Abstract
Nucleotide sequence of complete genome of a new isolate (KAN-6) of tomato leaf curl New Delhi virus (ToLCNDV) from Kanpur, Uttar Pradesh, India was determined. Sequence analysis indicated that it shared maximum identity to ToLCNDV isolates from pumpkin and ashgourd. Infectious clones of isolate KAN-6 along with two other ToLCNDV isolates (MOD-21 & FAI-19) obtained from potato fields of Modipuram and Faizabad, India were produced and used in symptom expression studies in N. benthamiana and potato plants through agro-inoculation. These isolates produced different symptoms both in N. benthamiana and potato. Severe symptoms of yellow mottling, downward curling and stunted growth were observed in N. benthamiana plants inoculated with KAN-6. MOD-21-inoculated plants also showed downward curling, stunted growth, but yellow mottling was observed only in older leaves whereas FAI-19-inoculated plants produced only downward curling symptoms. In case of potato, typical symptoms of apical leaf curl disease were observed in cultivar Kufri Pukhraj inoculated with MOD-21 and KAN-6 that are similar to those produced by virus-infected plants in the field. However, MOD-21 produced more prominent yellow mosaic symptoms as compared to KAN-6. FAI-19 produced only restricted yellow spots in Kufri Pukhraj. Only mild symptoms appeared in KAN-6 and no symptoms were observed in MOD-21- and FAI-19-inoculated Kufri Bahar plants which is known to show lowest seed degeneration under field conditions. Analysis of genomic components indicated that these isolates had 94.8–94.9% and 87.9–97.3% identity among them in DNA A and DNA B, respectively. The results of the study indicate the association of ToLCNDV isolates of different symptomatology with apical leaf curl disease of potato. This is also a first experimental demonstration of Koch’s postulate for a begomovirus associated with apical leaf curl disease of potato.Author names: Please confirm if the author names (Swarup Kumar Chakrabarti) are presented accurately and in the correct sequence (given name, middle name/initial, family name).Yes. It is correct
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02752-5.
Keywords: Begomovirus, Infectious clones, Symptoms, ToLCNDV, Potato
Introduction
The potato is susceptible to many diseases, some of which are widespread and others are localized. There are more than forty tuber transmitted viruses reported on potato of which eight are reported to cause economic losses in India. Apical leaf curl disease (ALCD) caused by tomato leaf curl New Delhi virus (ToLCNDV) is one among them and it is one of the most important viral diseases of potato in India (Jeevalatha et al. 2017a; Kreuze et al. 2020; Lakra 2002). The virus spreads to potato due to a change in planting date of potato which coincided with an increased whitefly population. Symptoms appear as curling/crinkling of apical leaves with distinct mosaic symptoms due to primary infection (Garg et al. 2001; Usharani et al. 2004). In case of secondary infection, the entire plant show severe leaf curling and stunting symptoms (Sohrab et al. 2013). Initially, sporadic incidence of the disease was reported in 1996 at Hisar in Haryana (Lakra 2003), later severe infection was observed in western UP and other parts of northern India (Chandel et al. 2010; Saha et al. 2014). About 40–75% infection in the Indo-Gangetic Plains and up to 100% infections from Hissar (Haryana) were observed with heavy yield losses in susceptible varieties (Lakra 2002; Venkatasalam et al. 2005). This virus is also reported from other parts of the country (Jeevalatha et al. 2013, 2014, 2017a). Its incidence is on the rise, menacing the cultivation of potato across the country.
Usharani et al. (2004) confirmed that the virus causing apical leaf curl disease in potato is a strain of ToLCNDV belonging to the genus Begomovirus within the family Geminiviridae. After a decade, diversity of ToLCNDV infecting potato from different parts of the country was studied based on coat protein gene and complete genome (Jeevalatha et al. 2017a). ToLCNDV, a bipartite begomovirus, is the most predominant virus in northern India, infecting elite cultivars of tomato (Kushwaha et al. 2015; Sahu et al. 2010). It also infects other crops such as okra, cucurbitaceous crops, eggplant and papaya (Chigurupati et al. 2012; Ito et al. 2008; Pratap et al. 2011; Raj et al. 2008). ToLCNDV is reported to be transmitted to potato from cucurbit crops grown nearby potato fields (Sohrab et al. 2013) and may be easily transmitted between tomato, cucurbits and potato. The component of bipartite begomoviruses known as DNA-A encodes for all factors required for virus replication, overcoming host defences, insect transmission, and control of gene expression; while DNA-B component encodes factors required for inter- and intra-cellular movement in host plants (Rojas et al. 2005; Moriones et al. 2017). Recombination and pseudo-recombination are very frequent phenomena occurring between as well as within species and genera of geminiviruses and are significant contributors to begomovirus evolution (Polston and Anderson 1997; Kumar and Chakraborty 2018) led to gain the ability to infect new hosts (Lefeuvre and Moriones 2015).
