Abstract
Imaging methods for hyperpolarized (HP) 13C agents must sample the evolution of signal from multiple agents with distinct chemical shifts within a very brief timeframe (typically <1 minute), which is challenging using conventional imaging methods. In this work, we compare two of the most commonly used HP spectroscopic imaging methods, spectral-spatial selective excitation and multi-echo chemical shift encoding (CSE, also referred to as IDEAL), for a typical preclinical HP [1-13C]pyruvate imaging scan at 7 T. Both spectroscopic encoding techniques were implemented and validated in HP experiments imaging enzyme phantoms and the murine kidney. SNR performance of these two spectroscopic imaging approaches was compared in numerical simulations and phantom experiments using a single-shot flyback EPI readout for spatial encoding. With identical effective excitation angles, the SNR of images acquired with spectral-spatial excitations and CSE were found to be effectively equivalent.
Keywords: Hyperpolarization, Carbon-13, Metabolic Imaging, Spectroscopic Imaging (MRSI)
Graphical Abstract
Introduction
Advances in dissolution dynamic nuclear polarization have enabled real-time MR imaging of metabolic processes that were previously undetectable with established imaging methods [1]. By far the most extensively studied hyperpolarized (HP) agent is [1-13C]pyruvate, which is used to assess metabolism in tumors, cardiac muscle and other organs [2]. Several promising HP agents are also being investigated for diverse applications, including [1,4-13C]fumarate for imaging necrosis [3], 13C-urea for perfusion [4], [2-13C]pyruvate for citric acid cycle metabolism [5], [1-13C]acetate for fatty acid metabolism [6], [1-13C]dehydroascorbate [7] and [1,3-13C]acetoacetate [8] for imaging redox potentials, and [2-13C]dihydroxyacetone [9] for measurement of gluconeogenic and glycolytic flux. Additionally, using multiple HP agents in a single experiment permits the simultaneous interrogation of different physiological processes [10].
Each HP metabolic agent produces a distinct set of downstream metabolites in tissue, requiring specialized MR methods to spectrally encode the corresponding chemical shifts that distinguish the biochemical fates of the HP 13C labels. Imaging of these signals complicates the MR methods needed in HP experiments, as the sequence must encode 3 spatial dimensions in addition to chemical shift. Further technical complexity is introduced by the dynamic evolution of HP signals over time, as this is often vital to quantitative modeling of HP substrate delivery and metabolism in vivo [11].
Various techniques for dynamic and single-timepoint spectroscopic imaging of HP substrates have been developed. These include, among others, traditional CSI [12] , EPSI [13, 14], multi-echo chemical shift encoding (CSE) [15] and spectral-spatial excitations coupled with spiral [16] , EPI [17], or EPSI [18] readouts. The choice of HP imaging method entails fundamental tradeoffs in spatial, spectral and temporal resolution. While traditional CSI can provide the greatest spectral resolution, it requires many excitations to encode a complete set of spectroscopic images. EPSI accelerates HP imaging by encoding spectral information along one spatial direction per excitation, and is therefore affected by coupling of spectral and spatial encoding parameters. CSE and single-band spectral-spatial excitation differ from traditional CSI and EPSI since they encode chemical shift in a manner that is decoupled from the method of spatial encoding. Multiple studies have evaluated the performance of EPSI [14], traditional CSI and spiral CSE [19] for HP imaging. Despite being two of the most widely used methods for HP MRSI, a direct quantitative comparison of spectral-spatial excitation and CSE imaging that fully accounts for the different HP magnetization usage between these methods has not been reported in the literature.
The CSE technique, which is often also referred to with the acronym IDEAL, is appealing for HP MRSI since it constrains chemical shift encoding in a way that is optimized for the specific set of chemical shifts known to be present in a given HP experiment [15]. However, there are limitations to this multi-echo CSE method when these chemical shifts occur over a very wide range of frequencies. In these cases, broadband excitation in the presence of slice selection gradients will result in large slice offsets for different chemical shifts, and the large excitation bandwidths necessary to cover the wide spectral range can require short RF pulses that exceed transmit power limitations. Spectral-spatial RF pulses can accomplish the task of spectral encoding during excitation if the pulses are appropriately designed to excite only a single metabolite at a time [20], which decouples spatial and spectral encoding and simplifies the imaging experiment. However, there are also difficulties in designing spectral-spatial pulses for selective excitation of individual metabolites within a wide range of chemical shifts due to spectral aliasing and slice position shifts. Given the close similarity in temporal, spatial and spectral resolution of single-band spectral-spatial excitation and CSE imaging, knowledge of the relative SNR performances of these encoding strategies is important in the selection of an appropriate imaging technique for a given HP MRI experiment. Such SNR comparisons are particularly valuable for maximizing the observability of products of metabolism in vivo, since these signals are often quite weak and represent the unique information provided by HP imaging.
The goal of this work was to compare single-band spectral-spatial excitations and multi-excitation CSE for 13C imaging on the Bruker preclinical MRI platform with spatial encoding via snapshot EPI readouts. For this purpose, we developed a flexible imaging sequence that enables both of these spectroscopic imaging methods. Through numerical simulation and phantom experiments, we quantitatively compare the performance of CSE and spectral-spatial excitations for imaging of HP [1-13C]pyruvate at 7 T.
Materials and Methods
All imaging experiments were performed on a 7 T small animal MRI system (Biospec 70/30 USR, Bruker Biospin MRI, Billerica, MA) running ParaVision 6.0.1 and equipped with a 72 mm inner diameter 1H/13C volume resonator (Bruker Biospin MRI). The BGA-12SHP imaging gradients are capable of a maximum gradient amplitude of 660 mT/m and a maximum slew rate of 4570 T/m/s. A 20 mm diameter miniflex coil (Rapid MRI International, Columbus, OH) was used for 13C signal reception with RF excitation from the volume resonator. All simulation and reconstruction was completed using custom-written MATLAB code. All code for the imaging method, experiment design and image reconstruction are publicly available online (https://github.com/mda-mrsl/PV6-HPMR, commit 609db2d). Animal experiment procedures were conducted with approval of our Institutional Animal Care and Use Committee.
