Abstract
CRISPR/Cas9 is a preferred genome editing tool and has been widely adapted to ranges of disciplines, from molecular biology to gene therapy. A key prerequisite for the success of CRISPR/Cas9 is its capacity to distinguish between single guide RNAs (sgRNAs) on target and homologous off-target sites. Thus, optimized design of sgRNAs by maximizing their on-target activity and minimizing their potential off-target mutations are crucial concerns for this system. Several deep learning models have been developed for comprehensive understanding of sgRNA cleavage efficacy and specificity. Although the proposed methods yield the performance results by automatically learning a suitable representation from the input data, there is still room for the improvement of accuracy and interpretability. Here, we propose novel interpretable attention-based convolutional neural networks, namely CRISPR-ONT and CRISPR-OFFT, for the prediction of CRISPR/Cas9 sgRNA on- and off-target activities, respectively. Experimental tests on public datasets demonstrate that our models significantly yield satisfactory results in terms of accuracy and interpretability. Our findings contribute to the understanding of how RNA-guide Cas9 nucleases scan the mammalian genome. Data and source codes are available at https://github.com/Peppags/CRISPRont-CRISPRofft.
Keywords: CRISPR/Cas9, sgRNA, On-target, Off-target, Deep learning
1. Introduction
CRISPR/Cas9 is a remarkable genome engineering technology and has a promising potential for genetic manipulation applications. In this system, a Cas9 endonuclease protein forms a complex with a single guide RNA (sgRNA) molecule and localizes to a target DNA sequence by sgRNA: genome DNA base-pairing rules [1], [2]. The target DNA sequence must be both complementary to the sgRNA and also contain a protospacer adjacent motif (PAM), which is required for compatibility with the Cas9 protein being used [3]. Using CRISPR/Cas9 system, DNA sequences with the endogenous genome and their functional outputs can be easily edited or modulated in almost any organism of choice [1]. However, variable activity across different sgRNA remains a significant limitation, which contributes to inconsistent sgRNA on-target activity [4]. The targeting specificity of Cas9 is regulated by the 20-nt guiding sequence of the sgRNA and the PAM adjacent to the target sequence in the genome [5]. Previous work has found that Cas9 off-target activity depends on both sgRNA sequence and experimental conditions [6], [7], [8], [9], [10], [11]. Albeit CRISPR/Cas9 has great potential for basic and clinical research, the determinants of on-target activity and the extent of off-target effects remain insufficiently understood. Therefore, it is crucial to develop computational algorithms for on- and off-target activity prediction, which can increase our knowledge of the mechanisms of this system and help maximize the safety of CRISPR-based approaches.
Presently, several deep learning-based methods have been explored for CRISPR sgRNA on-target prediction. Convolutional neural networks (CNNs) are attractive solutions for genomic sequence due to their capability of performing automatic and parallel feature extraction. For instance, DeepCRISPR [12], DeepCas9 [13], CNN_5layers [14] and DeepSpCas9 [15] are trained by CNN to predict sgRNA efficiency by automatically recognize sequence features. Zhang et al. developed a hybrid CNN-SVR (CNN with support vector regression) for sgRNA cleavage efficacy prediction, which uses a merged CNN as the front-end for extracting sequence features and an SVR as the back-end for regression [16]. Recurrent neural networks (RNNs) [17], particularly based on long short-term memory network (LSTM) [18] and gated recurrent unit (GRU) [19], are good at capturing interactions between distant elements along the sequence, thus widely used in natural language processing (NLP) and genomics field [20]. For example, DeepHF applied RNN to extract the dependence information of sgRNA and its biological features, thus achieving a satisfactory performance for on-target activity prediction [21]. To obtain a comprehensive sequence representation, CNN and RNN are cooperated for evaluating sgRNA on-target activity. For instance, C-RNNCrispr uses CNN for feature extraction and applies bi-directional GRU (BGRU) for modeling sequential dependencies of sgRNA sequences in both forward and backward directions [22]. AttnToCrispr_CNN uses CNN with an attention-based transformer module [23] to extract cell-specific gene property derived from biological network and gene expression profile for sgRNA efficiency prediction [24]. To the best of our knowledge, this is the first application of attention mechanism for the purpose of predicting sgRNA activity. On the other hand, deep learning has also been gradually applied for CRISPR sgRNA off-target prediction. For example, DeepCRISPR [12] and CNN_std [25] use CNNs to predict sgRNA specificity. AttnToMismatch_CNN applies CNN with a transformer to capture features from sgRNA-DNA sequence pair and the cell-line specific gene expression information for sgRNA off-targets prediction [24]. In CnnCrispr, bidirectional LSTM (BLSTM) [26] is also followed by CNN to predict off-target activity. CRISPR-Net utilizes RNN to quantify off-target activities with insertions or deletions between sgRNA and target DNA sequence pair [27]. Despite the progress made so far, there is still need for developing more accurate and interpretable methods for sgRNA on- and off-target activities prediction.
More recent works apply the attention mechanism in NLP [28] to produce interpretable results for deep learning models. Attention mechanism assigns different weights to individual positions of the input, so the model can focus on the most crucial features to achieve superior predictive power. In sequence analysis, attention values for individual site derived by embedding attention mechanism allow model to focus on the important locus that significantly contribute to the final predictions [29]. Previous studies showed that positions immediately upstream the PAM (PAM-proximal) are more important than the PAM-distal region for sgRNA activity prediction [30]. As mentioned, position of mismatches between sgRNA and target DNA depend on off-target mutations. Perfect base-pairing with 10 ~ 12 bp PAM-proximal determines Cas9 specificity, while multiple PAM-distal mismatches can be tolerated [30]. These motivate us to selectively discover relevant features from numerous features generated in the convolutional layers for sgRNA on- and off-target activity prediction. Hence, attention mechanism is a suitable strategy to facilitate the interpretation of Cas9 binding and cleavage patterns.
In this study, we introduce two attention-based deep learning frameworks, CRISPR-ONT and CRISPR-OFFT, to predict CRISPR/Cas9 sgRNA efficiency and specificity, respectively. CRISPR-ONT combines a deep CNN with an attention mechanism to build an accurate and interpretable model, which can effectively capture the intrinsic characteristics of Cas9-sgRNA binding and cleavage. The attention mechanism allows CRISPR-ONT to learn to pay attention to PAM-proximal regions of the sequence that convey more relevant information about cleavage efficacy. The permutation nucleotide importance analysis clearly indicates the relative importance of each position in the input sgRNA sequence, which helps to expand our understanding of the mechanism of CRISPR/Cas9. In addition, we visualize our CRISPR-ONT by converting the convolutional filters into sequence logos and reveal some important patterns for sgRNA efficiency. Similarly, CRISPR-OFFT combines CNN with an attention module to predict sgRNA off-target activity, in which features of different levels are extracted from sgRNA-DNA sequence pair. We applied the bootstrapping sampling to balance the samples in each batch since the labels of off-target datasets were highly imbalanced. Moreover, transfer learning was performed for small-size cell-line sgRNA specificity prediction. Experiments on public datasets demonstrate that both CRISPR-ONT and CRISPR-OFFT can perform competitively against other predictors.
2. Methods
2.1. Dataset
On-target dataset. In this work, we used six public datasets for model training, parameter tuning and comparison of our CRISPR-ONT with existing methods. Wang et al. performed a genome-scale screen to measure sgRNA activity for two highly specific SpCas9 variants (eSpCas9 and SpCas9-HF1) and wild-type SpCas9 (WT-SpCas9) in human cells [21]. The indel frequency of a sgRNA was calculated by:
| (1) |
After removing the non-edited sequences, the authors obtained indel rates of 58,616, 56,887 and 55,603 sgRNAs for these three nucleases, which we refer to as datasets SP, HF and WT, respectively. The corresponding log2 fold change of sgRNA counts between before and several days after treatment with CRISPR/Cas9 was calculated and normalized. Datasets Sniper-Cas9 [31], SpCas9-NG [32] and xCas9 [33] were curated from [34]. These SpCas9 variants have been shown both enhanced fidelity and altered or broadened PAM compatibilities [34]. Target sequences associated with low indel frequency (lower than 20%) were excluded from this study. After removing the redundancy, the number of datasets Sniper-Cas9, SpCas9-NG and xCas9 was 37,794, 30,585 and 37,738, respectively. Each entry in the datasets contains a 23-nt sgRNA and its normalized indel frequency. Dataset WT was used as a benchmark dataset during model selection phase. Other five datasets were utilized to evaluate the performance of our and the compared models. We randomly partitioned the sequences into training set and independent test set with 85% and 15% classes for each dataset. Experiments were performed under 10-fold cross-validation in the training phase. More details can be found in Supplementary Table S1.
