SUMMARY
Hepatocyte proliferation is the principal mechanism for generating new hepatocytes in liver homeostasis and regeneration. Recent studies have suggested that this ability is not equally distributed among hepatocytes but concentrated in a small subset of hepatocytes acting like stem cells, located around the central vein or distributed throughout the liver lobule and exhibiting active WNT signaling or high telomerase activity, respectively. These findings were obtained by utilizing components of these growth regulators as markers for genetic lineage tracing. Here, we used random lineage tracing to localize and quantify clonal expansion of hepatocytes in normal and injured liver. We found that modest proliferation of hepatocytes distributed throughout the lobule maintains the hepatocyte mass and that most hepatocytes proliferate to regenerate it, with diploidy providing a growth advantage over polyploidy. These results show that the ability to proliferate is broadly distributed among hepatocytes rather than limited to a rare stem cell-like population.
In Brief
Chen et al. investigate potential differences in how much hepatocytes in the different zones of the liver lobule contribute to hepatocyte replacement in normal and injured liver. They find that, in principle, location and extent of hepatocyte proliferation are broadly distributed, with specific effects of type of injury and ploidy.
Graphical abstract
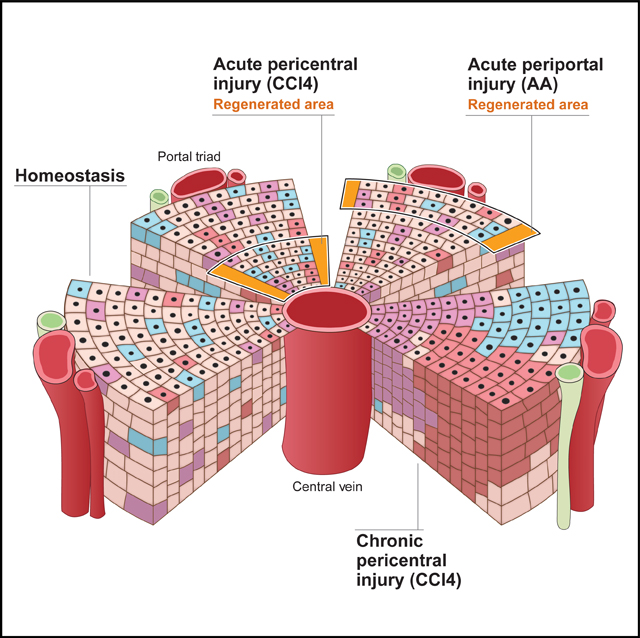
INTRODUCTION
Hepatocytes are unique among differentiated cells because they provide many vital functions and also can proliferate extensively, which allows efficient liver regeneration after injury. It is increasingly appreciated that hepatocytes are a heterogeneous cell population—depending on where they are located in the liver lobule, hepatocytes have different functions; they also differ in ploidy. Recently, hepatocyte heterogeneity was shown to extend to proliferation. In the normal liver, hepatocytes bordering on the central vein were found to harness WNT signaling to proliferate more than other hepatocytes, making them the main source of new hepatocytes (Wang et al., 2015). Another study showed that rare hepatocytes with high telomerase activity distributed throughout the liver lobule dominate both liver homeostasis and regeneration (Lin et al., 2018).
These findings suggest that the hepatocyte mass is maintained and restored by clonal expansion of a small subset of hepatocytes acting like stem cells. However, it is currently not known which of the 2 hepatocyte stem cell models is correct or if both are, and the relative contribution of other hepatocytes to liver homeostasis and regeneration has not been addressed. In addition, these findings were obtained using lineage tracing disrupting endogenous Axin2 or Tert genes, potentially altering how readily and extensively cells respond to growth signals (Minear et al., 2010; Sarin et al., 2005).
RESULTS
To address these questions, we used random lineage tracing—capturing a representative sample of all hepatocytes—to localize and quantify their clonal expansion in normal and injured liver. We permanently labeled hepatocytes throughout the liver lobule at a neutral genetic locus with 1 of 3 fluorophores by intravenously injecting AAV8-TBG-Cre into adult heterozygous Rosa26-Rainbow Cre reporter (R26Rrb/wt) mice (Figure 1A); the mice were housed under standard barrier conditions and any health problems were recorded (Table S1). Lineage tracing was active by 2 weeks after vector injection (Figures 1B and S1A). As previously reported for adeno-associated virus (AAV) capsids (Davidoff et al., 2003), including the AAV8 capsid (Greig et al., 2018), the labeling efficiency was higher in males than females and varied with weight (data not shown). Therefore, we doubled the dose in females and adjusted it to each mouse’s weight (Table S1). We used decreasing vector doses to induce high-, medium-, and low-density lineage tracing (Figure S1A; Table S1). The labeling efficiency gradually decreased along the central to portal axis, a characteristic of the AAV8 capsid (Bell et al., 2011; Figures 1B, S1A, and S1B). We used heterozygous mice to distinguish diploid from polyploid hepatocytes based on expression of 1 or ≥2 fluorophores, respectively. Accordingly, we found that lineage-traced hepatocytes expressing only 1 fluorophore were enriched for mononucleated cells, whereas those expressing ≥2 were enriched for binucleated cells (Figure S1C).
Figure 1. Hepatocyte Proliferation in Liver Homeostasis.