Infectious clones are used for characterizing the function of viral components in virus infections (Babu et al. 2018; Basu et al. 2018) and vector transmission (Pan et al. 2020). They also provide highly reproducible and simple inoculation method which facilitates host–virus interaction studies and crop breeding. This is especially useful for viruses that cannot be transmitted to host plants by mechanical inoculation (Pasin et al. 2019). Infectious clones are widely used in begomovirus studies including tomato leaf curl viruses (Jailani et al. 2016; Jyothsna et al. 2013; Malik et al. 2011; Qadir et al. 2019; Shafiq et al. 2019; Vinoth Kumar et al. 2015). In the present study, we cloned and sequenced a new isolate (KAN-6) of ToLCNDV from potato field of Kanpur, Uttar Pradesh, India. Infectious clones of KAN-6 and two other isolates (MOD-21 and FAI-19) of ToLCNDV were developed and infectivity studies were carried out in Nicotiana benthamiana and potato. In this study, association of tomato leaf curl New Delhi virus, a bipartite begomovirus, with apical leaf curl disease of potato was experimentally demonstrated. Thus, proving Koch’s postulate for the first time for a begomovirus associated with apical leaf curl disease of potato.
Materials and methods
Cloning of viral components
The leaf samples showing typical symptoms of potato apical leaf curl disease such as leaf curling, mosaic and stunting were collected from potato field in Kanpur and stored at – 80 °C until further use. Total DNA was extracted from the leaves using GenElute™ Plant Genomic DNA Miniprep kit (Sigma-Aldrich, Missouri, USA) following the manufacturer’s instructions. The concentration and quality of DNA were checked using a Nanodrop 2000 spectrophotometer (Thermoscientific, Leon-Rot, Germany). The viral DNA was amplified using the TempliPhi™ amplification kit (GE Healthcare) as follows: 80 ng of total DNA (2 µl) was added to 5 µl of the sample buffer, heated to 95 °C for 3 min to denature the DNA, chilled on ice, and combined with 5 µl of reaction buffer plus 0.2 µl of enzyme mix. The reaction mixtures were incubated for 18 h at 30 °C, followed by inactivation of the enzyme at 65 °C for 10 min. The resulting concatamers were subjected to digestion with Xba I enzyme to release the unit-length viral genome. Restricted products were checked in 1% agarose gel stained with Ethidium bromide and visualized under UV light. An approximately 2.7 kb band was observed after digestion with XbaI enzyme. The digested RCA product fragments of ≈ 2.7 Kb were gel purified using the MinElute gel extraction kit (Qiagen, Hilden, Germany). The eluted fragment was cloned in pUC18 vector at Xba I site following standard protocols (Sambrook et al. 1989). Colony PCR analysis was performed with CP gene-specific primers (Forward primer-5′-AAAGTCATGTGTGTTAGTGATGTTACC-3′ and Reverse primer-5′-TAGAAATAGATCCGGATTTTCAAAGTA-3′) and positive colonies showing an expected size of 491 bp amplicon were selected for DNA A analysis. Remaining negative colonies were used for DNA B analysis. Restriction digestion of the plasmids isolated from the colonies released an expected size of approx. 2.7 kb fragment.
Sequencing and analysis
The recombinant clones were sequenced using M13 forward and reverse primers and four internal primers (designed at approximate 500 bp interval) in the automated DNA sequencer (ABI PRISM ™ 3500 Genetic Analyzer from Applied Biosystems). The nucleotide sequences were assembled using DNAbaser to form contigs, the peaks were checked and manually edited. The complete nucleotide sequences are available in GenBank under the accession numbers, KX951455-KX951456. Multiple sequence alignments were made and per cent identity was calculated using Bioedit software. Phylogenetic dendrograms were generated using Neighbor joining algorithm of MEGA software version 4.0.2 and only bootstrap values > 80% are shown. Isolates, MOD-21 (KC874508 and KC874500) and FAI-19 (KC874505 and KC874497) reported earlier by Jeevalatha et al. (2017a) were used along with KAN-6. Genome organization of the tomato leaf curl New Delhi virus isolates of the present study is depicted in Supplementary Fig. 1.
Construction of infectious clones
To perform symptom expression studies, infectious clones were produced and artificially inoculated to N. benthamiana and potato plants through agro-infection. To develop partial dimers of DNA A components of MOD-21 and FAI-19, initially 2253 bp fragment was released from pUC18/DNA A recombinant plasmid by XbaI and EcoRI and ligated with plant transformation vector pRI101 digested with the same enzymes to develop 0.8 mer. Later, the complete DNA A (2740 bp) was released from pUC18/DNA A recombinant plasmid by digestion with XbaI enzyme and inserted at XbaI site pRI101-0.8 mer to get 1.8 mer tandem repeat of DNA A (pRI101-MA1.8 and pRI101-FA1.8). To develop partial dimers of DNA A of KAN-6 isolate, initially 1342 bp fragment was released from pUC18/DNA A recombinant plasmid by SacI and EcoRI and ligated with plant transformation vector pRI101 digested with the same enzymes to develop 0.4 mer. Later, the complete DNA A (2739 bp) was released from pUC18/DNA A recombinant plasmid by digestion with SacI enzyme and inserted at SacI site pRI101-0.4 mer to get 1.4 mer tandem repeat of DNA A (pRI101-KA1.4). The partial dimers of DNA B were developed by cloning ≈ 1800 bp fragment of DNA B released from pUC18-DNA B recombinant plasmid using XbaI and EcoRI enzymes and cloning in pRI101 to generate 0.6-mer of DNA B. Then, the full-length DNA B was released from pUC18-DNA B using XbaI enzyme and cloned at XbaI site of pRI101-0.6 mer to develop 1.6-mer tandem repeat of DNA B (pRI101-MB1.6, pRI101-FB1.6 and pRI101-KB1.6).
