Abstract
Extracellular vesicles derived from mammalian cells could be useful carriers for drug delivery systems (DDSs); however, with regard to clinical application, there are several issues to be overcome. Acerola (Malpighia emarginata DC.) is a popular health food. In this study, the feasibility of orally administered nucleic acid drug delivery by acerola exosome-like nanoparticles (AELNs) was examined. AELNs were recovered from acerola juice using an affinity column instead of ultracentrifugation. MicroRNA (miRNA) was sufficiently encapsulated in AELNs by 30-min incubation on ice and was protected against RNase, strong acid, and base treatments. The administration of an AELN/miRNA mixture in cells achieved downregulation of the miRNA’s target gene, and this mixture showed cytoplasmic localization. AELNs orally delivered small RNA to the digestive system in vivo. The target gene-suppressing effect in the small intestine and liver peaked 1 day after administration, indicating potential for use as an oral DDS for nucleic acid in the digestive system.
Keywords: acerola, nanovesicle, nucleic acid medicine, oral administration
Graphical abstract

Encapsulation of nucleic acids in acerola exosome-like nanovesicles (AELNs) had protective effects against RNase, acid, and base treatments. Kuroda and colleagues demonstrated that oral administration of AELNs enabled the functions of nucleic acids to be exerted in the digestive tract, and it is expected to become a new method for loading nucleic acids.
Introduction
Cells are in constant communication with each other for the performance of physiological activities. This communication is mediated primarily by extracellular vesicles, small transporters composed of lipid membranes.1 Normally, extracellular vesicles of the particular species in question carry out cell-to-cell signaling, but extracellular vesicles of another species can be used; for example, plant extracellular vesicles were shown to be able to carry out cell-to-cell signaling in mammalian cells.1, 2, 3, 4, 5
Mammalian exosomes can be docked and fused to the membrane of target cells to deliver exosome surface proteins and cytoplasm.6,7 Because the administration of exosomes does not induce deleterious immune responses and has a low risk of tumorigenesis, there is increasing interest in the use of exosomes as a vehicle for the in vivo delivery of microRNAs (miRNAs).8 In addition, therapeutic miRNA-loaded exosomes can be produced in large quantities by exosome-producing cells in vitro, allowing personalized therapy.9,10 These findings provide a rationale for the usefulness of exosomes as a drug delivery system (DDS).
Extracellular vesicles derived from edible plants are similar to mammalian exosomes due to their morphology and density and are called exosome-like nanovesicles (ELNs).11 They are composed of numerous lipids, RNAs, including miRNAs, and proteins.12,13 Upon the encapsulation of small interfering RNA (siRNA) in ELNs, siRNA was shown to be stable against physical stimuli such as sterilization, homogenization, and sonication.11,14, 15, 16 Several in vitro and in vivo studies reported that edible plant-derived ELNs accumulate in mammalian cells and have important functional effects on these cells.16 These findings suggest that plant ELNs may mediate mammalian cell-to-cell communication.15,17, 18, 19, 20
Acerola (Malpighia emarginata D.C.) fruit contains vitamin C, carotenoids, phenols, flavonoids, and anthocyanins,21 and it is widely used as a health food. It has various biological functions such as an antihyperglycemic effect,22 anticancer activity against lung cancer,23 and protective effects against genotoxicity induced by iron.24 In this study, we determined the feasibility of developing a DDS by the oral administration of nucleic acids using acerola ELNs (AELNs).
Results
The phenotypic characterization of nanovesicles derived from acerola juice
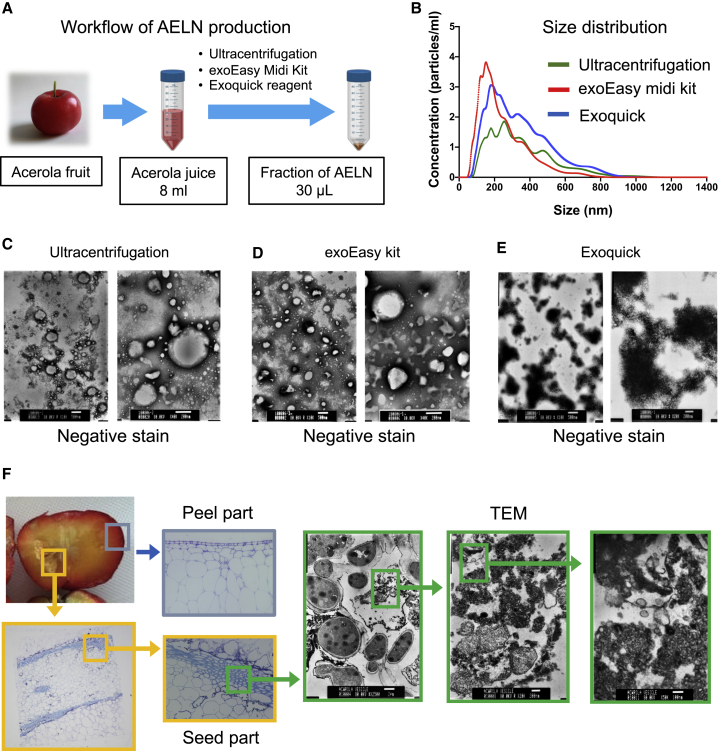
We compared the recovery of nanovesicles by several methods/tools (ultracentrifugation, exoEasy midi kit, ExoQuick). Using these approaches, it was possible to recover 9.28e+09, 1.2e+010, and 1.53e+010 nanoparticles from 8 mL of acerola juice (Figure 1A, n = 3). From the size distribution for these methods, the average diameters were 352 ± 180, 245 ± 132, and 340 ± 172 nm, respectively, as determined using a nanoparticle tracking assay (NTA) (Figure 1B). The vesicles showed a spherical shape, as revealed by electron microscopy. With the ultracentrifugation method, the obtained particles were not uniform in size (Figure 1C). With the exoEasy midi kit, the obtained particles were uniform in size (Figure 1D). Moreover, the ExoQuick method had the highest recovery rate (Figure S1), but the obtained particles were not uniform in size, and aggregated particles were also observed (Figure 1E). Among the three methods, the use of the exoEasy midi kit was thus the best option in terms of the quality of particles collected and the time required for collection.