Multi-Echo Chemical Shift Encoding Reconstruction
For gradient echo imaging, the MR signal within a volume containing K chemical shifts can be modeled as a sum of complex damped exponentials:
| [1] |
Here ρk is the relative complex amplitude of the signal arising from chemical species k, Δfk is its chemical shift and R2* is the spin-spin relaxation rate. The additional spatially-dependent phase shift ψ accounts for B0 field inhomogeneities present within the imaging volume (e.g. due to shimming or variations in material susceptibility). Assuming these field inhomogeneities can be demodulated from the observed signals and the chemical shifts are known, the complex signal values for a sequence that captures N gradient echoes for K species can be written in matrix form as:
| [2] |
A least-squares estimate for can be found using the Penrose-Moore pseudoinverse of A [15]:
| [3] |
Where H denotes the Hermitian transpose of a complex matrix. This chemical shift decomposition method may be applied to multi-echo MRI data in either image space or k-space [21]. In the case of k-space decomposition without field map demodulation, ρk represents the amplitude of a specific chemical signal at a given spatial frequency. Note that at least K echoes are required to decompose K chemical shifts, but the acquisition of more echoes may improve noise performance in the decomposed images for individual resonances. The careful selection of echo times is critical for CSE decomposition, since together with the known chemical shifts these echo times provide a basis for sampling the phase evolution of the chemical signals. In other words, the prior knowledge of chemical shifts present should be used to select echo times for which the phase angles of these signals are (at the very least) not static; when these phase angles are static, the condition number of A peaks and the system of equations described above are ill-posed.
The performance of CSE reconstruction for a given set of echo times and chemical shifts can be quantified through the condition number of A or through the effective number of signal averages (NSA) of individual chemical shifts [15]. While the condition number provides a summary metric of CSE performance for all resonances, it is often the case that specific chemical shifts are of interest while others may be present but optimizing the SNR of their corresponding images is not a priority. For example, in HP pyruvate imaging maximizing the SNR for products of in vivo metabolism—such as lactate, alanine and bicarbonate—is vital, whereas pyruvate-hydrate is often present from the moment of dissolution and provides little additional information. In these cases, the effective NSA can be used to assess noise performance for the specific chemical shifts of interest, calculated as the reciprocal of the diagonal element of the covariance matrix of A for that resonance:
| [4] |
In numerical and phantom experiments quantifying imaging method performance for HP [1-13C]pyruvate, six chemical shifts were simulated: lactate, pyruvatehydrate, alanine, pyruvate, urea and bicarbonate at 183.2, 179.5, 177, 171, 164 and 161 ppm (corresponding to +921, +642, +453, 0, −566 and −755 Hz at 7 T), respectively.
Sequence Design and Implementation
The template gradient-echo (FLASH) imaging method from Para Vision 6.0.1 was modified to permit gradient waveforms of arbitrary shape to be played out during excitation and readout. Gradient waveforms are stored in plain-text files with amplitude in units of mT/m and the dwell time of each point is determined by the gradient timing resolution of the system (here, 8 us). An optional variable TE delay between excitation and readout allows this sequence to be used for CSE [22]. In order to allow snapshot CSE image acquisition with flexible TE delays, each echo readout follows a separate excitation pulse. The inter-pulse delay is minimized to limit signal evolution across a single set of CSE echoes, and the delay between sets of CSE echoes may be set to control dynamic sampling of HP signal evolution.
Tools for single-shot flyback and symmetric EPI waveform design and image reconstruction were written in MATLAB R2017a (Mathworks, Natick, MA). The EPI waveforms designed with our method traverse in minimum time the k-space trajectory determined by the requested FOV, image matrix and readout bandwidth for the specified gyromagnetic ratio and gradient specifications (timing resolution, maximum amplitude and slew rate). All image data acquired and simulated in this work used the flyback EPI readout shown in Figure 1. Numerical simulations and phantom images for flyback and symmetric EPI trajectories on our preclinical MRI system are provided in supplementary materials.
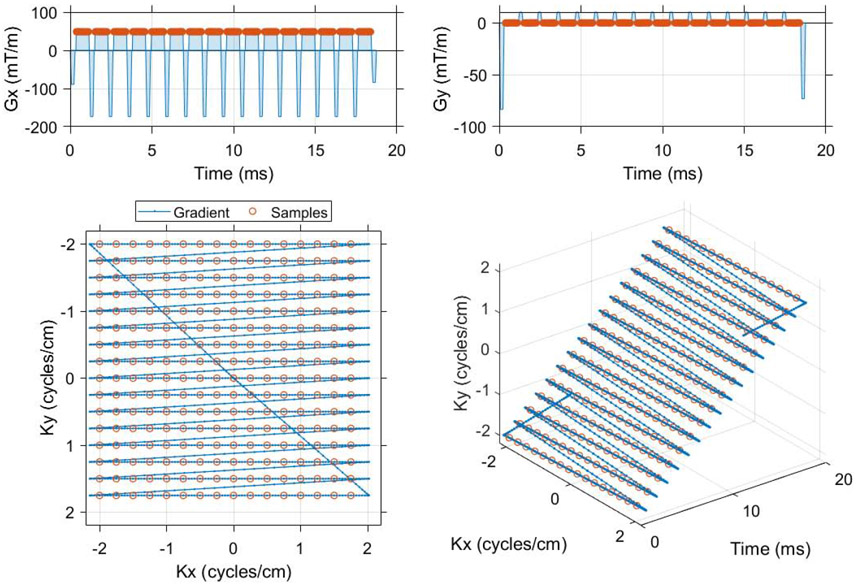
Figure 1.
The single shot EPI trajectory designed in MATLAB that was used to compare spectral encoding strategies for imaging of [1-13C]pyruvate and associated metabolites. Flyback and symmetrically sampled EPI waveforms were created as a function of matrix size, field of view and readout bandwidth (code available online, https://github.com/mda-mrsl/PV6-HPMR/blob/master/matlab/design/create2dEpi.m). This flyback EPI readout has a 16 x 16 matrix, 4 cm field of view and 20.833 kHz readout bandwidth (48 μs dwell time). The center of k-space is sampled 9.984 ms into the gradient waveform, which has a total duration of 18.76 ms and an echo spacing of 1.152 ms (868 Hz phase bandwidth).
The spectral-spatial RF pulse design toolbox written by Adam Kerr and Peder Larson [18] was adapted for use with this method. A flyback trapezoidal gradient was chosen with RF subpulses applied only during the unipolar gradient plateaus. The RF subpulses excite a 1 cm slice with a time-bandwidth factor of 3.5. The overall RF amplitude envelope was designed to impart a single on-resonance passband of at least 1 ppm (75.5 Hz) width separated from its spectral replicates by at least 12.3 ppm (930 Hz). The spatial and frequency response of the spectral-spatial RF pulse designed for HP pyruvate and lactate imaging in this work was validated by measuring one-dimensional spatial readouts along the slice direction in a gadolinium-doped water phantom with varying offsets between transmission and reception frequency. The spatial axis in this 1H measured spectral-spatial profile was corrected by multiplying it by the ratio of 1H to 13C gyromagnetic ratios for comparison with the 13C design Bloch simulation.
Images were reconstructed from EPI data by inverse 2DFT of points sampled in the readout gradient plateau. CSE signal decomposition was performed directly on k-space data prior to inverse 2DFT as described previously [21, 22], without demodulation of B0 field inhomogeneities. All 6 chemical shifts listed above were modeled as individual discrete peaks in this decomposition.