Agro-inoculation studies in N. benthamiana
The recombinant plasmids (MA, MB, FA, FB, KA and KB) were mobilized into Agrobacterium tumefaciens strain EHA105 through freeze–thaw method. Young seedlings (3–4 leaf stage) of Nicotiana benthamiana were inoculated with infectious clones by agro-infiltration method. Empty binary vector, pRI101 in A. tumefaciens was used for mock inoculation on control plants. The A. tumefaciens cultures were incubated at 28 °C and 200 rpm for 48 h (OD600 = 1) in Yeast Extract Mannitol (YEM) medium (pH 6.8) containing kanamycin (50 μg/ml) and rifampicin (20 μg/ml). Agrobacterium cells were harvested, resuspended in induction buffer [10 mM MES, pH 5.5, along with 200 µl of acetosyringone (ACS)] and incubated at 50 rpm for 3 h at room temperature. Then, the Agrobacterium cells were again harvested, resuspended in infiltration buffer (5 mM MES, pH 5.5) and used for agro-infiltration on N. benthamiana plants (Senthil Kumar and Mysore 2014). Three-to-four leaf stage plants were used and five plants were inoculated for each culture. The inoculated plants were maintained under 16/8 h light/dark periods, 18,000 lx, 28–30 °C, and 85% relative humidity for 3–4 weeks in the controlled growth chamber until they were scored for symptoms and analyzed for viral DNA (Kanakala et al. 2013). The experiment was repeated thrice for confirmation.
Agro-inoculation studies in potato
The potato cultivars, Kufri Pukhraj (susceptible) and Kufri Bahar (resistant) were agro-inoculated by tuber sprout method followed by stem injection. Germinating seed tubers were inoculated with Agrobacterium cells (mixture of DNA A and DNA B; 1:1 ratio) in a rotary shaker for 4 h after pricking around the sprouts and planted in sterile pot mixture. Then again 15 days after planting, the grown plants were inoculated with Agrobacterium cells through stem injection (Supplementary Fig. 2). Four replicates were maintained for each treatment and the experiment was repeated thrice. The inoculated plants were maintained under controlled condition along with control plants and observed periodically for symptom expression.
Viral DNA quantification
Viral DNA was quantified in agro-inoculated plants using qPCR assay with coat protein gene-specific primers. Total DNA was extracted from systemically infected N. benthamiana plant leaves collected at 10, 15, 20, and 25 days after inoculation (DAI) and potato plants at 30 days after planting (DAP)/30 days after first inoculation using GenElute™ Plant Genomic DNA Miniprep kit (Sigma-Aldrich, Missouri, USA) following the manufacturer’s instructions and the concentration and quality of DNA was checked using a Nanodrop 2000 spectrophotometer (Thermoscientific, Leon-Rot, Germany).The qPCR assays were carried out in an ABI PRISM HT7900 (Applied Biosystems) with the following thermal cycles: 50 °C for 2 min, 95 °C for 10 min; 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Each 15 µl reaction mixture contained 10 µl of Power SYBR Green PCR Master mix, 1.0 µl each of 5 µM forward and reverse primers, 1 µl of cDNA templates and 2 µl of nuclease free water. The primers used are PALCV-CPF: 5′-ACCGTCGTCCTACAGGATCTC-3′ and PALCV-CPR: 5′-GCTCGGTTCATTGTCAAACATGT-3′. The qPCR reactions were carried out in triplicates and average values were considered for quantification.
Results and discussion
Cloning and sequence analysis
The DNA A of isolate KAN-6 was 2739 bp long with a maximum of 96.0–96.8% identities to ToLCNDV isolate from pumpkin (JN129254) and ash gourd (JN208136). It shared 94.8% identity to MOD21 and FAI-19 and 94.9–95.2% identities to other ToLCNDV isolates from potato (Supplementary Table 1). The DNA B of KAN-6 isolate was 2693 bp long and had a maximum of 93.4% identity to DNA B of ToLCNDV isolates from pumpkin (AM286435) and ash gourd (JN208137). It shared 87.5–88.0% identity to DNA B of ToLCNDV isolates from potato (Supplementary Table 2). The per cent identities of these isolates at DNA A level to ToLCNDV is 93.3–96.8% which are above the threshold cut-off value 91% for species demarcation for begomoviruses based on DNA A (Brown et al. 2015). These results confirm that the begomovirus that infect potato is an isolate of ToLCNDV. It supports the report of Usharani et al. (2004) who initially found that the begomovirus infecting potato in India as ToLCNDV. These isolates (MOD-21, FAI-19 and KAN-6) showed about 5% variation in DNA A components. MOD-21 had more identity to the already reported ToLCNDV isolates from potato. While FAI-19 and KAN-6 shared more identity to ToLCNDV isolates from chilli and pumpkin/ash gourd, respectively.
