Abstract
Pseudomonas aeruginosa is a biofilm-forming opportunistic pathogen which causes chronic infections in immunocompromised patients and leads to high mortality rate. It is identified as a common coinfecting pathogen in COVID-19 patients causing exacerbation of illness. In our hospital, P. aeruginosa is one of the top coinfecting bacteria identified among COVID-19 patients. We collected a strong biofilm-forming P. aeruginosa strain displaying small colony variant morphology from a severe COVID-19 patient. Genomic and transcriptomic sequencing analyses were performed with phenotypic validation to investigate its adaptation in SARS-CoV-2 infected environment. Genomic characterization predicted specific genomic islands highly associated with virulence, transcriptional regulation, and DNA restriction-modification systems. Epigenetic analysis revealed a specific N6-methyl adenine (m6A) methylating pattern including methylation of alginate, flagellar and quorum sensing associated genes. Differential gene expression analysis indicated that this isolate formed excessive biofilm by reducing flagellar formation (7.4 to 1,624.1 folds) and overproducing extracellular matrix components including CdrA (4.4 folds), alginate (5.2 to 29.1 folds) and Pel (4.8–5.5 folds). In summary, we demonstrated that P. aeuginosa clinical isolates with novel epigenetic markers could form excessive biofilm, which might enhance its antibiotic resistance and in vivo colonization in COVID-19 patients.
Keywords: Pseudomonas aeruginosa, COVID-19, biofilm, coinfection, DNA methylation
Introduction
Coronavirus Disease 2019 (COVID-19) is a fatal lung infection caused by the novel coronavirus named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) which has influenced millions of people globally since its onset. Its pathogenicity, epidemiology and treatments have been extensively studied (Chen et al., 2020; Li et al., 2020; Shen et al., 2020). It causes severe tissue damage of lungs and multiple organs such as the heart, the liver, and the kidneys (Wang D. et al., 2020; Wang T. et al., 2020). Higher mortality rate was observed from COVID-19 patients with older age and underlying diseases (Liu et al., 2020; Wang T. et al., 2020; Zhou et al., 2020). Such viral lung infection weakens host immunity and alters the composition and functions of respiratory microbiota, predisposing hosts to bacterial coinfections (Hanada et al., 2018). Bacterial coinfection was one of the major causes of death in the influenza pandemics last century (Hers et al., 1958; Bisno et al., 1971; Brundage and Shanks, 2008; Gill et al., 2010; Cilloniz et al., 2012). About 30% of cases was found to have bacterial coinfection in the 2009 H1N1 influenza pandemic even with antibiotic treatment (Gill et al., 2010; Hanada et al., 2018). Bacteria coinfecting with SARS-CoV-2 have also been reported by several retrospective studies based on cases from different geographical regions (Hughes et al., 2020; Lansbury et al., 2020; Rawson et al., 2020; Vaughn et al., 2020; Zhu et al., 2020). The common coinfecting bacterial species include Haemophilus influenzae, Staphylococcus aureus, Klebsiella pneuminiae, Mycoplasma pneumonia, Streptococcus pneumoniae, and Pseudomonas aeruginosa (Lansbury et al., 2020; Zhu et al., 2020). According to Lansbury et.al, P. aeruginosa is the second most frequently detected pathogen in COVID-19 patients based on the data included in their study (Lansbury et al., 2020).
In our hospital, P. aeruginosa is one of the commonest coinfecting bacteria in COVID-19 patients. It is a biofilm-forming opportunistic pathogen causing life-threatening chronic infections in immunocompromised individuals with diseases like burn wounds, urinary tract infections, and respiratory infections (Gellatly and Hancock, 2013). Biofilm formation leads to higher antimicrobial resistance and lower virulence allowing chronic colonization of P. aeruginosa in the host (Gellatly and Hancock, 2013). Previous study showed that Respiratory Syncytial Virus (RSV) infection increases iron availability and promotes biofilm formation of coinfecting P. aeruginosa for prolonged colonization in the host (Hendricks et al., 2016). Fatal clinical case due to coinfection of P. aeruginosa and influenza virus has been also reported before (Su et al., 2019). P. aeruginosa adopts genetic modification in genes such as rpoN, lasR, fleQ, and mucA to reduce virulence, enhance biofilm formation, and increase antimicrobial resistance for long term colonization in CF patients (Smith et al., 2006; Marvig et al., 2015). Attenuation of its virulence by adaptive modification in lasR and mpl genes has also reported in P. aeruginosa isolated from ventilator-associated pneumonia patients (Wang et al., 2017). Although P. aeruginosa coinfection has been reported, its adaptive modification for prolonged colonization and invasion in COVID-19 patients still remains unclear.
In this study, we focused on a small colony variant (SCV) strain of P. aeruginosa isolated from sputum and bronchioalveolar lavage fluid (BALF) samples of a critical COVID-19 patient. Genomic and transcriptomic sequencing were performed using Illumina and PacBio sequencing analysis to investigate its genetic modification. Genome-wide DNA methylation analysis revealed that genes involved in its flagellar formation, exopolysaccharide (EPS) biosynthesis and quorum sensing systems were methylated. RNA sequencing analysis showed that expression of flagellar-related genes were significantly downregulated whereas EPS-related genes were overexpressed. More notably, expression of las and rhl quorum sensing genes were significantly elevated. This is opposite from previous studies that P. aeruginosa attenuates its quorum sensing systems in the biofilm for escaping from host immune clearance (Marvig et al., 2015; Wang et al., 2017). Phenotypic tests validated that the isolate expresses higher las and rhl quorum sensing systems and forms excessive biofilm by reduction of motility and overproduction of EPS. This study demonstrated that P. aeruginosa adopts complex genetic adaptations in the SARS-CoV-2 infected environment for higher antimicrobial resistance, persistent colonization and disease induction. Our findings contribute to the prognosis of disease development and treatment decision to manage P. aeruginosa coinfection in COVID-19 patients.
Materials and Methods
Strain Isolation
Two Pseudomonas aeruginosa small colony isolates were collected from sputum samples and bronchioalveolar lavage fluids (BALF) of a critically illed COVID-19 patient during routine clinical tests. Blood indices were recorded including counts of white blood cells (WBC), neutrophils (N), lymphocytes (L), and levels of interleukin-6 (IL-6), C-reactive protein (CRP), and Procalcitonin (PCT).
Ethical Statement
This work includes neither any identifiable human data nor direct participation of the patient. Bacterial samples were taken from routine microbiological tests of clinical respiratory samples. Blood indices were taken from the results of routine clinical blood examinations. This work is approved by the Ethics Committee of Shenzhen Third People’s Hospital, Second Hospital Affiliated to Southern University of Science and Technology [2020-184] and the Ethics Committee of Southern University of Science and Technology [20200069].
