ABSTRACT
Accumulating evidence indicates that the gut microbiota can promote or inhibit colonic inflammation and carcinogenesis. Promotion of beneficial gut bacteria is considered a promising strategy to alleviate colonic diseases including colitis and colorectal cancer. Interestingly, dietary polyphenols, which have been shown to attenuate colitis and inhibit colorectal cancer in animal models and some human studies, appear to reach relatively high concentrations in the large intestine and to interact with the gut microbial community. This review summarizes the modulatory effects of polyphenols on the gut microbiota in humans and animals under healthy and diseased conditions including colitis and colitis-associated colorectal cancer (CAC). Existing human and animal studies indicate that polyphenols and polyphenol-rich whole foods are capable of elevating butyrate producers and probiotics that alleviate colitis and inhibit CAC, such as Lactobacillus and Bifidobacterium. Studies in colitis and CAC models indicate that polyphenols decrease opportunistic pathogenic or proinflammatory microbes and counteract disease-induced dysbiosis. Consistently, polyphenols also change microbial functions, including increasing butyrate formation. Moreover, polyphenol metabolites produced by the gut microbiota appear to have anticancer and anti-inflammatory activities, protect gut barrier integrity, and mitigate inflammatory conditions in cells and animal models. Based on these results, we conclude that polyphenol-mediated alteration of microbial composition and functions, together with polyphenol metabolites produced by the gut microbiota, likely contribute to the protective effects of polyphenols on colitis and CAC. Future research is needed to validate the causal role of the polyphenol–gut microbiota interaction in polyphenols’ anti-colitis and anti-CAC effects, and to further elucidate mechanisms underlying such interaction.
Keywords: polyphenol, colitis, colitis-associated colorectal cancer, chemoprevention, gut microbiota, short-chain fatty acids, probiotics, 16S rRNA
Introduction
Colorectal cancer (CRC) is the second-leading cause of cancer death in men and women combined, accounting for 8–9% cancer mortality in the United States in 2020 (1). Chronic inflammation contributes to the etiology and pathogenesis of CRC. Inflammatory bowel diseases (IBDs), which include Crohn's disease and ulcerative colitis, significantly increase the risk of colitis-associated CRC (CAC) (2, 3). For instance, ulcerative colitis patients are found to have a 2.4-fold increased risk of CAC occurrence while patients with Crohn's disease develop CAC at a younger age compared with the healthy population (4). Mechanistically, IBD patients are exposed to excessive oxidative stress, which can promote DNA mutation and potentially lead to activation of oncogenes, disruption of tumor suppressor genes, and promotion of genomic instability (5, 6). Further, proinflammatory cytokines or lipid mediators are known to promote tumor progression via activating proliferative and antiapoptotic properties of premalignant cells and cancer cells in the tumor microenvironment (7–9).
Although tremendous effort has been made for developing effective therapies for CRC, the late-stage cancer has proven difficult to treat and has a low survival rate (10). Therefore, there is an urgent need to develop effective and relatively safe agents to inhibit CRC progression, potentially by attenuating chronic inflammation. To this end, polyphenols including anthocyanins, curcumins, resveratrol, and ellagic acid have been extensively studied as potential chemopreventive agents for CRC. In particular, many studies have documented the protective effects of polyphenols on colitis and CAC in clinically relevant disease models (11). Curcumin (12, 13) and epigallocatechin gallate (EGCG) (14, 15) have been shown to have anti-CRC effects in clinical patients, and resveratrol attenuated inflammation in ulcerative colitis patients (16).
Dietary polyphenols are produced by plants as part of their defense system. These compounds are the biggest group of phytochemicals with >8000 known structures and are rich in fruits such as berries, grapes, and pomegranates; vegetables such as broccoli; whole-grain products; and beverages such as tea, coffee, and wine (17). Structurally, they are characterized by phenolic moieties and can be roughly classified into phenolic acids, flavonoids, phenolic amides, and others such as stilbenes and lignans (18). The phenolic structure enables polyphenols to donate electrons/hydrogens to scavenge free radicals and thus possess strong antioxidant property, which may partially contribute to their antiaging, cardioprotective, and neuroprotective effects (17). Furthermore, polyphenols have also been shown to have strong anti-inflammatory and anticancer properties (19–21). In particular, many polyphenols inhibit NF-κB, a central transcription factor that regulates inflammation, cell growth, and cell survival, and plays critical roles in IBD and IBD-promoted CRC (22). Some polyphenols can inhibit the proinflammatory enzymes such as cyclooxygenase and 5-lipoxygenase, which are recognized targets for CRC prevention (23). Additionally, polyphenols such as flavonoids have been shown to modulate signaling kinases including phosphoinositide 3 kinase, Akt/protein kinase B (PKB), mitogen-activated protein kinase (MAPK), tyrosine kinase, and protein kinase C (24). A recent review by Alam et al. (25) summarized the anti-CRC mechanisms of polyphenols, including antiproliferation, proapoptosis, and anti-inflammation activities identified in cell-based studies.
Despite extensive studies showing anti-inflammatory and anticancer effects of polyphenols in cell and animal models, whether polyphenols can directly exert these activities in vivo has been questioned as these compounds are known to have poor bioavailability (26–28). For instance, polyphenols have been shown to exhibit anticancer or anti-inflammatory effects at 10 to >100 μM in cell-based studies (11, 29), but the concentrations of these compounds are often below a few micromoles in the plasma or serum of animals and humans (27, 30–32). Therefore, the observed antioxidant and anti-inflammatory effects in cells cannot fully explain the beneficial effects of these compounds manifested in the whole-body environment (11). Interestingly, emerging research shows that polyphenols appear to be bioavailable in the gut and able to modulate the gut microbiota (33–35). Since the gut microbiota are believed to play regulatory roles in colitis and CRC development (36, 37), we reason that polyphenols may alleviate colitis and inhibit cancer development, at least in part, via interacting with the gut microbial community. In this review, we summarize recent literature concerning the modulatory effect of polyphenols on the gut microbiota, analyze the interaction between polyphenols and the gut microbiota, and discuss the role of the gut microbiota in polyphenols’ protective effects on colitis and CAC. Additionally, we identify knowledge gaps in the field and propose potential future directions.
Current Status of Knowledge
The gut microbiota, intestinal inflammation, and CRC
Although the exact etiology of IBDs and CAC is not clear, over the years researchers have identified multiple factors contributing to the pathogenesis of these diseases, including genetic susceptibility (38), immune dysregulation (39), and barrier dysfunction (40). Additionally, recent evidence suggests that the interaction between the commensal gut microbiota and host immune system is also important to maintaining gut health, and disruption of such interplay may directly compromise gut health and result in the promotion of colitis and CAC. Herein we briefly discuss the relation between the gut microbiota and colitis or CAC, while the role of the gut microbiota in inflammation-induced cancer has been comprehensively reviewed by Elinav et al. (36).
IBD- and CAC-associated alteration of the gut microbiota
Studies in humans and animals have revealed potential links between the gut microbiota and CAC. In particular, patients with CRC and IBDs have altered gut microbial profile compared with healthy individuals. Specifically, CRC patients have been reported to have reduced relative abundance of butyrate producers including Roseburia and Lachnospiraceae bacterium A2, and increased abundance of Enterococcus,Escherichia/Shigella, and Streptococcus, which may contain opportunistic pathogen species (41). Additionally, in Crohn's disease patients, microbial dysbiosis is characterized by decreased abundance of Dialister invisus,Clostridium cluster XIVa, Faecalibacterium prausnitzii, and Bifidobacterium adolescentis and increased relative abundance of Ruminococcus gnavus compared with healthy subjects (42). In a study of ulcerative colitis patients, researchers found slightly decreased microbial richness and increased relative abundance of Actinobacteria and Proteobacteria in patients compared with their healthy twins (43). Consistent with these results in humans, research of murine CAC induced by azoxymethane (AOM) and dextran sulfate sodium (DSS) showed decreased microbial evenness in the DSS-induced colitis phase and increased relative abundance of Streptococcus luteciae, Lactobacillus hamster, Bacteroides uniformis, and Bacteroides ovatus in the carcinogenic phase (44). These results indicate that IBDs and CAC are associated with altered gut microbiota.
The interaction between the gut microbiota and colonic inflammation
Imbalanced immune responses including chronic inflammation and compromised immune system have been shown to induce microbial dysbiosis and even promote the growth of pathogenic bacteria that facilitate tumorigenesis. For instance, in the IL-10 knockout mouse model, Arthur et al. (45) revealed that intestinal inflammation changed the gut microbial composition, including the expansion of Escherichia coli NC101 pks, a genotoxic microbe that is shown to promote colonic carcinogenesis. Further, T-bet–deficient mice in the absence of adaptive immunity [recombination activating gene 2 (RAG2)-/-] are reported to develop spontaneous ulcerative colitis and significantly modulate the gut microbiota (46). Interestingly, the altered gut microbiota were colitogenic and transmitted colitis to T-bet–intact hosts through fostering litters and co-housing (46). Similarly, transmissible and colitogenic gut microbiota have been identified from mice deficient in caspase recruitment domain family member 9 (CARD9), a susceptibility factor for IBDs (47). Furthermore, in the 2,4,6-trinitrobenzene sulfonic acid (TNBS)–induced mouse model of Crohn's disease, ablation of proinflammatory TNF alleviated colonic inflammation and altered microbial composition compared with wild-type diseased mice (48). In addition, tumor-associated barrier defects have been shown to increase translocation of microbes or microbial products into the circulation, which, in turn, enhances inflammatory response and fosters cancer development (49). These results indicate mutual interactions between the gut microbiota and the host immune system.
The causal role of the gut microbiota in the development of colitis and CRC
Recent studies demonstrate the potential causal role of gut microbiota in colonic inflammation and CAC. For instance, fecal microbiota transplantation (FMT) of stool samples from CRC patients is found to promote AOM-induced high-grade dysplasia, multiplicity of macroscopic polyps, elevation of cytokines, and immune cell infiltration in mice compared with those fed stool samples from healthy volunteers (50). In the AOM-DSS–induced CAC model, Zackular et al. (51) showed that antibiotic treatment during the entire tumorigenesis or in the inflammation-mediated promotion phase diminished tumor burden, whereas antibiotics did not affect tumor development when given prior to AOM injection and stopped before DSS-induced colon inflammation. These results strongly indicate that the cross-talk between commensal bacteria and the host innate immune system during inflammation is critical for potentiating tumorigenesis (52). Consistently, Paramsothy et al. (53) performed an 8-wk FMT with stool samples from healthy donors to ulcerative colitis patients and observed significant improvement in steroid-free clinical remission and response rate in treated patients compared with those receiving FMT with the placebo controls. These results support the notion that the gut microbiota modulate colon inflammation and contribute to the inflammation-promoted colon tumorigenesis.
In addition to promoting inflammation, the gut microbiota may facilitate carcinogenesis through expanding pathogenic microbes or forming mucosal biofilm communities (5, 54). In particular, pathobionts including E. coli pks+(45), enterotoxigenic Bacteroides fragilis (55), and Fusobacterium nucleatum (56), which can be boosted during the inflammation process, have been shown to promote carcinogenesis by secreting mediators that increase DNA damage, reactive oxygen species production, cell proliferation, and immune cell recruitment. Interestingly, these pathobionts are enriched in colonic mucosal biofilms of familial adenomatous polyposis (57) and in sporadic CRC patients (58). Furthermore, the presence of mucus-invasive bacterial biofilms is more frequent in CRC patients, especially those with right-sided tumors, than healthy individuals (58). These existing biofilms are associated with enhanced cell proliferation (58) and polyamine metabolism in the colon tissue (59). Recently, Tomkovich et al. (60) demonstrated that mucus-invasive bacterial biofilms from both CRC patients and healthy participants are carcinogenic in different CRC models in mice.
Together, the above-cited studies indicate that the gut microbiota play significant roles in maintaining gut health and in developing colon inflammation and CRC. Therefore, modulation of the gut microbiota by chemopreventive agents may be an effective strategy for controlling colitis and inhibiting CAC progression. Specifically, bioactive compounds capable of cultivating a healthy microbial community that prevents or restores dysbiosis should be useful to maintain gut barrier functions, inhibit the growth of pathobionts, and alleviate colonic inflammation and carcinogenesis. In this regard, bioactive polyphenols are promising candidates. In the subsequent part of this review, we will discuss the interaction between dietary polyphenols and the gut microbiota, and how such interaction may contribute to attenuating colitis and preventing CAC.
The effect of polyphenols on the gut microbiota in healthy humans and animals
Although only a small portion of polyphenols appear to be absorbed in the upper gastrointestinal tract, up to 90–95% of consumed polyphenols are estimated to accumulate in the large intestine where microbes reside (1011–1012 bacteria/g of intestinal content) (33, 61–63). As a result, it is conceivable that polyphenols may interact with gut microbes in the colon. Indeed, polyphenols have been shown to modulate the gut microbiota in humans and animals. In this part, we summarize recent intervention studies where the effects of polyphenols on the gut microbiota have been evaluated in healthy humans and animals.
The impact of polyphenols on the gut microbiota in healthy humans
Recent clinical interventions have revealed that polyphenols are capable of modulating the gut microbiota in healthy humans, including elevation of potentially beneficial microbes (Table 1). In particular, intervention studies showed that fecal Bifidobacterium or Lactobacillus or both genera, compared with baseline, were enhanced as a result of supplementation of polyphenol-rich whole foods or polyphenol extracts including de-alcoholized red wine (64), polyphenol-rich green tea (66, 70), and blueberry drink (67). Further, healthy volunteers consuming 494 mg/d cocoa-derived flavanol drinks had increased fecal Bifidobacterium and Lactobacillus compared with those consuming 23 mg/d cocoa-derived flavanol drinks (65). Interestingly, in some studies, the increase in Lactobacillus or Bifidobacterium was reported to be inversely associated with the concentration of cholesterol or C-reactive protein, an inflammation marker (64, 65). Consistent with these intervention studies, the elevation of these 2 microbial genera was also observed in ex vivo experiments where fresh feces from healthy volunteers were cultivated with polyphenol extracts from dates (71), grape seeds (72), and sorghum bran (73).