Off-target dataset. The off-target data detected by GUIDE-seq, Digenome-seq, BLESS, etc were gathered from published studies [7], [9], [35], [36], [37], [38]. These datasets contain two human cell types, namely HEK 293 cell line and its derivatives, as well as K562t. Datasets HEK293T and K562 contain 18 and 12 sgRNAs, respectively. There are 656 positive data have been identified as off-targets among all 30 sgRNAs. Chuai et al. [12] obtained 152,577 possible sites across the whole genome using bowtie2 [39], with a maximum of 6 nucleotide mismatches. A previous study suggested that sgRNA off-target prediction using classification obtains superior performance than regression [12]. Therefore, we focus on sgRNA off-target prediction using binary classification. The labels of off-targets were represented with label 1, while the others of all non-off-targets were labeled by 0. We note that the whole dataset was highly unbalanced. The number of positive sample and negative sample of dataset HEK293T is 536 and 132378, respectively. For dataset K562, there are 20,199 negative samples while the number of positive samples is only 120. The ratio of positive and negative samples of these two datasets is nearly 1:233. Such a class label imbalance problem brings challenges in predictive model building. We applied bootstrapping sampling [40] to overcome this problem.
2.2. CRISPR-ONT for on-target efficiency prediction
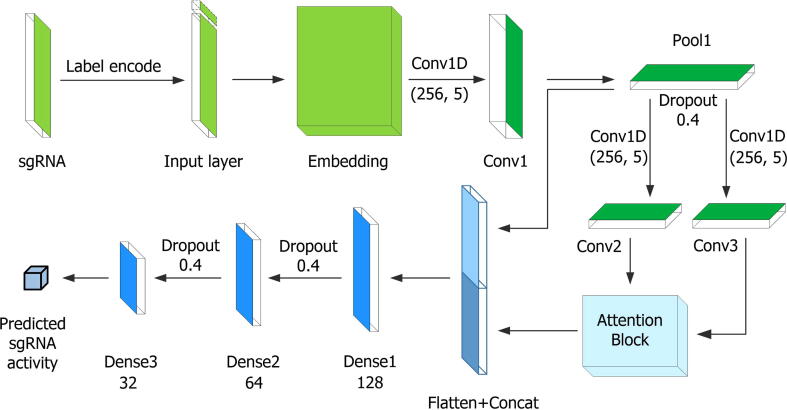
Fig. 1 illustrates the overall architecture of CRISPR-ONT. The input sequence is numerically encoded by using Tokenizer before being fed into our model. It first passes the encoded sgRNA sequence through an embedding layer. The encoded sequence is fed into a convolutional layer for feature extraction. Then, the feature maps are sent into the attention module. Afterwards, the outputs of the two modules are flatten and concatenated, subsequently being passed through three fully connected layers to make a final prediction. The detail regarding how CRISPR-ONT works step by step will be described in the following subsections.
Fig. 1.
Schematic illustration of CRISPR-ONT framework. The sgRNA sequence is label encoded and embedded as an input. After initial feature extraction, the encoded features are forwarded into both convolutional and attention modules for further processing. After being flatten, the features of these two modules are concatenated and passed through three fully connected layers to produce the final output. Dropout is used for model regularization to avoid overfitting. The output is a regression score that highly correlates with sgRNA on-target activity.
2.2.1. Embedding
The k-mer embedding has shown superior performance on sequential analysis [41]. We adopted k-mer embedding to represent the label encoded sequence. The 23-nt sgRNA was changed into a numerical sequence by using Tokenizer module in Keras (https://keras.io) before being fed into CRISPR-ONT. Each nucleotide in sgRNA sequence was denoted as an integer. We added a front padding of size 1 to the numerical sequence. Afterwards, we performed an embedding weight matrix of dimension k, which encodes each base into a vector of size k. The numerical sequence was mapped to a dense real-value spaceHere, L was 24 which represents the length of numerical sgRNA sequence with BOS (begin of sequence) token. is an embedding matrix, k is a hyperparameter corresponds to the embedding dimension by the embedding weight matrix . is computed by:
| (2) |
In the preliminary experiments, we evaluated model performance by varying the k-mer length from 7 to 120 and observed 44 achieved the best performance. The output of the embedding layer will be fed into the CNN layer. Supplementary Fig. S1 shows an example of embedding for a sgRNA sequence.
2.2.2. CNN feature extraction
The one-dimensional convolution (1D-CNN) layers were introduced to learn and scan a set of convolutional filters across its input and detects patterns of the embedding matrix . In the first convolutional layer (Conv1), we used 256 filters with kernel size of 5 to extract features from . To perform dimension reduction, one-dimensional average pooling operation (Pool1) was used, which computes mean value within a window whose size is 2 and with step size of 1. The second convolutional layer used two parallel convolution blocks (Conv2 and Conv3), each with identical filters and kernel size with Conv1. Rectified linear unit (ReLU) [42] activation function was applied in the convolution layers, which is defined as
| (3) |
The sgRNA features extracted after two rounds of convolution operation were concatenated with features of the first round of convolution-pooling operation, so that CRISPR-ONT can learn how sgRNA of different abstract levels determine its efficiency. Note that several convolutional layers stacked together can benefit for detecting motifs in different ranges. Besides, the parallel architecture can significantly reduce model complexity, which can help to combat potential overfitting. Similar ideas have been proposed in [43], [44].
2.2.3. Incorporation of the attention mechanism
Previous studies have shown that position immediately adjacent to PAM is critical for its activity [30], [45]. With this revelation, we assumed that not all features in sequences are equally decisive for sgRNA efficiency. In the field of neural machine translation, the attention mechanism is applied to capture long-range dependencies by training the network to learn which elements of the input sequence to focus on when predicting the output sequence [46]. In sequence analysis, the attention values for individual sites derived by the embedded attention mechanism allow the model to pay attention to those important sites that significantly contribute to the final results [47]. Here, we integrated an attention module into CRISPR-ONT to capture the positional importance of the input sequence, thus improving the predictive power and model interpretability at the level of nucleotide at specific position of sgRNA. The attention layer took the feature vectors after the second round of convolution operations (FConv2 and FConv3) as inputs, then computed a score reflecting whether the neural network shall pay attention to the sequence features at that position. In our early attempts to build the attention-based model, we compared with five attention scores including dot, general, concat, perceptron and add [48] as described in Formula (4) to choose the attentional architecture.
| (4) |
where and strand for the queries and keys metrics, respectively. , and were randomly initialized weight vectors. We applied a uniform distribution to initialize them. The add function works best among these functions. This observation is in accordance with a previous study, which shows that additive attention [46] achieves higher accuracy than multiplicative attention [48]. The detailed results will be presented in Section 3.1. Additive attention is a commonly used attention function which computes the compatibility function using a feed-forward network with a single hidden layer [23]. Hence, we implemented the additive attention to calculate attention scores in our CRISPR-ONT, thus efficiently capturing the internal correlations of the input sequence when predicting the output. It learns to assign different weights to the feature vector corresponding to individual positions and then computes their weighted average to improve prediction. This allows CRISPR-ONT to learn to focus on regions of the sequence that convey more relevant information about sgRNA on-target activity. Supplementary Fig. S2 depicts the attention architecture. Here, and denote features of the jth position in sgRNA sequence, with each dimension corresponding to a kernel in FConv2 and FConv3, respectively. The attentional vector score was then fed through the softmax layer to produce the predictive distribution formulates as:
| (5) |
where denotes attention weights, which reflects the similarity of and
| (6) |
Here, . Larger value of indicates the corresponding position is prior to sgRNA efficiency.
