Abstract
Advances in sequencing technology have led to the increased availability of genomes and metagenomes, which has greatly facilitated microbial pan-genome and metagenome analysis in the community. In line with this trend, studies on microbial genomes and phenotypes have gradually shifted from individuals to environmental communities.
Pan-genomics and metagenomics are powerful strategies for in-depth profiling study of microbial communities. Pan-genomics focuses on genetic diversity, dynamics, and phylogeny at the multi-genome level, while metagenomics profiles the distribution and function of culture-free microbial communities in special environments. Combining pan-genome and metagenome analysis can reveal the microbial complicated connections from an individual complete genome to a mixture of genomes, thereby extending the catalog of traditional individual genomic profile to community microbial profile. Therefore, the combination of pan-genome and metagenome approaches has become a promising method to track the sources of various microbes and decipher the population-level evolution and ecosystem functions.
This review summarized the pan-genome and metagenome approaches, the combined strategies of pan-genome and metagenome, and applications of these combined strategies in studies of microbial dynamics, evolution, and function in communities. We discussed emerging strategies for the study of microbial communities that integrate information in both pan-genome and metagenome. We emphasized studies in which the integrating pan-genome with metagenome approach improved the understanding of models of microbial community profiles, both structural and functional. Finally, we illustrated future perspectives of microbial community profile: more advanced analytical techniques, including big-data based artificial intelligence, will lead to an even better understanding of the patterns of microbial communities.
Keywords: Pan-genome, Metagenome, Integrated pan-genomics with metagenomics, Microbial community
1. Introduction
In recent years, genome analysis of microbial organisms has gradually shifted from focusing on single selected individuals or a few genomes to the large-scale comparative analysis of a set of related isolates. Since the gene pool of a species or community is typically much larger than that of any individual strain, the genetic dynamics and diversity of the genomes of different strains in the same species cannot be represented by a single individual genome. The genomic variation observed at the species and community level leads to the expansion of the pan-genome and metagenome concepts. Whole-genome sequencing lays a foundation for the powerful strategy of identifying core and accessory genes shared among close microbes through pan-genome. Pan-genome represents the entire gene repertoire of a group of isolates (e.g. strains from one species), which can characterize the dynamics and diversity of genomes in a given taxonomy, while individual genome usually only accounts for a small part of the pan-genome [1], [2]. The pan-genome approach is typically used to evaluate the microbial genetic composition in three ways: core genome profiling, accessory genome profiling, and specific genome profiling, revealing the characteristics of homology, diversity, and specialization between genomes [3], [4]. For example, the core genome found in all individuals is often used to evaluate the relatedness between strains in the same species [5], [6]. And the pan-genome analysis has revealed extensive horizontal transfer in microbial accessory genomes [7]. Pan-genome analysis has been used to study the genetic diversity of a group of related microbial genomes, including gene composition of individual strains, strain tracking, evolutionary impact, niche specialization, antimicrobial target screening, and diagnostic marker identification [5], [8], [9], [10], [11], [12], [13]. Although pan-genomics is highly informative in profiling microbial diversity and function, it still has limitations. The widely used pan-genome analysis methods [14], [15], such as PGAT [16], PGAP [17], and Panseq [18], provide a good strain-level pan-genome profile for isolates, but they cannot resolve the species relationships at the community level. Natural microbial communities have rich biodiversity, and these pan-genome analysis methods have limitations in studying the genetic variation and interaction of communities, hence fail to capture the dynamic behavior of microbial communities. Besides, when defining the size and content of pan-genome, these pan-genome analysis methods can only be applied to a limited number of species that cannot accurately reflect microbial ecology. Overcoming these issues requires new considerations at the community level, such as cross-community evaluation of pan-genome profile.
Metagenome refers to the entire genetic content of all microorganisms in a specific environment, which has been widely used to study microbial diversity in various habitats, such as air, soil, water, plants, and humans [19]. Metagenomics is a method to characterize the taxonomic and functional diversity of microbial communities by isolating genome sequences directly from the environment without prior cultivation [20]. Metagenomics can be used for taxonomic analysis and functional analysis to track community dynamics [21]. For example, it has been used to study the shifts in microbiome composition and function of the human body such as oral, skin, gut [22], [23]. Microbial composition analysis tools, such as MetaPhlAn2 [24] and Kraken2 [25], can characterize microbial community structure in the environment and human body by identifying microbial species and estimating their relative abundance. Also, HUMAnN2 [26] estimates the abundance of microbial pathways in terms of metagenomes to detect the metabolic potential of microbial communities. Metagenomics has great advantages in the taxonomic analysis at the species level, and some taxonomic studies have even reached the strain level [27], [28]. However, due to the lack of high-quality reference genomes, a higher taxonomic resolution is still challenging. For some microbial communities in complex environments such as marine sediments and soils, due to underrepresented reference genomes in databases, it is even difficult to distinguish microbes at the species level using metagenomics methods. Besides, metagenomics methods still have limitations in analyzing genomic heterogeneity, and additional efforts are needed to identify all accessory genes and their functions in the microbial communities. Thus, additional complete genomic information is needed to characterize the microbial community in detail.