A fair comparison of HP image SNR for a single spectrally-selective excitation and multi-excitation CSE imaging must properly account for the fraction of longitudinal magnetization depleted by each method. A variable excitation angle scheme was used in CSE experiments to provide a constant signal amplitude across echoes and deplete the longitudinal magnetization to a fraction specified by the desired effective excitation angle [23]. Individual excitation angles (θ) were calculated from a specified effective excitation angle (Θ) imparted by a series of N pulses as:
| [5] |
In this equation, Mz denotes the relative longitudinal magnetization present after each pulse, normalized to an initial value of 1 (θj=0 = 0) prior to the first pulse.
For chemical shift imaging, either a 6.6 ms flyback spectral-spatial RF pulse with a 1 cm slice width, 420 Hz passband and 930 Hz stopband was used (Figure 2), or CSE imaging was performed by acquiring 8 echoes with a TE shift of 0.52 ms (Figure 3). A 1.6 ms (5000 Hz) Shinnar Le-Roux calculated RF excitation with 1 cm slice width was used for CSE scans. The first TE for CSE imaging was 11.7 ms, and the TE for spectral-spatial excitation scans was 14 ms. The spectral-spatial pulse employed in this work was designed and tested specifically for selective imaging of pyruvate and lactate at 7 T, and is used in comparison to CSE imaging as a representative method for single-band excitation in HP MRI.
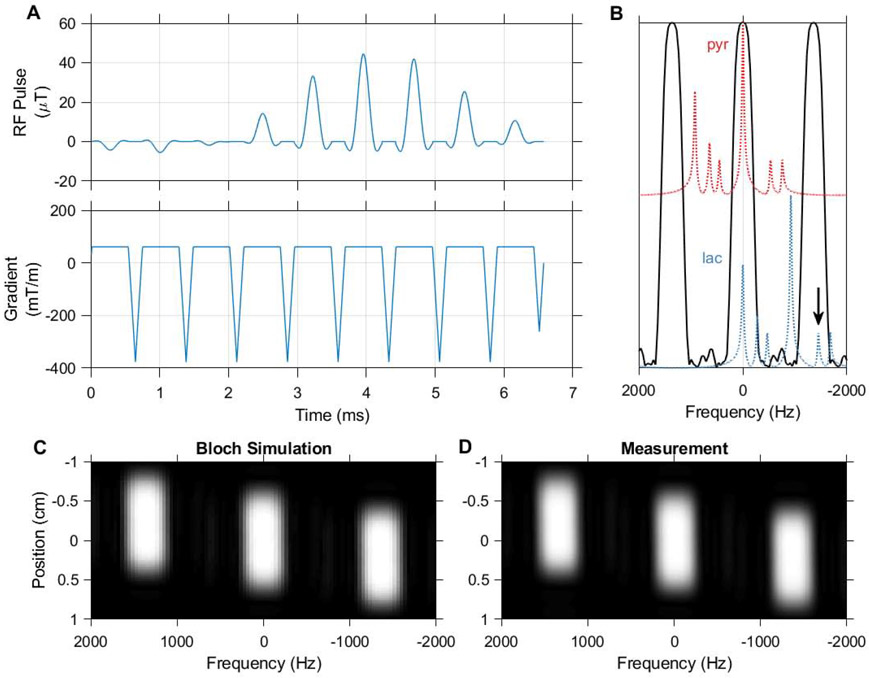
Figure 2.
The spectral-spatial pulse used in this work, designed using the spectral spatial RF pulse design package (https://github.com/LarsonLab/Spectral-Spatial-RF-Pulse-Design). The excitation waveform (A) consists of 9 subpulses with a total duration of 6.6 ms and slice thickness of 1 cm. The real RF B1 magnitude shown corresponds to a 90 degree excitation, and is scaled down to achieve lower excitation angles in HP experiments. A 420 Hz passband and 930 Hz stopband allow selective imaging of pyruvate and lactate at 7 T (B). 13C urea at −1680 Hz (arrow) falls within an aliased passband when the MRI center frequency is set to lactate, but can be presaturated when using this pulse for dynamic imaging. Bloch simulation of the transverse magnetization response to this pulse (C) agrees with measurement in a relaxed water phantom (D).
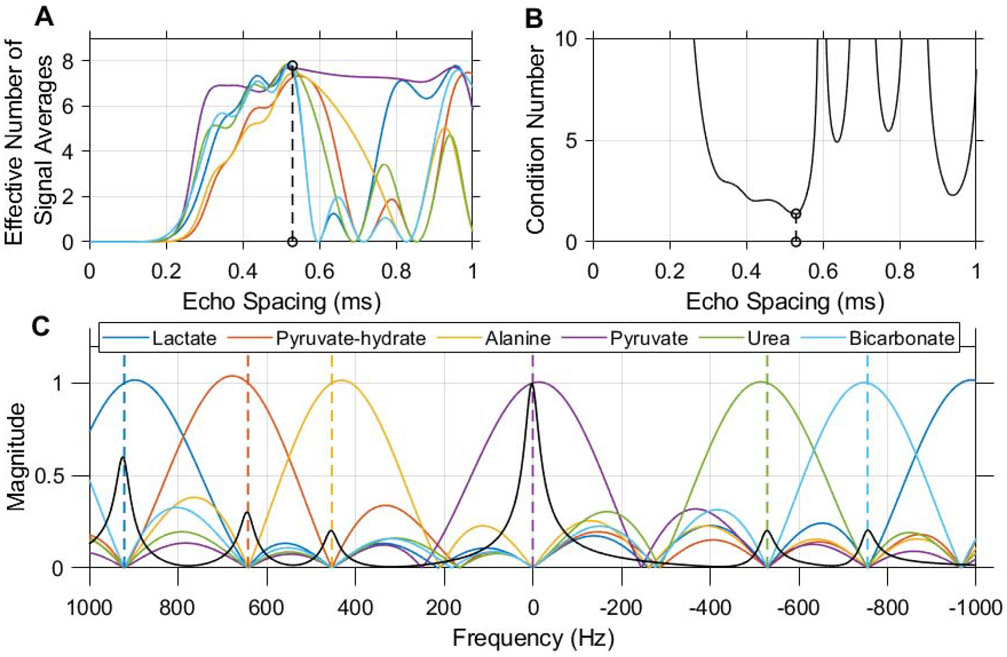
Figure 3.
An echo spacing of 0.52 ms with 8 echoes was chosen for multi-echo CSE of 13C urea, [1-13C]pyruvate and associated metabolites at 7 T. This echo spacing minimizes the condition number of the CSE phase coefficient matrix (B) and optimizes noise performance in the resulting individual chemical shift images (A). Without demodulation of B0 inhomogeneity, this CSE decomposition is simply a linear transform whose frequency response is characterized in the magnitude plot shown here (C). Dashed vertical lines indicate the center frequency of each resonance at 7 T when the scanner frequency is set to pyruvate. A synthetic spectrum depicting HP pyruvate and other expected 13C signals is plotted in black with the magnitude of the frequency response (C) in order to illustrate the potential for spectral misregistration of HP signals due to off-resonance effects.
