Abstract
With the development of molecular genetics and high-throughput sequencing technology, genotyping arrays consisting of large numbers of SNP have raised great interest in animal and plant research. However, the application of commercial chicken 600K SNP arrays has varied in different populations of egg-type chickens. Moreover, their genotyping cost is too high for large-scale population applications. Herein, we independently developed a custom Illumina 50K BeadChip, named PhenoixChip-I, for egg-type chickens based on SNP from 479 sequenced individuals in 7 lines. We filtered and selected SNP with stringent criteria, such as high polymorphism, genome coverage, design score, and priorities. Finally, a total of 43,681 effective SNP successfully genotyped were included on our custom array. Approximately 14K SNP were previously reported to be associated with important economic traits in egg-type chickens. Subsequently, we verified the applicability and efficiency of the PhenoixChip-I SNP array from many aspects, including evaluating its use scientific research (population structure analysis and genome-wide association study) and the poultry breeding industry (genomic selection). The findings in our study will play a crucial role in accelerating the genetic improvement of egg-type chickens.
Key words: genomic analysis, genotyping array, egg-type chicken, SNP
Introduction
With the development of molecular genetics and high-throughput sequencing technology, molecular markers—especially SNP—are now widely used in a variety of animals and plants. Genotyping arrays consisting of dense genome-wide SNP have become extremely valuable tools for genomic analyses, including genetic associations with qualitative and quantitative traits, detection of QTL, and signatures of selection (Matukumalli et al., 2009; Ramos et al., 2009). Moreover, it accelerates the development and application of genomic selection in animal and plant breeding and has become a new technical ground and bringing great changes in livestock and poultry breeding.
There are 2 commercial SNP chips for chickens, including an Illumina 60K SNP array (Illumina, San Diego) (Groenen et al., 2011) and an Axiom 600K genome-wide chicken array (Affymetrix, Inc., Santa Clara, CA) (Kranis et al., 2013). However, Illumina Co. stopped manufacturing the 60K SNP chip some time ago. Only the Affymetrix 600K SNP chip is currently available to both scientific researchers and breeding enterprises. The Affymetrix 600K SNP array comprised approximately 560K SNP originating from commercial broiler and layer lines and inbred lines. This array has a higher density than the Illumina 60K SNP chip, while its application in Chinese egg-type chickens did not achieve the desired effect. Our previous studies demonstrated that approximately half of the loci and one-third of SNP had low polymorphism in the pure line and F2 population (Yi et al., 2015; Liu et al., 2018a), respectively (Supporting Information File 1: Supplementary Figure 1). This meant its efficiency was very low. In addition, the high cost of genotyping further hindered its practical application in large-scale populations.
The original 60K SNP chip was based on the genome version of Gallus_gallus-2.1 (May 2006), and the Affymetrix 600K SNP array was based on Gallus_gallus_4.0 version (November 2011). In our research, the custom SNP array was based on the Gallus_gallus-6.0 (Mar. 2018) genome. Version 6.0 was improved relative to the former as it contained more genetic variants and fewer gap regions across chromosomes. The key point of the study was to design a moderate-density SNP array for egg-type chickens comprising SNP segregating at medium-to-high allele frequencies and covering all chromosomes in chickens of core breeding populations of high-yielding and characteristic breeds in China. Moreover, the cost of genotyping with the custom chip is lower than that with commercial SNP chips. It not only met the needs of scientific research but can also be applied on a larger scale to breeding industries.
Materials and methods
Animals and Whole Genome Sequencing
The 479 sequenced samples of 7 lines from this study came from 2 institutes in China, including Beijing Huadu Yukou Poultry Industry Co. Ltd., which is a leading layer breeding company in China, and the experimental station of China Agricultural University (detailed information is presented in Table 1).
Table 1.
Description of resource populations and sequencing strategy.
| Line | Breed | Sex | Number of individuals | Analysis type1 | Effective depth (X)2 |
|---|---|---|---|---|---|
| Line 1 | Rhode Island Red | Male | 92 | Individual | 7.38 |
| Line 2 | Rhode Island White | Male | 74 | Pool (9) | 22.75 |
| Line 3 | Rhode Island White | Male | 91 | Pool (9) | 21.95 |
| Line 4 | White Leghorn | Male | 60 | Pool (7) | 22.78 |
| Line 5 | White Leghorn | Male | 69 | Pool (7) | 21.11 |
| Line 6 | Houdan | Male | 68 | Pool (7) | 23.10 |
| Line 73 | Dong Xiang | Male | 25 | Pool (5) | 24.30 |
| Total | 479 |
The number of pools in the parentheses.