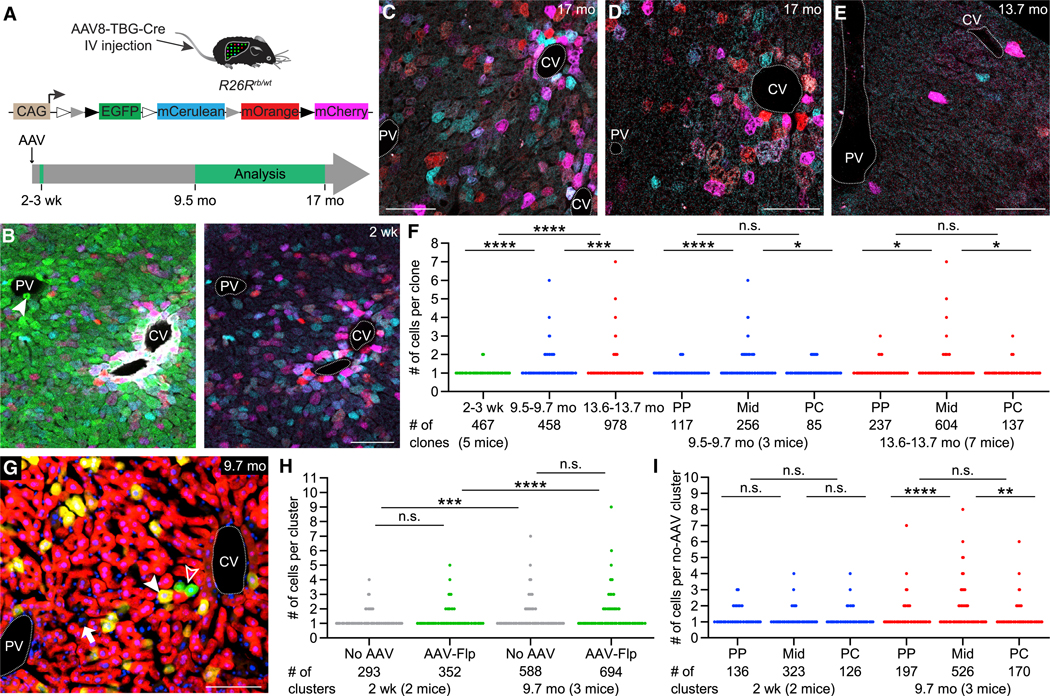
(A) Rosa26-Rainbow Cre reporter allele and lineage-tracing strategy. In the absence of Cre, R26Rrb/wt mice express EGFP in all cells. Hepatocyte-targeted AAV8-TBG-Cre mediates recombination at 1 set of the variant loxP sites, which deactivates EGFP and randomly activates mCerulean, mOrange, or mCherry expression. IV, intravenous; mo, months; wk, weeks; white triangle, loxN; gray triangle, lox2272; black triangle, loxP.
(B) Liver of a male R26Rrb/wt mouse 2 weeks after high-dose AAV8-TBG-Cre injection. Lineage-traced hepatocytes express mCerulean (light blue), mOrange (red), or mCherry (magenta) or a combination of these fluorophores. Hepatocytes and other liver cells not transduced by the AAV vector express only EGFP (green). Left image shows all 4 fluorophores; right image shows only the 3 Cre-activated fluorophores. Rare lineage-traced hepatocytes also express EGFP, presumably because 1 of the reporter alleles was not recombined. Portal tracts are identified by bile ducts (arrowhead) and pericentral hepatocytes by glutamine synthetase immunofluorescence (white). CV, central vein; PV, portal vein. Scale bar, 100μm.
(C–E) Livers of male R26Rrb/wt mice imaged at the indicated time points after high-dose (C), medium-dose (D), or low-dose (E) AAV8-TBG-Cre injection. Results are representative of 5–7 male and 4–6 female mice for each dose analyzed 9.5–17 months after AAV injection (Table S1). Scale bars, 100 μm.
(F) Number of hepatocytes per 3D clone in male and female R26Rrb/wt mice at the indicated time points after low-dose AAV8-TBG-Cre injection. 54–153 clones were analyzed in each mouse at 2 to 3 weeks, 142–167 clones at 9.5–9.7 months, and 103–181 clones at 13.6–13.7 months. PP, periportal; Mid, midlobular; PC, pericentral. Mann-Whitney U test; ****p < 0.0001; ***p < 0.001; *p < 0.05; n.s., not significant.
(G) Liver of a female R26RtdRFP/ZG mouse 9.7 months after co-injection of AAV8-TBG-Cre and AAV9-TTR-Flp. AAV8-TBG-Cre-lineage-traced hepatocytes express tdRFP (red) and AAV9-TTR-Flp-lineage-traced hepatocytes express EGFP (green; open arrowhead). Hepatocytes transduced with both AAV vectors express tdRFP and EGFP and appear yellow in the merged image (closed arrowhead), whereas hepatocytes not transduced with either AAV vector express neither tdRFP nor EGFP and are identified by large nuclei (DAPI staining, blue; arrow). Scale bar, 100 μm.
(H) Number of hepatocytes per 2D cluster not transduced with any AAV (no AAV) or transduced with AAV9-TTR-Flp (AAV-Flp) in 1 male and 1 female R26RtdRFP/ZG mouse 2 weeks and 1 male and 2 female mice 9.7 months after AAV8-TBG-Cre and AAV9-TTR-Flp co-injection. 99 and 194 no-AAV clusters and 117 and 235 AAV-Flp clusters were analyzed in male and female mice, respectively, at 2 weeks; 188–201 no-AAV and 206–275 AAV-Flp clusters were analyzed in each mouse at 9.7 months. Student’s t test on standardized cluster size (see STAR Methods and Figure S1M); ****p < 0.0001; ***p < 0.001.
(I) Number of hepatocytes per not transduced 2D cluster in male and female R26RtdRFP/ZG mice 2 weeks and 9.7 months after AAV8-TBG-Cre and AAV9-TTR-Flp co-injection (same mice as in H). 220 and 365 clusters were analyzed in each mouse at 2 weeks; 201–466 clusters were analyzed in each mouse at 9.7 months. Student’s t test on standardized cluster size (see STAR Methods and Figure S1N); ****p < 0.0001; **p < 0.01.
We started our investigation of hepatocyte proliferation in the normal liver by analyzing 2D clusters in mice after long-term lineage tracing. We found no or only modest expansion of labeled hepatocytes 9.5–17 months after AAV vector injection; we did not see large clusters or hepatocytes that streamed across the lobule (Figures 1C–1E). Moreover, the distribution of labeled hepatocytes appeared similar to that at 2 weeks after AAV vector injection, with denser labeling in the pericentral zone of the lobule (Figures 1B, S1A, and S1B). Had pericentral hepatocytes replenished the midlobular and periportal zones with new hepatocytes, the density of labeled cells in these zones should have increased.
Because 2D imaging of hepatocyte clusters might omit cells that lie beyond the plane of the section, we imaged 3D hepatocyte clones by confocal microscopy of liver tissue cleared using a modified CUBIC protocol (Susaki et al., 2015). This approach allowed us to image up to 150 μm deep through several hepatocyte layers, which confirmed absence of large clones or streaming of hepatocytes 13.5 or 17 months after injection of high (Video S1A) or medium (data not shown) AAV vector doses.
Next, we focused on the mice that received the low AAV vector dose. 2 to 3 weeks after injection, virtually all (457/467) labeled hepatocytes were single cells (Figure 1F; Video S1B). We rarely saw 2 differently colored clones next to each other (Figure S1D) and almost never 2 adjacent clones sharing a border (Video S1B). Few (10/467) clones that consisted of 2 cells expressing the same fluorophore were likely hepatocytes that proliferated between the time of AAV vector injection and analysis because they share a substantial cell border (Figure S1D) and because the probability of 2 different clones expressing the same fluorophore is only 1/3 or even less if cells are polyploid. Therefore, we used the mice that received the low AAV vector dose to quantify the size of individual clones.
