0–37.2) |
16.0 (0–47.2) |
27.0 (1.0–57.0) |
50.0 (30.0–69.0) |
57.3 (34.7–75.3) |
n.d. |
| Sperms with normal morphology | % | 0 (0–1.0) |
0 (0–6.0) |
2.0 (0–9.0) |
10.0 (2.0–19.1) |
12.0 (4.0–20.0) |
n.d. |
| FSH | IU/l | 13.7 (2.7–38.2) |
6.6 (1.9–22.8) |
5.2 (1.8–16.5) |
3.6 (1.5–8.3) |
2.8 (1.2–6.7) |
n.d. |
| LH | IU/l | 5.7 (2.1–12.0) |
4.6 (1.9–9.9) |
4.2 (1.8–8.4) |
3.6 (1.5–6.7) |
3.8 (1.8–7.2) |
n.d. |
| Total testosterone | nmol/l | 15.3 (7.7–28.4) |
16.6 (7.9–30.0) |
16.6 (8.5–30.3) |
16.5 (8.8–27.2) |
27.7 (15.4–46.3) |
n.d. |
All study subjects were recruited in Estonia. For each parameter, median and (5th–95th) percentile values are shown. Additional details in Supplementary file 1.
*Patients were subgrouped based on total sperm counts per ejaculate: azoospermia, no sperm; cryptozoospermia, sperm counts > 0–1 × 106; severe oligozoospermia, >1–10 × 106; moderate oligozoospermia, >10–39 × 106 (Punab et al., 2017).
†Male partners of pregnant women (Punab et al., 2017); eight men had sperm counts < 39 × 106; for four men, sperm analysis was not available.
‡Male cohort without fatherhood data (Grigorova et al., 2008); 47 men had sperm counts < 39 × 106; for nine men, sperm analysis was not available.
§REPROMETA study recruited and sampled couples after delivery of their newborn; details in Kikas et al., 2020; Pilvar et al., 2019.
n.d., not determined.
The partial AZFc deletions identified using the STS-based polymerase chain reaction (PCR) assays were gr/gr (n = 46), b2/b3 (n = 756), and b1/b3 (n = 1, reference case) (Table 2). A statistically significant excess of gr/gr deletions was detected in idiopathic male infertility patients (2.7%; n = 32/1190) compared to reference cases (1.2%; n = 14/1134) (Fisher’s exact test, p=0.016; odds ratio [OR] = 2.2 [95% confidence interval (CI) 1.2–4.2]) (Figure 1D, Supplementary file 2). The highest frequency of gr/gr deletion carriers (6.8%, n = 6/88) was detected in cryptozoospermia cases (sperm count > 0–1 × 106/ejaculate). However, in the reference group andrological parameters of men with or without the gr/gr deletion did not differ (Supplementary file 3). All 10 reference men with the gr/gr deletion and available andrological data were normozoospermic (220.3 [74.2–559.0] × 106 sperm/ejaculate). Also their other andrological parameters were within the normal range, overlapping with those of the subjects without a gr/gr deletion.
Table 2. Summary of the identified Y-chromosomal AZF deletion subtypes.
| Y-chromosomal rearrangements | Idiopathic male infertility patients (n) | Reference men (n) |
|---|---|---|
| All analyzed cases | 1190 | 1134 |
| Any AZFc gr/gr deletion | 32 (2.7%) | 14 (1.2%) |
| Fisher’s exact test, p=0.016; OR = 2.2 [95% CI 1.2–4.2] | ||
| Any AZFc b2/b3 deletion | 388 (32.6%) | 367 (32.4%) |
| Other type of AZF deletion | Loss of b2/b3 marker sY1191 (one case) | AZFc b1/b3 del (one case); partial AZFa del (one case) |
| No deletion | 769 (64.6%) | 751 (66.2%) |
| Simple partial AZFc deletions | ||
| Typical gr/gr deletion | 19/31 (61.3%) | 8/13 (61.5%) |
| Typical b2/b3 deletion* | 300/382 (78.5%) | 210/249 (84.3%) |
| AZFc partial deletion followed by b2/b4 duplication | ||
| gr/gr del + b2/b4 dupl† | 2/31 (6.5%) | 3/13 (23.1%) |
| Fisher’s exact test, p=0.144; OR = 0.2 [95% CI 0.0–1.6] | ||
| b2/b3 del + b2/b4 dupl*,† | 78/382 (20.4%) | 34/249 (13.7%) |
| Fisher’s exact test, p=0.026; OR = 1.6 [95% CI 1.0–2.4] | ||
| AZFc partial deletion and atypical genomic rearrangements‡ | ||
| gr/gr del + extra gene copies | 1 | 0 |
| b2/b3 del + extra gene copies | 3 | 4 |
| Complex events on the Y lineage R1a1-M458 with the preceding AZFc r2/r3 inversion | ||
| r2/r3 inv + gr/gr del | 8 | 2§ |
| r2/r3 inv + gr/gr del + b2/b4 dupl | 1 | 0 |
| r2/r3 inv + loss of marker sY1191 + secondary gene duplications¶ | 1 | 0 |
| r2/r3 inv + b2/b3 del + b2/b4 dupl | 0 | 1** |
| Carriers of any AZFc gr/gr deletion type without the preceding r2/r3 inversion | ||
| gr/gr del w/o detected r2/r3 inv | 23/1190 (1.9 %) | 12/1134 (1.1%) |
| Fisher’s exact test, p=0.090; OR = 1.8 [95% CI 0.9–3.7] | ||
*Deletion subtype analysis was carried out for cases with available sufficient quantities of DNA. REPROMETA subjects were excluded from the b2/b3 deletion subtype analysis and subsequent statistical testing due to missing andrological data.
†One or more amplicons of the retained ‘2xDAZ, 2xBPY2, 1xCDY1’ (gr/gr deletion) or ‘2xDAZ, 1xBPY2, 1xCDY1’ (b2/b3 deletion) genes.
‡Additional copies of DAZ, BPY2, and/or CDY1 genes inconsistent with the full ‘b2/b4’ duplication.
§Including one REPROMETA man without andrological data.
¶Detected gene copy numbers 6xDAZ, 4xBPY2, 3xCDY1; the obligate presence of r2/r3 inversion was defined based on Y-chromosomal phylogeny as the man carries Y lineage R1a1a1b1a1a1c-CTS11962.1 that was also identified in two cases with the r2/r3 inversion (Supplementary file 12).
**Man from ‘Partners of pregnant women’ cohort with sperm concentration 12 × 106/ml below normozoospermia threshold (15 × 106/ml) and sperm counts 39.4 × 106/ejaculate at the borderline of the lowest reference value (39.0 × 106/ejaculate).
The patient and the reference groups exhibited similar prevalence of b2/b3 deletions (388/1190, 32.6% vs 367/1134, 32.4%; Fisher’s exact test, p=0.8). No apparent clinically meaningful genetic effects on andrological parameters were observed in either of the study groups (Supplementary files 3 and 4).
Significant overrepresentation of Y lineage R1a1-M458 in gr/gr deletion carriers
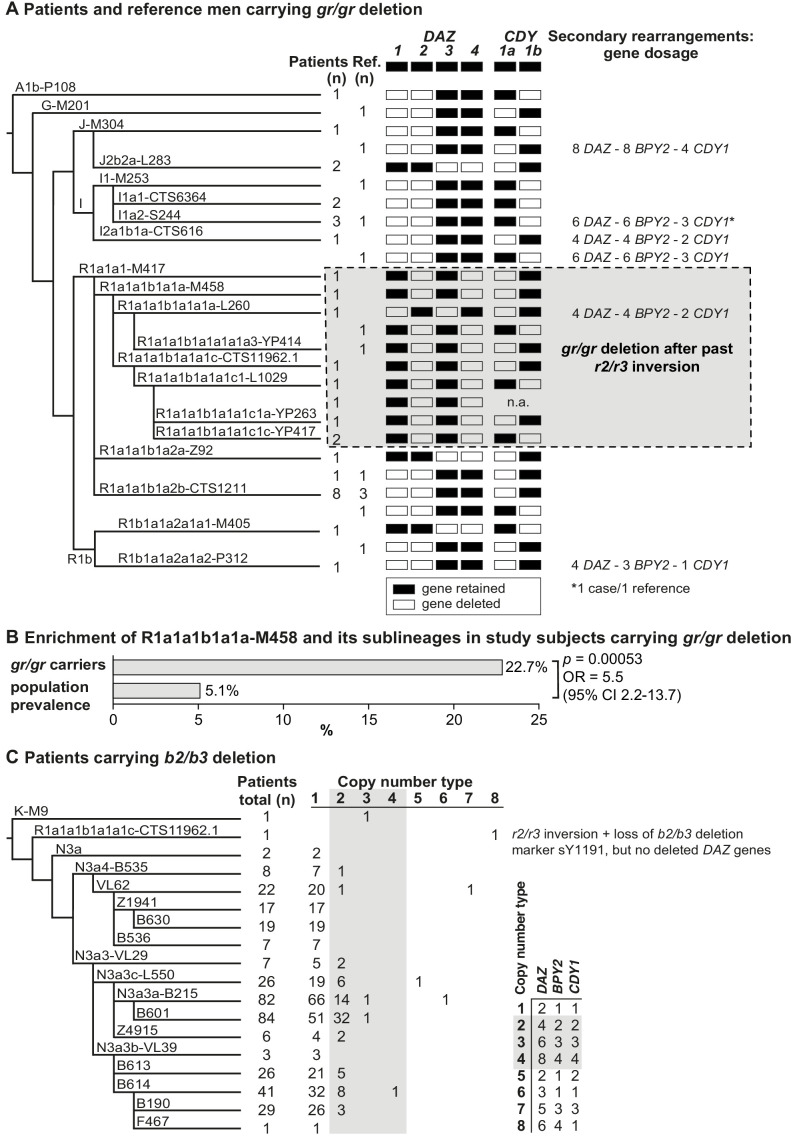
The Y-chromosomal haplogroups determined by typing phylogenetically informative markers in 31 patients and 13 reference men carrying a gr/gr deletion represented 20 different lineages (patients, 17; reference men, 10; Figure 2A, Supplementary file 5). Combining the phylogenetic context with the data on exact missing DAZ and CDY1 gene copies (see below) revealed that the gr/gr deletion events in 44 analyzed cases must have independently occurred at least 26 times. About two-thirds of these Y chromosomes belonged to haplogroup R1, whereas the rest represented A1b, G, I, and J lineages. Notably, there was a highly significant overrepresentation of Y chromosomes belonging to lineage R1a1-M458 in the gr/gr deletion carriers compared to the known Estonian population frequency (22.7% vs 5.1%; Fisher’s exact test, p=5.3×10−4, OR = 5.5 [95% CI 2.2–13.7]; Figure 2B, Supplementary file 5; Underhill et al., 2015).
