Abstract
Obstructive sleep apnea (OSA) is a common sleep-related respiratory disorder. Around the world, more and more people are suffering from OSA. Because of the limitation of monitor equipment, many people with OSA remain undetected. Therefore, we propose a sleep-monitoring model based on single-channel electrocardiogram using a convolutional neural network (CNN), which can be used in portable OSA monitor devices. To learn different scale features, the first convolution layer comprises three types of filters. The long short-term memory (LSTM) is used to learn the long-term dependencies such as the OSA transition rules. The softmax function is connected to the final fully connected layer to obtain the final decision. To detect a complete OSA event, the raw ECG signals are segmented by a 10 s overlapping sliding window. The proposed model is trained with the segmented raw signals and is subsequently tested to evaluate its event detection performance. According to experiment analysis, the proposed model exhibits Cohen's kappa coefficient of 0.92, a sensitivity of 96.1%, a specificity of 96.2%, and an accuracy of 96.1% with respect to the Apnea-ECG dataset. The proposed model is significantly higher than the results from the baseline method. The results prove that our approach could be a useful tool for detecting OSA on the basis of a single-lead ECG.
1. Introduction
Obstructive sleep apnea (OSA) is a major sleep-disordered breathing (SDB) syndrome that is an independent risk factor of coronary heart disease, hypertension, and arrhythmia [1]. According to the manual of the American Academy of Sleep Medicine (AASM) [2], OSA in adults is scored when there is a 90% or more reduction in the baseline of the oral and nasal respiration amplitude for 10 s or more, occuring during sleep. This condition is associated with repetitive airflow limitation and sleep fragmentation, decreasing the sleep time and degrading the sleep quality of the OSA patients [3]. OSA not only causes excessive daytime neurocognitive deficits, drowsiness, depression, fatigue, and heart stroke [4–6] but can also cause a brain stroke, high blood pressure, arrhythmias, myocardial infarction, and ischemia [7–9]. According to the AASM [2], polysomnography (PSG) is considered to be the gold standard for OSA detection, which is based on a comprehensive evaluation of the sleep signals [10]. PSG involves overnight recording of the patient and the measurement of many signals using the sensors attached to the body, e.g., an electroencephalogram (EEG), electromyogram (EMG), electrocardiogram (ECG), and electrooculogram (EOG), to monitor the respiratory effort and other biophysiological signals [1]. After collecting the PSG data, physicians inspect them using statistical tools to score the OSA events.
However, PSG has several disadvantages. First, patients need to sleep in the hospital for at least one night, which consumes a considerable amount of time and is expensive. Furthermore, many patients cannot sleep well in hospitals. Second, many electrodes have to be connected to the body of a patient. These electrodes will interrupt their sleep, which will result in the deviation of the measurement results. Therefore, it is important to develop methods that can reliably diagnose OSA with a few signals and that can be used at home. According to Mietus and Peng [11], the heart beat interval of patients fluctuates periodically during the occurrence and recovery of OSA. Zarei and Asl [12] indicated that significant changes in heart rate or abnormal activities of the heart may indicate OSA. Additionally, according to our clinical research, patients' compliance is very low when they wear the pressure transducer sensor to obtain the oral and nasal respiration. Patients often pull out the nasal cannula. Therefore, when compared with the ECG signal, nasal airflow data can be unstable due to lead falling off. Hence, in this study, we use ECG signals to detect OSA events.
Traditional visual OSA scoring is a very tedious and time-consuming process for a physician to conduct. Therefore, many alternative OSA detection methods have been developed [13]. These methods were based on biosignals such as the respiratory [14], snoring [15–17], SpO2 [8, 9, 18], and ECG [12, 19–24] signals, and many authors have obtained a high performance level in terms of OSA detection. However, almost all these methods involved data preprocessing, feature extraction, feature selection, and classification. Although feature extraction is essential to ensure good performance, this process requires considerable domain expertise and is particularly limited to high-dimensional data [25].
Deep learning is an attractive alternative because it can automatically learn and extract features from raw data and can be merged with a classification procedure. In particular, convolutional neural networks (CNNs), which are a popular deep-learning model, have gained considerable success owing to their excellent performance in various domains, including visual imagery [26], speech recognition [27], and text recognition [28]. CNNs have also been applied to biosignal classification problems. For example, in our previous study [29], a CNN can be used to score the sleep stages. Banluesombatkul et al. [30] used metalearning classify sleep stages. Piriyajitakonkij et al. [31] proposed a SleepPoseNet to recognize sleep postures. An event-related potential encoder network was applied to ERP-related tasks [32]. Wilaiprasitporn et al. [33] used a deep-learning approach to improve the performance of affective EEG-based person identification. Recently, some models based on CNNs have been employed to detect OSA. Urtnasan et al. [25] proposed a method for the automated detection of OSA from a single-lead ECG using a CNN. Ho et al. [10] developed an approach for OSA event detection using a CNN and a single-channel nasal pressure signal. Banluesombatkul et al. [34] used a CNN to extract ECG signal features and fully connected neural networks for OSA events detection. McCloskey et al. [35] used a CNN and wavelets to analyze the nasal airflow and detect the OSA events. However, most of these methods score OSA events by minute-by-minute analysis. According to the AASM ruler [2], OSA events occur in 10 s or more. Therefore, minute-by-minute analysis will lose some OSA events. At the same time, the duration of each OSA event is different. Multiple OSA events can occur as briefly within only a single minute (i.e., one epoch); at times, one OSA event can be prolonged over multiple epochs. Therefore, it is difficult to detect complete OSA events for these methods.