Figure 1.
Characterization of acerola-derived exosome-like nanovesicles (AELNs)
(A) Procedure of isolating AELNs from acerola juice. AELNs were recovered from 8 mL of acerola juice using three exosome recovery methods. (B) Every AELN collected by the three methods (green, ultracentrifugation; red, exoEasy; blue, ExoQuick; n = 3) was evaluated to determine the size distribution by nanoparticle tracking analysis (NTA). (C–E) Morphological evaluation of each AELN using negative staining by electron microscopy (C, ultracentrifugation; D, exoEasy kit; E, ExoQuick; scale bars, 500 nm [left panels], 200 nm [right panels]). (F) Localization of AELNs in acerola fruit. Enlarged photo of acerola fruit peel and seed part, and transmission electron microscope (TEM) analysis of the seed part.
To determine which tissue in the fruit AELNs were derived from, cross-sectional images of the acerola fruit obtained by transmission electron microscopy (TEM) were analyzed. As a result of carefully observing the vicinity of the peel and seed in the fruit, vesicles of several hundred nanometers similar to AELNs were observed in the space around the seed (Figure 1F, green boxes). However, it was not possible to clarify whether these were vesicles inside the cell or a substance released into the extracellular space.
Conjugation of exogenous miRNA with AELNs
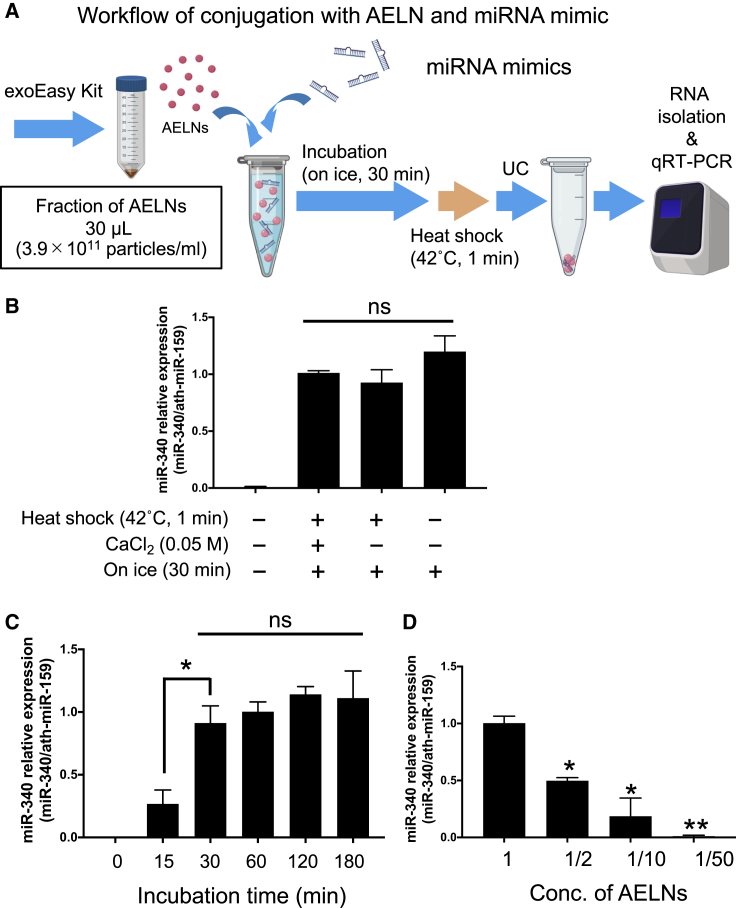
The miRNA conjugation efficiency of AELNs was examined by the following three methods. After mixing AELNs and hsa-miR-340, (1) first the cells were incubated on ice for 30 min, treated with calcium phosphate, and then treated at 42°C for 1 min (heat shock); (2) the heat shock treatment was omitted from the first method; and (3) only incubation for 30 min on ice was performed. The exosome fraction was isolated from the mixture by ultracentrifugation, and the amount of hsa-miR-340 was measured by quantitative real-time reverse transcriptase polymerase chain reaction (quantitative real-time RT-PCR) (Figure 2A). hsa-miR-340 could be detected to the same degree for the above three methods (Figure 2B). Sufficient conjugation of miRNA to AELNs occurred upon ice treatment, so we next attempted to optimize the treatment time. Upon comparison of the efficiency of conjugation between 15- and 30-min treatments, a significant difference was identified, but the efficiency did not change upon extending the incubation beyond 30 min (Figure 2C). Next, the optimum concentration of AELNs when incorporating nucleic acids was examined. AELNs serially diluted from a concentration of 3.9e+011 particles/mL were mixed with a fixed amount of nucleic acids, and ultracentrifugation was performed to collect the AELNs. Nucleic acids that could not bind to AELNs are present in the supernatant after ultracentrifugation. Nucleic acids that did not bind with AELNs were eliminated by removing the supernatant after ultracentrifugation, and the nucleic acid binding efficiency of AELNs was calculated (Figure S2). The amount of nucleic acids contained in the recovered AELNs peaked at the concentration of 3.9e+011 particles/mL, and the amount of encapsulated nucleic acids was also dependent on the concentration of AELNs (Figure 2D).
Figure 2.
Optimization of AELN and nucleic acid mixing
(A) Workflow of AELN and miRNA mixing. (1) A total of 30 μL of 1.2 × 1010 AELN particles and nucleic acid were prepared, (2) mixed by three methods (incubation on ice for 30 min plus heat shock at 42°C for 1 min, with or without 0.1 M CaCl2, and placing on ice for 30 min), (3) the ALEN-miRNA complex was pelleted by ultracentrifugation, and (4) 700 μL of QIAzol was added to the pellet to dissolve it, and then the nucleic acid (miRNA mimic bound to ALEN) was recovered. (5) miRNA was measured by quantitative real-time PCR (normalization by ath-miR-159). (B) Comparison of the methods of mixing AELNs and milk exosomes. The vertical axis represents the expression level of miR-340, which was standardized with ath-miR-159. (C) Comparing incubation time on ice, the vertical axis represents the expression level of miR-340, which was standardized with ath-miR-159. (D) Optimization of the mixing ratio of nucleic acids and AELNs (the amount of nucleic acids was constant). ∗p < 0.05, ∗∗p < 0.01. The vertical axis represents the expression level of miR-340, which was standardized with ath-miR-159.