Figure 2. Phylogenetic relationships and gene copies in study subjects with partial AZFc deletions.
(A) Y-chromosomal lineages indicated with typed terminal markers (left), deleted (white)/retained (black) DAZ and CDY1 gene copies (middle), and secondary rearrangements in the AZFc region (right) of idiopathic male factor infertility (n = 31) and reference cases (n = 13) carrying the gr/gr deletion. The human Y-chromosomal reference sequence has four DAZ and two CDY1 copies; the retained gene copies on each Y chromosome with a gr/gr deletion are shown as filled boxes. Chromosomes carrying atypical gr/gr subtypes with the loss of either the DAZ1/DAZ3 or DAZ2/DAZ4 gene pair due to complex genomic rearrangement combining the previous r2/r3 inversion with a subsequent gr/gr deletion are highlighted with a dashed gray square. (B) Enrichment of the Y-chromosomal lineage R1a1-M458 and its sub-lineages in study subjects carrying the gr/gr deletion in comparison to the Estonian general population (data from Underhill et al., 2015). Fisher’s exact test was used to test the statistical significance between the groups. (C) Y-chromosomal lineages indicated with typed terminal markers (left) and the copy number of the DAZ, BPY2, and CDY1 gene copies (right) determined for 382 idiopathic male factor infertility cases carrying the b2/b3 deletion. The light gray box denotes DAZ, BPY2, and CDY1 gene dosage consistent with full b2/b4 duplication(s). The legend for the deletion subtype is shown in the bottom right corner. Further information on the distribution of Y-chromosomal lineages in the carriers of AZFc partial deletions are provided in Supplementary files 5 and 6, and the AZFc rearrangement types are detailed in Figure 1—figure supplement 1 and Supplementary files 7 and 9–12. n, number; n.a., not available; Ref, reference cases.
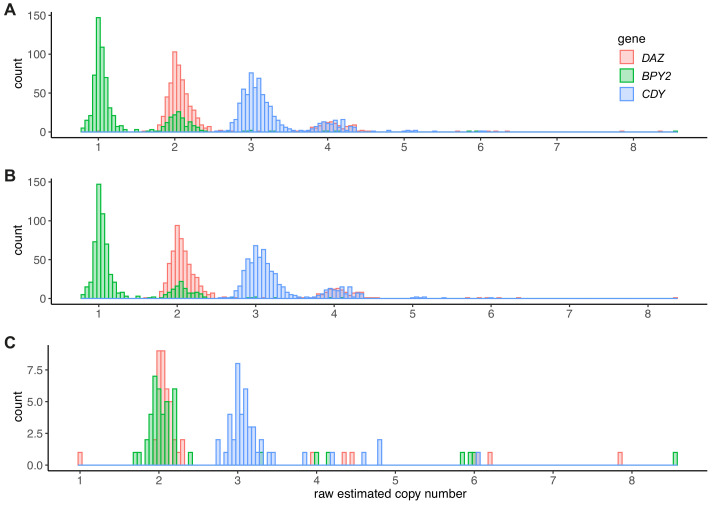
Figure 2—figure supplement 1. Histograms of estimated raw copy number values for DAZ, BPY2, and CDY genes by ddPCR.
Figure 2—figure supplement 2. Location of the identified exonic variants in the DAZ genes.
Nearly all (99.4%) Estonian cases with the b2/b3 deletion belonged to the Y haplogroup N3, in which this event is fixed (Repping et al., 2004; Fernandes et al., 2004). The most commonly detected sub-lineage was N3a3a-L550 (~51% of 436 typed chromosomes) and in total 15 different haplogroups that had diverged after the b2/b3 deletion event in the common ancestor of N3 were present in Estonian men (Figure 2C, Supplementary file 6). b2/b3 Y chromosomes representing non-N3 lineages were detected in two patients and three reference men. Lineage typing was possible for three of them, who carried either K-M9 (one patient) or R1a1a1b1a1a1c-CTS11962.1 (one patient and one reference case).
Increased prevalence of b2/b3 deletion followed by b2/b4 duplication in infertile men
The expected retained copy number of DAZ, BPY2, and CDY1 genes consistent with the typical gr/gr deletion, as determined by quantification using Droplet Digital PCR (ddPCR), was found in 37/44 (~84%) cases (Figure 2A, Figure 2—figure supplement 1, Table 2, Supplementary file 7). Three patients and three reference men carried a secondary b2/b4 duplication adding one or more amplicons of [two DAZ – two BPY2 – one CDY1] genes with no apparent effect on infertility status (Fisher’s exact test, p=0.34). Notably, four of six samples with secondary b2/b4 duplication events were identified in haplogroup I. This complex rearrangement has also been reported in the gnomAD SV database (v 2.1) in 114/5528 analyzed men from around the world with the prevalence of 3.5% in East Asians and 1.2% in Europeans (Supplementary file 8; Collins et al., 2020).
Similarly, 78.5% of patients and 84.3% reference men with the b2/b3 deletion presented gene dosage consistent with the typical deletion (Figure 2C, Figure 2—figure supplement 1, Table 2, Supplementary files 7 and 9). Indicative of recurrent secondary events, one or more b2/b4 duplications of [two DAZ – one BPY2 – one CDY1] genes were identified in 13 haplogroups, including non-N3 lineages K-M9 and R1a1a1b1a1a1c-CTS11962.1. In the gnomAD, 58/5115 men have been reported with this duplication, with the prevalence of 2.2% in East Asians and 0.9% in Europeans (Supplementary file 8). Although secondary b2/b4 duplications were detected with significantly higher prevalence in patients compared to the reference men (n = 78/382, 20.4% vs n = 35/249, 14.1%; Fisher’s exact test, p=0.026, OR = 1.57 [95% CI 1.02–2.42]), no consistent effect of increased gene copy number on andrological parameters was observed (Supplementary files 3 and 4). Reference men with b2/b4 duplication compared to subjects with no AZFc rearrangements showed a trend for lower follicle-stimulating hormone (FSH) (median 2.3 [5–95% range 1.4–7.5] vs 3.2 [1.3–7.1] IU/l; p<0.05) and luteinizing hormone (LH) (3.1 [1.7–5.0] vs 3.8 (1.7–7.2) IU/l; p<0.05). Additionally, in eight subjects with AZFc partial deletions, further atypical Y-chromosomal genomic rearrangements were detected, but also with no clear evidence for a phenotypic effect (Table 2, Supplementary file 7).
The data gathered from this analysis thus suggest that the dosage of DAZ, BPY2, and CDY1 genes does not play a major role in modulating the pathogenic effect of the gr/gr and b2/b3 deletions.
No specific DAZ or CDY1 gene copy is lost in men with spermatogenic failure
The deletion subtypes for b2/b3 and gr/gr carriers were identified by determining the genotypes of DAZ and CDY1 gene-specific paralogous sequence variants. The major b2/b3 deletion subtype in both patients (99.7%) and reference cases (98.1%) was the loss of DAZ3-DAZ4-CDY1a genes, whereas the most frequent gr/gr subtypes among all the deletion carriers were the loss of DAZ1-DAZ2-CDY1a (41.9%, 18/43 cases) and DAZ1-DAZ2-CDY1b (25.6%, 11/43 cases) combinations (Figure 2A, Supplementary files 10 and 11). The observed prevalence of the major gr/gr subtypes was concordant with the published data on other European populations (42.5% and 25.5%, respectively; Krausz et al., 2009). As these gr/gr deletion subtypes are prevalent in the reference group (total 11 of 13, 84.6%), their major role in spermatogenic impairment can be ruled out. As a novel insight, a subset of these Y chromosomes showed lineage-specific loss of some exon 7 subtypes of the retained DAZ4 gene (Figure 2—figure supplement 2; Supplementary file 11). All five exons 7Y in the DAZ4 gene were missing in the Y chromosomes with the DAZ1-DAZ2-CDY1a deletion that had occurred in sub-lineages of the R1a1a1b1a2 haplogroup (9/31 patients, 3/13 reference men), representing 12/18 DAZ1-DAZ2-CDY1a deletion carriers. The exon 7F in DAZ4 was lost in haplogroup I1 and its sub-lineages (5/31, 2/13), that is 7/11 individuals carrying the DAZ1-DAZ2-CDY1b deletion (Supplementary files 10 and 11). There was no evidence that loss of DAZ4 exons 7Y or 7F has any phenotypic consequences. Most likely, this observation reflects gene conversion events from DAZ3 to DAZ4 as the former lacks both, exons 7Y and 7F.
Taken together, our findings indicate that neither the loss of the DAZ1-DAZ2 nor the DAZ3-DAZ4 gene pair, combined with either a CDY1a or CDY1b gene, directly causes spermatogenic failure. Interestingly, no Y chromosomes were observed with fewer than two retained DAZ genes.