Numerical Simulation Experiments
The two primary spectral encoding methods enabled in our imaging method, multi-excitation CSE and spectrally-selective excitations, were compared in numerical simulations of HP experiments. Briefly, raw k-space data for a field strength of 7 T were synthesized for a numerical phantom consisting of 13C-urea and the 5 chemical shifts most commonly observed for in vivo imaging of [1-13C]pyruvate (listed above). For spectrally-selective excitations a perfect single-band excitation profile was assumed for the target metabolite. CSE data were synthesized by summation of complex k-space data for all metabolites at 8 echo times with an echo spacing chosen to optimize the performance of this method for these chemical shifts at 7 T (0.52 ms). The precise echo times for each spectral encoding technique matched those used in the phantom and small animal imaging experiments described below. Simulations were conducted for a range of effective excitation angles and T2* relaxation time constants, and repeated 100 times for each variance of complex zero-mean Gaussian noise in k-space.
All SNR values were calculated from magnitude image data and the standard deviation of a noise image, with correction for the Rayleigh distribution of magnitude noise [24]. In actual imaging experiments this noise image was obtained by acquiring a scan equivalent to the experimental scan but with negligible RF power [25].
Urea Phantom Experiments
13C urea doped with gadolinium was imaged using both spectral-spatial excitations and multi-echo CSE with various effective excitation angles. A single 10 mm NMR tube containing approximately 2 mL of 6 M 13C urea (Cambridge Isotope Laboratories, Tewksbury, MA) with 2 mM gadobutrol (Bayer Healthcare LLC, Whippany, NJ) was scanned in the center of the 4x4 cm2 FOV using the flyback EPI trajectory shown in Figure 1 for spatial encoding. For both spectral-spatial excitations and CSE imaging, data were collected for eight repetitions with nominal effective excitation angles of 10 to 90 degrees and a 10 second TR to allow full spin-lattice relaxation (the 13C urea T1 was measured to be approximately 1.6 seconds using a saturation recovery experiment). At each effective excitation angle, the eight measured SNR values for each encoding method were compared using the two-sided two-sample t-test (assuming equal variances). SNR differences were considered statistically significant when p < 0.05.
LDH Phantom Imaging
The spatial fidelity of CSE and single-band spectral-spatial excitations was tested in phantom experiments using lactate dehydrogenase (LDH) to catalyze the conversion of pyruvate to lactate [26]. The phantom consisted of a 50 mL conical tube containing gadolinium-doped water into which two 2 mL cryovials were inserted. One cryovial was prefilled with 1 mL of a solution containing 40 U/mL LDH (Worthington Biochemical, Lakewood, NJ) and 10 mM NADH (Sigma Aldrich, St. Louis, MO) dissolved in 0.2 M pH 7.6 TRIS buffer (Sigma Aldrich, St. Louis, MO), while the other cryovial was prefilled with 1 mL of the TRIS buffer solution. 1 mL of 20 mM HP [1-13C]pyruvate produced using an HyperSense Polarizer (Oxford Instruments, Abingdon, England) was injected into both cryovials simultaneously after the start of each dynamic acquisition, resulting in a final concentration of 20 U/mL LDH, 5 mM NADH and 10 mM pyruvate in the vial containing LDH solution.
A concentrated 13C urea phantom was used to set the MRI system center frequency and calibrate transmit power. All scans were acquired for 90 repetitions to sample the dynamics of pyruvate delivery and conversion to lactate. At the start of each repetition a 5-degree excitation was applied using the same RF pulse shape as CSE imaging to acquire a free induction decay prior to image data acquisition, and these slice-selective spectra were used to determine the frequencies of pyruvate and lactate for off-resonance correction of imaging data. A 1H RARE image was acquired after the HP 13C scan with the same FOV and slice position and used as a background for overlaying the 13C images.
In Vivo Kidney Imaging
Two healthy female BALB/c mice were imaged using the methods described above for LDH phantoms, with additional higher-order shimming applied to the 1 cm slice used for HP imaging. The 20 mm surface coil was placed over the right kidney and the imaging sequence was initiated prior to injecting 200 μL of 80 mM HP [1-13C]pyruvate via tail vein catheter. Each animal’s respiration was monitored continuously and anesthesia was initiated and maintained using 2-4% isoflurane delivered via nose cone.
Results
Sequence Design
The MATLAB tools written for design of flyback and symmetrically sampled EPI waveforms allow for flexibility in choice of readout trajectory as a function of matrix size, field of view and readout bandwidth. Figure 1 shows the flyback EPI trajectory used in phantoms and simulation experiments comparing spectral encoding strategies.
The imaging method developed in this work enables spectral-spatial excitations on a preclinical Bruker MRI system. Figure 2 shows the RF and gradient waveforms of the spectral-spatial pulse we have designed for selective imaging of HP pyruvate and lactate at 7 T. The magnetization profile of this pulse measured in a water phantom agrees with the profile obtained via the simulation tools of the spectral-spatial RF pulse design toolbox. The spectral profile of this pulse is suitable for selective imaging of pyruvate, however when the center frequency is set to lactate considerable excitation of urea occurs within a passband alias. Since urea is often used in these experiments only in thermally polarized phantoms to calibrate 13C RF power and frequency, this signal can be easily mitigated using a presaturation pulse.
This imaging sequence realizes CSE for HP imaging by acquiring multiple snapshot EPI datasets at equally spaced echo times. In general, the choice of echo spacing controls the performance of CSE decomposition for a given set of chemical shifts [27]. For the 6 chemical shifts considered in this work, a train of 8 echoes with 0.52 ms spacing was chosen to optimize noise performance and minimize the condition number of the CSE phase coefficient matrix. The plots in Figure 3 demonstrate how this echo spacing was chosen, and show the frequency response of CSE decomposition with these parameters. Without field map demodulation, CSE decomposition is a linear transform whose response to unit input signals at any frequency is shown in the magnitude response plot in Figure 3C.
Numerical Simulation Experiments
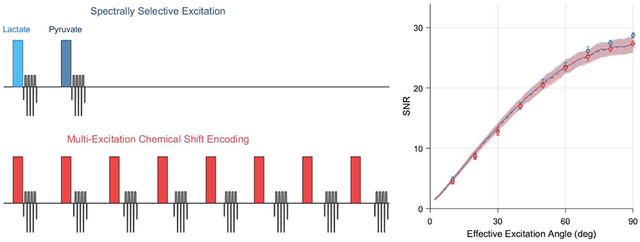
The performance of single-band spectrally selective excitations and multi-excitation CSE was compared in numerical simulations for various effective excitation angles, T2* relaxation times and levels of complex noise in the raw k-space data. The results of these simulations are summarized in Figure 4. Nearly identical theoretical SNR performance is seen with these two chemical shift encoding strategies for HP imaging, and at no set of simulation parameters were the SNRs of these two frequency encoding methods separated by more than one standard deviation about their respective means. Under the conditions of a typical preclinical HP imaging experiment, which is often conducted with dynamic imaging at low excitation angles to observe substrate uptake and metabolic conversion, we observed no significant difference in SNR performance between spectrally selective excitations and CSE decomposition in numerical simulation.