2.2.4. Features fusion and prediction
In order to integrate the features captured by the convolution-pooling module () and attention mechanism (), we employed a concatenation layer to concatenate all the values in and and then forward into the fully connected layers (Dense1, Dense2 and Dense3), among which the number of hidden layer units is 128, 64, 32, respectively. We used dropout [49] for model regularization with a 0.4 dropout rate.
| (7) |
where concat (⋅) and dense(⋅) denote concatenation and dense operations, respectively. represents feature map of the third dense layer () for the merged convolution-pooling and attention modules. was forwarded into an output layer which consists of one neuron corresponding to a regression score of sgRNA efficiency. The linear activation was applied in the final output layer to perform a linear transformation and made prediction of sgRNA cleavage efficacy. That is
| (8) |
where linear (⋅) represents the linear transformation.
2.3. CRISPR-OFFT for off-target activity prediction
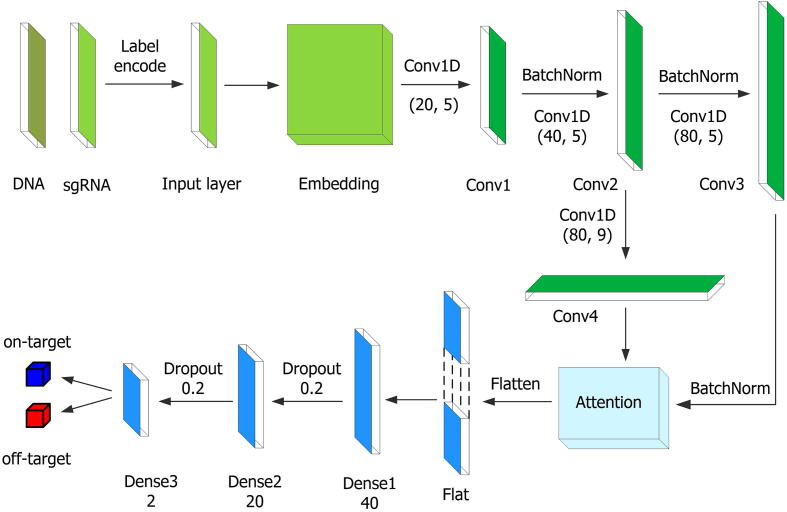
Fig. 2 depicts the network architecture of CRISPR-OFFT, which proceeds in six stages. (i) The pair-wise nucleotides in the particular position are encoded as integrals range from 0 to 15 according to the possible pairwise combinations of all single nucleotides (AA, AC, …, TG, TT). For example, an “AA” in sgRNA-DNA sequence pair is encoded as 0, and “TT” is represented as 15. Consequently, the sgRNA-DNA sequence pair is converted into an integral sequence with size of 23. Supplementary Fig. S3 illustrates how the encoding scheme encodes sgRNA-DNA sequence pair into an integral sequence with 23 (length of the sequence pair) in length and presents an example of how to encode a sgRNA-DNA sequence pair. (ii) Word2vec is applied to map the encoded sgRNA-DNA sequence pair to a dense real-valued high-dimensional space and being used as the input of CNN. Here, the dimension of word embedding is 100. (iii) 1D-CNN with 20 convolutional filters of size 5 with stride size 1 scans along the input sequence. ReLU is chosen as the activation function after each convolutional layer. The convolutional layers mentioned hereafter all use ReLU. Following the convolution and nonlinearity, batch normalization is applied to speed up convergence [50]. Then the extracted features are forwarded into another round of convolution and batch normalization operations, which consists of 40 convolutional filters of length 5. Subsequently, the extracted features are fed into the convolutional-batch normalization and convolutional modules, with 80 convolutional filters of length 5 and 80 convolutional filters of length 9, respectively. (iv) The outputs of these two modules are passed through the attention module for further processing. (v) After being flatten, the outputs of the attention module are fed into two consecutive fully connected layers, which consist 40 and 20 neurons, respectively. We use dropout for model regularization with a 0.2 dropout rate. (vi) The final output layer consists of two neurons that quantify propensity for on- and off-target sites. The two neurons are fully connected to the previous layer using softmax activation function, which is commonly used in the final output layer to the previous layer to distribute the probability throughout each of the output nodes. More details about the CRISPR-OFFT architecture can be found in Supplementary Notes Section S1.1.
Fig. 2.
The CRISPR-OFFT model architecture. The sgRNA-DNA sequence pair is encoded and embedded as an input. After initial feature extraction and two rounds of convolution operations, the encoded features are fed into both linear convolution and another convolution module. Then, the outputs of these two modules are combined to an attention module. Finally, the attention layer is followed by three fully connected layers to assess sgRNA off-target activity.
2.4. Model training and model selection
We implemented the proposed methods in Python 3.6.12 and Keras library 2.3.0 with a Tensorflow (2.2.0) backend. The training and testing processes were performed on a desktop computer with Intel (R) Xeon (R) CPU E5-2678 v3 @ 2.50 GHz, Ubuntu 20.04.1 LTS and 62.8 GB RAM. Four NVIDIA TITAN XP 12 GB of memory per GPU have been used to accelerate the training and testing process. We employed the Adamax [51] with MSE loss function to train our CRISPR-ONT. MSE loss function is formulated as follows:
| (9) |
where k is the number of testing samples, and represent measured sgRNA efficiency and predicted score, respectively. We aimed to optimize the parameters in the proposed CRISPR-ONT by minimizing the loss function in the training process. The distribution of the CRISPR-ONT network parameters such as the number and neurons per layer of CNNs were determined empirically. We applied grid search to adjust the following hyperparameters: dropout rate over the choice (0.2, 0.3, 0.4, 0.5), batch size over the choice (64, 128, 256) and number of epochs over the choice (50, 100, 150, 200). For all experiments, we used 10-fold cross-validation analysis to evaluate our models. The optimal hyperparameters were determined as: dropout rate was 0.4, epoch was 100 and batch size was 128. The learning rate was uniformly changed. We set its initial value of learning rate to 0.0005, and the value was reduced to 4/5 of the original value every 100 epochs. The lower limit of the learning rate was set to 1e-5.
Binary cross entropy (BCE) loss function is well used in binary-label neural networks, which is defined as
| (10) |
where N is the size of the batch. We used the Adam optimizer to train our CRISPR-OFFT for minimizing the BCE loss function. The learning rate was uniformly changed. An initial learning rate is set to 0.003 to CRISPR-OFFT and a reducing factor of 0.2 after 60 iterations. The lower limit of the learning rate was set to 1e-5. Similarly, the distribution of the CRISPR-OFFT network parameters was also determined empirically. Grid search was applied to adjust the following hyperparameters: dropout rate over the choice (0.2, 0.3, 0.4), batch size over the choice (64, 128, 256, 512) and number of epochs over the choice (30, 60, 90, 120). The optimal hyperparameters of CRISPR-OFFT were determined as: dropout rate was 0.2, epoch was 60 and batch size was 256.
2.5. Evaluation metrics
We quantitatively assessed the performance of the proposed CRISPR-ONT with two commonly used evaluation metrics including Spearman correlation coefficient (SCC) between predicted and measured on-target activity and the area under Receiver Operating Characteristic (AUROC). For a sample of size n, the n raw scores , are converted to ranks , . SCC is calculated by:
| (11) |
where represents the covariance of the rank variables, and donate the standard deviations of the rank variables. SCC was calculated using SciPy library (http://scipy.org). AUROC was calculated to comprehensively quantify the overall predictive ability. The value of AUROC is in 0.5 ~ 1, where a larger value means the model is predictive and robust. We used 0.5 AUROC as the baseline.
In addition, we assessed the performance of our CRISPR-OFFT using two evaluation metrics. The ROC curve is plotted with vertical axis shows sensitivity and horizontal axis shows 1-specificity, reflecting the relationship between sensitivity and specificity at different thresholds. The off-target datasets used in this study are extremely imbalance. ROC curve does not change due to the distribution of positive and negative samples. So, it is suitable for imbalanced binary classification. Precision recall (PR) curve is plotted with as the vertical axis and as the horizontal axis, which reflecting the trade-off between the Precision of the recognition of positive samples and the ability to cover positive examples. Area under the PR curve (PRAUC) closes to 1 represents the model is more predictive. Precision and Sensitivity are defined as follows:
| (12) |
| (13) |
where TP, FP and FN represent true positive, false positive and false negative, respectively.