In a phylogenetic analysis based on DNA A components, MOD-21 was grouped with already reported potato isolates of ToLCNDV from Meerut, Palampur and Happur. FAI-19 was grouped with an isolate of ToLCNDV from chilli in Tumkur. While the isolate, KAN-6 was grouped with ToLCNDV isolates from pumpkin and ashgourd. However, all the three isolates were found in the same cluster in which the ToLCNDV isolates from different crops were observed (Fig. 1a). Phylogenetic analysis using DNA B components showed that both MOD-21 and FAI-19 isolates were grouped with the ToLCNDV isolates reported earlier from potato, whereas the isolate KAN-6 was found to form another cluster with ashgourd and pumpkin isolates with 100% bootstrap support (Fig. 1b).
Fig. 1.
Phylogenetic dendrogram generated based on DNA A (a) and DNA B (b) components of ToLCNDV isolates from potato and other begomoviruses (isolates of this study are marked with violet color dots)
Symptoms expression in N. benthamiana
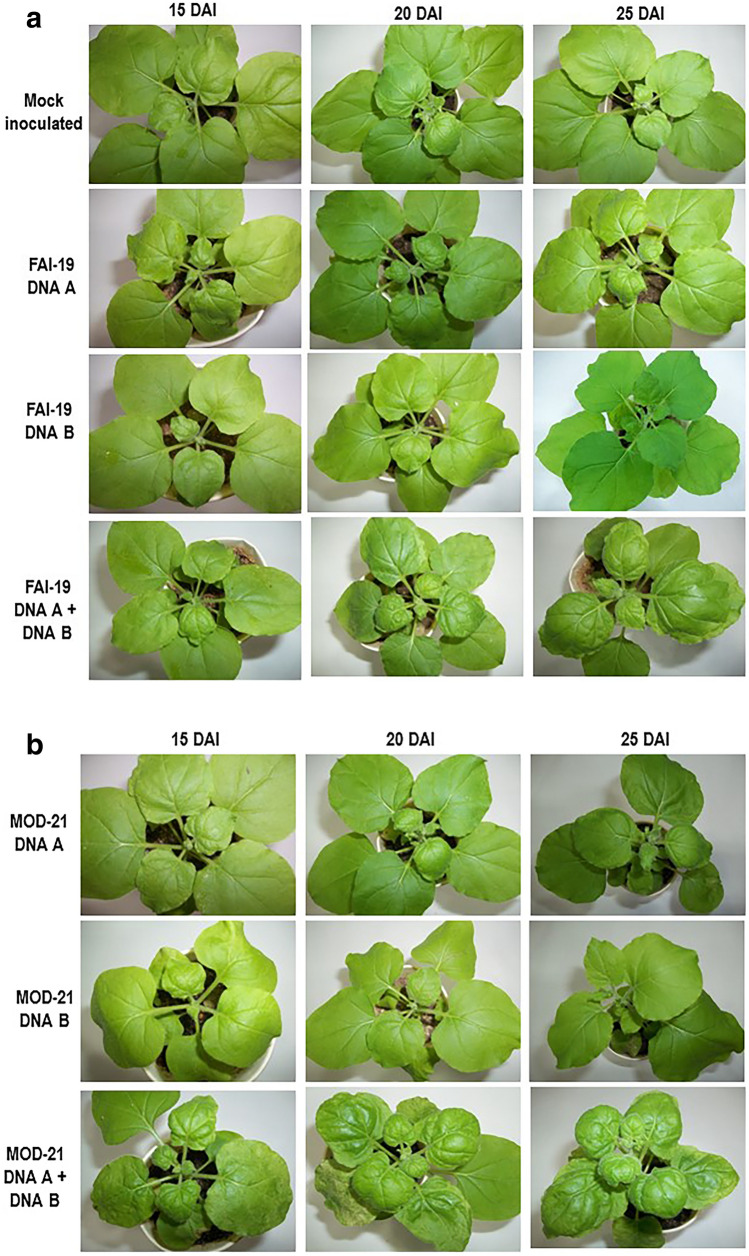
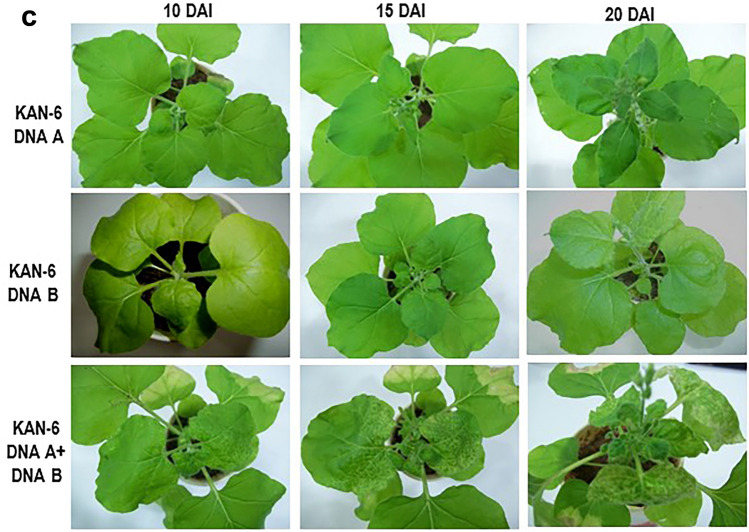
Among the three infectious clones, symptom expression started earlier in KAKB (KAN-6)-inoculated N. benthamiana plants (at 10 DAI). Symptoms appeared at 15 DAI in MAMB (MOD-21) and FAFB (FAI-19)-inoculated plants (Supplementary Table 3). The symptoms progressed faster in KAKB (KAN-6)-inoculated plants with prominent yellow mottling, small young leaves and severe downward curling of leaves. MAMB (MOD-21)-inoculated plants showed typical downward curling, stunted growth and yellow mottling in older leaves. FAFB (FAI-19) produced mild symptoms, i.e. merely downward curling of leaves was observed (Fig. 2a–c). In general, agro-inoculation of DNA A alone resulted in downward curling of leaves in N. benthamiana. This indicates DNA A can maintain itself and cause infection. However, symptoms were milder when compared to those plants co-inoculated with the DNA B component. In bipartite begomoviruses, it is generally the DNA B genes (movement protein) that lead to development of symptoms (Hou et al. 2000; Hussain et al. 2005; Kleinow et al., 2020) and hence, plants co-inoculated with DNA B produced severe symptoms.