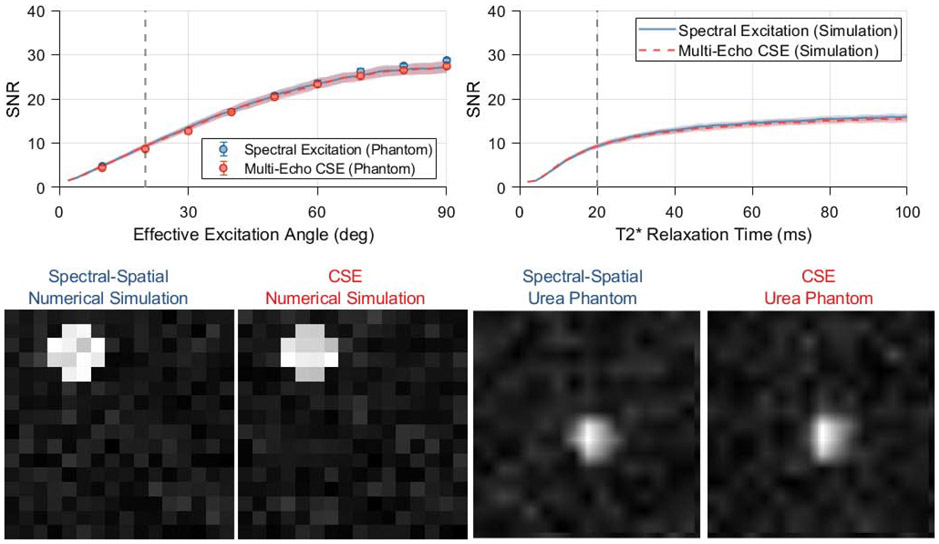
Figure 4.
Simulations comparing spectral encoding strategies for imaging of [1-13C]pyruvate and its associated metabolites at 7 T. The mean output SNRs for a single metabolite are summarized as a function of effective excitation angle and T2* relaxation time constant in the plots in the upper row. The lines and shaded regions depict one standard deviation about the mean for numerically simulated data. Vertical dashed lines indicate the standard value of each parameter that is used when plotting other parameters and displaying the numerical phantom images (lower left). Images from a thermal phantom experiment corresponding to 20 degree effective excitations are shown in the bottom right. SNR values were measured in eight repetitions of this phantom imaging experiment at various effective excitation angles, and are plotted with numerical simulation results in the upper left. Phantom data show a significantly lower SNR for multi-excitation CSE at effective excitation angles ≥70 degrees (* p<0.05, two-sample t-test), however the effect size is miniscule and not practically meaningful.
Urea Phantom Experiments
Images of the thermally polarized 13C urea phantom are shown in the bottom row of Figure 4, with the mean phantom SNR plotted as a function of effective excitation angle alongside numerical simulation data in the top left plot. These phantom images show 13C signal only for the on-resonance decomposed CSE chemical shift offset. Both chemical shift imaging methods show the expected sinusoidal trend in image SNR with increasing excitation angle, and these phantom data are congruent with the numerical simulation results for a T2* of 20 ms and k-space noise variance of 0.09. At an effective excitation angle of 20 degrees, the 13C urea images shown in the bottom row of Figure 4 exhibit SNRs of 8.6 ± 0.47 and 8.7 ± 0.50 (mean ± standard deviation) for spectral-spatial excitations and multi-excitation CSE, respectively.
The SNR obtained through multi-excitation CSE was statistically lower than spectral-spatial imaging for effective excitation angles of 70 degrees or greater. However, the magnitude of this difference was very small (<5%). This slight SNR difference is very unlikely to impact the decision of whether to use CSE for HP imaging in practice, especially since it was observed only for large effective excitation angles that are rarely used in HP imaging. Therefore, we conclude that both numerical and phantom simulation results demonstrate that the SNR performance of spectral-spatial and CSE imaging are effectively equivalent for HP pyruvate and lactate imaging on our preclinical MRI system.
LDH Phantom Imaging
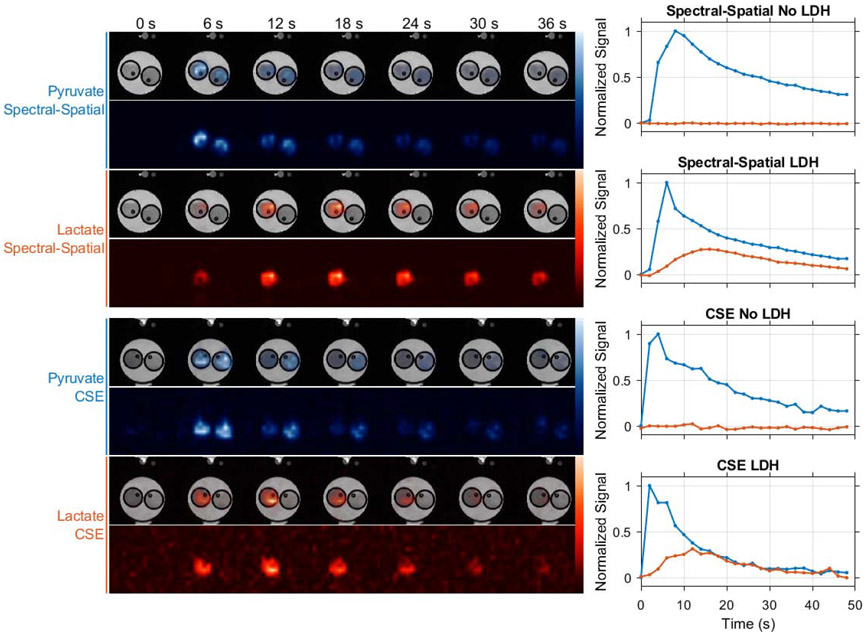
Experiments using LDH phantoms demonstrate that both chemical shift imaging strategies evaluated in this work can be used to selectively image HP pyruvate and lactate. As shown in Figure 5, similar HP imaging signal trends are seen in images acquired with spectral-spatial excitations and multi-excitation CSE. In these experiments, strong HP pyruvate signal is evident in both vials of our phantom, with lactate production observed only in the vial preloaded with the LDH enzyme and NADH cofactor. These results demonstrate that both chemical shift imaging methods provide spatial fidelity appropriate for small animal imaging when used with the snapshot EPI trajectory employed in this work.
