Abstract
Background: To determine the distribution and antimicrobial susceptibility pattern of pathogenic bacteria in patients with chronic cutaneous wounds on a national scale.
Methods: A retrospective study was conducted using the data recorded between January 1, 2018 and January1, 2020 in 195 hospitals across China. After screening the data, 815 patients with chronic wounds were finally analyzed. The data collected included information about the patients' general condition and local cutaneous wound assessments, especially microbial culture and antibiotic susceptibility tests. The analyses were performed using SPSS Version 26.
Results: The study included 815 patients (290 [35.6%] females; 63 [50–74] years). The most common causes of chronic cutaneous wounds were diabetes (183, 22.5%), infection (178, 21.8%), and pressure (140, 17.2%). Among these, 521(63.9%) samples tested yielded microbial growth, including 70 (13.4%) polymicrobial infection and 451 (86.6%) monomicrobial infection. The positive rate of microbial culture was highest in wound tissue of ulcers caused by infection (87.6%), followed by pressure (77.1%), diabetes (68.3%), and venous diseases (67.7%). Bates-Jensen wound assessment tool (BWAT) scores >25 and wounds that lasted for more than 3 months had a higher positive rate of microbial culture. BWAT scores >25 and wounds in the rump, perineum, and feet were more likely to exhibit polymicrobial infection. A total of 600 strains were isolated, of which 46.2% (277 strains) were Gram-positive bacteria, 51.3% (308 strains) were Gram-negative bacteria, and 2.5% (15 strains) were fungi. The most common bacterial isolates were Staphylococcus aureus (29.2%), Escherichia coli (11.5%), Pseudomonas aeruginosa (11.0%), Proteus mirabilis (8.0%), and Klebsiella pneumoniae (5.8%). The susceptibility tests showed that 116 cultured bacteria were Multidrug resistant (MDR) strains. The resistance rates of S. aureus were 92.0% (161/175) to penicillin, 58.3% (102/175) to erythromycin, and 50.9% (89/175) to clindamycin. Vancomycin was the most effective antibiotic (0% resistance rate) against all Gram-positive bacteria. Besides, the resistance rates of E. coli were 68.1% (47/69) to ampicillin, 68.1% (47/69) to ciprofloxacin, 60.9% (42/69) to levofloxacin. However, all the isolated Gram-negative bacteria showed low resistance rates to tigecycline (3.9%) and amikacin (3.6%).
Conclusions: The distribution of bacteria isolated from chronic cutaneous wounds varies with the BWAT scores, causes, duration, and the location of wounds. Multidrug resistance is a serious health issue, and therefore antibiotics used in chronic wounds must be under strict regulation. Our findings may help clinicians in making informed decisions regarding antibiotic therapy.
Keywords: pathogen, bacteria distribution, antibiotic resistance, multi drug resistant, chronic wounds
Introduction
Cutaneous wound healing is an incredibly complex and regulated process. A chronic cutaneous wound may develop when the wound healing process fails to progress in an orderly and timely manner (1, 2). The wound healing process can be delayed or stalled by a myriad of factors, including diabetes, skin infections, arterial and venous diseases, trauma, burn, pressure, and surgery (3–6). The number of patients developing chronic cutaneous wounds is rapidly increasing due to changing lifestyles and aging problems. Chronic cutaneous wounds present a major social and financial burden on both the individual patients and the entire healthcare system worldwide (1, 7).
Bioburden has been identified as one of the major barriers to wound healing (8). Colonization of the wound site by pathogens contributes substantially to the wound chronicity (9–11). Previous studies have shown that, in addition to primary skin infections, wounds caused by diabetes, pressure, venous diseases, and surgery (surgical site infections, SSIs) are more likely to be colonized by pathogenic bacteria (9, 12). Among them, SSIs represent about 15% of all nosocomial infections, and are extremely difficult to treat due to their resistance to multiple antibiotics (13, 14).
Several lines of evidence regarding the prevalence of pathogenic bacteria in chronic wounds have identified Staphylococcus aureus, Pseudomonas aeruginosa, Enterococcus faecalis, and Proteus mirabilis as the most prevalent bacteria in chronic wounds (9, 10, 15, 16). However, the distribution of pathogens depends on various factors, such as geographic location, causes of wounds, among others (17). China has a huge population living with chronic wounds that vary in types and causes (4). However, till date, no nationwide study has been done to investigate the distribution and antimicrobial susceptibility of pathogens in patients with chronic cutaneous wounds. To this end, this study aimed to investigate the distribution and antimicrobial susceptibility of pathogenic bacteria in patients with chronic cutaneous wounds in China. Our findings may help clinicians in making informed decisions regarding antibiotic therapy.
Materials and Methods
Study Identification and Data Extraction
Retrospective analysis of medical information downloaded from the WoundCareLog database and recorded between January 2018 and January 2020 was performed (18). Specifically, information on patients' general features and local cutaneous wounds was captured. The general features included the patient' name, gender, age, home address, hospital department, first admission time, complications, chief complaint, past medical history, and diagnosis. The information concerning local cutaneous wounds included wound classification, duration, wound location, wound photographs, among others. It should be noted that all wounds were classified according to their causes, such as diabetes, infection, pressure, etc. Among them, we defined “infection” as primary skin infections like erysipelas, impetigo, and scabies. Data on microbial culture and antimicrobial susceptibility tests were also gathered. The medical records in the WoundCareLog database were all uploaded by doctors and nurses in 195 cooperative hospitals across China following unified standards.
Inclusion and Exclusion Criteria
Patients diagnosed with chronic cutaneous wounds were initially included. Then patients with the following characteristics were excluded: duration of wounds less than 1 month; patients with incomplete information on general features or wounds; patients without records of microbial culture and antimicrobial susceptibility tests. These inclusion and exclusion criteria were applied in stages.
Swab Collection and Culture
Bacterial cultures and antimicrobial susceptibility tests were performed when patients presented with clinical signs of systemic or local infection, including fever, erythema, local warmth, serous exudate, discoloration of granulation tissue, and foul odor (19, 20). Samples were obtained from cutaneous wounds by trained nurses based on a standardized procedure (21). Before sample collection, wounds were cleansed with sterile normal saline. Excess saline was carefully removed using sterile gauze, the specimens were collected with a sterile swab by swabbing at the middle of the wounds for 5 s under sufficient pressure, and the swab was immediately inserted into a sterile tube and sent to the laboratory within 2 h. Swabs were streaked on MacConkey Agar (MCA), Blood Agar (BA) plates and incubated aerobically at 37°C and 5% CO2 for 24 h. Plates without bacterial growth were incubated for another 18–24 h.
Bacteria Identification
Identification of the purified isolated bacteria was performed using the VITEK MS automated system (bioMérieux, Marcy I'Etoile, France), VITEK 2 COMPACT System (bioMérieux, Marcy I'Etoile, France), or MALDI Biotyper System (Bruker Daltonics GmbH, Bremen, Germany) according to manufacturer's instructions.