TABLE 1.
Effects of polyphenol supplementation on the gut microbiota of healthy humans1
| Compound | Polyphenol dose | Study design | Molecular technique | Results | Reference |
|---|---|---|---|---|---|
| De-alcoholized red wine | 733.02 ± 23.61 mg gallic acid equivalent/d | Randomized, crossover, controlled study; 10 subjects (males), 20 d | DGGE-PCR, qPCR of selected bacterial 16S rRNA gene |
 Abundance of Fusobacteria, Bacteroidetes, Enterococcus, Blautia coccoides–Eubacterium rectale, Bifidobacterium, and Eggerthella lenta; Abundance of Fusobacteria, Bacteroidetes, Enterococcus, Blautia coccoides–Eubacterium rectale, Bifidobacterium, and Eggerthella lenta; Bifidobacterium correlate w/ Bifidobacterium correlate w/ cholesterol and C-reactive protein cholesterol and C-reactive protein |
(64) |
| Cocoa-derived flavanol drink | Low: 23 mg cocoa flavanols/d; high: 494 mg cocoa flavanols/d | Randomized, double-blind, crossover, controlled study; 22 subjects (12 males, 10 females), 4 wk | FISH targeting 16S rRNA |
 Fecal Bifidobacterium spp., Lactobacillus and Enterococcus spp. counts; Fecal Bifidobacterium spp., Lactobacillus and Enterococcus spp. counts;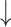 fecal Clostridia histolyticum counts in high-dosage compared with low-dosage group; fecal Clostridia histolyticum counts in high-dosage compared with low-dosage group; 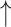 Lactobacilli correlates w/ Lactobacilli correlates w/ 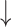 C-reactive protein C-reactive protein |
(65) |
| Green tea drink | 1000 mL/d | 10 subjects (4 males, 6 females), 10 d | T-RFLP, qPCR of selected bacterial 16S rRNA gene |
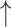 Relative abundance of Bifidobacterium spp. Relative abundance of Bifidobacterium spp. |
(66) |
| Wild blueberry powder drink | 375 mg anthocyanins, 127.5 mg chlorogenic acid/d + 4.5 g fiber/d | Randomized, crossover; 20 subjects (males), 6 wk | qPCR of selected bacterial 16S rRNA gene |
 Relative abundance of Bifidobacterium spp.; Relative abundance of Bifidobacterium spp.; relative abundance of Lactobacillus acidophilus relative abundance of Lactobacillus acidophilus
|
(67) |
| Palm date fruits | 750 mg polyphenol/d | Randomized, crossover, controlled study; 22 subjects (11 males, 11 females), 3 wk | FISH targeting 16S rRNA | No changes in biochemical or anthropometric parameters; no changes in microbial abundance | (68) |
| Freeze-dried cranberry powder (polyphenol components) | 30 g/d in animal-based diet (with matched controls) | Randomized, double-blind, crossover, controlled study; 11 subjects (7 males, 4 females), 5 d | 16S rRNA gene V4 region Illumina sequencing |
 Relative abundance of Bacteroidales, Anaerostipes, and Lachnospira; Relative abundance of Bacteroidales, Anaerostipes, and Lachnospira; relative abundance of Oribacterium and Clostridiales; relative abundance of Oribacterium and Clostridiales; urinary anthocyanins, phenolic acids; prevented animal-diet induced increase in fecal lithocholic acid and deoxycholic acid and decrease in acetic and butyric acid urinary anthocyanins, phenolic acids; prevented animal-diet induced increase in fecal lithocholic acid and deoxycholic acid and decrease in acetic and butyric acid |
(69) |
| Green tea liquid | 400 mL/d | 12 subjects (8 males, 4 females), 14 d | 16S rRNA gene V4-V5 region Illumina sequencing |
 α-Diversity; changed microbial composition; α-Diversity; changed microbial composition; relative abundance of (family) Lachnospiraceae and Bifidobactericeae; relative abundance of (family) Lachnospiraceae and Bifidobactericeae;  Bifidobacterium and SCFA-producing genus Roseburia, Feacalibacterium, Eubacterium, Blautia, Coprococcus, and Dorea; Bifidobacterium and SCFA-producing genus Roseburia, Feacalibacterium, Eubacterium, Blautia, Coprococcus, and Dorea; predicted microbial genes for LPS biosynthesis, glutathione metabolism, and glycosaminoglycan degradation predicted microbial genes for LPS biosynthesis, glutathione metabolism, and glycosaminoglycan degradation |
(70) |
1DGGE, denaturing gradient gel electrophoresis; FISH, fluorescence in situ hybridization; rRNA, ribosomal RNA; T-RFLP, terminal restriction fragment length polymorphism
In addition to specific bacterial taxa, a few studies examined the impact of polyphenol consumption on microbial metabolism and functions. For instance, in a controlled-feeding crossover study, researchers found that polyphenols from freeze-dried cranberry powder reversed the animal diet–induced increase in secondary bile acids and decrease in short-chain fatty acids (SCFAs) in feces (69). It is important to note that the increase in secondary bile acids and/or decrease in SCFAs are believed to be risk factors for CRC development (74, 75). In another study, using phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis of the 16S ribosomal RNA (rRNA) sequencing in fecal samples, Yuan et al. (70) reported that consumption of green tea was associated with decreased microbial genes encoding inflammation-associated pathways including LPS biosynthesis.
Despite relatively consistent modulatory effects by polyphenols on the gut microbiota in healthy humans (Table 1), Eid et al. (68) did not detect microbial changes in a clinical trial where healthy free-living subjects were given palm date as supplements for 3 wk. However, these researchers observed an increase in Bifidobacterium by polyphenol-rich palm date extract in the ex vivo incubation study (71). The discrepancy between ex vivo and in vivo results may result from confounding factors in the whole food such as dietary fibers. Indeed, when dividing volunteers into high- and low-fiber consumers, these investigators identified increased Bifidobacterium and Lactobacillus/Enterococcus counts in the low-fiber consumers. Additionally, this study used potentially biased and low-resolution techniques to detect gut microbes such as fluorescence in situ hybridization, qPCR, and denaturing gradient gel electrophoresis. With these approaches, researchers might overlook the response of potentially important microbes with relatively low abundances.
The impact of polyphenols on the gut microbiota in healthy animals
Consistent with clinical studies, animal studies also showed the increase in beneficial microbes including Lactobacillus and Bifidobacterium in response to polyphenol supplementation (Table 2). Specifically, supplementation of resveratrol (76), curcumin (77), hesperidin (78), anthocyanin-rich blackcurrant extracts (79, 80), water-soluble blueberry extracts (81), and aqueous extract of berry peels (82) increased the abundance of Lactobacillus and/or Bifidobacterium in the cecal or fecal contents in mice. However, supplementation of lowbush wild blueberry powder in male rats for 6 wk showed a decrease in relative abundance of Lactobacillus in the proximal colon contents (83).
TABLE 2.
Effects of polyphenol supplementation on the gut microbiota of healthy animals1
| Compound | Polyphenol dose | Study design | Molecular technique | Results | Reference |
|---|---|---|---|---|---|
| Curcumin | 0.5% wt/wt in diet | 129/SvEv mice, 20 wk | 16S rRNA gene V4 region Illumina sequencing | Prevented age-dependent species richness decrease; relative abundance of Lactobacillale (represented mainly by genus Lactobacillus); relative abundance of Lactobacillale (represented mainly by genus Lactobacillus);  relative abundance of Coribacteriales relative abundance of Coribacteriales
|
(77) |
| Resveratrol | 0.025% wt/wt in diet | Male CD-1 mice, 44 d | 16S rRNA gene V3-V4 region Illumina sequencing |
 Microbial richness measured by observed OTU and Shannon index; various enriched microbes identified by LEfSe Microbial richness measured by observed OTU and Shannon index; various enriched microbes identified by LEfSe |
(84) |
| Resveratrol | 1 mg/kg BW | Male F344 rats, 20 d | In vitro cultivation of cecum contents |
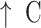 ecal counts of Bifidobacterium and Lactobacillus ecal counts of Bifidobacterium and Lactobacillus
|
(76) |
| Resveratrol | 100 mg/kg BW, gavage | Female BALBA/c mice, 5 d | 16S rRNA and qPCR gene V3-V4 region, Illumina sequencing | Tendency to increase the abundance of Akkermansia muciniphila and Ruminococcus gnavus | (89) |
| Resveratrol | 300 mg/kg BW, gavage | Male C57BL/6J mice, 16 wk | 16S/18S rRNA gene selected regions Illumina sequencing | Complete microbial separation from control;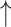 relative abundance of Bacteroides, Lachnospiraceae NK4A136 group, Blautia, Lachnoclostridium, Parabacteroides, and Ruminoclostridium 9 and relative abundance of Bacteroides, Lachnospiraceae NK4A136 group, Blautia, Lachnoclostridium, Parabacteroides, and Ruminoclostridium 9 and 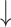 relative abundance of Firmicutes relative abundance of Firmicutes
|
(85) |
| Hesperidin | 100 and 200 mg/kg BW | Male Lewis rats, gavaged 3 times/wk for 4 wk | FISH targeting 16S rRNA |
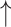 Total cecal bacteria count, total IgA-coated bacteria; Total cecal bacteria count, total IgA-coated bacteria; cecal relative abundance of Lactobacillus/Enterococcus and Staphylococcus spp.; cecal relative abundance of Lactobacillus/Enterococcus and Staphylococcus spp.; cecal relative abundance of C. coccoides/E. rectale cecal relative abundance of C. coccoides/E. rectale
|
(78) |
| Anthocyanin-rich blackcurrant extract | 40-g/kg diet (control diet matched for fiber content) | Male SD rats, 6 wk | qPCR of selected bacterial 16S rRNA gene |
 Cecal counts of Bacteroides-Prevotella-Porphyromonas group and Lactobacillus spp.; Cecal counts of Bacteroides-Prevotella-Porphyromonas group and Lactobacillus spp.; cecal counts of Bifidobacterium and Clostridium perfringens; cecal counts of Bifidobacterium and Clostridium perfringens; cecal acetate and butyrate level and cecal acetate and butyrate level and  cecal propionate level cecal propionate level |
(79) |
| Blackcurrant extract | 13.4 mg/kg BW, gavage 3 times weekly for 4 wk | Male SD rats | FISH targeting 16S rRNA |
 Cecal counts of Bifidobacterium and Lactobacillus; Cecal counts of Bifidobacterium and Lactobacillus; cecal counts of Bacteroides and Closteridia; cecal counts of Bacteroides and Closteridia; cecal enzymetic activity of B-glucosidase cecal enzymetic activity of B-glucosidase |
(80) |
| Water-soluble blueberry extract | 4 mL/kg BW, gavage | Female SD rats, 6 d | FISH targeting 16S rRNA |
 Cecal counts of Bifidobacterium and Lactobacillus Cecal counts of Bifidobacterium and Lactobacillus
|
(81) |
| Berry peel aqueous extract | 1.32 mg gallic acid equivalent/mL in drinking water | Male SD rats, 2 and 7 wk | In vitro cultivation of cecum contents |
 Cecal counts of Bifidobacterium, Lactobacilus, Enterobacteriaceae; Cecal counts of Bifidobacterium, Lactobacilus, Enterobacteriaceae; cecal acetate level after 7-wk compared with 2-wk supplementation cecal acetate level after 7-wk compared with 2-wk supplementation |
(82) |
| Green tea polyphenol | 0.5 and 1.5% wt/vol in drinking water | Female SD rats, 3 and 6 mo | 16S rRNA gene V3-V4 region, Illumina sequencing, shot gun metagenomics | Altered abundance of 10 gene orthologs after 3 mo, while 30 after 6 mo; altered relative abundance of 10 OTUs after 3 mo, 26 after 6 mo; α-diversity for 1.5% after 3 mo and 6 mo, α-diversity for 1.5% after 3 mo and 6 mo,  α-diversity for 0.5% after 6 mo, and α-diversity for 0.5% after 6 mo, and  α-diversity after 3 mo; dosage differentiated gut microbial community; 12 possible correlations between OTUs and clinical measurements α-diversity after 3 mo; dosage differentiated gut microbial community; 12 possible correlations between OTUs and clinical measurements |
(86) |
| Lowbush wild blueberry powder | 24.0 ± 5.2 mg anthocyanins/d + 4.5 g fiber/d | Male SD rats, 6 wk | Whole-genome sequencing |
 Relative abundance of Lactobacillus, Enterococcus, and Relative abundance of Lactobacillus, Enterococcus, and  Bifidobacterium/Coriobacteriaceae ratio; changes in 9% of KEGG Bifidobacterium/Coriobacteriaceae ratio; changes in 9% of KEGG |
(83) |
1BW, body weight; FISH, fluorescence in situ hybridization; KEGG, Kyoto Encyclopaedia of Gene and Genome; LEfSe, Linear discriminant analysis Effect Size; OTU, operational taxonomic unit; rRNA, ribosomal RNA; SD, Sprague-Dawley.
In addition to the changes in specific taxa, researchers have evaluated the impact of polyphenols on the richness and evenness (ɑ-diversity) of gut bacteria and overall gut microbial composition (beta-diversity). For example, supplementation of 0.5% curcumin was found to prevent the age-related reduction in gut microbial richness and maintained high ɑ-diversity until 30 wk of age, but did not change beta-diversity (77). The same increase in ɑ-diversity without affecting beta-diversity was also observed with supplementation of 0.025% resveratrol (84). In contrast, supplementation of resveratrol at 300 mg/kg body weight significantly changed microbial composition compared with control diets (85). Further, supplementation of green tea polyphenol (GTP) (86) significantly differentiated microbial composition from the controls. Interestingly, supplementation of GTP at 0.5% increased ɑ-diversity but at 1.5% decreased ɑ-diversity in female rats (86). These dose-dependent effects are not understood but may result from stronger promotion of specific gut microbes by the higher dosage of polyphenols, which consequently reduce the ɑ-diversity.