3. Results
3.1. Choices of attentional architectures for CRISPR-ONT
We first conducted extensive analysis to examine the performance of attention-based model with different alignment functions including dot, general, concat, perceptron and add and shed light on which function is best for the proposed attention architecture. We applied five alternatives to calculate the attention scores for predicting sgRNA efficiency on five independent datasets under 10-fold cross-validation. Overall, CRISPR-ONT_add achieved superior performance in terms of SCC on these datasets. Besides, it also showed higher AUROC scores on all datasets except for dataset SpCas9-NG (see details in Supplementary Table S2). Together, among different models, the attention model with add is best, as evaluated by SCC and AUROC.
3.2. Comparison of CRISPR-ONT and state-of-the-art sgRNA efficiency predictors
In order to access the predictive ability of CRISPR-ONT, we compared it with five state-of-the-art deep learning-based methods including attnToCrispr_CNN, DeepCas9, DeepSpCas9, DeepHF and C-RNNCrispr across five independent datasets. Prior to this, we briefly comment on some comparisons among these methods (Table 1). (1) CRISPR-ONT, AttnToCrispr_CNN and DeepHF apply k-mer computed using embedding method, while others use one-hot encoding to represent the nucleotides. (2) The architecture parameters are different among these methods. Specifically, DeepSpCas9 merges three 1D-CNN layers. DeepCas9 uses one 1D-CNN layer, whereas AttnToCrispr_CNN uses two layers of transformer-based 2D-CNNs. DeepHF uses a single BLSTM layer, whereas C-RNNCrispr uses a single 1D-CNN followed by a BGRU. CRISPR-ONT uses multiple layers of 1D-CNNs with attention mechanism to improve the interpretability. (3) Besides sgRNA sequences, attnToCrispr_CNN, DeepHF and C-RNNCrispr introduce biological features to improve performance, such as gene expression profile. Other methods such as CRISPR-ONT predict sgRNA on-target activity by considering sequence composition alone. For more details, see Supplementary Tables S3 to S8. We compared these models by considering sgRNA sequence. To make a fair comparison, we trained CRISPR-ONT based on the training data strictly consistent with other methods. Each dataset was randomly divided into training dataset and independent test dataset with the proportion of 85%:15%. The training process was performed under 10-fold cross-validation on each training dataset. In the testing stage, we tested the proposed CRISPR-ONT under the same condition with all the compared methods on each testing dataset.
Table 1.
Existing deep learning-based methods for CRISPR/Cas9 sgRNA efficiency prediction.
| Model | Encoding | Architecture | Sequence |
|---|---|---|---|
| CRISPR-ONT | embedding | 2 1D-CNNs, attention | sgRNA |
| AttnToCrispr_CNN | embedding | 2 2D-CNNs, transformer | sgRNA, gene expression |
| DeepCas9 | one-hot | 1D-CNN | sgRNA |
| DeepSpCas9 | one-hot | 3 1D-CNNs | sgRNA |
| DeepHF | embedding | BLSTM | melting temperatures, stem-loop |
| C-RNNCrispr | one-hot | 1D-CNN, BGRU | sgRNA, epigenetic features |
Note: BLSTM, bidirectional long short-term memory network; BGRU, bidirectional gate recurrent unit network.
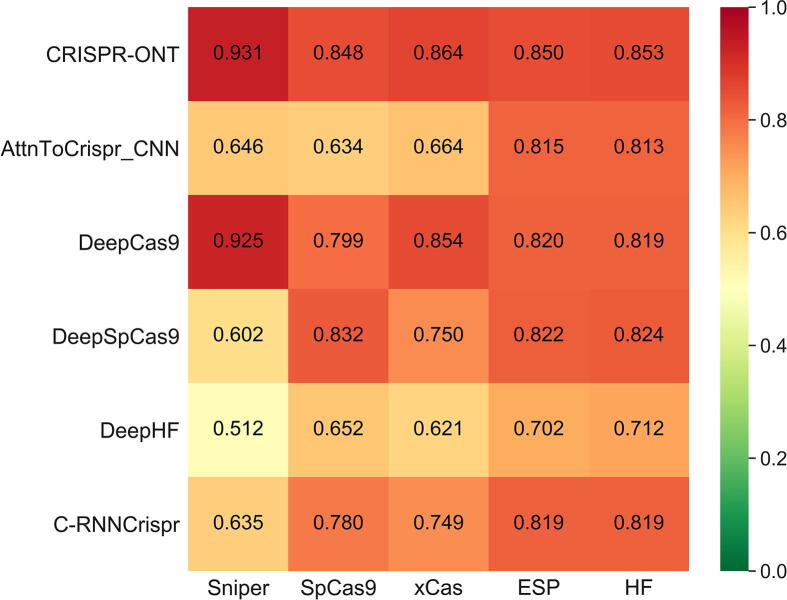
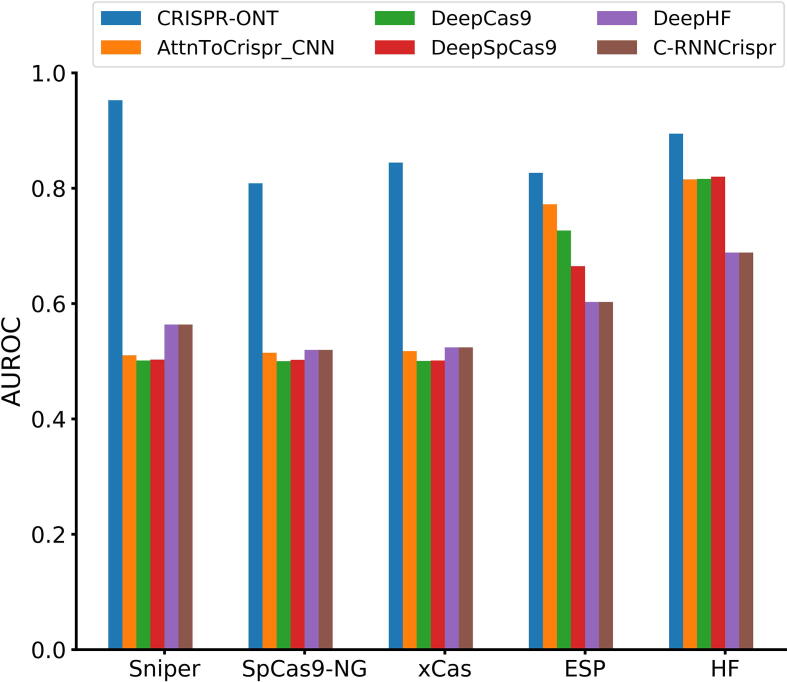
On the whole, CRISPR-ONT achieved the highest SCC, with increase by 2.6% on average compared with the second best DeepCas9 (Fig. 3). It is clear that CRISPR-ONT consistently outperformed others in terms of AUROC (Fig. 4). CRISPR-ONT achieved a mean AUROC of 0.865 while the second best attnToCrispr_CNN achieved 0.626 on the matched training, validation and test data split. This demonstrates the effectiveness of the proposed model for CRISPR/Cas9 sgRNA on-target activity prediction. Hence, we conclude that our CRISPR-ONT is competitive against other existing deep learning-based predictors.
Fig. 3.
Heatmap of SCC between CRISPR-ONT and other methods on various datasets under 10-fold cross-validation. The prediction models are placed vertically, whereas the test sets are arranged horizontally.
Fig. 4.
Comparison of AUROC of CRISPR-ONT and the state-of-the-art deep learning-based methods on five datasets under 10-fold cross-validation.
We further compared the running time of CNN integrates attention-based CRISPR-ONT, the CNN-RNN based C-RNNCrispr and the pure RNN-based DeepHF. Table 2 reports the training time for each predictor on five datasets over 10-fold cross-validation. We observed CRISPR-ONT gained superiority of running time than the best alternate method C-RNNCrispr. More specifically, we trained CRISPR-ONT using 4 NVIDIA TITAN XP GPUs, spending about 3.94 h for these datasets. Due to the internal structure of BLSTM, DeepHF needs more computational resource for model training. Under the same settings, DeepHF took about 10 times training time than CRISPR-ONT. As expected, the running time of C-RNNCrispr was somewhere in between. Therefore, the substantial difference in running time can be attributed to the efficiency of CRISPR-ONT over C-RNNCrispr and DeepHF, which helps it save computational resources, especially for large-scale prediction. Together, these observations suggest that CRISPR-ONT achieved superior performance in terms of accuracy and time complexity for sgRNA efficiency prediction.
Table 2.