Antimicrobial Susceptibility Test
The drug susceptibility tests were performed using the VITEK2 COMPACT System (bioMérieux, MarcyI'Etoile, France) according to manufacturer's instructions. The results were interpreted based on the guidelines of Clinical and Laboratory Standards Institute (CLSI) (22). Multidrug resistance (MDR) bacteria was defined as bacteria strains that exhibited non-susceptibility to at least one agent in three or more specified categories of antimicrobials (23).
BWAT Assessment
The status of the wounds was assessed using the Bates-Jensen wound assessment tool (BWAT) (24). Based on the medical data collected from the database, all wounds were rated based on 13 scored items listed in the instructions of BWAT, including wounds size, depth, edges, undermining, necrotic tissue type, necrotic tissue amount, exudate type, exudate amount, skin color surrounding the wound, peripheral tissue edema, peripheral tissue induration, granulation tissue, and epithelialization. The total score was determined by summing up the scores of the 13 items. A higher total score indicated a more severe wound status. All the wounds were independently rated by two researchers. The average value was adopted if the difference between the two scores was less than three; otherwise, the wound was rated by a third researcher who was more experienced.
Statistical Analysis
Data analyses were performed using SPSS Version 26 (IBM SPSS; Armonk, New York). The age of the patients was expressed as median and interquartile range. Categorical data, such as gender, the result of bacterial culture tests (positive/negative), and the types of bacteria, were presented as frequencies and proportions and were compared using χ2 tests or Fisher exact probability test. A p-value of less than 0.05 defined statistical significance.
Results
Demographics
A total of 38,380 medical records were analyzed, of which 9,617 patients with cutaneous wounds from 195 hospitals across China were identified. Out of which, 8,802 were excluded step by step according to the exclusion criteria, and 815 patients (290 [35.6%] females; median [interquartile range] age, 63 [50–74] years) from 195 hospitals (122 [62.6%] from southern China; 65 [33.3%] from northern China, and 8 [4.1%] from northwestern China) met the inclusion criteria. A flowchart with detailed information was outlined in Figure 1.
Figure 1.
Study selection flowchart.
In total, 450 (55.2%) patients were over 60 years old. The highest frequency of patients with chronic wounds was found in the age group of 60–80 years (40.9%). Data obtained showed that, most patients came from southern China (75.5%), followed by northern China (23.3%), and only 10 (1.2%) were from northwestern China. The top two most prevalent complications of the analyzed population were diabetes and high blood pressure (HBP), which affected 161 (19.8%) and 121 (14.8%) patients, respectively. The demographic features of the patients are listed in Table 1.
Table 1.
Participant demographics and clinical variables.
| Variable | Subgroup | Values, No. (%) |
|---|---|---|
| Age | 0–20 | 20 (2.5) |
| 21–40 | 96 (11.8) | |
| 41–60 | 249 (30.6) | |
| 61–80 | 333 (40.9) | |
| >80 | 117 (14.4) | |
| Gender | Female | 290 (35.6) |
| male | 525 (64.4) | |
| Geographical location | Southern China | 615 (75.5) |
| Northern China | 190 (23.3) | |
| Northwestern China | 10 (1.2) | |
| Complications | Diabetes | 161 (19.8) |
| HBP | 121 (14.8) | |
| CHD | 50 (6.1) | |
| Nephropathy | 13 (1.6) | |
| PVD | 71 (8.7) | |
| Cerebral infarction | 71 (8.7) | |
| Total | 815 (100) |
HBP, High Blood Pressure; CHD, Coronary Heart Disease; PVD, Peripheral Vascular Diseases.
Wound Information
Based on our analysis, chronic cutaneous wounds was caused by diabetes (183, 22.5%), infection (178, 21.8%), pressure (140, 17.2%), trauma (83, 10.2%), surgery (77, 9.4%), venous diseases (62, 7.6%), burn (34, 4.2%), arterial diseases (11, 1.3%), radiation (13, 1.6%), and malignant tumor (5, 0.6%), and other factors (29, 3.6%) including scar ulcers, toxicosis, and autoimmune diseases. The distribution of causes varied significantly in patients in different age groups (χ2 = 49.198, P < 0.001) (Figure 2). Other features of chronic cutaneous wounds, such as duration of wounds, BWAT scores, and location of the wounds are listed in Table 2.
Figure 2.
The distribution of different causes in patients of different age groups.
Table 2.
Features of patients' chronic cutaneous wounds.
| Variables | Subgroup | Values, No. (%) |
|---|---|---|
| Causes | Arterial disease | 11 (1.3) |
| Venous disease | 62 (7.6) | |
| Diabetes | 183 (22.5) | |
| Radiation | 13 (1.6) | |
| Infection | 178 (21.8) | |
| Burn | 34 (4.2) | |
| Trauma | 83 (10.2) | |
| Pressure | 140 (17.2) | |
| Surgery | 77 (9.4) | |
| Malignant Tumor | 5 (0.6) | |
| Others | 29 (3.6) | |
| Duration of wounds(months) | 1–3 | 457 (56.1) |
| 3–6 | 143 (17.5) | |
| 6–12 | 106 (13.0) | |
| >12 | 109 (13.4) | |
| BWAT scores | 1–25 | 362 (44.4) |
| 26–40 | 411 (50.4) | |
| >40 | 42 (5.2) | |
| wounds location | Head and neck | 25 (3.1) |
| Trunk | 156(19.1) | |
| Rump and perineum | 161 (19.7) | |
| Arm | 15 (1.8) | |
| Hand | 19 (2.3) | |
| Leg | 209 (25.6) | |
| Foot | 230 (28.2) | |
| Total | 815 (100) |
Microbial Culture
Herein, 63.9% (521 of 815) of samples analyzed yielded microbial growth. The positive rate of microbial culture was significantly higher in patients with cutaneous wounds that lasted for more than 3 months (χ2 = 8.765, P = 0.003). Also, the positive rate in BWAT scores > 25 was significantly higher than that of BWAT scores ≤ 25 (χ2 = 13.919, P < 0.001). Besides, the positive rate was highest in wound tissue of ulcers caused by infection (87.6%), followed by pressure (77.1%), diabetes (68.3%), and venous diseases (67.7%). Table 3 shows these results. Interestingly, a significant correlation between positive microbial culture and geographical location was observed. Participants from northern China exhibited higher positive rate than those from Southern China (χ2 = 5.099, P = 0.024). In total, 451 (86.6%) of the 521 wounds were monomicrobial infections, 70 (13.4%) wounds were polymicrobial infections (≥2 strains were isolated). Similarly, patients with BWAT score > 25 were more likely to have a polymicrobial infection (χ2 = 6.465, P = 0.011). Furthermore, we found that the anatomical sites of cutaneous wounds were related to types of infection. Compared to other locations, wounds in the rump, perineum and feet were more likely to have a polymicrobial infection (χ2 =9.897, P = 0.002).