According to Guilleminault et al. [36], there is a relation between the OSA events and heart rate variability. They indicated that the heart rate decelerates at the beginning of an OSA event and that it suddenly increases when normal breathing is resumed [36]. Because long short-term memory (LSTM) maintains internal memory and utilizes feedback connections to learn temporal information from sequences of inputs, in this study, we propose a new method for OSA detection using the CNN and LSTM. The LSTM [37] is used to learn these dependencies, such as the transition rules employed by physicians, to identify future OSA events from previous ECG epochs. To detect complete OSA events, a window overlapping method is required to accurately detect the OSA events, which can identify the start and end positions of the event. Therefore, the proposed method can alert for OSA events of long duration, which will reduce the rate of sudden death caused by OSA events [38].
This study is organized as follows: the datasets are presented in Section 2, and the methods are presented in Section 3. The experimental results and discussion are presented in Section 4, and Section 5 concludes this study.
2. Dataset and Preprocessing
The Apnea-ECG dataset [39], downloaded from https://www.physionet.org/content/apnea-ecg/1.0.0, was used to evaluate the proposed approach. The dataset comprises 70 PSG recordings, among which 35 are used in the training set and 35 are used in the test set. The training set was used to update the parameters of the proposed model, and the test set was used to perform independent performance assessments. Each recording contains a continuous digitized ECG signal, a set of apnea annotations (derived by human experts on the basis of the simultaneously recorded respiration and related signals), and a set of machine-generated QRS annotations. The sampling rate for the ECG was 100 Hz with a 12 bit resolution. The records contain variable lengths from 7 to 10 hours. The age of the subjects is between 27 and 63 years, and their weights are 35–135 kg.
First, according to Urtnasan et al. [25], a Chebyshev type-II band-pass filter (5–11 Hz) was used to remove undesirable noise from the single-lead ECG data. Second, the data were segmented into epochs (10 s long) to train the proposed model. Table 1 presents the distribution of all the epochs in the training and test sets. Abnormal epoch means an OSA event.
Table 1.
The number of normal epochs (NE) and abnormal epochs (AE).
| Training set | Test set | ||
|---|---|---|---|
| NE | AE | NE | AE |
| 210680 | 130050 | 213830 | 13102 |
3. Methods
3.1. Convolutional Neural Network
In this study, we used a one-dimensional (1D) CNN to classify the ECG signals. The CNN comprised convolutional, pooling, and fully connected layers. The net input of neuron j in layer l is defined as follows:
| (1) |
where Mj represents the selection of input maps, wj,i denotes the weight or the filter associated with the connection between neurons j and i, xil−1 is the output signal from neuron i in layer l − 1, bjl is the bias associated with neuron j in layer l, and (∗) denotes vector convolution. To acquire an output map, an activation function is required as follows:
| (2) |
When compared with other activation functions, a rectified linear unit (ReLU) exhibits robust training performance. Hence, in this study, we used ReLU as the activation function for the output maps, which can be expressed as follows:
| (3) |
After the convolutional layer, a pooling layer was placed, which was used to reduce the dimensions of the feature maps, network parameters, and the computational cost associated with successive layers using specific functions to summarize the subregions, such as by considering the average value or the maximum value. Additionally, the pooling layer allowed the CNN to learn features that were scale invariant or can be attributed to the orientation changes [40]. The pooling operation consisted of sliding a window across the previous feature map. Herein, max pooling was used after the convolutional layer was activated. Finally, a dense layer, which was generally used in the final stages of the CNN, was fully connected to the outputs of the previous layers.
3.2. Batch Normalization
During the training of a CNN, a change in the distribution of the inputs of each layer will affect the outputs of all the succeeding layers. This can result in difficulty when attempting to train models with saturated nonlinearities [41]. Therefore, batch normalization (BN) was used to solve this problem.
Suppose X = {x1, x2, ⋯, xd} is the input to a layer with dimension d. The corresponding minibatch is mb. The mean of all the inputs in the same minibatch can be expressed as follows:
| (4) |
The variance of the input in a minibatch can be expressed as follows:
| (5) |
Therefore, BN can be expressed as follows:
| (6) |
where , γ, and β are learnable parameters. The training efficiency of a CNN can be improved using BN. At the same time, BN helps the CNN to train faster and provides high accuracy [41].
3.3. Long Short-Term Memory
LSTM controls the cell state via three gates, i.e., a forgetting gate, an input gate, and an output gate. The output features obtained from the previous dense layer of a CNN layer are passed to the gate units. The memory cells constituting the LSTM update their states via the activation of each gate unit controlled to a continuous value between 0 and 1. The hidden state of the LSTM cell ht is updated after every t steps. The input gate, forget gate, and output gate can be written as shown in equations (7)–(9) [37], respectively.
| (7) |
| (8) |
| (9) |
where ° represents point-wise multiplication.