Fig. 2.
Symptom expression in N. benthamiana plants inoculated with infectious clones as labeled at different time intervals. a FAI-19, b MOD-21, c KAN-6
Symptoms expression in potato
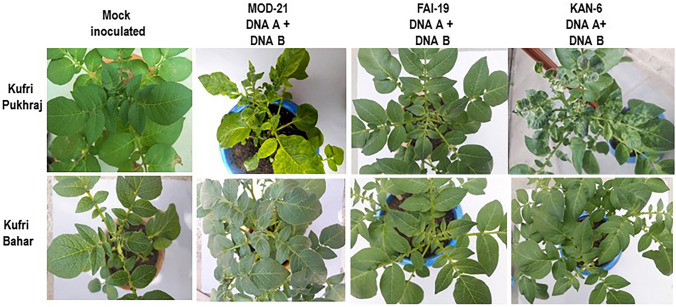
Symptoms appeared at 20 DAP or first inoculation in Kufri Pukhraj which is highly susceptible to the virus. Kufri Pukhraj plants showed typical symptoms of apical leaf curl disease, i.e. curling of leaves and mosaic symptoms with stunted growth that are similar to those occurred in virus-infected plants in the field (Fig. 3) upon inoculation with MOD-21 and KAN-6, whereas FAI-19 produced only restricted yellow spots. However, MOD-21 produced more prominent yellow mosaic symptoms which had bright yellow patches as compared to KAN-6. No symptoms were observed in Kufri Bahar after inoculation with MOD-21 and FAI-19 which is known to show lowest seed degeneration due to ToLCNDV (Lakra 2003) and no/mild symptoms of apical leaf curl disease in both field and under artificial inoculation (graft inoculation) in glass house (Jeevalatha et al. 2012, 2017b; Lakra 2003). Only mild yellow restricted spots appeared in KAN-6-inoculated Kufri Bahar plants at 20 DAP. However, later, mild symptoms appeared at 50 DAP in Kufri Bahar also. The presence or absence DNA A and DNA B components in young non-inoculated leaves of both tobacco and potato were confirmed by PCR analysis using DNA A- and DNA B-specific primers (data not-shown).
Fig. 3.
Symptom expression in potato cultivars, Kufri Pukhraj and Kufri Bahar inoculated with infectious clones (photographed 30 DAP/ 30 days after first inoculation)
So far, reports are available on complete genome sequencing of bipartite begomovirus, ToLCNDV from potato (Jeevalatha et al. 2017a; Usharani et al. 2004), but Koch’s postulate is not yet fulfilled. For the first time, in this study, we produced infectious clones of ToLCNDV isolates and studied symptom expression in potato to satisfy Koch’s postulate. Initially we tried with agro-infiltration in leaves and then stem injection. But these methods failed to produce symptoms in potato. Therefore, another method involving germinating tuber was used and combined with stem injection (Supplementary Fig. 2) which led to infection with infectivity of about 66.66–83.33%. Earlier reports also suggest that two to three doses of inoculation are required to agro-infect potato (Salaria et al. 2020).