Overall training time cost comparison of CRISPR-ONT, C-RNNCrispr and DeepHF on five datasets under 10-fold cross-validation.
| Model | Sinper | SpCas9 | xCas | ESP | HF |
|---|---|---|---|---|---|
| CRISPR-ONT | 2640 | 2145 | 2616 | 4093 | 2691 |
| C-RNNCrispr | 10,419 | 9288 | 10,354 | 16,140 | 15,643 |
| DeepHF | 24,017 | 19,314 | 24,275 | 37,899 | 36,292 |
3.3. Biological insights into CRISPR-ONT for sgRNA efficiency prediction
Next, we examined the contribution of attention mechanism. We explored whether CRISPR-ONT can learn the important positions in the protospacer, which have significant influences on sgRNA cleavage efficacy. If certain positions have great effects on sgRNA efficiency, then randomizing those nucleotides is expected to dramatically deteriorate the performance. We performed a permutation nucleotide importance analysis, systematically randomizing each position in testing sequences and examining the resulting effect on the outputs.
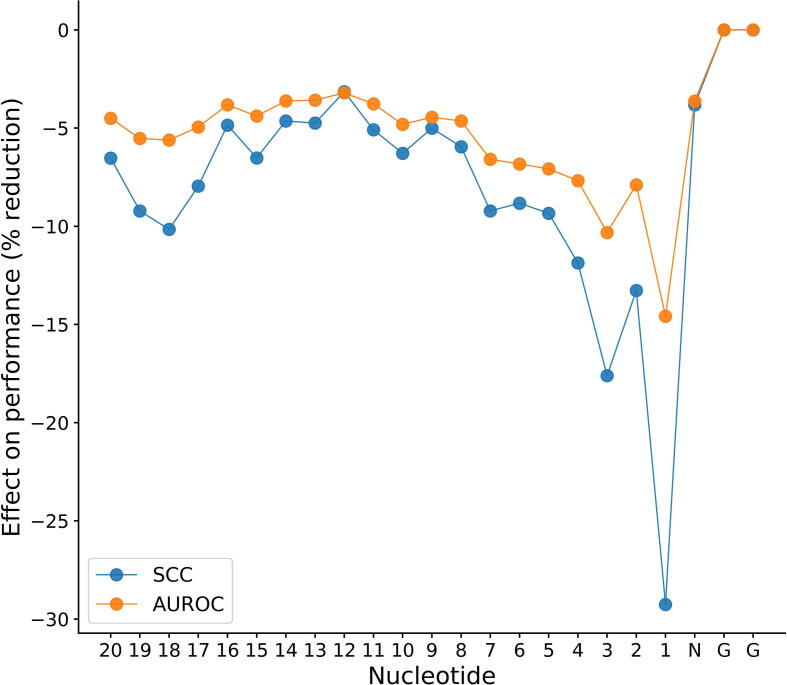
As depicted in Fig. 5, the nucleotide at 1 bp upstream of the PAM has the greatest effect on sgRNA activity, reducing SCC by 29.1% upon randomization. This observation is consistent with previous findings that nucleotides proximal to the PAM were the most predictive of efficiency [52]. Nucleotide positions 2, 3, 4 and 5 also showed effects, although weaker, reducing SCC by 12.78%, 17.41%, 11.92% and 9.41%, respectively. These observations are in accordance with a previous study, which corroborates that base pair in the first 5 nucleotides adjacent to the PAM (the core region) was found to be more important than pairing in the rest of the region [53]. The core region sequence largely determines target efficiency [3], [54]. These findings are also reminiscent of the previously observed nucleotides at 1 ~ 5 positions are crucial for the interaction with crRNA and subsequent being cut by Cas9 [53]. Intriguingly, we observed a second largest reduction at position 3 bp upstream of the PAM. This is likely caused by the factor that cleavage tend to be located at this position [55]. Chakrabarti et al. found that HNH usually cuts at position 3, while RuvC flexibly cuts at either 3, 4, 5 or even further upstream of the PAM [56]. Another study also demonstrated that 2~5 nucleotides upstream of the PAM are critical for the cleavage precision of the target site [57]. Interestingly, we also found 3 base pairs in the PAM distal region are also crucial for sgRNA activity, which in agreement with a previous finding [58]. Similar results were observed in terms of AUROC. Taken together, we conclude that CRISPR-ONT can reflect the local sgRNA efficiency characteristic, thus providing important biological insights into sgRNA cleavage efficacy.
Fig. 5.
Contribution of protospacer nucleotides to sgRNA efficiency. The effect of nucleotide randomization on dataset ESP is shown as reduction of SCC and AUROC. ‘1’ signifies position of the nucleotide with 1 base pair upstream of the NGG protospacer adjacent motif (PAM).
3.4. Visualization of sequence features learned by CRISPR-ONT
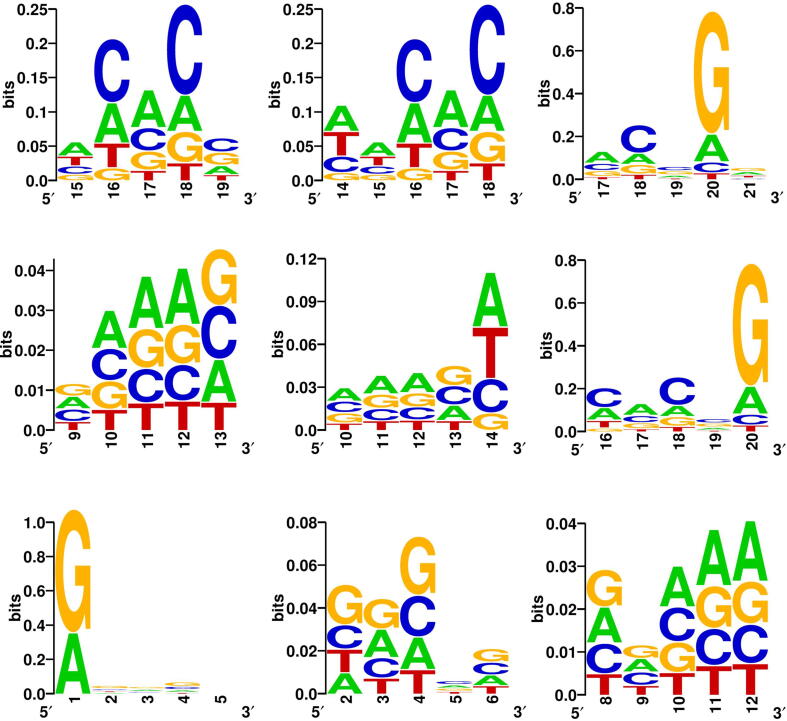
We further explore the ability of CRISPR-ONT to capture informative sequence features. Using the similar approach in previous studies [59], [60], [61], we visualized the specific sequence features from the first convolutional layer. Each filter in the first convolutional layer can be regarded as a specific sequence motif discriminator. The first convolutional layer of CRISPR-ONT contains 256 filters with kernel size of 5 to extract the sequence features. While feeding the encoded sgRNA sequence to the well-trained CRISPR-ONT, the filters of the first convolutional layer scan the 5-nt long subsequence from the beginning to the end of the input sequence with stride of 1. Each filter computes the 1D-CNN with the 5-nt subsequence and produces an activation value for this sequence fragment. If a 5-nt subsequence achieves a larger activation value for a specific filter, it means that this sequence fragment is more likely to be the pattern that the convolution unit tends to find. We fed all the test sequences of dataset ESP through the first convolutional layer of the model. For each filter, we extracted all 5-nt sequence fragments that activate the filter and use only activations that has the maximum value. After all the 5-nt subsequences were extracted, they were stacked and the nucleotide frequencies were counted to produce a position frequency matrix. Sequence logos of the corresponding subsequences from convolution units were then generated by WebLogo [62]. Fig. 6 depicts the top 9 highly activated convolution units that were most related to CRISPR/Cas9 sgRNA efficiency. Interestingly, T were disfavored at position 17 (4 bp upstream of the PAM), which also observed by Chuai et al. [12]. In addition, we observed many patterns are G-rich, indicating that G enrichment is likely to be an important feature. Moreover, C was found to be informative at position 18, which reflects that cleavage site usually resides 3 bp upstream of the PAM.
Fig. 6.
Examples of sequence motifs detected by first layer convolutional modules learned by CRISPR-ONT. X-axis represents the position of the protospacer sequence. Here, position 20 is referred to the nucleotide 1 bp upstream of the PAM.