Bacterial Strains and Growth Media
The isolates and all used P. aeruginosa strains and plasmids are listed in Table 1 . Cultures were grown in LB broth or ABTGC medium supplemented with 10% TSB unless stated. ABTGC medium contains 0.2% glucose, 0.2% casamino acids, 0.1% MgCl2, 0.1% CaCl2, 0.1% FeCl3, and 10% A10 medium consisting of 15.1mM (NH4)2SO4, 33.7mM Na2HPO4•2H2O, 22 mM KH2PO4, and 0.05mM NaCl. Three hundred μg/ml carbenicillin was used for selecting strains carrying pUCP22-cdrA::gfp plasmid. Colony morphology of the isolates was imaged by Olympus (Mshot, China) with 8X magnification.
Table 1.
Bacteria strains and plasmid used in this study.
| Strains or plasmids | Relevant genotype and/or characteristics | Reference |
|---|---|---|
| Pseudomonas aeruginosa PAO1 | Pseudomonas aeruginosa ATCC | (Hentzer et al., 2002) |
| Pseudomonas aeruginosa LYSZa7 | SCV Isolate 1 | This study |
| Pseudomonas aeruginosa LYSZa8 | SCV Isolate 2 | This study |
| PAO1ΔlasIΔrhlI | PAO1 lasI and rhlI double mutant | (Hentzer et al., 2003) |
| PAO1ΔlasIΔrhlI/PlasB::gfp | PAO1 lasI and rhlI mutant containing lasB-gfp(ASV) reporter fusion | (Hansen et al., 2012) |
| PAO1ΔlasIΔrhlI/PrhlA::gfp | PAO1 lasI and rhlI mutant containing rhlA-gfp(ASV) reporter fusion | (Hansen et al., 2012) |
| PAO1/PcdrA::gfp | PAO1 containing cdrA-gfp reporter fusion plasmid | This study |
| LYSZa7/PcdrA::gfp | Isolate 1 containing cdrA-gfp reporter fusion plasmid | This study |
| LYSZa8/PcdrA::gfp | Isolate 2 containing cdrA-gfp reporter fusion plasmid | This study |
| pUCP22-cdrA::gfp | Plasmid carrying cdrA::gfp | (Rybtke et al., 2012) |
Motility Assays
Swimming motility of the isolates and P. aeruginosa PAO1 was tested on 0.3% LB agar plates. Swarming motility of the strains was tested on 0.5% agar plates supplemented with 0.8% nutrient broth and 0.5% glucose. Overnight cultures were diluted to OD600nm of 0.01 and inoculated into the agar using sterile toothpicks for swimming while 2 µl of the inoculums were spotted onto the agar for swarming. The plates were incubated at 37°C statically for 14–16 h. Bacterial motility was imaged using Chemiluminescent (ChampChemiTM580, Sage, China).
Biofilm Formation Assay
Overnight cultures of the isolates and P. aeruginosa PAO1 were diluted to OD600nm 0.01 in fresh LB broth as innoculums. One hundred µl of the innoculums were aliquoted into 96-well microtiter plate in triplicates and incubated statically for 24 h at 37°C for biofilm formation. Biofilm was washed twice with ddH2O. One hundred twenty-five µl of 0.1% crystal violet (CV) was added to each well and incubated for 15 min at room temperature for staining. The wells were washed twice thoroughly with ddH2O and air-dried. CV stain was dissolved into 125 µl of 30% acetic acid. Relative biofilm biomass was quantified by measuring optical density of CV staining on a Tecan infinity pro200 microplate reader at 550 nm.
Quorum Sensing Inhibition Assay
Overnight cultures of the isolates, P. aeruginosa PAO1, PAO1ΔlasIΔrhlI, and quorum sensing (QS) reporter strains were diluted to OD600nm 0.01 in ABTGC medium with 10% TSB and cultured to OD600nm 0.8~1.0. Supernatants of the isolates, P. aeruginosa PAO1, and PAO1ΔlasIΔrhlI were collected by centrifugation at 12,000 g for 2 min and sterilized by filtration. Fifty µl of the supernatants were added to 50 µl of cultures of QS reporter strains respectively in triplicates. Green fluorescence and OD600nm were recorded for 16 h in Tecan infinity pro200 microplate reader for QS expression. The relative expression of las and rhl QS was quantified by GFP/OD.
Construction of Cyclic-Di-GMP Reporter Strains
P. aeruginosa PAO1 and the isolates were transformed with plasmid pUCP22-cdrA::gfp by electroporation. The transformants were selected on LB agar plates containing 300 µg/ml carbenicillin. The plasmid in the transformants was extracted and run on electrophoresis gel for confirmation.
Cyclic-Di-GMP Expression Assay
Overnight cultures of the cyclic-di-GMP reporter strains were diluted to OD600nm 0.01 in ABTGC medium with 10% TSB as inoculums. One hundred μl of inoculums were loaded into 96-well microplate in triplicates and allowed to grow to stationary phase with green fluorescence and OD600nm measured using a Tecan Infinite Pro2000 microplate reader. Relative cyclic-di-GMP levels expressed in the isolates and PAO1 was quantified by GFP/OD.
Genomic Extraction and Sequencing
The isolates were cultured to early stationary phase. Genomic DNA of the isolates was extracted by AxyPerp Bacterial Genomic DNA Miniprep Kit (Corning, New York, USA) and Mabio Bacterial DNA Extraction Mini Kits (Mabio), respectively, using manufacturer’s protocol. For short-read sequencing, PCR-free libraries of extracted genomic DNA were prepared by VAHTSTM PCR-Free DNA Library Prep Kit for Illumina® (Vazyme, China) following manufacturer’s protocol. Purified fragments were tagged with VAHTSTM DNA Adapters for Illumina® (Vazyme, China). Quality of the libraries were tested using qPCR and Agilent Technologies 2100 Bioanalyzer. Genomic sequencing was performed on Illumina HiSeq X platform for paired end reads of 150 bp. For long read sequencing, DNA was fragmented with G-tubes (Covaris) and end-repaired to prepare SMRTbell DNA template libraries according to the manufacturer’s specification (PacBio, Menlo Park, USA). Genomic sequencing was performed on the Pacific Biosciences RSII sequencer (PacBio, Menlo Park, USA) according to standard protocols.
Transcriptomic Extraction and Sequencing
The isolates and P. aeruginosa PAO1 were cultured to early stationary phase. Total RNA was extracted using Magen HiPure Universal RNA Mini kits (MCBio, China) according to the manufacturer’s instructions. Concentrations of RNA were measured by Qubit 2.0 (Thermo Fisher Scientific, MA, USA) and Nanodrop One (Thermo Fisher Scientific, MA, USA). RNA integrity was evaluated using Agilent 2100 system (Agilent Technologies, Waldbron, Germany). RNA libraries were prepared by NEB Next® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA) following manufacturer’s instruction. Ribosomal RNA were removed by Ribo-zero rRNA Removal Kit. cDNA was synthesized using NEB Next First Strand Synthesis Reaction Buffer. RNA sequencing was performed on Illumina NovaSeq 6000 platform for paired end reads of 150 bp.