Y lineage R1a1-M458 carries a fixed r2/r3 inversion predisposing to recurrent deletions
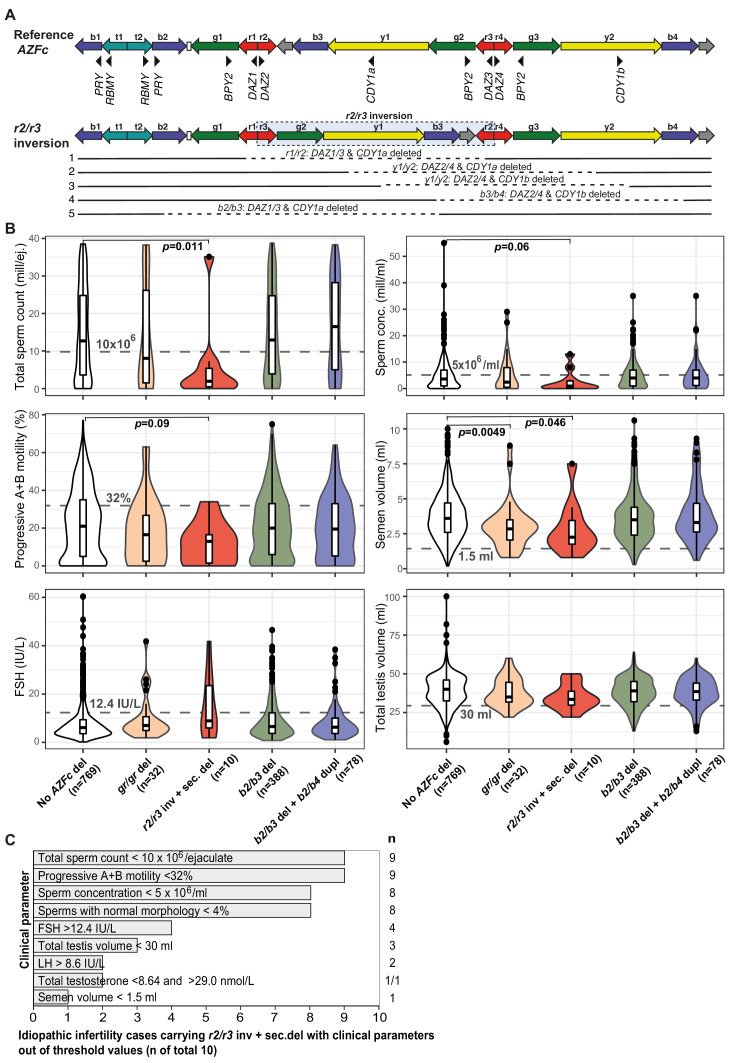
Novel atypical gr/gr and b2/b3 deletion subtypes with the loss of an unusual DAZ gene pair were identified (Figure 2A, Table 2, Supplementary files 10–12). Eight patients and two reference cases with a gr/gr deletion were missing DAZ2-DAZ4 genes. Loss of DAZ1-DAZ3 genes followed by a subsequent b2/b4 duplication event was identified in one infertile and one reference case with either gr/gr or b2/b3 deletion, respectively. All but one subject with this atypical pair of lost DAZ genes belonged to the Y haplogroup R1a1-M458 and its sub-lineages, significantly enriched in gr/gr deletion carriers (Figure 2B, Supplementary file 5). The most parsimonious explanation to explain the simultaneous deletion of either DAZ1-DAZ3 or DAZ2-DAZ4 genes is a preceding ~1.6 Mb long inversion between the r(ed)two and r(ed)three amplicons (Figure 3A). This new inverted structure might be more susceptible to recurrent deletions as it has altered the internal palindromic structure of AZFc region. In r2/r3 inversion chromosomes, the largest palindrome P1 is almost completely lost and the size of the palindrome P2 is greatly expanded by positioning the homologous g1/g2 segments in an inverted orientation. The r2/r3 inversion is consequently expected to destabilize the AZFc region as several long DNA amplicons with highly homologous DNA sequence are positioned in the same sequence orientation (b2, b3, and b4; g2 and g3; y1 and y3). Therefore, they are prone to non-allelic homologous recombination mediating recurrent deletions and duplications. Since these atypical deletion subtypes were identified only in a specific Y-chromosomal haplogroup, the detected r2/r3 inversion must have occurred only once in the common ancestor of R1a1-M458 sub-lineages. One patient with the loss of DAZ2-DAZ4 carried haplogroup R1a1a1-M417, an ancestral lineage to R1a1-M458 (Figure 2A). However, lineage R1a1a1-M417 is not fixed for this inversion since its other sub-lineage, R1a1a1b1a2, does not carry it and any subsequent inversion restoring the exact original AZFc structure is not credible. The more parsimonious explanation is that the inversion occurred in a sub-lineage of R1a1a1-M417 that has to be yet determined.
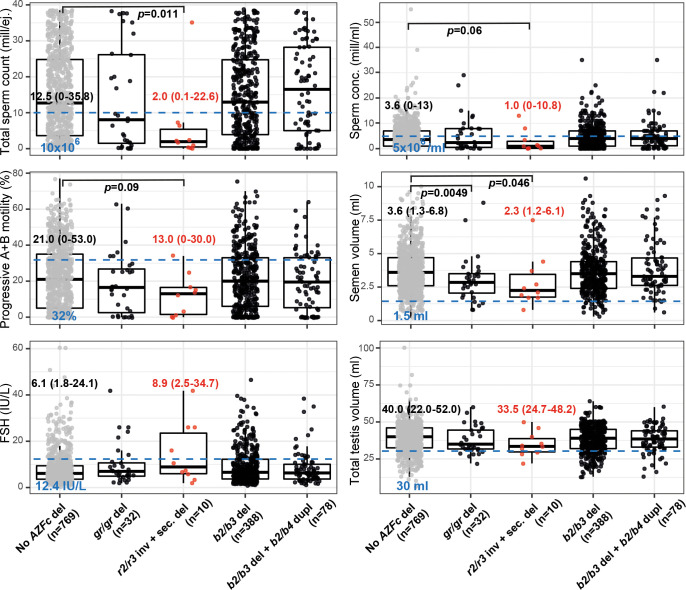
Figure 3. Complex structural variants at the Y-chromosomal lineage R1a1-M458 and their effect on andrological parameters.
(A) Schematic presentation of the Y chromosome with the r2/r3 inversion compared to the reference sequence. The r2/r3 inversion structure nearly destroys the large palindrome P1 and, consequently, destabilizes the AZFc region since several long DNA amplicons with highly similar DNA sequence (b2, b3, and b4; g2 and g3; y1 and y3) are positioned in the same sequence orientation. This structure promotes non-allelic homologous recombination mediating recurrent deletion and duplication events. The approximate regions removed by the identified gr/gr and b2/b3 deletions arising on the r2/r3 inverted Y chromosome are shown as dashed lines. (B) Distribution of andrological parameters in the idiopathic male factor infertility cases (total sperm counts 0–39 × 106) subgrouped based on the structure of the AZFc region. The pairwise Wilcoxon rank-sum test was applied to estimate the statistical difference between groups (Bonferroni threshold for multiple testing correction, p<1.0×10−3). Threshold values (shown in gray) for sperm parameters corresponding to severe spermatogenic failure are based on international guidelines (World Health Organization, 2010). For reproductive hormones, reference values of the laboratory service provider are shown. The empirical threshold for the total testis volume was based on routinely applied clinical criteria at the AC-TUH. For additional details, see Figure 3—figure supplements 1–3, Supplementary files 3 and 12. (C) The majority of idiopathic infertility cases carrying the r2/r3 inversion plus secondary AZFc partial deletions (total n = 10) exhibit severe oligoasthenoteratozoospermia (OAT) defined as extremely reduced sperm counts (<5 × 106/ml) and concentration (<10 × 106/ejaculate) combined with low fraction of sperms with normal morphology (<4% normal forms) and motility (<32% progressive motile spermatozoa). Reference values for andrological parameters have been applied as referred in (B). As total testis volume is mostly within the expected range, their infertility is not caused by intrinsic congenital testicular damage but rather due to severe spermatogenic failure per se. Del, deletion; inv, inversion; dupl, duplication; n, number; sec, secondary; mill, million; ej., ejaculate.
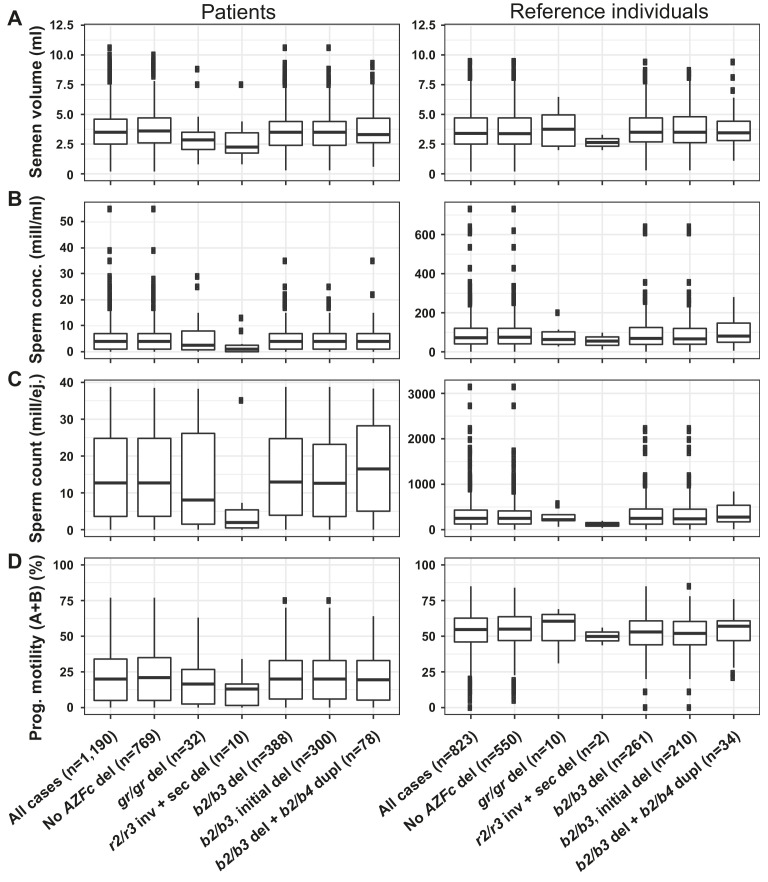
Figure 3—figure supplement 1. Distribution of seminal parameters in idiopathic male factor infertility cases with spermatogenic impairment and reference subjects.
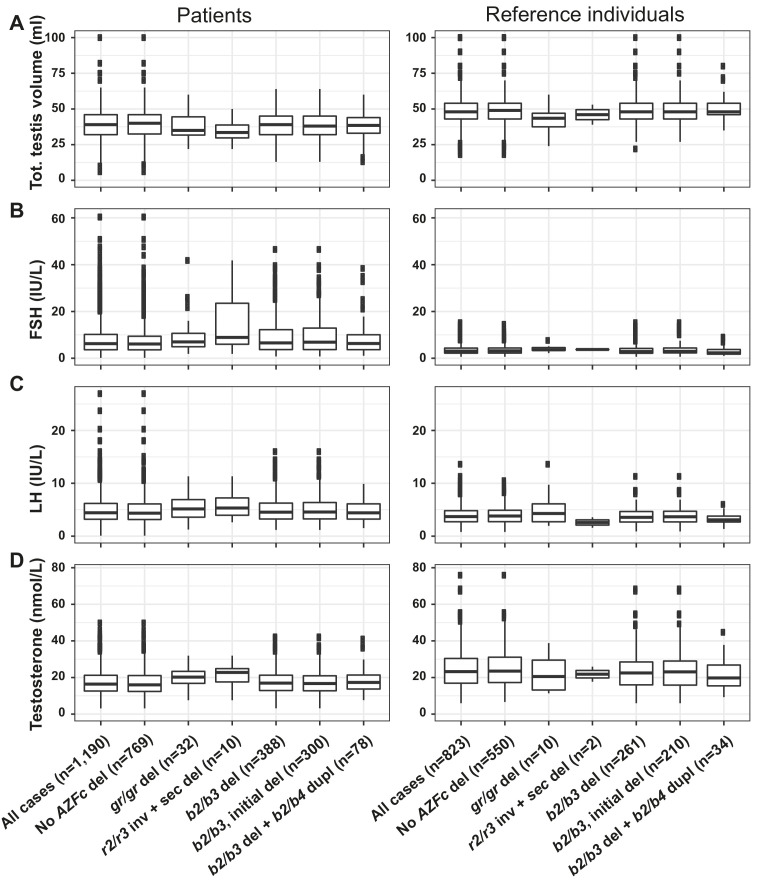
Figure 3—figure supplement 2. Distribution of hormonal and testicular parameters in idiopathic male factor infertility cases with spermatogenic impairment and reference individuals.
