Abstract
The bacterial Type VI secretion system (T6SS) is a nano-machine that delivers toxic effector proteins into target cells killing them. Here we show that the V. cholerae T6SS attacks members of the host commensal microbiota in vivo, facilitating the pathogen’s colonization of the murine gut. This microbial antagonistic interaction drives measurable changes in the pathogenicity of V. cholerae through enhanced intestinal colonization, expression of bacterial virulence genes, and activation of host innate immune genes. Because ablation of mouse commensals by this enteric pathogen correlated with more severe diarrheal symptoms, we conclude that antagonism toward the gut microbiota could improve the fitness of V. cholerae as a pathogen by elevating its transmission to new susceptible hosts.
One Sentence Summary:
Type 6 secretion-mediated killing of gut commensal microorganisms enhances virulence of Vibrio cholera and modulates host gene expression
Bacterial pathogens deploy numerous virulence factors that directly influence their pathobiology. However, little is known about the impact of pathogen interactions with the commensal microbiome on virulence and pathogen fitness. Vibrio cholerae, the causative agent of cholera (1, 2) assembles a dynamic organelle called Type VI secretion system (T6SS) that delivers toxic effector proteins to eukaryotic and prokaryotic cells (3, 4). The T6SS of V. cholerae can kill heterologous bacterial species in vitro, such as Escherichia coli (5), but it is also activated in vivo (within infected experimental animals) to kill V. cholerae cells that lack immunity to its toxic effectors (6, 7). Hence, we asked if the antibacterial activity of the T6SS is directed against gut commensals that share a niche with V. cholerae (6, 8). Here, we show that V. cholerae T6SS is used against commensal bacterial species and drives measurable changes in the pathogenicity of V. cholerae.
To test whether V. cholerae exerts antagonistic effects on the gut microbiota, we isolated several Gram-negative commensal bacteria from the small intestines of 5-day-old suckling mice (infant mice). All isolates were susceptible to T6SS killing by the V. cholerae 2740-80 strain, which expresses an active T6SS in vitro (9) (Table S1). Of the commensal strains isolated, we selected two E. coli strains that readily colonized the mouse small intestine, the niche V. cholerae also predominantly colonizes during infection. Although each strain was capable of colonizing the majority of animals within a mono-challenged group, when used as a mixture, two strains (WZ1-1 and WZ2-1) were found to reproducibly colonize all challenged animals (Figure S1). These two strains typically achieved a level of approximately 104–106 colony-forming units (CFU) per mouse small intestine after 24–48 hours of infection and were therefore used as a defined commensal microbiota in subsequent studies. To establish whether the V. cholerae T6SS could affect the gut colonization of these E. coli commensal strains, we inoculated groups of neonatal mice with the two E. coli strains, returned them to their dams for 24 hours, and then arbitrarily selected three groups of pups for challenge with 105 CFU of either V. cholerae WT (strain C6706 (8)), an isogenic T6SS-defective vipA mutant, or buffer control. E. coli intestinal load declined at 12 hours post V. cholerae WT challenge and was ~300-fold lower than the load in mice challenged with the T6SS mutant at 24 hours (Fig 1A. 5 ± 2 vs. 1200 ± 400 CFU/mouse, P < 0.05, WT and T6SS mutant treatment, respectively) or buffer (3840 ± 435 CFU/mouse). Surviving E. coli recovered from WT treated mice did not show a reproducible skew towards either WZ1-1 and WZ2-1 indicating that both strains were equally targeted for elimination in vivo. Because of its dependency on the T6SS vipA gene and the observation that C6706 is known to express T6SS genes in vivo during mouse infections (8), we conclude that the antibacterial activity directed against the E. coli commensal isolates is dependent on expression of V. cholerae T6SS within the mouse intestine.
Figure 1. T6SS facilitated V. cholerae (Vc) colonization of the mouse gut by eliminating T6SS-sensitive commensals.