Sequencing Data Analysis
For genomic sequencing reads, Illumina reads were trimmed with automatic adaptor trimming option and assembled into contigs using De Novo Assembly module of CLC Genomics Workbench 20 (Qiagen) with default parameters. Phylogenetic tree was drawn by libMUSCLE aligment mode of Parsnp package using complete genomes (Treangen et al., 2014). PacBio reads were assembled into complete genome using HGAP4 pipeline of SMRTLink software v9.0 with default settings. Multilocus sequence typing (MLST) and identification of antimicrobial resistance genes (85% identity and 60% minimal length) using LYSZa7 genome were performed on the Center of Genomic of Epidermiology webserver (Larsen et al., 2012; Zankari et al., 2012). Circular plot was drawn using BLAST Ring Image Generator (Alikhan et al., 2011). Genomic islands on LYSZa7 genome were predicted by IslandViewer 4 webserver (Bertelli et al., 2017). LYSZa7 genome was annotated by prokka v1.14.6 (-species Pseudomonas aeruginosa –metagenome –Kingdom bacterium) (Seemann, 2014). DNA methylation analysis was performed using Base Modification Analysis and Motif Analysis application of SMRTLink software v9.0. The complete genome sequence was uploaded into the SMRT portal as reference sequence. A default modification quality value (QV) score of 30 (correspond to a p-value of 0.001) was used to call the modified bases. Restriction modification system genes were predicted and assigned to identified recognition motifs, with REBASE (Roberts et al., 2010). Grep function in shell was used to match methylated genes and QS and biofilm related genes.
Illumina RNA sequencing reads of the isolates were preprocessed and analyzed using RNA analysis module of CLC Genomics Workbench 20 (Qiagen) with default settings. Differential gene expression was done using Empirical Analysis of DGE module using selection criteria of absolute fold change ≧4 and adjusted p-value <0.05, with P. aeruginosa PAO1 as reference. GO enrichment analysis of significantly regulated genes was performed on DAVID bioinformatics database v6.8 (Ma et al., 2012). PCoA plot and heatmap were drawn using vegan, ggplot2, and pheatmap packages in R 4.0.0 software.
Data Availability
All Illumina sequencing data used in this study could be found under BioProject No. PRJNA656063 and assembled genome of LYSZa7 could be found under BioProject No. PRJNA656096 on NCBI.
Results
Pseudomonas aeruginosa Coinfection and Immune Responses
In our hospital, P. aeruginosa is the third mostly identified coinfecting bacterium among COVID-19 patients. 5.1% (21/408) of all the COVID-19 patients was diagnosed with secondary infections, among which 23.8% (5/21, 4 critically illed and 1 severely illed patients) was infected with P. aeruginosa (unpublished data). Among all the P. aeruginosa isolates collected from these patients, two isolates collected from one critically illed patient exhibit small colony variant morphology with wrinkled edge and condensed extracellular matrix with stronger biofilm forming capacity ( Figures 1A, D ). We thus focused on these two isolates for further analysis. These two P. aeruginosa isolates were collected from routine clinical respiratory samples longitudinally with 10 days interval. The first isolate collected from sputum sample was named as LYSZa7 while the second was from BALF sample and named as LYSZa8. Counts of white blood cells (WBC), neutrophils (N), lymphocytes (L), and levels of interleukin-6 (IL-6), C-reactive protein (CRP) and Procalcitonin (PCT) were monitored as immune response indicators ( Figure 2 ). Two peaks were observed from levels of WBC, N and L whereas the isolates were collected over the period of the second peak ( Figure 2A ). CRP is an effective diagnostic marker to differentiate viral and mixed/bacterial infection in respiratory infections. Significantly higher levels of CRP (> 40 mg/L) could be detected from patients with bacterial and influenza viral coinfection comparing to that of patients with sole influenza viral infection (Ahn et al., 2011; Li et al., 2019). High levels of CRP observed from day 2 to day 23 suggested that there should be viral and bacterial coinfection in the patient’s respiratory system ( Figure 2B ). The emergence of P. aeruginosa aligned well with the peak of immune response indicators. Similar trend was observed from the levels of immune response indicators of the other 3 critically illed patients ( Figure S1 ). Thus, P. aeruginosa was highly associated with the bacterial coinfection induced in the patients during critical stage of COVID-19. We therefore further investigated the P. aeruginosa SCV isolates on their survival and adaptation in SARS-CoV-2 virus infected environment.
Figure 1.
Phenotypic tests. (A) Colony morphologies of P. aeruginosa PAO1, LYSZa7 and LYSZa8; (B) Cyclic-di-GMP levels in P. aeruginosa PAO1, LYSZa7 and LYSZa8 measured by cdrA::gfp reporter assay; (C) Swimming motility (left plate) and swarming motility(right plate) of P. aeruginosa PAO1, LYSZa7 and LYSZa8; (D) Biofilm formation of P. aeruginosa PAO1, LYSZa7, and LYSZa8 quantified by CV staining, **p-value <0.05; (E) las and rhl quorum sensing expression measured by PAO1ΔlasIΔrhlI/PlasB::gfp and PAO1ΔlasIΔrhlI/PrhlA::gfp reporter strains.
Figure 2.
Levels of the patient’s blood indices. (A) Counts of WBC, L and N during the course of hospitalization. The isolates were found over the period of the second immune peak. Red arrows indicate the isolation times of LYSZa7(day 21) and LYSZa8(day 31); (B) Levels of IL-6 (pg/ml), CRP (mg/L), and PCT (ng/ml) during the course of hospitalization. Orange arrows indicate the isolation times of LYSZa7(day 21) and LYSZa8(day 31).