Some studies explored the influence of polyphenols on gut microbial metabolism using metagenomic-based microbial functional predictions. For instance, long-term supplementation of GTP dose-dependently increased genes responsible for energy production and conversion (86). These modifications were accompanied by the increase in relative abundance of the phylum Bacteroidetes that possesses microbes capable of degrading resistant biopolymers (87). Moreover, supplementation of lowbush wild blueberries caused a 20% increase in the abundance of gene orthologs responsible for xenobiotic degradation and metabolism in male rats, including pathways of benzoate degradation, glycosaminoglycan degradation, and isoleucine/valine degradation (83). Further, supplementation of lowbush wild blueberries led to an 8-fold reduction in the number of genes assigned to bacterial invasion of epithelial cells compared with controls (83), which is consistent with the observation that blueberry husks inhibited DSS-induced microbial translocation (88). Overall, these data from animal studies are consistent with those observed in humans (70, 69).
Effects of polyphenol supplementation on the gut microbiota under diseased conditions including colitis and CAC
Many studies have documented the protective effects of polyphenols on colitis and CAC in clinically relevant disease models (11). In this section, we focus on the studies that evaluate the impact of polyphenols on the gut microbiota under diseased conditions, including colitis and CAC. Inflammation in the colon may cause outgrowth of pathogenic bacteria and induce microbial dysbiosis. As discussed earlier, some gut microbiota can play causal roles in the exacerbation of colitis and colon cancer, and consequently, gut microbial disturbance has been proposed as one of the promoters for colon carcinogenesis (36). Therefore, alleviating colitis symptoms by modulating gut microbiota or increasing the production of beneficial microbial metabolites is likely important for controlling colitis and chemoprevention of CAC (90).
Effects of polyphenols on the gut microbiota in the colitis models
Many studies have documented polyphenols’ modulatory effects on the gut microbiota in various experimental colitis models (Table 3). In the DSS-induced colitis model in rodents, resveratrol supplementation was found to increase fecal counts of potentially beneficial microbes including Lactobacillus and Bifidobacterium and decrease potentially pathogenic bacteria including E. coli and Enterobacteria compared with controls (76, 84). Interestingly, in the TNBS-induced colitis model, resveratrol restored the colitis-associated decrease in Akkermansia muciniphila and Ruminococcus gnavus (89), which are a mucin degrader (91, 92) and associated with Crohn disease (93), respectively. Resveratrol treatment also reversed the TNBS-caused increase in Bacteroides acidifaciens (89). Moreover, supplementation of nanoparticle curcumin increased the abundance of Clostridium cluster IV and XIVa compared with the control group in the DSS-induced colitis mice (94), and these microbes are known to contain butyrate producers (95). In a T-cell–dependent colitis model induced by the adoptive transfer of naive T cells into Rag1-deficient mice (Rag1-/-mice), quercetin supplementation decreased the relative abundance of the potentially pathogenic microbe E. coli (96). In addition, supplementation of grape seed extract increased the relative abundance of Bacteroides and Lactobacillus and decreased the relative abundance of F. prausnitzii in IL10-/-mice (97). Supplementation of polyphenol-containing whole food including Aronia berry, blueberry, and broccoli resulted in significant changes in specific microbes in the colitis-induced mice (98, 99).
TABLE 3.
Effects of polyphenol supplementation on the gut microbiota of animals with colitis1
| Compound | Polyphenol dose | Study design | Molecular technique | Results2 | Reference |
|---|---|---|---|---|---|
| Resveratrol | 1 mg/kg BW | Male F344 rats, DSS model, 25 d | In vitro cultivation of cecum contents |
 Cecal counts of Bifidobacterium and Lactobacillus compared with diseased control, while Cecal counts of Bifidobacterium and Lactobacillus compared with diseased control, while  cecal counts of Escherichia coli and Enterobacteria compared with diseased control cecal counts of Escherichia coli and Enterobacteria compared with diseased control |
(76) |
| Resveratrol | 100 mg/kg BW | Female BALB/c mice, TNBS model, 5 d | 16S rRNA and qPCR gene V3-V4 region, Illumina sequencing |
 Abundances of Akkermansia muciniphila and Ruminococcus gnavus; Abundances of Akkermansia muciniphila and Ruminococcus gnavus;  abundance of Bacteroides acidifaciens compared with diseased control, as well as various enriched microbes identified by LEfSe; abundance of Bacteroides acidifaciens compared with diseased control, as well as various enriched microbes identified by LEfSe; cecal acetate and isobutyrate level compared with diseased control cecal acetate and isobutyrate level compared with diseased control |
(89) |
| Resveratrol | 0.025% wt/wt in diet | Male CD-1 mice, DSS model, 44 d | 16S rRNA gene V3-V4 regions Illumina sequencing | Compared with diseased control,  microbial richness (Chao1 index) and separated gut microbiota; microbial richness (Chao1 index) and separated gut microbiota;  relative abundances of Bifidobacterium; relative abundances of Bifidobacterium;  relative abundances of Akkermansia, Biophila, Dorea, and Sutterella compared with diseased control, as well as various enriched microbes identified by LEfSe relative abundances of Akkermansia, Biophila, Dorea, and Sutterella compared with diseased control, as well as various enriched microbes identified by LEfSe |
(84) |
| Nanoparticle curcumin | 0.2% wt/wt in diet | Female BALB/c mice, DSS model, 18 d | qPCR of selected bacterial 16S rRNA gene, T-RFLP |
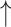 Relative abundances of Clostridium cluster IV and Clostridium subcluster XIV, while Relative abundances of Clostridium cluster IV and Clostridium subcluster XIV, while 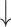 relative abundances of Clostridium cluster XI compared with diseased control; relative abundances of Clostridium cluster XI compared with diseased control; 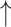 fecal butyrate level under both healthy and diseased conditions fecal butyrate level under both healthy and diseased conditions |
(94) |
| Chlorogenic acid | 1 mM in drinking water | Female C57BL/6 mice, DSS model, 15 d | qPCR and 16S rRNA gene V3 region Illumina sequencing | Microbial separation among healthy control group, DSS group and ChA + DSS group; enhanced DSS-induced reduction in ɑ-diversity; the ratio of Firmicutes to Bacteroidetes, relative abundance of Verrucomicrobia and Akkermansia compared with both healthy and diseased control the ratio of Firmicutes to Bacteroidetes, relative abundance of Verrucomicrobia and Akkermansia compared with both healthy and diseased control |
(101) |
| Salvianolic acid A | 4 and 8 mg/kg BW, intravenous injection | Male SD rats, DSS model, 7 d | 16S rRNA gene V3-V4 regions Illumina sequencing | Microbial separation among no-DSS, DSS and SAA + DSS group; 8 mg/kg dose  DSS-decreased ɑ-diversity; reversed DSS-decreased Firmicutes/Bacteroidetes; DSS-decreased ɑ-diversity; reversed DSS-decreased Firmicutes/Bacteroidetes;  relative abundance of Akkermansia spp., Bacillus spp., Blautia spp., Lactobacillus spp., Lachnoclostridium spp.; while relative abundance of Akkermansia spp., Bacillus spp., Blautia spp., Lactobacillus spp., Lachnoclostridium spp.; while  Bacteroides spp., Roseburia spp., and Ruminiclostridium spp., compared with healthy and diseased control Bacteroides spp., Roseburia spp., and Ruminiclostridium spp., compared with healthy and diseased control |
(100) |
| Quercetin | 10 mg/kg BW, gavage | C57BL/6 Rag1-/-mice, adoptive T-cell transfer model, every 3 d for 7 wk | qPCR of selected 16S rRNA gene, tissue LPS staining |
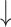 Firmicutes/Bacteroidetes ratio, relative abundance of Proteobacteria, Actinobacteria, segmented filamentous bacteria groups and E. coli;
Firmicutes/Bacteroidetes ratio, relative abundance of Proteobacteria, Actinobacteria, segmented filamentous bacteria groups and E. coli; 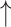 relative abundance of Bacteroides; relative abundance of Bacteroides; 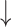 bacteria penetration in the basolateral surface of intestinal epithelial cells compared with diseased control bacteria penetration in the basolateral surface of intestinal epithelial cells compared with diseased control |
(96) |
| Quercetin aglycone/quercetin monoglycosides | 0.15% wt/wt in diet | Female ICR mice, DSS model, 14 d | 16S rRNA gene V4 region Illumina sequencing |
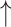 ɑ-Diversity compared with diseased control; ɑ-Diversity compared with diseased control; relative abundance of Firmicutes compared with diseased control relative abundance of Firmicutes compared with diseased control |
(102) |
| Blueberry and broccoli powder | 10% wt/wt in diet (whole food) | Male mdr1a-/-mice, 21 wk | qPCR of selected bacterial 16S rRNA gene | Both powder diets  relative abundance of Enterococcus spp., Lactobacillus spp., Clostridium perfringens, E. coli, while broccoli diet relative abundance of Enterococcus spp., Lactobacillus spp., Clostridium perfringens, E. coli, while broccoli diet  relative abundance of F. prausnitzii and total bacteria in mesenteric lymph nodes compared with diseased control relative abundance of F. prausnitzii and total bacteria in mesenteric lymph nodes compared with diseased control |
(98) |
| Aronia berry | 4.5% wt/wt in diet | C57BL/6 Rag1-/-mice, adoptive T-cell transfer model, 5 or 7 wk | 16S rRNA gene V4 region Illumina sequencing | Complete microbial separation among healthy control group, diseased group, and diseased supplementation group; ɑ-diversity and relative abundance of Firmicutes compared with diseased control ɑ-diversity and relative abundance of Firmicutes compared with diseased control |
(99) |
| Salvianolic acid B | 100 mg/kg BW | Female C57BL/6 mice, 14 d | 16S rRNA gene V3 region Illumina sequencing |
 ɑ-Diversity compared with diseased control; ɑ-Diversity compared with diseased control; predicted gene families for enzymes, predicted gene families for enzymes,  predicted cancer-related pathways and lipid metabolism pathways compared with diseased control; predicted cancer-related pathways and lipid metabolism pathways compared with diseased control; serum concentration of acetic acid, propionic acid, valerianic acid, isobutyric acid, and isovalerianic acid compared with diseased control serum concentration of acetic acid, propionic acid, valerianic acid, isobutyric acid, and isovalerianic acid compared with diseased control |
(103) |
| Grape seed extract | 1% wt/wt in diet | Female IL10-/- mice, 16 wk | qPCR of selected bacterial 16S rRNA gene |
 Relative abundance of Bacteroides and Lactobacillus; while Relative abundance of Bacteroides and Lactobacillus; while  Faecalibacterium prausnitzii compared with diseased control Faecalibacterium prausnitzii compared with diseased control |
(97) |
BW, body weight; ChA, chlorogenic acid; DSS, dextran sulfate sodium; LEfSe, Linear discriminant analysis Effect Size; mdr1a−/−,multidrug resistance gene deficient; Rag1-/-, recombination activating gene-1–deficient; rRNA, ribosomal RNA; SAA, salvianolic acid A; SAB, salvianolic acid B; SD, Sprague-Dawley; TNBS, 2,4,6-trinitrobenzenesulfonic acid; T-RFLP, terminal restriction fragment length polymorphism.
All studies showed that polyphenols alleviate experimental colitis.
In addition to documenting the changes in specific microbes, researchers have also reported the modulation of the microbial community, including ɑ- and beta-diversity, in response to polyphenol supplementation in murine colitis models (Table 3). For example, in the DSS-induced colitis models, administration of salvianolic acid A (100) and chlorogenic acid (101) induced significant separation of gut microbial composition among healthy control, colitis control, and polyphenol-supplemented colitis animals. Further, salvianolic acid A mitigated the colitis-induced decline in ɑ-diversity (100). The same salvage of ɑ-diversity has been observed as a result of supplementing quercetin aglycones/monoglycosides (102), resveratrol (84), polyphenol-rich Aronia berry (99), and salvianolic acid B (103) in murine colitis models. In contrast to the increase in ɑ-diversity, chlorogenic acid supplementation ameliorated experimental colitis and decreased ɑ-diversity compared with disease controls (101). Interestingly, the supplementation of chlorogenic acid markedly elevated Akkermansia, which is a recently identified mucin degrader and believed to have positive impacts on gut health (91, 92). Therefore, the substantial increase in this potentially beneficial microbe likely contributes to the uneven distribution of bacterial species that resulted in reduced ɑ-diversity (101). These observations suggest that the nature of the gut microbial alteration (e.g., expansion of a specific bacterium) may be more relevant to determining health outcomes than the overall changes in diversity measurements per se.
Since gut microbiota are considered a collective functional domain, it is critical to evaluate how polyphenols have impacts on the microbial functions besides the modulation of the microbial composition (104). To this end, quercetin has been shown to reduce colitis-induced bacterial penetration into the basolateral surface of colonic epithelial cells (96). A polyphenol-rich broccoli diet decreased translocation of total bacteria to the mesenteric lymph nodes in a mouse model of IBD (98). Moreover, supplementation of nanoparticle curcumin and resveratrol increased fecal/cecal butyrate concentration (89, 94), and salvianolic acid B elevated serum SCFAs in mice with colitis (103). These results indicate that polyphenol-mediated modulation of gut microbial functions, including suppression of epithelial invasion and increase in SCFA production, may contribute to the mitigation of colitis by these compounds (36, 105).
The effect of polyphenols on the gut microbiota in experimental CAC models
Many studies that investigated the effect of polyphenols on CAC development were carried out in the AOM-DSS model, which resembles histopathological and molecular profiles of CAC in humans (106). Briefly, AOM, a pre-carcinogen, is metabolized in the liver and consequently becomes an active mutagen that induces cancer-driving genetic mutations in the colon epithelial cells (107). Meanwhile, DSS induces colon inflammation that promotes colonic tumorigenesis via disrupting the gut barrier and forming fusible nanoparticles with medium-chain length fatty acids (108). In addition to the AOM-DSS model, IL-10 knockout (IL10-/-) mice coupled with AOM injection are also used to mimic CAC as IL-10 is a key anti-inflammatory regulator for IBD (109).