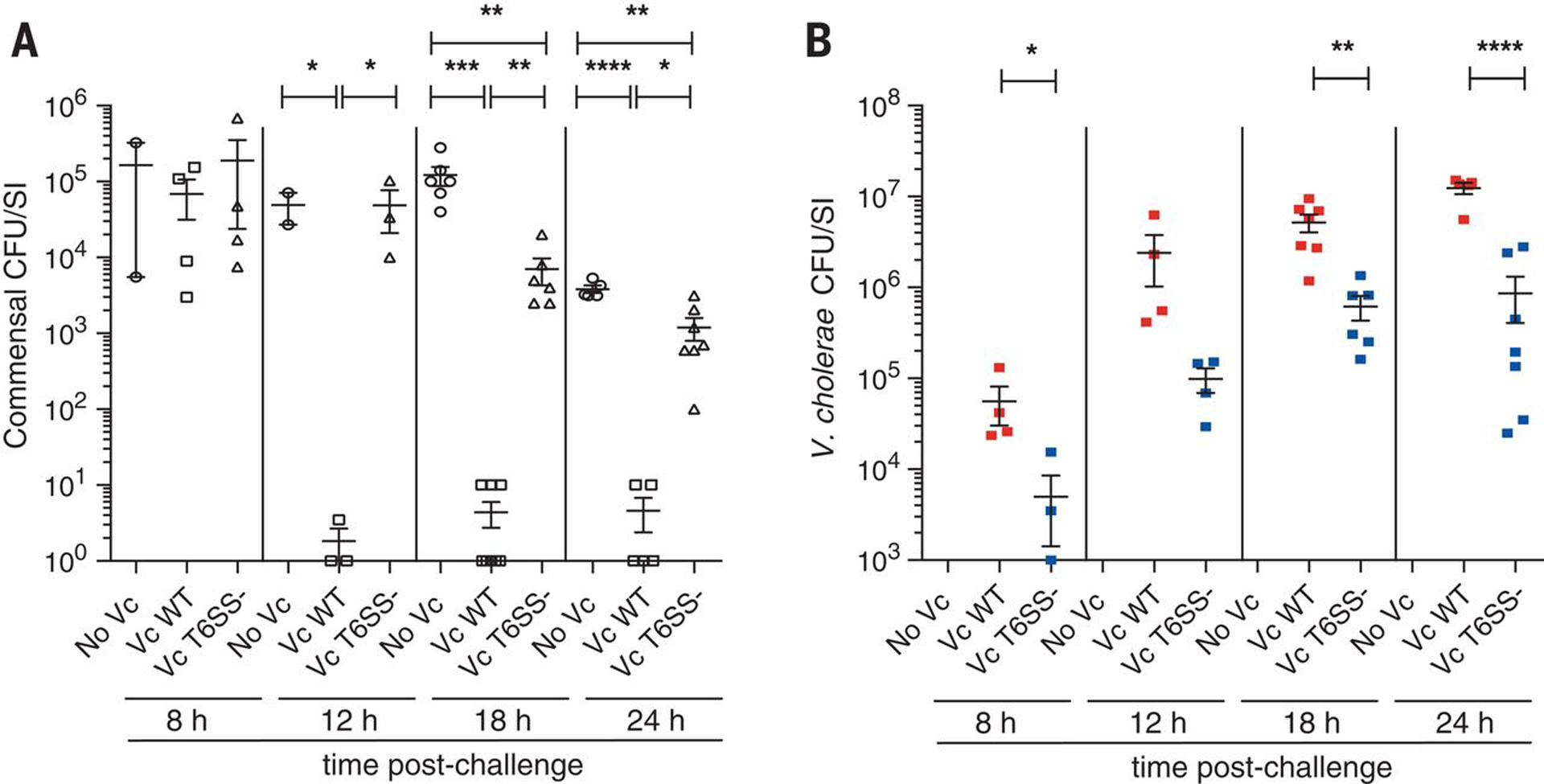
E. coli (A) and V. cholerae (B) CFU counts per SI homogenate collected from mice (n = 2–8) colonized first with commensal E. coli strains and subsequently challenged with 105 CFU of V. cholerae C6706 WT, T6SS-, or No Vc (buffer control). Every data point represents one mouse and error bars represent mean ± SEM of recovered CFUs. Statistical significance is shown based on an unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001).