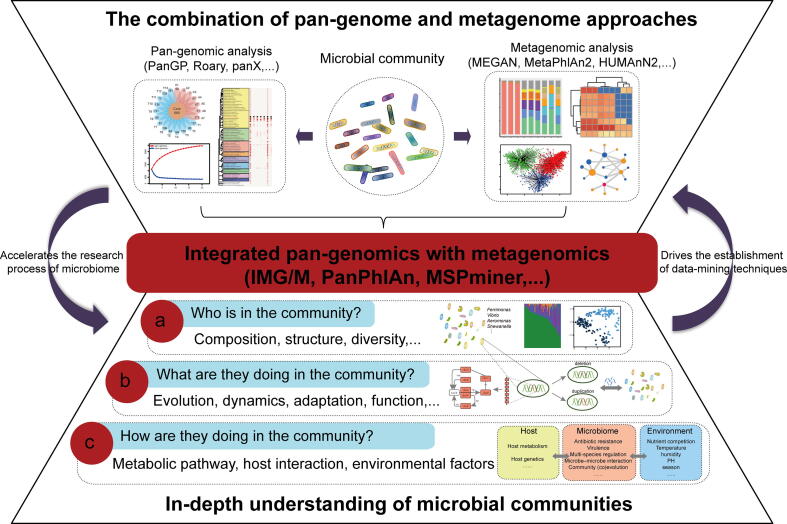
Pan-genomics and metagenomics have made independent breakthroughs in the study of microbial evolution and function in a given taxon and community respectively, but the limited number of cultivation-based microbes and the limited taxonomic resolution respectively restricts their development. Pan-genomics deciphers genomic heterogeneity and diversification of any given taxon, while metagenomics obtains the taxonomic and functional profiles in communities. Thus, combining complementary metagenome and pan-genome analysis strategies can break their limitations in the study of microbial communities, leading to an unprecedented opportunity to study the microbial interactions with their environments and hosts in communities. Recently, some microbiome studies have adopted this complementary strategy, and such integrated pan-genomics with metagenomics techniques have enabled researchers to study the diversity and dynamics of populations in microbial communities [29] (Fig. 1). Such a method takes advantage of both genome and metagenome to connect these two important genetic profiles to microbiome. For example, IMG/M [30] is a comparative data analysis system that integrates genomes and metagenomes. This system provides genome annotation such as COG clusters, Pfam, InterPro domains, and KEGG pathways. It allows comparative analysis of isolated genomes and metagenomes, including the determination of the phylogenetic and functional characteristics of individual genomes, as well as metabolic comparisons across microbial communities. PanPhlAn [31] is a tool that focuses on identifying the genetic composition of individual strains from metagenomes. This tool can characterize pan-genome and metagenome at the strain level, and determine the taxonomic profile, strain-level profile, functional profile, and phylogenetic profile from metagenomic samples. PanFP [32] is a method based on pan-genome reconstruction, which can describe the functions of KEGG Orthology, Gene Ontology, Pfam, TIGRFAMs of microbial community according to 16 s rRNA gene. Microbial communities can be explored not only by sequencing shotgun metagenome, but also by sequencing amplicon. The amplicon sequencing, such as 16S rRNA gene sequencing, is an economical and effective method for microbial abundance and diversity screening. At present, public databases have accumulated a large amount of amplicon survey data from different environments. Therefore, extending the application of amplicon survey to the pan-genome for the exploration of microbial communities is a cost-effective new method for studying complex ecosystems. Considering that 97.9% of the 16 s rRNA sequence divergence can be used as a representative of species boundaries [33], [34], pan-genome analysis with amplicon survey data may lead to hybrid genomes, so the application of amplicon survey to the pan-genome should pay close attention to the sequence identity thresholds.
Fig. 1.
The strategy that integrated pan-genomics with metagenomics for studying microbial diversity and dynamics in microbial communities. The genetic contents characterized in pan-genomes and metagenomes of the community can be collectively pooled to establish a community profile. The tools available for each workflow are shown in brackets.
Integrated pan-genome and metagenome approaches are critical to a system-level understanding of the relevant population-level genetic content of a particular habitat. In this review, we discussed the existing and new integration directions of pan-genomics and metagenomics, and highlighted the strategies and applications of such an integrative approach in the study of microbial community. This integrated strategy promotes the mechanism and model for the evolution and function of microbial communities to a higher level.
2. Microbial diversity and dynamics analyses by pan-genome approach
Microbial pan-genome analyses generally follow genomic homology-based strategies, including the assembly of individual genomes, independent annotation of genes, clustering of orthologous genes, phylogenetic relationships, gene composition, and functional characteristics. The pan-genome analysis focuses on the genomes of a group of species. The genomes can be a collection of sequences obtained by the whole-genome assembly after whole-genome sequencing, or the complete genomic sequences of a species downloaded from a public database. Series of microbial pan-genome analysis tools such as PanGP [35], Roary [36], and panX [37] have been developed for orthologous gene clustering, phylogenetic construction, and functional search. Pan-genome analysis has been carried out on the studies of diverse microbes such as Aeromonas [38], Shewanella [39] and others to handle functional dependencies and diversity among genomes. These studies have provided significant insights for the study of the evolutionary origins, niche adaptation, population structure, and relationship with health [40]. For example, Tettelin et al. characterized the pan-genome of Streptococcus agalactiae by using the genomes of eight strains [12]. This study reveals the genetic and functional diversity of Streptococcus agalactiae, in which housekeeping, regulatory cell envelope and transport genes dominate the core genome, while strain-specific genes consist of a series of dynamic genomic islands composed of atypical nucleotide. Kettler et al. revealed an open pan-genome of Prochlorococcus with a large number of different genes and tracked the diversity within and among newly discovered Prochlorococcus [41]. Besides, a pan-genome study focusing on population structure, genetic diversity and specific gene profiles associated with virulence and antibiotic resistance of Klebsiella pneumonia revealed a potential link between gene repertoire and disease [42]. These studies have shown that the size and composition of pan-genomes vary with species and their lifestyles, and this heterogeneity implies novel relationships, trends, and patterns in microbial pathogenicity, resistance, mobility, and taxonomy. In addition, these studies have detected and quantified microbial diversity that was not included in metagenome-based surveys, and many efforts are focusing on incorporating this pan-genomic view of microbial dynamics and diversity into metagenomics.