Studies have documented the impact of polyphenols on the gut microbiota in the CAC models (Table 4). Supplementation of isoliquirtigenin, a flavonoid extracted from traditional Chinese medicine licorice, increased the relative abundance of potentially beneficial microbes including Butyricicoccus, Clostridium, and Ruminococcus and decreased the relative abundance of IBD-associated bacteria including Escherichia, Enterococcus, and Helicobacteraceae, compared with the control group in the CAC mice (110). Importantly, the microbial composition of isoliquirtigenin-supplemented CAC mice was similar to that of healthy mice at the end of the 18-wk study. Similarly, 13-wk supplementation of EGCG increased the relative abundance of probiotics such as Lactobacillus, Bifidobacterium, and Ruminococcus compared with the CAC control mice (111). Like isoliquirtigenin, there was no significant difference in the relative abundance of the compared microbes between the healthy control and EGCG-supplemented CAC mice, except for Lactobacillus. Consistent with these studies, Chen et al. (112) observed greater gut microbial similarity between the healthy controls and the CAC mice that were supplemented with anthocyanin extracted from black raspberry than the CAC mice fed the control diet. In this study, supplementation of anthocyanin from black raspberry reversed AOM-DSS–caused decrease in beneficial microbes such as Eubacterium rectale, F. prausnitzii, and Lactobacillus, while it counteracted AOM-DSS–increased pathogenic microbes such as Campylobacter and Helicobacter pylori (112). Furthermore, supplementing anthocyanin-rich sausage also reduced the relative abundance of a proinflammatory microbe named Bilophila wadsworthia (113) in the CAC mice compared with the non-anthocyanin control (114). In contrast to the DSS model, AOM-DSS treatment increased ɑ-diversity, which was reversed by polyphenol supplementation (110, 111).
TABLE 4.
Effects of polyphenol supplementation on the gut microbiota of animals with colitis-associated colorectal cancer1
| Compound | Polyphenol dose | Animal model | Molecular technique | Results2 | Reference |
|---|---|---|---|---|---|
| Isoliquiritigenin | 30, 75, 150 mg/kg BW via gavage, 6 times weekly for 12 and 18 wk | Male Balb/c mice, AOM-DSS model | T-RFLP,16S rRNA gene V3-V5 region Illumina sequencing | Dose-specific effects on richness and ɑ-diversity; stabilized disease-shifted gut microbial structure; 150 mg/kg BW dosage relative abundance of Bacteriodes spp., Prevotella, Butyricicoccus, Clostridium, Ruminococcus,Lachnospiraceae, Rikenellaceae, while relative abundance of Bacteriodes spp., Prevotella, Butyricicoccus, Clostridium, Ruminococcus,Lachnospiraceae, Rikenellaceae, while  relative abundance of Helicobacteraceae,Turicibacter, and Enterococcus relative abundance of Helicobacteraceae,Turicibacter, and Enterococcus
|
(110) |
| Epigallocatechin gallate | 1% vol/vol in drinking water, gavage daily for 8 and 12 wk | Female FVB/N mice, AOM-DSS model | 16S rRNA gene V4 region pyrosequencing | Stabilized disease-shifted gut microbial structure; time-dependent effects; relative abundance of Lactobacillus,Clostridiaeae,Fusobacterium,Ruminococcus,Ochrobactrum,Veillonella,Desulfococcus,Prevotella,Enterobacteriaceae,Sulfurimonas, while relative abundance of Lactobacillus,Clostridiaeae,Fusobacterium,Ruminococcus,Ochrobactrum,Veillonella,Desulfococcus,Prevotella,Enterobacteriaceae,Sulfurimonas, while  relative abundance of Bacteroides,Anaerotruncus, and Faecalibaterium compared with diseased control relative abundance of Bacteroides,Anaerotruncus, and Faecalibaterium compared with diseased control |
(111) |
| Curcumin | 0.5% wt/wt in diet, 20 wk | 129/SvEv mice, IL10-/-, AOM model | 16S rRNA gene V4 region Illumina sequencing |
 Relative abundance of Lactobacillales,Bifidobacteriales,Erisipelotrichales, Coriobacteriales,Cyanobacteria YS2, and Lactobacillus, while Relative abundance of Lactobacillales,Bifidobacteriales,Erisipelotrichales, Coriobacteriales,Cyanobacteria YS2, and Lactobacillus, while  relative abundance of Clostridiales compared with diseased control; 19 predicted microbial gene categories were identified with no difference between healthy control and 0.5% supplementation group, but all differed from diseased control group relative abundance of Clostridiales compared with diseased control; 19 predicted microbial gene categories were identified with no difference between healthy control and 0.5% supplementation group, but all differed from diseased control group |
(77) |
| Anthocyanin | Anthocyanin (0.11%)-rich sausages, 21 wk | Male F344 rats, AOM-DSS model | 16S rRNA gene ION PGM next-generation sequencing |
 Relative abundance of Clostridiaceae, while Relative abundance of Clostridiaceae, while  relative abundance of Desulfovibrionaceae,Enterobacteriaceae (family) and Bilophila wadsworthia (genus) compared with diseased control fed matched sausage relative abundance of Desulfovibrionaceae,Enterobacteriaceae (family) and Bilophila wadsworthia (genus) compared with diseased control fed matched sausage |
(114) |
| Black raspberry anthocyanin extract | 7.0 μmol/g diet, 12 wk | C57BL/6J mice, AOM-DSS model | T-RFLP and qPCR of selected bacterial 16S rRNA gene |
 Desulfovibrio sp., Enterococcus spp.;
Desulfovibrio sp., Enterococcus spp.;  Eubacterium rectale,Faecalibacterium prausnitzii, and Lactobacillus, compared with diseased control; Eubacterium rectale,Faecalibacterium prausnitzii, and Lactobacillus, compared with diseased control; relative abundance of butyrate-producing bacteria and Neisseria; relative abundance of butyrate-producing bacteria and Neisseria;  Campylobacter,H. pylori,Bacteroides, and Prevotella compared with diseased control Campylobacter,H. pylori,Bacteroides, and Prevotella compared with diseased control |
(112) |
AOM, azoxymethane; BW, body weight; DSS, dextran sulfate sodium; FVB/N, Friend Virus B NIH Jackson, FVB; ION PGM, Ion Torrent™ Personal Genome Machine™ (PGM); rRNA, ribosomal RNA; T-RFLP, terminal restriction fragment length polymorphism.
All studies showed that polyphenols inhibited colitis-associated colorectal cancer in different animal models.
Similar to the beneficial effects observed in the AOM-DSS models, supplementation of curcumin was found to reduce tumor burden and caused favorable changes in gut microbiota in AOM-injected IL-10-/- mice (77). In this study, the gut microbial composition in curcumin-supplemented CAC mice was similar to that in healthy controls at the order level and showed increased relative abundance of Lactobacillus compared with the CAC controls. In the subsequent analysis with PICRUSt, the researchers found that the microbes in curcumin-supplemented, CAC-induced mice had a decreased relative abundance of genes involved in bacterial chemotaxis, motility protein synthesis, and arachidonic acid metabolism compared with the CAC mice fed the control diet.
To sum up, the above-cited studies indicate that polyphenols appear to restore colitis- or CAC-induced changes in gut microbes, including counteracting disease-induced decrease in probiotics and increase in potential pathogenic bacteria. As a result, polyphenol-supplemented animals often possess the microbial profiles resembling healthy control animals, indicating that polyphenols provide resistance to colitis- or CAC-induced dysbiosis.
Mechanisms underlying the interaction between polyphenols and the gut microbiota
While research in humans and animals indicates that polyphenols can selectively enhance specific gut microbes, the mechanisms underlying these modulatory effects are not well understood. Existing evidence suggests that polyphenols likely modulate gut microbes through their mutual interactions. On the one hand, polyphenols may affect the growth of bacteria via inhibiting nucleotide synthesis, chelating metabolically essential metal ions, interacting with cell membrane components, or facilitating electron transfer, which is critical to bacterial energy metabolism. On the other hand, gut microbes can catabolize polyphenols, which may, in turn, modulate the microbial community.
Polyphenols regulate the growth of gut bacteria via various distinct mechanisms
Polyphenols are known to have antibacterial properties, which are their originally recognized functions in the plant's defense system. Several studies have revealed polyphenols’ antibacterial activities using representative gut microbes including common intestinal pathogens (115–118). For instance, resveratrol at 1.56 μg/mL (6.8 μM) has been shown to inhibit biofilm formation of Fusobacterium nucleatum under anaerobic conditions (119). It is noteworthy that F. nucleatum, a commensal-turned pathogenic bacterium, has been reported to be elevated in the colonic biopsies of IBD patients (120) and is believed to promote colorectal carcinogenesis (121, 122). Moreover, polyphenols appear to selectively inhibit bacterial growth, probably because specific microbes possess distinct metabolism. For instance, tannic acid, under the anaerobic condition, inhibited the growth of intestinal bacteria such as Clostridium clostridiiforme, E. coli American Type Culture Collection (ATCC) 25,922 and Enterobacter cloacae ATCC 13,047, whereas Bifidobacterium infantis and Lactobacillus acidophilus remained unaffected (117). Since tannic acid is a strong chelator of iron, the inhibition of bacterial growth by this polyphenol is likely rooted in its deprivation of iron needed for bacterial metabolism, whereas neither Bifidobacterium infantis nor Lactobacillus acidophilus requires iron for growth (117). Additionally, although further validation is needed, mechanisms that were identified under aerobic conditions may also be applied to polyphenols’ anti-growth effects on anaerobes (123), including disruption of bacterial quorum sensing (124, 125), a universal communication pathway that also exists among gut microbes (126), inhibition of bacterial DNA and RNA synthesis (33, 127, 128), disruption of membrane function and permeability (129–131), or interference of bacterial biofilm formation (132, 133).
In contrast to antibacterial activities, polyphenols can promote the growth of gut bacteria via facilitating extracellular electron transfer (EET) or general anaerobic respiration in microbes. In the colon, flavin is believed to act as electron shuttles in the EET system to boost the growth of the intestinal anaerobes by accepting electrons from microbes and donating them to extracellular electron acceptors such as metal ions (134, 135). Like flavin, many natural polyphenols, in particular flavonoids with ortho-dihydroxyl groups on the B ring, possess such reversible redox behavior based on cyclic voltammetry scanning (136). Importantly, the gene orthologs responsible for EET are present in multiple intestinal microbes such as Clostridium perfringens, Enterococcus faecalis,Streptococcus dysgalactiae, Lactococcus spp., and Lactobacillus spp., making EET a possible target for polyphenols in the intestinal environment. Furthermore, besides serving as potential electron shuttles for EET, polyphenols can chelate redox-active metal ions such as iron and copper, which could be utilized in EET and therefore indirectly affect the growth of EET-based microbes (137). Alternatively, polyphenols with double bonds on the side chain, such as caffeic acids, may act as terminal electron acceptors to accelerate anaerobic respiration (138). To sum up, polyphenols can selectively affect the metabolism of individual microbes, which consequently modulates the microbial composition.
Gut microbiota extensively metabolize polyphenols and this interaction may, in turn, change microbial composition and metabolism
Accumulating research has shown that the gut microbiota can extensively metabolize polyphenols. In the food matrix, polyphenols are often in glycosylated forms, which limits their initial intestinal absorption (139). These polyphenol glycosides can be hydrolyzed into free polyphenols via small intestinal 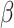 -glucosidase (140) or microbial enzymes such as α-rhamnosidase, β-glucosidase, and β-glucuronidase (141). Once deconjugated, free polyphenols can be readily metabolized by gut microbes and undergo catabolism into phenolic acids (141). The catabolic reactions include ester hydrolysis, ring-cleavage by microbial decarboxylase, demethylation by microbial demethylase, and reduction by microbial dehydroxylase or hydrogenase (34, 142). The resultant metabolites may be excreted into feces or absorbed for further colonocyte and liver metabolism. Representative metabolites of polyphenols by gut microbes include urolithin, equol, and hesperitin, which are metabolized from ellagitannins, isoflavones, and flavanones, respectively (34).
-glucosidase (140) or microbial enzymes such as α-rhamnosidase, β-glucosidase, and β-glucuronidase (141). Once deconjugated, free polyphenols can be readily metabolized by gut microbes and undergo catabolism into phenolic acids (141). The catabolic reactions include ester hydrolysis, ring-cleavage by microbial decarboxylase, demethylation by microbial demethylase, and reduction by microbial dehydroxylase or hydrogenase (34, 142). The resultant metabolites may be excreted into feces or absorbed for further colonocyte and liver metabolism. Representative metabolites of polyphenols by gut microbes include urolithin, equol, and hesperitin, which are metabolized from ellagitannins, isoflavones, and flavanones, respectively (34).
In addition to the formation of polyphenol catabolites, the interaction of polyphenols and the gut microbiota also results in enhanced production of SCFAs or their precursors. Such interaction has been observed via in vitro incubation of polyphenols with fecal samples (143, 144) and as a result of in vivo polyphenol supplementation (145–147). For instance, under in vitro settings, formation of SCFAs was observed to be accompanied with increased polyphenol catabolites and elevation of potential SCFAs producers including F. prausnitzii and cluster XIVa Clostridiales (144) as well as microbes that aid with SCFA production such as Bifidobacterium (143, 144). Interestingly, Parkar et al. (143) showed that the growth of Bifidobacterium was stimulated by polyphenol catabolites in the fermenta, but not by the original polyphenols. Therefore, polyphenols or their microbiota-produced metabolites may enrich SCFA producers to make SCFAs from sugar molecules in the culturing media. In addition, under the whole-body environment, polyphenols including EGCG have been shown to inhibit 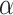 -amylase in the upper gastrointestinal tract, which likely results in an increase in fermentable substrates from the undigested starch. Consequently, the increased substrates become available to colonic SCFA producers, which enhances SCFA production (148, 149).
-amylase in the upper gastrointestinal tract, which likely results in an increase in fermentable substrates from the undigested starch. Consequently, the increased substrates become available to colonic SCFA producers, which enhances SCFA production (148, 149).
In summary, although the mechanism underlying polyphenols’ impact on gut microbes is not fully understood, the interaction of the 2 parties is essential for the mutual modulatory effects; specifically, polyphenols, which can reach relatively high concentrations in the gut, may selectively influence bacterial growth, while the process of gut microbe–involved metabolism of polyphenols may, in turn, change the microbial composition. This mutual interaction results in breaking down polyphenols to various metabolites, modulating the gut microbiota, and increasing the production of SCFAs (Figure 1).