Surprisingly, we found that V. cholerae C6706 exhibited enhanced colonization in in mice pre-colonized with E. coli commensals compared to its T6SS-defective mutant. At 8 hours post-challenge, V. cholerae WT showed an approximately 10-fold greater bacterial load than the T6SS mutant (Fig 1B. 6 × 104 vs. 5× 103 CFU/mouse, P < 0.05); and this ratio increased to approximately 15-fold 24 hours post-infection (1.2 × 107 vs. 8.6 × 105 CFU/mouse, Fig 1B, P < 0.001). To determine if T6SS was allowing colonization of V. cholerae strains that could not express a functional T6SS, we simultaneously introduced the WT strain (C6706) and an isogenic vipA T6SS mutant into mice pre-colonized with E. coli commensal strains. We observed no significant difference between WT and T6SS mutant colonization levels in these mice (Figure S2. competition index ~1.0), indicating that the WT strain could stimulate the colonization of the T6SS mutant presumably by deploying its functional T6SS to eliminate the E. coli commensals that were antagonizing the vipA mutant.
T6SS-dependent killing of E. coli commensals also caused changes in the transcription of pro-inflammatory factors known to be induced in response to V. cholerae infection (10). At 6 hours post-infection, we found that both IL6 and KC (IL6: interleukin 6, KC: chemokine (C-X-C motif) ligand 1) levels were 3.3 fold (10.7 ± 1.8 vs. 3.2 ± 0.7, P < 0.05) and 5.1 fold (10.8 ± 2.4 vs. 2.2 ± 0.3, P < 0.05) higher in intestinal extracts derived from mice infected with WT compared to the T6SS mutant strain (Fig 2A and B). These results indicate that during the early stage of infection, the induction of innate immune factors IL6 and KC is dependent on V. cholerae (C6706) possessing an active T6SS. If this early T6SS-dependent IL6/KC cytokine response depends on ablation of gut commensals, we reasoned that antibiotic treatment might suppress this response. We therefore eliminated much of the mouse gut microbiota by antibiotic treatment for two days, and tested whether mice challenged with either V. cholerae WT (C6706) or an isogenic T6SS mutant elicited detectable differences in cytokine expression levels. In the absence of streptomycin-sensitive gut commensals, no significant differences in IL6 or KC expression levels were observed between mice infected with V. cholerae WT and the T6SS mutant strain at 6 hours (1.5- and 1.0- fold, respectively) (Fig 2C). Thus, T6SS-mediated killing of commensal organisms correlates with the up-regulation of IL6 and KC during early stages of infection and this cytokine response is suppressed by antibiotic treatment.
Figure 2. V. cholerae WT induced a more acute host innate immune response compared to the T6SS mutant during early stages of infection.