3. Taxonomic profile and microbial diversity analyses by metagenome approach
Metagenomics has been utilized for the studies of changes in community organization and microbial inhabitants, resulting in the discovery of a remarkable amount of genomic diversity and the characterization of new bacterial members [43], [44]. Metagenomics analysis explores the entire genetic composition of the microbial community by sequencing and subsequent analysis of genomic information extracted directly from environmental samples. A series of metagenomics analysis tools have been proposed, such as MEGAHIT [45], MEGAN [46], and MetaPhlAn2 [24], allowing perform metagenomics assembly, taxonomy, and functional analysis. Analyses of microbiome composition and function in different sites of the human body including the skin, oral and gut show great differences in the microbial structure [47], [48]. For example, the taxonomic representation of bacteria on human skin include Staphylococcus, Micrococcus, and Corynebacterium [49], [50], while the dominant microbes in oral are Streptococci, Lactobacillus, and Fusobacterium [51], [52]. Also, the main components of microorganisms in the human gut are Bacteroides and Prevotella [53], [54]. These microbes in human body have coevolved with their hosts, which is also related to human health and disease [48], [55]. The composition of microbes in different hosts varies greatly, and there are dynamic changes under different environmental factors [48]. For example, Sonnenburg et al. revealed a seasonal cycle of intestinal microbiota corresponding to the enrichment of functions of the Hadza hunter-gatherers, especially Bacteroides, which varies with the season, especially between the dry and wet seasons [56]. Such a study reveals the succession of microbial communities with the season in human gut. In addition, studies on microbial communities in natural environments such as soil [57], deep-sea [58], and wastewater [59] have uncovered hundreds of microbes, new genes, and uncharacterized metabolism, revealing incredible microbial diversity and complexity. However, metagenomics analysis is still hindered by the scope of the reference genome, which is far from being able to cover all the microbial variability in the community. The metagenomics analysis alone cannot reveal the extent to which genes related to the ecology and adaptability of microbes are conserved in the phylogenetic clade.
4. Strategies to integrate pan-genome with metagenome
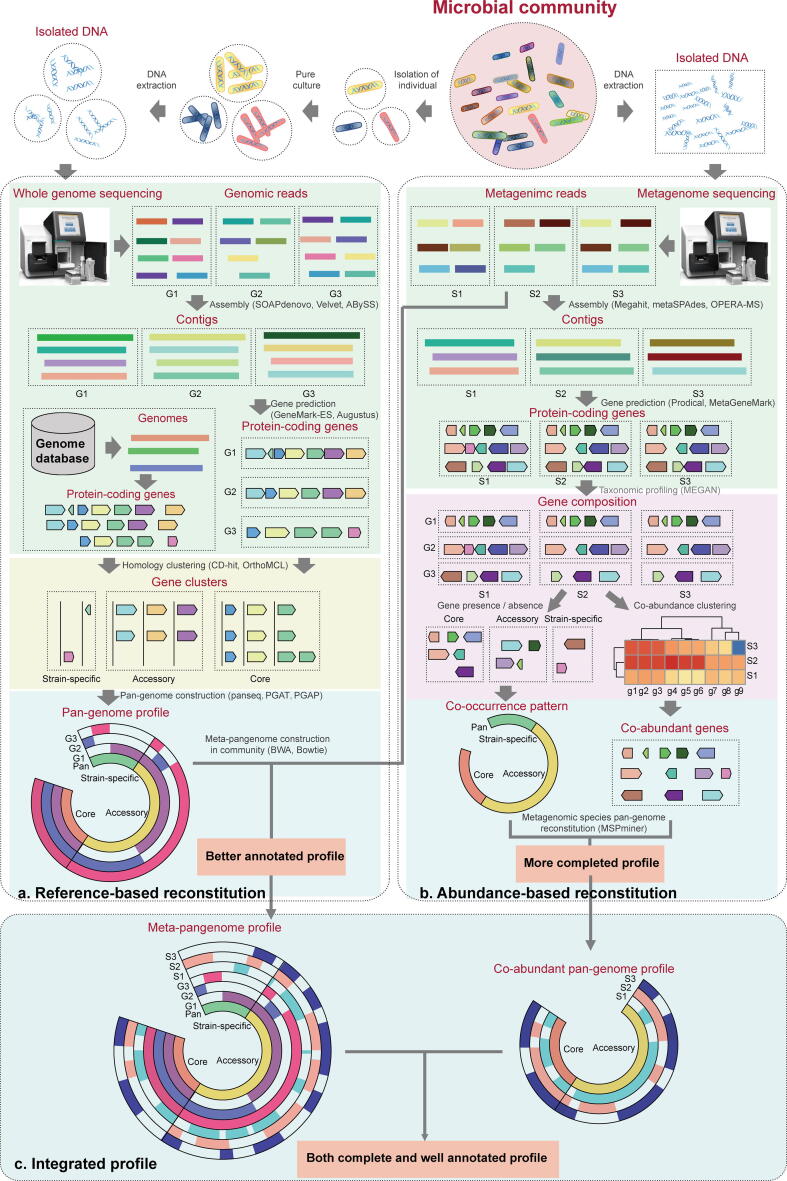
To take the next steps forward in understanding the basic biology of microbial communities, especially for human microbiota and environmental microbiota, the study of combining pan-genome and metagenome is necessary. Recently, new strategies that combine pan-genomics and metagenomics have emerged to characterize the strain-level variation and dynamics of microbial communities [29], [60] (Table 1). By linking pan-genomes and metagenomes with the appropriate resolution, the distribution of individual gene clusters across multiple microbial genomes can be extended to various environmental microbial communities [61] (Fig. 2a). This combination strategy first identifies gene clusters from environment-isolated genomes or existing genomes in the database, deduces the relatedness between genomes and constructs pan-genomes, and then in conjunction with metagenomes to track the abundance and prevalence of cross-environmental microbial genomes and individual genes by read recruitment. For example, the previous study by Delmont et al. characterized metagenome-assembled genomes according to the entire genetic content and linked genes with their environmental distribution patterns [61]. Kim et al. predicted core and strain-specific genes from metagenome-assembled genomes of multiple Bacillus species, and applied the core gene data to identify their properties in food microbial communities [62]. In addition, Farag et al. identified fragments of the Latescibacteria genomic by comparing the metagenome-assembled genome with the reference genome [63]. These studies target the core, accessory, or unique genomes from the microbial community to catalog the overall genomic and functional diversity of a specific environment. This pan-genome and metagenome combination strategy is based on the environmental connectivity of pan-genome and metagenome to recover the occurrence of populations or individual genes, which is not only applicable to genome-assembled genomes, but also for metagenome-assembled genomes. Metagenomic data recruiting pan-genomes requires low computationally demanding for high-resolution mapping, and provides a standard reference for comparison. However, these studies on the processing of metagenome data are based on the constructed pan-genome, which cannot be extended by using the abundant gene resources contained in the metagenome.