Table 3.
The distribution of common pathogenic bacteria in wounds of different causes.
| Value, n (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arterial | Venous | Diabetes | Radiation | Infection | Burn | Trauma | Pressure | Surgery | Malignant | Others | Total | |
| disease | disease | tumor | ||||||||||
| Total samples | 11 | 62 | 183 | 13 | 178 | 34 | 83 | 140 | 77 | 5 | 29 | 815 |
| Positive samples | 6 (54.5) | 42 (67.7) | 125 (68.3) | 8 (61.5) | 156 (87.6) | 18 (52.9) | 34 (41.0) | 108 (77.1) | 18 (23.4) | 3 (60.0) | 3 (10.3) | 521 (63.9) |
| Total strains | 8 | 47 | 145 | 8 | 173 | 19 | 34 | 139 | 20 | 4 | 3 | 600 |
| Polymicrobial infection | 2 (33.3) | 5 (11.9) | 16 (12.8) | 0 (0) | 16 (10.3) | 1 (5.6) | 0 (0) | 27 (25.0) | 2 (11.1) | 1 (33.3) | 0 (0) | 70 (11.7) |
| Monomicrobial infection | 4 (66.7) | 37 (88.1) | 109 (87.2) | 8 (100) | 140 (89.7) | 17 (94.4) | 34 (100) | 81 (75.0) | 16 (88.9) | 2 (66.7) | 3 (100) | 451 (75.2) |
| MDR | 0 (0) | 7 (14.9) | 31 (21.4) | 1 (12.5) | 38 (22.0) | 2 (10.5) | 7 (20.6) | 27 (19.4) | 2 (10.0) | 0 (0) | 1 (33.3) | 116 (19.3) |
| Gram-positive bacteria | 1 (12.5) | 22 (46.8) | 73 (50.3) | 6 (75.0) | 87 (50.3) | 13 (68.4) | 18 (52.9) | 47 (33.8) | 7 (35.0) | 1 (25.0) | 2 (66.7) | 277 (46.2) |
| S. aureus | 0 (0) | 18 (38.3) | 51 (35.2) | 4 (50.0) | 58 (33.5) | 7 (36.8) | 6 (17.6) | 25 (18.0) | 4 (20.0) | 0 (0) | 2 (66.7) | 175 (29.2) |
| MRSA | 0 (0) | 2 (4.3) | 20 (13.8) | 1 (12.5) | 22 (12.7) | 1 (5.3) | 3 (8.8) | 11 (7.9) | 1 (5.0) | 0 (0) | 1 (33.3) | 62 (10.3) |
| Enterococcus spp. | 0 (0) | 0 (0) | 8 (5.5) | 0 (0) | 5 (2.9) | 1 (5.3) | 4 (11.8) | 8 (5.8) | 1 (5.0) | 0 (0) | 0 (0) | 27 (4.5) |
| Gram-negative bacteria | 7 (87.5) | 23 (48.9) | 67 (46.2) | 2 (25.0) | 82 (47.4) | 6 (31.6) | 15 (44.1) | 89 (64.0) | 13 (65.0) | 3 (75.0) | 1 (33.3) | 308 (51.3) |
| E. coli | 1 (12.5) | 4 (8.5) | 11 (7.6) | 1 (12.5) | 18 (10.4) | 0 (0) | 0 (0) | 29 (20.9) | 5 (25.0) | 0 (0) | 0 (0) | 69 (11.5) |
A total of 600 bacterial strains were isolated from the 521 cases, 46.2% (277 strains) of which were Gram-positive bacteria, 51.3% (308 strains) were Gram-negative bacteria, and 2.5% (15 strains) were fungi. Samples from the wounds that lasted for more than 3 months mainly contained Gram-negative bacteria, whereas those from the wounds that lasted for less than 3 months mainly contained Gram-positive bacteria (Figure 3A). Besides, wounds caused by radiation and burn were mainly colonized by Gram-positive bacteria, whereas, wounds caused by arterial diseases, pressure, surgery, and malignant tumor were mainly colonized by Gram-negative bacteria. The distribution of common pathogenic bacteria in chronic wounds arising from different causes are listed in Table 3. However, there was no significant association between the distribution of pathogenic bacteria and the age or gender of patients (P = 0.527, 0.283, respectively).
Figure 3.
(A, B) The distribution of pathogens in wounds of different duration.
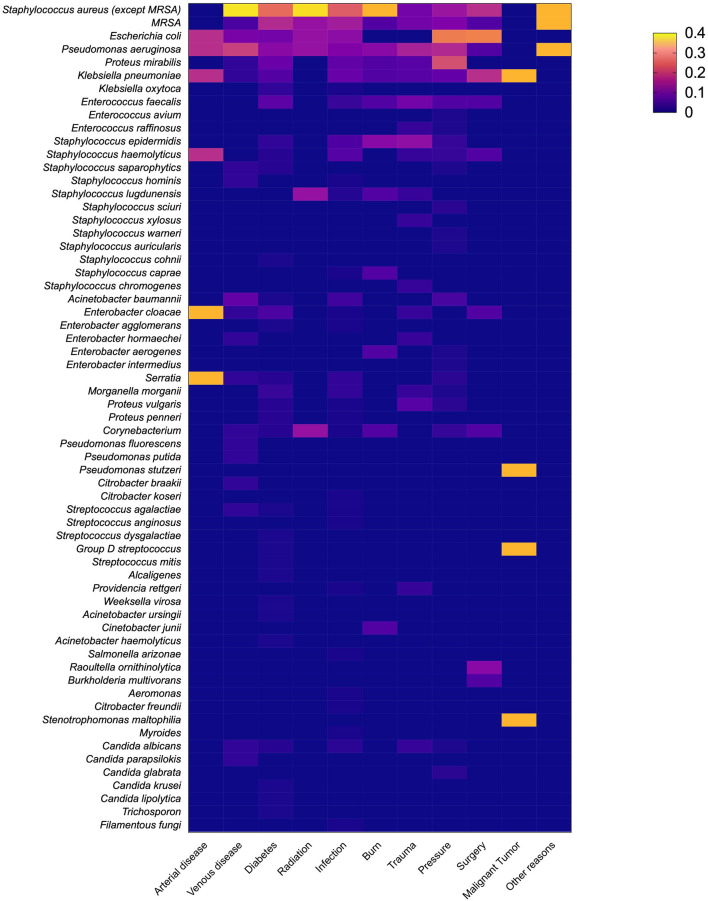
The most frequently isolated species were S. aureus (29.2%), followed by E. coli (11.5%), P. aeruginosa (11.0%), P. mirabilis (8.0%), and Klebsiella pneumoniae (5.8%). In the wounds that formed within 3 months, Gram-positive bacteria played a major role, and 32% of the infections involved S. aureus. E. coli (12%) was the most common Gram-negative bacteria. However, the wounds that lasted for more than 3 months showed different microbial composition. Gram-negative bacteria accounted for 56.3% of the infections, and P. aeruginosa (13%) was the most common Gram-negative bacteria (Figure 3B). The isolated fungi species were as follows: Candida albicans (8 strains), Candida parapsilokis (1 strain), Candida glabrata (2 strains), Candida krusei (1 strain), Candida lipolytica (1 strain), Trichosporon sp. (1 strain), and Filamentous fungi (1 strain). However, we did not include fungi in further comparative analysis because of possible bias caused by the small number of isolated fungal strains. The distribution of pathogens in wounds of different causes is presented in Figure 4.