Genomic and Epigenetic Characterization of P. aeruginosa SCV Isolates
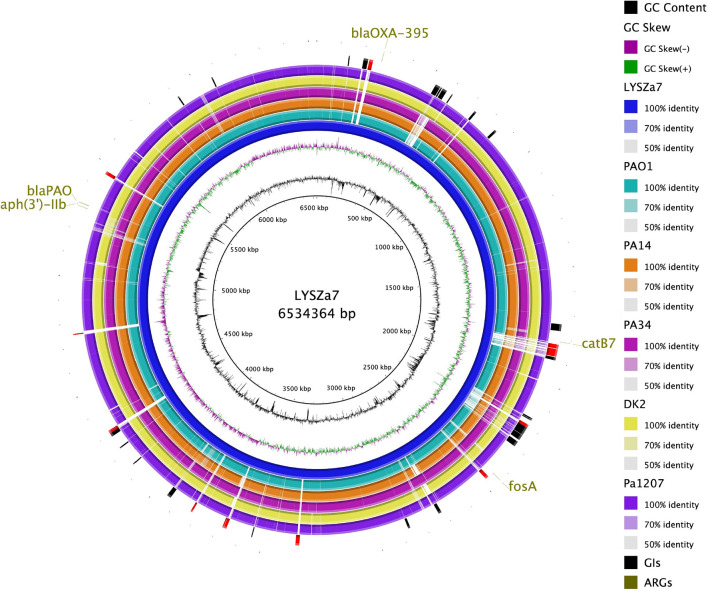
Genomic sequencing was performed using Illumina and PacBio sequencing platforms to investigate the genomic characteristics of the isolates. MLST analysis using Illumina contigs showed that both isolates are of P. aeruginosa ST1445. LYSZa7 and LYSZa8 are of same sequencing type with different evolving times. Thus, we put our focus solely on LYSZa7 genome. Its complete genome is 6,534,364 bp in length with an average GC of 66.21%. It carries several antimicrobial resistance genes (ARGs) including aph(3’)-IIb, blaOXA-395, blaPAO, fosA and catB7 against aminoglycoside, beta-lactam, fosfomycin, and phenicol drugs ( Figure 4 , outermost circle).
Figure 4.
Circular plot. From the innermost, Circle 1: GC content; Circle 2: GC Skew; Circle 3: P. aeruginosa LYSZa7; Circle 4: P. aeruginosa PAO1; Circle 5: P. aeruginosa PA14 (virulent clinical isolate); Circle 6: P. aeruginosa PA34 (strain isolated from Keratitis patient); Circle 7: P. aeruginosa DK2 (Strain isolated from Cystic Fibrosis patient); Circle 8: P. aeruginosa Pa1207 (strain isolated from Bacteremia patient); Circle 9: Genomic Islands predicted with specific GIs of LYSZa7 highlighted in red; Circle 10: Antimicrobial resistance genes (ARGs).
Phylogenetic tree was constructed by comparing LYSZa7 with 23 other P. aeruginosa genomes selected from NCBI Genbank to trace its evolutionary origin ( Table S1 ). LYSZa7 branched out individually with a closer distance to the virulent clinical strain PA14 while positioned further away from PAO1 reference strain and certain clinical strains such as DK2 isolated from cystic fibrosis patients ( Figure 3 ). Based on the phylogenetic tree, genome of LYSZa7 was compared with selected species including PAO1 reference strain and virulent clinical strains, PA14, PA 34, DK2, and Pa1207 isolated from patients with different diseases like cystic fibrosis, keratitis, and bacteremia to predict specific genomic islands (GIs) on LYSZa7 genome ( Figure 4 ). As seen from the circular plot, there are specific GIs present only on LYSZa7 genome ( Figure 4 , circle 9, highlighted in red). These GIs are involved in transposition, toxin transport, transcriptional regulation and especially DNA restriction-modification (R-M) system for survival, persistence, and invasion in the respiratory tract during coinfection ( Table S2 ).
Figure 3.

Phylogenetic tree constructed using whole genomes of LYSZa7 and 23 other P. aeruginosa laboratory, clinical and environmental strains selected from NCBI database. P. aeruginosa PAO1 was used as reference strain.
DNA methylation as part of DNA restriction-modification systems modifies the nucleotide without changing the genome sequence to regulate gene expression and control phenotypic traits in epigenetic perspective (Lobner-Olesen et al., 2005; Vasu and Nagaraja, 2013). Specific GIs for type I and type III DNA restriction-modification system were predicted on LYSZa7 genome. DNA methylation analysis indicated that genes related to las and rhl quorum sensing systems, alginate production and flagellar formation were methylated by two different methyltransferases from both R-M systems predicted ( Table 2 ). DNA sequencing analysis revealed the phylogeny, unique GIs and a particular pattern of DNA methylation of LYSZa7. Transcriptomic analysis was then performed to evaluate gene expression of the isolates.
Table 2.
Genes methylated in LYSZa7 comparing to P. aeruginosa PAO1 and motif information. Type: type of R-M system.
| Type | Methyltransferase | Motif | Genes methylated* |
|---|---|---|---|
| Type III | M.PaeZa7II | CCCGAG | algD algF algK fliC fliD lasB rhlB |
| Type I | M.PaeZa7I | AGGNNNNNPTGT | fliC |
*Grep function in shell was used to match methylated genes and QS and biofilm related genes.
Pre-Transcriptional and Transcriptional Regulations Lead to Excessive Biofilm Formation and Elevated Quorum Sensing Systems
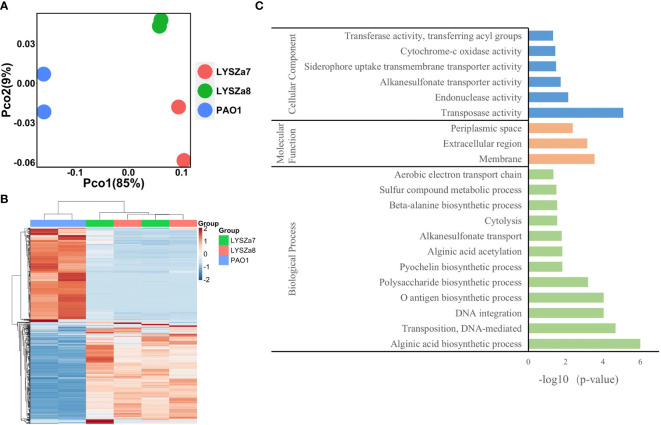
RNA sequencing was performed on Illumina sequencing platform and differential gene expression (DGE) in LYSZa7 and LYSZa8 was analyzed using P. aeruginosa PAO1 as reference to investigate transcriptional regulation during coinfection. We involved the second isolate here to learn the transcriptional versatility between two isolates. Clear separation between the isolates and PAO1 along PC1 (85%) on PCoA plot implied the significant differences in their transcriptomic profiles ( Figure 5A ). The proximity between clusters of the two isolates along both PC1 (85%) and PC2 (9%) implied the similarity in their gene expressions. Such differences among the transcriptomics profiles of PAO1 and the two isolates, and similarity between that of the two isolates could also be seen from heatmap plot ( Figure 5B ).
Figure 5.
Dissimilarity analysis of samples and Gene Orthology analysis of differentially expressed genes; (A) PCoA plot of P. aeruginosa PAO1, LYSZa7 and LYSZa8 by Bray Curtis dissimilarity; (B) Heatmap of PAO1, LYSZa7 and LYSZa8 based on euclidean clustering distance; (C) Gene Orthology Enrichment based on the significantly regulated genes (fold change ≧ 4, adjusted p-value < 0.05) in LYSZa7 comparing to those of PAO1.