Table 1.
Examples of the application of combining pan-genome and metagenome approaches at analyses of microbial communities.
| Object | Strategy | Time | Reference |
|---|---|---|---|
| Methanobrevibacter smithii | Identify bacteria co-occurring with M. smithii and construct the M. smithii pan-genome. | 2011 | [65] |
| Escherichia coli | Use metagenomic data to resolve microbial profiles at the strain level in complex communities. | 2016 | [31] |
| Bacillus | Perform a pan-genome analysis on genomes of different Bacillus species and reconstruct their core genes from the microbiome. | 2017 | [62] |
| Bacteroides | Use metagenome abundance of reference genes to identify different subgroups engrafted from human mothers to infants. | 2018 | [66] |
| Escherichia coli | Extract gene contents and construct the pan-genomic networks from large-scale metagenomic data | 2018 | [67] |
| Prochlorococcus | Generate a pan-genome and characterize the gene content in environmental samples through metagenomic read recruitment | 2018 | [61] |
| Proteobacteria | Binning co-abundant genes across metagenomic samples to reconstitute metagenomic species pan-genome | 2019 | [64] |
| Spiroplasma | Reconstruct and compare Spiroplasma genome from metagenome using metagenomics and pan-genomics analysis strategies. | 2019 | [68] |
| Aeromonas | Construct pan-genome of 29 Aeromonas and identify the virulence genes of Aeromonas from assembly metagenomes. | 2019 | [38] |
| Wolbachia | Identify genes of Wolbachia by pan-genomic analysis and combine metagenome-assembled genomes with reference genomes. | 2019 | [69] |
| Ruminococcus, Alistipes, Eubacterium. | Identify uncultured candidate bacterial species by reconstructing metagenome-assembled genomes from human gut microbiomes. | 2019 | [70] |
| Bacteroides | Reconstruct microbial genomes from yet-to-be-named species to expand the pan-genomes of human-associated microbes. | 2019 | [71] |
| Haemophilus parainfluenzae, Rothia | Construct pan-genome and study the degree of each gene in the healthy human mouth. | 2020 | [60] |
| Lactobacillus, Gardnerella. | Identify the totality of genes belonging to a species in multiple metagenomic samplings of a particular habitat. | 2020 | [29] |
| TM7 | Assembly metagenomes and construct pan-genome of tongue-specific TM7 clades and other host-associated TM7 genomes. | 2020 | [72] |
Fig. 2.
Scheme of integrative pan-genome with metagenome studies on microbial community. (a) The gene repertoire reconstitutes of microbial species based on metagenomic read recruitment. Using pan-genome from a set of isolated genomes as a reference, reads are recruited from metagenomes to quantify relative frequency of each gene sequence in community. (b) Binning co-abundant genes obtained from de novo assembly across metagenomic samples to reconstitute metagenomic species pan-genomes. Co-abundant core or accessory genes of microbial species co-occurrence in samples and yield co-abundance. (c) Strategy for integrating pan-genome construction and metagenomic read recruitment with co-abundant genes. The tools available for each step in these workflows are shown in brackets.
The gene repertoire reconstitutes of microbial species based on metagenomic read recruitment does not consider the distribution of co-abundant genes. When accessory or strain-specific genes are considered, this read recruitment strategy has poor discrimination. Some genes in the core genome, plasmids that move between strains or operons in the accessory genome, often tend to co-occur in the genomes of multiple samples. Binning these co-abundant genes provides a framework for characterizing cross-community pan-genome. The pan-genome based metagenome approach performs homologous clustering of all genes according to the gene composition and abundance of metagenomes to determine the core and variable genes in the community, and then constructs a cross-community pan-genome for genes with the same abundance (Fig. 2b). This pan-genome based metagenome approach is reference-free to bin co-abundant genes and reconstitute cross-community pan-genomes. For example, Plaza et al. proposed the MSPminer approach based on the strategy of co-abundant genes, extending the metagenome to pan-genome, which can not only distinguish species core genes, but also accessory genes of microbial species from human gut [64]. This MSPminer method delineates pan-genomes from metagenomics datasets without referring to genomes of isolated strains, and provides microbial population genetics for profiling species without reference genomes.
The strategy based on pan-genome construction and metagenomic read recruitment gives a good profile with complete gene annotation, while the strategy based on co-abundant genes provides a profile of all genes in the community. Combining these two strategies can obtain more accurate genomic information of the population (Fig. 2c), which will be more effective in studying microbial communities.