Nucleic acid-protective effect of AELNs
To examine the stability of nucleic acids in the AELN-nucleic acid complex, nucleic acid stability was compared among cases with miRNA alone, the miRNA-AELN complex, and the miRNA-human breast milk exosome complex, which were prepared and subjected to RNase, acid (HCl; pH 2.0), and base (NaOH; pH 10.0) treatments (Figure 3A). With miRNA alone, the degradation of nucleic acids was immediately observed upon RNase, acid, and base treatments. Nucleic acid degradation was also observed for the mixture of miRNA and milk exosomes for each treatment. However, for the mixture with AELNs, the degradation of nucleic acids was inhibited for each treatment (Figure 3B).
Figure 3.
Protective effect of AELNs on miRNA
(A) The procedure: (1) mixing of nucleic acids and AELNs on ice for 30 min, (2) collecting the supernatant upon ultracentrifugation, (3) treatment with RNase, acid (HCl; pH 2.0), or base (NaOH; pH 10.0), and (4) measurement of miRNA by quantitative real-time PCR. (B) Comparison of the miRNA protective effect is shown for miRNA alone (white bar), miRNA and AELN mixture (black bar), and miRNA and milk exosome mixture (gray bar). The vertical axis represents the expression level of miR-340, which was standardized with ath-miR-159. ∗p < 0.05, ∗∗p < 0.01.
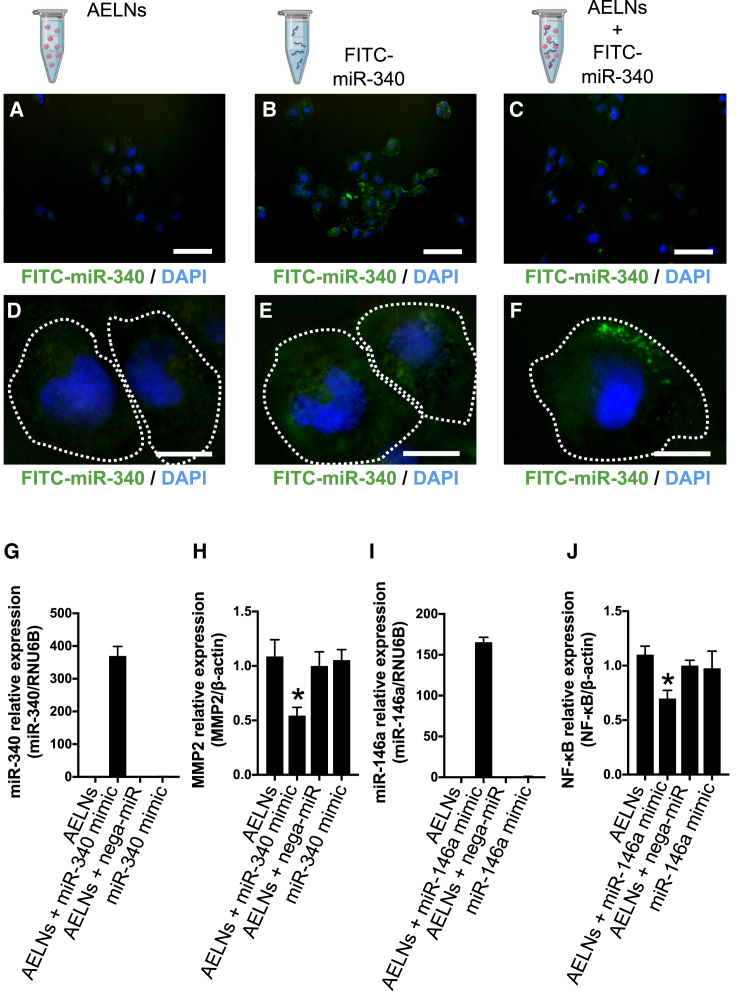
Uptake of the AELN-miRNA mimic complex into mammalian cells
Whether the AELN-miRNA complex was incorporated into mammalian cells was investigated using an in vitro system (Figures 4A–4F). Fluorescein isothiocyanate (FITC)-labeled miR-340 and the AELN complex was introduced into a cervical cancer cell line (SiHa) and then the fluorescent signal was observed under a fluorescence microscope after 24 h. The administration of AELNs alone did not produce any signals (Figures 4A and 4D). With the administration of FITC-miRNA alone, only an extracellular diffuse signal was observed (Figures 4B and 4E). However, the administration of the complex of FITC-miRNA and AELNs produced a granular signal in the cytoplasm (Figures 4C and 4F).
Figure 4.
Uptake of AELN and miRNA mixture into cells in in vitro study
(A–F) AELN alone (A and D), AELN and FITC-labeled miRNA mixture (B and E), and FITC-labeled miRNA alone (C and F) were introduced into SiHa cells. Double staining of FITC and nuclear staining (DAPI) was performed. (A)–(C) show weakly (original magnification, ×4; scale bars, 50 μm) magnified images, and (D)–(F) show strongly (original magnification, ×40; scale bars, 10 μm) magnified images. The white dashed lines in (D)–(F) indicate the cell boundaries. (G) Expression of hsa-miR-340 upon the introduction of AELNs, AELN and hsa-miR-340 mixture, hsa-miR-340, and negative control for hsa-miR-340 into SiHa cells. The vertical axis represents the expression level of hsa-miR-340, which was standardized by RNU6B. (H) hsa-miR-340 and AELNs were introduced into SiHa cells in the same manner as in (G), and the expression of MMP2, which is the target gene of hsa-miR-340, was evaluated. The vertical axis represents the expression level of MMP2, which was standardized by β-actin. (I) Expression of hsa-miR-146a upon the introduction of AELNs, AELN and hsa-miR-146a mixture, hsa-miR-146a, and negative control for hsa-miR-146a into NHDFs. The vertical axis represents the expression level of hsa-miR-146a, which was standardized by RNU6B. (J) hsa-miR-146a and AELNs were introduced into NHDFs in the same manner as in (I), and the expression of NF-κB, which is the target gene of hsa-miR-146a, was evaluated. The vertical axis represents the expression level of NFκB, which was standardized by β-actin. In (G)–(J), ∗p < 0.05.