We did not find large clones or streaming of hepatocytes 9.5–15 months after AAV vector injection when we imaged 5–23 fields (each containing at least 1 portal vein and 1 central vein and ~1,600 hepatocytes) from each of the left, median, right anterior, and right posterior lobe or the median or left lobe of each mouse (Videos S1C and S1D; Table S1). Specific analysis of 458 clones from at least 1 field per mouse at 9.5– 9.7 months showed that 84.1% of hepatocytes did not proliferate but remained single cells, 14.0% of hepatocytes divided once and gave rise to 2-cell clones, and 1.7% gave rise to 3-to 4-cell clones; only 1 (0.2%) clone was larger, consisting of 6 cells. Similarly, of 978 clones analyzed in mice at 13.6–13.7 months, 90.1% were single cells, 9.0% were 2-cell clones, 0.7% were 3-to 4-cell clones, and 0.2% consisted of 5–7 cells (Figure 1F; Videos S1C–S1E). Clones >2 cells were mostly located in the midlobular zone; pericentral and periportal clones almost exclusively consisted of 1 or 2 cells (Figure 1F). Hepatocytes proliferated more in females than in males, particularly in the midlobular zone, which was associated with higher baseline levels of CCND1 (Alvarado et al., 2016; Figures S1E–S1I). Clones ≥4 cells expressed only 1 fluorophore but at <1% were too rare to establish a growth advantage of diploid hepatocytes (Figure S1J). In accord, overall clonal expansion followed a Poisson distribution (Figure S1K), suggesting that hepatocyte proliferation occurs randomly in liver homeostasis.
To exclude that AAV vector transduction affected hepatocyte proliferation, we devised a lineage tracing strategy that let us compare the proliferation of transduced and untransduced cells. We intravenously co-injected adult Cre and Flp double-reporter (R26RtdRFP/ZG) mice with AAV8-TBG-Cre and AAV9-TTR-Flp at doses that activate Cre-dependent tdRFP in all but a few sparsely distributed hepatocytes and Flp-dependent EGFP in a small fraction of hepatocytes (Figures 1G and S1L; Table S1). Because Cre-and Flp-mediated reporter activation is very efficient in the normal liver (Schaub et al., 2018; Yanger et al., 2014), hepatocytes negative for both tdRFP and EGFP can be assumed to not have been transduced with either AAV vector. When we examined the number of hepatocytes in individual 2D clusters 2 weeks and 9.7 months after AAV injection, we found that hepatocytes not transduced with AAV vectors and hepatocytes labeled by AAV9-TTR-Flp expanded similarly at the 2 time points (Figures 1H and S1M), demonstrating that AAV transduction did not affect hepatocyte proliferation. Further analysis of the untransduced clusters confirmed our initial finding of modest hepatocyte proliferation in all 3 zones, with more proliferation occurring in the midlobular zone, and absence of streaming of hepatocytes (Figures 1I and S1N).
We also investigated clonal expansion of hepatocytes in the injured liver. After intravenously injecting low-or high-dose AAV8-TBG-Cre into adult R26Rrb/wt mice, we specifically injured their pericentral or periportal hepatocytes by intraperitoneal injection of 1 dose of carbon tetrachloride (CCl4) or 2 doses of allyl alcohol (AA), respectively (Figures S2A and S2B). We found that these acute injuries—depleting ~1/2 or ~1/4 of the hepatocyte mass, respectively—induced proliferation of hepatocytes adjacent to the injured area, leading to mostly 2-to 3-cell clones extending into the necrotic area (Figures 2A, 2B, and 2D). In addition to this localized repair, we saw compensatory proliferation of hepatocytes in zones further away from the injury (Figures 2A, 2B, 2D, S2C, and S2D).
Figure 2. Hepatocyte Proliferation in Liver Regeneration.
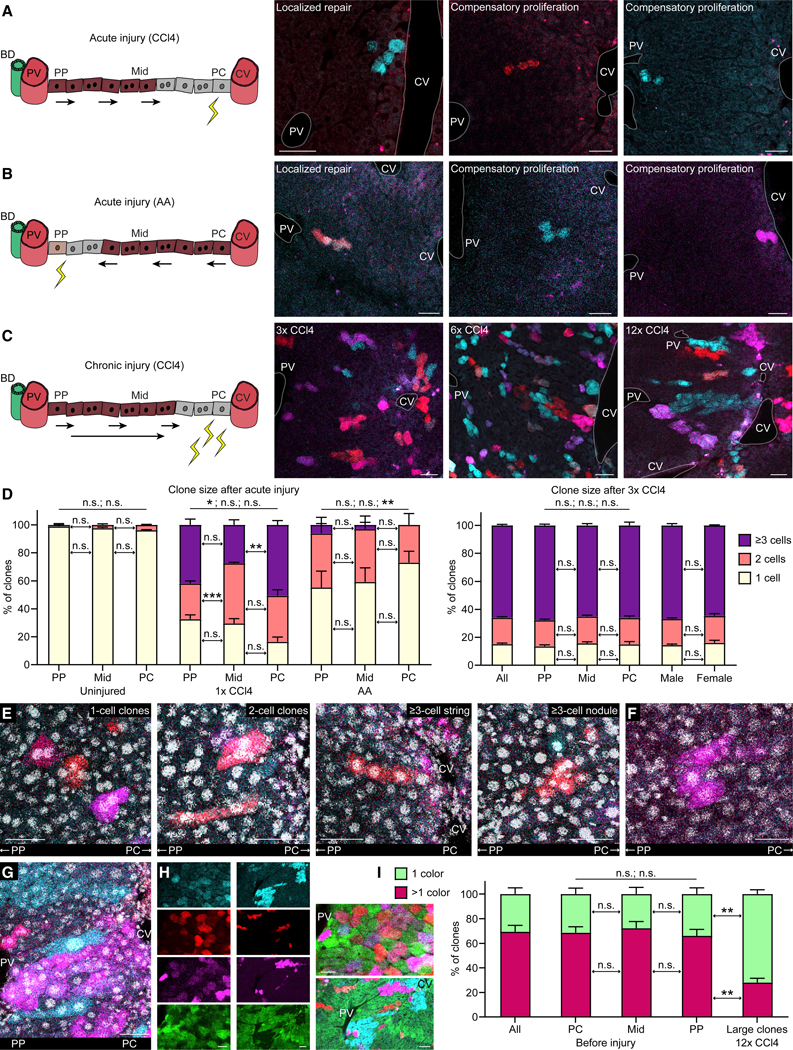
(A) Livers of male and female R26Rrb/wt mice injected with low-dose AAV8-TBG-Cre analyzed 2 weeks after 1 dose of CCl4. Lineage-traced hepatocytes express mCerulean (light blue) or mOrange (red). BD, bile duct. In the cartoons, healthy hepatocytes are brown, injured hepatocytes are gray, injury is represented by yellow zigzags, and arrows indicate the direction of hepatocyte clone expansion. Results are representative of 4 (2 male and 2 female) mice. An additional female mouse analyzed 5 days after the CCl4 dose showed similar results (data not shown). Scale bars, 50 μm.