Figure 5.
The spatial fidelity of both chemical shift imaging methods used in this work was evaluated in LDH enzyme phantom experiments. In these experiments only the vials seen on the left side of these images were preloaded with LDH, which catalyzed production of lactate when HP pyruvate was injected simultaneously into both vials. HP signal is plotted over time for a voxel at the center of each vial, showing similar substrate delivery and conversion trends for spectrally selective excitations and multi-excitation CSE imaging with lactate signal confined to the vial containing LDH. Note that due to variations in absolute polarization level and in the rate of imaging agent delivery by manual injection, this experiment is not suitable for quantitative SNR comparison of spectral-spatial and multi-excitation CSE imaging. Dynamic pyruvate (blue) and lactate (orange) images are therefore normalized to their respective maximum intensities.
In Vivo Kidney Imaging
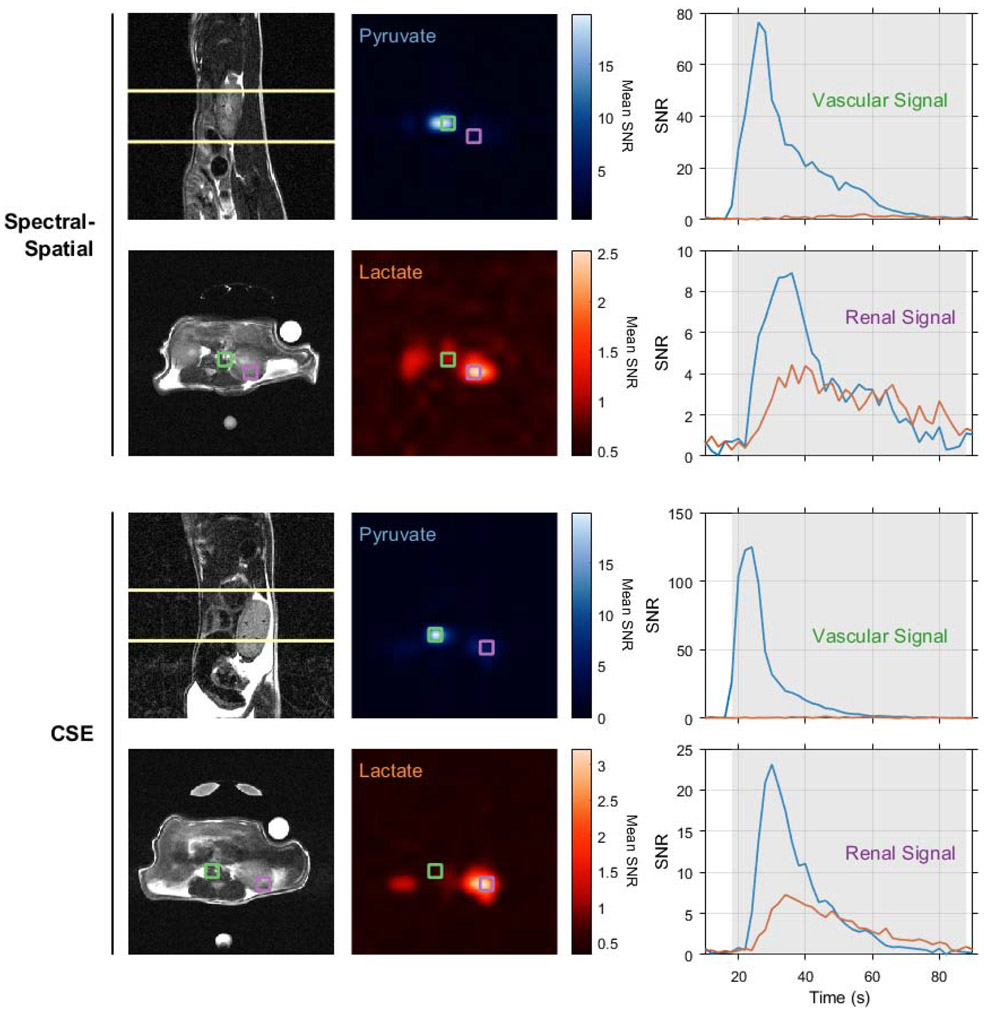
Both chemical shift imaging approaches compared in this work were tested in imaging renal metabolism in healthy BALB/c mice. Figure 6 shows the slice positions, time-averaged pyruvate and lactate images, and HP signal time courses for selected voxels from each experiment. As expected, both imaging strategies show a very intense pyruvate signal from the voxel containing the inferior vena cava and abdominal aorta, with weaker pyruvate and lactate observed in later time points from the kidney. While these animal experiments lack statistical power to provide any definitive conclusions regarding the relative performance of spectral-spatial excitation and multi-echo CSE for HP pyruvate imaging, they clearly show that each method resolves real-time metabolism in vivo.
Figure 6.
Renal pyruvate uptake and metabolism were imaged in healthy BALB/c mice using spectral-spatial excitation (upper panels) and multi-excitation CSE (lower panels). HP images in the central panel are bilinearly interpolated to the resolution of the axial proton images (lower left) and averaged over time. Native resolution voxels containing a large blood vessel and kidney parenchyma are outlined in green and purple, respectively. The HP signals for these vascular and renal voxels are plotted overtime on the right, and the shaded portions of these plots indicate the time points averaged in the HP images. All images depict a 4x4 cm2 FOV.
Discussion
In this work we developed an imaging method for preclinical HP 13C imaging and quantitatively compared the imaging approaches enabled in this pulse program. In comparing the two spectral encoding strategies available to us, multi-excitation CSE and spectrally selective excitations, we found in numerical simulations and confirmed in phantom measurements that both provide equivalent performance for HP imaging when the same amount of longitudinal magnetization is depleted. In phantom experiments we also demonstrate the spatial fidelity of the image encoding strategies enabled by our method in a system that recreates one of the enzymatic reactions this imaging technique seeks to quantify. We further demonstrate the feasibility of our method by imaging lactate production in the murine kidney in vivo.
MRI scan parameters must be set carefully to provide optimal performance for a given experiment. This is particularly important for HP MRI due to the non-renewable nature of the imaging signal and the relatively low amplitude of signals produced via metabolic conversion in vivo. Quantitative evaluation of methods for encoding position and chemical shift is therefore critical for HP MRI. Previous work by Durst, et al. compared various chemical shift readout methods for HP 13C imaging on a clinical 3 T system in extensive simulation, phantom and animal experiments [19]. While this study included a thorough comparison of traditional CSI, EPSI and multi-echo CSE imaging using spiral readouts, spectrally selective excitations were not considered for encoding chemical shift nor was EPI used for spatial encoding. A separate study by Schulte, et al. focused on the evaluation of spectral-spatial excitation imaging at 3 T with a 15 degree excitation angle on pyruvate and saturation of downstream metabolites [16]. This study included a SNR comparison of this specific spectral-spatial imaging strategy to CSE imaging with seven 10 degree excitations in a rat tumor model. Greater mean peak SNR was observed for pyruvate using CSE compared to spectral-spatial imaging. The opposite result was seen for lactate and other downstream metabolites, where 90 degree spectral-spatial excitations provided superior SNR than multi-excitation CSE imaging. These results are consistent with ours, since the effective excitation angle for all metabolites in their method of CSE imaging (≈26 degrees) is greater than the 15 degree spectral-spatial excitation angle applied to pyruvate, but much lower than the 90 degree spectral-spatial excitation angle applied to downstream metabolites.