The average effective depth after mapping to the chicken genome.
This line was from the experimental station of China Agricultural University, and all other lines were from Beijing Huadu Yukou Poultry Industry Co., Ltd.
Genomic DNA was extracted from whole blood samples using the TIANamp Genomic DNA Kit (Tiangen Biotech [Beijing] Co., Ltd., China) and then measured for quality using a NanoDrop 2000 (Thermo Fisher Scientific Inc. Waltham, MA). For each line, equal amounts of qualified genomic DNA from 5–12 individuals were used to make DNA pools except line 1, which was sequenced individually for each chicken. All libraries were constructed as per the manufacturer's protocol of Illumina performed on an Illumina X Ten platform with 150 bp paired-end reads (Illumina Inc.).
SNP Discovery and Collection
Candidate SNP in custom SNP arrays came from 2 important sources. The first part was detection from sequencing samples. Before aligning the raw Illumina sequence reads to the chicken whole reference genome (Gallus_gallus6.0), low-quality and adapter reads were filtered out using the NGS QC Toolkit (v2.3.3) with default parameters (Patel and Jain, 2012). After quality control, mapping reads to the reference genome was conducted by the Burrows-Wheeler Alignment tool (v0.7.15) (Li and Durbin, 2009). In addition, SAMtools (v1.3.1) and the Picard package (v1.119) (Li et al., 2009) were used to convert the file format and sort the mapped reads, respectively. The raw variant calling was performed in line with the pipeline of recommendations made in the Genome Analysis Toolkit (v3.7) Best Practices documentation (McKenna et al., 2010). In addition, we filtered raw SNP per line with stringent criteria: 1) QUAL >60; 2) DP > 5; 3) QD > 5.0; 4) FS < 60.0; 5) MQ > 40; 6) MQRankSum > −12.5; and 7) ReadPosRankSum > −8.0. We deleted SNP if more than 3 of them were clustered in a 10-bp window. Finally, eligible SNP in each line were retained for further custom SNP array analysis.
The second part was collected from an online SNP database, including genome-wide association studies (GWAS)–related SNPs, candidate genes, and QTL regions (https://www.animalgenome.org/cgi-bin/QTLdb/GG/index), which were previously reported to be associated with some important economic traits (such as egg production, feed efficiency, egg quality, and disease resistance) in egg-type chickens. These curated SNP were mostly detected by GWAS using Illumina 60K or Affymetrix 600K SNP chips in our previous studies (Liu et al., 2011; Yi et al., 2015; Sun et al., 2015a; Sun et al., 2015b; Yuan et al., 2015a; Yuan et al., 2015b; Yuan et al., 2017; Liu et al., 2018a; Liu et al., 2018b), while candidate genes originated from searching a large number of reported studies. For trait-related QTL regions, we reorganized different QTL with overlapping regions and excluded QTL with positions having wide CI. Only significant QTL (P value < 0.05 supplied by the chicken QTL database file) were retained in the following analysis.
Selection Strategy of the Final SNP List
To ensure good polymorphism of SNP on the chip, SNP shared by 7 sequencing lines (a total of 1,846,003 SNP) were screened first, while only 64,396 SNP remained after low minor allele frequency (MAF < 0.05) was eliminated, and these loci were regarded as the backbone core sites. Second, considering the importance of line 1, which occupies more than 40% of the Chinese layer market share, we selected 7,304,179 SNP that distinguished line 1 from at least 1 other line. At the same time, these SNP were also converted to overlap with the abovementioned candidate genes and QTL regions. Thus, 845,896 of 4,290,088 loci (filtering MAF < 0.05) associated with egg production, feed efficiency, quality of eggs, and disease resistance were treated as the second priority, and the remaining SNP and SNP in the database were used to fill the gaps where the first 2 categories failed to cover. In addition, 1,355 important trait-related loci as obligatory SNP were added to the SNP array design because all SNP from the abovementioned GWAS underwent deduplication and conversion of the different versions. Next, all of the aforementioned SNP were submitted to Illumina Co., and SNP with a score greater than 0.8 were reserved through the Illumina XT platform. Apart from the Illumina design score and the trait priority, other parameters were also considered, including the estimated MAF, the average spacing of neighboring SNP along each chromosome (which determines the number of SNP for chromosomes), whole genome-wide coverage, and the presence of neighboring SNP within 10 bp. Any SNP known to be connected with a patent was removed from the list. Finally, SNP in the 50K SNP array were selected with an even distribution across each chromosome in the whole chicken genome using a custom Perl script. For example, in chromosome 1, the SNP are distributed as shown.