(B) Liver of a female R26Rrb/wt mouse injected with low-dose AAV8-TBG-Cre analyzed 2 weeks after 2 doses of AA. mCerulean and mOrange double-positive hepatocytes appear orange. Small white and magenta dots are autofluorescent cellular debris. Results are representative of 2 (1 male and 1 female) mice. Additional 3 (2 male and 1 female) mice analyzed 1 week after the last AA dose showed similar results (data not shown). Scale bars, 50 μm.
(C) Livers of 3 male R26Rrb/wt mice injected with high-dose AAV8-TBG-Cre analyzed 2 weeks after 3, 6, or 12 doses of CCl4. Results are representative of 5 (3 male and 2 female) mice for 3 doses and 2 (1 male and 1 female) mice each for 6 doses and 12 doses. Additional 3, 1, and 2 male and female mice analyzed 3–6 days after the last of 3, 6, and 12 doses of CCl4 showed similar results (data not shown). Scale bars, 50 μm.
(D) Left graph shows percentage of 3D clones with the indicated number of hepatocytes in 3 (1 male and 2 female) uninjured R26Rrb/wt mice injected with low-dose AAV8-TBG-Cre (mice also shown in Figure 1F), 2 (1 male and 1 female) R26Rrb/wt mice injected with low-dose and 1 female mouse injected with medium-dose AAV8-TBG-Cre, all injured with 1 dose of CCl4, and 3 (2 male and 1 female) R26Rrb/wt mice injected with low-dose AAV8-TBG-Cre and injured with 2 doses of AA. Livers were analyzed 2 weeks after the single CCl4 dose or 1 week after the last AA dose. 46–99 clones from the midlobular zone and 21–69 clones each from the periportal and pericentral zones were analyzed in each mouse. Right graph shows percentage of 3D clones with the indicated number of hepatocytes in 8 (4 male and 4 female) R26Rrb/wt mice injected with high-dose AAV8-TBG-Cre analyzed 4–6 days or 2 weeks after 3 doses of CCl4. 76–180 clones from the midlobular zone and 32–102 clones each from the periportal and pericentral zones were analyzed in each mouse. In mice receiving AA, liver lobules lacking injured portal areas were excluded from analysis (see STAR Methods). Results are means + SEM. Student’s t test; ***p < 0.001; **p < 0.01; *p < 0.05. Differences between 1-, 2-, and ≥3-cell clones in periportal and pericentral zones are indicated by p values above bars.
(E) Characteristic types of hepatocyte clones in a male R26Rrb/wt mouse injected with high-dose AAV8-TBG-Cre analyzed 4 days after 3 doses of CCl4, including single large (hypertrophied) binucleated hepatocytes, hepatocytes that divided once and generated 2-cell clones, or hepatocytes that proliferated more than once, generating ≥3 cells that either streamed along the portal to central axis or formed a nodule. Nuclei are identified by DAPI staining (white). Results are representative of 2 (1 male and 1 female) mice. Scale bars, 50 μm.
(F) mCherry (magenta)-expressing clone consisting of several small hepatocytes with small nuclei and 2 large hepatocytes with large nuclei in a male R26Rrb/wt mouse injected with low-dose AAV8-TBG-Cre analyzed 3 days after 6 doses of CCl4. Nuclei are identified by DAPI staining (white). Scale bar, 50 μm.
(G) mCherry (magenta)-expressing clone consisting of many small hepatocytes that covers the distance from the portal vein to the central vein in a female R26Rrb/wt mouse injected with high-dose AAV8-TBG-Cre analyzed 4 days after 12 doses of CCl4. Adjacent to this clone is an mCerulean (light blue)-expressing 1-cell clone consisting of a large hepatocyte stretching along the portal to central axis. Nuclei are identified by DAPI staining (white). Scale bar, 50 μm.
(H) Columns of small images show the fluorophores mCerulean (light blue), mOrange (red), mCherry (magenta), and EGFP (green) separately; large images show them merged. Left column and related merged image show hepatocytes in a male R26Rrb/wt mouse 2 weeks after high-dose AAV8-TBG-Cre injection before initiation of liver injury. Many hepatocytes express more than 1 fluorophore, i.e., are polyploid. Scale bar, 25 μm. Right column and related merged image show hepatocyte clones in a male R26Rrb/wt mouse injected with high-dose AAV8-TBG-Cre analyzed 4 days after 12 doses of CCl4. Many large clones consist of diploid hepatocytes as indicated by expression of only 1 fluorophore. Scale bar, 50 μm.
(I) Percentages of diploid-enriched (1 color) and polyploid (>1 color) hepatocyte clones in 4 (2 male and 2 female) R26Rrb/wt mice analyzed 2 weeks after high-dose AAV8-TBG-Cre injection and in 4 (2 male and 2 female) R26Rrb/wt mice injected with high-dose AAV8-TBG-Cre analyzed 4 days or 2 weeks after 12 doses of CCl4. In injured mice, only large clones consisting of ≥12 cells were analyzed. Results are means + SEM. Welch’s t test; **p < 0.01. Differences between 1 color and >1 color in pericentral and periportal zones are indicated by p values above bars.
Next, we modeled chronic liver injury by repeating CCl4 application as soon as hepatocyte regeneration was complete (Figures S2E and S2F). After 3 doses of CCl4, we found that 84.9% ± 2.2% of lineage-traced hepatocytes in all zones divided, with 65.9% ± 2.2% dividing at least twice, giving rise to clones consisting of ≥3 cells (Figures 2C and 2D; Video S1F). Hepatocyte proliferation was similar between males and females and uniform across the lobule (Figures 2C and 2D). Clone morphology ranged from single enlarged cells—characteristic of hepatocyte hypertrophy, as after partial hepatectomy (Miyaoka et al., 2012)—to clones consisting of several small cells that streamed along the portal to central axis or formed nodules (Figure 2E; Video S1F). Although many 1-cell clones were enlarged after either chronic or acute injury (54.6% ± 2.0% of 69–102 clones each in 2 males and 2 females for 3 × CCl4, 28.0% ± 2.0% of 25–30 clones each in 1 male and 2 females for 1 × CCl4, and 35.2% ± 4.7% of 32–48 clones each in 2 males and 1 female for AA), proliferation was the predominant mechanism of hepatocyte regeneration (Figure 2D).