To clarify whether the complex functioned in the SiHa cells, the amount of miRNA contained in the complex and the target gene-suppressing effect of the miRNA were examined. The expression of miR-340 was increased and the expression level of matrix metalloproteinase-2 (MMP2), its target, was decreased in SiHa cells treated with miR-340 and the AELN complex (AELNs + miR-340 mimic) compared with the control (AELNs + nega-miR) (Figures 4G and 4H). Additionally, we analyzed the expression of four genes that are not targets of miR-340. As a result, it was evaluated that none of the four genes showed any change in expression due to the addition of the AELNs + miR-340 mimic (Figure S3). Moreover, the introduction of the mixture of AELNs and hsa-miR-146a into normal human dermal fibroblasts (NHDFs) showed the upregulation of hsa-miR-146a and downregulation of nuclear factor κB (NF-κB), the target of miR-146a (Figures 4I and 4J).
The efficiency of gene transfer into luciferase-expressing reporter cells was also compared between AELNs and HiPerFect reagent, which is a liposome-based gene transfer reagent. It was found that, when the luciferase siRNA was introduced into cells overexpressing luciferase, the method using HiPerFect reagent suppressed luciferase activity more than that using AELNs (Figure S4).
In vivo pharmacodynamics of the orally administered AELN complex
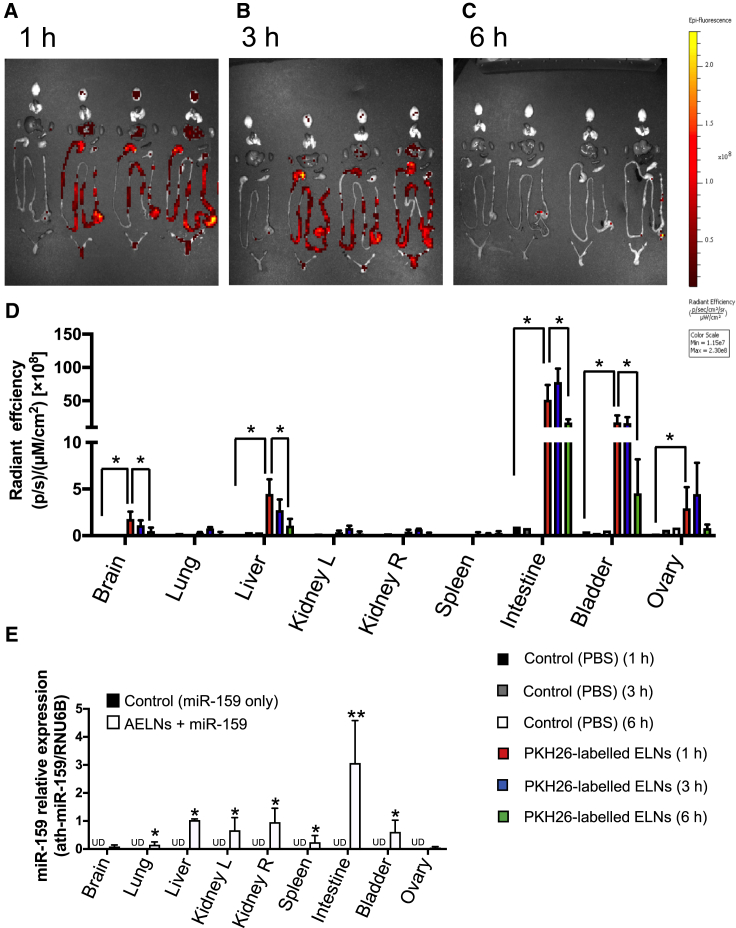
First, fluorescently labeled AELNs were orally administered and then observed using an in vivo imaging system (IVIS). One hour after the oral administration of PKH26-labeled AELNs to wild-type mice, fluorescence signals were strongly detected in the intestine, liver, and bladder (Figure 5A). Interestingly, a weak signal was also observed in the brain at the same time (Figure 5A). After 3 h, the signal of each organ decreased continuously and, after 6 h, no signal was detected (Figures 5B–5D).
Figure 5.
Distribution of labeled AELNs after oral administration
(A–C) Uptake of labeled AELNs by each organ. Left shows the signal in IVIS, while right shows the quantified degree of uptake. Oral ingestion of labeled AELNs 1 h later (A), 3 h later (B), and 6 h later (C) is shown. (D) Quantitative data for uptake of labeled AELNs by each organ. The black, gray, and white bars show PBS administration (control), and the red, blue, and green bars show PKH-26-labeled AELN administration at 1, 3, and 6 h later, respectively. (E) Detection level of ath-miR-159 upon the oral ingestion of ath-miR-159 alone or a mixture of AELNs and atr-miR-159. The vertical axis represents the expression level of ath-miR-159, which was standardized by RNU6B. The black bar shows the administration of ath-miR-159, while the white bar shows the administration of the AELN and ath-miR-159 mixture. ∗p < 0.05, ∗∗p < 0.01.
Next, to monitor the pharmacodynamics of nucleic acids, an experiment involving the administration of exogenous miRNA (ath-miR-159) was performed. One hour after the oral administration of the AELN and miR-159 mimic complex to wild-type mice, the expression of miR-159 was detected in lung and kidney, in addition to other organs (intestinal tract, liver, brain, bladder), as confirmed by the localization of AELNs (Figure 5E).
Functional analysis of the orally administered ADEN-siRNA complex
Luciferase transgenic mice were used to observe the pharmacodynamics of an orally administered mixture of AELNs and siRNA for luciferase. Twenty-four hours after the oral administration of this mixture, the luciferase signal was observed by IVIS (Figures 6A–6D). The whole-body observation revealed that the intensity of the luciferase signal decreased, while the observation for each organ revealed that the intensity of the luciferase signal in the digestive tract decreased (Figures 6E and 6F).
Figure 6.