Figure 3—figure supplement 3. Distribution of andrological parameters in the idiopathic male factor infertility cases (total sperm counts 0–39 × 106) subgrouped based on the structure of the AZFc region.
Based on the Y-chromosomal phylogenetic data, one additional patient was identified as an obligate carrier of the r2/r3 inversion as his Y chromosome represents the lineage R1a1a1b1a1a1c-CTS11962.1 that was also identified in two cases with the r2/r3 inversion. This patient exhibited signs of unusual deletion and duplication events in the AZFc region as he carried six DAZ, four BPY2, and three copies of the CDY1 gene (Figure 2B, Table 2, Supplementary file 9).
Among the analyzed 2324 men, 13 cases with the complex AZFc rearrangement combining r2/r3 inversion with a subsequent deletion (from here on referred to as 'r2/r3 inversion plus deletion' for simplicity), represented 0.6% (Table 3). Considering the reported population prevalence of R1a1-M458 lineage in Estonians (5.1%; Underhill et al., 2015), the estimated number of subjects representing this Y lineage in the study group was ~119. Thus, approximately one in ten chromosomes with the r2/r3 inversion had undergone a subsequent deletion event (13/119, 11%).
Table 3. Enrichment of the AZFc r2/r3 inversion followed by a partial AZFc deletion in men with severe spermatogenic failure.
| AZFc r2/r3 inversion + AZFc partial deletion | ||||
|---|---|---|---|---|
| Group | All (n) | Estimated non-carriers (n) | Detected carriers (n) | % of carriers in the (sub)group |
| a. Full study group | ||||
| All analyzed study subjects | 2324 | 2311 | 13 | 0.6% |
| Study subjects with sperm counts | 2000 | 1988 | 12 | 0.6% |
| Subjects stratified based on total sperm counts per ejaculate | ||||
| Sperm counts 0–10 × 106 | 524 | 515 | 9 | 1.7% |
| Sperm counts > 10 × 106 | 1476 | 1473 | 3 | 0.2% |
| Fisher’s exact test, p=6.0×10−4, OR = 8.6 [95% CI 2.3–31.8] | ||||
| b. Carriers of the Y lineage R1a1a-M458* | ||||
| In all analyzed study subjects | 119 | 106 | 13 | 11.0% |
| In study subjects with sperm counts | 102 | 90 | 12 | 11.8% |
| Subjects stratified based on total sperm counts per ejaculate | ||||
| Sperm counts 0–10 × 106 | 27 | 18 | 9 | 33.7% |
| Sperm counts > 10 × 106 | 75 | 72 | 3 | 4.0% |
| Fisher’s exact test, p=3.0×10−4, OR = 12.0 [95% CI 2.9–48.9] | ||||
*Expected number of Y lineage R1a1-M458 in each subgroup was estimated using the known Estonian population prevalence 5.1% (Underhill et al., 2015).
r2/r3 inversion promotes recurrent deletions that lead to severe oligoasthenoteratozoospermia
Idiopathic infertility cases carrying the r2/r3 inversion plus deletion in the AZFc region (n = 10) exhibited extremely low sperm counts compared to subjects without any AZFc deletions (median 2.0 vs 12.5 × 106/ejaculate; Wilcoxon test, nominal p=0.011) (Figure 3B, Figure 3—figure supplements 1–3, Supplementary file 3). Nine of 10 men showed severe spermatogenic failure (total sperm counts <10 × 106/ejaculate), either azoospermia (n = 1), cryptozoospermia (n = 3), or severe oligozoospermia (n = 5) (Supplementary files 11 and 12). They also showed consistently the poorest sperm concentration (median 1.0 × 106/ml) and progressive motility (13%), as well as the lowest semen volume (2.3 ml) compared to the rest of analyzed infertile men. The data suggests that extreme oligoasthenoteratozoospermia (OAT) observed in these subjects was due to the severely affected process of spermatogenesis, whereas their testicular volume and hormonal profile were within the typical range of male factor infertility cases (Figure 3C).
When all the men with andrological data (n = 2000) were stratified based on sperm counts, there was a highly significant enrichment of r2/r3 inversion plus deletion in men with severe spermatogenic failure (sperm counts 0–10 × 106) compared to the rest (1.7% vs 0.2%, Fisher’s exact test, p=6.0×10−4, OR = 8.6 [95% CI 2.3–31.8]; Table 3). The estimated number of phenotyped subjects representing the Y haplogroup R1a1-M458 with the fixed r2/r3 inversion was 102 (based on population prevalence 5.1%; Underhill et al., 2015). Among carriers of this Y lineage, 33.7% of men with sperm counts 0–10 × 106 (9/27), but only 4.0% with sperm counts of >10 × 106 (3/72) had undergone a subsequent AZFc partial deletion (Fisher’s exact test, p=3.0×10−4, OR = 12.0 [95% CI 2.9–48.9]).
Only three reference cases carried a Y chromosome with the r2/r3 inversion plus deletion. At the time of phenotyping, all three subjects were younger (aged 18, 21, and 23 years) than the variant carriers in the idiopathic infertility group (median 32.4, range 26–51 years) (Supplementary file 12). The only reference subject with this complex AZFc rearrangement, but unaffected sperm analysis was the youngest (18 years). Notably, another reference man (23 years) with andrological data would actually be classified, based on WHO guidelines (World Health Organization, 2010), as an oligozoospermia case (sperm concentration 12 × 106/ml vs threshold 15 × 106/ml). Also, his total sperm counts (39.4 × 106/ejaculate) represented a borderline value.
Sequence diversity of the retained DAZ, BPY2, and CDY genes is extremely low and has no detectable effect on sperm parameters
The re-sequenced retained DAZ1-4, BPY2, and CDY1-2 genes were characterized by extremely low nucleotide variability in all Y-chromosomal lineages and deletion subtypes (Supplementary file 13). For 476 samples (gr/gr, n = 40; b2/b3, n = 436), re-sequenced for the >94 kb region using Illumina MiSeq, a total of 42 variants were identified with median 0.8 variants/kb and maximum two variants per individual. Most of them were previously undescribed (Giachini et al., 2008), singletons (Jobling and Tyler-Smith, 2017), and/or non-coding SNVs/short indels (Lu et al., 2011; Supplementary file 14). The CDY2a-CDY2b genes harbored only one variable site, whereas DAZ1-DAZ2 carried 24 or 26 SNVs/indels. Most variants appeared paralogous as both the reference and alternative alleles were identified. Among the four detected missense variants, CDY1b p.T419N was fixed in all three CDY1b copies present on the Y chromosome with the b2/b3 deletion plus b2/b4 duplications that represented an oligozoospermia case. However, the effect of this conservative substitution is unclear.
There was thus no evidence that the sequence variation in DAZ, BPY2, and CDY genes has any effect on infertility related parameters in the subjects examined.
Discussion
We conducted a comprehensive investigation of partial deletion subtypes of the Y-chromosomal AZFc region in 2324 Estonian men, approximately half with idiopathic spermatogenic impairment (n = 1190) in comparison to the reference group (n = 1134). Importantly, 2000 men had undergone full and uniformly conducted andrological workup at a single clinical center, facilitating fine-scale genotype–phenotype analysis. Previously, no study had undertaken re-sequencing of the retained DAZ, BPY2, and CDY genes along with the assessment of the Y haplogroup, dosage, and retained/deleted genes in the gr/gr or b2/b3-deleted chromosomes in both infertile men and controls. Concordant with the reports from other European populations, the gr/gr, but not the b2/b3 deletion, is a risk factor for spermatogenic impairment in Estonian men with >2-fold increased susceptibility to infertility (Figure 1D). However, the gathered data on the large group of reference men in the current study demonstrated the existence of Y chromosomes carrying a gr/gr deletion without any documented effect on andrological parameters (Supplementary file 3). As a novel finding, the study uncovered complex AZFc rearrangements within a specific Y haplogroup, R1a1-M458 and its sub-lineages, causing severe spermatogenic failure in the majority of carriers (Figure 3, Table 3). This Y lineage has undergone a ~1.6 Mb r2/r3 inversion in the AZFc region that has disrupted the structure of the palindromes P1 and P2, promoting subsequent recurrent deletions and consequently, severely impaired the process of spermatogenesis.
Consistent with key early observations (Rozen et al., 2012; Krausz et al., 2009; Machev, 2004), this study supports the recurrent nature and high subtype diversity of the AZFc partial losses that are currently considered jointly under the umbrella term ‘gr/gr deletions’. The 44 detected gr/gr deletions in our study sample were estimated to have originated independently at least 26 times across the Y phylogenetic tree and include seven different combinations of DAZ and CDY1 gene losses. Apparently, there is a substantial undescribed heterogeneity in the spread and structure of gr/gr deletions that in turn contributes to the phenotypic variability of the genetic effects. Unexpectedly, one in four Estonian gr/gr deletion carriers belonged to the Y-chromosomal haplogroup R1a1-M458 (and its sub-lineages) (Figure 2A,B; 22.7% vs 5.1% reported as the Estonian population frequency; Underhill et al., 2015). Notably, a previous study has reported a significant enrichment of the haplogroup R1a (ancestral lineage to the R1a1-M458) among gr/gr-deleted chromosomes in the Polish population (Rozen et al., 2012), which has a high prevalence, 25%, of R1a1-M458 (Underhill et al., 2015; Figure 4A, Supplementary file 15). All the Estonian gr/gr cases and also additional b2/b3 deletion chromosomes representing this Y lineage carried unusual retained DAZ gene pairs (DAZ1-DAZ3 or DAZ2-DAZ4) in combination with either CDY1a or CDY1b gene copy (Figure 2). These complex AZFc rearrangements were best explained by a preceding (and apparently fixed in R1a1-M458) ~1.6 Mb inversion between the homologous r2 and r3 amplicons, followed by recurrent secondary partial AZFc deletions (Figure 3A). The latter are facilitated by large ampliconic segments positioned in the same orientation. Inversions in the AZFc region are not uncommon, but none of the previously described inversions is expected to substantially disrupt the core palindromic structure of the AZFc region (Figure 1—figure supplement 1; Repping et al., 2004; Machev, 2004). In contrast, the r2/r3 inversion disrupts the structure of palindrome P1 and expands the size of the P2 palindrome more than twofold (Figure 3A). The critical role of intact P1–P2 palindromes in the AZFc structure is supported by the observation that no Y chromosomes have been described with a single DAZ gene copy, whereas the inverted DAZ gene pairs form the ‘heart’ of both P1 and P2. In future studies, long-read sequencing technologies should be applied to determine the detailed genomic structure of the AZFc region in the R1a1-M458 chromosomes and the exact chromosomal breakpoints of the identified r2/r3 inversion and secondary deletion events in oligozoospermia patients.