Viral DNA quantification
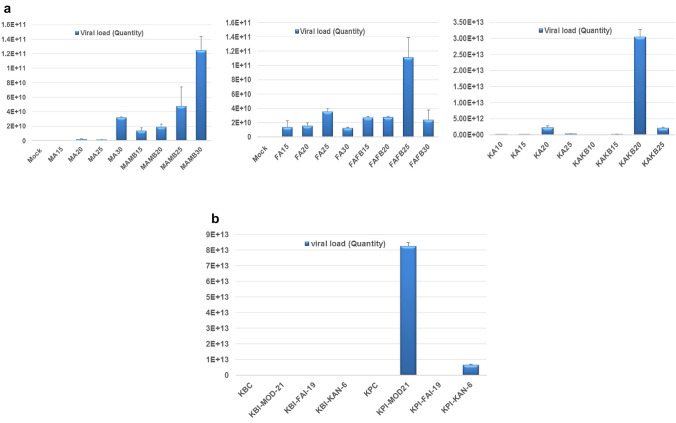
Differences in the levels of viral DNA were observed among the infectious clones. In N. benthamiana, an increase in viral DNA concentration with the progression of time was noticed in case of MOD-21. But in case of FAI-19 and KAN-6, the viral DNA concentration increased initially and then reduced at later stages (30 DAI in FAI-19 and 25 DAI in KAN-6). Viral DNA concentration was maximum at 25 DAI in FAI-19 and 20 DAI in KAN-6. The concentration of viral DNA was higher when both DNA A and DNA B constructs were co-inoculated and it was observed in all the three infectious clones (Fig. 4a). In potato, the viral DNA was very low (2.09 × 102 to 2.8 × 103 copies in 20 ng of total DNA) in Kufri Bahar inoculated with all three isolates (Fig. 4b). Maximum viral DNA was observed in Kufri Pukhraj inoculated with MOD-21 (8.25 × 1013) followed by KAN-6 (6.64 × 1012) and FAI-19 (4.8 × 106). The results of the study supports the earlier findings that Kufri Bahar produce no/mild symptom with very low virus load (Jeevalatha et al. 2017b).
Fig. 4.
Quantitative PCR analysis of viral DNA in agro-infiltrated N. benthamiana at 15, 20, 25 and 30 DAI (a); MA-MOD-21 DNA A, MB-MOD-21 DNA B, MAMB- MOD-21 DNA A + DNA B, FA- FAI-19 DNA A, FB-FAI-19 DNA B, FAFB- FAI19 DNA A + DNA B, KA-KAN-6 DNA A, KB-KAN-6 DNA B, KAKB-KAN-6 DNA A + DNA B and potato plants (b); Kufri Bahar and Kufri Pukhraj plants inoculated with both DNA A and DNA B components of MOD-21, FAI-19 and KAN-6 (quantified at 30 DAP or 30 days after first inoculation). The error bar represents the standard deviation of the three replicates
Conclusion
Infectious clones of three ToLCNDV isolates (KAN-6, MOD-21 and FAI-19) produced dissimilar symptoms in both tobacco and potato, indicating the association of ToLCNDV isolates of varying symptomatology with apical leaf curl disease of potato. The infectious clones of this study could serve as study material to explore the changes in the genomic components that led to infection in potato and also the agro-inoculation method standardized in this study will be useful in screening of hybrids and germplasms for ToLCNDV resistance.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. Genome organization of ToLCNDV isolates of the present study (JPG 91 KB)
Supplementary Fig. 2. Steps involved in agro-inoculation of potato with infectious clones (JPG 143 KB)
Acknowledgements
This study was supported by Indian Council of Agricultural Research, New Delhi, India.
Author contributions
AJ—conceptualization, AJ and PK—sample processing, cloning and sequencing of KAN-6 isolate, AJ—development of infectious clones AJ and AK—agro-inoculation experiments in N. benthamiana, VG, SS, AJ and RK—agro-inoculation experiments in potato, AJ, PK, SS and RK—qPCR analysis, AJ, SS, RK and SKC helped in preparing and editing the manuscript.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the author.
Footnotes
Contributor Information
Arjunan Jeevalatha, Email: A.Jeevalatha@icar.gov.in.
G. Vanishree, Email: Vanishree.G@icar.gov.in
Sundaresha Siddappa, Email: Sundaresha.S@icar.gov.in.
Ravinder Kumar, Email: Ravinder.Kumar2@icar.gov.in.
Priyanka Kaundal, Email: priyanka_kaundal@yahoo.co.in.
Ashwani Kumar, Email: aryan.verma07@gmail.com.
Swarup Kumar Chakrabarti, Email: skc_cpri@yahoo.co.in.
References
- Babu KSD, Guria A, Karanthamalai J, Srikakulam N, Kumari K, Sharma P, Chandran SA, Barnabas AD, Tennyson J, Pandi G. DNA methylation suppression by bhendi yellow vein mosaic virus. Epigenomes. 2018;2:7. [Google Scholar]
- Basu S, Kushwaha N, Singh AK, Sahu PP, Vinoth Kumar R, Chakraborty S. Dynamics of a geminivirus-encoded pre-coat protein and host RNA-dependent RNA polymerase 1 in regulating symptom recovery in tobacco. J Exp Bot. 2018;69:2085–2102. doi: 10.1093/jxb/ery043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JCF, Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, Idris A, Malathi VG, Marin DP, Rivera-Bustamante R, Ueda S, Varsani A. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol. 2015;160:1593–1619. doi: 10.1007/s00705-015-2398-y. [DOI] [PubMed] [Google Scholar]
- Chandel RS, Banyal DK, Singh BP, Malik K, Lakra BS. Integrated management of whitefly Bemisia tabaci (Gennadius) and potato apical leaf curl virus in India. Potato Res. 2010;53:129–139. [Google Scholar]
- Chigurupati P, Sambasiva Rao KRS, Jain RK, Mandal B. Tomato leaf curl New Delhi virus is associated with Pumpkin Leaf Curl: A New Disease in Northern India. Indian J Virol. 2012;23:42–45. doi: 10.1007/s13337-011-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg ID, Paul-Khurana SM, Kumar S, Lakra BS. Association of a geminivirus with potato apical leaf curl in India and its immuno-electron microscopic detection. J Indian Potato Assoc. 2001;28:227–232. [Google Scholar]