Figure 4.
The distribution of pathogens in wounds of different causes. Different colors indicate the percentage of each pathogen in wounds of different etiologies.
In this study, 22.2% (116 of 521) of the patients developed MDR bacterial colonization, and 116 MDR bacterial strains were cultivated. Among the different causes of chronic cutaneous wounds, MDR bacteria were more likely to be found in wounds caused by infection (22.0%, 38 of 173). The wounds that lasted for more than 3 months had a significantly higher incidence rate of MDR bacterial strains than those that lasted for less than 3 months (χ2 = 4.911, P = 0.027). Apart from Methicillin-resistant Staphylococcus aureus (MRSA), which was isolated from 62 patients (53.4%), other MDR bacterial species were not common. None of the patients was colonized with vancomycin-resistant enterococci (VRE). Among the other 54 patients colonized with MDR bacterial strains, the most common genera were P. mirabilis (15, 12.9%), P. aeruginosa (13, 11.2%), Acinetobacter baumannii (9, 7.8%), Morganella morganii (6, 5.2%), Staphylococcus epidermidis (5, 4.3%), E. coli (4, 3.4%), and Proteus vulgaris (2, 1.7%).
Antimicrobial Susceptibility Test
Details of the antimicrobial resistance pattern of the isolated Gram-positive and Gram-negative bacteria are shown in Tables 4, 5. S. aureus showed high resistance rates to penicillin (92.0%), erythromycin (58.3%), and clindamycin (50.9%). The MRSA strains were 100% resistant to oxacillin, followed by penicillin (95%), erythromycin (61.3%), clindamycin (54.8%), and moxifloxacin (32.3%). In contrast, MDR S. epidermidis exhibited 100% resistance rates to penicillin, followed by erythromycin (80.0%), clindamycin (80.0%), and levofloxacin (32.3%), but no resistance to moxifloxacin (0%) (Figure 5A). Meanwhile, MDR E. faecalis showed high resistance rates to tetracycline (79.2%), quinupristin/dalfotristin (70.8%), and Gentamicin (54.2%). Vancomycin was the most effective antibiotic (0% resistance rate) against all of the Gram-positive bacteria.
Table 4.
Drug resistance patterns of Gram-positive pathogenic bacteria.
| Value, No. (%) | |||||
|---|---|---|---|---|---|
| Antibiotics | S. aureus | E. faecalis | S. epidermidis | S. haemolyticus | Streptococcus spp. |
| (n = 175) | (n = 24) | (n = 20) | (n = 16) | (n = 8) | |
| Penicillin | 161 (92.0) | 6 (25.0) | 19 (95.0) | 12(75) | 1 (12.5) |
| Oxacillin | 61 (34.9) | 0 (0) | 16 (80.0) | 11 (68.8) | 0 (0) |
| Ampicillin | 25 (14.3) | 5 (20.8) | 0 (0) | 2 (12.5) | 0 (0) |
| Erythromycin | 102 (58.3) | 12 (50.0) | 15 (75.0) | 12(75) | 5 (62.5) |
| Clindamycin | 89 (50.9) | 7 (29.2) | 12 (60.0) | 11 (68.8) | 8 (100) |
| Moxifloxacin | 17 (9.7) | 2 (8.3) | 3 (15.0) | 8 (50) | 0 (0) |
| Levofloxacin | 29 (16.6) | 11 (45.8) | 8 (40.0) | 9 (56.3) | 4 (50.0) |
| Ciprofloxacin | 31 (17.7) | 11 (45.8) | 6 (30.0) | 11 (68.8) | 0 (0) |
| Tetracycline | 45 (25.7) | 19 (79.2) | 7 (35.0) | 4 (25.0) | 1 (12.5) |
| Rifampicin | 10 (5.7) | 2 (8.3) | 2 (10.0) | 5 (31.3) | 0 (0) |
| Gentamicin | 31 (17.7) | 13 (54.2) | 0 (0) | 8 (50.0) | 0 (0) |
| Cotrimoxazole | 22 (12.6) | 2 (8.3) | 6 (30.0) | 4 (25) | 1 (12.5) |
| Ceftriaxone | 7 (4.0) | 1 (4.2) | 0 (0) | 0 (0) | 0 (0) |
| Cefoxitin | 15 (8.6) | 0 (0) | 0 (0) | 2 (12.5) | 0 (0) |
| Quinupristin/dalfotristin | 1 (0.6) | 17 (70.8) | 2 (10.0) | 1 (6.3) | 0 (0) |
| Amoxil | 13 (7.4) | 2 (8.3) | 0 (0) | 2 (12.5) | 0 (0) |
| Vancomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Table 5.
Drug resistance patterns of Gram-negative pathogenic bacteria.