Application of integrated approach for the in-depth understanding of microbial ecology and evolution
As already shown in Table 1, dozens of studies have been conducted based on the combination of pan-genome and metagenome approaches to analyze microbial communities. These studies have used pan-genomics and metagenomics methods to generate a union of genes from species found in their habitats. In the study of Bacillus [62] and Prochlorococcus [61], pan-genome was analyzed, and then the gene content was characterized by metagenomic read recruitment. This read recruitment method is suitable for species with high-quality reference genomes, but not for unknown components in the microbial community. Many studies, such as Wolbachia [69], Ruminococcus [70], and Bacteroides [71] study, recovered metagenome-assembled genomes from metagenomic samples, resulting in the entire genome of the species from samples collected in a given environment. These studies use the metagenomic binning strategy, which employs sequence composition characteristics and differential coverage statistics of contigs to reconstruct highly complete genomes. Then the pan-genome of genomes assembled by metagenomes were analyzed. This metagenomic binning strategy depends on the quality of the assembled genome and benefits the most abundant organisms. The study of Proteobacteria [64], binning co-abundance genes to reconstruct pan-genomes across communities. This co-occurrence model-based approach has advantages in extending the pan-genome analysis to uncultured microbial groups, but ignores the information of microbial genomes. Apart from proving that the integrated approaches are indeed feasible, these studies have also answered several key questions regarding microbial ecology and evolution, included but not limited to: taxonomic profile and diversity assessment of the community, microbial niche adaptation, microbial evolution, functional activities, and interaction networks of the community, etc.
Taxonomic profile and diversity: Natural microbial communities are usually highly diverse at multiple taxonomic levels. Metagenomics-based taxonomic profile of microbial communities has limitations in capturing information that is crucial for the accurate description of individuals, such as variation at the strain level. In the genomes of different Escherichia coli strains, the genes encoding toxins vary slightly, which can contribute to their different pathogenicity [73]. Therefore, the strain-level profile is needed, especially the gene content based on pan-genome, to identify diversity in uncultured or unknown organisms. For example, Peng et al. integrated the pan-genomes of Escherichia coli in conjunction with metagenomes for a comprehensive exploration of genetic diversity [67]. The applications of this integrated pipeline in five pathogenic strains of Escherichia coli and 760 human gut microbiomes revealed extensive genetic diversity of Escherichia coli within both isolates and the human gut microbial population. Such diversity would be undetected in a pan-genome only approach. In addition, Zou et al. constructed 1520 non-redundant genomes from more than 6000 healthy human intestinal bacteria [74]. Pan-genome analysis of 38 important species revealed the diversity and specificity of functional enrichment between their core genome and dispensable genome [74]. In addition to human gut, the integrated approaches of pan-genome and metagenome have been applied to the study of food microbiome, revealing significant differences in the genomic signature of Bacillus, and identifying Bacillus species in the food microbiome [62]. Julie et al. de novo reconstructed Wolbachia genomes from single mosquitoes and compared these four Wolbachia metagenome-assembled genomes through a pan-genome strategy [69]. By linking the genes with their abundance in the metagenome, new viral genes in the Wolbachia metagenome were revealed. These studies enable a higher-resolution description of gene pool and population structure of environmental microbes. However, the use of pan-genome to capture the taxonomic profiles and discover biomarkers of microbial communities still has certain limitations. The number of sequenced genomes is insufficient for diverse species and cannot be extended to other species in the community.
Although the sizes of most eukaryotic genomes are very large, it is still possible to assemble the genomes of eukaryotes, such as fungi, from metagenomes. Although there are more and more reports recovering eukaryotic genomes from metagenomes [75], [76], to our knowledge, these genomes assembled from eukaryotic metagenomes are susceptible to contamination by bacterial and archaeal genome fragments. Therefore, applying pan-genomics and metagenomics strategies in the community may be an effective way to identify eukaryotes.
Microbial niche adaptation: Microbes occupy different environmental niches in complex communities and interact with the surrounding environment. The combining measure of pan-genomics and metagenomics provides access to the microbial niche partitioning of microbial populations adapting to different habitats. In the pan-genomes of many bacteria such as Methanobrevibacter smithii [65] and Pantoea ananatis [77], genes existing in the accessory genome and species-specific or strain-specific genes usually participate in the process of niche adaptation. To clarify the ecology of the microbial population in the community, Delmont et al. revealed detailed pan-genomic characteristics related to occurrence patterns of Prochlorococcus populations in different marine communities [61]. This study revealed differential environmental patterns of Prochlorococcus isolates belonging to the same phylogenetic clade and identified a set of core genes that appeared on the hypervariable genomic islands, which were associated with subtle fitness trends. For these genes maintained within their niche boundaries of Prochlorococcus populations, the metagenomic recruitment approach alone does not provide access to them. Furthermore, Utter et al. combined pan-genome and metagenome to investigate the habitat adaptation and cultivar diversity of oral bacterial populations in different habitats of tongue dorsum, buccal mucosa, and supragingival plaque [78]. This study showed the different habitat-specific abundance of three distinct subgroups of Haemophilus parainfluenzae and revealed species-level taxonomy and habitat preferences of genus Rothia [78]. These results demonstrate the power of pan-genome and metagenome integration in the interpretation of microbial ecology and adaptation across communities, as well as genomic subgroups represent the niche partitioning. Because of the limited coverage of environmental metagenomes and genomes, it is still challenging to characterize all accessory genes of a given population in the environment.