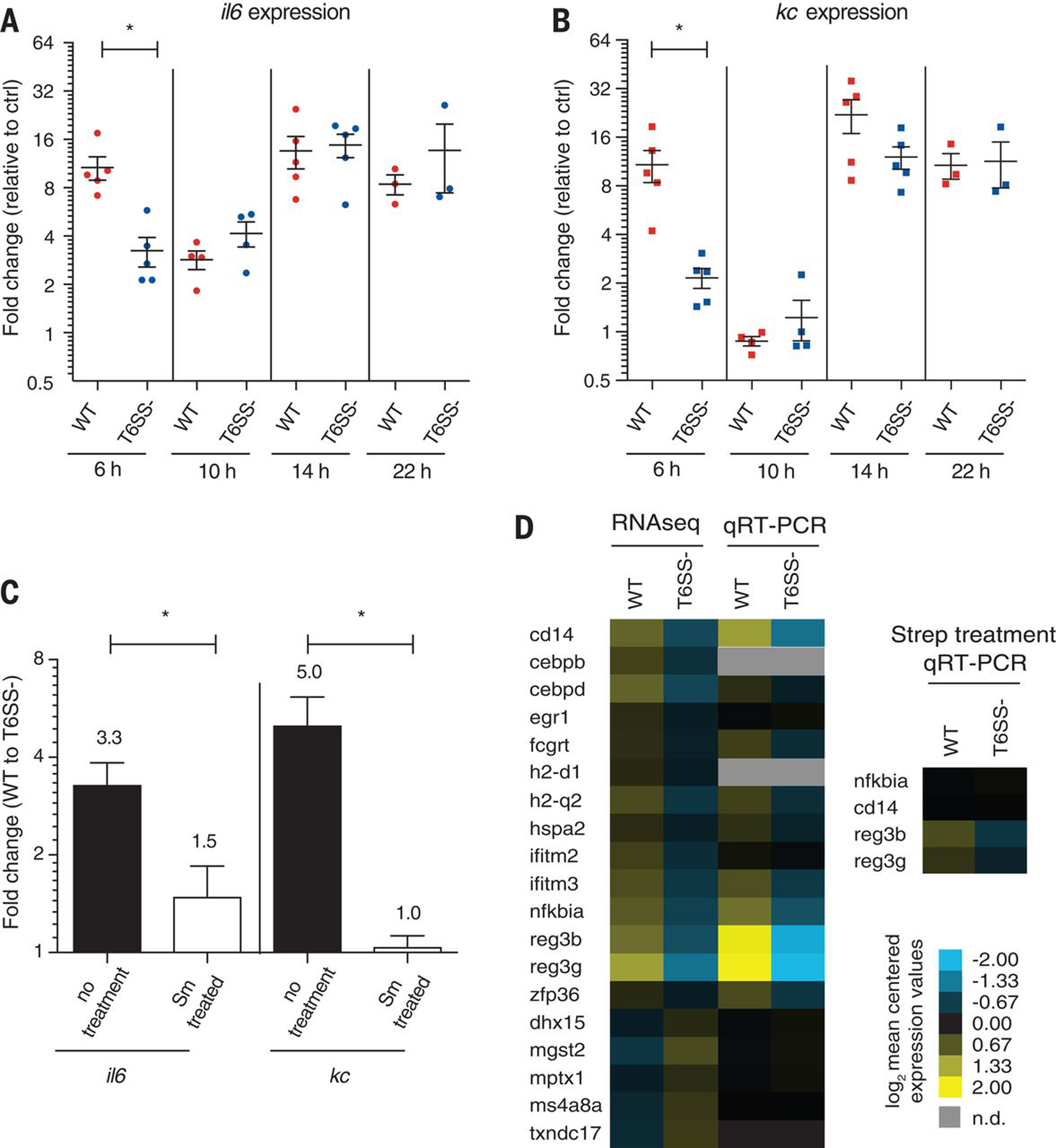
Transcript levels of inflammation markers IL6 (A) and KC (B) were measured by qRT-PCR from WT or T6SS- infected mice (n = 3–5) and are expressed as fold change with respect to No Vc (buffer) control. (C) IL6 and KC transcript levels were measured in streptomycin (Sm) treated or untreated mice. Error bars represent mean ± SEM of fold changes. Statistical significance is shown based on an unpaired t test (*P < 0.05). (D) Heatmap of host innate immune response transcripts levels measured by RNASeq and confirmed by qRT-PCR in WT or T6SS- infected mice (each group n = 5).