| Value, No. (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antibiotics | E. coli | P. aeruginosa | P. mirabilis | Klebsiella spp. | Enterobacter spp. | A. baumannii | Serratia spp. | M. morganii |
| (n = 69) | (n = 66) | (n = 48) | (n = 40) | (n = 19) | (n = 15) | (n = 11) | (n = 10) | |
| Ampicillin | 47 (68.1) | 23 (34.8) | 30 (62.5) | 18 (45.0) | 8 (42.1) | 8 (53.3) | 4 (36.4) | 3 (30) |
| Ciprofloxacin | 47 (68.1) | 9 (13.6) | 31 (64.6) | 16 (40.0) | 5 (26.3) | 7 (46.7) | 0 (0) | 3 (30) |
| Levofloxacin | 42 (60.9) | 10 (15.2) | 17 (35.4) | 14 (35.0) | 5 (26.3) | 7 (46.7) | 0 (0) | 1 (10) |
| Cefazolin | 30 (43.5) | 15 (22.7) | 35 (72.9) | 18 (45.0) | 13 (68.4) | 10 (66.7) | 8 (72.7) | 7 (70) |
| Ceftriaxone | 28 (40.6) | 18 (27.3) | 29 (60.4) | 12 (30.0) | 5 (26.3) | 6 (40.0) | 1 (9.1) | 0 (0) |
| Ceftazidime | 12 (17.4) | 3 (4.5) | 9 (18.8) | 6 (15.0) | 4 (21.1) | 8 (53.3) | 0 (0) | 0 (0) |
| Cefuroxime | 17 (24.6) | 3 (4.5) | 14 (29.2) | 8 (20.0) | 6 (31.6) | 4 (26.7) | 4 (36.4) | 1 (10) |
| Cefotaxime | 7 (10.1) | 3 (4.5) | 6 (12.5) | 4 (10.0) | 4 (21.1) | 0 (0) | 1 (9.1) | 0 (0) |
| Cefepime | 11(15.9) | 4 (6.1) | 11 (22.9) | 2 (5.0) | 3 (15.8) | 5 (33.3) | 0 (0) | 0 (0) |
| Cotrimoxazole | 28 (40.6) | 17 (25.8) | 18 (37.5) | 24 (60.0) | 6 (31.6) | 7 (46.7) | 2 (18.2) | 5 (50) |
| Gentamicin | 18 (26.1) | 8 (12.1) | 16 (33.3) | 12 (30.0) | 2 (10.5) | 7 (46.7) | 1 (9.1) | 1 (10) |
| Piperacillin | 18 (26.1) | 6 (9.1) | 6 (12.5) | 4 (10.0) | 3 (15.8) | 3 (20.0) | 2 (18.2) | 0 (0) |
| Imipenem | 1 (1.4) | 15 (22.7) | 9 (18.8) | 2 (5.0) | 0 (0) | 5 (33.3) | 0 (0) | 8 (80) |
| Meropenem | 0 (0) | 9 (13.6) | 1 (2.1) | 2 (5.0) | 1 (5.3) | 4 (26.7) | 0 (0) | 0 (0) |
| Aztreonam | 10 (14.5) | 16 (24.2) | 8 (16.7) | 5 (12.5) | 4 (21.1) | 9 (60.0) | 0 (0) | 0 (0) |
| Tobramycin | 10 (14.5) | 15 (22.7) | 9 (18.8) | 5 (12.5) | 3 (15.8) | 3 (20.0) | 0 (0) | 1 (10) |
| Tetracycline | 8 (11.6) | 7 (10.6) | 6 (12.5) | 1 (2.5) | 1 (5.3) | 0 (0) | 3 (27.3) | 0 (0) |
| Amikacin | 0 (0) | 6 (9.1) | 3 (6.3) | 0 (0) | 0 (0) | 2 (13.3) | 0 (0) | 0 (0) |
| Nitrofurantoin | 0 (0) | 8 (12.1) | 18 (37.5) | 11 (27.5) | 0 (0) | 1 (6.7) | 0 (0) | 6 (60) |
| Tigecycline | 0 (0) | 10 (15.2) | 2 (4.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Amoxil | 5 (7.2) | 5 (7.6) | 3 (6.3) | 0 (0) | 0 (0) | 2 (13.3) | 4 (36.4) | 1 (10) |
Figure 5.

Drug resistance patterns of MDR bacteria. (A) Drug resistance patterns of MDR Gram-positive bacteria. (B) Drug resistance patterns of MDR Gram-negative bacteria. The color and size of the bubbles indicate the percentage of bacteria that are resistant to certain antibiotics. The blank indicates no resistance.
The resistance rates of E. coli were 68.1% to ampicillin, 68.1% to ciprofloxacin, and 60.9% to levofloxacin. For P. aeruginosa, 34.8% of isolates were resistant to ampicillin, 27.3% to ceftriaxone, and 25.8% were resistant to cotrimoxazole. P. mirabilis showed high resistance rates to cefazolin (72.9%), ciprofloxacin (64.6%), and ampicillin (62.5%). However, all the isolated Gram-negative bacteria showed low resistance rates to tigecycline (3.9%) and amikacin (3.6%). In addition, we analyzed antibiotic resistance rates of MDR Gram-negative bacteria, and the results revealed that 49 MDR Gram-negative strains were isolated, which accounted for 15.9% of the Gram-negative bacteria. MDR E. coli showed 100% resistance rates to ampicillin, ceftriaxone, and gentamicin whereas MDR Pseudomonas aeruginosa, Proteus mirabilis, Proteus vulgaris, and Morganella morganii were highly resistant to imipenem (80–100%) (Figure 5B).
Discussion
Consistent with previous studies (4, 7), this study showed that the most common causes of chronic cutaneous wounds in China were diabetes (22.5%), infection (22.5%), pressure (17.2%), and trauma (10.2%). Comparatively, the primary causes of chronic wounds in western countries are diabetes, venous diseases, pressure, and surgery (12, 25–27). The differences in the etiological characteristics of east and west are, at least in part, a consequence of the differences in economic development, healthcare systems, as well as living and eating habits. Notably, chronic cutaneous wounds have become a major challenge worldwide, and therefore there is an urgent need to develop more effective treatment options.
Microbial infection is the most common challenge to wound healing. A wound is considered infected if the bacteria exceeds a threshold of 105 per gram of wound tissue (28, 29). Chronic wounds are normally colonized by a large collection of pathogenic bacteria that are more likely to form biofilms, and directly contribute to delayed wound healing (9, 30–32). Bacteria that commonly colonize wounds include S. aureus, P. aeruginosa, and E. coli. These bacterial species usually exert a damaging effect on wound healing (15, 33). Many other species of bacteria in chronic wounds have also been reported, including Enterobacter cloacae (17, 34–36), Citrobacter sp. (37, 38), Peptostreptococcus sp. (39), Flavobacter sp. (40), Serratia sp. (41, 42), and Candida sp. (43–45).
In the present study, the positive rate of microbial culture (63.9%) was much lower than reported by Howell-Jones (82%) and Kassam (91.4%) in western countries (30, 46), and this could be owing to the fact that many patients use antibiotics for self-treatment before seeking medical attention, thus reducing the total positive rate of microbial culture.
Consistent with other studies (9, 32), the results of the present study revealed that chronic cutaneous wounds contained mainly Gram-negative bacteria. The most common species were S. aureus (29.2%), E. coli (11.5%), P. aeruginosa (11.0%), P. mirabilis (8.0%), and K. pneumoniae (6.7%). These results were fall in line with the study by Calina et al. (12), which focused on surgical site infections. Meanwhile, we observed a variation in the bacterial species depending on the causes of wounds. Contrary to previous studies (47), wounds caused by diabetes, radiation, trauma, and burn were mainly colonized by Gram-positive bacteria, whereas those caused by vascular diseases, pressure, surgery and malignant tumor were mainly infected with Gram-negative bacteria. The bacteria in chronic wounds mostly form a polymicrobial environment, which provides a suitable environment for genetic exchange between different bacteria and contribute to antibiotic resistance (30). However, the present study illustrated the majority of wound infections were monomicrobial infections (75.2%). In addition, wounds in the rump, perineum, and feet were more likely to form polymicrobial infections than wounds in other body parts.