Applying additional CCl4 doses promoted the expansion of hepatocyte clones throughout the lobule. After 6 doses of CCl4, we found large clones (~7 cells in 2D) in the periportal and midlobular zones that converged to form regenerative patches (Figures 2C and S2G). Most hepatocytes in these clones were small, but large cells could also be found within a clone, suggesting that hepatocytes can switch from proliferation to hypertrophy (Figure 2F). After 12 doses of CCl4, we saw very large clones (~20 cells in 2D), some of which covered the distance from the portal vein to the central vein (Figures 2C, 2G, and S2H). In addition, we saw large clones in the midlobular zone, both right next to and further away from the pericentral zone (Figure S2H; Video S1G). 1-or 2-cell clones were rare but could still be found; the cells were large and elongated (Figure 2G). However, most lineage-traced hepatocytes divided markedly after 6 or 12 doses of CCl4 (Figures 2C, 2G, S2G, and S2H).
Diploid hepatocytes proliferated more efficiently than polyploid hepatocytes—in normal liver, most (69.4% ± 10.3%) clones, independent of size or location, were polyploid, whereas after 12 doses of CCl4, most (71.9% ± 7.0%) large clones were diploid (Figures 2H and 2I).
DISCUSSION
In summary, our random lineage tracing of hepatocytes in the normal liver does not show predominant proliferation of pericentral hepatocytes, leading to large hepatocyte patches streaming toward the periportal zone, as was shown by lineage tracing of Axin2-expressing hepatocytes (Wang et al., 2015). We did not find that the dose of tamoxifen used for Axin2 lineage tracing significantly induces clonal expansion of hepatocytes (Figure S2I), suggesting genetic or environmental differences, e.g., Axin2 haploinsufficiency or diet and housing, as potential reasons for the discrepant results. In accord with results from lineage tracing of hepatocytes exhibiting high Tert expression (Lin et al., 2018), we found that hepatocyte proliferation occurs throughout the liver lobule, with a slight predominance of the midlobular zone. Our results differ in how much each lineage-traced hepatocyte proliferates: high-Tert lineage tracing showed an average clone size of 4.2 cells at 6 months, with 64% of clones being >2 cells; expansion of lineage-traced hepatocytes from 3%–5% to 30% suggests an average clone size of 6–10 cells at 1 year (Lin et al., 2018). We found an average clone size of 1.1 cells at 13.6–13.7 months, with 9.9% of lineage-traced hepatocytes proliferating and 0.9% of clones being >2 cells (average clone size of 3.9 cells). To confirm that our strategy captured a representative sample of all hepatocytes, we ascertained that it was not biased against cells highly expressing Tert or Axin2 (Figures S2J–S2N). Our results show that modest proliferation of hepatocytes in all zones maintains the hepatocyte mass, which is consistent with most hepatocytes being long lived as suggested by isotope labeling (Arrojo E Drigo et al., 2019).
Our findings were similar for how the hepatocyte mass is restored in the injured liver. Acute loss of pericentral or periportal hepatocytes is repaired by proliferation of adjacent hepatocytes, aided by proliferation of hepatocytes located further away from the injury. Repeated pericentral injury leads to clonal expansion of as many as 85% of hepatocytes in all zones. As clones derived from midlobular hepatocytes moving into the pericentral injury zone continue to be depleted, periportal hepatocytes give rise to large clones that stream along the portal to central axis (Font-Burgada et al., 2015). Although diploidy does not have a significant effect on hepatocyte proliferation in the normal liver, it affords a selective growth advantage in the chronically injured liver (Wilkinson et al., 2019). Diploid clones may derive from pre-existing diploid hepatocytes or form by ploidy reversal of polyploid hepatocytes (Duncan et al., 2010).
Collectively, our findings show that the ability to proliferate is not concentrated in rare stem cell-like hepatocytes but distributed among hepatocytes in liver homeostasis and regeneration, with type of injury and ploidy affecting location and extent of proliferation. Because hepatocytes are constantly exposed to exogenous and endogenous toxins, it is conceivable that this principle of broad distribution of proliferation evolved to reinforce regeneration and minimize cancer risk.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Holger Willenbring (holger.willenbring@ucsf.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were housed under standard barrier conditions and experiments were approved by the Institutional Animal Care and Use Committee at UCSF. Mice were bred in mating pairs as 1 male with 1 female, or trios of 1 male with 2 females. All mice were housed at ambient room temperature in groups of up to 5 mice; remaining males of each litter were housed singly. All mice were immune competent, not involved in previous procedures, and drug and test naive. Food (diets specified in Table S1) and water was provided ad libitum. R26Rrb/rb mice (Red-Horse et al., 2010) (gift from Irving Weissman, Stanford University) maintained on a C57BL/6 background were bred to wild-type C57BL/6J mice (Jackson Laboratory, Stock# 000664) to generate R26Rrb/wt mice. R26RtdRFP/ZG mice were generated by removing the Cre reporter from R26RNZG/NZG mice (Yamamoto et al., 2009) (Jackson Laboratory, Stock# 012429) as previously reported (Schaub et al., 2018), and crossing the resulting R26RZG/ZG mice with R26RtdRFP/tdRFP mice (Luche et al., 2007). R26RtdRFP/ZG mice were maintained on a mixed C57BL/6 and FVB background.
METHOD DETAILS
AAV vector injection and dosing
All AAV vectors were diluted in PBS with 5% sorbitol and 100 μL were intravenously injected through the tail vein. For high, medium, and low-density lineage tracing, 8–15-week-old male or female R26Rrb/wt mice were injected with AAV8-TBG-Cre at 1.01 × 109 viral genomes per gram body weight (vg/g), 3.35 × 108 vg/g, and 5.58 × 107 vg/g or 2.01 × 109 vg/g, 6.70 × 108 vg/g, and 1.12 × 108 vg/g, respectively. Liver tissue was collected for analysis 2–3 weeks after AAV vector injection, or mice were maintained for up to 17 months. Injury protocols were started 2–4 weeks after AAV vector injection. To induce dual labeling of hepatocytes, 12-week old male or female R26RtdRFP/ZG mice were injected with 6.09 × 109 vg/g or 1.00 × 1010 vg/g AAV8-TBG-Cre and 3.48 × 108 vg/g or 4.00 × 108 vg/g AAV9-TTR-Flp, respectively. Liver tissue was collected for analysis 2 weeks after AAV vector injection, or mice were maintained for 9.7 months.