FIGURE 1.
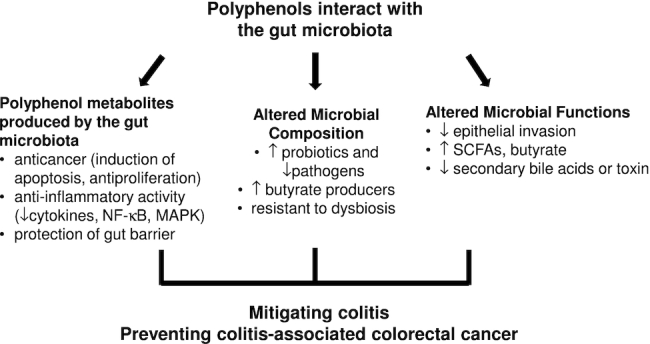
Roles of the polyphenol–gut microbiota interaction in polyphenols’ protective effects against colitis and colitis-associated colorectal cancer. The interaction between polyphenols and the gut microbiota can modulate gut microbial composition and functions. Specifically, polyphenols may promote butyrate producers and suppress pathogenic bacteria, which may lead to the increase in SCFAs and suppression of epithelial invasiveness, respectively. Additionally, gut microbes metabolize polyphenols into absorbable metabolites, which have been shown to have anticancer and anti-inflammatory activities and provide protection of gut barrier functions. These activities contribute to polyphenols’ disease-alleviation effects in animals and humans. MAPK, mitogen-activated protein kinase.
Roles of the polyphenol–gut microbiota interaction in polyphenols’ anti-colitis and anti-CAC effect
The mutual interaction between gut microbiota and polyphenols likely plays key roles in polyphenols’ protective effects on colitis and CAC, as summarized in Figure 1. On the one hand, polyphenols can alter the composition and functions of gut microbiota, which may improve gut health. On the other hand, the metabolites generated via gut microbiota–mediated catabolism of polyphenols can offer anticancer and anti-inflammatory effects, protect gut barrier integrity, and therefore play significant roles in anti-colitis and anti-CAC actions.
The role of polyphenols’ modulation of the gut microbiota in the mitigation of colitis and prevention of CAC
Based on the above-cited literature, polyphenol supplementation appears to be capable of increasing the level of known probiotics under both healthy and diseased conditions, including Lactobacillus and Bifidobacterium (64–67, 70–73, 76–84, 111, 112), the emerging probiotic Akkermansia (89, 101, 100), and potential butyrate producers such as F. prausnitzii, E. rectale (112), Clostridium cluster IV and XIVa (94), and Butyricicoccus (110). Further, polyphenols can reduce some opportunistic pathogens and proinflammatory microbes such as E. coli (76, 96, 98) and B. wadsworthia (114). Indeed, there is ample evidence showing that Lactobacillus (lactic acid bacteria) and Bifidobacterium attenuate colitis and inhibit the development of CRC including CAC. In particular, several lactic acid bacteria have been shown to inhibit CRC by inducing cancer cell apoptosis, anticancer immune response, and epigenetic modification of tumor suppressor genes (150). Jacouton et al. (151) found that Lactobacillus casei BL23 inhibited CAC development, reduced colonic IL-22 concentration, and increased caspase-7, -9, and Bik expression to promote apoptosis in the AOM-DSS–induced CAC model in mice. Moreover, Mizuno et al. (152) demonstrated that co-cultured Lactobacillus species including L. casei inhibits the growth of potentially pathogenic E. coli in vitro. With regard to Bifidobacterium spp., multiple clinical trials and animal studies have demonstrated their anti-colitis effects in the form of fermented milk products (153), synbiotics (154, 155), or single strains (156), while their anti-CAC effects were observed in animal models either alone (157) or mixed with other Lactobacillus strains (158, 159). Interestingly, there appears to be cross-feeding between lactic acid bacteria and butyrate producers, as the latter microbes utilize lactate produced by the former bacteria to produce butyrate, which is known to inhibit colon cancer development (160). These results, together with the observation that polyphenol supplementation promotes these “beneficial” microbes, strongly suggest that polyphenols likely have prebiotic activities contributing to polyphenols’ anti-colitis and anti-CAC actions.
In addition to its enhancing effect on probiotics, animal studies have revealed that polyphenol supplementation reverses disease-associated changes in the gut microbial profile. Thus, the gut microbial composition in polyphenol-treated disease animals often resembles that in healthy ones (77, 110–112). These results indicate that polyphenols can stabilize the gut microbial community and make it resistant to disease-caused dysbiosis.
Additionally, polyphenol consumption leads to functional changes in microbial metabolism, which is relevant to the attenuation of the diseases. For instance, polyphenols have been shown to increase energy production and conversion (86), elevate production of SCFAs (69, 82) and beta-glucosidase activity (80), and decrease epithelial invasion (83) and secondary bile acid production (69, 161) as well as toxins secreted by pathogenic bacteria (162).
Anticancer effects of polyphenol metabolites generated by the gut microbiota
Since gut microbiota substantially metabolize polyphenols, studies have been conducted to investigate the potential anticancer effects of gut microbe–mediated polyphenol metabolites. Specifically, Wang et al. (163) identified baicalein as a major metabolite of baicalin from Scutellaria baicalensis extract and showed that, compared with the parental compound, baicalein exhibited enhanced antiproliferation and proapoptotic effects, and increased cell cycle arrest at S phase in colon cancer cells. Consistently, baicalein more strongly inhibited tumor growth than baicalin in a xenograft nude mouse model (163). Similarly, metabolites of ellagitannin and ellagic acid by gut microbes decreased the number and size of colonospheres formed by Caco-2 cells and primary colon tumor cells (164), inhibited cancer cell proliferation and induced cell cycle arrest at the G2M and S phase (165). Moreover, 8-prenylnaringenin, a microbial metabolite of isoxanthohumol, has been shown to inhibit colon cancer cell growth, and reduce H2O2-induced DNA damage and cell invasiveness (166). In addition, metabolites of quercetin from 7 bacteria strains showed dose-dependent antiproliferation effects on HCT-116 cells (167). Interestingly, the quercetin metabolites generated from different bacteria strains showed differential antiproliferation potency, suggesting potentially distinct polyphenol metabolites produced by different microbes. Additionally, 3,4-dihydroxyphenylacetic acid and 3-(3,4-dihydroxyphenyl)-propionic acid, which are microbe-produced metabolites of quercetin and chlorogenic acid, respectively, decreased cyclooxygenase-2 expression and modulated CumOOH-induced DNA damage in colon adenoma cells (168). These results indicate that polyphenol metabolites produced by the gut microbiota may directly contribute to polyphenols’ anticancer effects.
However, not all microbially made polyphenol metabolites showed superior anticancer effects compared with the original compounds. For instance, after in vitro fermentation, antiproliferation effectiveness of palm date polyphenol-rich extract decreased by >2 fold in Caco-2 cells (71). Although the nature of the reduction remained unknown, it might result from the reduction in originally active compounds or the formation of inactive metabolites.
Regardless of the positive or negative impact of metabolites on cancer compared with the parental compounds, the above-mentioned studies strongly suggest that analyses of polyphenol metabolites, together with microbiota alteration and disease outcomes, are needed to understand the role of polyphenols in their modulation of gut microbes and anticancer mechanisms.
Anti-inflammatory activities and protective effects on gut barrier integrity by polyphenol metabolites synthesized by the gut microbiota
Some polyphenol metabolites have been shown to have anti-inflammatory activities in vitro and in vivo. For instance, 3,4-dihydroxyphenylpropionic acid (hydrocaffeic acid), which is a major gut microbe–producing metabolite of caffeic acid and chlorogenic acid in the colon, has been shown to inhibit LPS-stimulated TNF-α, IL-1β, and IL-6 in peripheral blood mononuclear cells isolated from healthy volunteers (169). In another study, Larrosa et al. (170) reported that hydrocaffeic acid attenuated DSS-induced colitis and cytokines including TNF-α, IL-1β, and IL-8 in rats. Further, these investigators showed that hydrocaffeic acid and other polyphenol metabolites, such as dihydroxyphenyl acetic and hydroferulic acid, effectively inhibited IL-1β–stimulated prostaglandin E2 (PGE2) in CCD-18 colon fibroblasts. Furthermore, ferulaldehyde, a water-soluble microbial degradation product of polyphenols and having high concentrations in human urine after red wine and chocolate consumption (171, 172), was reported to prolong the survival and attenuate inflammatory response in a murine LPS-induced septic shock model (173). In addition, Tucsek et al. (174) demonstrated that ferulaldehyde suppressed LPS-induced reactive oxygen and nitrogen species and activation of NF-κB via blocking MAPK pathways in macrophages.
Emerging evidence shows that polyphenol metabolites may be protective to gut barrier functions. Urolithin A (UroA), a major microbe-producing metabolite of polyphenolics from berries and pomegranate fruits, has been shown to have anti-inflammatory, antioxidative, and antiaging activities (175). Recently, Singh et al. (176) reported that UroA and its analogue protected gut barrier functions by upregulation of epithelial tight junction proteins such as zona occludens 1 (ZO-1), occluding, and claudin 4 in colon epithelial cells. Mechanistic studies indicate that the increase in tight junction proteins was through activation of aryl hydrocarbon receptor (AhR) and nuclear erythroid 2-related factor 2 (Nrf2)–dependent pathways (176). Consistently, treatment with UroA and its analogue attenuated TNBS-induced colitis and gut barrier dysfunction, and inhibited proinflammatory cytokines in mice (176). These data therefore provide direct evidence that microbial polyphenol metabolites protect gut barrier integrity, in addition to anti-inflammatory effects.
To sum up, polyphenol-mediated resistance to disease-associated microbial dysbiosis, expansion of potential beneficial microbes, and inhibition of pathogenic microbes likely play significant roles in the attenuation of colonic inflammation and maintaining gut barrier integrity. Additionally, polyphenol catabolites and SCFAs as a result of the polyphenol–microbiota interaction may mitigate colitis via anti-inflammation and protection of barrier functions, and inhibit the development of colonic carcinogenesis through various anticancer mechanisms. Together, the polyphenol–gut microbiota interaction may contribute to polyphenols’ protective effects against colitis and CAC (Figure 1).
Knowledge gaps in the understanding of the role of gut microbial modulation in polyphenols’ health benefits—challenges and opportunities
Despite existing evidence suggesting the prebiotic role of polyphenols in maintaining gut health, little research has been conducted to establish the causative link between polyphenols’ modulation of microbiota and disease-alleviation effects. In particular, polyphenols are known to have anticancer and anti-inflammatory effects, which may lead to the mitigation of colitis and prevention of colitis-caused dysbiosis. In other words, modulation of gut microbes may be a consequence, rather than a cause, of polyphenol-mediated beneficial effects. To address whether the polyphenol–gut microbiota interaction is critical to this compound-mediated protection against diseases, an effective approach is to test polyphenols’ impact on diseases in germ-free animals or in conventional animals treated with pan-antibiotics that deplete gut bacteria. The lack of or weakening of protective effects in animals without the gut microbiota will support a critical role of the polyphenol–gut microbiota interaction in disease prevention (177). Moreover, FMT may be used to examine the potential impact of modulated gut microbiota on polyphenol-mediated disease prevention (178, 179). Recently, in the TNBS-induced colitis model, Alrafas et al. (89) observed that transferring fecal materials from resveratrol-fed and colitis-induced mice, rather than those from control diet–fed colitis mice, ameliorated colitis symptoms in gut bacteria–depleted mice. Although this observation supports the causality of resveratrol-changed gut microbes in anti-colitis activities, the results are somewhat confounded with FMT using feces from resveratrol-fed diseased mice. Further research is needed to test the anti-colitis effects of fecal samples from resveratrol-fed healthy mice. In addition, utilization of polyphenols as prebiotics may be another approach to validate key microbes responsible for alleviation of diseases (180) and the causal role of the polyphenol–gut microbiota interaction in attenuation of colitis and CAC. In these studies, gene knockout or other molecular approaches may be applied to elucidate in-depth mechanisms underlying polyphenol–gut microbiota interactions at the molecular level (181, 182).
In addition to the necessity of establishing a causative role of polyphenol–gut microbiota interactions in protection against diseases, research using cutting-edge technologies is needed to overcome current challenges in gut microbiota analyses to further understand the role of the gut microbiota in polyphenol-mediated health benefits. First, polyphenol-altered gut microbes have often been identified at the genus, family, and phylum level. The lack of taxonomic determination of microbes at the species or strain level hinders accurate interpretation of microbial functions and therefore health outcomes. This concern is underscored by the observation that different strains of Lactococcus lactis are found to show varied ability to inhibit CAC development (183). Nevertheless, with the recent advances in microbiome analysis (184), it is foreseeable that rigorous identification of species or strains will be achieved through optimized maker gene amplification methods (185), and advanced algorithms for sequencing error correction (186) and taxonomic classification (187). Second, even with accurate classification of a microbe's identity, it may still be difficult to determine its function in vivo. This is because individual microbes may acquire new functions through horizontal gene transfer among each other (188), and may carry out specific functions under unique host conditions, such as the E. coli pks strain, which promotes colon carcinogenesis only under inflammatory conditions (45). Further, the gut microbial community has developed functional redundancy to improve resilience and resistance to environmental perturbations (189). As a result, alteration of the microbial profile does not necessarily mean functional changes in the community. Indeed, realization of functional redundancy and adaptive changes in gut microbiota has urged researchers to shift focus to the functional analysis of gut microbes, including targeted microbial functional outcomes such as SCFA production (89, 94, 103), host response to bacterial translocation such as LPS in the circulation (190, 191), exploratory metagenomic predictions (83, 86), and extensive metabolomic analyses of polyphenols and microbial metabolites. Finally, to translate the observations in animal models to clinical use in humans, it is important to consider differences in gut bacterial compositions between animals and humans (192, 193). Fortunately, despite the low percentage of shared genes between mouse and human gut microbiota, the mouse gut microbiome is functionally similar to its human counterparts (194). In the future, investigation of shared microbial functional outcomes between animals and humans and utilization of humanized animals inoculated with human gut microbiota may help obtain clinically relevant data for the translation of basic discovery to humans (195, 196).