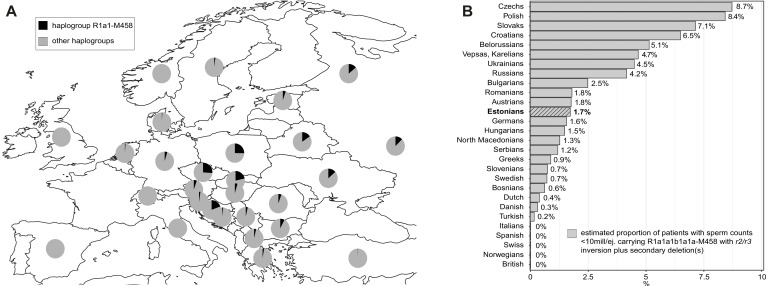
Figure 4. The prevalence of the Y-chromosomal haplogroup R1a1-M458 carrying a fixed r2/r3 inversion.
(A) Geographical distribution of haplogroup R1a1-M458 and its sub-lineages in Europe. Pie charts indicate populations, with the black sector showing the proportion of R1a1-M458 according to Underhill et al., 2015. (B) The estimated proportion of subjects among idiopathic cases with severe spermatogenic failure (sperm counts 0–10 × 106/ejaculate) carrying R1a1-M458 Y lineage (and its sub-lineages) chromosomes that have undergone a subsequent partial AZFc deletion. The prevalence was estimated using reported population frequencies of R1a1-M458, including Estonians (Underhill et al., 2015) and data available in the current study for Estonian men with spermatogenic failure. Estonians are shown in bold and with a striped filling. For full details, see Supplementary file 15.
As a likely scenario, the r2/r3 inversion plus deletion may predispose to spermatogenic impairment through substantial destabilization of the intra-chromosomal structure affecting meiotic recombination and chromosomal segregation. The removal of some specific genetic factor(s) being responsible for the phenotypic outcome seems a less likely explanation as the gr/gr deletion locations are variable, and so far, no reproducible associations with the exact deleted regions or specific gene copies have been identified. However, recent reports have uncovered an abundance of Y-chromosomal non-coding RNAs and their potential functional involvement in spermatogenesis (Johansson et al., 2019). The AZFc region contains at least one multicopy family of non-coding RNA genes, TTTY4 (Testis-Specific Transcript, Y-Linked 4) with testis-enriched expression. These genes are located in the three g1–g3 duplicons flanking the P1 and P2 palindromic ‘hearts’. The phenotypic consequences of TTTY4 copy number changes are still to be studied. Among 12 Estonian subjects carrying the AZFc r2/r3 inversion plus deletions and with available data for sperm counts, nine cases exhibited severe spermatogenic failure, two cases had moderate oligozoospermia, and only one case (aged 18 years) was normozoospermic (Figure 3B,C). This represented ~8- to 9-fold enrichment of this complex rearrangement among men with severely reduced sperm counts (0–10 × 106; Table 3). This genetic effect was observed specifically on the effectiveness of spermatogenesis, whereas the measurements of bitesticular volume and reproductive hormone levels did not stand out among the rest of analyzed infertile men. Unfortunately, none of the patients carrying the r2/r3 inversion plus deletion had undergone a testicular biopsy during their infertility workup. The histopathological pattern of germ cell abnormalities among these cases remains to be investigated in follow-up studies. This knowledge would facilitate understanding of the consequences this Y-chromosomal rearrangement on spermatogenesis and so maximize the benefit of molecular diagnostics in evidence-based clinical management decisions.
To our knowledge, no other Y-lineage-specific risk variants for spermatogenic impairment have been reported so far. Previously, the DAZ2–DAZ4 deletion had been shown as a high-risk factor for male infertility in the Tunisian population, but the Y haplogroups of those subjects were not investigated (Ghorbel et al., 2016). The survival of such a high-risk lineage in the population seems at first sight surprising, but may be accounted for by its possible age-specific effects on spermatogenesis, which may be exacerbated by the recent general decline in sperm count (Andersson et al., 2008). In the past, this lineage may not have been disadvantageous. The possible age-related progressive worsening of the reproductive phenotype among r2/r3 inversion plus deletion carriers should be investigated in follow-up, ideally longitudinal, studies of sufficiently large numbers of patients to make robust conclusions.
This study outcome has notable clinical implications for the improvement of molecular diagnostics and reducing the proportion of idiopathic male factor infertility cases. In Northern and Central Europe, the prevalence of R1a1-M458 haplogroup carrying the r2/r3 inversion ranges from ~1% in the Netherlands and Denmark to ~2–5% in Austria, Hungary, Germany, Baltics, and most Balkan countries, whereas it is widespread in Slavic populations and carried by 12–26% of men (Figure 4A, Supplementary file 15; Underhill et al., 2015). In non-European populations, the R1a1-M458 Y chromosomes are virtually non-existent (for details, see Underhill et al., 2015). However, in some European populations, recurrent secondary AZFc partial deletions on Y chromosomes representing the R1a1-M458 haplogroup (and its sub-lineages) may potentially explain from 0.3% up to ~9% of cases presenting severe spermatogenic impairment (sperm counts < 10 million per ejaculate) (Figure 4B). Further studies in other populations and large samples of patients and normozoospermic controls are required to fully establish the value of extending the current recommended testing of Y-chromosomal deletions by including the analysis of this novel Y-lineage-specific pathogenic AZFc rearrangement.
The evidence from the literature has shown that the increased prevalence of either gr/gr or b2/b3 deletions in infertility cases appears to be population-dependent (Bansal et al., 2016a; Bansal et al., 2016b). It can be speculated that also in other populations some specific Y lineages may carry AZFc structural variants that in combination with partial deletions (or other rearrangements) predispose to chromosomal instability in the complex process of spermatogenesis involving multiple well-coordinated cell divisions. So far, the largest study of the Y-chromosomal phylogeny of gr/gr deletion carriers included 152 infertile subjects representing seven countries with different population genetic structures (Krausz et al., 2009). However, the number of cases per population was low and the study included only 17 fertile men. Also, the study did not include fine-scale analysis of Y sub-lineages and the retained gene content. Long-range re-sequencing of the whole AZFc region in large numbers of men would be the preferred approach to uncover its structural complexity. Additional pathogenic AZFc rearrangements may also exist among Estonian infertile men. Even after omitting the cases carrying a gr/gr deletion at the r2/r3 inversion background, a non-significant enrichment of the remaining gr/gr deletion chromosomes can be observed in patients compared to reference men (1.9 vs 1.1%, p<0.1; Table 2).
In addition to the main finding, our deep re-sequencing dataset revealed that neither the dosage, sequence variation, nor exact copy of the retained DAZ, BPY2, and CDY1 gene showed any detectable effect on spermatogenic parameters. All chromosomes with AZFc partial deletions exhibit extremely low overall sequence variation of the retained DAZ, BPY2, and CDY genes. This observation is consistent with previous reports showing low levels of genetic diversity of the human Y chromosome (Jobling and Tyler-Smith, 2017) and suggesting that novel variants may be rapidly removed by active gene conversion among Y-chromosomal duplicate genes or selective constraint (Hallast et al., 2013; Rozen et al., 2003; Trombetta and Cruciani, 2017). Among the re-sequenced 382 chromosomes with b2/b3 deletions, no pathogenic mutations were detected in the single retained BPY2 and CDY1 gene copies. At the same time, the high rates of large structural rearrangements and copy number variation in the Y chromosome are well established, contrasting with low levels of sequence variation (Teitz et al., 2018; Shi et al., 2019). One in five or six Estonian Y chromosomes with gr/gr and b2/b3 deletions had undergone secondary rearrangements with no apparent effect on tested andrological parameters and fertility potential (Table 2). In the literature, the data about the effects of secondary duplications after an initial AZFc partial deletion on sperm parameters are inconclusive. Some studies have suggested increased pathogenicity (Lu et al., 2011; Yang et al., 2010; Lin et al., 2007; Ye et al., 2013; Yang et al., 2015), whereas others have reported neutral or even positive effects on spermatogenesis (Krausz et al., 2009; Giachini et al., 2008; Lo Giacco et al., 2014; Noordam et al., 2011). However, further copy number reductions in this genomic region appear to be very rare – none of the 44 gr/gr or 631 b2/b3 deletion carriers were identified with further reductions beyond what is expected from the initial deletion.
The current study is the largest and most detailed to date in terms of both the number of patients with spermatogenic impairment and reference samples with available andrological data from a single population, and detailed characterization of the genetic diversity of the AZFc region and phylogenetic background of the Y chromosomes. Yet, the total number of identified cases with the r2/r3 inversion followed by a deletion was relatively small and also inadequate to reach the statistical power in association testing with andrological parameters. Follow-up replication studies utilizing sample cohorts from populations with high(er) R1a1-M458 frequency (e.g. Polish, Czech) should be undertaken to confirm the prevalence and significance of the identified risk variant. The biggest challenge of such studies is the availability of sufficiently large sample collections of both patients and reference cases with andrological data. The identification of men with R1a1-M458 Y chromosomes and characterization of subsequent deletion subtypes only require standard inexpensive laboratory techniques such as PCR, restriction fragment length polymorphism (RFLP) analysis, and Sanger sequencing.