Because of the difference in recombination across different chromosomes (ICGSC, 2004; Groenen et al., 2009; Megens et al., 2009), SNP were spaced based on similar recombination rates regardless of chromosomes in the genome, with spacing ranging between 6,000 bp for the microchromosomes and 20,000 bp for the macrochromosomes (GGA1 to GGA5) (Supporting Information File 1: Supplementary Table 1).
Validation of 50K Array for Studying Population Structure
To evaluate the performance of the 50K SNP chip, we genotyped 185 individuals to study population structure from 10 breeds, including Rhode Island Red (20 individuals), Rhode Island White (20 individuals), White Leghorn (19 individuals), Houdan (19 individuals), Beijing You (19 individuals), Dongxiang (20 individuals), Silkie (20 individuals), Tibetan (20 individuals), dwarf (20 individuals), and yellow broiler (8 individuals). The genotyping and original quality control were performed by Beijing Compass Biotechnology Co., Ltd. (http://www.kangpusen.com/, China) using the standard protocol for Infinium XT BeadChips supplied by Illumina Co. Then, principal component analysis was implemented using GCTA software (v1.26.0) (Yang et al., 2011) after filtering the MAF of SNP less than 0.01. We drew the plot of the first 2 components using R (https://www.r-project.org/). In addition, to better understand the genetic structure of these individuals, we constructed a neighbor-joining phylogenetic tree based on an identical by state distance matrix by the SNPhylo package (version 20160204) (Lee et al., 2014) and displayed with FigTree software (v1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/).
SNP Array for GWAS
To determine the capacity of the layer 50K SNP assay to map traits, we performed a GWAS for egg weight in a population of 680 Rhode Island Red hens at 36 wk of age. The genotyping was the same as that described previously. Quality control was conducted by PLINK (v1.90) software (Purcell et al., 2007) with including sample call rate > 97%, Geno < 0.05, MAF > 0.01, and HWE < 1e-06. A final total of 38,627 SNP and 673 individuals remained for the following association analyses.
A univariate GWAS based on a linear mixed model was carried out using the GEMMA (v0.94) package (Zhou and Stephens, 2012). The detailed method of GWAS was described in our previous studies (Liu et al., 2018b). Moreover, the significant thresholds were calculated from the number of independent SNP markers and linkage disequilibrium blocks. Therefore, the threshold P-value of genome-wide significance was 8.1e-06 (0.05 of 6,174), and the suggestive significance was set at 1.62e-4 (1 of 6,174).
Application of 50K SNP Array for Genomic Prediction in Layer
Genomic selection has become a very important technology in the area of animal breeding, especially in cattle (Schaeffer, 2006), and has tremendous potential in other species, such as poultry, pig, and beef cattle. To evaluate the efficiency of the 50K SNP array for Genomic selection, we compared the accuracy of genomic prediction for economic traits by the single-step method with the traditional prediction method in the study. The pedigree and phenotype data of a pure-line layer across 3 generations (G1–G3) were supplied by Beijing Huadu Yukou Poultry Industry Co., Ltd. In addition, 2,977 chickens across 3 generations were selected for genotyping by the 50K SNP chip. The criteria of genotyping quality control were as follows: sample call rate >0.97, MAF >0.01, GENO <0.05, and MIND <0.05. Finally, 2,950 individuals and 38,413 SNP were retained for the genomic analyses.
The genetic assessment of 3 economic traits, including BW and egg weight at 28 wk of age and total egg number from onset to 38 wk, was analyzed based on the pedigree best linear unbiased prediction (PBLUP) and single-step genomic best linear unbiased prediction (SSGBLUP) models. In addition, the accuracy of predicting offspring and prediction bias were calculated on the aforementioned two models using chickens in G1 ∼ G2 as the training population and individuals in G3 as the validation population. Detailed information of the model and analyses was previously described (Yan et al., 2018). In short, the predictive ability was calculated as the correlation between the predicted genomic estimated breeding values and the corresponding phenotypes, which were corrected for fixed effects, including birth year, batch, and sex. The bias was defined as the regression coefficients of corrected phenotypes with predicted genomic estimated breeding values.