To gain a more comprehensive understanding of the T6SS-dependent modulation of the host immune response during early stages of infection, we harvested the small intestine of mice at 6 hours post V. cholerae WT (C6706) or T6SS mutant infection to extract host RNA for RNA-Seq. We found 135 differentially expressed host genes; 14 of the 19 genes involved in host immune function were up-regulated in WT compared to T6SS mutant infected mice (Table S2), including several known or predicted targets of the NF-κB pathway (cd14, nf-kbia, fcgrt, erg1, and cebpd) and regenerating islet-derived protein 3 gamma and beta (reg3γ and reg3β), which were among the most highly induced genes (16.8 and 13.1-fold, respectively) (Fig 2D, Table S3). Thus, WT V. cholerae triggered a greater innate immune transcriptional response than the T6SS mutant. We wondered if innate immune response observed in these experiments were driven by release of Microbe-Associated Molecular Patterns (MAMPs), bacterial molecules recognized by the host innate immune system (11). Consistent with this hypothesis, we found that V. cholerae T6SS-dependent killing of E. coli releases MAMPs in vitro (Figure S3A and S3B).
To test whether disease symptoms might be modulated by the T6SS-mediated killing of commensal species, mice were gavaged with 108 CFU of V. cholerae WT (C6706), its isogenic vipA T6SS mutant, or buffer, and tested for fluid accumulation (FA ratio, indicative of diarrhea) after 22 hours. The FA ratio was highest for the WT challenged group (0.0875 ± 0.0025), and was significantly lower (0.0749 ± 0.0026) for the T6SS mutant group (Fig 3A, P < 0.01). We also monitored the bacterial load in the SI and colon of the infected mice. In both organs, WT exhibited 3–9 fold higher CFU than the T6SS mutant (Fig 3B, P < 0.01). Furthermore, elimination of host gut commensals with antibiotic treatment abolished the observed T6SS-dependent enhancement of the FA ratio and bacterial load for animals infected with WT versus the T6SS mutant were (Fig 3C and D).
Figure 3. V. cholerae T6SS enhances the development of diarrhea disease symptoms.
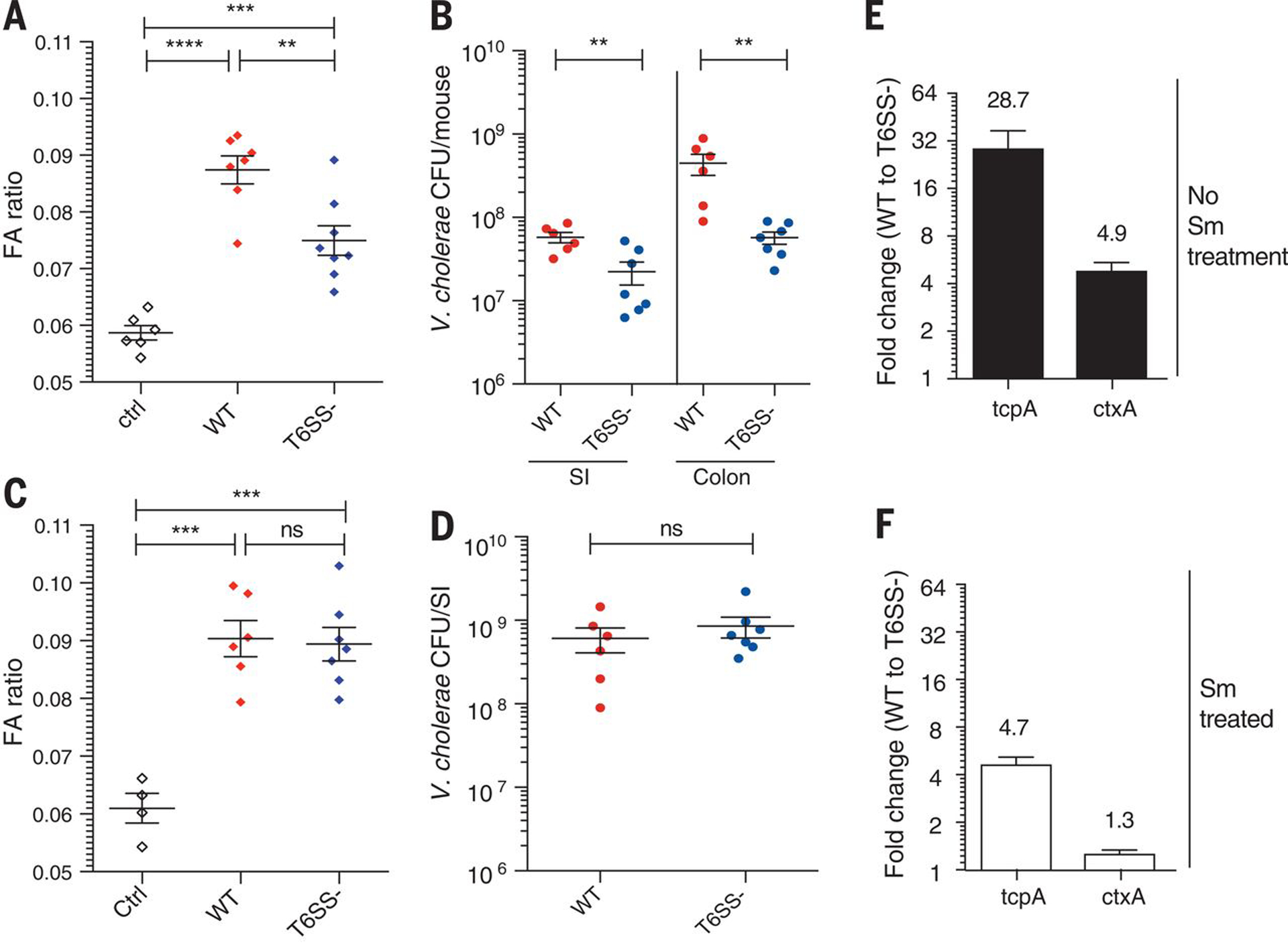