3.5. Comparison of CRISPR-OFFT and existing sgRNA off-target predictors
To evaluate the predictive ability of CRISPR-OFFT, we compared it with four computational off-target predictors including CFD, CNN_std, AttnToMismatch_CNN and CnnCrispr on the above off-target datasets. For each cell line, we randomly selected 20% of the data as the independent test set. The remaining 80% of data for two cell lines were merged as the training data to train CRISPR-OFFT and tune the hyperparameters during cross-validation procedure. Notably, the labels of the whole dataset were highly imbalanced, so we applied the bootstrapping sampling algorithm to the mini-batches for model training. For any given dataset, the training procedure is as follows: (i) the negative samples of each dataset were randomly divided into n parts, where , N is the number of negative samples and 256 is the mini-batch size. (ii) 256 samples were randomly selected with replacement from positive data in the training set and being combined with the negative sample in (i) to construct a mini-batch training dataset. (iii) Repeat step (ii) N times to get a total of N balanced mini-batch training set. A similar idea has been adopted in previous studies [12], [16]. This strategy ensures that mini-batches make the ratio of positive and negative examples 1:1 in the training data, thus avoiding gradient update instability and substantially improving the predictive power.
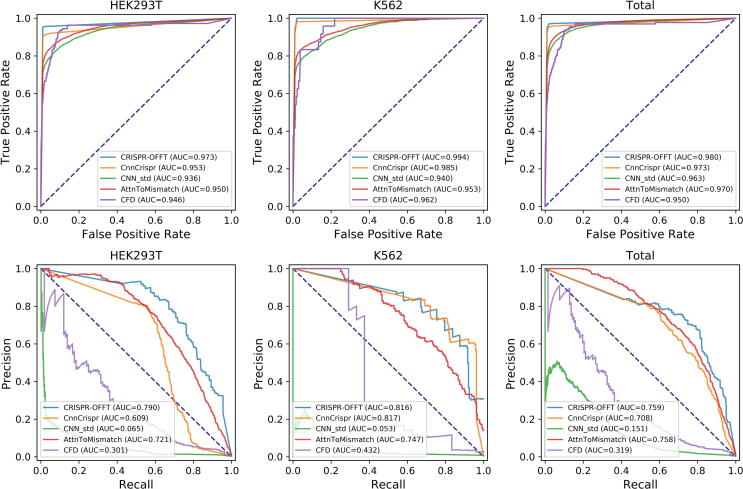
When evaluated on datasets HEK293T, K562 and Total, CRISPR-OFFT achieved substantially superior performance compared to others, reaching AUROCs of 0.973, 0.994 and 0.980, respectively (Fig. 7). This implies CRISPR-OFFT achieves the highest true positive rate among all predictors when the false positive rate is fixed. Besides, it can also be observed that CRISPR-OFFT performs better than other methods on datasets HEK293T and Total in PRAUC, with values of 0.790 and 0.759, respectively. CRISPR-OFFT achieved comparable performance with CnnCrispr on dataset K562, with PRAUCs of 0.816 and 0.817, respectively, reflecting that CRISPR-OFFT can dramatically decrease the false positives. In summary, our CRISPR-OFFT outperforms the others on specific cell lines.
Fig. 7.
Performance comparison of CRISPR-OFFT and different computational models in terms of ROC curves (up) and Precision-Recall curves (bottom) on three independent datasets.
3.6. Testing the generalizability of CRISPR-OFFT
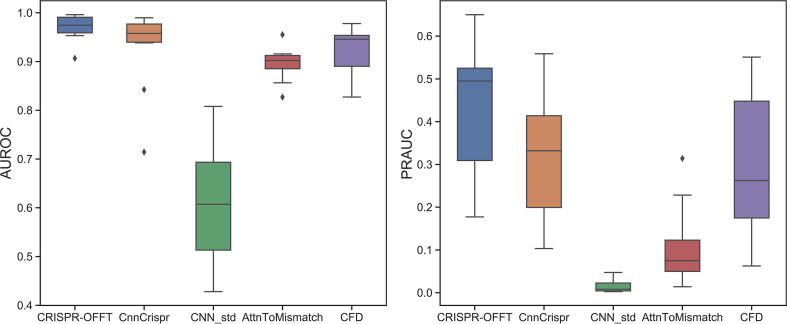
To examine the generalizability of CRISPR-OFFT for off-target prediction, we tested the compared models on the above datasets under 10-fold cross-validation. We randomly split all 30 sgRNAs into 10 groups for testing, each contained 3 sgRNAs. For each test set, we chose 27 sgRNAs from total 30 sgRNAs as a training set, which have no overlaps with current selected testing sgRNAs. We applied the bootstrapping method as mentioned in Section 3.7 to eliminate data imbalance of positive samples and negative samples. Fig. 8 shows the distribution of AUROCs and PRAUCs for CRISPR-OFFT and other methods. Overall, we observed that our CRISPR-OFFT consistently outperforms others. We obtained mean AUROCs of 0.970, 0.927, 0.610, 0.896 and 0.922 for CRISPR-OFFT, CnnCrispr, CNN_std, AttnToMismatch_CNN and CFD, respectively. Besides, the above methods yielded PRAUCs of 0.440, 0.317, 0.015, 0.107 and 0.300, respectively. On average, CRISPR-OFFT achieved 4% and 12% higher than the second best CnnCrispr. These observations indicated that CRISPR-OFFT has better generalizability than other tools.
Fig. 8.
Performance comparison of CRISPR-OFFT and different computational models in terms of AUROC (left) and PRAUC (right) on dataset under 10-fold cross-validation. AttnToMismatch: AttnToMismatch_CNN.
3.7. Advantages of transfer learning for small-size cell line sgRNAs off-target prediction
In CRISPR/Cas9 sgRNA specificity analysis, a most common issue is that the number of valid off-target data is limited and small. Training a fully deep network structure with small number of data may lead to overfitting, which is the main cause of low performance and generalization ability. Transfer learning [63] is a good way to tackle this challenge where the learned parameters of well-trained model on a large dataset are shared to the targeted network model. Hence, the parameters’ transfer of the pre-trained model may provide the new target model a powerful feature extraction ability and reduce computation cost.
We applied a transfer learning strategy to enhance the predictive power of CRISPR-OFFT for unseen sgRNAs. More narrowly, we randomly selected 15 sgRNAs from datasets HEK293T and K562 to construct a benchmark dataset for model pre-training. The remaining 15 sgRNAs were randomly partitioned into 3 groups, each group contains 5 sgRNAs. The training and test data for each dataset were generated in the same way as described in Section 3.7. We first trained our CRISPR-OFFT on the benchmark for feature extraction. During fine-tuning, we optimized the BCE loss function, only updating the weight parameters in the last two fully connected layers (862 free parameters) again with the remaining sgRNAs. For any given sgRNA of interest, the training process proceeds in four stages. (i) Pre-train the CRISPR-OFFT with the benchmark dataset for 20 epochs. (ii) Freeze the convolution, attention and the first fully connected layers. (iii) Train the last two fully connected layers of the model with the training data from the sgRNAs of interest for another 50 epochs. (iv) Evaluate the model on the test data.
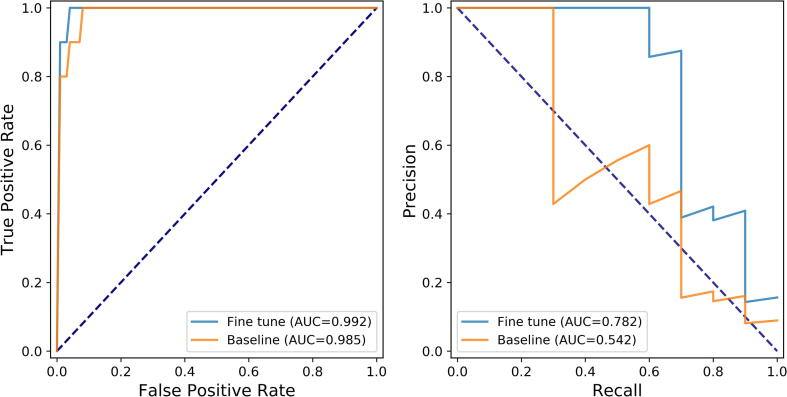
In order to estimate the effect of transfer learning, we compared two strategies: training CRISPR-OFFT from scratch based on new sgRNAs alone and transferring the well-trained CRISPR-OFFT to fit new sgRNAs data via fine-tune. As depicted in Fig. 9, fine tune improved the predictive power compared to model training from scratch, leading to 0.7% and 24% improvements on average in terms of AUROC and PRAUC, respectively. These observations indicated that transfer learning precedes the naive training way in terms of sgRNA-specific prediction.