Results and discussion
SNP Discovery
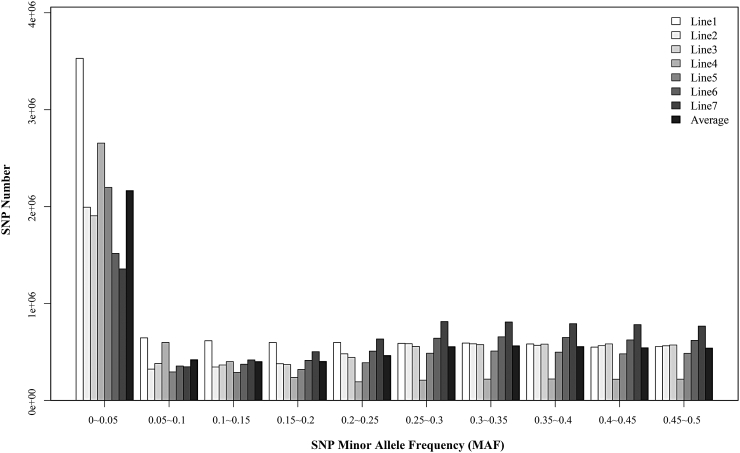
The sequencing strategy for each line is displayed in Table 1, and the effective depth was defined as the read depth calculated from the reads with mapping quality greater than 20. The results showed that the average effective depth ranged from 21.11x to 24.3x in pools and was 7.38x in individual sequencing, which was sufficient for further analysis of genetic variation detection. Then, SNP sequencing detection was carried out separately within each line, and the combined results are displayed in Table 2 after primary quality control with Genome Analysis Toolkit software. The total number of segregating SNP identified in the 7 lines was 13.31 Mb and ranged from 5.17 Mb to 8.86 Mb per line. Line 1 has more SNP than any other line because of the larger number of individual sequences. Interestingly, line 7 has fewer individuals, but it contains more variations, which may be because it is a local Chinese breed and suffers less intense artificial selection. The SNP MAF spectrum of each strain and average MAF across 7 lines are displayed in Figure 1. Among them, SNP with MAF greater than 0.25 occupied almost the same proportion in different lines, while half of the SNP (51.3%) in line 4 had low allele frequencies (MAF < 0.05).
Table 2.
SNP identified in the 7 sequencing populations.
| Lines | Line 1 | Line 2 | Line 3 | Line 4 | Line 5 | Line 6 | Line 7 |
|---|---|---|---|---|---|---|---|
| Line 1 | 8,857,074 | 5,588,202 | 5,719,096 | 3,888,326 | 4,403,317 | 4,705,196 | 5,388,358 |
| Line 2 | 6,389,088 | 5,336,579 | 3,216,878 | 3,604,678 | 3,826,008 | 4,337,184 | |
| Line 3 | 6,336,080 | 3,202,261 | 3,607,286 | 3,813,167 | 4,311,237 | ||
| Line 4 | 5,171,152 | 3,816,265 | 3,585,181 | 3,335,626 | |||
| Line 5 | 5,950,902 | 3,998,969 | 3,739,701 | ||||
| Line 6 | 6,361,263 | 4,018,880 | |||||
| Line 7 | 7,225,068 | ||||||
| Total | 13,309,506 |
The diagonal represents the number of SNP detected in each line. The upper triangle represents the number of SNP shared between any pair of lines.
Figure 1.
The SNP MAF spectrum of 7 lines. The MAF of line 1 was calculated by PLINK. The others were estimated by directly counting the number of reads for each allele. Average represented the mean MAF across 7 lines. Abbreviation: MAF minor allele frequency.
We compared SNP detected in our project with publicly available SNP from the NCBI database (ftp://ftp.ncbi.nih.gov/snp/organisms/archive/chicken_9031/VCF/). Approximately 89.65% of our SNP (13.31 Mb) were validated in the SNP database (19.38 Mb). However, many fewer SNP (11,651) were detected from chromosome 16 in our study because of the complex structure and partial presentation of this chromosome in the current genome version (Gallus_gallus-6.0). In addition, the distribution of SNP in sex-linked chromosome Z appeared extremely uneven. This finding was in accordance with previous studies that reported lower recombination rates and reduced genetic variations on chromosome Z than on autosomes (Sundstrom et al., 2004).