In summary, we have undertaken a comprehensive study of the carriers of AZFc partial gr/gr and b2/b3 deletions and uncovered high levels of structural variation in the AZFc locus, but low sequence diversity of the coding genes within the region. As a major finding, we discovered a large inversion specific to the Y lineage R1a1-M458 that represents a hotspot for subsequent AZFc partial deletions. Men carrying Y chromosomes with this complex rearrangement have >10 fold increased risk of severe spermatogenic failure, but the consequences of this risk could potentially be alleviated by early identification of the variant carriers and facilitating the storage of their sperm samples. Our study results thus have the potential to improve clinical diagnostics and management of idiopathic impaired spermatogenesis in a significant fraction of men originating from Northern and Central European populations.
Materials and methods
Study subjects
Patients with idiopathic spermatogenic impairment (n = 1190) were recruited at the Andrology Centre at Tartu University Hospital (AC-TUH) in 2003–2015 (PI: M. Punab). Included cases showed reduced sperm counts (<39 × 106/ejaculate) in at least two consecutive semen analyses (World Health Organization, 2010). Recruitment and sampling, semen analyses, hormone assays, and definition of idiopathic cases have previously been described in detail (Punab et al., 2017). Men with known causes of male infertility detected during routine diagnostic workup were excluded, for example cryptorchidism, testicular cancer, orchitis/epididymitis, mumps orchitis, testis trauma, karyotype abnormalities, and complete Y-chromosomal microdeletions. The final idiopathic infertility group included 104 azoospermia (no sperm), 88 cryptozoospermia (sperm counts > 0–1 × 106/ejaculate), 319 severe oligozoospermia (1–10 × 106), and 679 moderate oligozoospermia (10–38 × 106) cases (Table 1, Supplementary file 1).
The reference sample of Estonian men (n = 1134) comprised healthy young men (n = 499) and subjects with proven fatherhood (n = 635) (Table 1, Supplementary file 1). The cohort of ‘Estonian young men’ (n = 499) was recruited at the AC-TUH in 2003–2004 (PI: M. Punab), representing a healthy male group with median age 18.6 (17.2–22.9) years at the time of recruitment (Grigorova et al., 2008). The subgroup of ‘Partners of pregnant women’ (n = 324) includes male partners of pregnant women, recruited in 2010–2014 at the Tartu University Hospital and the West Tallinn Central Hospital (Punab et al., 2017). Eight hundered and ten men (of 823) in these subgroups underwent sperm analysis.
The subgroup of ‘REPROMETA proven fathers’ (n = 311) was recruited in 2006–2011 at the Women's Clinic at Tartu University Hospital during the REPROMETA study (PI: M. Laan), originally designed to collect mother–father–placenta trios at delivery to investigate genetics of pregnancy complications (Kikas et al., 2020; Pilvar et al., 2019). In this study, the REPROMETA fathers represented reference men with proven fertility. Only self-reported age and body mass index (BMI) data were available for this subgroup.
All men, who had turned to Andrology Centre, Tartu University Hospital (AC-TUH) due to idiopathic infertility (n = 1190), as well as the participants of the 'Estonian young men' cohort (n = 499) and the subgroup ‘Partners of pregnant women' (n = 324) were offered complete routine andrological workup. The subjects were examined by specialist andrologists at the AC-TUH, who had received respective training in clinical assessment and standardized andrological workup, locally and in collaboration with other European Andrology Academy (EAA)-accredited centers. Also, anthropometric parameters were documented during clinical examination. Details are described in Punab et al., 2017.
Physical examination for the assessment of genital pathology and testicular size (orchidometer; made of birch wood, Pharmacia and Upjohn, Denmark) was performed with the patients in standing position. The total testis volume is the sum of right and left testicles. The position of the testicles in the scrotum, pathologies of the genital ducts (epididymitis and ductus deference), and the penis, urethra, presence, and, if applicable, grade of varicocele were registered for each subject.
For 2000 study subjects, sperm analysis was performed, whereas 13 reference cases did not agree with this procedure. Semen samples were obtained by patient masturbation, and semen analysis was performed in accordance with the World Health Organization (WHO) recommendations. In brief, after ejaculation, the semen was incubated at 37°C for 30–40 min for liquefaction. Semen volume was estimated by weighing the collection tube with the semen sample and subsequently subtracting the predetermined weight of the empty tube assuming 1 g = 1 ml. For assessment of the spermatozoa concentration, the samples were diluted in a solution of 0.6 mol/l NaHCO3% and 0.4% (v/v) formaldehyde in distilled water. The spermatozoa concentration was assessed using the improved Neubauer haemocytometers.
Genomic DNA was extracted from EDTA-blood. After blood draw in the morning, serum and plasma fractions were separated immediately for hormone measurements (FSH, LH, testosterone). All laboratory analyses and routine genetic testing (karyotyping, Y-chromosomal microdeletions) were performed at the United Laboratories of Tartu University Hospital according to the established clinical laboratory guidelines. Detailed methodology and reference values for hormonal levels are available by the service provider: https://www.kliinikum.ee/yhendlabor/analueueside-taehestikuline-register.
Genotyping Y-chromosomal microdeletions
All study subjects (n = 2324) were typed for complete AZFa (loss of markers sY84 and sY86), AZFb (sY127 and sY134), AZFc (sY254 and sY255), and partial AZFc deletions gr/gr (sY1291), b2/b3 (sY1191), and b1/b3 (sY1161, sY1191, and sY1291) following established PCR primers (Krausz et al., 2014; Lin et al., 2006; Supplementary file 16). The multiplex PCR contained the final concentrations of 1× PCR buffer B1 (Solis Biodyne, Estonia), 2.5 mM MgCl2, 2.5 mM dNTP, 2 µM PCR primers for STS markers sY1291 and sY1201, 3 µM PCR primers for STS markers sY1191, sY1206, and sY1161 (Supplementary file 16), 1U FIREPol DNA polymerase (Solis Biodyne), and 10 ng of template genomic DNA per reaction. The following PCR conditions were used: for 5 min at 95°C, followed by 32 cycles of 30 s at 95°C, 30 s at 63°C and 1 min at 72°C, final extension of 10 min at 72°C and a 4°C hold. The presence/absence of PCR products in a reaction were checked on 2% agarose gel. Lack of amplification of STS marker sY1291 (but presence of all others) was used to determine the gr/gr deletion and lack of sY1191 the b2/b3 deletion.
Re-sequencing of retained DAZ, BPY2, and CDY genes using Illumina MiSeq
Re-sequencing of the exonic regions of the retained AZFc genes (according to Ensembl release 84, RRID:SCR_002344) in 476 cases with either gr/gr or b2/b3 deletions targeted in total 94,188 bp per subject. CDY, BPY2, and DAZ genes were amplified using 8, 10, or 26 PCR primer pairs, respectively (Supplementary files 17 and 18). The presence of all amplicons was confirmed using gel electrophoresis. Amplicons were pooled in equimolar concentrations, barcoded per sample, and sequenced (250 bp reads, paired-end) on Illumina MiSeq (RRID:SCR_016379) with at least 40× coverage. BWA (v0.7.15, RRID:SCR_010910) (Li and Durbin, 2009) was implemented to map the sequencing reads to a modified human genome reference (GRChg38), where CDY1a, CDY2a, BPY2a and either DAZ3-DAZ4 (gr/gr carriers) or DAZ1-DAZ2 (b2/b3 carriers) remained unchanged, but the sequences of other CDY, BPY2, and DAZ gene copies were replaced with 'Ns'. SNVs and indels were identified using GATK HaplotypeCaller (v3.7, RRID:SCR_001876), with a minimum base quality 20 and outputting all sites (McKenna et al., 2010). Y-chromosomal phylogenetic markers were called using bcftools (v1.8, RRID:SCR_005227), with minimum base quality 20, mapping quality 20 and defining ploidy as 1.
Re-sequencing included 31 patients and nine reference men with AZFc gr/gr deletions. Six men carrying gr/gr deletions were not analyzed due to DNA limitations (two cases) or unavailable andrological data (four cases). The analysis of b2/b3 deletion carriers included 382 patients (haplogroup N3: n = 380; non-N3, n = 2) and 54 'Partners of pregnant women’ (N3: n = 53; non-N3, n = 1).
Analysis of variant effects from the illumina MiSeq dataset
Variant effect prediction was performed using the Variant Effect Predictor tool (VEP, https://www.ensembl.org/Tools/VEP, Ensembl release 99, RRID:SCR_007931) (McLaren et al., 2016). The Combined Annotation Dependent Depletion (CADD) score ≥ 20, that is, including variants among the top 1% of deleterious variants in the human genome, was considered indicative of potential functional importance of identified SNVs in the coding regions (Rentzsch et al., 2019).
Y-chromosomal haplogroup typing
Y lineages of the gr/gr samples were defined using 14 markers included in the re-sequencing, plus 34 additional markers determined by Sanger sequencing or restriction fragment length polymorphism (RFLP) analysis (Supplementary files 18 and 19). The b2/b3-deletion carriers were typed for Y marker N3-M46 (Tat) (Zerjal et al., 1997). The sub-lineages of the re-sequenced haplogroup N3 samples were defined in more detail using 16 phylogenetic markers from the Illumina MiSeq dataset, following established nomenclature (Ilumäe et al., 2016; Karmin et al., 2015). For the other haplogroups, nomenclature according to the International Society of Genetic Genealogy (ISOGG, version 14.14) was followed.
The R1a1a1b1a1a-lineage-specific phylogenetic marker M458 (rs375323198, A > G polymorphism, GRCh38 genomic coordinate: chrY: 22220317), indicating the carrier status of r2/r3 inverted Y chromosome was amplified using the following conditions: PCR contained the final concentrations of 1× PCR buffer B1 (Solis Biodyne), 2.5 mM MgCl2, 2.5 mM dNTP, 10 µM forward and reverse PCR primers for M458 (see Supplementary file 19 for primer sequences), 1U FIREPol DNA polymerase (Solis Biodyne OÜ), and 10 ng of template genomic DNA per reaction. The following PCR conditions were used: for 5 min at 95°C, followed by 32 cycles of 30 s at 95°C, 30 s at 52°C and 1 min at 72°C, final extension of 10 min at 72°C and a 4°C hold. The presence of the M458 marker in derived state (instead of the ancestral allele ‘A’ presence of allele ‘G’ at position 147) was determined using Sanger sequencing.