Fig. 9.
Impact of transfer learning via fine tune on AUROC (left) and PRAUC (right). Baseline corresponds to training the CRISPR-OFFT from scratch only with the sgRNAs of interest, and fine tune corresponds to pre-training the CRISPR-OFFT on the benchmark dataset followed by updating the fully connected layers on sgRNAs of interest.
4. Discussion
Accurate prediction of CRISPR/Cas9 sgRNA on-target activity is a major goal in genetic manipulation research. Besides, the potential homologous off-target binding and cleavage are important concerns for this system and they are highly depending on the selection of sgRNA. In this study, we developed two attention-based CNN frameworks, namely CRISPR-ONT and CRISPR-OFFT, for CRISPR/Cas9 sgRNA efficiency and specificity prediction, respectively. Both the proposed algorithms use CNNs frameworks to extract the contextual sequence features and have built-in attention modules to focus on the specific part of the input to help extract interpretable Cas9 binding sgRNA patterns. Comprehensive tests on public datasets showed the superior performance of the proposed methods.
The architecture of CRISPR-ONT allows it to combine features of different abstraction levels extracting from the one round convolution-pooling and two rounds convolution-pooling modules with attention for boosting the predictive power for sgRNA efficiency prediction. Furthermore, dropout regularization was applied to combat overfitting of CRISPR-ONT. We compared various alignments functions and showed attention with add function is superior for sgRNA on-target prediction. Given the black box nature of deep learning, we try to interpret our CRISPR-ONT through three aspects. First, to embed the explainability in our framework, we introduced the attention mechanism, which has been widely applied in deep learning to indicate important positions in the input. As demonstrated earlier, using attention module makes model pay attention to some more important segments in the sequence. It can also be understood that the influence of nucleotides differs at different positions of sgRNA sequence on its efficiency, therefore, applying the attention module may contribute to the prediction accuracy. Second, we applied the in-depth permutation nucleotide importance analysis to analyze the effect of nucleotide at particular position for sgRNA efficiency, thus revealing the underlying biological mechanisms of our model. Third, we interpret our model by converting the convolutional filters into sequence logos to find the informative sequence patterns for sgRNA activity. We visually inspected the convolution units of the first convolutional layer of the pre-trained CRISPR-ONT to find the important sequence patterns that can affect the sgRNA efficiency. In fact, these three approaches are complementary to each other in generating biological insights from CRISPR-ONT framework. To sum up, CRISPR-ONT is a computational tool for accurate sgRNA target efficacy prediction which has the potential to provide novel biological hypotheses. We also showed that CRISPR-ONT is competitive with the state-of-the-art methods when tested against prokaryotes (Supplementary Table S9). Additionally, our work could be applied for CRISPR/Cas12a system by adjusting the input shape of the model.
In spite of widely used of CRISPR/Cas9 for genome editing, general principles that govern genome-wide off-target activity remain largely unknown. We proposed an attention-based CNN architecture named CRISPR-OFFT to predict sgRNA off-target activity. CRISPR-OFFT automatically trains the sequence features of sgRNA-DNA pairs, and embeds the trained word vector matrix into CNN with attention mechanism. Extensive tests on public datasets demonstrated that CRISPR-OFFT outperforms other methods in terms of AUROC and PRAUC. More importantly, we applied transfer learning via fine tune to train CRISPR-OFFT by taking advantage of data from 15 sgRNAs from total off-target dataset and their commonalities. To our knowledge, this is the first study for investigating the effect of transfer learning for sgRNA off-target activity prediction. Applying this strategy, CRISPR-OFFT achieved clearly superior performance than training from scratch. This is not surprising given that there are commonalities across sgRNAs. Hence, we conclude that the representations learned from other sgRNAs are effectively transferred and exploited for the target dataset. When transfer learning is applied, fewer annotations are needed to achieve satisfactory performance. Our results suggest that there is promise in the idea of sharing information between tasks and between cell lines for off-target prediction and may help overcome the limitations of training deep learning models on small size datasets.
Generalization is a drawback of deep learning-based CRISPR/Cas9 activity and specificity prediction methods, namely a model only preforms well in a specific dataset but not in an unseen dataset [64]. These techniques are data-driven, however, there are several challenges for achieving sufficient unbiased training dataset to adequately fit the on- and off-target prediction models. On the one hand, data heterogeneity is a common issue for on-target prediction. Datasets from different cell types, organisms, and platforms are heterogeneous and could not be simply mixed [65]. Feature learning on heterogeneous datasets may improve the performance by accounting for more influencing factors, such as energetic or epigenetic factors [16]. For example, Chari et al. identified epigenetic status to be an additional modulator of sgRNA activity considering DNase-seq and H3K4 trimethylation data [66]. Although adding epigenetic information may improve model accuracy, it also leads to decreasing the generalizability compared to sequence-only models. This is because epigenetic features are highly variable cross-species and depend on the cell type and cell state, restricting the application of these methods for species-specific and cell type-specific predictions [67]. They hence opted for considering only sequence information for their sgRNA Scorer and sgRNA Scorer 2.0 methods [66], [67]. On the other hand, compared to the datasets for on-target prediction, the amount of off-target raw data might be less sufficient [12]. Additionally, data imbalance [68] is a common pitfall for off-target prediction. A majority of the available sgRNA off-target data are detected via high-throughput sequencing. For each target site, the homologous off-target sequences with cleavage efficiency can be genome-wide detected. The detected homologous sgRNAs are defined as positive data, and all possible nucleotide mismatch loci are regarded as negative data [12], [16], [65], [69]. The homologous sgRNA target sites with undetected cleavage outnumber that of the detective ones. Deep learning models trained on imbalanced data tend to achieve high accuracies for the majority class. But the learning models generally perform worse for the minority class, which is noteworthy in this case. These issues could be mitigated by applying proper normalization and regularization techniques to penalize and reduce data-associated noise and biases. Continuous efforts are required to further improve the accuracy and robustness methods for CRISPR/Cas9 on- and off-target predictions.
Our future work will focus on prediction of sgRNA off-target activities with insertions or deletions of sgRNA-DNA pairs. Currently, the proposed CRISPR-OFFT only uses sgRNA-DNA pairs to evaluate CRISPR/Cas9 sgRNA specificity. A previous study has shown that DNA sequences contain insertions (DNA bulge) and deletions (RNA bulge) are an integral part of the CRISPR system, which are important for comprehensive off-target analysis [70]. Abadi et al. proposed a machine learning-based method named CRISTA for CRISPR cleavage propensity prediction and found that DNA/RNA bulges are predictive features for boosting the performance [16]. Recently, a RNN-based CRISPR-Net has been developed to predict off-target activities with insertions and deletions from sgRNA-DNA pairs [27]. However, the improvement in the predictive power comes at the expense of training time due to the RNN-based model structure. Therefore, it is still room for developing more accurate and faster deep learning-based models for CRISPR/Cas9 specificity by considering both DNA and RNA bulges, which is deserved to be explored in future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 61872396), the Natural Science Foundation of Guangdong Province (No. 2014A030308014) and the STU Scientific Research Foundation for Talents (No. NTF20032).
Author’s contributions
All authors contributed to the project design. GSZ and TZ wrote the analysis source code, analyzed the data. GSZ drafted the full manuscript. ZMD and XXD critically revised the final manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.001.
Contributor Information
Zhiming Dai, Email: daizhim@mail.sysu.edu.cn.