SNP Selection and Array Design
Owing to the limitation of the Infinium XT chip containing up to 50 thousand SNP sites, we selected and filtered SNP with multiple steps as described in the materials and methods section. We removed SNP with an Illumina design score less than 0.8 to increase the successful conversion rate on the array. Moreover, 2 design types of SNP can be included in the array: Infinium I (A/T or C/G), requiring 2 bead types per SNP, and Infinium II (A/C; A/G; T/C; T/G), requiring only 1 bead type per marker (Gunderson, 2009). We preferred to choose SNP of the Infinium II type to maximize the number of genotyped polymorphisms. In addition, considering the difference in recombination previously reported (ICGSC, 2004), we divided chromosomes into 4 types based on the decay of linkage disequilibrium (Supporting Information File 1: Supplementary Figure 2), which was calculated in line 1 populations using Affymetrix 600K SNP genotyping data (Liu et al., 2018a). The results indicated that microchromosomes (chromosomes 10–28) had relatively higher recombination than macrochromosomes, which agreed with results of previous studies (Megens et al., 2009). Thus, the average spacing of SNP on different chromosomes varied as per the size of the chromosome (Supporting Information File 1: Supplementary Table 1). Finally, the SNP lists containing 43,681 effective SNP on the Illumina XT BeadChip were retained for versatile analysis because some SNP failed to successfully convert. The summary of functional SNP in the 50K array (named PheonixChip-I) is shown in Table 3. A total of 14.7K functional SNP related to important economic traits were designed on our custom array as per the selection strategy in which 13,189 SNP were from the traits-related QTL in SNP database, and 156 SNP from candidate genes, and 1,355 SNP from our previous GWAS results.
Table 3.
Functional SNP selected for the custom 50K array.
| SNPs on the chip | Number |
|---|---|
| Original total number of SNP | 50,000 |
| Total effective SNP | 43,681 |
| SNP in trait-related QTL | 13,189 |
| SNP detected by GWAS | 1,355 |
| SNP related with candidate genes | 156 |
Abbreviation: GWAS, genome-wide association study.
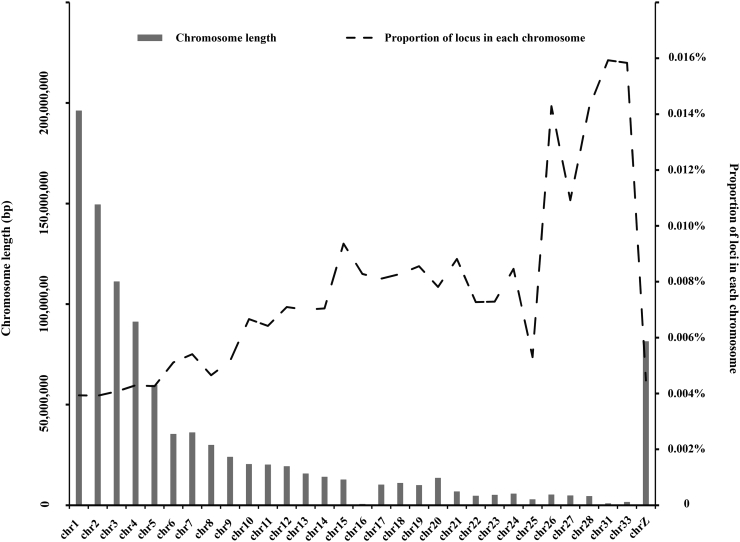
The proportion of SNP in each chromosome is presented in Figure 2. The results indicated that microchromosomes had higher SNP density than the others, which could better cover and represent the chromosomes. Fewer SNP were assigned to chromosome 16 and chromosome Z because even the better assembled genome (Gallus_gallus-6.0) contained some underrepresented regions. Moreover, the higher GC content or pieces of chromosome repeats might result in the absence of SNP in those gaps (ICGSC, 2004). Therefore, we updated the custom 50K SNP chip for the future version to improve the performance.
Figure 2.
The proportion of SNP in each chromosome. The left ordinate represents the length of the chromosome, and the right ordinate indicates the proportion of SNP in each chromosome.