Fluid accumulation (FA) ratio (A) and V. cholerae CFU count per SI (B) of either WT or T6SS- treated mice. FA (C) ratio and V. cholerae CFU (D) measured in mice pre-treated with streptomycin (Sm). tcpA and ctxA transcript levels were measured in no streptomycin (Sm) mice (E) or streptomycin (Sm) treated (F) at 6 hours post infection. Statistical significance is shown based on an unpaired t test (NS, not significant; **P < 0.01; ***P < 0.005; ****P < 0.001).
The V. cholerae ToxT regulatory cascade is known to enhance diarrheal responses and bacterial loads in vivo through its control of the ctx and tcp virulence operons (12–15). Thus we asked whether T6SS-mediated killing of commensal organisms could release a commensal-derived signal that activates the ToxT regulatory cascade. We found that tcpA and ctxA transcript levels in bacteria recovered from infected mice were much higher in WT compared to its T6SS mutant during early stages of infection (Fig S5). This difference significantly dropped when mice where pre-treated with streptomycin (Fig 3E and 3F) indicating that T6SS-dependent killing of streptomycin-sensitive commensal bacteria can activate V. cholerae virulence gene expression during early stages of infection. Because control experiments showed that T6SS-mediated killing E. coli on laboratory media did activate tcpA or ctxA transcription in vitro (Fig. S4) we conclude that the host is driving these T6SS-dependent changes in virulence gene expression.
Genome analysis has recently revealed that variant strains of the 7th pandemic El Tor V. cholerae clade are now responsible for the vast majority of cholera cases in South Asia, Africa, and Haiti (12). Because these variant strains cause more severe diarrhea in cholera victims, we wondered whether they showed higher levels of T6SS expression. Transcriptome analysis showed that variant strains H1 (isolated early in the Haitian cholera epidemic) and MDC126 (isolated in Bangladesh in 2008 (12)) show 4- to 24-fold (average of 6-fold) greater T6SS gene transcript levels than two earlier 7th pandemic strains (N16961 isolated in Bangladesh in 1971 (13) and C6706 isolated in Peru in 1991) (Fig. 4 and Auxiliary File S1). Variant strains also displayed more T6SS-dependent killing activity in vitro (Fig. S6). Thus, the enhanced T6SS expression in V. cholerae variant strains may improve their fitness through T6SS-mediated killing of commensal Gram-negative microbiota or even enteric pathogens, such as pathogenic E. coli that share an upper intestinal nitch with V. cholerae and a similar epidemiological distribution (14).