Determination of DAZ, BPY2, and CDY gene dosage and gene types
The Bio-Rad QX 200 Droplet Digital PCR system (RRID:SCR_019707) was used to quantify the copy numbers of the retained DAZ, BPY2, and CDY genes for 44 gr/gr deletion carriers (31 cases, 13 reference men with sperm analysis data) and 631 b2/b3 carriers (382 cases/249 reference men) (Figure 2—figure supplement 1). The PCR primers and probes were designed using Primer3plus (version 2.4.2, RRID:SCR_003081), PCR were performed according to the recommendations in the Droplet Digital PCR Application Guide (Bio-Rad, U.S.) (Supplementary file 20) and described in Shi et al., 2019. A XQ200 Droplet Reader was used to measure the fluorescence of each droplet and QuantaSoft software (v1.6.6.0320; Bio-Rad) to cluster droplets into distinct fluorescent groups. The copy number of each gene was determined by calculating the ratio of target (unknown – DAZ, BPY2, or CDY) and reference (single-copy SRY gene) concentration. ddPCR for each gene were performed once for every sample. For samples carrying the gr/gr deletion, if the copy number obtained differed from the expected (two copies of DAZ and BPY2, three copies of CDY), then the ddPCR reaction was repeated. For b2/b3 carriers, typing was repeated for all samples not carrying the two most typical copy numbers (2-1-3 or 4-2-4 copies of DAZ, BPY2, and CDY genes, respectively). Additionally, a total of 5% of random samples were replicated. If the copy number estimates between replicates differed by 0.8 or more, then a third replicate was performed, and the final copy number was calculated as average of the two closest replicates.
The re-sequencing data of the DAZ genes covered nine paralogous sequence variants that were used to determine the retained gene copies in the gr/gr and b2/b3 deletion carriers (Supplementary file 21). For the validation of the DAZ gene copy mapping approach, at least five gr/gr carriers were additionally typed for published SNV combinations differentiating the DAZ gene copies (Machev, 2004; Fernandes et al., 2002). The retained CDY1 gene was identified according to Machev, 2004.
Genetic association testing with andrological parameters
Statistical testing for the associations between AZFc gr/gr or b2/b3 deletions and andrological parameters was conducted using RStudio (version 1.2.1335, RRID:SCR_000432), and data were visualised using ggplot2 (version 3.2.1, RRID:SCR_014601) (Wickham, 2009). Differences in continuous clinical variables between groups were compared using the non-parametric pairwise Wilcoxon rank-sum test.
Genetic association with the carrier status of b2/b3 deletion and its subtypes was also tested using linear regression analyses adjusted for age. For sperm parameters abstinence time and for total testosterone levels, BMI estimates were additionally used as cofactors. Natural log transformation was used to achieve an approximately normal distribution of values. In all cases (except total sperm counts), the applied transformation resulted in a close-to-normal distribution of values. For the linear regression analyses, statistical significance threshold after correction for multiple testing was estimated p<1.0×10−3 (six tests × eight independent parameters).
Acknowledgements
We thank all the patients for making this study possible. The clinical team at the Andrology Centre, Tartu University Hospital is thanked for the professional phenotyping and assistance in patient recruitment over many years. Mart Adler and Eve Laasik are specifically acknowledged for the management of the Androgenetics Biobank, and all ML team members are thanked for their contributions to the DNA extractions. Our work was supported by the Wellcome Trust (098051). This work was supported by Estonian Research Council Grants PUT1036 (PH and LK), PUT181 (MP, KP, and OP), IUT34-12 (LK, MG, KR, and ML), PRG1021 (MP, KR, and ML), IUT24-1 (RF and SR), and MOBTT53 (SR). Establishment of the cohort of Estonian infertility subjects was also supported by the EU through the European Regional Development Fund, project HAPPY PREGNANCY, no. 3.2.0701.12–004 (ML, MP, and KR).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Pille Hallast, Email: pille.hallast@ut.ee.
Maris Laan, Email: maris.laan@ut.ee.
George H Perry, Pennsylvania State University, United States.
George H Perry, Pennsylvania State University, United States.
Funding Information
This paper was supported by the following grants:
Estonian Research Council PUT1036 to Pille Hallast, Laura Kibena.
Estonian Research Council PUT181 to Margus Punab, Olev Poolamets, Kristjan Pomm.
Wellcome Trust 098051 to Pille Hallast, Laura Kibena, Elena Arciero, Yali Xue, Chris Tyler-Smith.
Estonian Research Council IUT34-12 to Laura Kibena, Marina Grigorova, Kristiina Rull, Maris Laan.
Estonian Research Council IUT24-1 to Siiri Rootsi, Rodrigo Flores.
Estonian Research Council MOBTT53 to Siiri Rootsi.
European Regional Development Fund 3.2.0701.12-004 to Margus Punab, Kristiina Rull, Maris Laan.
Estonian Research Council PRG1021 to Margus Punab, Kristiina Rull, Maris Laan.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing - original draft, Project administration, Writing - review and editing.
Data curation, Formal analysis, Validation, Investigation, Methodology, Writing - review and editing.
Resources, Data curation, Supervision, Writing - review and editing.
Investigation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Data curation, Writing - review and editing.
Resources, Supervision, Writing - review and editing.
Resources, Supervision, Funding acquisition, Writing - review and editing.
Resources, Formal analysis, Supervision, Funding acquisition, Investigation, Writing - original draft, Writing - review and editing.
Ethics
Human subjects: The study was approved by the Ethics Review Committee on Human Research of the University of Tartu, Estonia (permissions 146/18, 152/4, 221/T-6, 221/M-5, 272/M-13, 267M-13, 286M-18, 288M-13), and sequencing/genotyping was approved at the Wellcome Sanger Institute under WTSI HMDMC 17/105. Written informed consent for evaluation and use of their clinical data for scientific purposes was obtained from each person prior to recruitment. All procedures and methods have been carried out in compliance with the guidelines of the Declaration of Helsinki.
Additional files
(a) Y haplogroup distribution of the Estonian patients with idiopathic spermatogenic impairment and reference cases carrying gr/gr deletions.
(b) Enrichment of Y-chromosomal lineage R1a1a1b1a1a-M458 in men carrying gr/gr deletion.
Data availability
Illumina MiSeq re-sequencing data are available through the European Genome-phenome Archive (EGA, https://www.ebi.ac.uk/) under the accession number: EGAS00001002157.
The following dataset was generated:
Hallast P, Kibena L, Punab M, Laan M, Xue Y, Tyler-Smith C. 2017. Resequencing candidate genes for male spermatogenic impairment. EGA. EGAS00001002157
References
- Andersson AM, Jørgensen N, Main KM, Toppari J, Rajpert-De Meyts E, Leffers H, Juul A, Jensen TK, Skakkebaek NE. Adverse trends in male reproductive health: we may have reached a crucial 'tipping point'. International Journal of Andrology. 2008;31:74–80. doi: 10.1111/j.1365-2605.2007.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASRM Diagnostic evaluation of the infertile male: a committee opinion. Fertility and Sterility. 2015;103:e18–e25. doi: 10.1016/j.fertnstert.2014.12.103. [DOI] [PubMed] [Google Scholar]
- Bansal SK, Jaiswal D, Gupta N, Singh K, Dada R, Sankhwar SN, Gupta G, Rajender S. Gr/gr deletions on Y-chromosome correlate with male infertility: an original study, meta-analyses, and trial sequential analyses. Scientific Reports. 2016a;6:19798. doi: 10.1038/srep19798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal SK, Gupta G, Rajender S. Y chromosome b2/b3 deletions and male infertility: a comprehensive meta-analysis, trial sequential analysis and systematic review. Mutation Research/Reviews in Mutation Research. 2016b;768:78–90. doi: 10.1016/j.mrrev.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Colaco S, Modi D. Genetics of the human Y chromosome and its association with male infertility. Reproductive Biology and Endocrinology. 2018;16:14. doi: 10.1186/s12958-018-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L, Taylor KD, Lin HJ, Rich SS, Post WS, Chen YI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME, Genome Aggregation Database Production Team. Genome Aggregation Database Consortium A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Palmer MJ, Tanton C, Gibson LJ, Jones KG, Macdowall W, Glasier A, Sonnenberg P, Field N, Mercer CH, Johnson AM, Wellings K. Prevalence of infertility and help seeking among 15 000 women and men. Human Reproduction. 2016;31:2108–2118. doi: 10.1093/humrep/dew123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho CMB, Zuccherato LW, Fujisawa M, Shirakawa T, Ribeiro-Dos-Santos AKC, Santos SEB, Pena SDJ, Santos FR. Study of AZFc partial deletion gr/gr in fertile and infertile japanese males. Journal of Human Genetics. 2006;51:794–799. doi: 10.1007/s10038-006-0024-2. [DOI] [PubMed] [Google Scholar]
- Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, Rajpert De Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M, Barros A, Vogt PH. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Molecular Human Reproduction. 2002;8:286–298. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. The American Journal of Human Genetics. 2004;74:180–187. doi: 10.1086/381132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel M, Baklouti-Gargouri S, Keskes R, Chakroun N, Sellami A, Fakhfakh F, Ammar-Keskes L. Gr/gr-DAZ2-DAZ4-CDY1b deletion is a high-risk factor for male infertility in tunisian population. Gene. 2016;592:29–35. doi: 10.1016/j.gene.2016.07.050. [DOI] [PubMed] [Google Scholar]
- Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the italian population. Human Genetics. 2008;124:399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- Grigorova M, Punab M, Ausmees K, Laan M. FSHB promoter polymorphism within evolutionary conserved element is associated with serum FSH level in men. Human Reproduction. 2008;23:2160–2166. doi: 10.1093/humrep/den216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, Balaresque P, Bowden GR, Ballereau S, Jobling MA. Recombination dynamics of a human Y-chromosomal palindrome: rapid GC-biased gene conversion, multi-kilobase conversion tracts, and rare inversions. PLOS Genetics. 2013;9:e1003666. doi: 10.1371/journal.pgen.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilumäe AM, Reidla M, Chukhryaeva M, Järve M, Post H, Karmin M, Saag L, Agdzhoyan A, Kushniarevich A, Litvinov S, Ekomasova N, Tambets K, Metspalu E, Khusainova R, Yunusbayev B, Khusnutdinova EK, Osipova LP, Fedorova S, Utevska O, Koshel S, Balanovska E, Behar DM, Balanovsky O, Kivisild T, Underhill PA, Villems R, Rootsi S. Human Y chromosome haplogroup N: a Non-trivial Time-Resolved phylogeography that cuts across language families. The American Journal of Human Genetics. 2016;99:163–173. doi: 10.1016/j.ajhg.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. Human Y-chromosome variation in the genome-sequencing era. Nature Reviews Genetics. 2017;18:485–497. doi: 10.1038/nrg.2017.36. [DOI] [PubMed] [Google Scholar]