Effect of siRNA upon oral administration
(A–D) Using luciferase transgenic mice, PBS (A and B) or a mixture of AELNs and siRNA for luciferase (C and D) was orally administered and observed 1 h later. Luciferase signals in the whole body (A and C) and in each organ (B and D) were observed by IVIS. The luciferase signal of the whole body (E) and the luciferase signal of the digestive tract and other organs (F) observed by IVIS were quantified. Black and white bars indicate PBS and a mixture of AELN and siRNA for luciferase, respectively. ∗p < 0.05.
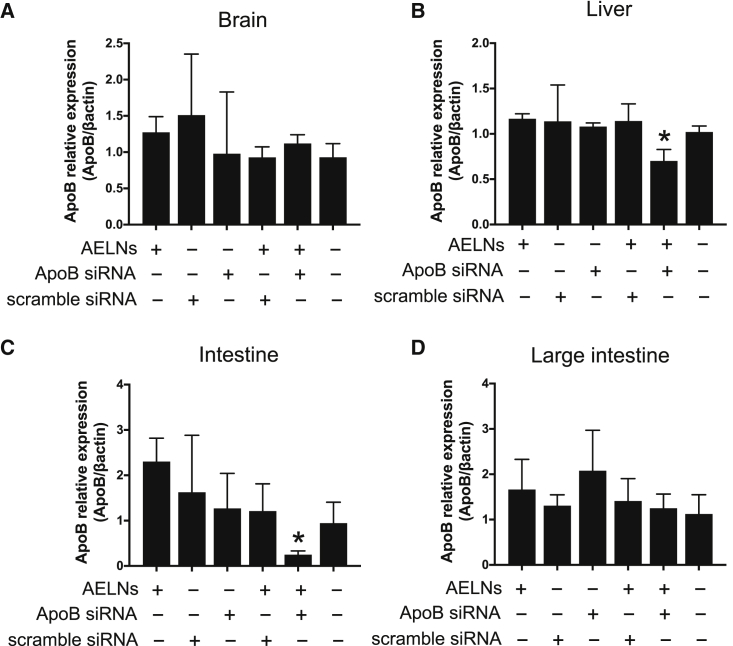
Upon the oral administration of AELNs mixed with siRNA for apolipoprotein B (Apob) to normal mice, the expression of Apob was suppressed in the liver and digestive tract (Figures 7A–7D), with a particularly pronounced effect in the small intestine (Figure 7C). However, the effect of siRNA in the large intestine was unclear (Figure 7D).
Figure 7.
Effect of siRNA upon oral administration
Normal mice were used to test the oral administration of siRNA targeting Apob that was not overexpressed. (A–D) Twenty-four hours after the oral administration of AELNs alone, scramble siRNA alone, Apob siRNA alone, a mixture of AELNs and Apob siRNA, and a mixture of AELNs and scramble siRNA, Apob expression levels in brain (A), liver (B), intestine (C), and (D) large intestine were observed. Vertical axis represents the expression level of Apob normalized to β-actin. ∗p < 0.05.
Discussion
In this study, we demonstrated the feasibility of developing a DDS involving the oral administration of AELNs. AELNs have the following key features: (1) nucleic acids encapsulated in AELNs are protected against RNase, acid, and base treatments, with greater protection than that of milk exosomes; and (2) AELNs can encapsulate any nucleic acids without the use of special reagents and enable the functions of the nucleic acids to be exerted in the digestive tract via oral administration.
It has been reported that gene transfer via exosomes has several advantages compared with viral vector-mediated gene transfer and has the potential to be introduced into gene therapy.25 As an example of gene transfer via exosomes, exosomes in which the surface membrane expressed the Apo-A1/CD63 complex were shown to enhance the efficacy of gene knockdown in a mouse model of hepatocellular carcinoma.26 It has been reported that exosome-mediated delivery is peptide-dependent and that the efficiency of delivery is similar to that of traditional transfection reagents.27
As a method for loading nucleic acids on ELNs, optimization by electroporation has been carried out. At that time, biological characteristics such as the surface charge of the ELNs were retained.17
One factor enabling ELNs to be taken up by cells is the ability of macrophages to actively take up the synthesized nanovesicles.28, 29, 30 Macrophages and edible plant-derived ELNs (EPDENs) have been shown to coexist in the lamina propria of the small and large intestine.12 Mu et al.12 showed that EPDENs exert different biological effects on the expression of genes with anti-inflammatory and antioxidant effects and activation of the Wnt/TCF4 signaling pathway.
In this study, we show that nucleic acids are incorporated into AELNs by incubation on ice without special reagents and without electroporation. It was confirmed that the charge of AELNs was negative, while when the localization of colloidal gold-labeled nucleic acids and AELNs was observed by electron microscopy, gold colloidal particles were observed on the surface of AELNs (Figure S5). From the above results, we predict that the nucleic acid is not encapsulated in the AELN, but that the nucleic acid is attached to the surface of the AELN by some physical action other than the electric charge.
Furthermore, we show that the oral administration of siRNA targeted to AELNs and Apob enabled the siRNA’s functions to be exerted in the digestive system (Figure 6). Most of the AELN-nucleic acid complex is excreted from the digestive tract after oral administration. Part of this complex was absorbed from the intestinal tract and distributed to the liver and spleen, and it was then hematogenously distributed throughout the body. In vitro epithelial and vascular endothelial models were constructed to analyze the dynamics and stability of AELN in the intestine, and permeation experiments of epithelial and endothelial layers were performed. It was revealed that some nucleic acids conjugated with AELNs are taken up by the small intestinal epithelium and vascular endothelium, but some pass through the cell layer (Figure S6). From this, it is considered that the orally administered AELN-nucleic acid complex is taken into the body (intravascular) from the intestinal tract, flows through the bloodstream, and reaches the nucleic acids and each organ in vivo.
It is suggested that the function in the liver occurs via delivery from the digestive tract via the portal vein, and the function in the digestive tract is due to the use of macrophages derived from there. Since the analysis of the expressed antigens and internal structure of AELNs has not been performed in this analysis, it cannot be clarified whether they have an affinity for a specific organ. We were able to confirm the localization in the small intestine, liver, brain, and other areas, but we would like to consider in the future whether we are targeting specific tissues depending on the nature of AELNs.