Xianhua Dai, Email: issdxh@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hsu P., Lander E., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science:1258096. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12(10):982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 9.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D., Kim S., Kim S., Park J., Kim J.-S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26(3):406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19(1) doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue L.i., Tang B., Chen W., Luo J. Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J Chem Inf Model. 2019;59(1):615–624. doi: 10.1021/acs.jcim.8b00368. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Zhang J. Prediction of sgRNA on-target activity in bacteria by deep learning. BMC Bioinf. 2019;20:517. doi: 10.1186/s12859-019-3151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H.K., Kim Y., Lee S., Min S., Bae J.Y., Choi J.W. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci Adv. 2019;5(11):eaax9249. doi: 10.1126/sciadv.aax9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abadi S., Yan W.X., Amar D., Mayrose I., Xu H. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol. 2017;13(10):e1005807. doi: 10.1371/journal.pcbi.1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengio Y., Simard P., Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5(2):157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 18.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 19.Jozefowicz R, Zaremba W, Sutskever I. An empirical exploration of recurrent network architectures. 2015.
- 20.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep learning. [Google Scholar]
- 21.Wang D., Zhang C., Wang B., Li B., Wang Q., Liu D. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-12281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devikanniga D., Raj J.S., R Classification of osteoporosis by artificial neural network based on monarch butterfly optimisation algorithm. Healthcare Technol Lett. 2018;5:70–75. doi: 10.1049/htl.2017.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. arXiv preprint arXiv:1706.03762; 2017.
- 24.Liu Qiao, He Di, Xie Lei, Segata Nicola. Prediction of off-target specificity and cell-specific fitness of CRISPR-Cas System using attention boosted deep learning and network-based gene feature. PLoS Comput Biol. 2019;15(10):e1007480. doi: 10.1371/journal.pcbi.1007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J, Wong K-C. Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics 2018; 34:i656–i63. [DOI] [PMC free article] [PubMed]
- 26.Haapaniemi Emma, Botla Sandeep, Persson Jenna, Schmierer Bernhard, Taipale Jussi. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 27.Lin Jiecong, Zhang Zhaolei, Zhang Shixiong, Chen Junyi, Wong Ka‐Chun. CRISPR-Net: a recurrent convolutional network quantifies CRISPR off-target activities with mismatches and indels. Adv Sci. 2020;7(13):1903562. doi: 10.1002/advs.v7.1310.1002/advs.201903562. [DOI] [Google Scholar]
- 28.Li J, Luong M-T, Jurafsky D. A hierarchical neural autoencoder for paragraphs and documents. arXiv preprint arXiv:1506.01057; 2015.
- 29.Hu Y, Wang Z, Hu H, Wan F, Chen L, Xiong Y, et al. ACME: pan-specific peptide-MHC class I binding prediction through attention-based deep neural networks-附件. Bioinformatics 2019. [DOI] [PubMed]
- 30.Jiang Wenyan, Bikard David, Cox David, Zhang Feng, Marraffini Luciano A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Jungjoon K., Jeong Euihwan, Lee Joonsun, Jung Minhee, Shin Eunji, Kim Young-hoon. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimasu Hiroshi, Shi Xi, Ishiguro Soh, Gao Linyi, Hirano Seiichi, Okazaki Sae. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361(6408):1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Johnny H., Miller Shannon M., Geurts Maarten H., Tang Weixin, Chen Liwei, Sun Ning. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Nahye, Kim Hui Kwon, Lee Sungtae, Seo Jung Hwa, Choi Jae Woo, Park Jinman. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat Biotechnol. 2020;38(11):1328–1336. doi: 10.1038/s41587-020-0537-9. [DOI] [PubMed] [Google Scholar]
- 35.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frock Richard L, Hu Jiazhi, Meyers Robin M, Ho Yu-Jui, Kii Erina, Alt Frederick W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran F. Ann, Cong Le, Yan Winston X., Scott David A., Gootenberg Jonathan S., Kriz Andrea J. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Xiaoling, Wang Yebo, Wu Xiwei, Wang Jinhui, Wang Yingjia, Qiu Zhaojun. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33(2):175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 39.Langmead Ben, Salzberg Steven L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalal Z., Matas J., Mikolajczyk K. 2010 IEEE computer society conference on computer vision and pattern recognition. IEEE; 2010. Pn learning: Bootstrapping binary classifiers by structural constraints; pp. 49–56. [Google Scholar]
- 41.Lopez MM, Kalita J. Deep Learning applied to NLP. arXiv preprint arXiv:1703.03091; 2017.
- 42.Krizhevsky A., Sutskever I., Hinton G. ImageNet classification with deep convolutional neural networks. Adv Neural Inform Process Syst. 2012;25 [Google Scholar]
- 43.Kim Y. Convolutional neural networks for sentence classification. arXiv preprint arXiv:1408.5882; 2014.
- 44.Zhang Sai, Hu Hailin, Zhou Jingtian, He Xuan, Jiang Tao, Zeng Jianyang. Analysis of ribosome stalling and translation elongation dynamics by deep learning. Cell Syst. 2017;5(3):212–220.e6. doi: 10.1016/j.cels.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Kim Hui K, Song Myungjae, Lee Jinu, Menon A Vipin, Jung Soobin, Kang Young-Mook. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14(2):153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 46.Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473; 2014.
- 47.Hu Y, Wang Z, Hu H, Wan F, Chen L, Xiong Y, et al. ACME: pan-specific peptide-MHC class I binding prediction through attention-based deep neural networks. Bioinformatics 2019; 35:4946–54. [DOI] [PubMed]
- 48.Luong M-T, Pham H, Manning CD. Effective approaches to attention-based neural machine translation. arXiv preprint arXiv:1508.04025; 2015.
- 49.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 50.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167; 2015.
- 51.Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980; 2014.
- 52.Dhanjal Jaspreet Kaur, Dammalapati Samvit, Pal Shreya, Sundar Durai. Evaluation of off-targets predicted by sgRNA design tools. Genomics. 2020;112(5):3609–3614. doi: 10.1016/j.ygeno.2020.04.024. [DOI] [PubMed] [Google Scholar]
- 53.O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res 2015; 43:3389–404. [DOI] [PMC free article] [PubMed]
- 54.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shou Jia, Li Jinhuan, Liu Yingbin, Wu Qiang. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell. 2018;71(4):498–509.e4. doi: 10.1016/j.molcel.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 56.Chakrabarti Anob M., Henser-Brownhill Tristan, Monserrat Josep, Poetsch Anna R., Luscombe Nicholas M., Scaffidi Paola. Target-specific precision of CRISPR-mediated genome editing. Mol Cell. 2019;73(4):699–713.e6. doi: 10.1016/j.molcel.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemos Brenda R., Kaplan Adam C., Bae Ji Eun, Ferrazzoli Alexander E., Kuo James, Anand Ranjith P. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci U S A. 2018;115(9):E2040–E2047. doi: 10.1073/pnas.1716855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo J., Chen W., Xue L., Tang B. Prediction of activity and specificity of CRISPR-Cpf1 using convolutional deep learning neural networks. BMC Bioinf. 2019;20:332. doi: 10.1186/s12859-019-2939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alipanahi Babak, Delong Andrew, Weirauch Matthew T, Frey Brendan J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33(8):831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 60.Seo S, Oh M, Park Y, Kim S. DeepFam: deep learning based alignment-free method for protein family modeling and prediction. Bioinformatics 2018; 34:i254–i62. [DOI] [PMC free article] [PubMed]
- 61.Trabelsi A, Chaabane M, Ben-Hur A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 2019;35:i269–i77. [DOI] [PMC free article] [PubMed]
- 62.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bengio Y. Proceedings of ICML workshop on unsupervised and transfer learning. 2012. Deep learning of representations for unsupervised and transfer learning; pp. 17–36. [Google Scholar]
- 64.Haeussler Maximilian, Schönig Kai, Eckert Hélène, Eschstruth Alexis, Mianné Joffrey, Renaud Jean-Baptiste. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1) doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Jun, Zhang Xiuqing, Cheng Lixin, Luo Yonglun. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 2020;17(1):13–22. doi: 10.1080/15476286.2019.1669406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chari Raj, Mali Prashant, Moosburner Mark, Church George M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12(9):823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chari Raj, Yeo Nan Cher, Chavez Alejandro, Church George M. sgRNA Scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6(5):902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao Y, Chuai G, Yu W, Qu S, Liu Q. Data imbalance in CRISPR off-target prediction. Brief Bioinform 2020;21:1448–54. [DOI] [PubMed]
- 69.Listgarten Jennifer, Weinstein Michael, Kleinstiver Benjamin P., Sousa Alexander A., Joung J. Keith, Crawford Jake. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018;2(1):38–47. doi: 10.1038/s41551-017-0178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.