Microbial evolution: The increasing availability of microbial genomes and metagenomes provides new opportunities for studying the evolutionary relationship of microbial populations. The evolutionary relationships among species constructed based on pan-genome, combined with their distribution in the community, can reveal their subtle evolutionary traces more effectively. The isolates closely related to the population of Prochlorococcus showed a subtle distribution gradient, leading to this difference involving gene clusters that affect the fitness among close members [61]. This pan-genome based identification method, in conjunction with the metagenomics analysis, can perform the taxonomic assignment with high resolution. A multi-omics study that combines pan-genomics, metagenomics, and phylogenomics, has been applied in investigating the ecology and evolution of Saccharibacteria (TM7). This study showed that TM7 has six monophyletic clades related to plaque or tongue [72]. In this analysis, tongue-specific TM7 clades are classified into other host-associated TM7 genomes, while plaque-specific TM7 clades are more closely related to environmental TM7 genomes [72]. The evolution of TM7 indicated a resemblance between the dental plaque and the non-host environment. In addition, Moran et al. used short-read mapping to estimate the abundance of genes in the pan-genome of B. uniformis in an environmental population, thereby identifying transmission events in B. uniformis population [66]. With the combination of genome-resolved metagenomics and pan-genomics, the Wolbachia genome was reconstructed from metagenome, revealing a diverse set of mobile genetic elements [69]. Incorporating pan-genome and metagenome has been applied in the context of individual microbes and microbial communities, and enables predictions of microevolution and phylogenetic resolution in microbial communities, especially the concept of horizontal gene transfer in communities. These studies have achieved more accurate phylogenetic characteristics and contributed to a more powerful analysis of the evolutionary process. However, there still exist certain limitations in using pan-genome to identify the phylogenetic profile of microbial communities. Due to the different rates of evolution of different species, species for pan-genomic phylogeny need to be carefully selected and evaluated in the community.
Functional activities and interaction networks: When combined with pan-genomics and metagenomics, the measurement of functional activity of microbial communities is more effective because it highlights the respective pathways of high abundance, low abundance, and core and accessory genes to understand how microbes interact in various environments and hosts. Typically, there are a considerable number of genes with unknown functions in the metagenome. Hence, restricting metagenomic analysis to genes with functional annotations will ignore a large proportion of genes. As a solution, the strategy of clustering and analyzing metagenome-assembled genes through pan-genome approach has been proposed. For example, combining pan-genomics with metagenome-based taxonomic and functional profile revealed a set of core genes that have high sequence diversity related to sugar metabolism in Prochlorococcus [61]. The pan-genome and metatranscriptome studies of Lactobacillus sakei revealed the homolactic and heterolactic pathways of fermenting various carbohydrates [79]. The combination of pan-genomics and metagenomics can also reveal changes in the functional activity of Aeromonas in response to different packages. For example, a series of virulence genes existed in low abundance in the community, and increased during the storage period, some of which were significant correlated with Aeromonas abundance [38]. This complementary method studies specificity of functional enrichment in microbial genomes and the diversity of core and accessory genes in community, expands the microbial functional profile, and can track the functional dynamic changes in ecological adaptation.
To understand the interaction and emergency properties of the community, interaction and network must be performed on the community. The co-abundant patterns can show how particular organisms or genes in a system occur together and vary with environmental factors [64]. By extending the pan-genome of pathogenic Escherichia coli strains to uncultured Escherichia coli, a pan-genomic network was derived from in human gut microbiomes, showing adjacency pattern among genes [67]. The detailed patterns of a particular community structure can be represented as a network, which is amenable to predicting metabolic interactions between microorganisms and hosts. These studies have also improved the understanding of how microbes, gene families and functions are distributed.
5. Conclusion and future perspectives of microbial community profile
The diversity of microorganisms at the individual level indicates the significance of cataloging a pan-genome in the community, which can be used in subsequent applications in metagenomics. Metagenomics constitutes an effective approach for studying intact microbial communities, especially when incorporating pan-genomics. In this review, we summarized the applications of the pan-genome approaches to metagenomics, for comprehensively describing the microbial communities in specific habitats and conditions. We discussed the application of the integration of pan-genomics and metagenomics in the community to characterize the genetic content in a specific environment and obtain ecologically meaningful views of different ecosystems. Extending the concept of pan-genome to incorporate metagenome has advanced our understanding of microbial diversity and metabolism.
Pan-genomics and metagenomics are complementary. Pan-genomics provides unique insights for a given taxon of microbial genomes, while metagenomics identifies the composition and metabolic patterns of microorganisms in the environment. Metagenomes allow estimating the abundance of variation of specific genes in environmental populations, thus avoiding the limitations of cultivation and genomic assembly. Extending from the traditional pan-genome concept to metagenome, it overcomes many biases related to whole-genome sequencing and provides a comprehensive description of the microbial genetic diversity in specific habitats. The combination of pan-genome and metagenome strategy can not only estimate the abundance and distribution of gene clusters in the environment, but also link them with the distribution of microbial populations, thereby providing a solution for the expansion of genome-wide conventional analysis. In addition, to clarify the basic biology of microbial communities, broader pan-genome and metagenome studies should include the regulatory relationship profile, signal transduction and interactions between microbes.