Array for Studying Population Structure
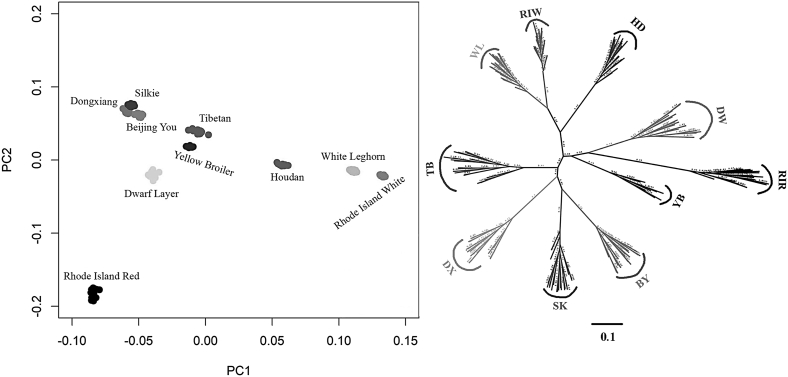
It is of great significance to study population structure regardless of evolutionary analysis or GWAS because the stratification of population may lead to more false positives in the results (Xu and Shete, 2005; Price et al., 2010). We carried out principal component analysis and phylogenetic tree analyses using the genotype data to investigate the population stratification in 185 individuals of 10 breeds to evaluate the performance of the 50K SNP array. Figure 3 shows that samples originating from the same breed were clustered together. Three local Chinese breeds, including Silkie, Beijing You, and Dongxiang, appeared to be clustered tightly from both the principal component analysis and phylogenetic tree results, while international breeds (Rhode Island White, White Leghorn, and Houdan) were in close proximity in the phylogenetic tree suggesting that they shared ancestry. Interestingly, the genetic distance of Rhode Island Red was remote relative to exotic breeds but relatively close to cultivated strains (dwarf and yellow Broiler) in China. We speculated that it originated from crossing the Asian (Red Malay and Asiatic native stock) and European (Leghorn) breeds and also from geographical isolation more than 10 yr ago (Muir et al., 2008; Elferink et al., 2012). In addition, Tadano et al previously reported that different lines of Rhode Island Red were separated clearly (Tadano et al., 2007), although they were from the same breed. Another explanation could be ascertainment bias because the SNP were selected from Rhode Island Red and Rhode Island White. Our results of population structure analyses agree with those of previous studies (Elferink et al., 2012; Kranis et al., 2013).
Figure 3.
Population genetic structure analysis based on the custom 50K SNP array. The left panel shows principal component analysis (PCA), and the right panel displays a neighbor-joining phylogenetic tree. Abbreviations: BY, Beijing You; DW, dwarf; DX, Dongxiang chicken; HD, Houdan; RIR, Rhode Island Red; RIW, Rhode Island White; SK, Silkie; TB, Tibetan; WL, White Leghorn; YB, yellow broiler.
Application of Association and Genomic Selection Analyses
Before the genome-wide association analysis, we treated the first 5 principal components as covariates and included them in a univariate linear mixed model as fixed effects to control the population stratification. The Manhattan plot for egg weight at 36 wk of age is shown in Supplementary Figure 3 (Supporting information File 1). The results showed that a 650-kb region spanning between 168.97 and 169.62 Mb on GGA1 was significantly associated with egg weight (Supporting Information File 2). Several genes, including fibronectin type III domain containing 3A, calcium-binding protein 39-like, lymphocytic leukemia 7, and MIR15A, were annotated using the online Ensembl database (http://asia.ensembl.org). The aforementioned genes around the significant region were also reported to be related to egg weight in studies based on the commercial 600K SNP array (Yi et al., 2015; Liu et al., 2018b).
For genomic selection, the basic information of individuals used for genetic evaluation across 3 generations is shown in Table 4. The results of descriptive statistics and variance component estimation for BW at 28 wk of age, egg weight at 28 wk of age, and total egg number from onset to 38 wk are supplied in Supporting Information File 3. For all traits, the estimates of heritability by PBLUP were smaller than those by SSGBLUP because of the relatively higher residual variance. These results were in agreement with those of a previous study (Yan et al., 2018). Moreover, the predictive ability of genotyped and ungenotyped individuals by the 2 models is displayed in Table 5. Generally, the single-step method (SSGBLUP) yields higher predictive ability than PBLUP. In particular, the improvements in SSGBLUP were 52.6, 76.5, and 72.7% higher than those in PBLUP for BW at 28 wk of age, egg weight at 28 wk of age, and total egg number from onset to 38 wk in genotyped candidates, respectively. The increased predictive ability for these traits is higher than that in a previous study because of the large-scale reference population.