Liver injury protocols
CCl4 was intraperitoneally injected as a 1:4 dilution with corn oil. A dose of 1 μL/g body weight was used to induce acute pericentral injury. A dose of 0.5 μL/g every 3–4 days for a total of 3, 6, or 12 doses was used to induce chronic injury. Liver tissue was collected for analysis 2 days, 3–6 days, or 2 weeks after the last dose. AA was intraperitoneally injected as a 1% solution with sodium chloride 0.9% to induce acute periportal injury. Mice received 0.030 μL/g body weight followed 3 days later by 0.046 μL/g (< 20 g), 0.060 μL/g (20–30 g), or 0.076 μL/g (30–40 g). Liver tissue was collected for analysis 1 day, 1 week, or 2 weeks after the last dose. Untreated controls were analyzed 2–3 weeks after AAV injection. Tamoxifen was intraperitoneally injected as a 20 mg/mL solution in corn oil at a dose of 160 μg/g body weight 2–3 weeks after AAV injection. Liver tissue was collected for analysis 4 weeks later. Untreated controls were analyzed 7 weeks after AAV injection. Protocols were established in C57BL/6 and mixed C57BL/6 and FVB mice.
Tissue collection, processing and imaging
Livers were perfused with 4% paraformaldehyde (PFA), cut into 0.5 cm-thick pieces, and further fixed in 4% PFA for 4–8 hours at 4°C (R26Rrb/wt mice) or overnight (all other mice). Tissues for paraffin embedding were fixed overnight in 10% formalin at 4°C or room temperature (samples for RNAscope). PFA-fixed tissues were washed in PBS and sectioned into 150 μm-to 300 μm-thick sections using a vibratome. Sections from the median or left lobes were used for analysis of clonal expansion during homeostasis and after CCl4 injury. Additionally, sections from each of the left, median, right anterior, and right posterior lobe were used for analysis of clonal expansion during homeostasis in a subset of mice (Table S1). Sections from the right (anterior and posterior) lobe were used to analyze clonal expansion after AA injury because injury was more severe in this lobe. Sections from the median, left, or right anterior lobes were used to assess clone size in control mice 2–3 weeks after AAV injection. Vibratome sections were stained with rabbit anti-glutamine synthetase antibody at 1:2,000 dilution, which was detected with Alexa Fluor-647-conjugated donkey anti-rabbit antibody at 1:250 dilution. For 3D imaging, vibratome sections were cleared using a modified CUBIC protocol (Susaki et al., 2015; Susaki and Ueda, 2016) where incubation in Reagent 1A (10% weight Triton X-100, 5% weight N,N,N’,N’-Tetrakis(2-hydroxypropyl)ethylenediamine, 10% weight urea, and 25 mM sodium chloride dissolved in ultrapure water) was done for 4 days to 1 week at 4°C, followed by staining with 1 μg/mL DAPI in a 1:1 solution of Reagent 2 (25% weight urea, 50% weight sucrose, 15% weight ultrapure water, and 10% weight triethanolamine):PBS overnight at 4°C, and incubation in Reagent 2 at room temperature for 3 hours to overnight. Cleared sections were mounted with Reagent 2 for imaging. In addition, fixed tissues were cryoprotected in 30% sucrose and embedded in OCT (Tissue-Tek) for cryosectioning. 7 μm-thick cryosections from R26RtdRFP/ZG mice were stained with rabbit anti-glutamine synthetase antibody and chicken anti-GFP antibody at 1:2,000 and 1:1,000 dilution, respectively, which were detected with Alexa Fluor-647-conjugated donkey anti-rabbit antibody and Alexa Fluor-488-conjugated donkey anti-chicken antibody, both at 1:250 dilution. In addition, these sections were stained with rabbit anti-DsRed antibody, chicken anti-GFP antibody, and mouse anti-glutamine synthetase antibody at 1:500, 1:1,000, and 1:1,000 dilution, respectively, which were detected with Alexa Fluor-555-conjugated donkey anti-rabbit antibody, Alexa Fluor-488-conjugated donkey anti-chicken antibody, and M.O.M. Immunodetection Kit with Alexa Fluor 647 streptavidin conjugate, all at 1:250 dilution. To assess Tert and Axin2 expression in R26Rrb/wt mice, 5 μm-thick sections of formalin-fixed paraffin-embedded livers were incubated with Tert or Axin2 RNAscope probe and signal was amplified and detected with RNAscope 2.5 HD Reagent Kit-RED. Sections were subsequently incubated with rabbit anti-DsRed antibody at 1:25 dilution to detect mOrange-and mCherry-expressing clones. Anti-DsRed antibody signal was amplified by sequential incubation with donkey anti-rabbit biotin antibody and Alexa Fluor 647 streptavidin conjugate, both at 1:250 dilution. Endogenous biotin, biotin receptors, and streptavidin binding sites were blocked with Streptavidin/Biotin Blocking Kit. Sections of formalin-fixed paraffin-embedded livers of C57/B6J mice were stained with rabbit anti-CCND1 antibody or mouse anti-PCNA antibody at 1:100 or 1:16,000 dilution, respectively, followed by 3,3’-Diaminobenzidine detection. Sections of formalin-fixed paraffin-embedded livers were used for H&E staining. Both immunohistochemistry and H&E staining were performed at Peninsula Histopathology Laboratory. Thick vibratome sections were imaged on a confocal microscope. Confocal image stacks were acquired with a step size of 4 μm along the z axis. Thin cryosections and formalin-fixed paraffin-embedded liver sections were imaged on a fluorescence microscope or a confocal microscope. See Table S1 for numbers of fields imaged per sample. Fluorescent signal from each fluorophore was acquired separately, pseudo-colored, and the brightness and contrast optimized before combining into 1 RGB image using Fiji (Schindelin et al., 2012). Micrographs were prepared using Adobe Creative Suite and were cropped to focus on an area covering a representative half lobule containing 1 portal tract and 1 central vein, or to focus on a clone of interest. Exceptions are Figures S1A, S2A, S2B, and S2E–S2H, which were not cropped. Micrograph series featured in Video S1 were cropped to show a representative field or clone of interest.