The cell states and hidden states can be expressed using equations (10) and (11), respectively.
| (10) |
| (11) |
The CNN and LSTM can be used as backpropagation algorithms to update the parameters of the model during training.
4. Experiments
4.1. Statistical Evaluation Methods
In this study, we use the kappa coefficient (KP) [42], which is a robust statistical measure of the inter-rater agreement, to evaluate the performance of our method. Additionally, the total accuracy (TAC), sensitivity (SE), specificity (SP), positive predictive value (PPV), and negative predictive value (NPV) were calculated according to an epoch-by-epoch analysis as follows:
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
where TP, TN, FP, and FN denote the true positive, true negative, false positive, and false negative, respectively. We implement our experiments on a workstation with a GeForce GTX2060 GPU in a Windows environment. The TensorFlow framework is used to train the proposed model.
4.2. The Proposed Deep Model Architecture and Parameters
To build an optimal OSA detection architecture, we need to understand the characteristics of the input data. The sampling rate of the ECG was 100 Hz, and the 10 s input dimension was 1000. To extract different scale features, we need to set up different size filters. Therefore, experiments are implemented while varying the filters size of the convolution layer to identify the optimal parameters for automated OSA detection. According to existing study [25, 29], we design a network model, which contains a convolution, BN, pooling, dropout, and dense layer, as shown in Figure 1. N denotes the number of the filters. The parameters and results are shown in Table 2. From Table 2, we can see that model_2 performs best and model_1 is the second. However, the parameters of model_2 are large than those of model_1. For portable OSA devices or real-time OSA analysis systems, model_1 is more appropriate. Therefore, model_1 is used to learning the features representation of ECG. To learn the transition rules of OSA, LSTM is used. The proposed model contains the BN, convolutional, pooling, LSTM, and dense layer, as shown in Figure 2.
Figure 1.
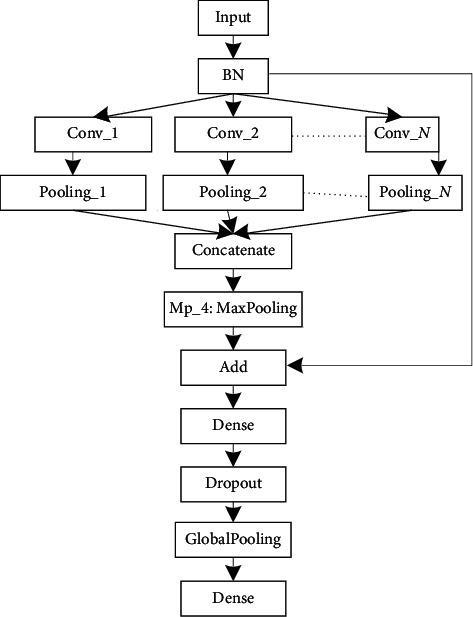
Schematic of the proposed CNN model for the automated detection of OSA.
Table 2.
The parameters and TACs of the different models.
| Name | N | Layer | Units | Size | Stride | TAC (%) |
|---|---|---|---|---|---|---|
| Model_1 | 3 | Cn_1 | 24 | 125 × 1 | 1 × 1 | 94.832 |
| Cn_2 | 24 | 15 × 1 | 1 × 1 | |||
| Cn_3 | 24 | 5 × 1 | 1 × 1 | |||
| Model_2 | 4 | Cn_1 | 24 | 125 × 1 | 1 × 1 | 94.835 |
| Cn_2 | 20 | 100 × 1 | 1 × 1 | |||
| Cn_3 | 24 | 15 × 1 | 1 × 1 | |||
| Cn_4 | 24 | 5 × 1 | 1 × 1 | |||
| Model_3 | 4 | Cn_1 | 24 | 125 × 1 | 1 × 1 | 93.92 |
| Cn_2 | 20 | 50 × 1 | 1 × 1 | |||
| Cn_3 | 20 | 15 × 1 | 1 × 1 | |||
| Cn_4 | 20 | 5 × 1 | 1 × 1 | |||
| Model_4 | 3 | Cn_1 | 24 | 100 × 1 | 1 × 1 | 94.78 |
| Cn_2 | 24 | 15 × 1 | 1 × 1 | |||
| Cn_3 | 24 | 5 × 1 | 1 × 1 | |||
| Model_5 | 2 | Cn_1 | 30 | 125 × 1 | 1 × 1 | 90.4 |
| Cn_2 | 30 | 15 × 1 | 1 × 1 |
Figure 2.
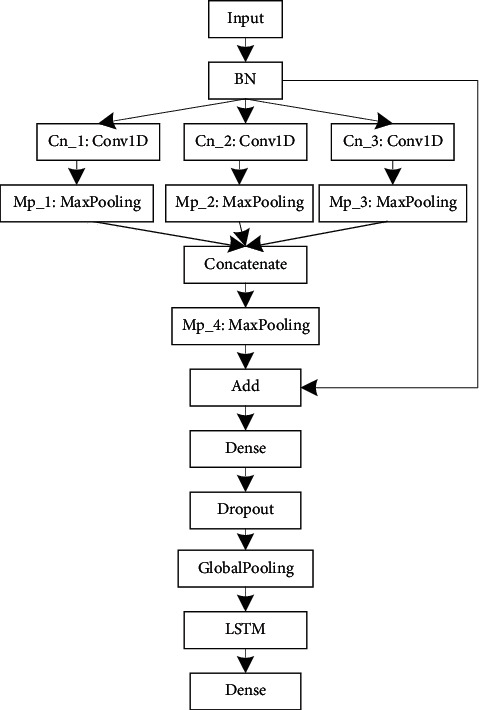
Architecture of the proposed model.