Results of DGE between LYSZa7 and LYSZa8 aligned with the dissimilarity analysis showing only minor variation in their gene expression profiles was observed. Only 39 genes were significantly downregulated in LYSZa8 comparing to LYSZa7 with mostly low mean counts ( Table S3 ). None of the genes participate in biofilm formation and quorum sensing systems. This indicated that the isolates proliferated rather stably in the respiratory system during coinfection.
DGE of PAO1 was compared with that of LYSZa7 since there was only minimal difference between LYSZa7 and LYSZa8. In total, 481 genes were differentially regulated in LYSZa7 ( Table S4 ). Among which, 38 genes were involved in biofilm formation and virulence regulation of the isolate ( Table 3 ). Gene ontology analysis also demonstrated enrichment of pathways essential for biofilm formation and virulence such as polysaccharide/alginate biosynthesis and O antigen biosynthesis pathways ( Figure 5C ). In P. aeruginosa, the second messenger, cyclic-di-GMP promotes biofilm formation while it is controlled by two groups of enzymes, phosphodiesterase (PDE) for its degradation and diguanylate cyclase (DGC) for biosynthesis (Kulasakara et al., 2006). As cdrA is positively regulated by cyclic-di-GMP, overexpression of cdrA reflects an increase in cyclic-di-GMP biosynthesis (Borlee et al., 2010). In LYSZa7, expression of cdrA (PA4625) gene was upregulated for 4.41 folds indicating an accumulation of cyclic-di-GMP. cdrA::gfp reporter assay confirmed the excessive production of cyclic-di-GMP in both LYSZa7 and LYSZa8 ( Figure 1B ). Such cyclic-di-GMP accumulation is probably caused by the downregulation of the PDE gene, arr, which decreased for 318.93 folds. Cyclic-di-GMP in turn bound to the I-site on PA2771 inducing self-inhibition of this DGC and resulted in its downregulation (Hoffman et al., 2005; Chen et al., 2016) ( Table 3 ). As cyclic-di-GMP promotes biofilm formation, we tested biofilm formation of LYSZa7 and LYSZa8 by CV staining assay. Results showed that LYSZa7 and LYSZa8 are indeed strong biofilm-formers as they formed significantly more biofilm comparing to the reference PAO1 strain ( Figure 1D ).
Table 3.
Significantly regulated genes involved in virulence and biofilm formation in LYSZa7 comparing to P. aeruginosa PAO1 (fold change ≧ 2, FDR adjusted p-value < 0.05); means: normalized mean counts of the sample groups.
| Feature ID | Product | Fold change | Adjusted p-value | PAO1 Means | LYSZa7 Means |
|---|---|---|---|---|---|
| alg44 | alginate biosynthesis protein Alg44 | 11.17 | 3.14E-47 | 26.0 | 277.9 |
| alg8 | alginate biosynthesis protein Alg8 | 7.87 | 8.78E-57 | 53.3 | 389.8 |
| algA | phosphomannose isomerase/guanosine 5’-diphospho-D-mannose pyrophosphorylase | 29.12 | 1.16E-75 | 47.9 | 1352.3 |
| algD | GDP-mannose 6-dehydrogenase AlgD | 19.91 | 7.43E-60 | 56.3 | 1076.1 |
| algE | Alginate production outer membrane protein AlgE precursor | 15.83 | 8.60E-64 | 25.1 | 393.2 |
| algF | alginate o-acetyltransferase AlgF | 19.26 | 5.84E-56 | 14.5 | 322.3 |
| algI | alginate o-acetyltransferase AlgI | 5.19 | 2.60E-23 | 47.9 | 226.6 |
| algJ | alginate o-acetyltransferase AlgJ | 17.14 | 2.28E-46 | 4.8 | 182.5 |
| algK | alginate biosynthetic protein AlgK precursor | 14.33 | 8.76E-48 | 16.2 | 250.3 |
| algL | poly(beta-d-mannuronate) lyase precursor AlgL | 14.69 | 1.29E-58 | 21.6 | 321.1 |
| algX | alginate biosynthesis protein AlgX | 20.54 | 1.60E-59 | 10.8 | 308.2 |
| arr | aminoglycoside response regulator | -318.93 | 1.25E-49 | 229.6 | 2.5 |
| flgL | flagellar hook-associated protein type 3 FlgL | -8.39 | 1.14E-72 | 1,319.7 | 173.3 |
| fliC | flagellin type B | -7.44 | 0.00E+00 | 33,619.2 | 6865.2 |
| fliD | flagellar capping protein FliD | -1,624.06 | 8.80E-207 | 6,412.3 | 5.7 |
| PA2771 | diguanylate cyclase with a self-blocked I-site, Dcsbis | -17.32 | 2.07E-06 | 348.4 | 20.4 |
| PA4625 | cyclic diguanylate-regulated TPS partner A, CdrA | 4.41 | 4.60E-17 | 971.8 | 5208.9 |
| pelD | PelD | 5.55 | 5.54E-35 | 104.8 | 561.9 |
| pelE | PelE | 4.80 | 6.62E-20 | 65.3 | 280.4 |
| pelF | PelF | 5.00 | 8.20E-27 | 88.6 | 414.8 |
| pilA | type 4 fimbrial precursor PilA | -961.48 | 1.61E-221 | 29,476.2 | 40.7 |
| waaL | O-antigen ligase, WaaL | -8.00 | 3.94E-10 | 545.0 | 71.8 |
| wbpA | UDP-N-acetyl-d-glucosamine 6-Dehydrogenase | -1,275.14 | 1.09E-199 | 6,270.7 | 7.2 |
| wbpB | UDP-2-acetamido-2-deoxy-d-glucuronic acid 3-dehydrogenase, WbpB | -1,832.88 | 2.71E-164 | 3,815.0 | 4.8 |
| wbpD | UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid N-acetyltransferase, WbpD | -2,685.85 | 6.53E-163 | 1,683.3 | 3.4 |
| wbpE | UDP-2-acetamido-2-dideoxy-d-ribo-hex-3-uluronic acid transaminase, wbpE | -2,156.03 | 6.33E-173 | 4,611.0 | 4.8 |
| wbpG | LPS biosynthesis protein WbpG | -2,523.51 | 3.59E-141 | 4,083.4 | 4.2 |
| wbpH | probable glycosyltransferase WbpH | -1,978.78 | 4.72E-166 | 2,238.8 | 3.7 |
| wbpI | UDP-N-acetylglucosamine 2-epimerase WbpI | -2,058.27 | 6.90E-147 | 3,268.5 | 4.3 |
| wbpJ | probable glycosyl transferase WbpJ | -1,336.39 | 9.86E-162 | 1,523.7 | 4.3 |
| wbpK | probable NAD-dependent epimerase/dehydratase WbpK | -1,800.55 | 2.03E-149 | 1,150.5 | 3.7 |
| wbpL | glycosyltransferase WbpL | -1,912.92 | 3.51E-164 | 1,218.6 | 3.8 |
| wzx | O-antigen translocase | -638.58 | 3.01E-98 | 435.9 | 3.4 |
| wzy | B-band O-antigen polymerase | -6,615.39 | 2.54E-138 | 757.6 | 3.2 |
| wzz | O-antigen chain length regulator | -655.23 | 7.50E-152 | 1,113.7 | 4.5 |
| lasB | elastase LasB | 2.60 | 7.02E-06 | 4,484.2 | 15730.4 |
| rhlA | rhamnosyltransferase chain A | 2.74 | 1.41E-14 | 592.2 | 1887.9 |
| rhlB | rhamnosyltransferase chain B | 3.05 | 5.51E-16 | 294.8 | 940.1 |
Cyclic-di-GMP promotes biofilm formation through various mechanisms including reducing motility and enhancing biosynthesis of exopolysaccharides. Decrease in expression of genes involved in flagellar formation, fliCD and flgL, impaired swimming and swarming motility of the isolates ( Table 3 , Figure 1C ). Moreover, expression of alg and pel genes for exopolysaccharide biosynthesis raised significantly for 4.8 to 29.12 folds ( Table 3 ). Methylation of fliCD genes and algDFK genes indicated that these genes were regulated at pre-transcriptional level as well ( Table 2 ). Reduction in motility and overproduction of exopolysaccharides, alginate and Pel, are major factors leading to excessive biofilm formation by the isolates for prolong colonization.