Replication, randomization and exclusion
Experiments were replicated independently once (Figures 1F (9.5–9.7 and 13.6–13.7 months), 1G–1I, 2B, 2F, 2G, S1C, S1D, S1E–S1H (9.5–9.7 and 13.6–13.7 months), S1I–S1N, S2C, and S2I–S2N; Videos S1A, S1C–S1E, and S1G), twice (Figures 2C (6 x and 12 x CCl4), 2D (uninjured and AA), 2H, 2I, S1B (2–3 weeks), S2A, S2B, and S2D–S2H), or at least three times (Figures 1B, 1C–1E (time points > 1 year), 1F (2–3 weeks), 2A, 2C (3 x CCl4), 2D (1 x and 3 x CCl4), 2E, S1A, S1B (13.5–17 months), and S1E–S1H (2–3 weeks); Videos S1B and S1F). For each series of experiments, attempts at replication were successful. Mice excluded from the analysis shown in Figures 1 and S1 due to spontaneous development of liver pathologies are indicated in Table S1. Labeling efficiency with AAV8-TBG-Cre varied moderately with the virus production lot; an effort was made to inject the same lot into all mice within the same experiment. Extent of injury with AA showed variability between mice and within the same liver (Figure S2B); in Figures 2B, 2D, and S2D, analysis focused on fields that had autofluorescent cellular debris (2 of 11 mice were excluded because fields with cellular debris were not found in the sections analyzed). Mice from the same litter and sex were randomly assigned to different experimental conditions, and each condition had at least 1 male and 1 female mouse. Researchers were not blinded when analyzing results. No statistical methods were used to predetermine sample size.
QUANTIFICATION AND STATISTICAL ANALYSIS
To generate the graphs in Figures 1F, S1E–S1H, S1J, S1K, and S2I, all 3D clones from 1–14 fields per sample were analyzed. Only clones completely within the tissue sample were analyzed. Number of cells per clone was determined based on clone morphology and nuclear staining with DAPI. Clones containing 2 mononucleated cells were distinguished from binucleated single-cell clones based on whether the cell shape appeared to be 2 cells and whether the 2 nuclei were separated by at least half a nuclear diameter or in separate planes. Hepatocyte hypertrophy was estimated from nuclear DAPI staining and cell size; a clone was considered hypertrophied if it had a nucleus or cell diameter at least 1/3 larger than neighboring hepatocytes in the same zone. Clones containing a cell located within 4 cell distances from the portal vein or bile duct were classified as periportal; clones containing a cell within 2 cell distances from the central vein were classified as pericentral; clones not meeting either criterion were classified as midlobular. Because the periportal and pericentral zones are smaller, clones were counted from additional fields to increase the number of clones in these zones. Number of new hepatocytes per 3D clone was calculated by subtracting the number of cells in each clone by 1, following the assumption that all clones are initially single cells. Dividing the total number of new hepatocytes by the number of 3D clones analyzed in each mouse or at each time point gives the average number of new hepatocytes per 3D clone as shown in Figures S1F and S1K and Table S1. To generate the graphs in Figures 2D and 2I, at least 3 fields were analyzed from 1 vibratome section of R26Rrb/wt mice injected with high-or medium-dose AAV8-TBG-Cre and subjected to CCl4 injury. 18–48 fields were analyzed from 1–7 vibratome sections of R26Rrb/wt mice injected with low-dose AAV8-TBG-Cre and subjected to CCl4 or AA injury. To generate the graphs in Figures 1H, 1I, S1M, and S1N, at least 10 10x fields of stained cryosectioned tissue were analyzed; each of the liver lobes of each mouse are represented in the fields analyzed. Hepatocytes were considered to be in the same 2D cluster if they shared a cell border. Clusters containing a cell located within 4 cell distances from the portal vein were classified as periportal, and clusters containing 1 glutamine synthetase-positive cell (usually within 2 cell distances from the central vein) were classified as pericentral; clusters not meeting either criterion were classified as midlobular. To account for differences in no-AAV cluster size across the lobule due to zonation of AAV8 transduction, cluster sizes were standardized by Z score calculated as Z = (x – μ)/σ where x is the number of cells per 2D cluster and μ and σ are the average and standard deviation, respectively, of the number of cells per no-AAV or AAV-Flp cluster in mice of the corresponding sex (Figures 1H and S1M) or in the corresponding lobular location in mice of the corresponding sex (Figures 1I and S1N) 2 weeks after AAV injection. To generate the graphs in Figures S2L and S2N, 3–6 fields were analyzed to assess Tert and Axin2 expression in all hepatocytes; 76–108 fields were analyzed to assess Tert and 25–35 fields were analyzed to assess Axin2 expression in lineage-traced clones.
Data were tabulated and analyzed in Microsoft Excel or GraphPad Prism. In Figures 1F, S1E, S1G, S1H, S1J, and S2I a similar number of clones were analyzed from each mouse within each group. Data from all mice in the group were pooled and Mann-Whitney U tests were used to compare different groups. Standardized Z score data from all mice in each group in Figures 1H, 1I, S1M, and S1N were pooled and Student’s 2-sided unpaired t tests were used to compare different groups. χ2 tests were used to determine goodness of fit to a Poisson distribution in Figure S1K. Student’s 2-sided unpaired t tests or 2-sided unpaired t tests with Welch’s correction were used to compare groups shown in Figures 2D, 2I, S1B, S1C, and S2N. A p value < 0.05 was considered statistically significant. Statistical parameters are shown in figures and figure legends. Error bars in the figures show SEM to illustrate how precisely the data define the mean; in the main text the mean ± SD is given to show the variability. No methods were used to determine whether data met assumptions of statistical approach. Graphs were generated in Prism. In Figures 1F, 1H, 1I, S1E, S1G, S1H, S1M, and S1N width distribution of points is proportionate to the number of points at each y axis value and represents data distribution. In Figures S1J and S2I minimizing overlap of individual data points is prioritized over representing shape of data distribution.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-glutamine synthetase | BD Transduction Laboratories | Cat# 610517; RRID AB_397879 |
| Mouse monoclonal anti-PCNA | Cell Signaling | Cat# 2586; RRID AB_2160343 |
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970; RRID AB_300798 |
| Donkey polyclonal anti-Chicken IgY, Alexa Fluor 488 | Jackson ImmunoResearch | Cat# 703–545-155; RRID AB_2340375 |
| Donkey polyclonal anti-Rabbit IgG, Biotin | Jackson ImmunoResearch | Cat# 711–065-152; RRID AB_2340593 |
| Donkey polyclonal anti-Rabbit IgG, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31572; RRID AB_162543 |