In the present study, the number of MDR bacterial isolates was 116 from 116 cases, with an occurrence rate of 19.3%. Several studies have reported a 10–59% occurrence rate of MDR bacterial strains in chronic wounds (48–50). We cultivated 62 MRSA strains, accounting for 53.4% of all MDR strains and 35.4% of the S. aureus strains. Our findings were consistent with other studies reporting that MRSA take up about 40% of the S. aureus strains in SSIs (12, 51). S. aureus strains were the most common pathogens in chronic cutaneous wounds and exhibited a high frequency of resistance to antibiotics. Studies have demonstrated that S. aureus usually forms biofilms in chronic wounds, thereby causing drug-resistance (52). Similar to findings by Shittu et al. (53) and Shah et al. (54), no strains of S. aureus, including those in the MRSA group, showed resistance to vancomycin in our study. However, high levels of resistance to tetracycline (79.2%), quinupristin/dalfotristin (70%), gentamicin (54.2%), and erythromycin (50.0%) were found in Enterococcus spp.
In this study, E. coli (11.5%) were the most common Gram-negative bacteria in chronic wounds. However, Wong et al. (15) and Gadepalli et al. (47) both reported that P. aeruginosa (14.8–16.7%) were the most common Gram-negative bacteria in chronic wounds. Another study indicated that P. aeruginosa were more likely to colonize deeper layers of tissue (55). However, all samples were swab cultures instead of deep tissue cultures in the present study, which might have contributed to the differences in the results. We also found that the resistance rates of E. coli to ampicillin, ciprofloxacin, and levofloxacin were high (>60%), with no resistant to meropenem, amikacin, nitrofurantoin, and tigecycline.
In addition, we found a lower resistance rate (<5%) of Klebsiella spp., Enterobacter spp., and Serratia spp.to imipenem, but M. morganii showed a high resistant rate to imipenem (80%). Gram-negative bacteria have been shown to be highly susceptible to amikacin and meropenem (56, 57). However, the resistivity of A. baumannii to amikacin and meropenem were 13.3 and 26.7%, respectively. The lowest resistivity of Gram-negative was found in tigecycline (3.9%) and amikacin (3.6%). Study elsewhere have reported similar findings (58).
An epidemiological study in 2008 indicated that about 78% of patients with chronic cutaneous wounds in China received antimicrobial treatment (7), which is a higher proportion compared to western countries (59, 60). Admittedly, the overuse and misuse of antibiotics is a global problem, which directly contributes to the spreading of antibiotic resistance, especially in China.
Evolving antibiotic resistance has prompted the judicious use of systemic antimicrobials, particularly in treating local infections, such as cutaneous wounds. The use of topical antimicrobials to manage chronic wounds is necessary for controlling wounds infection and even the formation of bacterial biofilms. Topical antibiotic treatments like polymyxin B, silver sulfadiazine are preferred over systemic antibiotic treatments for infected wounds, and antibiotics should be stopped once the wound is clean (61–63). A retrospective study from Hammond et al. (64) indicated that triple antibiotic (polymyxin B, neomycin, bacitracin) ointments can significantly reduce biofilms produced by S. aureus and P. aeruginosa isolates in burn wounds. It is noteworthy that the use of topical antibiotics can reduce the amount of systemic antibiotics and delay the occurrence of drug resistance. Therefore, antibiotics, especially systemic antibiotics for chronic wounds treatments, must be used under strict control. Furthermore, for chronic wounds, there is a need to perform microbial culture and antibiotics susceptibility tests prior to prescribing antibiotics.
Our study does have some limitations. Firstly, despite being a non-invasive and widely used method, swabs might provide a less truly status of bacterial colonization in the wounds as compared to puncture or tissue biopsy samples if not operated properly. The results of bacteria culture can be affected by colonizing organisms by improper collection, making it difficult to define that this bacterium is infecting wounds or just colonizing them. Secondly, due to the retrospective study design, we were unable to determine the sources of infections (community or hospital acquired) with insufficient information in our database, which was important for epidemiological purposes and impact directly in the antimicrobial resistance rates. Thirdly, as a large-sample research, the data of our study were gathered from 195 hospitals across the country, and the susceptibility profile for some antimicrobials were still reported for those microorganisms showing intrinsic resistance to these agents in a few hospitals. These data were also included in our study which need to be analyzed carefully. Therefore, we suggest these factors should be taken into account in future studies.
However, our findings may help clinicians to establish informed guidelines regarding antibiotic therapy for patients with chronic cutaneous wounds, with the aim to control the infections more effectively and to avoid the overuse and misuse of antibiotics.
Conclusion
In summary, we analyzed data on the distribution and antimicrobial susceptibility tests of pathogenic bacteria isolated from chronic cutaneous wounds of patients in China. Collectively, we recommend that the antibiotics used in the treatment of chronic wounds should be under strict regulation. Furthermore, there is a need to perform microbial culture and antibiotics susceptibility tests for bacterial isolates from chronic wounds before prescribing antibiotics. Our findings may guide clinicians in making informed decisions regarding antibiotic treatment for patients with chronic wounds.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
HG, SL, and JT: concept and design. HG, WD, BG, and YL: drafting of the manuscript. HG, MJ, DZ, and YL: statistical analysis. YA, JD, and YN: administrative, technical, or material support. SL: supervision. All authors: acquisition, analysis, or interpretation of data, and critical revision of the manuscript for important intellectual content.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.609584/full#supplementary-material
References
- 1.Han G, Ceilley R. Chronic wound healing: a review of current management and treatments. Adv Ther. (2017) 34:599–610. 10.1007/s12325-017-0478-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. (2009) 17:763–71. 10.1111/j.1524-475X.2009.00543.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones RE, Foster DS, Longaker MT. Management of chronic wounds-2018. JAMA. (2018) 320:1481–2. 10.1001/jama.2018.12426 [DOI] [PubMed] [Google Scholar]