Conclusions
Existing evidence indicates that polyphenols can elevate probiotics capable of alleviating colitis and inhibiting CAC, decrease opportunistic pathogenic or proinflammatory microbes, reverse disease-induced dysbiosis, and change microbial functions such as increasing butyrate formation. Polyphenol metabolites produced by the gut microbiota have anticancer and anti-inflammatory activities and are protective to gut barrier integrity. Future research is needed to establish causation between polyphenols’ disease-preventive effects and modulation of the gut microbiota in germ-free mice or with an FMT approach. The polyphenol–gut microbiota interaction should be further delineated by performing microbial functional analysis with metabolomic, transcriptomic, and proteomic approaches (197). Ultimately, these different lines of research should achieve the goal of personalized nutrition and precision medicine (198), such as designing specific polyphenol–microbe synbiotics or adjustment of polyphenol consumption based on the individual's microbial profile and baseline physiological status for optimal disease alleviation.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—YZ and QJ: are responsible for the design, writing, and final content and read and approved the final version of the manuscript.
Notes
Supported by a Purdue Center for Cancer Research Phase I grant and a research grant from California Table Grape Commission.
Author disclosures: The authors report no conflicts of interest.
Abbreviations used: AOM, azoxymethane; ATCC, American Type Culture Collection; CAC, colitis-associated colorectal cancer; CRC, colorectal cancer; DSS, dextran sulfate sodium; EET, extracellular electron transfer; EGCG, epigallocatechin gallate; FMT, fecal microbiota transplantation; GTP, green tea polyphenol; IBD, inflammatory bowel disease; PICRUSt, phylogenetic investigation of communities by reconstruction of unobserved states; RAG, recombination activating gene; TNBS, 2,4,6-trinitrobenzene sulfonic acid; UroA, urolithin A.
Contributor Information
Yiying Zhao, Department of Nutrition Science, Purdue University, West Lafayette, IN, USA.
Qing Jiang, Department of Nutrition Science, Purdue University, West Lafayette, IN, USA.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA A Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Lakatos PL, Lakatos L. Risk for colorectal cancer in ulcerative colitis: changes, causes and management strategies. World J Gastroenterol. 2008;14(25):3937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lutgens MW, van Oijen MG, van der Heijden GJ, Vleggaar FP, Siersema PD, Oldenburg B. Declining risk of colorectal cancer in inflammatory bowel disease: an updated meta-analysis of population-based cohort studies. Inflamm Bowel Dis. 2013;19(4):789–99. [DOI] [PubMed] [Google Scholar]
- 4. Sobczak M, Wlazłowski M, Zatorski H, Sałaga M, Fichna J. Current overview of colitis-associated colorectal cancer. Open Life Sci. 2014;9(11):1022–9.. doi: 10.2478/s11535-014-0345-7. [Google Scholar]
- 5. Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–14 e5. [DOI] [PubMed] [Google Scholar]
- 6. Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2009;6(5):297–305. [DOI] [PubMed] [Google Scholar]
- 7. Kraus S, Arber N. Inflammation and colorectal cancer. Curr Opin Pharmacol. 2009;9(4):405–10. [DOI] [PubMed] [Google Scholar]
- 8. Fantini MC, Pallone F. Cytokines: from gut inflammation to colorectal cancer. Curr Drug Targets. 2008;9(5):375–80. [DOI] [PubMed] [Google Scholar]
- 9. Cathcart MC, Lysaght J, Pidgeon GP. Eicosanoid signalling pathways in the development and progression of colorectal cancer: novel approaches for prevention/intervention. Cancer Metastasis Rev. 2011;30(3–4):363–85. [DOI] [PubMed] [Google Scholar]
- 10. DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–71. [DOI] [PubMed] [Google Scholar]
- 11. Nunez-Sanchez MA, Gonzalez-Sarrias A, Romo-Vaquero M, Garcia-Villalba R, Selma MV, Tomas-Barberan FA, Garcia-Conesa MT, Espin JC. Dietary phenolics against colorectal cancer—from promising preclinical results to poor translation into clinical trials: pitfalls and future needs. Mol Nutr Food Res. 2015;59(7):1274–91. [DOI] [PubMed] [Google Scholar]
- 12. Carroll RE, Benya RV, Turgeon DK, Vareed S, Neuman M, Rodriguez L, Kakarala M, Carpenter PM, McLaren C, Meyskens FL. Phase IIa clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prev Res. 2011;4(3):354–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Z-Y, Shi C-B, Wen H, Li F-L, Wang B-L, Wang J. Upregulation of p53 expression in patients with colorectal cancer by administration of curcumin. Cancer Invest. 2011;29(3):208–13. [DOI] [PubMed] [Google Scholar]
- 14. Shimizu M, Fukutomi Y, Ninomiya M, Nagura K, Kato T, Araki H, Suganuma M, Fujiki H, Moriwaki H. Green tea extracts for the prevention of metachronous colorectal adenomas: a pilot study. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3020–5. [DOI] [PubMed] [Google Scholar]
- 15. Hoensch H, Groh B, Edler L, Kirch W. Prospective cohort comparison of flavonoid treatment in patients with resected colorectal cancer to prevent recurrence. World J Gastroenterol. 2008;14(14):2187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samsami-kor M, Daryani NE, Asl PR, Hekmatdoost A. Anti-inflammatory effects of resveratrol in patients with ulcerative colitis: a randomized, double-blind, placebo-controlled pilot study. Arch Med Res. 2015;46(4):280–5. [DOI] [PubMed] [Google Scholar]
- 17. Pandey KB, Rizvi SI. Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev. 2009;2(5):270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsao R. Chemistry and biochemistry of dietary polyphenols. Nutrients. 2010;2(12):1231–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez R, Ballester I, Lopez-Posadas R, Suarez MD, Zarzuelo A, Martinez-Augustin O, Sanchez de Medina F. Effects of flavonoids and other polyphenols on inflammation. Crit Rev Food Sci Nutr. 2011;51(4):331–62. [DOI] [PubMed] [Google Scholar]
- 20. Niedzwiecki A, Roomi MW, Kalinovsky T, Rath M. Anticancer efficacy of polyphenols and their combinations. Nutrients. 2016;8(9):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Y, Zheng J, Li Y, Xu DP, Li S, Chen YM, Li HB. Natural polyphenols for prevention and treatment of cancer. Nutrients. 2016;8(8):515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10(3):181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams RJ, Spencer JP, Rice-Evans C. Flavonoids: antioxidants or signalling molecules?. Free Radic Biol Med. 2004;36(7):838–49. [DOI] [PubMed] [Google Scholar]
- 25. Alam MN, Almoyad M, Huq F. Polyphenols in colorectal cancer: current state of knowledge including clinical trials and molecular mechanism of action. Biomed Res Int. 2018;2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scalbert A, Morand C, Manach C, Remesy C. Absorption and metabolism of polyphenols in the gut and impact on health. Biomed Pharmacother. 2002;56(6):276–82. [DOI] [PubMed] [Google Scholar]
- 27. Clifford MN, van der Hooft JJJ, Crozier A. Human studies on the absorption, distribution, metabolism, and excretion of tea polyphenols. Am J Clin Nutr. 2013;98(6):1619s–30s. [DOI] [PubMed] [Google Scholar]
- 28. Hanske L, Loh G, Sczesny S, Blaut M, Braune A. Recovery and metabolism of xanthohumol in germ-free and human microbiota-associated rats. Mol Nutr Food Res. 2010;54(10):1405–13. [DOI] [PubMed] [Google Scholar]
- 29. Nunes S, Danesi F, Del Rio D, Silva P. Resveratrol and inflammatory bowel disease: the evidence so far. Nutr Res Rev. 2018;31(1):85–97. [DOI] [PubMed] [Google Scholar]
- 30. Czank C, Cassidy A, Zhang Q, Morrison DJ, Preston T, Kroon PA, Botting NP, Kay CD. Human metabolism and elimination of the anthocyanin, cyanidin-3-glucoside: a 13C-tracer study. Am J Clin Nutr. 2013;97(5):995–1003. [DOI] [PubMed] [Google Scholar]
- 31. Boocock DJ, Faust GE, Patel KR, Schinas AM, Brown VA, Ducharme MP, Booth TD, Crowell JA, Perloff M, Gescher AJ. Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1246–52. [DOI] [PubMed] [Google Scholar]
- 32. Patel KR, Brown VA, Jones DJ, Britton RG, Hemingway D, Miller AS, West KP, Booth TD, Perloff M, Crowell JA. Clinical pharmacology of resveratrol and its metabolites in colorectal cancer patients. Cancer Res. 2010;70(19):7392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cardona F, Andres-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuno MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. 2013;24(8):1415–22. [DOI] [PubMed] [Google Scholar]
- 34. Espin JC, Gonzalez-Sarrias A, Tomas-Barberan FA. The gut microbiota: a key factor in the therapeutic effects of (poly)phenols. Biochem Pharmacol. 2017;139:82–93. [DOI] [PubMed] [Google Scholar]
- 35. Hervert-Hernández D, Goñi I. Dietary polyphenols and human gut microbiota: a review. Food Rev Int. 2011;27(2):154–69. [Google Scholar]
- 36. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–71. [DOI] [PubMed] [Google Scholar]
- 37. Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6):954–64. [DOI] [PubMed] [Google Scholar]
- 38. Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. 2012;30(4):330–3. [DOI] [PubMed] [Google Scholar]
- 39. Arseneau KO, Tamagawa H, Pizarro TT, Cominelli F. Innate and adaptive immune responses related to IBD pathogenesis. Curr Gastroenterol Rep. 2007;9(6):508–12. [DOI] [PubMed] [Google Scholar]
- 40. Atreya R, Neurath MF. IBD pathogenesis in 2014: molecular pathways controlling barrier function in IBD. Nat Rev Gastroenterol Hepatol. 2015;12(2):67–8. [DOI] [PubMed] [Google Scholar]
- 41. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60(5):631–7. [DOI] [PubMed] [Google Scholar]
- 43. Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Dore J, Raedler Aet al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141(1):227–36. [DOI] [PubMed] [Google Scholar]
- 44. Liang X, Li H, Tian G, Li S. Dynamic microbe and molecule networks in a mouse model of colitis-associated colorectal cancer. Sci Rep. 2015;4:4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers ABet al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JMet al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22(6):598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jones-Hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS One. 2015;10(3):e0119441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KEet al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKKet al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621. [DOI] [PubMed] [Google Scholar]
- 51. Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4(6):e00692–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11(1):9–20. [DOI] [PubMed] [Google Scholar]
- 53. Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy Ret al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet North Am Ed. 2017;389(10075):1218–28. [DOI] [PubMed] [Google Scholar]
- 54. Sears CL, Garrett WS, Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15(3):317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, Huso DL, Brancati FL, Wick E, McAllister F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu XQ, Shields CED, Hechenbleikner EM, Huso DLet al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Welch JLM, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev Met al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111(51):18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EMet al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, Pope JL, Gauthier J, Sun XL, Muhlbauer Met al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest. 2019;129(4):1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Olthof MR, Hollman PC, Katan MB. Chlorogenic acid and caffeic acid are absorbed in humans. J Nutr. 2001;131(1):66–71. [DOI] [PubMed] [Google Scholar]
- 62. Saura-Calixto F, Serrano J, Goñi I. Intake and bioaccessibility of total polyphenols in a whole diet. Food Chem. 2007;101(2):492–501. [Google Scholar]
- 63. Hollman PC, de Vries JH, van Leeuwen SD, Mengelers MJ, Katan MB. Absorption of dietary quercetin glycosides and quercetin in healthy ileostomy volunteers. Am J Clin Nutr. 1995;62(6):1276–82. [DOI] [PubMed] [Google Scholar]
- 64. Queipo-Ortuno MI, Boto-Ordonez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, Estruch R, Cardona Diaz F, Andres-Lacueva C, Tinahones FJ. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr. 2012;95(6):1323–34. [DOI] [PubMed] [Google Scholar]
- 65. Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am J Clin Nutr. 2011;93(1):62–72. [DOI] [PubMed] [Google Scholar]
- 66. Jin JS, Touyama M, Hisada T, Benno Y. Effects of green tea consumption on human fecal microbiota with special reference to Bifidobacterium species. Microbiol Immunol. 2012;56(11):729–39. [DOI] [PubMed] [Google Scholar]
- 67. Vendrame S, Guglielmetti S, Riso P, Arioli S, Klimis-Zacas D, Porrini M. Six-week consumption of a wild blueberry powder drink increases bifidobacteria in the human gut. J Agric Food Chem. 2011;59(24):12815–20. [DOI] [PubMed] [Google Scholar]
- 68. Eid N, Osmanova H, Natchez C, Walton G, Costabile A, Gibson G, Rowland I, Spencer JP. Impact of palm date consumption on microbiota growth and large intestinal health: a randomised, controlled, cross-over, human intervention study. Br J Nutr. 2015;114(8):1226–36. [DOI] [PubMed] [Google Scholar]
- 69. Rodriguez-Morato J, Matthan NR, Liu J, de la Torre R, Chen CO. Cranberries attenuate animal-based diet-induced changes in microbiota composition and functionality: a randomized crossover controlled feeding trial. J Nutr Biochem. 2018;62:76–86. [DOI] [PubMed] [Google Scholar]