The detailed parameters of the proposed model are presented in Table 3. This table shows the number of filters, the size, and stride in each convolution layer, the size and stride of the kernel in each pooling layer, and the output size of each layer, including the LSTM layer. The batch size is 30, the training epoch is 100, and the learning rate is 0.1. Figure 3 shows the learning results in terms of accuracy and loss obtained as the number of epochs is varied. The results show that the accuracy and loss reach stable values after several iterations of learning when applied to the validation dataset. Figure 4 shows the filter morphology and training time with each training epoch. From Figure 4(a), we can see that, after 90 training epochs, the morphology of the filter almost does not change. Figure 4(b) indicates that the speed of model training is fast.
Table 3.
The parameters of the proposed model.
| Layer | Layer type | Units | Unit type | Size | Stride | Output size |
|---|---|---|---|---|---|---|
| Input | 1000 × 1 | |||||
| BN | 1000 × 1 | |||||
| Cn_1 | Convolutional | 24 | ReLU | 125 × 1 | 1 × 1 | 876 × 24 |
| Cn_2 | Convolutional | 24 | ReLU | 15 × 1 | 1 × 1 | 986 × 24 |
| Cn_3 | Convolutional | 24 | ReLU | 5 × 1 | 1 × 1 | 996 × 24 |
| Mp_1 | Max pooling | 24 | 2 × 1 | 1 × 1 | 438 × 24 | |
| Mp_2 | Max pooling | 24 | 2 × 1 | 1 × 1 | 493 × 24 | |
| Mp_3 | Max pooling | 24 | 2 × 1 | 1 × 1 | 498 × 24 | |
| Concatenate | 24 | 1429 × 24 | ||||
| Mp_4 | Max pooling | 24 | 3 × 1 | 1 × 1 | 476 × 24 | |
| Add | Add | 24 | 1000 × 24 | |||
| Dense | Fully connected | 48 | LeakyReLU | 1000 × 48 | ||
| Dropout | Dropout | 1000 × 48 | ||||
| Gp | Global pooling | 48 × 1 | ||||
| LSTM | LSTM | 64 × 1 | ||||
| Dense | Fully connected | 2 | Softmax | 2 |
Figure 3.
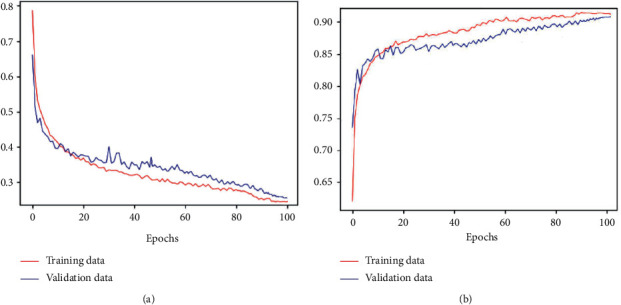
Accuracy and loss of the proposed model for automated OSA detection. (a) Loss curve. (b) Accuracy curve.
Figure 4.
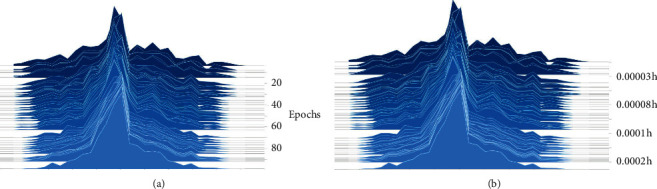
Filters morphology and training time with each epoch. (a) Filter morphology. (b) Training time.
4.3. Performance Results
Table 4 presents the performances of the proposed model for the automated detection of OSA from a single-lead ECG signal. When applied to the test dataset, we obtained a KP of 0.92, an SE of 96.1%, an SP of 96.2%, a TAC of 96.1%, a PPV of 97.6%, and an NPV of 93.8%. As can be seen, the proposed model performed very well for the detection of OSA.
Table 4.
The performances of the proposed model for automated detection of OSA.
| NE | AE | KP | SE (%) | SP (%) | TAC (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|
| NE | 202460 | 4940 | 0.92 | 96.1 | 96.2 | 96.1 | 97.6 | 93.8 |
| AE | 8220 | 125110 |
From Table 4, we can observe that 3.9% of the AEs were misclassified as NEs and that 3.8% of the NEs were misclassified as AEs. According to our research, these misclassifications could have been caused by two probable reasons. One reason is that a transition epoch from NE to AE or AE to NE is difficult to classify. For example, Figure 5 shows a transition epoch from NE to AE, whereas Figure 6 shows a transition epoch from AE to NE. A skilled physician would be able to classify these epochs based on the contextual information. However, the proposed model does not use the contextual information to score OSA, making it unable to distinguish the transition epochs. The other reason may be that the proposed model finds it difficult to score the artifact epochs. The ECG signals can be polluted by unwanted noise signals, including body movement. Figure 7 shows a polluted ECG epoch. Because the artifact epochs are few and varied, the proposed model was unable to learn the distributions of all the artifact epochs. Therefore, it is difficulty for the proposed model to detect the OSA of artifact epochs. In this case, the usage of handcrafted features seems to be considerably robust.