- 4.Cheng B, Jiang Y, Fu X, Hao D, Liu H, Liu Y, et al. Epidemiological characteristics and clinical analyses of chronic cutaneous wounds of inpatients in China: prevention and control. Wound Repair Regen. (2020) 28:623–30. 10.1111/wrr.12825 [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Liang H, Clarke E, Jackson C, Xue M. Inflammation in chronic wounds. Int J Mol Sci. (2016) 17:2085. 10.3390/ijms17122085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong J, Tian M, Song F, Tang J, Liu Y, Wu M, et al. Epidemiological investigation of vascular etiological examinations in the diagnosis and treatment of lower-extremity ulcers in China. Wound Repair Regen. (2020) 28:532–8. 10.1111/wrr.12810 [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Huang S, Fu X, Liu H, Ran X, Lu S, et al. Epidemiology of chronic cutaneous wounds in China. Wound Repair Regen. (2011) 19:181–8. 10.1111/j.1524-475X.2010.00666.x [DOI] [PubMed] [Google Scholar]
- 8.Rhoads DD, Wolcott RD, Sun Y, Dowd SE. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci. (2012) 13:2535–50. 10.3390/ijms13032535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahim K, Saleha S, Zhu X, Huo L, Basit A, Franco OL. Bacterial contribution in chronicity of wounds. Microb Ecol. (2017) 73:710–21. 10.1007/s00248-016-0867-9 [DOI] [PubMed] [Google Scholar]
- 10.Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, Moser C, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. (2008) 46:2717–22. 10.1128/JCM.00501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg SR, Diegelmann RF. What makes wounds chronic. Surg Clin North Am. (2020) 100:681–93. 10.1016/j.suc.2020.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Calina D, Docea AO, Rosu L, Zlatian O, Rosu AF, Anghelina F, et al. Antimicrobial resistance development following surgical site infections. Mol Med Rep. (2017) 15:681–8. 10.3892/mmr.2016.6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulon M, Haamann F, Peters C, Schablon A, Nienhaus A. MRSA prevalence in. European healthcare settings: a review. BMC Infect Dis. (2011) 11:138. 10.1186/1471-2334-11-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweizer ML, Chiang HY, Septimus E, Moody J, Braun B, Hafner J, et al. Association of a bundled intervention with surgical site infections among patients undergoing cardiac, hip, or knee surgery. JAMA. (2015) 313:2162–71. 10.1001/jama.2015.5387 [DOI] [PubMed] [Google Scholar]
- 15.Wong SY, Manikam R, Muniandy S. Prevalence and antibiotic susceptibility of. bacteria from acute and chronic wounds in Malaysian subjects. J Infect Dev Ctries. (2015) 9:936–44. 10.3855/jidc.5882 [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Pan H, Leng W, Lei X, Chen L, Liang Z. Distribution of microbes and drug susceptibility in patients with diabetic foot infections in Southwest China. J Diabetes Res. (2018) 2018:9817308. 10.1155/2018/9817308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. (2008) 8:43. 10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong W, Nie LJ, Wu MJ, Xie T, Liu YK, Tang JJ, et al. WoundCareLog APP – A new application to record wound diagnosis and healing. Chin J Traumatol. (2019) 22:296–9. 10.1016/j.cjtee.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haesler E, Swanson T, Ousey K, Carville K. Clinical indicators of wound infection and biofilm: reaching international consensus. J Wound Care. (2019) 28:s4–12. 10.12968/jowc.2019.28.Sup3b.S4 [DOI] [PubMed] [Google Scholar]
- 20.Gardner SE, Frantz RA, Doebbeling BN. The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen. (2001) 9:178–86. 10.1046/j.1524-475x.2001.00178.x [DOI] [PubMed] [Google Scholar]
- 21.Gjodsbol K, Skindersoe ME, Christensen JJ, Karlsmark T, Jorgensen B, Jensen AM, et al. No need for biopsies: comparison of three sample techniques for wound microbiota determination. Int Wound J. (2012) 9:295–302. 10.1111/j.1742-481X.2011.00883.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassim A, Omuse G, Premji Z, Revathi G. Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: a cross-sectional study. Ann Clin Microbiol Antimicrob. (2016) 15:21. 10.1186/s12941-016-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 24.Harris C, Bates-Jensen B, Parslow N, Raizman R, Singh M, Ketchen R. Bates-Jensen wound assessment tool: pictorial guide validation project. J Wound Ostomy Continence Nurs. (2010) 37:253–9. 10.1097/WON.0b013e3181d73aab [DOI] [PubMed] [Google Scholar]
- 25.Valencia IC, Falabella A, Kirsner RS, Eaglstein WH. Chronic venous insufficiency and venous leg ulceration. J Am Acad Dermatol. (2001) 44:401–21; quiz 22–4. 10.1067/mjd.2001.111633 [DOI] [PubMed] [Google Scholar]
- 26.Richmond NA, Maderal AD, Vivas AC. Evidence-based management of common chronic lower extremity ulcers. Dermatol Ther. (2013) 26:187–96. 10.1111/dth.12051 [DOI] [PubMed] [Google Scholar]
- 27.Frykberg RG, Banks J. Challenges in the treatment of chronic wounds. Adv Wound Care. (2015) 4:560–82. 10.1089/wound.2015.0635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trengove NJ, Stacey MC, McGechie DF, Mata S. Qualitative bacteriology and leg ulcer healing. J Wound Care. (1996) 5:277–80. 10.12968/jowc.1996.5.6.277 [DOI] [PubMed] [Google Scholar]
- 29.Stojadinovic A, Carlson JW, Schultz GS, Davis TA, Elster EA. Topical advances. in wound care. Gynecol Oncol. (2008) 111:S70–80. 10.1016/j.ygyno.2008.07.042 [DOI] [PubMed] [Google Scholar]
- 30.Howell-Jones RS, Wilson MJ, Hill KE, Howard AJ, Price PE, Thomas DW. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J Antimicrob Chemother. (2005) 55:143–9. 10.1093/jac/dkh513 [DOI] [PubMed] [Google Scholar]
- 31.Mudge EJ. Recent accomplishments in wound healing. Int Wound J. (2015) 12:4–9. 10.1111/iwj.12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu YK, Cheng NC, Cheng CM. Biofilms in chronic wounds: pathogenesis and diagnosis. Trends Biotechnol. (2019) 37:505–17. 10.1016/j.tibtech.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 33.Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. (2015) 13:605–13. 10.1586/14787210.2015.1023291 [DOI] [PubMed] [Google Scholar]
- 34.Mokracka J, Koczura R, Pawlowski K, Kaznowski A. Resistance patterns and integron cassette arrays of Enterobacter cloacae complex strains of human origin. J Med Microbiol. (2011) 60:737–43. 10.1099/jmm.0.027979-0 [DOI] [PubMed] [Google Scholar]
- 35.Wu M, Ruan H, Huang Y, Liu C, Ni P, Ye J, et al. Bacteriological investigation of chronic wounds in a specialized wound healing department: a retrospective analysis of 107 cases. Int J Low Extrem Wounds. (2015) 14:178–82. 10.1177/1534734615572825 [DOI] [PubMed] [Google Scholar]