P. aeruginosa possesses several virulence factors for invasion and disease induction in hosts, such as quorum sensing systems, siderophore production and lipopolysaccharide (LPS) (Hentzer et al., 2003). Transcription of genes responsible for LPS biosynthesis (waaL, wbp, wzx/y/z) was greatly inhibited in LYSZa7 impairing LPS biosynthesis ( Table 3 ). More notably, expression of quorum sensing genes, lasB and rhlAB, increased for 2.6, 2.74, and 3.05 folds, respectively, in LYSZa7 ( Table 2 ). We thus collected the supernatants of the reference PAO1 strain, the isolates and quorum sensing mutant to test the expression of lasB and rhlA genes using lasB::gfp and rhlA::gfp reporter assays. Results clearly showed that the expression of lasB and rhlA genes was much higher in the isolates comparing to PAO1 and the mutant ( Figure 1E ). This is opposite from previous studies reporting that P. aeruginosa attenuated quorum sensing systems during colonization in the respiratory systems (Smith et al., 2006; Wang et al., 2017). Methylation of lasB and rhlB genes indicated that regulation of quorum sensing systems started pre-transcriptionally. Our results suggested that P. aeruginosa isolate remodels its biofilm forming capacity and dynamically adjusts its virulence to adapt to the SARS-CoV-2 infected environment in COVID-19 patients.
Discussion
In this study, we characterized two P. aeruginosa SCVs isolated from respiratory samples of one critical COVID-19 patient on genomic, transcriptomic, and phenotypic levels for their adaptation and underlying mechanisms causing bacterial coinfection. The two isolates are of same sequence typing with relatively stable transcriptomic profiles. Characterization of LYSZa7 genome indicated that the isolates carry specific GIs for R-M systems and may result in DNA methylation. N6-methyl adenine (m6A) methylation was observed on genes involved in flagellar formation, alginate biosynthesis and quorum sensing systems based on the results of epigenetic analysis. RNA-seq analysis indicated that expression of 38 genes for biofilm formation and virulence were differentially regulated. Overlapped genes between epigenetic analysis and transcriptomic analysis showed that gene regulation may occur at both pre-transcriptional and transcriptional levels. We demonstrated here that the isolates reduced motility and increased exopolysaccharides biosynthesis to enhance biofilm formation, and reinforced las and rhl quorum sensing systems to colonize and persist in the COVID-19 patient.
Bacterial coinfection is complex process which has been frequently observed from patients with respiratory viral infections such as influenza pneumonia and resulted in high mortality rate. Viral infection damages tissues along respiratory track and regulates immune cells/cytokines resulting in dysbiosis of microbiome and facilitate bacterial colonization (McCullers, 2014; Bakaletz, 2017). In this study, fluctuated levels of immune cells (WBC, N, L) and blood indices (CRP and PCT) were recorded. CRP has been reported as an indicator of bacterial coinfection in viral pneumonia patients with a cut off value of more than 40 mg/L for mixed coinfection and lower for sole viral infection (Korppi and Kroger, 1993; Haran et al., 2013). With this as a reference, there should be viral and bacterial coinfection in the patient as the peak value of CRP reached more than 80 mg/L. Although there was no direct evidence to prove P. aeruginosa to be the causative pathogen, the emerging time of the isolates inferred a high correlation of this bacterium with bacterial coinfection in the patients based on the clinical data collected from all critical patients infected by P. aeruginosa ( Figures 2 , S1 ).
Previous studies revealed that viral infections facilitate bacterial colonization and promote biofilm formation. Influenza virus activates TGF-β and increase synthesis of cellular adhesins for enhancing attachment of group A Streptococcus to host cells (Li et al., 2015). RSV infection in the airway of CF patient activates antiviral interferon signaling which leads to higher iron availability and enhances P. aeruginosa biofilm formation (Hendricks et al., 2016). We observed similar phenomenon in this COVID-19 case that P. aeruginosa survived as small colony variant with enhanced biofilm formation in SARS-CoV-2 infected environment, by attenuating flagellar formation and enhancing exopolysaccharide production. In addition, phenotypic tests of P. aeruginosa isolates from the other three critically illed patients indicated obvious increases in biofilm formation in the later isolates as well (data not shown), suggesting elevated biofilm formation as a general traits evolved during coinfection with SARS-CoV-2 virus. We here put our focus solely on LYSZa7 and LYSZa8 as they exhibited small colony variant morphology and showed the strongest biofilm forming capacity among all the P. aeruginosa isolates collected.
Moreover, as reported previously, coinfecting pathogens either form biofilm with compromised virulence to escape from immune clearance or dispersed from biofilm gaining higher virulence for disease induction. Secretion of virulent factors upon biofilm dispersion in the coinfecting bacteria, such as Streptococcus pneumoniae and Staphylococcus aureus, upon influenza virus infection enabled a transition from non-invasive colonization to secondary bacterial infection in the patients (Pettigrew et al., 2014; Reddinger et al., 2016). It is also well-known that P. aeruginosa attenuates its virulence in the biofilm to escape from host immune clearance for chronic colonization (Smith et al., 2006). A recent research demonstrated that damaged mucus prevents P. aeruginosa biofilm dispersion and enhances the expression of virulence pathways by regulating motility, quorum sensing systems, and production of siderophore and toxins (Wheeler et al., 2019). These suggest that the virulence of P. aeruginosa biofilm is dependent on pathological changes in host tissues and variation in environmental conditions in the niches. We, for the first time, reported here that P. aeruginosa could express higher virulence in SARS-CoV-2 infected respiratory system by upregulating las and rhl quorum sensing systems. SARS-CoV-2 virus causes tissue damages including diffuse alveolar damage and shedding of epithelial cells of alveolar (Martines et al., 2020). Such defects in tissues and weakened host immunity after viral infection probably give chance to P. aeruginosa to elevate its virulence.