Figure 5.
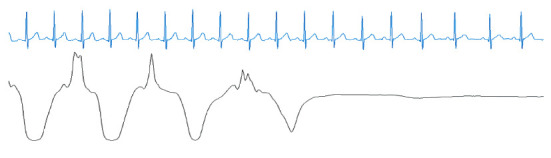
A transition epoch from an NE to an AE. Blue denotes the ECG signal, and black denotes the nasal airflow signal.
Figure 6.
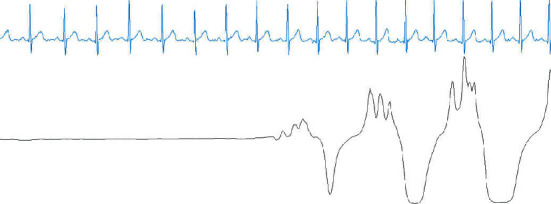
A transition epoch from an AE to an NE. Blue denotes the ECG signal, and black denotes the nasal airflow signal.
Figure 7.
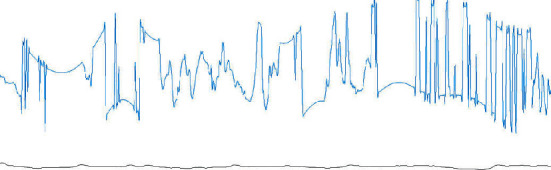
An ECG artifact epoch. Blue denotes the ECG signal, and black denotes the nasal airflow signal.
4.4. Benefits of Long Short-Term Memory
The major advantage associated with the usage of LSTM is that it can be trained to learn long-term dependencies, including the transition rules that are used by the physicians to identify the next possible OSA event(s) from a sequence of ECG epochs. To validate the usefulness of LSTM, we removed the LSTM layer from the model (Figure 2) and then reimplemented the experiment. This test was named CNN_1. Table 5 shows the comparison results, where we can see that the proposed model (CNN + LSTM) results in a gain of 1.3% over the TAC of CNN_1. In addition, KP increased by 0.03 when LSTM was added, verifying our assumption.
Table 5.
Comparison of classification performances.
| Model | KP | TAC (%) |
|---|---|---|
| CNN_1 | 0.89 | 94.8 |
| CNN + LSTM | 0.92 | 96.1 |
Figure 8 shows an example of the NE ECG signal. When the proposed method (CNN + LSTM) is used, the epoch is classified as an NE. However, when CNN_1 is used, this epoch is scored as an OSA event. The reason for OSA misclassification is that the heart rate is slow at the center of this epoch. According to a previously conducted study [11], the heart rate decelerates when OSA occurs. Therefore, CNN_1 learned this feature. However, from Figure 8, we can observe that the heart rate changes very little. At the same time, the heart rates of previous epochs are similar to those of this epoch. However, because the LSTM learns long-term dependencies, the CNN + LSTM method accurately detects the epoch, which is the benefit associated with the usage of LSTM.
Figure 8.
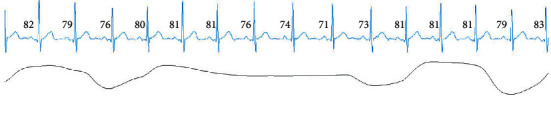
A normal ECG epoch.
4.5. OSA Detection
As mentioned previously, long OSA is dangerous because it can lead to sudden death. To identify long OSA, the window overlapping method can be used to detect the start and end positions of an OSA event. In this way, long OSA can be detected Figure 9 shows that the proposed model can detect complete OSA events from the ECG signals. From the nasal airflow signal, we can observe that the OSA events detected by our model have been accurately identified.
Figure 9.
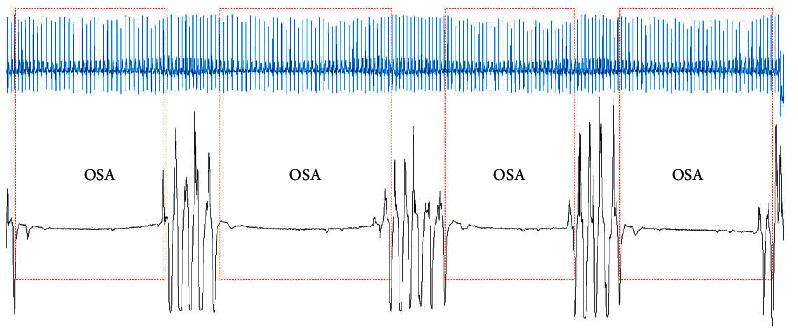
The start and end positions of multiple OSA events.
4.6. Comparison of the Proposed Method with Existing Studies
The comparison of various methods of automatic OSA detection is difficult because different datasets, feature sets, and classifiers are used in different studies. For ensuring a fair comparison with existing studies, Table 6 shows the classification performances of different methods based on single-lead ECG signals. From Table 6, we can observe that the proposed model achieved better performance when compared with those achieved in the previous studies. More importantly, our method can be used in conjunction with wearable medical devices, which is very important for home OSA monitoring.
Table 6.