Moreover, it cannot be denied that there are fruits that are more suitable than acerola as exosome-like nanoparticles. We compared the collection method and collection efficiency with the lemons and grapefruits used in the analysis of this area (data not shown). Compared to acerola, lemons and grapefruits had thicker skins and were rich in pectin, making it very difficult to grind the fruits. Furthermore, the extracted fruit juice was also highly viscous, and when it was stored 4°C, a precipitate would form, making it difficult to recover exosome-like nanoparticles with a kit such as the exoEasy kit. In summary, it was found that acerola is very easy to recover exosome-like nanoparticles, and the vesicle recovery rate per fruit is also good.
Conclusions
With regard to drug discovery, nucleic acids have the advantages of being easier to design than proteins and small molecules and they have a low cost. We have established a method that allows some specific types of nucleic acids to be conveniently packaged in AELNs and administered orally. Because this approach involves delivery directly to the digestive tract, which has thus far been considered difficult, rather than systemic administration, it is expected to become a new method for loading nucleic acids.
Materials and methods
Ethics
All animal experiments were conducted in compliance with institutional guidelines of the Animal Experimental Center of Tokyo Medical University/Animal Biosafety Level-II Laboratory for Use of Animals. The experimental protocols were approved by the Institutional Animal Care and Use Committee of Tokyo Medical University (approval no. R2-0084).
Cell culture
Human cervical cancer cells SiHa (human papillomavirus type 16 [HPV16]+) were purchased from the American Type Culture Collection (ATCC, USA). NHDFs were purchased from Lonza (Basel, Switzerland). The cells were maintained using DMEM (Gibco, Invitrogen, Carlsbad, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified atmosphere containing 5% CO2, and the adherent cells were harvested by trypsinization. NHDFs were either passaged (passages 1–5) for expansion or subjected to analyses for in vitro experiments.
Purification of AELNs
Acerola juice was obtained from Nichirei Biosciences (Saitama, Japan). The ELNs derived from acerola juice were extracted using an exoEasy maxi kit (QIAGEN, Valencia, CA, USA) and ExoQuick reagent (System Biosciences, Mountain View, CA, USA), in accordance with the manufacturers’ protocols.
In the methods using an exoEasy maxi kit, 8 mL of acerola juice was filtered through a 0.45-μm polyvinylidene fluoride (PVDF) filter (Millipore, Billerica, MA, USA), supplemented with an equal volume of buffer XBP, mixed well, transferred into an exoEasy spin column, and subjected to treatment in line with the manufacturer’s instructions. A total of 800 μL of buffer XE was added, followed by centrifugation for 5 min at 5,000 × g, after which the flowthrough was concentrated by ultracentrifugation at 100,000 × g using a TLA-110 rotor 1 (Beckman Coulter, Brea, CA, USA) for 70 min at 4°C to change the elution buffer XE to 30 μL of phosphate-buffered saline (PBS).
In the methods using ExoQuick reagent, 8 mL of acerola juice was filtered using a 0.45-μm PVDF filter, and 2 mL of ExoQuick reagent was added and mixed thoroughly by inverting. After refrigeration for 12 h, the mixture was centrifuged at 1,500 × g for 30 min and all supernatant was removed by aspiration, after which 30 μL of PBS was added for resuspension.
Characterization of AELNs according to density and size
NTA measurements were performed using the NanoSight LM10 system (Malvern, Herrenberg, Germany) equipped with a blue laser (405 nm). AELNs were illuminated by the laser and their movement under Brownian motion was recorded in 90-s sample videos, which were analyzed with NTA 2.0 analytical software (Malvern Panalytical). All samples were diluted with PBS to reach a particle concentration suitable for analysis with NTA (1 × 108 to 2.5 × 109 particles/mL). The capture settings (shutter and gain) and analysis settings were set manually in accordance with the manufacturer’s instructions. All analysis settings were kept constant within each experiment. NTAs were averaged within each sample across the video replicates and then averaged across samples to provide total nanoparticle concentrations.
TEM
For electron microscopy analysis, AELNs were prepared, fixed with 4% paraformaldehyde and 4% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at the incubation temperature, and placed in a refrigerator to lower their temperature to 4°C. The samples were adsorbed to a 400-mesh carbon-coated grid and immersed in 2% phosphotungstic acid solution (pH 7.0) for 30 s. The samples were then observed using a transmission electron microscope (JEM-1200EX; JEOL, Tokyo, Japan) at an acceleration voltage of 80 kV.
Complex of nucleic acids with AELNs
AELNs (3.9 × 1011 particles/mL) and miRNA mimics (100 pmol, hsa-miR-340, hsa-miR-146a; Applied Biosystems, Carlsbad, CA, USA) were mixed in 50 μL of PBS with 0.1 M CaCl2. After incubation on ice for 30–120 min, the mixture was heated at 42°C for 45 s, centrifuged at 100,000 × g for 70 min, and all supernatant was removed by aspiration, after which 30 μL of PBS was added for resuspension, followed by chilling on ice until use. For the complex formation of AELNs and nucleic acids other than the first mixing experiment, each mixture was simply incubated for 30 min on ice without CaCl2 and the heat shock step was omitted. As a control experiment, human breast milk exosome (Cosmo Bio, Tokyo, Japan) was used as a substitute for AELNs to form a complex with nucleic acids in the same procedure.
Nuclease, strong acid, and strong base degradation assay
RNase, strong acid, and strong base digestion was performed by mixing the purified AELN-miRNA complex with RNase (6.25 μg/mL; QIAGEN), HCl (pH 2.0; Fujifilm Wako Pure Chemical, Tokyo, Japan), or NaOH (pH 10; Fujifilm Wako Pure Chemical), incubated at 37°C for 1 h, and then dissolved with 700 μL of QIAzol lysis reagent (QIAGEN) to estimate the amount of miRNA mimic that remained undegraded.
Uptake of the AELN-miRNA complex into cell lines
To visualize the uptake of the AELN-miRNA complex into cells, miR-340 mimic was labeled using a Label IT siRNA Tracker FITC kit (Mirus Bio, Madison, WI, USA), in accordance with the manufacturer’s instructions. NHDFs were seeded on a glass coverslip (Matsunami Glass, Osaka, Japan) and cultured with a complex of AELNs and FITC-labeled miR-340 mimic. Twenty-four hours after incubation, NHDFs were fixed in 2% paraformaldehyde for 15 min and washed twice with cold PBS. DAPI (Abbott Diagnostics, Lake Forest, IL, USA) was used for nuclear staining. Analyses were performed with a fluorescence microscope (Biozero BZ-8000; Keyence, Tokyo, Japan).