The simulation and phantom experiments presented here demonstrate the relative performance of these imaging techniques on our preclinical 7 T MRI system. Different results could be observed if the same imaging methods were evaluated on a system operating at another field strength, or with different gradient capabilities that would require redesign of gradient waveforms. The spectral-spatial pulse shown in Figure 2 is used here as a representative implementation of single-band chemically selective excitation. Alternate spectral-spatial pulses used for selective excitation of individual chemical shifts may affect our results due to disparities in the minimum echo time and effective slice profile. The CSE image reconstruction method used in this work does not correct for B0 field inhomogeneities, and our simulation results therefore represent the optimal performance expected from CSE imaging with a homogenous field map or with perfect correction of these effects. As in the work presented here, the 13C imaging techniques available should be evaluated thoroughly to compare their performance on the specific hardware that is to be used in a given experiment.
When comparing single-band spectral-spatial excitations to multi-echo CSE for spectral encoding, there are several practical distinctions worth noting. The longer chemical shift encoding time generally required in CSE increases the chance of artifacts from physiological motion in these images. Depending on the organ of interest, these motion effects could be mitigated by physiological gating of the image acquisition [28], however this gating can place strict limitations on the ability to dynamically measure HP signal evolution. These motion effects on both spectral-spatial excitation and multi-excitation CSE imaging warrant further study. In the context of multislice imaging, the faster encoding time of spectral-spatial excitations can permit greater volumetric coverage and superior temporal resolution than multi-excitation CSE. For single-band spectral-spatial imaging, reliable signal excitation depends on proper setting of the frequency offset so that the chemical shift of interest falls in the passband of the pulse’s frequency response. If set incorrectly, the chemical shift may occur at the edge of the passband and experience a reduced excitation angle, or outside the passband and not be detectable. This can be particularly problematic for HP imaging with narrowband pulses since this frequency offset must be set before the signals of interest are present. For CSE imaging, the wide bandwidth of the RF pulse used is typically sufficient to consistently excite all chemical shifts present, however the constant excitation gradient will impart positional shifts in the slice direction that vary across chemical shift offsets [29]. This effect can become significant when using CSE with thick slices [30] and when decomposing images for chemical shifts separated by large frequency offsets. Although we have focused on comparing the performance of a multi-echo CSE spectroscopic readout method with spectroscopic excitation for imaging of HP pyruvate and lactate, these two approaches are not mutually exclusive. Multi-band excitation pulses can easily be used to control the chemical content of signals encoded with CSE [28] or other spectroscopic readouts such as EPSI [18]. This combination of chemical shift encoding in both excitation and readout provides methodological flexibility that is very useful in studies of novel HP preparations [31].
In conclusion, we have demonstrated that multi-excitation CSE (IDEAL) and spectral-spatial imaging provide equivalent SNR performance for preclinical HP imaging of [1-13C]pyruvate when each method consumes equal amounts of magnetization. The imaging method we developed in this work provides flexibility in the choice of encoding methods for position and chemical shift and is readily adaptable for use in various hyperpolarized imaging experiments. The source code for our numerical simulations, imaging method and reconstruction of images acquired with the techniques presented in this work available online (https://github.com/mda-mrsl/PV6-HPMR). Readers who conduct hyperpolarized imaging experiments on Bruker MRI systems are invited to adapt this method for their needs.
Supplementary Material
Highlights:
HP MRI requires specialized imaging methods for measuring metabolism in vivo
Selective excitation and multi-echo imaging readouts can encode HP chemical shift
A framework that supports both these imaging strategies permits their comparison
When equal HP magnetization is consumed, equivalent SNR is obtained
Acknowledgments
This work was supported in part by the National Institutes of Health (R01-DK105346, R01-CA211150, P30-CA016672) and the Cancer Prevention and Research Institute of Texas (RP170366). The content is solely the responsibility of the authors and does not necessarily represent the official views of these agencies.
The authors gratefully acknowledge Jorge De La Cerda and Charles Kingsley for their assistance with animal handling and preparation, and Reshmi Patel and Karen Wang for their assistance in editing this manuscript. This research was performed in partial fulfillment of the requirements for the PhD degree from The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences.
Abbreviations:
- CSE
(multi-echo) Chemical Shift Encoding
- HP
Hyperpolarized
- LDH
Lactate Dehydrogenase
- NSA
Number of Signal Averages
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR, Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research, Neoplasia, 13 (2011) 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Comment A, Merritt ME, Hyperpolarized magnetic resonance as a sensitive detector of metabolic function, Biochemistry, 53 (2014) 7333–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gallagher FA, Kettunen MI, Hu DE, Jensen PR, Zandt RI, Karlsson M, Gisselsson A, Nelson SK, Witney TH, Bohndiek SE, Hansson G, Peitersen T, Lerche MH, Brindle KM, Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors, Proc Natl Acad Sci U S A, 106 (2009) 19801–19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].von Morze C, Larson PE, Hu S, Keshari K, Wilson DM, Ardenkjaer-Larsen JH, Goga A, Bok R, Kurhanewicz J, Vigneron DB, Imaging of blood flow using hyperpolarized [(13)C]urea in preclinical cancer models, J Magn Reson Imaging, 33 (2011) 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schroeder MA, Atherton HJ, Ball DR, Cole MA, Heather LC, Griffin JL, Clarke K, Radda GK, Tyler DJ, Real-time assessment of Krebs cycle metabolism using hyperpolarized 13C magnetic resonance spectroscopy, Faseb j, 23 (2009) 