Comparison of performances of different methods.
| Input | Author | Method | TAC (%) | SE (%) | SP (%) |
|---|---|---|---|---|---|
| ECG | Jafari [43] | Handcrafted features, SVM | 94.8 | 94.1 | 95.4 |
| Chen et al. [44] | Handcrafted features, SVM | 82.1 | 83.2 | 80.2 | |
| Urtnasan et al. [25] | CNN | 96 | 96 | 96 | |
| Banluesombatkul et al. [34] | CNN | 79.45 | 77.6 | 80.1 | |
| Zarei and Asl [12] | Handcrafted features, SVM | 94.63 | 94.43 | 94.77 | |
| Tripathy [45] | Handcrafted features, kernel extreme learning machine | 76.37 | 78.02 | 74.64 | |
| Hassan and Haque [46] | Handcrafted features, RUSboot | 88.88 | 87.58 | 91.49 | |
| Hassan [47] | Handcrafted features, AdaBoost | 87.33 | 81.99 | 90.72 | |
| Our method | CNN | 96.1 | 96.1 | 96.2 |
5. Conclusions
In this study, we developed an automated OSA event detection method using a CNN, where the feature extraction and selection processes were not required. The proposed method detected the start and end positions of the OSA events based on the overlapping epochs in the ECG signal dataset. Our method automatically extracted the time-invariant features from raw ECG signals without utilizing any handcrafted features. The proposed approach is robust and completely automated, and the method can be easily adapted to other physiological signal analyses and prediction problems. The TAC and KP of the proposed model applied to the single-channel ECG reached 96.1% and 0.92, respectively. The experimental results showed that the proposed method could accurately score the OSA events and that it achieved comparable performance with other state-of-the-art studies. More importantly, our method can prevent sudden death from OSA, which is important for the patients who are severely affected by OSA.
There are some limitations associated with our CNN method. First, the proposed model can only detect OSA and normal events but not hypopnea events. Although hypopnea is not as serious as OSA, it is still prevalent in sleep-disordered breathing patients. Second, it is difficult to score transition epochs using our method. In the future, we will improve the discrimination ability of our method for AEs and NEs. In addition, the automated anomaly detection of ECG based on the CNN, which is important to rapidly assess the quality of the ECG data, will be studied.
Acknowledgments
This work was supported in part by the Henan Provincial Key Science and Technology Research Projects under Grant nos. 202102210127 and 212102210142, in part by the National Science Foundation of China under Grant no. 61973177, in part by the Henan Key Laboratory of Smart Lighting, in part by Henan International Joint Laboratory of Behavior Optimization Control for Smart Robots, in part by the Programme of Henan Innovative Research Team of Cooperative Control in Swarm-based Robotics, in part by the Award Plan for Tianzhong Scholars of Huanghuai University in 2019, in part by Zhumadian Artificial Intelligence & Medical Engineering Technical Research Centre, and in part by Zhumadian Industrial Innovation and Development Research Major Project under Grant no. 2020ZDA06.
Data Availability
The Apnea-ECG dataset, downloaded from https://www.physionet.org/content/apnea-ecg/1.0.0, was used to evaluate our proposed approach.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Young T., Evans L., Finn L., Palta M. Estimation of the clinically diagnosed proportion of sleep apnea syndrome in middle-aged men and women. Sleep. 1997;20(9):705–706. doi: 10.1093/sleep/20.9.705. [DOI] [PubMed] [Google Scholar]
- 2.Iber C., Ancoli-Israel S., Chesson A. L., et al. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Darien, IL, USA: American Academy of Sleep Medicine; 2007. pp. 17–48. [Google Scholar]
- 3.Engleman H. M., Douglas N. J. Sleep·4: sleepiness, cognitive function, and quality of life in obstructive sleep apnoea/hypopnoea syndrome. Thorax. 2004;59(7):618–622. doi: 10.1136/thx.2003.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung D. W., Hwang S. H., Lee Y. J., Jeong D.-U., Park K. S. Apnea-hypopnea index prediction using electrocardiogram acquired during the sleep-onset period. IEEE Transactions on Biomedical Engineering. 2017;64(2):295–301. doi: 10.1109/tbme.2016.2554138. [DOI] [PubMed] [Google Scholar]
- 5.Dixon J. B., Schachter L. M., O’Brien P. E. Predicting sleep apnea and excessive day sleepiness in the severely obese. Chest. 2003;123(4):1134–1141. doi: 10.1378/chest.123.4.1134. [DOI] [PubMed] [Google Scholar]
- 6.Ghahjaverestan N. M., Masoudi S., Shamsollahi M., et al. Coupled hidden markov model-based method for apnea bradycardia detection. IEEE Journal of Biomedical and Health Informatics. 2016;20(2):527–538. doi: 10.1109/JBHI.2015.2405075. [DOI] [PubMed] [Google Scholar]
- 7.Flemons W. W., Remmers J. E., Gillis A. M. Sleep apnea and cardiac arrhythmias: is there a relationship? American Review of Respiratory Disease. 1993;148(3):618–621. doi: 10.1164/ajrccm/148.3.618. [DOI] [PubMed] [Google Scholar]