- 70. Yuan X, Long Y, Ji Z, Gao J, Fu T, Yan M, Zhang L, Su H, Zhang W, Wen Xet al. Green tea liquid consumption alters the human intestinal and oral microbiome. Mol Nutr Food Res. 2018;62(12):1800178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Eid N, Enani S, Walton G, Corona G, Costabile A, Gibson G, Rowland I, Spencer JP. The impact of date palm fruits and their component polyphenols, on gut microbial ecology, bacterial metabolites and colon cancer cell proliferation. J Nutr Sci. 2014;3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou L, Wang W, Huang J, Ding Y, Pan Z, Zhao Y, Zhang R, Hu B, Zeng X. In vitro extraction and fermentation of polyphenols from grape seeds (Vitis vinifera) by human intestinal microbiota. Food Function. 2016;7(4):1959–67. [DOI] [PubMed] [Google Scholar]
- 73. Ashley D, Marasini D, Brownmiller C, Lee JA, Carbonero F, Lee SO. Impact of grain sorghum polyphenols on microbiota of normal weight and overweight/obese subjects during in vitro fecal fermentation. Nutrients. 2019;11(2):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Onc. 2014;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–72. [DOI] [PubMed] [Google Scholar]
- 76. Larrosa M, Yanez-Gascon MJ, Selma MV, Gonzalez-Sarrias A, Toti S, Ceron JJ, Tomas-Barberan F, Dolara P, Espin JC. Effect of a low dose of dietary resveratrol on colon microbiota, inflammation and tissue damage in a DSS-induced colitis rat model. J Agric Food Chem. 2009;57(6):2211–20. [DOI] [PubMed] [Google Scholar]
- 77. McFadden RM, Larmonier CB, Shehab KW, Midura-Kiela M, Ramalingam R, Harrison CA, Besselsen DG, Chase JH, Caporaso JG, Jobin Cet al. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention. Inflamm Bowel Dis. 2015;21(11):2483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Estruel-Amades S, Massot-Cladera M, Perez-Cano FJ, Franch A, Castell M, Camps-Bossacoma M. Hesperidin effects on gut microbiota and gut-associated lymphoid tissue in healthy rats. Nutrients. 2019;11(2):324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Paturi G, Butts CA, Monro JA, Hedderley D. Effects of blackcurrant and dietary fibers on large intestinal health biomarkers in rats. Plant Foods Hum Nutr. 2018;73(1):54–60. [DOI] [PubMed] [Google Scholar]
- 80. Molan AL, Liu ZJ, Kruger M. The ability of blackcurrant extracts to positively modulate key markers of gastrointestinal function in rats. World J Microbiol Biotechnol. 2010;26(10):1735–43. [Google Scholar]
- 81. Molan AL, Lila MA, Mawson J, De S. In vitro and in vivo evaluation of the prebiotic activity of water-soluble blueberry extracts. World J Microbiol Biotechnol. 2009;25(7):1243–9. [Google Scholar]
- 82. da Silva-Maia JK, Batista AG, Correa LC, Lima GC, Bogusz Junior S, Marostica Junior MR. Aqueous extract of berry (Plinia jaboticaba) byproduct modulates gut microbiota and maintains the balance on antioxidant defense system in rats. J Food Biochem. 2019;43(2):e12705. [DOI] [PubMed] [Google Scholar]
- 83. Lacombe A, Li RW, Klimis-Zacas D, Kristo AS, Tadepalli S, Krauss E, Young R, Wu VC. Lowbush wild blueberries have the potential to modify gut microbiota and xenobiotic metabolism in the rat colon. PLoS One. 2013;8(6):e67497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li F, Han Y, Cai X, Gu M, Sun J, Qi C, Goulette T, Song M, Li Z, Xiao H. Dietary resveratrol attenuated colitis and modulated gut microbiota in dextran sulfate sodium-treated mice. Food Function. 2020;11(1):1063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang P, Li D, Ke W, Liang D, Hu X, Chen F. Resveratrol-induced gut microbiota reduces obesity in high-fat diet-fed mice. Int J Obes. 2020;44(1):213–25. [DOI] [PubMed] [Google Scholar]
- 86. Wang J, Tang L, Zhou H, Zhou J, Glenn TC, Shen CL, Wang JS. Long-term treatment with green tea polyphenols modifies the gut microbiome of female Sprague-Dawley rats. J Nutr Biochem. 2018;56:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol. 2011;2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hakansson A, Branning C, Molin G, Adawi D, Hagslatt ML, Jeppsson B, Nyman M, Ahrne S. Blueberry husks and probiotics attenuate colorectal inflammation and oncogenesis, and liver injuries in rats exposed to cycling DSS-treatment. PLoS One. 2012;7(3):e33510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Alrafas HR, Busbee PB, Nagarkatti M, Nagarkatti PS. Resveratrol modulates the gut microbiota to prevent murine colitis development through induction of Tregs and suppression of Th17 cells. J Leukoc Biol. 2019;106(2):467–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, Dolara P. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res. 2009;53(8):1044–54. [DOI] [PubMed] [Google Scholar]
- 91. Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Applied and Environmental Microbiology. 2008;74(5):1646–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr. 2018;63(1):33–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A. 2019;116(26):12672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ohno M, Nishida A, Sugitani Y, Nishino K, Inatomi O, Sugimoto M, Kawahara M, Andoh A. Nanoparticle curcumin ameliorates experimental colitis via modulation of gut microbiota and induction of regulatory T cells. PLoS One. 2017;12(10):e0185999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ju S, Ge Y, Li P, Tian X, Wang H, Zheng X, Ju S. Dietary quercetin ameliorates experimental colitis in mouse by remodeling the function of colonic macrophages via a heme oxygenase-1-dependent pathway. Cell Cycle. 2018;17(1):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang H, Xue Y, Zhang H, Huang Y, Yang G, Du M, Zhu MJ. Dietary grape seed extract ameliorates symptoms of inflammatory bowel disease in IL 10‐deficient mice. Mol Nutr Food Res. 2013;57(12):2253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Paturi G, Mandimika T, Butts CA, Zhu S, Roy NC, McNabb WC, Ansell J. Influence of dietary blueberry and broccoli on cecal microbiota activity and colon morphology in mdr1a(-/-) mice, a model of inflammatory bowel diseases. Nutrition. 2012;28(3):324–30. [DOI] [PubMed] [Google Scholar]
- 99. Pei R, Martin DA, Valdez JC, Liu J, Kerby RL, Rey FE, Smyth JA, Liu Z, Bolling BW. Dietary prevention of colitis by aronia berry is mediated through increased Th17 and Treg. Mol Nutr Food Res. 2018;63:(5):e1800985. doi: 10.1002/mnfr.201800985. [DOI] [PubMed] [Google Scholar]
- 100. Wang K, Yang Q, Ma Q, Wang B, Wan Z, Chen M, Wu L. Protective effects of salvianolic acid a against dextran sodium sulfate-induced acute colitis in rats. Nutrients. 2018;10(6):791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang Z, Wu X, Cao S, Cromie M, Shen Y, Feng Y, Yang H, Li L. Chlorogenic acid ameliorates experimental colitis by promoting growth of Akkermansia in mice. Nutrients. 2017;9(7):677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hong Z, Piao M. Effect of quercetin monoglycosides on oxidative stress and gut microbiota diversity in mice with dextran sodium sulphate-induced colitis. Biomed Res Int. 2018;2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wu XY, Wang LJ, Tang L, Wang L, Cao SY, Wu Q, Zhang Z, Li L. Salvianolic acid B alters the gut microbiota and mitigates colitis severity and associated inflammation. J Funct Foods. 2018;46:312–9.. doi: 10.1016/j.jff.2018.04.068. [Google Scholar]
- 104. Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18:2–4. [DOI] [PubMed] [Google Scholar]
- 105. Gomes SD, Oliveira CS, Azevedo-Silva J, Casanova M, Barreto J, Pereira H, Chaves S, Rodrigues L, Casal M, Corte-Real Met al. The role of diet related short-chain fatty acids in colorectal cancer metabolism and survival: prevention and therapeutic implications. Curr Med Chem. 2018;27(24):4087–108. [DOI] [PubMed] [Google Scholar]
- 106. Thaker AI, Shaker A, Rao MS, Ciorba MA. Modeling colitis-associated cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS). J Vis Exp. 2012;; 67:e4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Takahashi M, Wakabayashi K. Gene mutations and altered gene expression in azoxymethane-induced colon carcinogenesis in rodents. Cancer Sci. 2004;95(6):475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Laroui H, Ingersoll SA, Liu HC, Baker MT, Ayyadurai S, Charania MA, Laroui F, Yan Y, Sitaraman SV, Merlin D. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS One. 2012;7(3):e32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kanneganti M, Mino-Kenudson M, Mizoguchi E. Animal models of colitis-associated carcinogenesis. J Biomed Biotechnol. 2011;2011:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wu M, Wu Y, Deng B, Li J, Cao H, Qu Y, Qian X, Zhong G. Isoliquiritigenin decreases the incidence of colitis-associated colorectal cancer by modulating the intestinal microbiota. Oncotarget. 2016;7(51):85318–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang X, Ye T, Chen WJ, Lv Y, Hao Z, Chen J, Zhao JY, Wang HP, Cai YK. Structural shift of gut microbiota during chemo-preventive effects of epigallocatechin gallate on colorectal carcinogenesis in mice. World J Gastroenterol. 2017;23(46):8128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Chen L, Jiang B, Zhong C, Guo J, Zhang L, Mu T, Zhang Q, Bi X. Chemoprevention of colorectal cancer by black raspberry anthocyanins involved the modulation of gut microbiota and SFRP2 demethylation. Carcinogenesis. 2018;39(3):471–81. [DOI] [PubMed] [Google Scholar]
- 113. Feng Z, Long W, Hao B, Ding D, Ma X, Zhao L, Pang X. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. 2017;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fernandez J, Garcia L, Monte J, Villar CJ, Lombo F. Functional anthocyanin-rich sausages diminish colorectal cancer in an animal model and reduce pro-inflammatory bacteria in the intestinal microbiota. Genes. 2018;9(3):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Duda-Chodak A. The inhibitory effect of polyphenols on human gut microbiota. J Physiol Pharmacol. 2012;63(5):497–503. [PubMed] [Google Scholar]
- 116. Bialonska D, Kasimsetty SG, Schrader KK, Ferreira D. The effect of pomegranate (Punica granatum L.) byproducts and ellagitannins on the growth of human gut bacteria. J Agric Food Chem. 2009;57(18):8344–9. [DOI] [PubMed] [Google Scholar]
- 117. Chung KT, Lu Z, Chou MW. Mechanism of inhibition of tannic acid and related compounds on the growth of intestinal bacteria. Food Chem Toxicol. 1998;36(12):1053–60. [DOI] [PubMed] [Google Scholar]
- 118. Puupponen-Pimia R, Nohynek L, Hartmann-Schmidlin S, Kahkonen M, Heinonen M, Maatta-Riihinen K, Oksman-Caldentey KM. Berry phenolics selectively inhibit the growth of intestinal pathogens. J Appl Microbiol. 2005;98(4):991–1000. [DOI] [PubMed] [Google Scholar]
- 119. He Z, Huang Z, Zhou W, Tang Z, Ma R, Liang J. Anti-biofilm activities from resveratrol against Fusobacterium nucleatum. Front Microbiol. 2016;7:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Tahara T, Shibata T, Kawamura T, Okubo M, Ichikawa Y, Sumi K, Miyata M, Ishizuka T, Nakamura M, Nagasaka Met al. Fusobacterium detected in colonic biopsy and clinicopathological features of ulcerative colitis in Japan. Dig Dis Sci. 2015;60(1):205–10. [DOI] [PubMed] [Google Scholar]
- 121. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Barbieri R, Coppo E, Marchese A, Daglia M, Sobarzo-Sanchez E, Nabavi SF, Nabavi SM. Phytochemicals for human disease: an update on plant-derived compounds antibacterial activity. Microbiol Res. 2017;196:44–68. [DOI] [PubMed] [Google Scholar]
- 124. Huber B, Eberl L, Feucht W, Polster J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z Naturforsch C. 2003;58(11-12):879–84. [DOI] [PubMed] [Google Scholar]
- 125. Truchado P, Gimenez-Bastida JA, Larrosa M, Castro-Ibanez I, Espin JC, Tomas-Barberan FA, Garcia-Conesa MT, Allende A. Inhibition of quorum sensing (QS) in Yersinia enterocolitica by an orange extract rich in glycosylated flavanones. J Agric Food Chem. 2012;60(36):8885–94. [DOI] [PubMed] [Google Scholar]
- 126. Papenfort K, Bassler BL. Quorum sensing signal-response systems in gram-negative bacteria. Nat Rev Microbiol. 2016;14(9):576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Plaper A, Golob M, Hafner I, Oblak M, Šolmajer T, Jerala R. Characterization of quercetin binding site on DNA gyrase. Biochem Biophys Res Commun. 2003;306(2):530–6. [DOI] [PubMed] [Google Scholar]
- 128. Huang YH, Huang CC, Chen CC, Yang KJ, Huang CY. Inhibition of Staphylococcus aureus PriA helicase by flavonol kaempferol. Protein J. 2015;34(3):169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Siriwong S, Thumanu K, Hengpratom T, Eumkeb G. Synergy and mode of action of ceftazidime plus quercetin or luteolin on Streptococcus pyogenes. Evid Based Complement Alternat Med. 2015;2015:759459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yi S, Wang W, Bai F, Zhu J, Li J, Li X, Xu Y, Sun T, He Y. Antimicrobial effect and membrane-active mechanism of tea polyphenols against Serratia marcescens. World J Microbiol Biotechnol. 2014;30(2):451–60. [DOI] [PubMed] [Google Scholar]
- 131. Sirk TW, Friedman M, Brown EF. Molecular binding of black tea theaflavins to biological membranes: relationship to bioactivities. J Agric Food Chem. 2011;59(8):3780–7. [DOI] [PubMed] [Google Scholar]
- 132. Kurzbaum E, Iliasafov L, Kolik L, Starosvetsky J, Bilanovic D, Butnariu M, Armon R. From the Titanic and other shipwrecks to biofilm prevention: the interesting role of polyphenol-protein complexes in biofilm inhibition. Sci Total Environ. 2019;658:1098–105. [DOI] [PubMed] [Google Scholar]