| Donkey polyclonal anti-Rabbit IgG, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID AB_2536183 |
| Rabbit monoclonal anti-CCND1 | Thermo Fisher Scientific | Cat# MA5–14512; RRID AB_10985779 |
| Rabbit polyclonal anti-DsRed | Takara Bio | Cat# 632496; RRID AB_10013483 |
| Rabbit polyclonal anti-glutamine synthetase | Abcam | Cat# ab49873; RRID AB_880241 |
| Bacterial and Virus Strains | ||
| AAV8-TBG-Cre | Penn Vector Core | AV-8-PV1091 |
| AAV9-TTR-Flp | Vector Biolabs | Custom order |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Allyl alcohol | Sigma-Aldrich | Cat# 240532 |
| Carbon tetrachloride | Sigma-Aldrich | Cat# 319961 |
| N,N,N’,N’-Tetrakis(2-hydroxypropyl)ethylenediamine | Tokyo Chemical Industry (TCI) | Cat# T0781 |
| Paraformaldehyde | VWR International | Cat# 100504–858 |
| D-Sorbitol | Sigma-Aldrich | Cat# S1876 |
| Triethanolamine | Sigma-Aldrich | Cat# 90279 |
| Triton X-100 | Sigma-Aldrich | Cat# X100 |
| Urea | Thermo Fisher Scientific | Cat# 15505035 |
| Critical Commercial Assays | ||
| H&E and immunohistochemical stainings | Peninsula Histopathology Laboratory | N/A |
| M.O.M. Immunodetection Kit | Vector Laboratories | Cat# BMK-2202 |
| RNAscope 2.5 HD Reagent Kit-RED | Advanced Cell Diagnostics (ACD) | Cat# 322350 |
| RNAscope Probe-Mm-Axin2 | Advanced Cell Diagnostics (ACD) | Cat# 400331 |
| RNAscope Probe-Mm-Tert | Advanced Cell Diagnostics (ACD) | Cat# 313441 |
| Streptavidin, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# S21374 |
| Streptavidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP-2002 |
| Experimental Models: Organisms/Strains | ||
| R26Rrb/rb mice | Red-Horse et al., 2010 | N/A |
| R26RtdRFP/tdRFP mice | Luche et al., 2007 | N/A |
| R26RZG/ZG mice | Schaub et al., 2018 | N/A |
| C57BL/6J mice | Jackson Laboratory | Stock# 000664 |
| Software and Algorithms | ||
| Creative Suite | Adobe | CC 2017 |
| Excel | Microsoft | Office 16 |
| Fiji | Schindelin et al., 2012 | N/A |
| Prism | GraphPad | Version 8.1.2 |
| Other | ||
| BX51 fluorescence microscope | Olympus | N/A |
| TCS SP5 confocal microscope | Leica | N/A |
| VT1000 S vibrating-blade microtome | Leica | Cat# 14047235613 |
Highlights.
Random lineage tracing provides a representative sample of all hepatocytes
Liver homeostasis relies on modest proliferation of hepatocytes in all zones
The burden of proliferation in liver regeneration is distributed among hepatocytes
Chronic injury reveals differences in hepatocyte proliferation caused by ploidy
ACKNOWLEDGMENTS
F.C. was supported by NIH T32 DK060414 and the Jane Coffin Childs Memorial Fund for Medical Research, H.Y.L. by an Eli and Edythe Broad Regeneration Medicine and Stem Cell Fellowship, and H.W. by NIH R01 AA026578, CIRM DISC2–10088, and NIH P30 DK026743. The authors thank Donghui Wang (UCSF Preclinical Therapeutics Core) for tail vein injection, Tara Friedrich (UCSF Liver Center) and Jan Tchorz (Novartis) for discussion, and Pamela Derish (UCSF Department of Surgery) for manuscript editing.
Footnotes
DECLARATION OF INTERESTS
H.W. is co-founder and advisory board member of Ambys Medicines. The authors declare no competing interests.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets/code.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2019.11.001.
REFERENCES
- Alvarado TF, Puliga E, Preziosi M, Poddar M, Singh S, Columbano A, Nejak-Bowen K, and Monga SP (2016). Thyroid hormone receptor β agonist induces b-catenin-dependent hepatocyte proliferation in mice: implications in hepatic regeneration. Gene Expr. 17, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrojo E Drigo R, Lev-Ram V, Tyagi S, Ramachandra R, Deerinck T, Bushong E, Phan S, Orphan V, Lechene C, Ellisman MH, and Hetzer MW (2019). Age mosaicism across multiple scales in adult tissues. Cell Metab. 30, 343–351.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P, Wang L, Gao G, Haskins ME, Tarantal AF, McCarter RJ, Zhu Y, Yu H, and Wilson JM (2011). Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab 104, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff AM, Ng CY, Zhou J, Spence Y, and Nathwani AC (2003). Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood 102, 480–488. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, and Grompe M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, Taniguchi K, Nakagawa H, Valasek MA, Ye L, et al. (2015). Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig JA, Nordin JML, Draper C, McMenamin D, Chroscinski EA, Bell P, Gray JT, Richman LK, and Wilson JM (2018). Determining the minimally effective dose of a clinical candidate AAV vector in a mouse model of Crigler-Najjar syndrome. Mol. Ther. Methods Clin. Dev 10, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Nascimento EM, Gajera CR, Chen L, Neuhöfer P, Garbuzov A, Wang S, and Artandi SE (2018). Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, and Fehling HJ (2007). Faithful activation of an extra-bright red fluorescent protein in “knock-in” Crereporter mice ideally suited for lineage tracing studies. Eur. J. Immunol 37, 43–53. [DOI] [PubMed] [Google Scholar]
- Minear S, Leucht P, Jiang J, Liu B, Zeng A, Fuerer C, Nusse R, and Helms JA (2010). Wnt proteins promote bone regeneration. Sci. Transl. Med 2, 29ra30. [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, and Miyajima A. (2012). Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol 22, 1166–1175. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, and Krasnow MA (2010). Coronary arteries form by developmental reprogramming of venous cells. Nature 464, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, and Artandi SE (2005). Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436, 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, Karns RA, Chen F, Rezvani M, Luu HY, et al. (2018). De novo formation of the biliary system by TGFb-mediated hepatocyte trans differentiation. Nature 557, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, and Ueda HR (2016). Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol 23, 137–157. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, and Ueda HR (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc 10, 1709–1727. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, and Nusse R. (2015). Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 524, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson PD, Delgado ER, Alencastro F, Leek MP, Roy N, Weirich MP, Stahl EC, Otero PA, Chen MI, Brown WK, and Duncan AW (2019). The polyploid state restricts hepatocyte proliferation and liver regeneration in mice. Hepatology 69, 1242–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, and Goldhamer DJ (2009). A multifunctional reporter mouse line for Cre-and FLP-dependent lineage analysis. Genesis 47, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, Pikarsky E, and Stanger BZ (2014). Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