The strategy of integrating the pan-genome into the metagenome has led to the discovery of remarkable genomic diversity and the characterization of novel membership in communities. However, for genomes with incomplete metagenome assembly, this strategy should always be used with caution, as it may reduce the quality of genome repositories. Due to the limited coverage of metagenomes and genomes in the environmental communities, coupled with the inherent complexity of assembly algorithms, determining the characteristics of accessory genes in the community is still a challenging task. The integration of pan-genomes derived from metagenomes needs to be careful to avoid deviations caused by spurious expansion of gene content. Efforts need to be made to improve the accuracy of assembly, pan-genome and metagenome analysis tools, and integrate them into the study of microbial communities. In addition, future research on pan-genomes and metagenomes should not be limited to those gene sets with coding functions. The regulatory regions of genomes also have a decisive influence on microbial characteristics. The focus of future research should expand the definition of pan-genome to include non-coding sequences that may perform regulatory or other important functions. This combination strategy also can be applied to broader taxonomic groups as well as other eukaryotes such as fungus in communities. Besides, integrated amplicon-based taxonomies and pan-genomes will give new insights into complex ecosystems. Moreover, the rapid development of microbial single-cell sequencing reveals the intra-population structure or host-microbe interactions of complex microbial communities [80], and short-reading and assembly-based strategies may not be effective. The method that combines microbial single-cell sequencing, pan-genomics and metagenomics will allow us to understand complex dynamics of population, gene expression and metabolic functions of the microbial genomes and their communities.
With recent technological advances, the integration of pan-genomics and metagenomics data with other complex omics data has become increasingly popular. Applying more advanced analytical techniques to large-scale microbial datasets, including big-data analysis based on artificial intelligence, such as meta-analysis of large metagenomic datasets by machine learning and metagenomic signatures analysis by deep learning, will result in an even better understanding of the patterns and dynamics of microbial communities.
CRediT authorship contribution statement
Chaofang Zhong: Writing - original draft, Visualization. Chaoyun Chen: Visualization. Lusheng Wang: Visualization, Funding acquisition. Kang Ning: Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was partially supported by the National Key Research and Development Program of China [2018YFC0910502], the National Natural Science Foundation of China [32071465, 31871334, 31671374, 61972329] and the GRF grant CityU [11210119].
Contributor Information
Lusheng Wang, Email: cswangl@cityu.edu.hk.
Kang Ning, Email: ningkang@hust.edu.cn.
References
- 1.Muzzi A., Masignani V., Rappuoli R. The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov Today. 2007;12(11-12):429–439. doi: 10.1016/j.drudis.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Snipen L., Almoy T., Ussery D.W. Microbial comparative pan-genomics using binomial mixture models. BMC Genomics. 2009;10:385. doi: 10.1186/1471-2164-10-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mira A. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol. 2010;13(2):45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 4.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15(6):589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Lefebure T., Stanhope M.J. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8(5):R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy C.G.P., Fitzpatrick D.A. Pan-genome analyses of model fungal species. Microb Genom. 2019;5(2) doi: 10.1099/mgen.0.000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingstone PG, Morphew RM, Whitworth DE, Genome Sequencing and Pan-Genome Analysis of 23 Corallococcus spp. Strains Reveal Unexpected Diversity, With Particular Plasticity of Predatory Gene Sets. Frontiers in Microbiology, 2018. 9(3187). [DOI] [PMC free article] [PubMed]
- 8.Vernikos G., Medini D., Riley D.R., Tettelin H. Ten years of pan-genome analyses. Curr Opin Microbiol. 2015;23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Sugawara M., Epstein B., Badgley B.D., Unno T., Xu L., Reese J. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 2013;14(2):R17. doi: 10.1186/gb-2013-14-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J.-N., Kim Y., Jeong Y., Roe J.-H., Kim B.-G., Cho B.-K. Comparative genomics reveals the core and accessory genomes of streptomyces species. J Microbiol Biotechnol. 2015;25(10):1599–1605. doi: 10.4014/jmb.1504.04008. [DOI] [PubMed] [Google Scholar]
- 11.Donati C., Hiller N.L., Tettelin H., Muzzi A., Croucher N.J., Angiuoli S.V. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11(10):R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Auria G. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics. 2010;11:181. doi: 10.1186/1471-2164-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao J. A brief review of software tools for pangenomics. Genomics Proteomics Bioinf. 2015;13(1):73–76. doi: 10.1016/j.gpb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zekic T., Holley G., Stoye J. Pan-genome storage and analysis techniques. Methods Mol Biol. 2018;1704:29–53. doi: 10.1007/978-1-4939-7463-4_2. [DOI] [PubMed] [Google Scholar]
- 16.Brittnacher M.J. PGAT: a multistrain analysis resource for microbial genomes. Bioinformatics. 2011;27(17):2429–2430. doi: 10.1093/bioinformatics/btr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, et al., PGAP: pan-genomes analysis pipeline. Bioinformatics, 2012. 28(3): p. 416-8. [DOI] [PMC free article] [PubMed]
- 18.Laing C. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinf. 2010;11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Council, N.R., The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. 2007, Washington, DC: The National Academies Press. 170. [PubMed]
- 20.Rondon M.R. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66(6):2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Human Microbiome Project, C., Structure, function and diversity of the healthy human microbiome. Nature, 2012. 486(7402): p. 207-14. [DOI] [PMC free article] [PubMed]
- 22.Turnbaugh P.J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 23.Kong H.H. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truong D.T. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 25.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzosa E.A. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dilthey A.T. Strain-level metagenomic assignment and compositional estimation for long reads with MetaMaps. Nat Commun. 2019;10(1):3066. doi: 10.1038/s41467-019-10934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goltsman D.S.A. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28(10):1467–1480. doi: 10.1101/gr.236000.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma B, France M, and Ravel J, Meta-Pangenome: At the Crossroad of Pangenomics and Metagenomics. The Pangenome, 2020: p. 205. [PubMed]
- 30.Chen IA et al., IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res, 2019. 47(D1): p. D666-D677. [DOI] [PMC free article] [PubMed]
- 31.Scholz M. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13(5):435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 32.Jun S.R. PanFP: pangenome-based functional profiles for microbial communities. BMC Res Notes. 2015;8:479. doi: 10.1186/s13104-015-1462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton R.J. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol. 2007;73(22):7169–7176. doi: 10.1128/AEM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L.-X. Accurate and complete genomes from metagenomes. Genome Res. 2020;30(3):315–333. doi: 10.1101/gr.258640.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y et al., PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics, 2014. 30(9): p. 1297-9. [DOI] [PMC free article] [PubMed]
- 36.Page A.J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding W, Baumdicker F, and Neher RA, panX: pan-genome analysis and exploration. Nucleic Acids Res, 2018. 46(1): p. e5. [DOI] [PMC free article] [PubMed]
- 38.Zhong C. Comprehensive analysis reveals the evolution and pathogenicity of aeromonas, viewed from both single isolated species and microbial communities. mSystems. 2019;4(5) doi: 10.1128/mSystems.00252-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong C. Pan-genome analyses of 24 Shewanella strains re-emphasize the diversification of their functions yet evolutionary dynamics of metal-reducing pathway. Biotechnol Biofuels. 2018;11(1):193. doi: 10.1186/s13068-018-1201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deschamps P et al., Pangenome Evidence for Extensive Interdomain Horizontal Transfer Affecting Lineage Core and Shell Genes in Uncultured Planktonic Thaumarchaeota and Euryarchaeota. Genome Biology and Evolution, 2014. 6(7): p. 1549-1563. [DOI] [PMC free article] [PubMed]
- 41.Kettler G.C. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3(12):e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt K.E. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riesenfeld C.S., Schloss P.D., Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38(1):525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 44.Integrative, H.M.P.R.N.C., The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe, 2014. 16(3): p. 276-89. [DOI] [PMC free article] [PubMed]
- 45.Li D et al., MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 2015. 31(10): p. 1674-6. [DOI] [PubMed]
- 46.Huson D.H. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koren O. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Supplement_1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costello E.K. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fredricks D.N. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 2001;6(3):167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 50.Grice E.A. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dewhirst F.E. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng F. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015;18(3):296–306. doi: 10.1016/j.chom.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Costea P.I. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3(1):8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu G.D. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clemente J.C. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smits S.A. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357(6353):802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daniel R. The soil metagenome – a rich resource for the discovery of novel natural products. Curr Opin Biotechnol. 2004;15(3):199–204. doi: 10.1016/j.copbio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Mason O.U. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 2014;8(7):1464–1475. doi: 10.1038/ismej.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo J. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017;123:468–478. doi: 10.1016/j.watres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 60.Utter D et al., Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. 2020. [DOI] [PMC free article] [PubMed]
- 61.Delmont TO, Eren AM, Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ, 2018. 6: p. e4320. [DOI] [PMC free article] [PubMed]
- 62.Kim Y. Pan-genome analysis of Bacillus for microbiome profiling. Sci Rep. 2017;7(1):10984. doi: 10.1038/s41598-017-11385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farag I.F., Youssef N.H., Elshahed M.S., Löffler F.E. Global distribution patterns and pangenomic diversity of the candidate phylum “Latescibacteria” (WS3) Appl Environ Microbiol. 2017;83(10) doi: 10.1128/AEM.00521-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plaza Onate F et al., MSPminer: abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics, 2019. 35(9): p. 1544-1552. [DOI] [PMC free article] [PubMed]
- 65.Hansen E.E. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A. 2011;108(Supplement_1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yassour M. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24(1):146–154.e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng Y et al., MetaPGN: a pipeline for construction and graphical visualization of annotated pangenome networks. Gigascience, 2018. 7(11). [DOI] [PMC free article] [PubMed]
- 68.Yeoman CJ, et al., Genome-resolved insights into a novel Spiroplasma symbiont of the Wheat Stem Sawfly (Cephus cinctus). PeerJ, 2019. 7: p. e7548. [DOI] [PMC free article] [PubMed]
- 69.Reveillaud J. The Wolbachia mobilome in Culex pipiens includes a putative plasmid. Nat Commun. 2019;10(1):1051. doi: 10.1038/s41467-019-08973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almeida A. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasolli E. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaiber A et al., Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. bioRxiv, 2020: p. 2020.04.29.069278. [DOI] [PMC free article] [PubMed]
- 73.Karch H., Tarr P.I., Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005;295(6-7):405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 74.Zou Y. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37(2):179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greshake B. Potential and pitfalls of eukaryotic metagenome skimming: a test case for lichens. Mol Ecol Resour. 2016;16(2):511–523. doi: 10.1111/1755-0998.12463. [DOI] [PubMed] [Google Scholar]
- 76.Saary P, Mitchell A, Finn R, Estimating the quality of eukaryotic genomes recovered from metagenomic analysis. 2019. [DOI] [PMC free article] [PubMed]
- 77.De Maayer P. Analysis of the Pantoea ananatis pan-genome reveals factors underlying its ability to colonize and interact with plant, insect and vertebrate hosts. BMC Genomics. 2014;15(1):404. doi: 10.1186/1471-2164-15-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Utter DR, et al., Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity. bioRxiv, 2020: p. 2020.05.01.072496. [DOI] [PMC free article] [PubMed]
- 79.Kim K.H. Genomic and metabolic features of Lactobacillus sakei as revealed by its pan-genome and the metatranscriptome of kimchi fermentation. Food Microbiol. 2020;86 doi: 10.1016/j.fm.2019.103341. [DOI] [PubMed] [Google Scholar]
- 80.Woyke T., Doud D.F.R., Schulz F. The trajectory of microbial single-cell sequencing. Nat Methods. 2017;14(11):1045–1054. doi: 10.1038/nmeth.4469. [DOI] [PubMed] [Google Scholar]