- 8.Wang D., Wong K. K., Rowsell L., Don G. W., Yee B. J., Grunstein R. R. Predicting response to oxygen therapy in obstructive sleep apnoea patients using a 10-minute daytime test. European Respiratory Journal. 2018;51(1):p. 1701587. doi: 10.1183/13993003.01587-2017. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y.-Y., Wu H.-T., Hsu C.-A., Huang P.-C., Huang Y.-H., Lo Y.-L. Sleep apnea detection based on thoracic and abdominal movement signals of wearable piezoelectric bands. IEEE Journal of Biomedical and Health Informatics. 2017;21(6):1533–1545. doi: 10.1109/jbhi.2016.2636778. [DOI] [PubMed] [Google Scholar]
- 10.Ho C. S., Heenam Y., Seok K. H., et al. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Computers in Biology and Medicine. 2018;100:123–131. doi: 10.1016/j.compbiomed.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Mietus J. E., Peng C. K. Detection of obstructive sleep apnea from cardiac interbeat interval time series. Computers in Cardiology. 2000;27:753–756. [Google Scholar]
- 12.Zarei A., Asl B. M. Automatic detection of obstructive sleep apnea using wavelet transform and entropy-based features from single-lead ECG signal. IEEE Journal of Biomedical and Health Informatics. 2019;23(3):1011–1021. doi: 10.1109/jbhi.2018.2842919. [DOI] [PubMed] [Google Scholar]
- 13.Faizal W. M., Ghazali N. N. N., Badruddin I. A., et al. A review of fluid-structure interaction simulation for patients with sleep related breathing disorders with obstructive sleep. Computer Methods and Programs in Biomedicine. 2019;180 doi: 10.1016/j.cmpb.2019.105036.105036 [DOI] [PubMed] [Google Scholar]
- 14.Lakhan P., Ditthapron A., Banluesombatkul N., et al. Deep neural networks with weighted averaged overnight airflow features for sleep apnea-hypopnea severity classification. Proceedings of the 2018 IEEE Region 10 Conference; October 2018; Jeju Island, Korea. pp. 0441–0445. [Google Scholar]
- 15.Rachel K., Christian G. Obstructive sleep apnea syndrome. Clinics in Chest Medicine. 2017;31(2):187–201. doi: 10.1016/j.ccm.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Erdenebayar U., Park J.-U., Jeong P., Lee K.-J. Obstructive sleep apnea screening using a piezo-electric sensor. Journal of Korean Medical Science. 2017;32(6):893–989. doi: 10.3346/jkms.2017.32.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui J., Liang L., Lijuan S. Acoustic analysis of snoring in the diagnosis of obstructive sleep apnea syndrome: a call for more rigorous studies. Journal of Clinical Sleep Medicine JCSM: Official Publication of the American Academy of Sleep Medicine. 2015;11(7):765–771. doi: 10.5664/jcsm.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutta S., Cheng Q. Modeling of oxygen saturation and respiration for sleep apnea detection. Proceedings of the 50th Asilomar Conference on Signals, Systems and Computers; November 2016; Pacific Grove, CA, USA. pp. 1636–1640. [Google Scholar]
- 19.Cheng C., Kan C., Yang H. Heterogeneous recurrence analysis of heartbeat dynamics for the identification of sleep apnea events. Computers in Biology and Medicine. 2016;75:10–18. doi: 10.1016/j.compbiomed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Sharma H., Sharma K. K. An algorithm for sleep apnea detection from single-lead ECG using Hermite basis functions. Computers in Biology and Medicine. 2016;77:116–124. doi: 10.1016/j.compbiomed.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Viswabhargav C. S. S., Tripathy R. K., Acharya U. R. Automated detection of sleep apnea using sparse residual entropy features with various dictionaries extracted from heart rate and EDR signals. Computers in Biology and Medicine. 2019;108:20–30. doi: 10.1016/j.compbiomed.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Varon C., Caicedo A., Testelmans D., Buyse B., Van Huffel S. A novel algorithm for the automatic detection of sleep apnea from single-lead ECG. IEEE Transactions on Biomedical Engineering. 2015;62(9):2269–2278. doi: 10.1109/tbme.2015.2422378. [DOI] [PubMed] [Google Scholar]
- 23.Manish S., Shreyansh A., Rajendra A. U. Application of an optimal class of antisymmetric wavelet filter banks for obstructive sleep apnea diagnosis using ECG signals. Computers in Biology and Medicine. 2018;100:100–113. doi: 10.1016/j.compbiomed.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Lei W., Youfang L., Jing W. A RR interval based automated apnea detection approach using residual network. Computer Methods and Programs in Biomedicine. 2019;176:93–104. doi: 10.1016/j.cmpb.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Urtnasan E., Park J.-U., Joo E.-Y., Lee K.-J. Automated detection of obstructive sleep apnea events from a single-lead electrocardiogram using a convolutional neural network. Journal of Medical Systems. 2018;42(6) doi: 10.1007/s10916-018-0963-0. [DOI] [PubMed] [Google Scholar]