- 36.Hosny AEM, Rasmy SA, Aboul-Magd DS, Kashef MT, El-Bazza ZE. The increasing threat of silver-resistance in clinical isolates from wounds and burns. Infect Drug Resist. (2019) 12:1985–2001. 10.2147/IDR.S209881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. (2018) 7:37. 10.1186/s13756-018-0324-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thanganadar Appapalam S, Muniyan A, Vasanthi Mohan K, Panchamoorthy R. A study on isolation, characterization, and exploration of multiantibiotic-resistant bacteria in the wound site of diabetic foot ulcer patients. Int J Low Extrem Wounds. (2019). 10.1177/1534734619884430. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Charles PG, Uckay I, Kressmann B, Emonet S, Lipsky BA. The role of anaerobes in diabetic foot infections. Anaerobe. (2015) 34:8–13. 10.1016/j.anaerobe.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 40.Kienzle N, Muller M, Pegg S. Chryseobacterium in burn wounds. Burns. (2001) 27:179–82. 10.1016/s0305-4179(00)00087-5 [DOI] [PubMed] [Google Scholar]
- 41.Fernandez AL, Adrio B, Martinez Cereijo JM, Martinez Monzonis MA, El-Diasty MM, Alvarez Escudero J. Clinical study of an outbreak of postoperative mediastinitis caused by Serratia marcescens in adult cardiac surgery. Interact Cardiovasc Thorac Surg. (2020) 30:523–7. 10.1093/icvts/ivz312 [DOI] [PubMed] [Google Scholar]
- 42.Mamishi S, Mahmoudi S, Naserzadeh N, Hosseinpour Sadeghi R, Haghi Ashtiani MT, Bahador A, et al. Antibiotic resistance and genotyping of gram-negative bacteria causing hospital-acquired infection in patients referred to Children's Medical Center. Infect Drug Resist. (2019) 12:3377–84. 10.2147/IDR.S195126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farjah MH, Farahpour MR. Efficacy of topical platelet-rich plasma and chitosan co-administration on Candida albicans-infected partial thickness burn wound healing. Burns. (2020) 46:1889–95. 10.1016/j.burns.2020.05.019 [DOI] [PubMed] [Google Scholar]
- 44.Zmuda HM, Mohamed A, Raval YS, Call DR, Schuetz AN, Patel R, et al. Hypochlorous acid-generating electrochemical scaffold eliminates Candida albicans biofilms. J Appl Microbiol. (2020) 129:776–86. 10.1111/jam.14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alam F, Catlow D, Di Maio A, Blair JMA, Hall RA. Candida albicans enhances meropenem tolerance of Pseudomonas aeruginosa in a dual-species biofilm. J Antimicrob Chemother. (2020) 75:925–35. 10.1093/jac/dkz514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kassam NA, Damian DJ, Kajeguka D, Nyombi B, Kibiki GS. Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a Tertiary Hospital, northern Tanzania. BMC Res Notes. (2017) 10:757. 10.1186/s13104-017-3092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadepalli R, Dhawan B, Sreenivas V, Kapil A, Ammini AC, Chaudhry R. A clinico-microbiological study of diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care. (2006) 29:1727–32. 10.2337/dc06-0116 [DOI] [PubMed] [Google Scholar]
- 48.Mendo-Lopez R, Jasso L, Guevara X, Astocondor AL, Alejos S, Bardossy AC, et al. Multidrug-resistant microorganisms colonizing lower extremity wounds in patients in a Tertiary Care Hospital, Lima, Peru. Am J Trop Med Hyg. (2017) 97:1045–8. 10.4269/ajtmh.17-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandemir O, Akbay E, Sahin E, Milcan A, Gen R. Risk factors for infection of the diabetic foot with multi-antibiotic resistant microorganisms. J Infect. (2007) 54:439–45. 10.1016/j.jinf.2006.08.013 [DOI] [PubMed] [Google Scholar]
- 50.Cataldo MC, Bonura C, Caputo G, Aleo A, Rizzo G, Geraci DM, et al. Colonization of pressure ulcers by multidrug-resistant microorganisms in patients receiving home care. Scand J Infect Dis. (2011) 43:947–52. 10.3109/00365548.2011.591821 [DOI] [PubMed] [Google Scholar]
- 51.Mohamed N, Wang MY, Le Huec JC, Liljenqvist U, Scully IL, Baber J, et al. Vaccine development to prevent Staphylococcus aureus surgical-site infections. Br J Surg. (2017) 104:e41–54. 10.1002/bjs.10454 [DOI] [PubMed] [Google Scholar]
- 52.Banu A, Hassan MMN, Rajkumar J, Srinivasa S. Spectrum of bacteria associated. with diabetic foot ulcer and biofilm formation: a prospective study. Australas Med J. (2015) 8:280–5. 10.4066/Amj.2015.2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis. (2006) 6:125. 10.1186/1471-2334-6-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahmohammadi MR, Nahaei MR, Akbarzadeh A, Milani M. Clinical test to detect mecA and antibiotic resistance in Staphylococcus aureus, based on novel biotechnological methods. Artif Cells Nanomed Biotechnol. (2016) 44:1464–8. 10.3109/21691401.2015.1041639 [DOI] [PubMed] [Google Scholar]
- 55.Fazli M, Bjarnsholt T, Kirketerp-Moller K, Jorgensen B, Andersen AS, Krogfelt KA, et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol. (2009) 47:4084–9. 10.1128/Jcm.01395-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekhar S, Vyas N, Unnikrishnan M, Rodrigues G, Mukhopadhyay C. Antimicrobial susceptibility pattern in diabetic foot ulcer: a pilot study. Ann Med Health Sci Res. (2014) 4:742–5. 10.4103/2141-9248.141541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugandhi P, Prasanth DA. Microbiological profile of bacterial pathogens from diabetic foot infections in tertiary care hospitals, Salem. Diabetes Metab Syndr. (2014) 8:129–32. 10.1016/j.dsx.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 58.Alhussain FA, Yenugadhati N, Al Eidan FA, Al Johani S, Badri M. Risk factors, antimicrobial susceptibility pattern and patient outcomes of Pseudomonas aeruginosa infection: a matched case-control study. J Infect Public Health. (2021) 14:152–7. 10.1016/j.jiph.2020.11.010 [DOI] [PubMed] [Google Scholar]
- 59.Howell-Jones RS, Price PE, Howard AJ, Thomas DW. Antibiotic prescribing for chronic skin wounds in primary care. Wound Repair Regen. (2006) 14:387–93. 10.1111/j.1743-6109.2006.00144.x [DOI] [PubMed] [Google Scholar]
- 60.Hernandez R. The use of systemic antibiotics in the treatment of chronic wounds. Dermatol Ther. (2006) 19:326–37. 10.1111/j.1529-8019.2006.00091.x [DOI] [PubMed] [Google Scholar]
- 61.Stevens DL, Bisno AL, Chambers HF, Dellinger EP, Goldstein EJC, Gorbach SL, et al. Executive summary: practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the infectious diseases society of America. Clin Infect Dis. (2014) 59:147–59. 10.1093/cid/ciu444 [DOI] [PubMed] [Google Scholar]
- 62.Schultz GS, Sibbald RG, Falanga V, Ayello EA, Dowsett C, Harding K, et al. Wound bed preparation: a systematic approach to wound management. Wound Repair Regen. (2003) 11:S1–28. 10.1046/j.1524-475x.11.s2.1.x [DOI] [PubMed] [Google Scholar]
- 63.Tang J, Guan H, Dong W, Liu Y, Dong J, Huang L, et al. Application of compound polymyxin B ointment in the treatment of chronic refractory wounds. Int J Low Extrem Wounds. (2020). 10.1177/1534734620944512. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 64.Hammond AA, Miller KG, Kruczek CJ, Dertien J, Colmer-Hamood JA, Griswold JA, et al. An in vitro biofilm model to examine the effect of antibiotic ointments on biofilms produced by burn wound bacterial isolates. Burns. (2011) 37:312–21. 10.1016/j.burns.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.