Table 4.
Information on individuals across 3 generations in this study.
| Category | G1 | G2 | G3 | Total |
|---|---|---|---|---|
| Sires | 759 | 551 | 610 | 1,920 |
| Dams | 3,662 | 3,710 | 3,919 | 11,291 |
| Genotyped sires | 88 | 110 | 92 | 290 |
| Genotyped dams | 680 | 1,104 | 903 | 2,687 |
Table 5.
Predictive ability and bias of different models for genotyped and ungenotyped individuals.
| Traits | Genotyped |
Ungenotyped |
||
|---|---|---|---|---|
| PBLUP | SSGBLUP | PBLUP | SSGBLUP | |
| BW28 | 0.19(0.709)1 | 0.29(0.827) | 0.20(0.866) | 0.22(0.840) |
| EW28 | 0.17(0.549) | 0.30(0.685) | 0.27(1.119) | 0.31(1.119) |
| EN38 | 0.22(0.857) | 0.38(0.900) | 0.21(1.350) | 0.25(1.244) |
Abbreviations: BW28, BW at 28 wk of age; EN38: total egg number from onset to 38 wk; EW28, egg weight at 28 wk of age.
The value is predictive ability, and the value in parentheses is predictive bias.
All of the aforementioned results further verify the reliability and efficiency of applying the 50K SNP chip in association studies and genomic selection.
Conclusion
In our study, we independently developed the Illumina 50K BeadChip PheonixChip-I for egg-type chickens based on next-generation sequence technology and an online SNP database. In addition, we verified the applicability and efficiency of this SNP array in many aspects, including scientific research and breeding industries. The findings in our study play a crucial role in accelerating the genetic improvement of egg-type chickens.
Acknowledgments
This work was funded in part by grants from the China Agriculture Research Systems (CARS-40), Programs for Changjiang Scholars and Innovative Research in University (IRT_15R62), and Chinese Universities Scientific Fund (2018QC030). We are grateful for the help of members of the Beijing Engineering Research Center of Layer and Beijing Compass Biotechnology Co., Ltd.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2021.101044.
Disclosures
The authors declare no conflicts of interest.
Supplementary data
References
- Elferink M.G., Megens H.J., Vereijken A., Hu X., Crooijmans R.P., Groenen M.A. Signatures of selection in the genomes of commercial and non-commercial chicken breeds. PLoS One. 2012;7:e32720. doi: 10.1371/journal.pone.0032720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M.A., Megens H.-J., Zare Y., Warren W.C., Hillier L.W., Crooijmans R.P., Vereijken A., Okimoto R., Muir W.M., Cheng H.H. The development and characterization of a 60K SNP chip for chicken. BMC Genomics. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M.A., Wahlberg P., Foglio M., Cheng H.H., Megens H.J., Crooijmans R.P., Besnier F., Lathrop M., Muir W.M., Wong G.K., Gut I., Andersson L. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson K. Whole-genome genotyping on bead arrays. Methods Mol. Biol. 2009;529:197–213. doi: 10.1007/978-1-59745-538-1_13. [DOI] [PubMed] [Google Scholar]
- ICGSC Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Kranis A., Gheyas A.A., Boschiero C., Turner F., Yu L., Smith S., Talbot R., Pirani A., Brew F., Kaiser P., Hocking P.M., Fife M., Salmon N., Fulton J., Strom T.M., Haberer G., Weigend S., Preisinger R., Gholami M., Qanbari S., Simianer H., Watson K.A., Woolliams J.A., Burt D.W. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics. 2013;14:59. doi: 10.1186/1471-2164-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-H., Guo H., Wang X., Kim C., Paterson A.H. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:62. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome S., Project Data Processing The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li D., Liu J., Chen S., Qu L., Zheng J., Xu G., Yang N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PLoS One. 2011;6:e28600. doi: 10.1371/journal.pone.0028600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sun C., Yan Y., Li G., Shi F., Wu G., Liu A., Yang N. Genetic variations for egg quality of chickens at late laying period revealed by genome-wide association study. Sci. Rep. 2018;8:10832. doi: 10.1038/s41598-018-29162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sun C., Yan Y., Li G., Wu G., Liu A., Yang N. Genome-wide association analysis of age-Dependent egg weights in chickens. Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matukumalli L.K., Lawley C.T., Schnabel R.D., Taylor J.F., Allan M.F., Heaton M.P., O'Connell J., Moore S.S., Smith T.P., Sonstegard T.S., Van Tassell C.P. Development and characterization of a high density SNP genotyping assay for cattle. PloS One. 2009;4:e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens H.J., Crooijmans R.P., Bastiaansen J.W., Kerstens H.H., Coster A., Jalving R., Vereijken A., Silva P., Muir W.M., Cheng H.H., Hanotte O., Groenen M.A. Comparison of linkage disequilibrium and haplotype diversity on macro- and microchromosomes in chicken. BMC Genetics. 2009;10:86. doi: 10.1186/1471-2156-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir W.M., Wong G.K., Zhang Y., Wang J., Groenen M.A., Crooijmans R.P., Megens H.J., Zhang H., Okimoto R., Vereijken A., Jungerius A., Albers G.A., Lawley C.T., Delany M.E., MacEachern S., Cheng H.H. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17312–17317. doi: 10.1073/pnas.0806569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a Toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Zaitlen N.A., Reich D., Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Reviews. Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A.M., Crooijmans R.P., Affara N.A., Amaral A.J., Archibald A.L., Beever J.E., Bendixen C., Churcher C., Clark R., Dehais P., Hansen M.S., Hedegaard J., Hu Z.L., Kerstens H.H., Law A.S., Megens H.J., Milan D., Nonneman D.J., Rohrer G.A., Rothschild M.F., Smith T.P., Schnabel R.D., Van Tassell C.P., Taylor J.F., Wiedmann R.T., Schook L.B., Groenen M.A. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006;123:218–223. doi: 10.1111/j.1439-0388.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- Sun C., Lu J., Yi G., Yuan J., Duan Z., Qu L., Xu G., Wang K., Yang N. Promising loci and genes for Yolk and ovary weight in chickens revealed by a genome-wide association study. PLoS One. 2015;10:e0137145. doi: 10.1371/journal.pone.0137145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Qu L., Yi G., Yuan J., Duan Z., Shen M., Qu L., Xu G., Wang K., Yang N. Genome-wide association study revealed a promising region and candidate genes for eggshell quality in an F2 resource population. BMC Genomics. 2015;16:565. doi: 10.1186/s12864-015-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström H., Webster M.T., Ellegren H. Reduced variation on the chicken Z chromosome. Genetics. 2004;167:377–385. doi: 10.1534/genetics.167.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadano R., Nishibori M., Nagasaka N., Tsudzuki M. Assessing genetic diversity and population structure for commercial chicken lines based on forty microsatellite analyses. Poult. Science. 2007;86:2301–2308. doi: 10.3382/ps.2007-00233. [DOI] [PubMed] [Google Scholar]
- Xu H., Shete S. Effects of population structure on genetic association studies. BMC Genetics. 2005;6(Suppl 1):S109. doi: 10.1186/1471-2156-6-S1-S109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Wu G., Liu A., Sun C., Han W., Li G., Yang N. Genomic prediction in a nuclear population of layers using single-step models. Poult. Sci. 2018;97:397–402. doi: 10.3382/ps/pex320. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Shen M., Yuan J., Sun C., Duan Z., Qu L., Dou T., Ma M., Lu J., Guo J., Chen S., Qu L., Wang K., Yang N. Genome-wide association study dissects genetic architecture underlying longitudinal egg weights in chickens. BMC Genomics. 2015;16:746. doi: 10.1186/s12864-015-1945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Chen S., Shi F., Wu G., Liu A., Yang N., Sun C. Genome-wide association study reveals putative role of gga-miR-15a in controlling feed conversion ratio in layer chickens. BMC Genomics. 2017;18:699. doi: 10.1186/s12864-017-4092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Sun C., Dou T., Yi G., Qu L., Qu L., Wang K., Yang N. Identification of promising Mutants associated with egg production traits revealed by genome-wide association study. PLoS One. 2015;10:e0140615. doi: 10.1371/journal.pone.0140615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Wang K., Yi G., Ma M., Dou T., Sun C., Qu L.J., Shen M., Qu L., Yang N. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet. Sel. Evol. 2015;47:82. doi: 10.1186/s12711-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.