Besides quorum sensing systems, lipopolysaccharide is also a key virulence factor of P. aeruginosa inducing inflammation in the host (Pier, 2007). It has been reported that alginate, Psl and Pel share the same precursors, mannose-1-phosphate, with LPS (Ma et al., 2012). The increase in the exopolysaccharide of the isolates was probably due to the biosynthetic flux towards alginate and Pel leading to a reduction in LPS biosynthesis. Thus, LPS is probably not a major contributor to virulence during coinfection. Moreover, results of in vitro antimicrobial susceptibility tests indicated that the isolates were susceptible to various antibiotics, but rather resistant to continuous clinical antibiotic treatments and persisted in patients’ respiratory systems. Such observation suggested that increased drug resistance is predominantly due to biofilm formation which shields P. aeruginosa from antimicrobial attack. Our results suggested that P. aeruginosa adopts unique adaptations in COVID-19 pneumoniae patients for survival and disease induction. Future studies on a larger scale are necessary to provide further support of such adaptation observed here.
This study demonstrated that P. aeruginosa, as a top coinfecting pathogen, could form more biofilm and elevate quorum sensing systems for prolonged colonization and bacterial coinfection induction during critical stage of COVID-19 pneumonia. Understanding these genetic adaptations of P. aeruginosa would greatly contribute to the prognosis of disease development and treatment scheme decision to manage secondary infections induced by P. aeruginosa in COVID-19 patients.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author Contributions
JQ and ZC contributed to data analysis and manuscript preparation. YuL, XD, SH, JL, YuZ, ZJ, and YiZ contributed to data collection, experimental design, and operation. CZ, YaL, YiL, LL, and LY contributed to study design and manuscript verification. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Guangdong Natural Science Foundation for Distinguished Young Scholar [2020B1515020003] and Start-up Grants [Y01416106 and Y01416206] from the Southern University of Science and Technology (SUSTech) to LY, Science and Technology Program of Shenzhen [JCYJ20190809144005609] and Guangdong Basic and Applied Basic Research Foundation [2020A1515010586] to JQ, the Guangzhou Municipal Science and Technology Bureau [Project No. 201607020044] to CZ, Grant from Bill & Melinda Gates Foundation to LL, Natural Science Fund of Guangdong Province [2020A1515010316] to YaL and Guangdong Basic and Applied Basic Research Foundation[2019A1515110640] to YiZ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has appeared online previously as a preprint on BioRxiv (doi: https://doi.org/10.1101/2020.08.05.238998).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.641920/full#supplementary-material
Blood index levels of the other 3 critically illed patients. (A) Blood index levels of patient 2: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 12) and last P. aeruginosa isolate (day 48); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 12) and last P. aeruginosa isolate (day 48); (B) Blood index levels of patient 3: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 17) and last P. aeruginosa isolate (day 39); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 17) and last P. aeruginosa isolate (day 39); (C) Blood index levels of patient 4: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 31) and last P. aeruginosa isolate (day 43); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 31) and last P. aeruginosa isolate (day 43).
References
- Ahn S., Kim W. Y., Kim S. H., Hong S., Lim C. M., Koh Y., et al. (2011). Role of procalcitonin and C-reactive protein in differentiation of mixed bacterial infection from 2009 H1N1 viral pneumonia. Influenza Other Respir. Viruses 5 (6), 398–403. 10.1111/j.1750-2659.2011.00244.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakaletz L. O. (2017). Viral-bacterial co-infections in the respiratory tract. Curr. Opin. Microbiol. 35, 30–35. 10.1016/j.mib.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Laird M. R., Williams K. P., Simon Fraser University Research Computing G. Lau B. Y., Hoad G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45 (W1), W30–WW5. 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno A. L., Griffin J. P., Van Epps K. A., Niell H. B., Rytel M. W. (1971). Pneumonia and Hong Kong influenza: a prospective study of the 1968-1969 epidemic. Am. J. Med. Sci. 261 (5), 251–263. 10.1097/00000441-197105000-00004 [DOI] [PubMed] [Google Scholar]
- Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., Parsek M. R. (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75 (4), 827–842. 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage J. F., Shanks G. D. (2008). Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg. Infect. Dis. 14 (8), 1193–1199. 10.3201/eid1408.071313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu S., Liu C., Huang Y., Chi K., Su T., et al. (2016). Dcsbis (PA2771) from Pseudomonas aeruginosa is a highly active diguanylate cyclase with unique activity regulation. Sci. Rep. 6, 29499. 10.1038/srep29499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395 (10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloniz C., Ewig S., Menendez R., Ferrer M., Polverino E., Reyes S., et al. (2012). Bacterial co-infection with H1N1 infection in patients admitted with community acquired pneumonia. J. Infect. 65 (3), 223–230. 10.1016/j.jinf.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly S. L., Hancock R. E. W. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67 (3), 159–173. 10.1111/2049-632X.12033 [DOI] [PubMed] [Google Scholar]
- Gill J. R., Sheng Z. M., Ely S. F., Guinee D. G., Beasley M. B., Suh J., et al. (2010). Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch. Pathol. Lab. Med. 134 (2), 235–243. 10.5858/134.2.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada S., Pirzadeh M., Carver K. Y., Deng J. C. (2018). Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 9, 2640. 10.3389/fimmu.2018.02640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. K., Rau M. H., Johansen H. K., Ciofu O., Jelsbak L., Yang L., et al. (2012). Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. Isme J. 6 (1), 31–45. 10.1038/ismej.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran J. P., Beaudoin F. L., Suner S., Lu S. (2013). C-reactive protein as predictor of bacterial infection among patients with an influenza-like illness. Am. J. Emerg. Med. 31 (1), 137–144. 10.1016/j.ajem.2012.06.026 [DOI] [PubMed] [Google Scholar]
- Hendricks M. R., Lashua L. P., Fischer D. K., Flitter B. A., Eichinger K. M., Durbin J. E., et al. (2016). Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. U S A 113 (6), 1642–1647. 10.1073/pnas.1516979113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M., Riedel K., Rasmussen T. B., Heydorn A., Andersen J. B., Parsek M. R., et al. (2002). Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148 (Pt 1), 87–102. 10.1099/00221287-148-1-87 [DOI] [PubMed] [Google Scholar]
- Hentzer M., Wu H., Andersen J. B., Riedel K., Rasmussen T. B., Bagge N., et al. (2003). Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22 (15), 3803–3815. 10.1093/emboj/cdg366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers J. F., Masurel N., Mulder J. (1958). Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet 2 (7057), 1141–1143. 10.1016/s0140-6736(58)92404-8 [DOI] [PubMed] [Google Scholar]
- Hoffman L. R., D’Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., Miller S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436 (7054), 1171–1175. 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- Hughes S., Troise O., Donaldson H., Mughal N., Moore L. S. P. (2020). Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 26(10), 1395–1399. 10.1016/j.cmi.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korppi M., Kroger L. (1993). C-Reactive Protein in Viral and Bacterial Respiratory-Infection in Children. Scand. J. Infect. Dis. 25 (2), 207–213. 10.3109/00365549309008486 [DOI] [PubMed] [Google Scholar]