- Johansson MM, Pottmeier P, Suciu P, Ahmad T, Zaghlool A, Halvardson J, Darj E, Feuk L, Peuckert C, Jazin E. Novel Y-Chromosome long Non-Coding RNAs expressed in human male CNS during early development. Frontiers in Genetics. 2019;10:891. doi: 10.3389/fgene.2019.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G, Krausz CI. European association of urology working group on male, european association of urology guidelines on male infertility: the 2012 update. European Urology. 2012;62:324–332. doi: 10.1016/j.eururo.2012.04.048. [DOI] [PubMed] [Google Scholar]
- Karmin M, Saag L, Vicente M, Wilson Sayres MA, Järve M, Talas UG, Rootsi S, Ilumäe AM, Mägi R, Mitt M, Pagani L, Puurand T, Faltyskova Z, Clemente F, Cardona A, Metspalu E, Sahakyan H, Yunusbayev B, Hudjashov G, DeGiorgio M, Loogväli EL, Eichstaedt C, Eelmets M, Chaubey G, Tambets K, Litvinov S, Mormina M, Xue Y, Ayub Q, Zoraqi G, Korneliussen TS, Akhatova F, Lachance J, Tishkoff S, Momynaliev K, Ricaut FX, Kusuma P, Razafindrazaka H, Pierron D, Cox MP, Sultana GN, Willerslev R, Muller C, Westaway M, Lambert D, Skaro V, Kovačevic L, Turdikulova S, Dalimova D, Khusainova R, Trofimova N, Akhmetova V, Khidiyatova I, Lichman DV, Isakova J, Pocheshkhova E, Sabitov Z, Barashkov NA, Nymadawa P, Mihailov E, Seng JW, Evseeva I, Migliano AB, Abdullah S, Andriadze G, Primorac D, Atramentova L, Utevska O, Yepiskoposyan L, Marjanovic D, Kushniarevich A, Behar DM, Gilissen C, Vissers L, Veltman JA, Balanovska E, Derenko M, Malyarchuk B, Metspalu A, Fedorova S, Eriksson A, Manica A, Mendez FL, Karafet TM, Veeramah KR, Bradman N, Hammer MF, Osipova LP, Balanovsky O, Khusnutdinova EK, Johnsen K, Remm M, Thomas MG, Tyler-Smith C, Underhill PA, Willerslev E, Nielsen R, Metspalu M, Villems R, Kivisild T. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Research. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikas T, Inno R, Ratnik K, Rull K, Laan M. C-allele of rs4769613 near FLT1 represents a High-Confidence placental risk factor for preeclampsia. Hypertension. 2020;76:884–891. doi: 10.1161/HYPERTENSIONAHA.120.15346. [DOI] [PubMed] [Google Scholar]
- Kohn TP, Kohn JR, Owen RC, Coward RM. The prevalence of Y-chromosome microdeletions in Oligozoospermic men: a systematic review and Meta-analysis of european and north american studies. European Urology. 2019;76:626–636. doi: 10.1016/j.eururo.2019.07.033. [DOI] [PubMed] [Google Scholar]
- Krausz C, Giachini C, Xue Y, O'Bryan MK, Gromoll J, Rajpert-de Meyts E, Oliva R, Aknin-Seifer I, Erdei E, Jorgensen N, Simoni M, Ballescà JL, Levy R, Balercia G, Piomboni P, Nieschlag E, Forti G, McLachlan R, Tyler-Smith C. Phenotypic variation within european carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. Journal of Medical Genetics. 2009;46:21–31. doi: 10.1136/jmg.2008.059915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Hoefsloot L, Simoni M, Tüttelmann F, European Academy of Andrology. European Molecular Genetics Quality Network EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2:5–19. doi: 10.1111/j.2047-2927.2013.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Casamonti E. Spermatogenic failure and the Y chromosome. Human Genetics. 2017;136:637–655. doi: 10.1007/s00439-017-1793-8. [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genetics. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YW, Hsu CL, Yen PH. A two-step protocol for the detection of rearrangements at the AZFc region on the human Y chromosome. Molecular Human Reproduction. 2006;12:347–351. doi: 10.1093/molehr/gal038. [DOI] [PubMed] [Google Scholar]
- Lin YW, Hsu LC, Kuo PL, Huang WJ, Chiang HS, Yeh SD, Hsu TY, Yu YH, Hsiao KN, Cantor RM, Yen PH. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in han chinese in Taiwan. Human Mutation. 2007;28:486–494. doi: 10.1002/humu.20473. [DOI] [PubMed] [Google Scholar]
- Lo Giacco D, Chianese C, Sánchez-Curbelo J, Bassas L, Ruiz P, Rajmil O, Sarquella J, Vives A, Ruiz-Castañé E, Oliva R, Ars E, Krausz C. Clinical relevance of Y-linked CNV screening in male infertility: new insights based on the 8-year experience of a diagnostic genetic laboratory. European Journal of Human Genetics. 2014;22:754–761. doi: 10.1038/ejhg.2013.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhang F, Yang H, Xu M, Du G, Wu W, An Y, Qin Y, Ji G, Han X, Gu A, Xia Y, Song L, Wang S, Jin L, Wang X. Additional genomic duplications in AZFc underlie the b2/b3 deletion-associated risk of spermatogenic impairment in han chinese population. Human Molecular Genetics. 2011;20:4411–4421. doi: 10.1093/hmg/ddr369. [DOI] [PubMed] [Google Scholar]
- Machev N. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. Journal of Medical Genetics. 2004;41:814–825. doi: 10.1136/jmg.2004.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The ensembl variant effect predictor. Genome Biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordam MJ, Westerveld GH, Hovingh SE, van Daalen SK, Korver CM, van der Veen F, van Pelt AM, Repping S. Gene copy number reduction in the azoospermia factor c (AZFc) region and its effect on total motile sperm count. Human Molecular Genetics. 2011;20:2457–2463. doi: 10.1093/hmg/ddr119. [DOI] [PubMed] [Google Scholar]
- Olesen IA, Andersson AM, Aksglaede L, Skakkebaek NE, Rajpert-de Meyts E, Joergensen N, Juul A. Clinical, genetic, biochemical, and testicular biopsy findings among 1,213 men evaluated for infertility. Fertility and Sterility. 2017;107:74–82. doi: 10.1016/j.fertnstert.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Pilvar D, Reiman M, Pilvar A, Laan M. Parent-of-origin-specific allelic expression in the human placenta is limited to established imprinted loci and it is stably maintained across pregnancy. Clinical Epigenetics. 2019;11:94. doi: 10.1186/s13148-019-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punab M, Poolamets O, Paju P, Vihljajev V, Pomm K, Ladva R, Korrovits P, Laan M. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Human Reproduction. 2017;32:18–31. doi: 10.1093/humrep/dew284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nature Genetics. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. A family of human Y chromosomes has dispersed throughout northern eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, Oates RD, Silber SJ, Ardlie K, Page DC. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. The American Journal of Human Genetics. 2012;91:890–896. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Louzada S, Grigorova M, Massaia A, Arciero E, Kibena L, Ge XJ, Chen Y, Ayub Q, Poolamets O, Tyler-Smith C, Punab M, Laan M, Yang F, Hallast P, Xue Y. Evolutionary and functional analysis of RBMY1 gene copy number variation on the human Y chromosome. Human Molecular Genetics. 2019;28:2785–2798. doi: 10.1093/hmg/ddz101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Stahl PJ, Masson P, Mielnik A, Marean MB, Schlegel PN, Paduch DA. A decade of experience emphasizes that testing for Y microdeletions is essential in american men with azoospermia and severe oligozoospermia. Fertility and Sterility. 2010;94:1753–1756. doi: 10.1016/j.fertnstert.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Teitz LS, Pyntikova T, Skaletsky H, Page DC. Selection has countered high mutability to preserve the ancestral copy number of Y chromosome amplicons in diverse human lineages. The American Journal of Human Genetics. 2018;103:261–275. doi: 10.1016/j.ajhg.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta B, Cruciani F. Y chromosome palindromes and gene conversion. Human Genetics. 2017;136:605–619. doi: 10.1007/s00439-017-1777-8. [DOI] [PubMed] [Google Scholar]
- Tüttelmann F, Werny F, Cooper TG, Kliesch S, Simoni M, Nieschlag E. Clinical experience with azoospermia: aetiology and chances for spermatozoa detection upon biopsy. International Journal of Andrology. 2011;34:291–298. doi: 10.1111/j.1365-2605.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Poznik GD, Rootsi S, Järve M, Lin AA, Wang J, Passarelli B, Kanbar J, Myres NM, King RJ, Di Cristofaro J, Sahakyan H, Behar DM, Kushniarevich A, Sarac J, Saric T, Rudan P, Pathak AK, Chaubey G, Grugni V, Semino O, Yepiskoposyan L, Bahmanimehr A, Farjadian S, Balanovsky O, Khusnutdinova EK, Herrera RJ, Chiaroni J, Bustamante CD, Quake SR, Kivisild T, Villems R. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. European Journal of Human Genetics. 2015;23:124–131. doi: 10.1038/ejhg.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009. [DOI] [Google Scholar]
- World Health Organization . WHO Laboratory Manual for the Examination and Processing of Human Semen. Geneva: World Health Organization; 2010. [Google Scholar]
- Yang Y, Ma M, Li L, Su D, Chen P, Ma Y, Liu Y, Tao D, Lin L, Zhang S. Differential effect of specific gr/gr deletion subtypes on spermatogenesis in the chinese han population. International Journal of Andrology. 2010;33:745–754. doi: 10.1111/j.1365-2605.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- Yang B, Ma YY, Liu YQ, Li L, Yang D, Tu WL, Shen Y, Dong Q, Yang Y. Common AZFc structure may possess the optimal spermatogenesis efficiency relative to the rearranged structures mediated by non-allele homologous recombination. Scientific Reports. 2015;5:10551. doi: 10.1038/srep10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JJ, Ma L, Yang LJ, Wang JH, Wang YL, Guo H, Gong N, Nie WH, Zhao SH. Partial AZFc duplications not deletions are associated with male infertility in the yi population of Yunnan Province, China. Journal of Zhejiang University. Science. B. 2013;14:807–815. doi: 10.1631/jzus.B1200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhövel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C. Genetic relationships of asians and northern europeans, revealed by Y-chromosomal DNA analysis. American Journal of Human Genetics. 1997;60:1174–1183. [PMC free article] [PubMed] [Google Scholar]