- 133. Najarzadeh Z, Mohammad-Beigi H, Nedergaard Pedersen J, Christiansen G, Sonderby TV, Shojaosadati SA, Morshedi D, Stromgaard K, Meisl G, Sutherland Det al. Plant polyphenols inhibit functional amyloid and biofilm formation in Pseudomonas strains by directing monomers to off-pathway oligomers. Biomolecules. 2019;9(11):659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, Ajo-Franklin CM, Portnoy DA. A flavin-based extracellular electron transfer mechanism in diverse gram-positive bacteria. Nature. 2018;562(7725):140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Khan MT, Duncan SH, Stams AJM, van Dijl JM, Flint HJ, Harmsen HJM. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6(8):1578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hsueh C-C, Wu C-C, Chen B-Y. Polyphenolic compounds as electron shuttles for sustainable energy utilization. Biotechnol Biofuels. 2019;12(1):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Singh M, Arseneault M, Sanderson T, Murthy V, Ramassamy C. Challenges for research on polyphenols from foods in Alzheimer's disease: bioavailability, metabolism, and cellular and molecular mechanisms. J Agric Food Chem. 2008;56(13):4855–73. [DOI] [PubMed] [Google Scholar]
- 138. Hess V, Gonzalez JM, Parthasarathy A, Buckel W, Muller V. Caffeate respiration in the acetogenic bacterium Acetobacterium woodii: a coenzyme a loop saves energy for caffeate activation. Appl Environ Microbiol. 2013;79(6):1942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chen L, Cao H, Xiao J, Polyphenols: absorption, bioavailability, and metabolomics, Polyphenols: properties, recovery, and applications. Cambridge (UK): Woodhead Publishing; 2018. pp. 45–67. [Google Scholar]
- 140. Németh K, Plumb GW, Berrin J-G, Juge N, Jacob R, Naim HY, Williamson G, Swallow DM, Kroon PA. Deglycosylation by small intestinal epithelial cell β-glucosidases is a critical step in the absorption and metabolism of dietary flavonoid glycosides in humans. Eur J Nutr. 2003;42(1):29–42. [DOI] [PubMed] [Google Scholar]
- 141. Aura A-M. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem Rev. 2008;7(3):407–29. [Google Scholar]
- 142. Selma MV, Espin JC, Tomas-Barberan FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. 2009;57(15):6485–501. [DOI] [PubMed] [Google Scholar]
- 143. Parkar SG, Trower TM, Stevenson DE. Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe. 2013;23:12–9. [DOI] [PubMed] [Google Scholar]
- 144. Tomas-Barberan F, Garcia-Villalba R, Quartieri A, Raimondi S, Amaretti A, Leonardi A, Rossi M. In vitro transformation of chlorogenic acid by human gut microbiota. Mol Nutr Food Res. 2014;58(5):1122–31. [DOI] [PubMed] [Google Scholar]
- 145. Unno T, Hisada T, Takahashi S. Hesperetin modifies the composition of fecal microbiota and increases cecal levels of short-chain fatty acids in rats. J Agric Food Chem. 2015;63(36):7952–7. [DOI] [PubMed] [Google Scholar]
- 146. Zdunczyk Z, Juskiewicz J, Estrella I. Cecal parameters of rats fed diets containing grapefruit polyphenols and inulin as single supplements or in a combination. Nutrition. 2006;22(9):898–904. [DOI] [PubMed] [Google Scholar]
- 147. Haraguchi T, Kayashima T, Okazaki Y, Inoue J, Mineo S, Matsubara K, Sakaguchi E, Yanaka N, Kato N. Cecal succinate elevated by some dietary polyphenols may inhibit colon cancer cell proliferation and angiogenesis. J Agric Food Chem. 2014;62(24):5589–94. [DOI] [PubMed] [Google Scholar]
- 148. Happy O. Molecular interaction and inhibitory potential of selected plant-derived polyphenolic compounds with human α-amylase. Int J Comput Biol Bioinform. 2016;1:21–6. [Google Scholar]
- 149. Xiao J, Kai G, Ni X, Yang F, Chen X. Interaction of natural polyphenols with alpha-amylase in vitro: molecular property-affinity relationship aspect. Mol Biosyst. 2011;7(6):1883–90. [DOI] [PubMed] [Google Scholar]
- 150. Zhong L, Zhang X, Covasa M. Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J Gastroenterol. 2014;20(24):7878–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Jacouton E, Chain F, Sokol H, Langella P, Bermudez-Humaran LG. Probiotic strain Lactobacillus casei BL23 prevents colitis-associated colorectal cancer. Front Immunol. 2017;8:1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Mizuno K, Mizuno M, Yamauchi M, Takemura AJ, Medrano Romero V, Morikawa K. Adjacent-possible ecological niche: growth of Lactobacillus species co-cultured with Escherichia coli in a synthetic minimal medium. Sci Rep. 2017;7(1):12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ishikawa H, Akedo I, Umesaki Y, Tanaka R, Imaoka A, Otani T. Randomized controlled trial of the effect of bifidobacteria-fermented milk on ulcerative colitis. J Am Coll Nutr. 2003;22(1):56–63. [DOI] [PubMed] [Google Scholar]
- 154. Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'Neil DA, Macfarlane GT. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54(2):242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ishikawa H, Matsumoto S, Ohashi Y, Imaoka A, Setoyama H, Umesaki Y, Tanaka R, Otani T. Beneficial effects of probiotic bifidobacterium and galacto-oligosaccharide in patients with ulcerative colitis: a randomized controlled study. Digestion. 2011;84(2):128–33. [DOI] [PubMed] [Google Scholar]
- 156. Osman N, Adawi D, Ahrne S, Jeppsson B, Molin G. Modulation of the effect of dextran sulfate sodium-induced acute colitis by the administration of different probiotic strains of Lactobacillus and Bifidobacterium. Dig Dis Sci. 2004;49(2):320–7. [DOI] [PubMed] [Google Scholar]
- 157. Kim SW, Kim HM, Yang KM, Kim SA, Kim SK, An MJ, Park JJ, Lee SK, Kim TI, Kim WHet al. Bifidobacterium lactis inhibits NF-kappaB in intestinal epithelial cells and prevents acute colitis and colitis-associated colon cancer in mice. Inflamm Bowel Dis. 2010;16(9):1514–25. [DOI] [PubMed] [Google Scholar]
- 158. Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Hontecillas R. Immunoregulatory mechanisms underlying prevention of colitis-associated colorectal cancer by probiotic bacteria. PLoS One. 2012;7(4):e34676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Mendes MCS, Paulino DS, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol. 2018;24(18):1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Han Y, Haraguchi T, Iwanaga S, Tomotake H, Okazaki Y, Mineo S, Moriyama A, Inoue J, Kato N. Consumption of some polyphenols reduces fecal deoxycholic acid and lithocholic acid, the secondary bile acids of risk factors of colon cancer. J Agric Food Chem. 2009;57(18):8587–90. [DOI] [PubMed] [Google Scholar]
- 162. Tombola F, Campello S, De Luca L, Ruggiero P, Del Giudice G, Papini E, Zoratti M. Plant polyphenols inhibit VacA, a toxin secreted by the gastric pathogen Helicobacter pylori. FEBS Lett. 2003;543(1-3):184–9. [DOI] [PubMed] [Google Scholar]
- 163. Wang CZ, Zhang CF, Chen L, Anderson S, Lu F, Yuan CS. Colon cancer chemopreventive effects of baicalein, an active enteric microbiome metabolite from baicalin. Int J Oncol. 2015;47(5):1749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Nunez-Sanchez MA, Karmokar A, Gonzalez-Sarrias A, Garcia-Villalba R, Tomas-Barberan FA, Garcia-Conesa MT, Brown K, Espin JC. In vivo relevant mixed urolithins and ellagic acid inhibit phenotypic and molecular colon cancer stem cell features: a new potentiality for ellagitannin metabolites against cancer. Food Chem Toxicol. 2016;92:8–16. [DOI] [PubMed] [Google Scholar]
- 165. Gonzalez-Sarrias A, Nunez-Sanchez MA, Garcia-Villalba R, Tomas-Barberan FA, Espin JC. Antiproliferative activity of the ellagic acid-derived gut microbiota isourolithin A and comparison with its urolithin A isomer: the role of cell metabolism. Eur J Nutr. 2017;56(2):831–41. [DOI] [PubMed] [Google Scholar]
- 166. Allsopp P, Possemiers S, Campbell D, Gill C, Rowland I. A comparison of the anticancer properties of isoxanthohumol and 8-prenylnaringenin using in vitro models of colon cancer. Biofactors. 2013;39(4):441–7. [DOI] [PubMed] [Google Scholar]
- 167. Zhang Z, Peng X, Zhang N, Liu L, Wang Y, Ou S. Cytotoxicity comparison of quercetin and its metabolites from in vitro fermentation of several gut bacteria. Food Funct. 2014;5(9):2152–6. [DOI] [PubMed] [Google Scholar]
- 168. Miene C, Weise A, Glei M. Impact of polyphenol metabolites produced by colonic microbiota on expression of COX-2 and GSTT2 in human colon cells (LT97). Nutr Cancer. 2011;63(4):653–62. [DOI] [PubMed] [Google Scholar]
- 169. Monagas M, Khan N, Andres-Lacueva C, Casas R, Urpi-Sarda M, Llorach R, Lamuela-Raventos RM, Estruch R. Effect of cocoa powder on the modulation of inflammatory biomarkers in patients at high risk of cardiovascular disease. Am J Clin Nutr. 2009;90(5):1144–50. [DOI] [PubMed] [Google Scholar]
- 170. Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, Dolara P. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res. 2009;53(8):1044–54. [DOI] [PubMed] [Google Scholar]
- 171. Rios LY, Gonthier MP, Remesy C, Mila L, Lapierre C, Lazarus SA, Williamson G, Scalbert A. Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am J Clin Nutr. 2003;77(4):912–8. [DOI] [PubMed] [Google Scholar]
- 172. Gonthier MP, Cheynier V, Donovan JL, Manach C, Morand C, Mila I, Lapierre C, Remesy C, Scalbert A. Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J Nutr. 2003;133(2):461–7. [DOI] [PubMed] [Google Scholar]
- 173. Radnai B, Tucsek Z, Bognar Z, Antus C, Mark L, Berente Z, Gallyas F, Sumegi B, Veres B. Ferulaldehyde, a water-soluble degradation product of polyphenols, inhibits the lipopolysaccharide-induced inflammatory response in mice. J Nutr. 2009;139(2):291–7. [DOI] [PubMed] [Google Scholar]
- 174. Tucsek Z, Radnai B, Racz B, Debreceni B, Priber JK, Dolowschiak T, Palkovics T, Gallyas F, Sumegi B, Veres B. Suppressing LPS-induced early signal transduction in macrophages by a polyphenol degradation product: a critical role of MKP-1. J Leukoc Biol. 2011;89(1):105–11. [DOI] [PubMed] [Google Scholar]
- 175. Espin JC, Larrosa M, Garcia-Conesa MT, Tomas-Barberan F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid-Based Compl Alt. 2013. doi: Artn 270418. 10.1155/2013/270418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, Hiwale AA, Saiyed T, Patel P, Vijay-Kumar Met al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lundberg R, Toft MF, August B, Hansen AK, Hansen CHF. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes. 2016;7(1):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Clavel T, Lagkouvardos I, Blaut M, Stecher B. The mouse gut microbiome revisited: from complex diversity to model ecosystems. Int J Med Microbiol. 2016;306(5):316–27. [DOI] [PubMed] [Google Scholar]
- 180. Neville BA, Forster SC, Lawley TD. Commensal Koch's postulates: establishing causation in human microbiota research. Curr Opin Microbiol. 2018;42:47–52. [DOI] [PubMed] [Google Scholar]
- 181. Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, Curry KP, Renner SW, Greenwalt A, Ryan EP. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota-and butyrate-dependent manner. Cancer Discov. 2014;4(12):1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Sivaprakasam S, Gurav A, Paschall A, Coe G, Chaudhary K, Cai Y, Kolhe R, Martin P, Browning D, Huang L. An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis. 2016;5(6):e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Darby TM, Owens JA, Saeedi BJ, Luo L, Matthews JD, Robinson BS, Naudin CR, Jones RM. Lactococcus Lactis Subsp. cremoris is an efficacious beneficial bacterium that limits tissue injury in the intestine. iScience. 2019;12:356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald Det al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–22. [DOI] [PubMed] [Google Scholar]
- 185. Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter Ret al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;34(9):942. [DOI] [PubMed] [Google Scholar]
- 186. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Liu L, Chen XW, Skogerbo G, Zhang P, Chen RS, He SM, Huang DW. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics. 2012;100(5):265–70. [DOI] [PubMed] [Google Scholar]
- 189. Louca S, Polz MF, Mazel F, Albright MB, Huber JA, O'Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2(6):936–43. [DOI] [PubMed] [Google Scholar]
- 190. Gonzalez-Sarrias A, Romo-Vaquero M, Garcia-Villalba R, Cortes-Martin A, Selma MV, Espin JC. The endotoxemia marker lipopolysaccharide-binding protein is reduced in overweight-obese subjects consuming pomegranate extract by modulating the gut microbiota: a randomized clinical trial. Mol Nutr Food Res. 2018;62(11):1800160. [DOI] [PubMed] [Google Scholar]
- 191. Gonzalez-Sarrias A, Nunez-Sanchez MA, Avila-Galvez MA, Monedero-Saiz T, Rodriguez-Gil FJ, Martinez-Diaz F, Selma MV, Espin JC. Consumption of pomegranate decreases plasma lipopolysaccharide-binding protein levels, a marker of metabolic endotoxemia, in patients with newly diagnosed colorectal cancer: a randomized controlled clinical trial. Food Function. 2018;9(5):2617–22. [DOI] [PubMed] [Google Scholar]
- 192. Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research?. Dis Models Mechanisms. 2015;8(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75(1):149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Xiao L, Feng Q, Liang SS, Sonne SB, Xia ZK, Qiu XM, Li XP, Long H, Zhang JF, Zhang DYet al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33(10):1103. [DOI] [PubMed] [Google Scholar]
- 195. Gootenberg DB, Turnbaugh PJ. Companion animals symposium: humanized animal models of the microbiome. J Anim Sci. 2011;89(5):1531–7. [DOI] [PubMed] [Google Scholar]
- 196. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. O'Keefe SJD, Li JV, Lahti L, Ou JH, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder Eet al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]