Assay for miRNA expression by quantitative real-time RT-PCR
To assess the levels of miRNAs in cells, tissues, and the AELN-miRNA complex, miRNAs were isolated using the miRNeasy mini kit (QIAGEN), in accordance with the manufacturer’s recommendations. The samples were dissolved with 700 μL of QIAzol lysis reagent (QIAGEN). After incubation for 2 min, 1 μL of 1 nM ath-miR-159 (Hokkaido System Science, Hokkaido, Japan) was added to each aliquot as a spike control for losses in preparation, followed by vortexing for 30 s and incubation on ice for 10 min. Subsequent cartridge filtration was performed in accordance with the manufacturer’s instructions.
Quantitative real-time RT-PCR was carried out on a 7900HT thermal cycler (Applied Biosystems, Bedford, MA, USA), using the manufacturer’s recommended program. TaqMan miRNA assays were used to determine the levels of individual miRNAs (hsa-miR-340, hsa-miR-146a; Applied Biosystems). Using SDS2.2 software (Applied Biosystems), cells and the AELN-miRNA complex were run in duplicate and standardized to RNU6B and ath-miR-159, respectively.
Assay for mRNA expression by quantitative real-time RT-PCR
The expression levels of MMP2 and NF-κB in SiHa cells and NHDFs were determined by quantitative real-time RT-PCR using Assay-on-Demand primers and TaqMan Universal PCR master mix reagent (Applied Biosystems). β-Actin (ACTB) was used as a normalization control. The following primers and probes were used: MMP2 (Hs01548727_m1), NF-κB (NFKB1: Hs00765730_m1), and ACTB (Hs01060665_g1).
Oral administration of PKH26-labeled AELNs
The AELNs were incubated with 2 μM PKH26 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at 25°C, and washed four times using Amicon Ultra-0.5 (100 kDa; Millipore, Billerica, MA, USA). C57BL/6J mice (Japan SLC, Shizuoka, Japan) were maintained in the experimental animal facility of Tokyo Medical University. They were kept under a 12-h light/12-h dark cycle at 22°C–24°C. Standard laboratory feed (FR-1, 30 kGy; Funabashi Farm, Chiba, Japan) and tap water were given ad libitum. The care and handling of the mice conformed to the guidelines for animal research of the National Institutes of Health. The Institutional Animal Care and Use Committee approved the experimental protocols. The mice were randomly divided into four groups (n = 3 per group): oral administration of a sample with a total volume of 200 μL (including PKH26-labeled AELNs derived from 2 mL of acerola juice; 3e+009 particles) was via a feeding tube. The fluorescence intensity was measured using an IVIS (PerkinElmer, Waltham, MA, USA) at 1, 3, and 6 h after oral administration, and the Living Image software (PerkinElmer, Waltham, MA, USA) was used to measure the fluorescence intensity in each organ.
Oral administration of the AELN-nucleic acid complex
C57BL/6 female mice (8 weeks old, n = 3−4 per cohort) were used to measure the oral administration of the ath-miR-159 (Hokkaido System Science) or ApoB siRNA (Ajinomoto Bio-Pharma, Osaka, Japan).
AELNs derived from 2 mL of acerola juice (3e+009 particles) and synthetic ath-miR-159 or ApoB siRNA were mixed in 200 μL of PBS. After a 12-h fast, oral administration was conducted with an AELN-nucleic acid complex with a total volume of 200 μL (including 10 mg/kg synthetic ath-miR-159 or the ApoB siRNA-AELN complex) via a feeding tube (Natsume Seisakusho, Tokyo Japan). The mice treated with synthetic ath-miR-159 or the ApoB siRNA-AELN complex were scarified, and each organ was collected at 1 or 24 h after oral administration. The expression of miR-159 or the ApoB gene in each organ was measured by quantitative real-time RT-PCR.
Luciferase transgenic mice, which had luciferase cDNA driven under a cytomegalovirus immediate early (CMV-IE) promoter (8 weeks old, n = 3−4 per cohort), were used to measure changes in luciferase activity after the oral administration of luciferase siRNA (Ajinomoto Bio-Pharma, Osaka, Japan). After a 12-h fast, oral administration with a sample with a total volume of 200 μL (including 10 mg/kg luciferase siRNA-AELN complex) was performed via a feeding tube. The mice treated with the synthetic luciferase siRNA-AELN complex were sacrificed, each organ was collected at 24 h after oral administration, and the luciferase activity was measured by an IVIS (PerkinElmer).
Statistical analyses
Data are expressed as mean ± SD. Two treatment groups were compared using a Student’s t test. Multiple group comparisons were performed by ANOVA. GraphPad Prism version 5c for Macintosh (GraphPad, La Jolla, CA, USA) was used for statistical analyses. Results were considered statistically significant when p <0.05.
Acknowledgments
We would like to Edanz for editing the English text of a draft of this manuscript and Akio Ishikawa and Rei Kashiwagi for experimental assistance. This study was supported in part by the Private University Strategic Research-Based Support Project (S1511011) from the Ministry of Education, Culture, Sports, Science, and Technology (Tokyo, Japan).
Author contributions
T.U., Y.M., T.O., and M.K. designed the research and wrote the paper. T.U., S.O., K.K., K.N., and S.T. performed the research and analyzed the in vitro data. T.U., M.T., and K.S. performed the research and analyzed the in vivo data.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.03.006.