2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jensen PR, Peitersen T, Karlsson M, in 't Zandt R, Gisselsson A, Hansson G, Meier S, Lerche MH, Tissue-specific Short Chain Fatty Acid Metabolism and Slow Metabolic Recovery after Ischemia from Hyperpolarized NMR in Vivo*, in: J Biol Chem, 2009, pp. 36077–36082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bohndiek SE, Kettunen MI, Hu DE, Kennedy BW, Boren J, Gallagher FA, Brindle KM, Hyperpolarized [1-13C]-ascorbic and dehydroascorbic acid: vitamin C as a probe for imaging redox status in vivo, J Am Chem Soc, 133 (2011) 11795–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].von Morze C, Ohliger MA, Marco-Rius I, Wilson DM, Flavell RR, Pearce D, Vigneron DB, Kurhanewicz J, Wang ZJ, Direct assessment of renal mitochondrial redox state using hyperpolarized (13) C-acetoacetate, Magn Reson Med, 79 (2018) 1862–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moreno KX, Satapati S, DeBerardinis RJ, Burgess SC, Malloy CR, Merritt ME, Real-time detection of hepatic gluconeogenic and glycogenolytic states using hyperpolarized [2-13C]dihydroxyacetone, J Biol Chem, 289 (2014) 35859–35867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wilson DM, Keshari KR, Larson PE, Chen AP, Hu S, Van Criekinge M, Bok R, Nelson SJ, Macdonald JM, Vigneron DB, Kurhanewicz J, Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo, J Magn Reson, 205 (2010) 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bankson JA, Walker CM, Ramirez MS, Stefan W, Fuentes D, Merritt ME, Lee J, Sandulache VC, Chen Y, Phan L, Chou PC, Rao A, Yeung SC, Lee MH, Schellingerhout D, Conrad CA, Malloy C, Sherry AD, Lai SY, Hazle JD, Kinetic Modeling and Constrained Reconstruction of Hyperpolarized [1-13C]-Pyruvate Offers Improved Metabolic Imaging of Tumors, Cancer Res, 75 (2015) 4708–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Golman K, in 't Zandt R, Thaning M, Real-time metabolic imaging, Proc Natl Acad Sci U S A, 103 (2006) 11270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen YF, Pauly JM, Nelson SJ, Vigneron DB, Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C, J Magn Reson, 187 (2007) 357–362. [DOI] [PubMed] [Google Scholar]
- [14].Yen YF, Kohler SJ, Chen AP, Tropp J, Bok R, Wolber J, Albers MJ, Gram KA, Zierhut ML, Park I, Zhang V, Hu S, Nelson SJ, Vigneron DB, Kurhanewicz J, Dirven HA, Hurd RE, Imaging considerations for in vivo 13C metabolic mapping using hyperpolarized 13C-pyruvate, Magn Reson Med, 62 (2009) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Reeder SB, Brittain JH, Grist TM, Yen YF, Least-squares chemical shift separation for (13)C metabolic imaging, J Magn Reson Imaging, 26 (2007) 1145–1152. [DOI] [PubMed] [Google Scholar]
- [16].Schulte RF, Sperl JI, Weidl E, Menzel MI, Janich MA, Khegai O, Durst M, Ardenkjaer-Larsen JH, Glaser SJ, Haase A, Schwaiger M, Wiesinger F, Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1-13C] pyruvate using spectral-spatial excitation, Magn Reson Med, 69 (2013)1209–1216. [DOI] [PubMed] [Google Scholar]
- [17].Lupo JM, Chen AP, Zierhut ML, Bok RA, Cunningham CH, Kurhanewicz J, Vigneron DB, Nelson SJ, Analysis of hyperpolarized dynamic 13C lactate imaging in a transgenic mouse model of prostate cancer, Magn Reson Imaging, 28 (2010) 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Larson PE, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB, Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging, J Magn Reson, 194 (2008) 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Durst M, Koellisch U, Frank A, Rancan G, Gringeri CV, Karas V, Wiesinger F, Menzel MI, Schwaiger M, Haase A, Schulte RF, Comparison of acquisition schemes for hyperpolarised (1)(3)C imaging, NMR Biomed, 28 (2015) 715–725. [DOI] [PubMed] [Google Scholar]
- [20].Cunningham CH, Chen AP, Lustig M, Hargreaves BA, Lupo J, Xu D, Kurhanewicz J, Hurd RE, Pauly JM, Nelson SJ, Vigneron DB, Pulse sequence for dynamic volumetric imaging of hyperpolarized metabolic products, J Magn Reson, 193 (2008) 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brodsky EK, Holmes JH, Yu H, Reeder SB, Generalized k-space decomposition with chemical shift correction for non-Cartesian water-fat imaging, Magn Reson Med, 59 (2008) 1151–1164. [DOI] [PubMed] [Google Scholar]
- [22].Wiesinger F, Weidl E, Menzel MI, Janich MA, Khegai O, Glaser SJ, Haase A, Schwaiger M, Schulte RF, IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate, Magn Reson Med, 68 (2012) 8–16. [DOI] [PubMed] [Google Scholar]
- [23].Nagashima K, Optimum pulse flip angles for multi-scan acquisition of hyperpolarized NMR and MRI, J Magn Reson, 190 (2008) 183–188. [DOI] [PubMed] [Google Scholar]
- [24].Henkelman RM, Measurement of signal intensities in the presence of noise in MR images, Med Phys, 12 (1985) 232–233. [DOI] [PubMed] [Google Scholar]
- [25].NEMA MS 1-2008 (R2014) Determination of Signal-to-Noise Ratio (SNR) in Diagnostic Magnetic Resonance Imaging, in, 2008. [Google Scholar]
- [26].Walker CM, Lee J, Ramirez MS, Schellingerhout D, Millward S, Bankson JA, A catalyzing phantom for reproducible dynamic conversion of hyperpolarized [1-13C]-pyruvate, PLoS One, 8 (2013) e71274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brodsky EK, Chebrolu VV, Block WF, Reeder SB, Frequency response of multipoint chemical shift-based spectral decomposition, J Magn Reson Imaging, 32 (2010) 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sigfridsson A, Weiss K, Wissmann L, Busch J, Krajewski M, Batel M, Batsios G, Ernst M, Kozerke S, Hybrid multiband excitation multiecho acquisition for hyperpolarized (13) C spectroscopic imaging, Magn Reson Med, 73 (2015) 1713–1717. [DOI] [PubMed] [Google Scholar]
- [29].Yang S, Lee J, Joe E, Lee H, Choi YS, Park JM, Spielman D, Song HT, Kim DH, Metabolite-selective hyperpolarized (13)C imaging using extended chemical shift displacement at 9.4T, Magn Reson Imaging, 34 (2016) 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grist JT, McLean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, Patterson I, Slough R, Gill AB, Chhabra A, Eichenberger R, Laurent MC, Comment A, Gillard JH, Coles AJ, Tyler DJ, Wilkinson I, Basu B, Lomas DJ, Graves MJ, Brindle KM, Gallagher FA, Quantifying normal human brain metabolism using hyperpolarized [1-(13)C]pyruvate and magnetic resonance imaging, Neuroimage, 189 (2019) 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marco-Rius I, Cao P, von Morze C, Merritt M, Moreno KX, Chang GY, Ohliger MA, Pearce D, Kurhanewicz J, Larson PE, Vigneron DB, Multiband spectral-spatial RF excitation for hyperpolarized [2-(13) C]dihydroxyacetone (13) C-MR metabolism studies, Magn Reson Med, 77 (2017) 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.