- Kulasakara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3’-5’)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U.S.A. 103 (8), 2839–2844. 10.1073/pnas.0511090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury L., Lim B., Baskaran V., Lim W. S. (2020). Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 81 (2), 266–275. 10.1016/j.jinf.2020.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 (4), 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Ren A. H., Wang X. S., Fan X., Zhao Y., Gao G. F., et al. (2015). Influenza viral neuraminidase primes bacterial coinfection through TGF-beta-mediated expression of host cell receptors. Proc. Natl. Acad. Sci. U.S.A. 112 (1), 238–243. 10.1073/pnas.1414422112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., He L., Li S., He W., Zha C., Ou W., et al. (2019). Combination of procalcitonin and C-reactive protein levels in the early diagnosis of bacterial co-infections in children with H1N1 influenza. Influenza Other Respir. Viruses 13 (2), 184–190. 10.1111/irv.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. (2020). Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 382 (13), 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Chen Y., Lin R., Han K. (2020). Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J. Infect. 80 (6), e14–ee8. 10.1016/j.jinf.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobner-Olesen A., Skovgaard O., Marinus M. G. (2005). Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8 (2), 154–160. 10.1016/j.mib.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Ma L., Wang J., Wang S., Anderson E. M., Lam J. S., Parsek M. R., et al. (2012). Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14 (8), 1995–2005. 10.1111/j.1462-2920.2012.02753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines R. B., Ritter J. M., Matkovic E., Gary J., Bollweg B. C., Bullock H., et al. (2020). Pathology and Pathogenesis of SARS-CoV-2 Associated with Fatal Coronavirus Disease, United States. Emerg. Infect. Dis. 26 (9), 2005–2015. 10.3201/eid2609.202095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig R. L., Sommer L. M., Molin S., Johansen H. K. (2015). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47 (1), 57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- McCullers J. A. (2014). The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 12 (4), 252–262. 10.1038/nrmicro3231 [DOI] [PubMed] [Google Scholar]
- Pettigrew M. M., Marks L. R., Kong Y., Gent J. F., Roche-Hakansson H., Hakansson A. P. (2014). Dynamic Changes in the Streptococcus pneumoniae Transcriptome during Transition from Biofilm Formation to Invasive Disease upon Influenza A Virus Infection. Infect. Immun. 82 (11), 4607–4619. 10.1128/IAI.02225-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier G. B. (2007). Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 297 (5), 277–295. 10.1016/j.ijmm.2007.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson T. M., Moore L. S. P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., et al. (2020). Bacterial and fungal co-infection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 71(9):2459–2468. 10.1093/cid/ciaa530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddinger R. M., Luke-Marshall N. R., Hakansson A. P., Campagnari A. A. (2016). Host Physiologic Changes Induced by Influenza A Virus Lead to Staphylococcus aureus Biofilm Dispersion and Transition from Asymptomatic Colonization to Invasive Disease. mBio 7 (4), e01235–16. 10.1128/mBio.01235-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Vincze T., Posfai J., Macelis D. (2010). REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, D234–D2D6. 10.1093/nar/gkp874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtke M. T., Borlee B. R., Murakami K., Irie Y., Hentzer M., Nielsen T. E., et al. (2012). Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78 (15), 5060–5069. 10.1128/AEM.00414-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 (14), 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. (2020). Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 323(16):1582–1589. 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. E., Buckley D. G., Wu Z., Saenphimmachak C., Hoffman L. R., D’Argenio D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 103 (22), 8487–8492. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I. C., Lee K. L., Liu H. Y., Chuang H. C., Chen L. Y., Lee Y. J. (2019). Severe community-acquired pneumonia due to Pseudomonas aeruginosa coinfection in an influenza A(H1N1)pdm09 patient. J. Microbiol. Immunol. 52 (2), 365–366. 10.1016/j.jmii.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Treangen T. J., Ondov B. D., Koren S., Phillippy A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15 (11), 524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77 (1), 53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn V. M., Gandhi T., Petty L. A., Patel P. K., Prescott H. C., Malani A. N., et al. (2020). Empiric Antibacterial Therapy and Community-onset Bacterial Co-infection in Patients Hospitalized with COVID-19: A Multi-Hospital Cohort Study. Clin. Infect. Dis. ciaa1239. 10.1093/cid/ciaa1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. (2020). Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 323 (11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen Y. Q., Salido M. M., Kohli G. S., Kong J. L., Liang H. J., et al. (2017). The rapid in vivo evolution of Pseudomonas aeruginosa in ventilator-associated pneumonia patients leads to attenuated virulence. Open Biol. 7 (9), 170029. 10.1098/rsob.170029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., et al. (2020). Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 395 (10228), e52. 10.1016/S0140-6736(20)30558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler K. M., Carcamo-Oyarce G., Turner B. S., Dellos-Nolan S., Co J. Y., Lehoux S., et al. (2019). Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat. Microbiol. 4 (12), 2146–2154. 10.1038/s41564-019-0581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 (11), 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395 (10229), 1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. J., Ge Y. Y., Wu T., Zhao K. C., Chen Y., Wu B., et al. (2020). Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 285, 198005. 10.1016/j.virusres.2020.198005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blood index levels of the other 3 critically illed patients. (A) Blood index levels of patient 2: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 12) and last P. aeruginosa isolate (day 48); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 12) and last P. aeruginosa isolate (day 48); (B) Blood index levels of patient 3: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 17) and last P. aeruginosa isolate (day 39); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 17) and last P. aeruginosa isolate (day 39); (C) Blood index levels of patient 4: Upper panel: Counts of WBC, L and N during the course of hospitalization. Red arrows indicate the isolation times of first P. aeruginosa isolate (day 31) and last P. aeruginosa isolate (day 43); Lower panel: Levels of IL-6(pg/mL), CRP (mg/L) and PCT (ng/mL) during the course of hospitalization. Orange arrows indicate the isolation times of first P. aeruginosa isolate (day 31) and last P. aeruginosa isolate (day 43).
Data Availability Statement
All Illumina sequencing data used in this study could be found under BioProject No. PRJNA656063 and assembled genome of LYSZa7 could be found under BioProject No. PRJNA656096 on NCBI.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .