Abstract
Actinobacteriophages are viruses that infect bacterial hosts in the phylum Actinobacteria. More than 17,000 actinobacteriophages have been described and over 3,000 complete genome sequences reported, resulting from largescale, high-impact, integrated research-education initiatives such as the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Sciences (SEA-PHAGES) program. Their genomic diversity is enormous; actinobacteriophages comprise many architecturally mosaic genomes with distinct DNA sequences. Their genome diversity is driven by the highly dynamic interactions between phages and their hosts, and prophages can confer a variety of systems that defend against attack by genetically distinct phages; phages can neutralize these defense systems by coding for counter-defense proteins. These phages not only provide insights into diverse and dynamic phage populations but also have provided numerous tools for mycobacterial genetics. A case study using a three-phage cocktail to treat a patient with a drug-resistant Mycobacterium abscessus suggests that phages may have considerable potential for the therapeutic treatment of mycobacterial infections.
Keywords: bacteriophage, mycobacterium, phage therapy, genomics
INTRODUCTION
Actinobacteriophages are viruses that infect bacteria in the phylum Actinobacteria. The Actinobacteria encompass a large group of gram-positive bacteria found both terrestrially and aquatically and are prominent components of soil microbiomes (1). Many species are pathogens of humans or animals, although the Actinobacteria are especially rich in bacteria that produce commercially useful antibiotics (2). The Actinobacteria span six major classes, of which the class Actinobacteria is perhaps the largest and the most important, including the pathogenic Mycobacteria and the antibiotic-producing Streptomyces. Most (but not all) have characteristically high G+C% genomes. The bacteriophages of these fascinating and important bacteria can reveal key insights into viral diversity and evolution while also providing tools for genetic analysis and clinical utility (3).
Actinobacteriophage:
bacteriophage that infects bacterial hosts in the phylum Actinobacteria
Phage(s):
the plural “phages” refers to more than one type of phage; the plural “phage” refers to many phage of the same type
The phages of the mycobacteria have been the primary focus among the actinobacteriophages, which were first characterized in the 1950s (4). A key motivation for their characterization was their potential utility for typing Mycobacterium tuberculosis, although the relatively fast-growing nonpathogenic M. smegmatis was often used as a surrogate for phage isolation and characterization (5, 6). This strategy has well stood the test of time. Of the 10,000 individual phages isolated using M. smegmatis strain mc2155, over 1,800 have been sequenced and annotated; this remains the best-characterized collection of phages on any single bacterial host strain (7). New types of phages infecting M. smegmatis continue to be discovered, but finding new genomes and genes is much less common than it was even a few years ago. As interest has turned to phages of other actinobacterial hosts, it is becoming clear that the diversity of phages of many other actinobacteria is at least as great as that of the mycobacteriophages. Moreover, the comparative genomics of phages of closely related bacteria provides new insights into pathways of their evolution.
Several reviews on mycobacteriophages have described different aspects of their genomics and utilities in the past few years (8–12). The last review on mycobacteriophages published by Annual Reviews was 10 years ago (8). At that time, the 70 completely sequenced mycobacteriophage genomes provided marvelous insights into these creatures. With 1,800 sequenced mycobacteriophages and 1,200 sequenced phages of other actinobacterial hosts, the landscape of diversity has changed considerably (7). Moreover, this remarkable collection has provided new insights into the microbial dynamics that drive the evolution of an enormously diverse viral population and ways in which they might find therapeutic potential. These newer findings are the focus of this review.
ACTINOBACTERIOPHAGE GENOMICS
Integrated Research-Education Programs for Phage Discovery
The past 10 years have seen a dramatic increase in the number of sequenced actinobacteriophage genomes (a greater than 40-fold increase), resulting from both enhanced simplicity and reduced sequencing costs, together with the development of integrated research-education programs (enrolling students are sometimes referred to as phage hunters). The first of these was the Phage Hunters Integrating Research and Education (PHIRE) program starting in 2002, which provided authentic research experiences to undergraduate and high school students with a local focus in Pittsburgh, PA (13, 14). In 2008, the Howard Hughes Medical Institute–supported Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Sciences (SEA-PHAGES) program was launched, in which participating institutions offer a phage discovery and genomics course for early-career undergraduates (15). SEA-PHAGES is an example of an Inclusive Research Education Community (Figure 1) in which a centralized programmatic infrastructure supports implementation at each component college and university (16). This enables implementation at a large scale. At the time of writing (fall 2019), there are 147 participating institutions—ranging from community colleges to R1 research universities—and over 5,000 enrolled students.
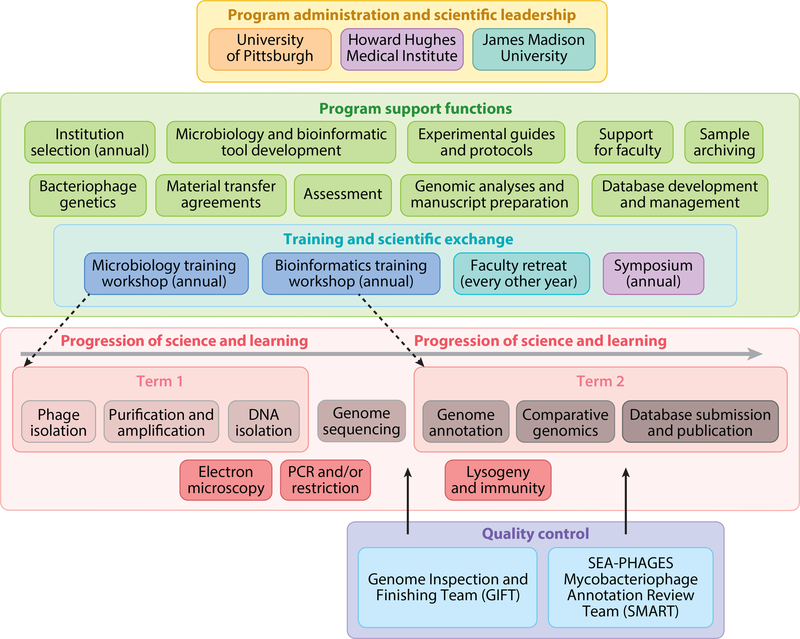
Figure 1.
Organization and structure of the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Sciences (SEA-PHAGES) program. SEA-PHAGES program administrators (yellow box, top) oversee support components critical to program implementation (green box, upper middle). The typical two-term course structure (pink box, lower middle) includes phage isolation through comparative genomics; additional characterization includes electron microscopy and PCR/restriction analysis. Sequence and annotation quality control are shared with SEA-PHAGES faculty teams.
The outcomes of the SEA-PHAGES program are impressive, with clear evidence of enhanced student persistence in science and strong contributions to bacteriophage discovery and genomics (16). Students collect environmental samples, extract with a simple buffer, and use either direct plating or enrichment to discover phages that form plaques on a specific bacterial host. The substantial diversity of the phage population enhances the prospects of students identifying a novel phage (i.e., one that has not been previously described), which they then name and characterize (16). The phage is purified and amplified, and the virion morphology is determined by electron microscopy. Phage genomic DNA is extracted, analyzed by restriction and gel electrophoresis, and sequenced. The genome is then analyzed bioinformatically, annotated, compared to other phage genomes, and submitted to GenBank. Detailed protocols for all steps are available at https://phagesdb.org and https://seaphages.org. Typically, the program is implemented as a two-term research laboratory class targeted at first-year undergraduate students (~4 h/week), with microbiology and bioinformatics being the foci of the first and second terms, respectively (16). The combination of rich scientific discovery and involvement of prospective researchers lacking prior technical expertise or content knowledge not only makes the overall platform fully inclusive but also lends it to implementation in many configurations. One such alternative implementation is the Mycobacteriophages Genetic Course, an intensive 2-week workshop in Durban, South Africa, from 2008 to 2018.
Bacterial Hosts and Host Ranges
As of November 2019, the total number of isolated actinobacteriophages was 17,323, of which 3,055 are fully sequenced. These bacterial host strains span 14 genera, 70 species, and 110 individual strains (Supplemental Table 1). The representation of phages isolated on these various strains is heterogenous and ranges from ~1,800 sequenced phages of M. smegmatis to fewer than a half dozen for about 50% of the represented species (Supplemental Table 1). However, there are 50 or more sequenced phages for seven of the genera. In general, there is substantial diversity of the phages for all the bacterial species, with the notable exception of Cutibacterium acnes (formerly Propionibacterium acnes), for which the phage diversity appears to be quite restricted (17, 18); although few phages of Propionibacterium species such as P. freudenreichii have been described, these seem to be more varied (19).
The advantage of constraining phage characterization to bacteria within a single phylum is that it enhances the prospects of learning about their evolutionary pathways. In general, the host ranges of these phages are narrow and typically do not extend to other host genera—with some exceptions (20)—and commonly do not extend to other species within a genus (21); phages may also discriminate between strains within a single species (21, 22). It is thus not surprising that phages isolated on strains of one actinobacterial genus typically are not related to phages isolated on other actinobacterial genera, although this is discussed further below.
Actinobacteriophage Virion Morphologies
All of the actinobacteriophages examined to date contain double-stranded DNA (3). Until recently, all were also tailed phages, with examples of siphoviruses, myoviruses, and podoviruses (23–25) (Figure 2). Curiously, of the more than 1,000 mycobacteriophages that have been examined by electron microscopy (https://phagesdb.org), all of these are siphoviruses and myoviruses; no podoviruses have been identified (3). Moreover, the mycobacteriophage myoviruses are confined to a single genomic group (Cluster C). The reason for the lack of mycobacteriophage podoviruses is unclear but likely reflects a physical constraint imposed by the complex cell wall that includes a mycobacterial outer membrane composed of mycolic acids (26). Podoviruses have been described for other Actinobacteria such as Arthrobacter (24), so it is unlikely that their absence from mycobacteriophages reflects a lack of evolutionary opportunity (Figure 2). The vast majority of the actinobacteriophages have isometric heads—with diameters ranging from 40 nm to 80 nm—although some have prolate heads, with length:width ratios ranging from 2.5:1 to 4:1 (27). Prolate-headed phages have been described for Mycobacterium, Microbacterium, and Gordonia hosts and may reflect evolutionary opportunities to expand capsid volume and thus genome length, facilitating acquisition of additional genomic segments (Figure 2).
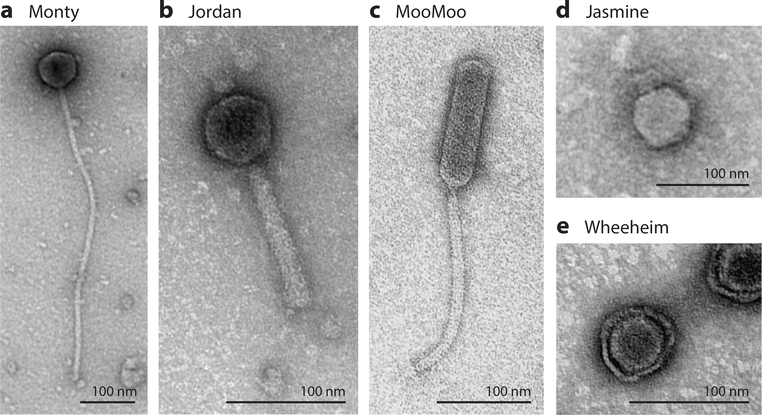
Figure 2.
Actinobacteriophage virion morphologies. Electron microscope images are shown for phages (a) Monty, (b) Jordan, (c) MooMoo, (d) Jasmine, and (e) Wheeheim. Monty and MooMoo are examples of siphoviral morphotypes with long flexible tails, but MooMoo has a prolate (elongated) head, whereas Monty has an isometric head. Jordan, Jasmine, and Wheeheim are exmplaes of myoviral, podoviral, and tectiviral morphotypes.
Recently, nontailed phages have been described for both Rhodococcus opacus (phage Toil) and Streptomyces scabiei (phages Forthebois and Wheeheim), which have lipid-containing virions and are members of the Tectiviridae (28, 29) (Figure 2). Although these are not closely related to each other or to Tectiviridae of Escherichia coli (e.g., phage PRD1) or Bacillus thuringiensis (e.g., phage Bam35), all of these share common features in addition to their capsid compositions. They have similar genome sizes (14–18 kbp), all have terminal proteins covalently attached to their genomes, and all have short (24–110 bp) inverted terminal repeats.
Groupings into Clusters, Subclusters, and Singletons
Bacteriophage genomes are characteristically architecturally mosaic with the mosaic units often being single genes (23, 30, 31). Consequently, genomic comparisons identify numerous examples of gene homologs (often sharing relatively low amino acid sequence identities) in otherwise unrelated genomes and flanked by unrelated sequences (3). It has been proposed that this arises predominantly through nonsequence-directed illegitimate recombination events and selection for function, rather than sequence-directed events (30, 31); transposition and site-specific recombination processes likely contribute to this mosaicism (9). Homologous recombination (between common sequences) plays a large role in reassembling gene combinations but does not directly create new gene boundaries. Not surprisingly, genomic mosaicism substantially confounds phage taxonomy, creating fuzzy divisions between different groups of phages, and it is likely that there is an underlying continuum of diversity, albeit with unequal sampling and unequal representation (23). Thus, although it has proven useful to place actinobacteriophages into groups, they are groupings of convenience more than a reflection of inviolable biological divisions (23, 25).
Cluster:
group of bacteriophages related to each other and sharing at least 35% of genes with at least one other cluster member
Subcluster:
subset of phages more closely related to each other than to other cluster members
Singleton:
phage with no other close relatives
The sequencing of the first few mycobacteriophage genomes revealed that they often share little or no nucleotide sequence similarity and thus could be readily placed into distinct clusters (i.e., Clusters A, B, C, etc.) (30, 32–34). Some of these clusters have distinct subgroups (e.g., differing in Average Nucleotide Identity values) and can be divided into subclusters; phages with no close relatives are referred to as singletons (33, 34). Initially, phages were placed in the same cluster if their genomes shared nucleotide similarity spanning greater than 50% of their genome lengths, a convenient threshold value that was rarely encountered (33, 34). With greatly increased numbers of sequenced actinobacteriophage genomes, this threshold has presented challenges and has been revised such that cluster membership requires an average of 35% shared genes (25). This can be readily determined by sorting actinobacteriophage predicted gene products into groups of related sequences (phamilies or phams) that are displayed using the program Phamerator (https://phamerator.org) (35). A tool is available at http://phagesdb.org to calculate the proportion of pairwise shared phams and to determine predicted cluster designations for newly identified phages (7).
A common nomenclature has been deployed for cluster designation of all of the actinobacteriophages regardless of their host. The total number of mycobacteriophage clusters is currently 29, and they are labeled Clusters A–Z and AA–AC (together with 10 singletons). Phages of other hosts are designated with two-letter blocks that can be increased as needed. Further expansion of host and phage discovery will likely require three-letter blocks, and so on. Table 1 shows a list of current cluster assignments according to host.
Table 1.
Actinobacteriophage cluster allocations and designations
| Host genus | Cluster allocation (number)a | Current designationsb,c | Number of phagesd |
|---|---|---|---|
| Mycobacterium | A-Z, AA-AJ (36) | A-Z, AA-AC (29); 10 singletons | 1,795 |
| Arthrobacter | AK-AZ, FA-FZ (42) | AK-AZ, FA-FG (23); 7 singletons | 270 |
| Streptomyces | BA-BT (20) | BA-BO (15); 12 singletons | 217 |
| Propionibacterium | BU-BZ (6) | BU-BX (4) | 55 |
| Rhodococcus | CA-CC, CE-CP (15) | CA-CC (3); 13 singletons | 55 |
| Gordonia | CD, CQ-DZ (36)e | CDf, CQ-DW (32); 9 singletons | 282 |
| Microbacterium | EA-EM, GA-GM (26) | EA-EM, GA-GC (16); 7 singletons | 229 |
| Corynebacterium | EN-EZ (13) | EN-EP (3); 2 singletons | 21 |
Blocks of letters are allocated to phages isolated on a particular host genus.
Current cluster designations assigned are shown, with the number of singletons.
Designations and assignments as of September 2019.
Numbers of sequenced phages isolated on that host genus.
There is not a strict segregation of phages into new clusters based on isolation host; e.g., some phages isolated on Gordonia group in Cluster A, which is predominantly populated by mycobacteriophages.
The first phage isolated in Cluster CD was originally described as a Rhodococcus phage, but it and all other Cluster CD phages have been confirmed as Gordonia phages.
Actinobacteriophage Lifestyles
The actinobacteriophages can be generally grouped into those that are obligatorily lytic and those that are temperate, the latter defined as those forming visible plaques on a bacterial lawn but that also form stable lysogens. The former often are referred to simply as lytic phages and produce characteristically clear plaques on a bacterial lawn. The temperate phages form visibly turbid plaques reflecting the two outcomes of infection: lytic growth for phage replication and lysis, and formation of stable lysogens that are immune to superinfection [as described for the lambda prototype (36)]. Lytic and temperate phages can often be distinguished bioinformatically, with temperate phages commonly coding for repressor and integrase genes. Phage lifestyles correlate closely with cluster designation, and phages within a cluster have similar properties (7). Nonetheless, within clusters of temperate phages, it is not uncommon for some naturally occurring isolates to be lytic, having lost the ability to establish lysogeny (37); whether these clear plaque variants were present in the initial environmental sample or were selected during isolation is unresolved.
Phamily:
group of mycobacteriophage genes related to each other, according to amino acid sequence relatedness
Of the current 125 actinobacteriophage clusters, 51 (40%) are temperate and 74 (60%) are lytic (7). However, this distribution varies considerably depending on the host. For Mycobacterium and Gordonia phages, 50% of clusters are temperate, whereas 40% and 30% of the Streptomyces and Arthrobacter clusters, respectively, are temperate. However, among the Microbacterium phages, only a lone singleton is temperate (https://phagesdb.org); the other 16 clusters and 6 singletons are all lytic. The reason for the dearth of Microbacterium temperate phages is unclear, but it appears to be a genus-specific rather than a species-specific phenomenon, as the phages were isolated on ten different strains corresponding to nine different species of Microbacterium (7) (Supplemental Table 1).
The designation of phage lifestyle is sometimes ambiguous. Plaque morphology alone can be misleading, as some temperate phages form lysogens at relatively low frequencies [for Cluster G phages it is typically <5% (38)], and the plaques are not evidently turbid unless incubated for extended periods of time. However, lysogeny can be readily observed by survival of 1–5% of cells following infection, whereas for a lytic phage, the survivors are typically phage-resistant mutants arising at frequencies of <10−6. Bioinformatically, repressors cannot always be readily predicted, although a divergently transcribed pair of small genes with DNA binding motifs (equivalent to cI and cro in lambda) is remarkably common. Integrase genes can be easily identified—and many tyrosine-integrase and serine-integrase members are present—although some temperate phages encode a partitioning system for extrachromosomal maintenance rather than an integrase (39) (Figure 3).
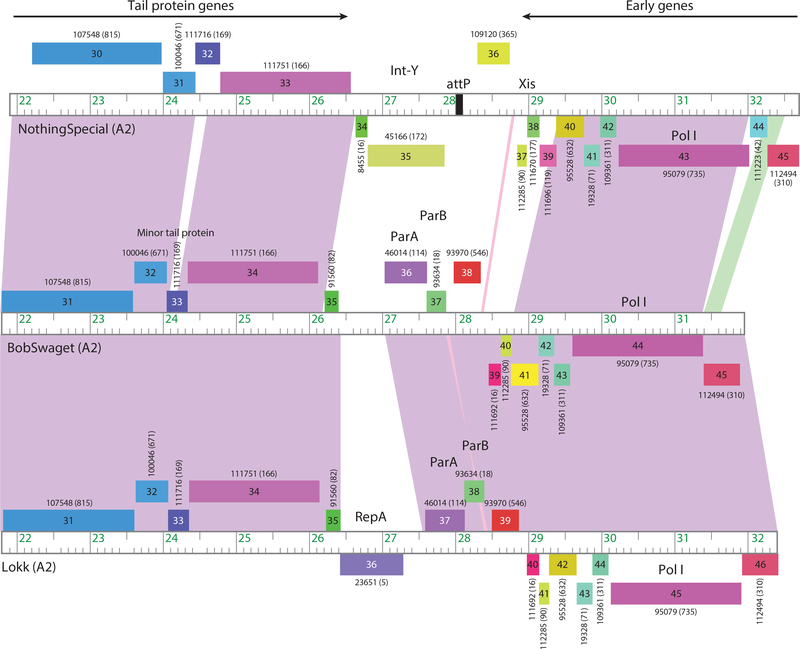
Figure 3.
Temperate phage systems for prophage maintenance. The central parts of three Cluster A2 phage genomes are shown that vary in their prophage maintenance mechanisms. Rightward- and leftward-transcribed genes are shown as boxes above and below the genome markers, respectively, with gene numbers shown in the boxes. Numbers above or below the genes indicate their assignment into gene phamilies, with the numbers of phamily members shown in parentheses. Pairwise nucleotide similarity is indicated as spectrum-colored shading between genomes, with violet reflecting closest similarity. The ends of the structural gene operons (tail protein genes) and the early gene operons are indicated. Between these, phage NothingSpecial has an integration cassette including a tyrosine-integrase (Int-Y) and attP site, whereas phages BobSwaget and Lokk have partitioning cassettes encoding parAB genes. Lokk also codes for a RepA-like protein for extrachromosomal prophage replication, whereas BobSwaget likely replicates using an alternative mechanism.
Curiously, no lytically replicating phages of the gut actinobacteria Bifidobacteria have been described. However, genome comparisons have identified several different prophages, some of which are mitomycin C inducible (40). These prophages—integrated in the host dnaJ2 gene—undergo excision, replication, and viral assembly following induction and presumably are fully competent to replicate lytically, although permissive hosts have yet to be identified. Interestingly, several of the phages have a putative phase variation shufflon system that generates variations in a tail-associated receptor binding protein (40).
Overall Genomic Diversity
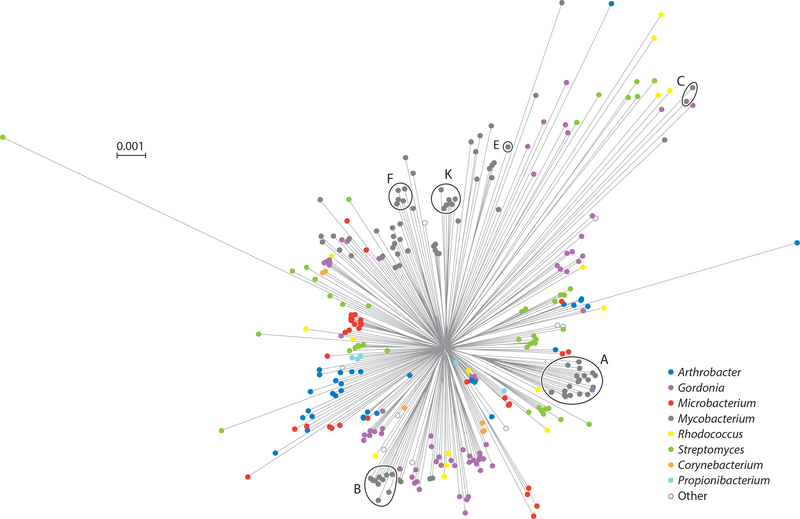
The genomic diversity of the actinobacteriophages is substantial. It is notable that there are only two known instances of phages with exactly the same genome sequences being isolated twice—at different locations and times, ruling out the possibility of cross contamination (although there are several examples of students working at adjacent benches isolating the same phage!). All the other phages are different from each other, with differences ranging from a few nucleotide substitutions and insertion/deletion of one or more genes within otherwise near-identical genomes, to shared segments of very close nucleotide similarities spanning near 50% genome length, to sharing of few if any amino acid sequence motifs. A network phylogeny based on shared gene content (Figure 4) reflects the overall diversity, with generally long branch lengths between phages of different clusters, subclusters, and singletons.
Figure 4.
A network phylogeny of actinobacteriophages. A randomly chosen phage from each subcluster and nonsubclustered cluster together with the singletons were compared by their gene contents and the relationships displayed using SplitsTree (132). Each phage node is indicated by a colored circle indicating the genus of the bacterial host used for isolation. Clusters containing 100 or more individual phages are circled and the cluster indicated.
There are striking differences in the representations of different cluster members. For example, of the ~1,800 sequenced actinobacteriophages, over 600 (35%) are in Cluster A, and the intracluster variation is substantial with 20 different subclusters (7) (Figure 4); six clusters (A, B, C, E, F, and K)—all mycobacteriophages—have more than 100 individual members (Figure 4). Cluster A phages are also relatively isolated, sharing few genes with other mycobacteriophages, notwithstanding one subcluster (A15) that is solely composed of Gordonia phages (Figure 4). In contrast, there are nine clusters with six or fewer members and ten singletons. This heterogenous representation confounds predictions for the total genetic diversity, and ongoing sampling of individual phages only rarely reveals either new mycobacteriophage singletons or relatives of extant singletons. The large portion of isolated but unsequenced mycobacteriophages in the collection has been explored using a Deconvolution Of Genomes by En Masse Sequencing (DOGEMS) strategy in which pools of phages are sequenced and deconvoluted by PCR, identifying one new singleton (Kumao) and several members of poorly represented clusters. This strategy has also been useful for sampling all the phages (20–40) isolated by a given class of SEA-PHAGES students, when only one or two have been fully sequenced.
Gordonia phages represent 41 clusters+singletons from 382 sequenced phages, whereas 1,800 sequenced mycobacteriophages span 39 clusters+singletons (Table 1). Thus, the diversity of Gordonia phages is at least that of the mycobacteriophages. Also, the Gordonia phage sampling is somewhat less heterogenous and more even within clusters, with the largest Gordonia cluster having only 44 genomes (~12%). Although there are fewer phages of Arthrobacter, Microbacterium, and Streptomyces, these all have one cluster (AK, EA, and BD, respectively) that is overrepresented relative to the others. It is noteworthy that whereas the mycobacteriophages were predominantly isolated on a single host strain (M. smegmatis mc2155), the Gordonia phages were isolated on 16 strains representing nine different species (7, 25). Likewise, the phages of Arthrobacter, Microbacterium, and Streptomyces also were isolated on multiple strains/species (5/4, 10/9, and 31/25, respectively). In general, there is not a close correlation between cluster assignment and the host strain used for isolation (24). The implications of this are substantial, as it suggests that it will be necessary to characterize much larger numbers of phages on different host species within a genus to understand diversity. Moreover, the mapping of phages isolated on different hosts to the same cluster could indicate the relative ease with which host range preferences change during their evolution. For example, currently there are five Gordonia phages grouped in Cluster CW, but these were isolated on G. malaquae, G. neofelifaecis, and G. rubripertincta, and these phages may be able to transition between these hosts quite readily. Finally, this also suggests that the spectrum of phages infecting hosts within the genus Mycobacterium is vastly greater than those isolated using the single strain M. smegmatis mc2155 as a host, and deeper exploration using other mycobacterial strains is warranted.
Evolutionary Perspectives
What drives the enormous bacteriophage diversity? Genome mosaicism likely arises by either replication errors or recombination occurring between sequences lacking extended sequence similarity (31, 41). Phage-encoded RecET-like systems may mediate these events (41–43) and are present in genomes in Clusters G, I, N, P, T, DC, DE, DW, EP, AO, AR, AS, BV, BW, CV CY, CZ, and DC; these phages represent most of the bacterial hosts, with the notable exception of Streptomyces. Reassortment of genes can be mediated by RecA-like proteins, and these can also be phage encoded (30). Many actinobacterial hosts also have nonhomologous end joining (NHEJ) systems that may facilitate nonsequence-dependent DNA rearrangements, and some phages code for Ku-like proteins playing roles in NHEJ-mediated recircularization of genome ends following DNA injection during infection (44). Together, these mechanisms create highly mosaic genomes in which it is common for boundaries between shared and nonshared nucleotide sequences to coincide with gene boundaries (see Figure 3). These likely arise from illegitimate recombination events coupled with selection for gene function.
The dynamic effects of phage resistance and phage coevolution can drive phage host-range evolution and thus contribute to the overall diversity (21). Phages migrate across this diverse bacterial landscape, pursuing numerous routes across species- and strain-rich environments, sampling different segments of the larger gene pool. Reconstructing these pathways is currently challenging because of the dearth of phage genome sequences for the vast majority of bacterial genera, species, and strains. However, an illuminating example is provided by examination of phage Patience, an M. smegmatis phage with G+C% content (50.3%) substantially below its host (67%). The Patience codon usage preferences are widely different from M. smegmatis, making it quite likely that it recently migrated from moderate G+C% hosts into the Mycobacterium neighborhood (45). Proteomic characterization suggests that highly expressed genes are more rapidly evolving to have codon usage profiles corresponding to those of the host (45).
Comparison of the rates of gene acquisition/loss and nucleotide divergence suggests that there are at least two distinct evolutionary modes for bacteriophages (46). The two modes differ in the rates of horizontal gene exchange relative to nucleotide distance, with a high gene content flux (HGCF) mode in which gene exchange is tenfold greater than in the low gene content flux (LGCF) mode. Lytic phages predominantly use the LGCF mode, whereas temperate phages distribute between the HGCF and LGCF modes (46). The basis for the difference between the two modes is unclear, but it suggests that prophages offer a reservoir of genetic information undergoing frequent exchange. It is noteworthy that phages of different bacterial hosts also vary in their use of the HGCF and LGCF modes (46). Because both overall diversity and evolutionary modes can vary substantially, different perspectives on phage evolution can easily arise depending on which parts of the phage world are examined (23, 47).
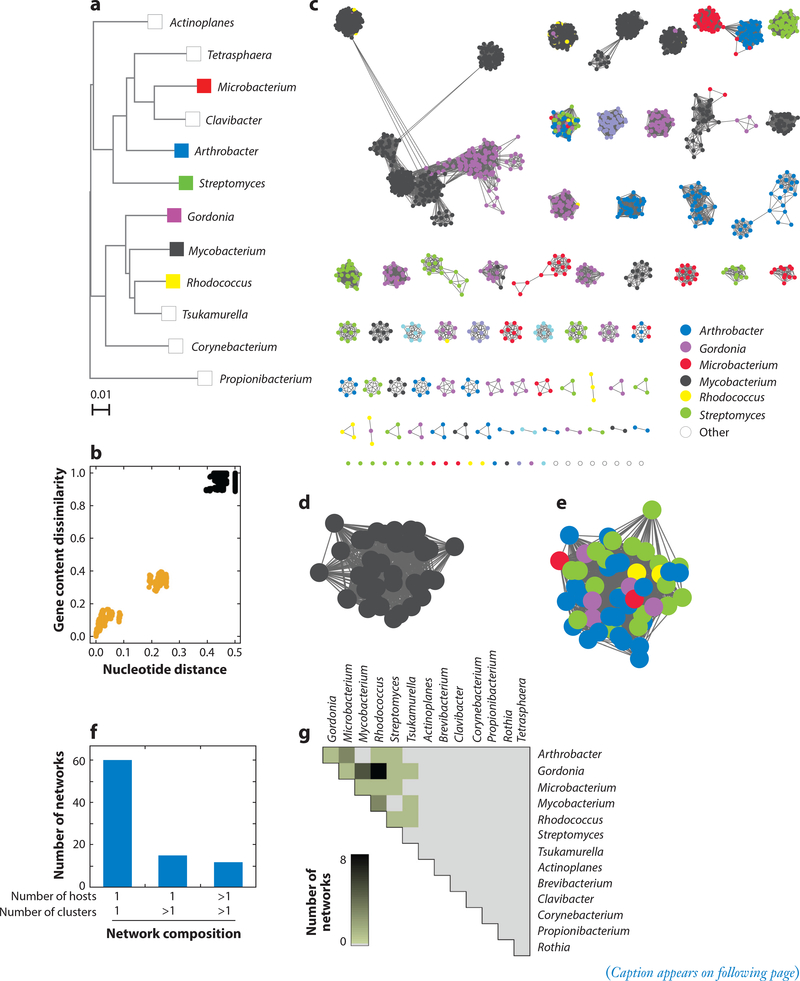
Inclusion of a variety of hosts from different bacterial genera facilitates examination of how phages share genetic information relative to their hosts (Figure 5). In general, pairwise comparison of phages shows that phages within a cluster have greater shared gene content and more similar nucleotide sequences than with other phages (as illustrated for Cluster L phages in Figure 5b) (46). Networks can then be constructed with connections between pairs of phages that meet threshold values for both nucleotide distance relationships and shared gene content (Figure 5c). Using thresholds that approximate previously determined cluster designations, 87 networks are produced (Figure 5c). (This approach has the advantage of avoiding the binary decisions in cluster designation, i.e., whether a phage is grouped in a particular cluster or not.) Many of the individual networks contain phages of a single cluster and bacterial host (e.g., Cluster L, mycobacteria; Figure 5d), whereas others have phages of multiple hosts (Figure 5e). Almost 70% of the networks consist of a single cluster from a single bacterial host (Figure 5f), but ~16% contain phages of more than one cluster/singleton, and ~14% have phages both of more than one cluster and from more than one host (Figure 5f). Although the total number of such networks is still relatively small, it is clear that phages of phylogenetically closely related hosts (Figure 5a) are more likely to be in shared networks and are thus sharing their genes at a greater rate than with phages of more distantly related hosts (Figure 5g). Phage genomes also exchange genes with their hosts, and it is common to find genes thought of as bacterial in bacteriophage genomes (30).
Figure 5.
Heterogeneous genomic diversity of actinobacteriophages. (a) Phylogenetic tree of several actinobacterial host genera. (b) Representative genomic similarity plot comparing gene content dissimilarity and nucleotide distance (46) between phages in Cluster L (n = 38) and not in Cluster L (n = 2,384). Each data point represents a pairwise comparison involving a phage within Cluster L and another phage within (gold) or without (black) Cluster L. (c) Genome networks (n = 87) for all sequenced actinobacteriophages (n = 2,422). A node represents a phage genome and is colored according to its host genus. Two nodes connected by an edge reflect phages with an intracluster genomic relationship, having gene content dissimilarity <0.89 and nucleotide distance <0.42 (46). A network consists of a group of phages that contain at least one edge to another phage in the group and no edges to any phages outside of the group. (d) Enlarged representative network from panel c containing phages (n = 38) from a single cluster (Cluster L) and a single host genus (Mycobacterium). (e) Enlarged representative network from panel c containing phages (n = 52) from multiple clusters (Clusters AM, AU, BI, CC, DJ, and EL and Singleton RosaAsantewaa) and multiple host genera. (f) Histogram reflecting phage diversity based on the composition of the networks in panel c. The number of clusters and host genera represented within each network were quantified, and the number of networks containing the indicated number of clusters and host genera were reported. For this analysis, singletons were treated as clusters. (g) Heatmap reflecting phage relationships within networks from panel c by unique host genera (n = 14). Within each network, host genera pairs connected by an edge were identified. For each possible host genera pair in the database, the number of networks containing at least one edge connecting the two host genera was quantified and represented by a color spectrum. Panel a adapted with permission from Reference 24.
Genome Architecture and Gene Expression
Given the genetic diversity, it is not surprising that these phages have a multitude of different genome organizations. Nonetheless, there are common themes in gene arrangements and expression profiles. All the phages have virions containing linear genomic DNA, but a variety of different types of virion DNA termini are observed (3, 7, 48). The two most common are cohesive ends with short single-stranded extensions—all the actinobacteriophages have 3′ extensions—and circularly permuted (and presumably terminally redundant) ends reflecting pac-type packaging systems. However, some have short inverted repeats and covalently linked proteins (e.g., Cluster BO, FD), and others have long direct repeats (e.g., Cluster BE, BF) that can be up to 11-kbp long.
Phage genes are generally densely packed, with the protein-coding and transfer RNA genes accounting for greater than 95% of the genome span. As such, the genes are organized into operons with little or no space between individual open reading frames, and it is common for translation start and stop codons of adjacent genes to overlap. With the exception of the Myoviridae (e.g., Clusters AA, AO, AR, C, DO), the virion structural genes are organized into an apparently single operon of closely linked genes with well-conserved synteny; a noteworthy departure is phage Marvin (Cluster S) in which some of the tail genes are displaced into the right part of the genome (49). All 15 Cluster S phages share this organization, suggesting that this rearrangement is not a recent event at all. The genomes are oriented for convenience, with the virion structural genes at the left part of the genome and transcribed rightward (32). Typically, the 15–25-virion structural genes span 15–25 kbp of the genome; thus, in the smallest genomes (~15 kbp), there are few nonstructural genes. In contrast, the larger siphoviral genomes (e.g., Microbacterium phage PauloDiaboli, 192 kbp) have a large number of nonstructural genes, most of which are of unknown function. Indeed, the main difference between small and large genomes of the siphoviruses is the number of these nonvirion structure and assembly genes (50). Overall, only ~30% of the phage genes have assigned functions.
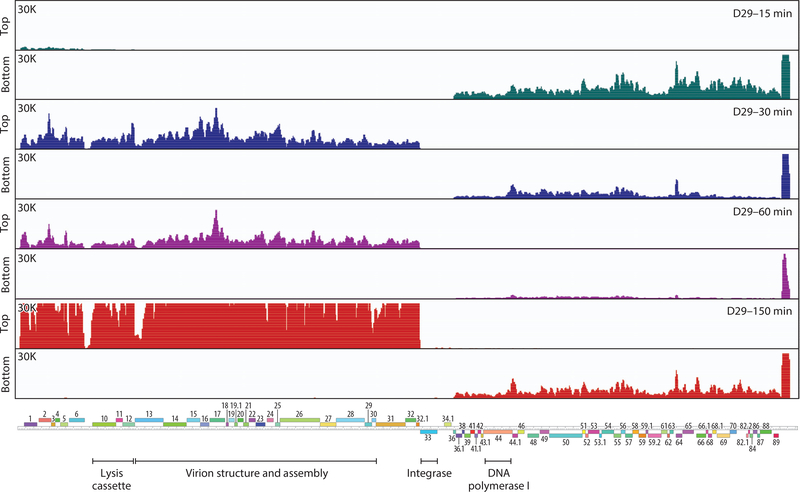
Transcription profiles have been described for several mycobacteriophages during growth and lysogeny, and common themes emerge (37, 39, 51–55). During lytic growth, there are typically two temporally separable patterns, designated as early and late, with the early genes expressed in the first 30 minutes of infection and the late gene expression beginning shortly after and continuing until lysis, typically 2.5–3 hours after infection (Figure 6). RNA sequencing profiles at earlier times show little evidence of a separable immediate early set of genes, but these could be easily overlooked, especially if expressed at low levels (Figure 6). It is also noteworthy that at late times a substantial signal from the early genes remains, either from ongoing expression or stability of early transcripts. Early genes usually correspond to nonvirion structure and assembly genes (located in the rightmost parts of the genomes), and the virion structural genes are expressed late (Figure 6).
Figure 6.
Transcription of the D29 genome. Strand-specific RNA sequencing analysis of D29 (Multiplicity of Infection = 3) infected Mycobacterium smegmatis me2 155. Time points after adsorption are indicated on the upper right: 15 min (teal), 30 min (blue), 60 min (purple), and 150 min (red). At the left are scale maxima and indications of top or bottom strand. The D29 map is shown at the bottom. Figure adapted from Reference 37.
The late genes are highly expressed, and the transcripts are among the most abundant in the cell (Figure 6). Late gene transcription initiates upstream of the structural genes in Cluster A phages (Figure 6) but can also start at the right end of the genome and proceed through cos (52) in the circularized genome. For some phages (e.g., Cluster G), additional promoters may be located within the body of the late operon (56). In Cluster A phages, there is a highly abundant transcript at the extreme right end of the genome (37, 51) within a region lacking protein-coding genes; although several small RNAs have been described (37), their roles are uncertain (Figure 6). There are likely other small noncoding RNAs expressed in the actinobacteriophages that warrant further examination.
Lysogenic gene expression patterns show that several phage genes may be expressed in addition to the repressor. Usually these are located near the center of the genome (i.e., close to an attachment junction of the prophage). Some of these have been shown to be involved in viral defense and are described in more detail below. For Cluster A and Cluster G phages the operator sites used to repress lytic gene expression have been identified and overlap early lytic promoters (38, 56). These have characteristics of sigA-like promoters with canonical −10 and −35 sequences (38, 57, 58). Unfortunately, little is known about initiation of late gene expression or its regulation in any of the actinobacteriophages. Late transcription does not start at sigA-like promoters and is likely dependent on transcriptional activators expressed in early lytic growth. Characterizing late expression is of interest given their high levels of transcriptional activity.
The Cluster A phages have unusual regulatory systems with multiple (25–35) asymmetric 13–14-bp repressor-binding sites located throughout the genomes in small intergenic spaces and in one orientation relative to the direction of transcription (27, 59, 60). These are referred to as stoperators because repressor binding leads to interruption of transcription (59). Immune specificity is conferred by repressor binding preferences for the operator and stoperator sites, and evolution of immunity involves multiple and complex selection factors (55). The immunity system is not constrained to mycobacteriophages but is also found in some phages of Gordonia (25) and Streptomyces (61).
PHAGE-HOST DYNAMICS
Host-Mediated Actinobacteriophage Defense Mechanisms
Most mycobacterial strains do not have CRISPR-Cas systems including M. smegmatis and M. abscessus. M. tuberculosis does have a CRISPR-like array, although its functionality is unclear (62) and there has yet to be a report of a sequence match between a CRISPR spacer and a phage protospacer (Figure 7). It is plausible that there is a reservoir of M. tuberculosis-specific phages yet to be described, assuming the CRISPR system is still functional. Clearly the M. tuberculosis CRISPR-Cas system was active at some point, and spacer sequence variation has been used extensively for spoligotyping strains (63). Restriction-modification (R-M) systems have been described for Mycobacterium (64–68), Microbacterium (69, 70), and Arthrobacter (71–75) strains and for many Streptomyces strains (e.g., 76, 77). It is likely there are many R-M systems yet to be discovered, playing important roles in viral defense. It is notable that there are substantial deviations in genome composition including tetranucleotide usage that likely reflect phage responses to host R-M systems (78).
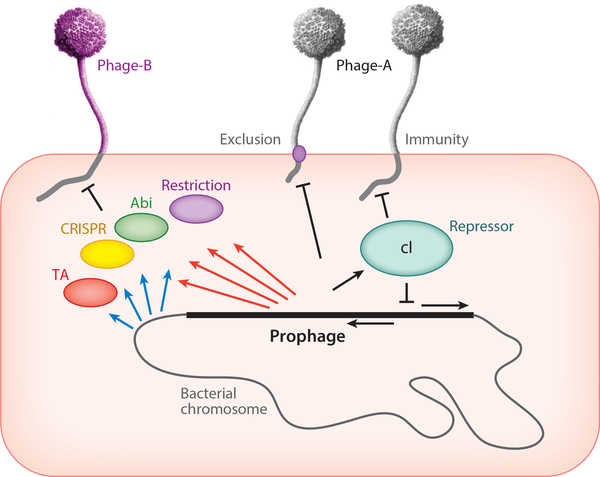
Figure 7.
Phage-host dynamics. A lysogenic cell is depicted carrying a prophage integrated into the bacterial chromosome (not to scale). The prophage genome is derived from Phage-A and encodes a repressor protein (cI) that shuts down lytic genes of both the integrated prophage and superinfecting Phage-A particles. Some prophages may express membrane proteins that prevent superinfection by the same phage (Phage-A) or closely related phages. The bacterial chromosome may express a variety of systems to defend against viral attack (blue arrows), including restriction, various abortive infection systems (abi), CRISPR-Cas, and toxin-antitoxin (TA) systems. Prophages can express analogous systems (red arrows) that defend against infection by heterotypic (i.e., unrelated) phages, such as Phage-B.
Many phage defense abortive infection systems have been described (79, 80), and bioinformatic analyses have predicted many new viral defense systems in bacteria (81–83). Toxin-antitoxin (TA) systems are also involved in phage defense, including both Type II and Type III systems (84, 85) (Figure 7). Experimental evidence for the involvement of TA systems for phage defense in the actinobacteria is sparse, but it is noteworthy that M. tuberculosis contains a vast number of TA systems (>80), many of which are likely active (86). These TA pairs could have been selected for in an M. tuberculosis environmental recent ancestor—defending against phage attack—similar to the CRISPR-Cas systems.
Prophage-Mediated Viral Defense
Interestingly, temperate phages are themselves rich reservoirs of viral defense systems (Figure 7). This should not be unexpected, as temperate phages are abundant in the environment and prophages can be acquired at relatively high frequencies, presenting gifts of protection against attack from other viruses (53, 87). The prophage-encoded defense genes are expressed lysogenically and are typically located near the repressor and integrase genes at the center of the viral genome (53). These are often hypervariable parts of the genome, reflecting exchange of these segments among phages. Unlike repressor-mediated superinfection immunity, which is homotypic and prevents reinfection by the same or very closely related phages, these defense systems are typically heterotypic and defend against unrelated phages (53, 88, 89) (Figure 7). Furthermore, they sometimes act with remarkable specificity, defending against only a single known phage (53, 88); in general this specificity appears much tighter than for the host-encoded resistance mechanisms (81). The mechanisms and bases for the targeting specificity are largely unknown, although characterization of defense escape mutants reveals genes required for targeting (88, 89). The genes are typically dispensable for lytic growth, reflecting defense mechanisms acting through abortive infection rather than direct inhibition of phage growth per se (53, 88, 89). Defense systems have been described in mycobacterial Cluster N and I phages and Gordonia Cluster CV phages (53, 88, 89) but are likely present in many of the temperate phages. It is noteworthy that defense genes can be discovered by their lysogenic expression, but identifying the targeted phages requires a very large collection of phages known to infect the same host strain.
Phages compete with other phages not only through lysogeny but also by exclusion in lytic growth. Lytically growing phages can exclude other phages from superinfection by a variety of small membrane-localized proteins (90) or by inhibition of host-encoded functions required for infection. In one example, the Cluster F phage Fruitloop gp52 protein is expressed in early lytic growth and binds to and inactivates the host DivIVA (Wag31) protein, excluding infection by the Cluster B phage Rosebush, which requires DivIVA for efficient infection (91); it is remarkably specific, and most other Cluster B phages are not excluded. Fruitloop 52 is not required for lytic growth and has no other known function. It is plausible that many of the small genes replete throughout phages genomes play similar roles.
Phage-Encoded Counter-Defense Systems
Phages can avoid or escape defense mechanisms either by mutationally changing their host preferences to infect a different bacterium or by acquiring counter-defense systems. Antirestriction and anti-CRISPR phage genes have been described for Proteobacteria phages (92–94), and it is likely that there are counter-defense systems for other host-encoded defenses. Additionally, counter-defense mechanisms can facilitate escape from prophage-mediated systems. In one intriguing example, phage Tweety is targeted by the prophage-mediated defenses of phages Phrann and MichelleMyBell (53), and Tweety escape mutants can be readily isolated. The mutations are in gene 54, and Tweety gp54 has an unusual organization with 40–48 tetrapeptide repeats flanked by unique N- and C-terminal regions (53, 95). The escape mutants alter the repeat number and often the sequence of the repeat units (53), and different mutants tune gp54 for specificity against either the Phrann or MichelleMyBell systems. Tweety 54 is not required for defense targeting and has no other known function other than to neutralize the prophage-mediated defense systems (53).
Protection from host defenses that target the phage genome directly can be countered by chemical modification of the phage DNA, and many types have been described (96, 97). Some of the actinobacteriophages that contain modified DNA bases can display resistance to restriction in vitro. A well-characterized example is the presence of 7-deazaguanine modifications in several Cluster B phages (98).
TOOLS AND APPLICATIONS FOR ACTINOBACTERIOPHAGES
Genetic Tools
Mycobacteriophages have contributed many tools used widely in mycobacterial genetics (9, 99). Notable among these are the integration-proficient vectors that use phage integration systems to insert plasmids into the host chromosome (38, 50, 95, 100–102). These have the advantages of integrating at a specific chromosome location, efficient transformation, and the ability to form stable single-copy recombinants. They are ideal for complementation studies and have been developed to use multiple different attB loci (9). Mycobacteriophage-derived recombination systems have been exploited to develop recombineering systems for constructing mutants and recombinants of M. smegmatis and M. tuberculosis (42, 43, 103), and they have been adapted for efficient manipulation of the phage genomes themselves (104–106). Phage repressors have been adapted as selectable markers for use in antibiotic-resistant strains (107, 108), phage expression systems have been characterized (58), and phage partitioning systems have been described that promote plasmid stability (39). There are numerous other potential genetic tools awaiting development.
Exploitation of mycobacteriophages as vectors to efficiently deliver DNA to host cells has been advanced through the construction of shuttle phasmids that grow as plasmids in E. coli and as phages in mycobacteria (109, 110). These have been used to deliver transposons (111) and allelic exchange substrates (112) to a variety of mycobacterial species, as well as reporter genes such as luciferase and gfp (113, 114).
Clinical Tools
Several diagnostic tools using mycobacteriophages have been described including the FastPlaque assay (115, 116) that takes advantage of M. tuberculosis replication of phage D29 and recombinant phages that deliver luciferase or fluorescent reporter genes (113,117). Reporter phages have shown considerable potential for drug susceptibility testing of M. tuberculosis in clinical specimens (118–121) but have yet to be widely implemented. Rapid diagnosis of M. tuberculosis drug susceptibility profiles remains a clinically important issue, and reporter phages continue to show considerable promise (119, 122).
The idea of using phages therapeutically to control mycobacterial infections has been widely contemplated, especially in response to the emergence of multiple drug resistance in tuberculosis (10,123) (see sidebar titled Phage Therapy: Fact or Fiction?). On the one hand, phages have been described that efficiently kill lab strains of M. tuberculosis, but on the other hand, there are key questions about access to the bacteria given their intracellular nature and presence in granulomas (124). In the first therapeutic effort to date, a three-phage cocktail was used to treat a 15-year-old patient with cystic fibrosis, a bilateral lung transplant, and a disseminated and highly drug-resistant M. abscessus infection (22). The patient showed substantial improvement after intravenous and topical phage administration, progressing from palliative care to a normal routine. Although this is just a single case study, the possibility of using phages more broadly is worth further consideration. The success of this case required identification of specific phages effective in infecting and killing the specific clinical isolate of M. abscessus (22). Few phages among those isolated on M. smegmatis met this requirement, and Bacteriophage Recombineering of Electroporated DNA (BRED)-engineering and host-range evolution were required to assemble a three-phage cocktail (22, 125). There is substantial variation in phage susceptibility among M. abscessus clinical isolates, and similar case studies will also require personalization of the phages (22, 126). Nonetheless, advances in understanding phage host range determinants and in genome engineering and synthetic biology could facilitate a broad-based approach for controlling mycobacterial infections.
PHAGE THERAPY: FACT OR FICTION?
The idea of using bacteriophages therapeutically was clearly in French-Canadian Felix d’Herelle’s mind shortly after discovering them a hundred years ago. Without knowing truly what a bacteriophage was, d’Herelle figured that something capable of killing bacteria in the lab might also do so in a patient, and he tested the idea in 1919. This caught the attention of the American writer Sinclair Lewis, who published the classic novel Arrowsmith in 1925. Martin Arrowsmith, an aspiring young doctor, uses phages to try to control an outbreak of bubonic plaque on a fictional Caribbean island. Arrowsmith was awarded the Pulitzer Prize in 1926, but Lewis refused it. However, in 1930 he accepted the Nobel Prize in Literature, the only Nobel Award associated with phage therapy!
These early themes reflect the challenges and opportunities for phage therapy today. In d’Herrelle’s first patients, the phages were effectively matched to the infectious strain in the lab, personalizing the therapy, similar to two recent successful applications (22, 126). Martin Arrowsmith, however, was conflicted on mass administration of phages to control plague, in the absence of a more thorough understanding of the phages, a challenge that we have yet to live up to.
Interestingly, clinical isolates of M. tuberculosis are genomically more homogenous than other mycobacteria, and a simple phage cocktail may be effective against a wide spectrum of strains. Early animal model studies suggest that phages might be useful against M. tuberculosis infections (127–129), but clinical interventions in humans have yet to be reported. An alternative application is the prophylactic administration of phages to interfere with tuberculosis transmission, and recent studies in mice suggest this may be effective (130). Aerosol administration for pulmonary infections may be attractive, and delivery systems have been compared and evaluated (131).
SUMMARY AND FUTURE PROSPECTS
Over the past 10 years we have seen substantial advances in our understanding of actinobacteriophage diversity, glimpses into the complexities of the dynamic interactions between phages and their hosts, and the first attempt at therapeutic use. A fuller comprehension of genomic diversity will require both a deeper study of phages using current bacterial hosts and substantial expansion to include many more hosts in the actinobacterial phylum. Fortunately, the development of integrated research-educational communities provides a means by which this could be achievable (16). A major challenge remains in determining the functions of the ~200,000 actinobacteriophage genes of unknown function. A substantial portion are likely involved in determining phage-host dynamics; conferring prophage-mediated viral defenses; exclusion during lytic growth; or counter-defenses to neutralize restriction, CRISPR-Cas, and other resistance mechanisms (53, 91). The specificity within these dynamic interactions complicates disentangling these activities, and large collections of characterized phages provide enormously powerful tools for doing so. Finally, although the first successful application of mycobacteriophages to control a drug-resistant M. abscessus infection had a positive outcome, the potential for clinical application awaits critical evaluation (22).
BRED:
Bacteriophage Recombineering on Electroporated DNA
Supplementary Material
SUMMARY POINTS.
Actinobacteriophages encompass enormous genetic diversity.
Integrated research-education programs such as the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Sciences (SEA-PHAGES) program are powerful models for advancing both student engagement and scientific discovery.
There are multiple modes of bacteriophage evolution involving varying degrees of horizontal genetic exchange.
Over 70% of actinobacteriophage genes are of unknown function.
Phages compete with other phages through both exclusion mechanisms in lytic growth and prophage-mediated viral defense in lysogeny.
Actinobacteriophages are a rich source of genes for tool development to advance genetics of mycobacteria and other actinobacteria.
Bacteriophage Recombineering of Electroporated DNA (BRED) engineering is an effective tool for constructing recombinant phages.
A case study suggests there may be potential for therapeutic use of phages to treat mycobacterial infections.
FUTURE ISSUES.
Our understanding of phage genetic diversity has yet to scratch the surface, and intensive efforts are required to broadly map viral genomes.
Determining the functions of the many hundreds of thousands of phage-encoded genes is critical but requires new strategies and approaches.
Phages play critical roles in microbial dynamics, but prophage-mediated viral defense systems, exclusion systems, counter-defense systems, and counter-exclusion mechanisms are likely highly prevalent in phage genomes but greatly underexplored.
The opportunities and challenges in the therapeutic use of phages for mycobacterial infections warrant active investigation.
ACKNOWLEDGMENTS
Many thanks to Travis Mavrich for data and design of Figure 5 and to Elizabeth Amarh, Bekah Dedrick, Krista Freeman, Becky Garlena, Christian Gauthier, Carlos Guerrero, Debbie Jacobs-Sera, Ching Chung Ko, Travis Mavrich, Matthew Montgomery, Welkin Pope, Dan Russell, Bailey Smith, and Katie Wetzel for comments on the manuscript. This work was supported by grants from the National Institute of Health (GM131729) and the Howard Hughes Medical Institute (#GT12053). The actinobacteriophage collection reflects contributions of over 400 SEA-PHAGES faculty and over 30,000 student phage hunters, and special thanks go to them. I would also like to thank Steve Cresawn at James Madison University and David Asai, Vic Sivanathan, Billy Biederman, and Danielle Heller at the Howard Hughes Medical Institute for their wonderful partnership in SEA-PHAGES program administration.
Footnotes
DISCLOSURE STATEMENT
G.F.H. is an Editorial Committee Member of the Annual Review of Virology and a fellow for the American Society of Microbiology and the American Association for the Advancement of Science. He is also a paid consultant for Tessera Therapeutics and Janssen Inc.
LITERATURE CITED
- 1.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, et al. 2016. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev 80:1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durand GA, Raoult D, Dubourg G. 2019. Antibiotic discovery: history, methods and perspectives. Int. J. Antimicrob. Agents 53:371–82 [DOI] [PubMed] [Google Scholar]
- 3.Hatfull GF. 2018. Mycobacteriophages. Microbiol. Spectr 6. 10.1128/microbiolspec.GPP3-0026-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Froman S, Will DW, Bogen E. 1954. Bacteriophage active against Mycobacterium tuberculosis I. Isolation and activity. Am. J. Pub. Health 44:1326–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murohashi T, Tokunaga T, Mizuguchi Y, Maruyama Y. 1963. Phage typing of slow-growing mycobacteria. Am. Rev. Respir. Dis 88:664–69 [DOI] [PubMed] [Google Scholar]
- 6.Mizuguchi Y. 1984. Mycobacteriophages. In The Mycobacteria: A Sourcebook, ed. Kubica GP, Wayne LG, pp. 641–62. New York: Marcel Dekker [Google Scholar]
- 7.Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatfull GF. 2010. Mycobacteriophages: genes and genomes. Annu. Rev. Microbiol 64:331–56 [DOI] [PubMed] [Google Scholar]
- 9.Hatfull GF. 2012. The secret lives of mycobacteriophages. Adv. Virus Res 82:179–288 [DOI] [PubMed] [Google Scholar]
- 10.Hatfull GF. 2014. Mycobacteriophages: windows into tuberculosis. PLOS Pathog 10:e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatfull GF. 2015. Dark matter of the biosphere: the amazing world of bacteriophage diversity. J. Virol 89:8107–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNerney R, Traore H. 2005. Mycobacteriophage and their application to disease control. J. Appl. Microbiol 99:223–33 [DOI] [PubMed] [Google Scholar]
- 13.Hatfull GF. 2010. Bacteriophage research: gateway to learning science. Microbe 5:243–50 [Google Scholar]
- 14.Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. 2006. Teaching scientific inquiry. Science 314:1880–81 [DOI] [PubMed] [Google Scholar]
- 15.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, et al. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5:e01051–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanauer DI, Graham MJ, Sea P, Betancur L, Bobrownicki A, et al. 2017. An inclusive Research Education Community (iREC): impact of the SEA-PHAGES program on research outcomes and student learning. PNAS 114:13531–36Presents the SEA-PHAGES program, with evidence for scientific and educational impact.
- 17.Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, et al. 2012. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio 3:e00279–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ybazeta G, Graham J, Trifkovic J, Giroux L, Saleh M, et al. 2018. Complete genome sequences of a diverse group of 13 Propionibacterium acnes bacteriophages isolated from urban raw sewage. Genome Announc 6:e00224–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng L, Marinelli LJ, Grosset N, Fitz-Gibbon ST, Bowman CA, et al. 2018. Complete genomic sequences of Propionibacterium freudenreichii phages from Swiss cheese reveal greater diversity than Cutibacterium (formerly Propionibacterium) acnes phages. BMC Microbiol 18:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson ZA, Tucci J, Seviour RJ, Petrovski S. 2015. Lysis to kill: evaluation of the lytic abilities, and genomics of nine bacteriophages infective for Gordonia spp. and their potential use in activated sludge foam biocontrol. PLOS ONE 10:e0134512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, et al. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedrick R, Guerrero-Bustamante C, Garlena RA, Russell DA, Ford K, et al. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med 25:730–33Describes the first therapeutic use of bacteriophages for a mycobacterial infection.
- 23.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, et al. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4:e06416.Shows evidence supporting an underlying continuum of viral diversity.
- 24.Klyczek KK, Bonilla JA, Jacobs-Sera D, Adair TL, Afram P, et al. 2017. Tales of diversity: genomic and morphological characteristics of forty-six Arthrobacter phages. PLOS ONE 12:e0180517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pope WH, Mavrich TN, Garlena RA, Guerrero-Bustamante CA, Jacobs-Sera D,et al.2017. Bacteriophages of Gordonia spp. display a spectrum of diversity and genetic relationships. mBio 8:e01069–17Shows comparative genomics of Gordonia phages and revisits cluster definitions.
- 26.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. PNAS 105:3963–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, et al. 2011. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLOS ONE 6:e16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill JJ, Wang B, Sestak E, Young R, Chu KH. 2018. Characterization of a novel Tectivirus phage toil and its potential as an agent for biolipid extraction. Sci. Rep 8:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caruso SM, deCarvalho TN, Huynh AB, Morcos G, Kuo N, et al. 2019. A novel genus of actinobacterial Tectiviridae. Viruses 11:1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–82 [DOI] [PubMed] [Google Scholar]
- 31.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. PNAS 96:2192–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatfull GF, Sarkis GJ. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol 7:395–405 [DOI] [PubMed] [Google Scholar]
- 33.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, et al. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLOS Genet 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, et al. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol 397:119–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinform. 12:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ptashne M. 1987. A Genetic Switch. Oxford/Cambridge: Blackwell Science/Cell Press [Google Scholar]
- 37.Dedrick RM, Mavrich TN, Ng WL, Hatfull GF. 2017. Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives. BMC Microbiol 17:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broussard GW, Oldfield LM, Villanueva VM, Lunt BL, Shine EE, Hatfull GF. 2013. Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches. Mol. Cell 49:237–48Describes a new model for the regulation and mechanism of lysogenic establishment.
- 39.Dedrick RM, Mavrich TN, Ng WL, Cervantes Reyes JC, Olm MR, et al. 2016. Function, expression, specificity, diversity and incompatibility of actinobacteriophage parABS systems. Mol. Microbiol 101:625–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrich TN, Casey E, Oliveira J, Bottacini F, James K, et al. 2018. Characterization and induction of prophages in human gut-associated Bifidobacterium hosts. Sci. Rep 8:12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinsohn JT, Radman M, Petit MA. 2008. The λ red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLOS Genet 4:e1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147–52 [DOI] [PubMed] [Google Scholar]
- 43.van Kessel JC, Hatfull GF. 2008. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol. Microbiol 67:1094–107 [DOI] [PubMed] [Google Scholar]
- 44.Pitcher RS, Brissett NC, Doherty AJ. 2007. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol 61:259–82 [DOI] [PubMed] [Google Scholar]
- 45.Pope WH, Jacobs-Sera D, Russell DA, Rubin DH, Kajee A, et al. 2014. Genomics and proteomics of mycobacteriophage patience, an accidental tourist in the Mycobacterium neighborhood. mBio 5:e02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mavrich TN, Hatfull GF. 2017. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol 2:17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory AC, Solonenko SA, Ignacio-Espinoza JC, LaButti K, Copeland A, et al. 2016. Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genom 17:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes. MethodsMol. Biol 1681:109–25 [DOI] [PubMed] [Google Scholar]
- 49.Mageeney C, Pope WH, Harrison M, Moran D, Cross T, et al. 2012. Mycobacteriophage Marvin: a new singleton phage with an unusual genome organization. J. Virol 86:4762–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pope WH, Jacobs-Sera D, Best AA, Broussard GW, Connerly PL, et al. 2013. Cluster J mycobacteriophages: intron splicing in capsid and tail genes. PLOS ONE 8:e69273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halleran A, Clamons S, Saha M. 2015. Transcriptomic characterization of an infection of Mycobacterium smegmatis by the cluster A4 mycobacteriophage Kampy. PLOS ONE 10:e0141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Pinches RS, Cornely K, Hatfull GF. 2019. Mycobacteriophage ZoeJ: a broad host-range close relative of mycobacteriophage TM4. Tuberculosis 115:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, et al. 2017. Prophagemediated defence against viral attack and viral counter-defence. Nat. Microbiol 2:16251.Reports a variety of prophage-encoded systems for defense against heterotypic vial attack.
- 54.Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. 2013. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol. Microbiol 88:577–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mavrich TN, Hatfull GF. 2019. Evolution of superinfection immunity in cluster A mycobacteriophages. mBio 10:e00971–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villanueva VM, Oldfield LM, Hatfull GF. 2015. An unusual phage repressor encoded by mycobacteriophage BPs. PLOS ONE 10:e0137187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nesbit CE, Levin ME, Donnelly-Wu MK, Hatfull GF. 1995. Transcriptional regulation of repressor synthesis in mycobacteriophage L5. Mol. Microbiol 17:1045–56 [DOI] [PubMed] [Google Scholar]
- 58.Oldfield LM, Hatfull GF. 2014. Mutational analysis of the mycobacteriophage BPs promoter Pr reveals context-dependent sequences for mycobacterial gene expression. J. Bacteriol 196:3589–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown KL, Sarkis GJ, Wadsworth C, Hatfull GF. 1997. Transcriptional silencing by the mycobacteriophage L5 repressor. EMBO J 16:5914–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganguly T, Bandhu A, Chattoraj P, Chanda PK, Das M, et al. 2007. Repressor of temperate mycobacteriophage L1 harbors a stable C-terminal domain and binds to different asymmetric operator DNAs with variable affinity. Virol. J 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith MC, Hendrix RW, Dedrick R, Mitchell K, Ko CC, et al.2013. Evolutionary relationships among actinophages and a putative adaptation for growth in Streptomyces spp. J. Bacteriol 195:4924–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He L, Fan X, Xie J. 2012. Comparative genomic structures of Mycobacterium CRISPR-Cas. J. Cell Biochem 113:2464–73 [DOI] [PubMed] [Google Scholar]
- 63.Sola C 2015. Clustured regularly interspersed short palindromic repeats (CRISPR) genetic diversity studies as a mean to reconstruct the evolution of the Mycobacterium tuberculosis complex. Tuberculosis 95(Suppl. 1):S159–66 [DOI] [PubMed] [Google Scholar]
- 64.Rado TA, Bates JH, Fitzhugh JK. 1976. Evidence for host-dependent modification and restriction of bacteriophage DNA in Mycobacterium tuberculosis. J. Gen. Virol 30:91–97 [DOI] [PubMed] [Google Scholar]
- 65.Jones WD Jr., Greenberg J. 1977. Host modification and restriction with a mycobacteriophage isolated from a pseudolysogenic Mycobacterium chelonei. J. Gen. Microbiol 99:389–95 [DOI] [PubMed] [Google Scholar]
- 66.Crawford JT, Cave MD, Bates JH. 1981. Evidence for plasmid-mediated restriction-modification in Mycobacterium avium intracellulare. J. Gen. Microbiol 127:333–38 [DOI] [PubMed] [Google Scholar]
- 67.Shankar S, Tyagi AK. 1993. MchAI and MchAII, two class-II restriction endonucleases from Mycobacterium chelonei. Gene 132:119–23 [DOI] [PubMed] [Google Scholar]
- 68.Shankar S, Tyagi AK. 1993. Purification and characterization of restriction endonuclease Mgol from Mycobacterium gordonae. Gene 131:153–54 [DOI] [PubMed] [Google Scholar]
- 69.Striebel HM, Schmitz GG, Kaluza K, Jarsch M, Kessler C. 1990. MamI, a novel class-II restriction endonuclease from Microbacterium ammoniaphilum recognizing 5′-GATNNf.NNATC-3′. Gene 91:95–100 [DOI] [PubMed] [Google Scholar]
- 70.Striebel HM, Seeber S, Jarsch M, Kessler C.1996. Cloning and characterization of the MamI restriction-modification system from Microbacterium ammoniaphilum in Escherichia coli. Gene 172:41–46 [DOI] [PubMed] [Google Scholar]
- 71.Rexer BU, Jarsch M, Sagmeister C, Gluck B, Berger G, Kessler C. 1988. AsnI: a novel class II restriction endonuclease from Arthrobacter sp., strain N-CM, recognizing 5′-AT/TAAT-3′. FEBS Lett 235:241–46 [DOI] [PubMed] [Google Scholar]
- 72.Polisson C, Morgan RD. 1990. AciI, a unique restriction endonuclease from Arthrobacter citreus which recognizes 5′ CCGC 3′. Nucleic Acids Res 18:5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polisson C, Robinson D. 1992. ApoI, a unique restriction endonuclease from Arthrobacter protophormiae which recognizes 5′ RAATTY 3′. Nucleic Acids Res 20:2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Degtyarev S, Kolyhalov AA, Rechkunova NI, Abdurashitov MA. 1992. AcsI, a new restriction endonuclease from Arthrobacter citreus 310 recognizing 5′-Pu decreases AATTPy-3′. Nucleic Acids Res 20:3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grigaite R, Maneliene Z, Janulaitis A. 2002. AarI, a restriction endonuclease from Arthrobacter aurescens SS2–322, which recognizes the novel non-palindromic sequence 5′-CACCTGC(N)4/8–3′. Nucleic Acids Res 30:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arrand JR, Myers PA, Roberts RJ. 1978. A new restriction endonuclease from Streptomyces albus G. J. Mol. Biol 118:127–35 [DOI] [PubMed] [Google Scholar]
- 77.Yu H, Liu G, Zhao G, Hu W, Wu G, et al. 2018. Identification of a conserved DNA sulfur recognition domain by characterizing the phosphorothioate-specific endonuclease SprMcrA from Streptomyces pristinaespiralis. Mol. Microbiol 110:484–97 [DOI] [PubMed] [Google Scholar]
- 78.Siranosian B, Perera S, Williams E, Ye C, de Graffenried C, Shank P. 2015. Tetranucleotide usage highlights genomic heterogeneity among mycobacteriophages. F1000Res 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chopin MC, Chopin A, Bidnenko E. 2005. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol 8:473–79 [DOI] [PubMed] [Google Scholar]
- 80.Salmond GP, Fineran PC. 2015. A century of the phage: past, present and future. Nat. Rev. Microbiol 13:777–86 [DOI] [PubMed] [Google Scholar]
- 81.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, et al. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, et al. 2018. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol 3:90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, et al. 2019. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574:691–95 [DOI] [PubMed] [Google Scholar]
- 84.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. PNAS 106:894–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob. Genet. Elements 3:e26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLOS Genet 5:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, et al. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10:2854–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gentile GM, Wetzel KS, Dedrick RM, Montgomery MT, Garlena RA, et al. 2019. More evidence of collusion: a new prophage-mediated viral defense system encoded by mycobacteriophage Sbash. mBio 10:e00196–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Montgomery MT, Guerrero-Bustamante CA, Dedrick RM, Jacobs-Sera D, Hatfull GF. 2019. Yet more evidence of collusion: a new viral defense system encoded by Gordonia phage CarolAnn. mBio 10:e02417–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cumby N, Edwards AM, Davidson AR, Maxwell KL. 2012. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J. Bacteriol 194:5012–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ko CC, Hatfull GF. 2018. Mycobacteriophage Fruitloop gp52 inactivates Wag31 (DivIVA) to prevent heterotypic superinfection. Mol. Microbiol 108:443–60Describes how gp52 of phage Fruitloop confers exclusion by inactivation of the host DivIVA protein.
- 92.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. 2008. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J. Mol. Biol 375:720–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberts GA, Stephanou AS, Kanwar N, Dawson A, Cooper LP, et al. 2012. Exploring the DNA mimicry of the Ocr protein of phage T7. Nucleic Acids Res 40:8129–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. 2007. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology 153:2711–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weigele P, Raleigh EA. 2016. Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev 116:12655–87 [DOI] [PubMed] [Google Scholar]
- 97.Khudyakov IY, Kirnos MD, Alexandrushkina NI, Vanyushin BF. 1978. Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology 88:8–18 [DOI] [PubMed] [Google Scholar]
- 98.Hutinet G, Kot W, Cui L, Hillebrand R, Balamkundu S, et al. 2019. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat. Commun 10:5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hatfull GF. 1994. Mycobacteriophage L5: a toolbox for tuberculosis. ASM News 60:255–60 [Google Scholar]
- 100.Lee MH, Pascopella L, Jacobs WR Jr., Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. PNAS 88:3111–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2008. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J. Bacteriol. 190:2172–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, et al. 2011. Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4. PLOS ONE 6:e26750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Kessel JC, Hatfull GF. 2008. Mycobacterial recombineering. Methods Mol. Biol 43 5:203–15 [DOI] [PubMed] [Google Scholar]
- 104.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat. Rev. Microbiol 6:851–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marinelli LJ, Hatfull GF, Piuri M. 2012. Recombineering: a powerful tool for modification of bacteriophage genomes. Bacteriophage 2:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marinelli LJ, Piuri M, Hatfull GF. 2019. Genetic manipulation of lytic bacteriophages with BRED: bacteriophage recombineering of electroporated DNA. Methods Mol. Biol 1898:69–80Describes how recombineering can be applied for efficient genetic manipulation of phage genomes.
- 107.Donnelly-Wu MK, Jacobs WR Jr., Hatfull GF. 1993. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol. Microbiol 7:407–17 [DOI] [PubMed] [Google Scholar]
- 108.Petrova ZO, Broussard GW, Hatfull GF. 2015. Mycobacteriophage-repressor-mediated immunity as a selectable genetic marker: Adephagia and BPs repressor selection. Microbiology 161:1539–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jacobs WR Jr., Tuckman M, Bloom BR. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532–35 [DOI] [PubMed] [Google Scholar]
- 110.Jacobs WR Jr., Snapper SB, Tuckman M, Bloom BR. 1989. Mycobacteriophage vector systems. Rev. Infect. Dis 11(Suppl. 2):S404–10 [DOI] [PubMed] [Google Scholar]
- 111.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, et al. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. PNAS 94:10961–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bardarov S, Bardarov S Jr., Pavelka MS Jr., Sambandamurthy V, Larsen M, et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–17 [DOI] [PubMed] [Google Scholar]
- 113.Jacobs WR Jr., Barletta RG, Udani R, Chan J, Kalkut G, et al. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819–22 [DOI] [PubMed] [Google Scholar]
- 114.Piuri M, Jacobs WR Jr., Hatfull GF. 2009. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLOS ONE 4:e4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wilson SM, al-Suwaidi Z, McNerney R, Porter J, Drobniewski F. 1997. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat. Med 3:465–68 [DOI] [PubMed] [Google Scholar]
- 116.Albay A, Kisa O, Baylan O, Doganci L. 2003. The evaluation of FASTPlaqueTB™ test for the rapid diagnosis of tuberculosis. Diagn. Microbiol. Infect. Dis 46:211–15 [DOI] [PubMed] [Google Scholar]
- 117.Sarkis GJ, Jacobs WR Jr., Hatfull GF. 1995. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol. Microbiol 15:1055–67 [DOI] [PubMed] [Google Scholar]
- 118.Jain P, Hartman TE, Eisenberg N, O’Donnell MR, Kriakov J, et al. 2012. ϕ2GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J. Clin. Microbiol 50:1362–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Donnell MR, Pym A, Jain P, Munsamy V, Wolf A, et al. 2015. A novel reporter phage to detect tuberculosis and rifampin resistance in a high-HIV-burden population. J. Clin. Microbiol 53:2188–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rondon L, Urdaniz E, Latini C, Payaslian F, Matteo M, et al. 2018. Fluoromycobacteriophages can detect viable Mycobacterium tuberculosis and determine phenotypic rifampicin resistance in 3–5 days from sputum collection. Front. Microbiol 9:1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Banaiee N, Bobadilla-Del-Valle M, Bardarov S Jr., Riska PF, Small PM, et al. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol 39:3883–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, et al. 2016. Dual-reporter mycobacteriophages (ϕ2 DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 7:e01023–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hatfull GF, Vehring R. 2016. Respirable bacteriophage aerosols for the prevention and treatment of tuberculosis. In Drug Delivery Systems for Tuberculosis Prevention and Treatment, ed. Hickey AJ, Misra A, Fourie PB, pp. 277–92. Chichester, UK: Wiley & Sons [Google Scholar]
- 124.Azimi T, Mosadegh M, Nasiri MJ, Sabour S, Karimaei S, Nasser A. 2019. Phage therapy as a renewed therapeutic approach to mycobacterial infections: a comprehensive review. Infect. Drug Resist 12:2943–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, et al. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLOS ONE 3:e3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, et al. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother 61:e00954–17Shows use of personalized phage cocktails for treatment of an Acinetobacter infection.
- 127.Sula L, Sulova J, Stolcpartova M. 1981. Therapy of experimental tuberculosis in guinea pigs with mycobacterial phages DS-6A, GR-21 T, My-327. Czech Med 4:209–14 [PubMed] [Google Scholar]
- 128.Trigo G, Martins TG, Fraga AG, Longatto-Filho A, Castro AG, et al. 2013. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLOS Negl. Trop. Dis 7:e2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koz’min-Sokolov BN, Vabilin. 1975. [Effect of mycobacteriophages on the course of experimental tuberculosis in albino mice]. Probl. Tuberk 4:75–79 (In Russian) [PubMed] [Google Scholar]
- 130.Carrigy NB, Larsen SE, Reese V, Pecor T, Harrison M, et al. 2019. Prophylaxis of Mycobacterium tuberculosis H37Rv infection in a preclinical mouse model via inhalation of nebulized bacteriophage D29. Antimicrob. Agents Chemother 63:e00871–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carrigy NB, Chang RY, Leung SSY, Harrison M, Petrova Z, et al. 2017. Anti-tuberculosis bacteriophage D29 delivery with a vibrating mesh nebulizer, jet nebulizer, and soft mist inhaler. Pharm. Res 34:2084–96 [DOI] [PubMed] [Google Scholar]
- 132.Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.