Figure 4. Constitutive transcriptional up-regulation of T6SS genes in recent El Tor variant strains.
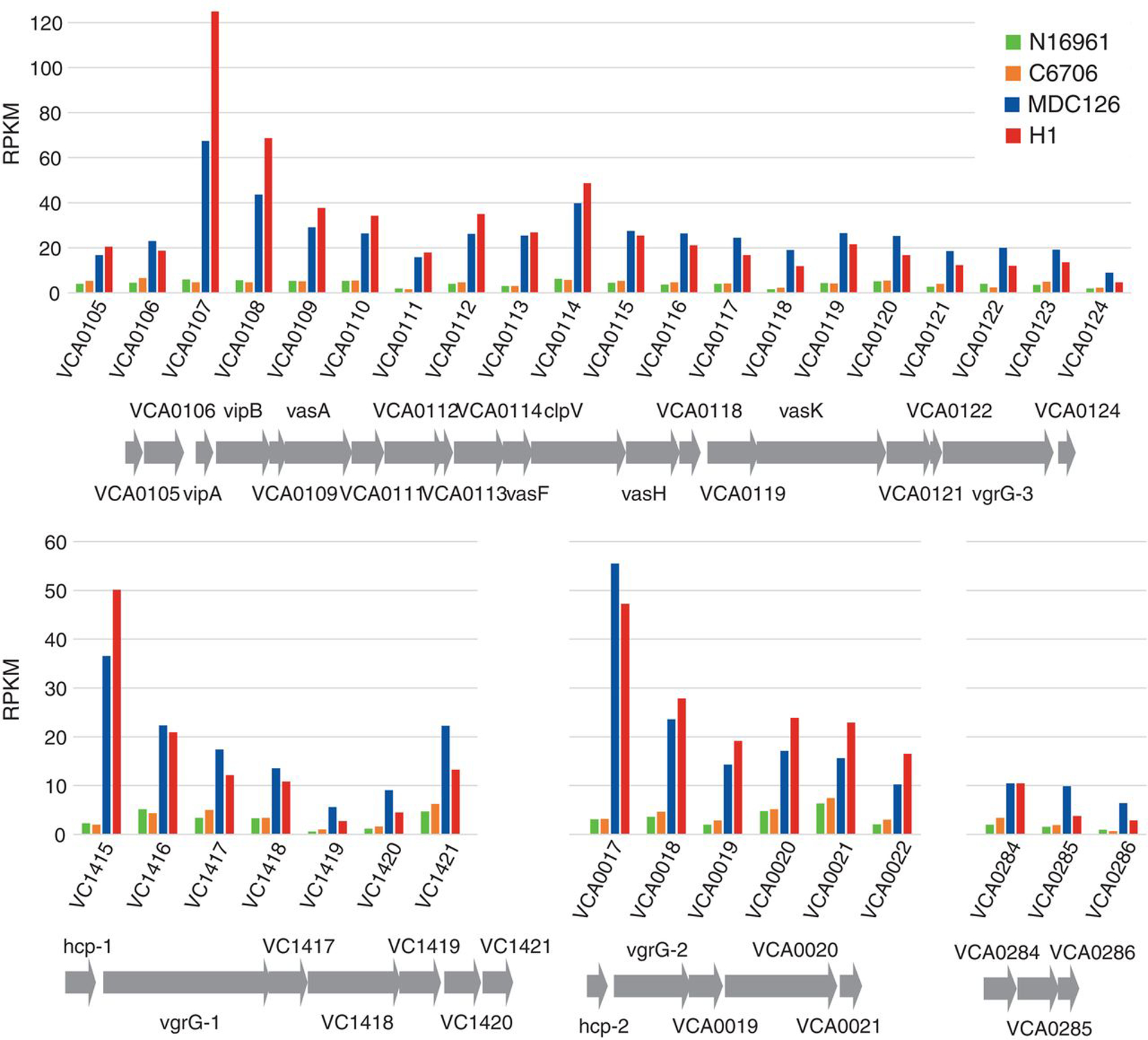
The transcriptomes of past seventh pandemic El Tor strains N16961 (1971) and C6706 (1991) are compared to those of recent variant strains MDC126 (2008) and H1 (2010) within T6SS-related operons in both chromosomes I and II. The relative expression of T6SS core/accessory, hcp, vgrG and effector/immunity genes are normalized as RPKM units
T6SS-mediated antagonistic behavior has been previously observed between enteric pathogens such as Salmonella typhimurium and Shigella sonnei and gut commensal organisms (15, 16). Although secreted molecules such as autoinducers have been shown to activate or repress the expression of T6SS and virulence factors (17–19) it is unclear whether microbial antagonism affects inter-species communication in vivo and pathogen fitness. Our results suggest that microbial antagonism may also change the interaction of an enteric pathogen with its host through altering its expression of virulence determinants as well as driving host innate immune responses. In sum, our data support a working model (Fig. 7S) that provides a framework for how V. cholerae might use an antibacterial mechanism to clear its target niche of inhibitory competitors and simultaneously enhance disease symptom-associated transmission.