- 26.Krizhevsky A., Sutskever I., Hinton G. E. Advances in Neural Information Processing Systems. Cambridge, MA, USA: The MIT Press; 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 27.Abdel-Hamid O., Mohamed A. R., Jiang H., et al. Applying convolutional neural networks concepts to hybrid NN-HMM model for speech recognition. Proceedings of the 2012 IEEE International Conference on Acoustics, Speech and Signal Processing; March 2012; Kyoto, Japan. pp. 4277–4280. [Google Scholar]
- 28.Wang T., Wu D. J., Coates A., Ng A. Y. End-to-end text recognition with convolutional neural networks. Proceedings of the 21st International Conference on Pattern Recognition; November 2012; Tsukuba, Japan. pp. 3304–3308. [Google Scholar]
- 29.Zhang J., Wu Y. A new method for automatic sleep stage classification. IEEE Transactions on Biomedical Circuits and Systems. 2017;11(5):1097–1110. doi: 10.1109/tbcas.2017.2719631. [DOI] [PubMed] [Google Scholar]
- 30.Banluesombatkul N., Ouppaphan P., Leelaarporn P., et al. MetaSleepLearner: a pilot study on fast adaptation of bio-signals-based sleep stage classifier to new individual subject using meta-learning. IEEE Journal of Biomedical and Health Informatics. 2020:p. 1. doi: 10.1109/JBHI.2020.3037693. [DOI] [PubMed] [Google Scholar]
- 31.Piriyajitakonkij M., Warin P., Lakhan P., et al. SleepPoseNet: multi-view learning for sleep postural transition recognition using UWB. IEEE Journal of Biomedical and Health Informatics. 2020:p. 1. doi: 10.1109/JBHI.2020.3025900. [DOI] [PubMed] [Google Scholar]
- 32.Ditthapron A., Banluesombatkul N., Ketrat S., Chuangsuwanich E., Wilaiprasitporn T. Universal joint feature extraction for P300 EEG classification using multi-task autoencoder. IEEE Access. 2019;7:68415–68428. doi: 10.1109/access.2019.2919143. [DOI] [Google Scholar]
- 33.Wilaiprasitporn T., Ditthapron A., Matchaparn K., Tongbuasirilai T., Banluesombatkul N., Chuangsuwanich E. Affective EEG-based person identification using the deep learning approach. IEEE Transactions on Cognitive and Developmental Systems. 2020;12(3):486–496. doi: 10.1109/tcds.2019.2924648. [DOI] [Google Scholar]
- 34.Banluesombatkul N., Rakthanmanon T., Wilaiprasitporn T. Single channel ECG for 0bstructive sleep apnea severity detection using a deep learning approach. Proceedings of the 2018 IEEE Region 10 Conference; October 2018; Jeju Island, Korea. pp. 2011–2016. [Google Scholar]
- 35.McCloskey S., Haidar R., Koprinska I., Jeffries B. Detecting hypopnea and obstructive apnea events using convolutional neural networks on wavelet spectrograms of nasal airflow. Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining; June 2018; Melbourne, Australia. pp. 361–372. [DOI] [Google Scholar]
- 36.Guilleminault C., Winkle R., Connolly S., Melvin K., Tilkian A. Cyclical variation of the heart rate in sleep apnoea syndrome. The Lancet. 1984;323(8369):126–131. doi: 10.1016/s0140-6736(84)90062-x. [DOI] [PubMed] [Google Scholar]
- 37.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M., Li L., Fowler D., et al. Causes of sudden death in patients with obstructive sleep apnea. Journal of Forensic Sciences. 2013;58(5):1171–1174. doi: 10.1111/1556-4029.12220. [DOI] [PubMed] [Google Scholar]
- 39.Goldberger A. L., Amaral L. A. N., Glass L., et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2003;101(23):e215–e220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 40.LeCun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 41.Ioffe S., Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proceedings of the 32nd International Conference on Machine Learning; July 2015; Lille, France. pp. 448–456. [Google Scholar]
- 42.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20(1):37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 43.Jafari A. Sleep apnoea detection from ECG using features extracted from reconstructed phase space and frequency domain. Biomedical Signal Processing and Control. 2013;8(6):551–558. doi: 10.1016/j.bspc.2013.05.007. [DOI] [Google Scholar]
- 44.Chen L., Zhang X., Song C. An automatic screening approach for obstructive sleep apnea diagnosis based on single-lead electrocardiogram. IEEE Transactions on Automation Science and Engineering. 2015;12(1):106–115. doi: 10.1109/tase.2014.2345667. [DOI] [Google Scholar]
- 45.Tripathy R. K. Application of intrinsic band function technique for automated detection of sleep apnea using HRV and EDR signals. Biocybernetics and Biomedical Engineering/Polish Academy of Sciences. Institute of Biocybernetics and Biomedical Engineering. 2018;38:136–144. [Google Scholar]
- 46.Hassan A. R., Haque M. A. An expert system for automated identification of obstructive sleep apnea from single-lead ECG using random under sampling boosting. Neurocomputing. 2017;235:122–130. doi: 10.1016/j.neucom.2016.12.062. [DOI] [Google Scholar]
- 47.Hassan A. R. Computer-aided obstructive sleep apnea detection using normal inverse Gaussian parameters and adaptive boosting. Biomedical Signal Processing and Control. 2016;29:22–30. doi: 10.1016/j.bspc.2016.05.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Apnea-ECG dataset, downloaded from https://www.physionet.org/content/apnea-ecg/1.0.0, was used to evaluate our proposed approach.