Supplemental information
References
- 1.Rutter B.D., Innes R.W. Extracellular vesicles as key mediators of plant-microbe interactions. Curr. Opin. Plant Biol. 2018;44:16–22. doi: 10.1016/j.pbi.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Szempruch A.J., Sykes S.E., Kieft R., Dennison L., Becker A.C., Gartrell A., Martin W.J., Nakayasu E.S., Almeida I.C., Hajduk S.L., Harrington J.M. Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell. 2016;164:246–257. doi: 10.1016/j.cell.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svennerholm K., Park K.S., Wikström J., Lässer C., Crescitelli R., Shelke G.V., Jang S.C., Suzuki S., Bandeira E., Olofsson C.S., Lötvall J. Escherichia coli outer membrane vesicles can contribute to sepsis induced cardiac dysfunction. Sci. Rep. 2017;7:17434. doi: 10.1038/s41598-017-16363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ionescu M., Zaini P.A., Baccari C., Tran S., da Silva A.M., Lindow S.E. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. USA. 2014;111:E3910–E3918. doi: 10.1073/pnas.1414944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou Y., Zhai Y., Feng L., Karimi H.Z., Rutter B.D., Zeng L., Choi D.S., Zhang B., Gu W., Chen X. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe. 2019;25:153–165.e5. doi: 10.1016/j.chom.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Begne M., Lu B., Han X., Hagen F.K., Hand A.R., Melvin J.E., Yates J.R. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT) J. Proteome Res. 2009;8:1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheshomi H., Matin M.M. Exosomes and their importance in metastasis, diagnosis, and therapy of colorectal cancer. J. Cell. Biochem. 2018 doi: 10.1002/jcb.27582. [DOI] [PubMed] [Google Scholar]
- 8.Hu G., Drescher K.M., Chen X.M. Exosomal miRNAs: Biological properties and therapeutic potential. Front. Genet. 2012;3:56. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katakowski M., Buller B., Zheng X., Lu Y., Rogers T., Osobamiro O., Shu W., Jiang F., Chopp M. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno S., Takanashi M., Sudo K., Ueda S., Ishikawa A., Matsuyama N., Fujita K., Mizutani T., Ohgi T., Ochiya T. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woith E., Fuhrmann G., Melzig M.F. Extracellular vesicles-connecting kingdoms. Int. J. Mol. Sci. 2019;20:E5695. doi: 10.3390/ijms20225695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu J., Zhuang X., Wang Q., Jiang H., Deng Z.B., Wang B., Zhang L., Kakar S., Jun Y., Miller D., Zhang H.G. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014;58:1561–1573. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Zhuang X., Mu J., Deng Z.B., Jiang H., Zhang L., Xiang X., Wang B., Yan J., Miller D., Zhang H.G. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013;4:1867. doi: 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y., Gao J., He Y., Jiang L. Plant extracellular vesicles. Protoplasma. 2020;257:3–12. doi: 10.1007/s00709-019-01435-6. [DOI] [PubMed] [Google Scholar]
- 15.Woith E., Melzig M.F. Extracellular vesicles from fresh and dried plants-simultaneous purification and visualization using gel electrophoresis. Int. J. Mol. Sci. 2019;20:E357. doi: 10.3390/ijms20020357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rome S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019;10:529–538. doi: 10.1039/c8fo02295j. [DOI] [PubMed] [Google Scholar]
- 17.Lukasik A., Brzozowska I., Zielenkiewicz U., Zielenkiewicz P. Detection of plant miRNAs abundance in human breast milk. Int. J. Mol. Sci. 2017;19:E37. doi: 10.3390/ijms19010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao J., Feng S., Wang X., Long K., Luo Y., Wang Y., Ma J., Tang Q., Jin L., Li X., Li M. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ. 2018;6:e5186. doi: 10.7717/peerj.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X., Odenthal M., Fries J.W. Exosomes as miRNA carriers: Formation-function-future. Int. J. Mol. Sci. 2016;17:E2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z., Yu S., Li M., Gui X., Li P. Isolation of exosome-like nanoparticles and analysis of microRNAs derived from coconut water based on small RNA high-throughput sequencing. J. Agric. Food Chem. 2018;66:2749–2757. doi: 10.1021/acs.jafc.7b05614. [DOI] [PubMed] [Google Scholar]
- 21.Mezadri T., Fernández-Pachón M.S., Villaño D., García-Parrilla M.C., Troncoso A.M. [The acerola fruit: composition, productive characteristics and economic importance] Arch. Latinoam. Nutr. 2006;56:101–109. [PubMed] [Google Scholar]
- 22.Hanamura T., Mayama C., Aoki H., Hirayama Y., Shimizu M. Antihyperglycemic effect of polyphenols from Acerola (Malpighia emarginata DC.) fruit. Biosci. Biotechnol. Biochem. 2006;70:1813–1820. doi: 10.1271/bbb.50592. [DOI] [PubMed] [Google Scholar]
- 23.Nagamine I., Akiyama T., Kainuma M., Kumagai H., Satoh H., Yamada K., Yano T., Sakurai H. Effect of acerola cherry extract on cell proliferation and activation of ras signal pathway at the promotion stage of lung tumorigenesis in mice. J. Nutr. Sci. Vitaminol. (Tokyo) 2002;48:69–72. doi: 10.3177/jnsv.48.69. [DOI] [PubMed] [Google Scholar]
- 24.Horta R.N., Kahl V.F.S., Sarmento M. da S., Nunes M.F.S., Porto C.R.M., Andrade V.M., Ferraz Ade.B., Silva J.D. Protective effects of acerola juice on genotoxicity induced by iron in vivo. Genet. Mol. Biol. 2016;39:122–128. doi: 10.1590/1678-4685-GMB-2015-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niidome T., Huang L. Gene therapy progress and prospects: Nonviral vectors. Gene Ther. 2002;9:1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 26.Liang G., Kan S., Zhu Y., Feng S., Feng W., Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomedicine. 2018;13:585–599. doi: 10.2147/IJN.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 28.He C., Yin L., Tang C., Yin C. Multifunctional polymeric nanoparticles for oral delivery of TNF-α siRNA to macrophages. Biomaterials. 2013;34:2843–2854. doi: 10.1016/j.biomaterials.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 29.Coco R., Plapied L., Pourcelle V., Jérôme C., Brayden D.J., Schneider Y.J., Préat V. Drug delivery to inflamed colon by nanoparticles: Comparison of different strategies. Int. J. Pharm. 2013;440:3–12. doi: 10.1016/j.ijpharm.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Collnot E.M., Ali H., Lehr C.M. Nano- and microparticulate drug carriers for targeting of the inflamed intestinal mucosa. J. Control. Release. 2012;161:235–246. doi: 10.1016/j.jconrel.2012.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.