Supplementary Material
Acknowledgements:
We thank Dr. Michaela Gack and Jessica Chiang for assistance with luciferase assays, and for a kind gift of reporter. We thank all Mekalanos lab members for helpful comments. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-01845 to J.J.M.. The authors declare no competing financial interests. All authors helped design and analyze experiments; W.Z., F.C. and W.R. performed experiments, and W.Z., F.C., W.R. and J.J.M. wrote the paper. All data generated or analyzed during this study are included in this published article (and its supplementary materials).
Footnotes
Supplementary Materials:
Materials and Methods
Fig S1-S7
Table S1-S5
Reference (20–25)
Auxiliary File S1. Transcriptome data of Vibrio cholerae strains
References and Notes
- 1.Mutreja A et al. , Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB, Cholera. Lancet 379, 2466–2476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pukatzki S et al. , Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103, 1528–1533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ, Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S, The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107, 19520–19524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, Waldor MK, Mekalanos JJ, Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmann V et al. , Bile Salts Modulate the Mucin-Activated Type VI Secretion System of Pandemic Vibrio cholerae. PLoS Negl Trop Dis 9, e0004031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandlik A et al. , RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basler M, Ho BT, Mekalanos JJ, Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AL, Patimalla B, Camilli A, Vibrio cholerae-induced inflammation in the neonatal mouse cholera model. Infect Immun 82, 2434–2447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman MA, Sundelin T, Nielsen JT, Erbs G, MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4, 139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin CS et al. , The origin of the Haitian cholera outbreak strain. N Engl J Med 364, 33–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidelberg JF et al. , DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris AM et al. , Shifting prevalence of major diarrheal pathogens in patients seeking hospital care during floods in 1998, 2004, and 2007 in Dhaka, Bangladesh. Am J Trop Med Hyg 79, 708–714 (2008). [PMC free article] [PubMed] [Google Scholar]
- 15.Sana TG et al. , Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proceedings of the National Academy of Sciences of the United States of America 113, E5044–E5051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ, Shigella sonnei Encodes a Functional T6SS Used for Interbacterial Competition and Niche Occupancy. Cell Host & Microbe 21, 769–776.e763 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Higgins DA et al. , The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Hsiao A et al. , Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borgeaud S, Metzger LC, Scrignari T, Blokesch M, The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Miller VL, Mekalanos JJ, A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170, 2575–2583 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon R, Priefer U, Puhler A, A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech 1, 784–791 (1983). [Google Scholar]
- 22.Angelichio MJ, Spector J, Waldor MK, Camilli A, Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun 67, 3733–3739 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baselski V, Briggs R, Parker C, Intestinal fluid accumulation induced by oral challenge with Vibrio cholerae or cholera toxin in infant mice. Infect Immun 15, 704–712 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Spandidos A, Wang H, Seed B, PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 40, D1144–1149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