- Hou YM, Sanders R, Ursin VM, Gilbertson RL. Transgenic plants expressing geminivirus movement proteins: abnormal phenotypes and delayed infection by Tomato mottle virus in transgenic tomatoes expressing the Bean dwarf mosaic virus BV1 or BC1 proteins. Mol Plant Microbe Interact. 2000;13:297–308. doi: 10.1094/MPMI.2000.13.3.297. [DOI] [PubMed] [Google Scholar]
- Hussain M, Mansoor S, Iram S, Fatima AN, Zafar Y. The nuclear shuttle protein of tomato leaf curl New Delhi virus is a pathogenicity determinant. J Virol. 2005;79:4434–4439. doi: 10.1128/JVI.79.7.4434-4439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Sharma P, Kittipakorn K, Ikegami M. Complete nucleotide sequence of a new isolate of tomato leaf curl New Delhi virus infecting cucumber, bottle gourd and muskmelon in Thailand. Arch Virol. 2008;153:611–613. doi: 10.1007/s00705-007-0029-y. [DOI] [PubMed] [Google Scholar]
- Jailani AAK, Kumar A, Mandal B, Sivasudha T, Roy A. Agroinfection of tobacco by croton yellow vein mosaic virus and designing of a replicon vector for expression of foreign gene in plant. Virus Dis. 2016;27:277–286. doi: 10.1007/s13337-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeevalatha A, Tomar G, Shandil RK, Sharma NN, Venkatasalam EP, Sharma S, Baswaraj R, Kumar R, Sagar V, Chakrabarti SK, Singh BP. Screening methodology developed for apical leaf curl disease. CPRI News Lett. 2012;48:p1. [Google Scholar]
- Jeevalatha A, Kaundal P, Venkatasalam EP, Chakrabarti SK, Singh BP. Uniplex and duplex PCR detection of geminivirus associated with potato apical leaf curl disease in India. J Virol Methods. 2013;193:62–67. doi: 10.1016/j.jviromet.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Jeevalatha A, Singh BP, Kaundal P, Kumar R, Raigond B. RCA-PCR: a robust technique for the detection of tomato leaf curl New Delhi virus-potato at ultra low virus titre. Potato J. 2014;41:76–80. [Google Scholar]
- Jeevalatha A, Chakrabarti SK, Sharma S, Sagar V, Malik K, Raigond B, Singh BP. Diversity analysis of tomato leaf curl New Delhi virus—[potato], causing apical leaf curl disease of potato in India. Phytoparasitica. 2017;45:33–43. [Google Scholar]
- Jeevalatha A, Sundaresha S, Kumar A, Kaundal P, Guleria A, Sharma S, Nagesh M, Singh BP. An insight into differentially regulated genes in resistant and susceptible genotypes of potato in response to tomato leaf curl New Delhi virus-[potato] infection. Virus Res. 2017;232:22–33. doi: 10.1016/j.virusres.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Jyothsna P, Haq QM, Singh P, et al. Infection of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl Microbiol Biotechnol. 2013;97:5457–5471. doi: 10.1007/s00253-012-4685-9. [DOI] [PubMed] [Google Scholar]
- Kanakala S, Jyothsna P, Shukla R, Tiwari N, Veer BS, Swarnalatha P, Krishnareddy M, Malathi VG. Asymmetric synergism and heteroencapsidation between two bipartite begomoviruses, tomato leaf curl New Delhi virus and tomato leaf curl Palampur virus. Virus Res. 2013;174:126–136. doi: 10.1016/j.virusres.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Kleinow T, Happle A, Kober S, Linzmeier L, Rehm TM, Fritze J, Buchholz PCF, Kepp G, Jeske H, Wege C. Phosphorylations of the Abutilon mosaic virus movement protein affect its self-interaction, symptom development, viral DNA accumulation, and host range. Front Plant Sci. 2020;11:1155. doi: 10.3389/fpls.2020.01155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze JF, Souza-Dias JAC, Jeevalatha A, Figueira AR, Valkonen JPT, Jones RAC. Viral diseases in potato. In: Campos H, Ortiz O, editors. The potato crop. Cham: Springer; 2020. pp. 389–430. [Google Scholar]
- Kumar RV, Chakraborty S. Evolution and emergence of geminiviruses: reasons and consequences. In: Gaur RK, Khurana SMP, Dorokhov YL, editors. Plant viruses: diversity, interaction and management. Boca Raton: CRC Press; 2018. pp. 97–116. [Google Scholar]
- Kushwaha N, Singh AK, Basu S, Chakraborty S. Differential response of diverse solanaceous hosts to tomato leaf curl New Delhi virus infection indicates coordinated action of NBS-LRR and RNAi-mediated host defense. Arch Virol. 2015;160:1499–1509. doi: 10.1007/s00705-015-2399-x. [DOI] [PubMed] [Google Scholar]
- Lakra BS. Leaf curl: a threat to potato crop in Haryana. J Mycol Plant Pathol. 2002;32:367. [Google Scholar]
- Lakra BS. Potato apical leaf curl begomovirus-symptoms, appraisal of a scale and losses in potato crop. J Indian Potato Assoc. 2003;30:119–120. [Google Scholar]
- Lefeuvre P, Moriones E. Recombination as a motor of host switches and virus emergence: geminiviruses as case studies. Curr Opin Virol. 2015;10:14–19. doi: 10.1016/j.coviro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Malik AH, Briddon RW, Mansoor S. Infectious clones of Tomato leaf curl Palampur virus with a defective DNA B and their pseudo-recombination with Tomato leaf curl New Delhi virus. Virol J. 2011;8:173. doi: 10.1186/1743-422X-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriones E, Praveen S, Chakraborty S. Tomato leaf curl New Delhi virus: an emerging virus complex threatening vegetable and fiber crops. Viruses. 2017;9(10):264. doi: 10.3390/v9100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L-L, Chi Y, Liu C, Fan Y-Y, Liu S-S. Mutations in the coat protein of a begomovirus result in altered transmission by different species of whitefly vectors. Virus Evol. 2020;6(1):14. doi: 10.1093/ve/veaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin F, Menzel W, Daros J-A. Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnol J. 2019;17:1010–1026. doi: 10.1111/pbi.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polston JE, Anderson PL. The emergence of whitefly transmitted geminiviruses in tomato in the Western Hemisphere. Plant Dis. 1997;81:1358–1369. doi: 10.1094/PDIS.1997.81.12.1358. [DOI] [PubMed] [Google Scholar]
- Pratap D, Kashikar AR, Mukherjee SK. Molecular characterization and infectivity of a Tomato leaf curl New Delhi virus variant associated with newly emerging yellow mosaic disease of eggplant in India. Virol J. 2011;8:305. doi: 10.1186/1743-422X-8-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir R, Khan ZA, Monga D, Khan JA. Diversity and recombination analysis of Cotton leaf curl Multan virus: a highly emerging begomovirus in northern India. BMC Genomics. 2019;20:274. doi: 10.1186/s12864-019-5640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj SK, Snehi SK, Khan MS, Singh R, Khan AA. Molecular evidence for association of Tomato leaf curl New Delhi virus with leaf curl disease of papaya (Carica papaya L.) in India. Australas Plant Dis Notes. 2008;3:152–155. [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol. 2005;43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- Saha A, Saha B, Saha D. Molecular detection and partial characterization of a begomovirus causing leaf curl disease of potato in sub-Himalayan West Bengal, India. J Environ Biol. 2014;35:601–606. [PubMed] [Google Scholar]
- Sahu PP, Rai NK, Chakraborty S, Singh M, Chandrappa PH, Ramesh B, Chattopadhyay D, Prasad M. Tomato cultivar tolerant to tomato leaf curl New Delhi virus infection induces virus-specific short interfering RNA accumulation and defence-associated host gene expression. Mol Plant Pathol. 2010;11:531–544. doi: 10.1111/j.1364-3703.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaria N, Siddappa S, Thakur K, Tomar M, Goutam U, Sharma N, Sood S, Bhardwaj V, Singh B. Solanum tuberosum (CYCLING DOF FACTOR) CDF1.2 allele: a candidate gene for developing earliness in potato. S Afr J Bot. 2020;132:242–248. [Google Scholar]
- Sambrook J, Fritschi EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Senthil Kumar M, Mysore KS. Tobacco rattle virus based virus induced gene silencing in Nicotiana bethamiana. Nat Protoc. 2014;9:1549–1562. doi: 10.1038/nprot.2014.092. [DOI] [PubMed] [Google Scholar]
- Shafiq M, Ahmad M, Nisar A, et al. Molecular characterization and phylogenetic analysis of tomato leaf curl Palampur virus, a bipartite begomovirus, associated with Cucumis sativus L. in Pakistan. 3 Biotech. 2019;9:204. doi: 10.1007/s13205-019-1727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrab SS, Karim S, Varma A, Abuzenadah AM, Chaudhary AG, Mandal B. Role of sponge gourd in apical leaf curl disease of potato in Northern India. Phytoparasitica. 2013;41:403–410. [Google Scholar]
- Usharani KS, Surendranath B, Paul-Khurana SM, Garg ID, Malathi VG. Potato leaf curl—a new disease of potato in northern India caused by a strain of Tomato leaf curl New Delhi virus. Plant Pathol. 2004;53:235. [Google Scholar]
- Venkatasalam EP, Singh S, Gawande SJ, Malathi VG (2005) Detection of whitefly transmitted geminivirus associated with potato apical leaf curl by serological and molecular tools. In: Proceedings of the Annual Meeting of Indian Society of Plant Pathologists and Centenary Symposium on Plant Pathology, 7/8 April, Central Potato Research Institute, Shimla, p 18.
- Vinoth Kumar R, Singh AK, Singh AK, Yadav T, Basu S, Kushwaha N, Chattopadhyay B, Chakraborty S. Complexity of begomovirus and betasatellite populations associated with chilli leaf curl disease in India. J Gen Virol. 2015;96:3143–3158. doi: 10.1099/jgv.0.000254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Genome organization of ToLCNDV isolates of the present study (JPG 91 KB)
Supplementary Fig. 2. Steps involved in agro-inoculation of potato with infectious clones (JPG 143 KB)