Abstract
Mitochondria maintain a distinct pool of ribosomal machinery, including tRNAs and tRNAs activating enzymes, such as mitochondrial tyrosyl-tRNA synthetase (YARS2). Mutations in YARS2, which typically lead to the impairment of mitochondrial protein synthesis, have been linked to an array of human diseases including optic neuropathy. However, the lack of YARS2 mutation animal model makes us difficult to elucidate the pathophysiology underlying YARS2 deficiency. To explore this system, we generated YARS2 knockout (KO) HeLa cells and zebrafish using CRISPR/Cas9 technology. We observed the aberrant tRNATyr aminoacylation overall and reductions in the levels in mitochondrion- and nucleus-encoding subunits of oxidative phosphorylation system (OXPHOS), which were especially pronounced effects in the subunits of complex I and complex IV. These deficiencies manifested the decreased levels of intact supercomplexes overall. Immunoprecipitation assays showed that YARS2 bound to specific subunits of complex I and complex IV, suggesting the posttranslational stabilization of OXPHOS. Furthermore, YARS2 ablation caused defects in the stability and activities of OXPHOS complexes. These biochemical defects could be rescued by the overexpression of YARS2 cDNA in the YARS2KO cells. In zebrafish, the yars2KO larva conferred deficient COX activities in the retina, abnormal mitochondrial morphology, and numbers in the photoreceptor and retinal ganglion cells. The zebrafish further exhibited the retinal defects affecting both rods and cones. Vision defects in yars2KO zebrafish recapitulated the clinical phenotypes in the optic neuropathy patients carrying the YARS2 mutations. Our findings highlighted the critical role of YARS2 in the stability and activity of OXPHOS and its pathological consequence in vision impairments.
Keywords: mitochondrial tyrosyl-tRNA synthetase, oxidative phosphorylation, animal disease model, vision function, retina
Abbreviations: DMEM, Dulbecco’s modified eagle medium; EHC, enzyme histochemistry; GCL, ganglion cell layer; H&E, hematoxylin and eosin; INL, inner nuclear layer; IPL, inner plexiform layer; KO, knockout; MLASA, myopathy, lactic acidosis, and sideroblastic anemia; mtDNA, mitochondrial DNA; OCR, oxygen consumption rate; ONL, outer nuclear layer; OPL, outer plexiform layer; OXPHOS, oxidative phosphorylation system
Defects in mitochondrial protein synthesis are associated with a wide spectrum of human diseases, with a diversity of etiologies, ages of onset, involved organ systems and clinical presentations (1, 2, 3). Mitochondrial translation deficiencies resulted from defects in two rRNAs or 22 tRNAs, encoded by mitochondrial DNA (mtDNA), or alterations in the components of mitochondrial translation machinery (ribosomal proteins, ribosomal assembly proteins, aminoacyl-tRNA synthetases, tRNA-modifying enzymes, tRNA methylating enzymes, initiation, elongation, and termination factors), encoded by nuclear genes (3, 4, 5, 6, 7, 8, 9). In particular, all 19 mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs) were synthesized in the cytosol and subsequently imported into mitochondria. These mt-aaRSs are a group of enzymes that catalyze a two-step reaction where they first activate the cognate amino acid with ATP to form an aminoacyl-adenylate, then subsequently transfer the aminoacyl group of the aminoacyl-adenylate to the bound tRNA for protein synthesis (8, 9, 10). In addition to their central roles in the translation, these aminoacyl-tRNA synthetases may have other functions, as in the case of cytoplasmic aminoacyl-tRNA synthetases (11, 12, 13). Defects in nuclear genes encoding mt-aaRSs have emerged as an important cause of mitochondrial disorders linked to diverse clinical presentations, usually with an early onset and transmitted as autosomal recessive traits (8, 9, 10, 14, 15, 16). Remarkably, the pathologies linked to defects in mt-aaRSs display a marked bias for the central nervous system, including the leukodystrophy, encephalopathy, Perrault syndrome, sensorineural deafness, and visual impairment (17, 18, 19, 20, 21, 22, 23). However, the pathophysiology of these disease-linked mt-aaRS mutations, especially the pleiotropic and tissue-specific effects, remains poorly understood.
Mutations of YARS2 gene encoding mitochondrial tyrosyl-tRNA synthetase were responsible for the myopathy, lactic acidosis, and sideroblastic anemia (MLASA) with mitochondrial respiratory chain complex deficiencies (24, 25, 26, 27, 28, 29, 30). Recently, we demonstrated that the YARS2 p.Gly191Val mutation contributed to the phenotypic expression of Leber's hereditary optic neuropathy (LHON)-associated mtDNA mutations or deafness-associated tRNASer(UCN) 7511A > G mutation (31, 32). The primary defects in these YARS2 mutations were the deficient aminoacylation of tRNATyr (30, 31, 32). The aberrant tRNATyr metabolism impaired mitochondrial translation, especially for the synthesis of polypeptides with high content of tyrosine codon such as ND4, ND5, ND6, and CO2 in cell lines carrying YARS2 mutations (24, 29, 31, 32). Strikingly, cell lines carrying the YARS2 mutations exhibited marked reductions in the nucleus-encoding mitochondrial proteins: NDUFS3 and NDUFB8 [subunits of NADH:ubiquinone oxidoreductase (complex I)] and COX10 [subunit of cytochrome c oxidase (complex IV)] (24, 29, 32). These implied that YARS2 may play an important role in the biogenesis of oxidative phosphorylation (OXPHOS) system (1). To test this hypothesis, we used CRISPR/Cas9 genomic editing approach in HeLa cells to produce the targeted deletion in YARS2 gene. To further confirm the defects of YARS2 knockout (KO) (YARS2KO) in the HeLa cells, we transferred a plasmid carrying the full-length YARS2 cDNA into the YARS2KO HeLa cells. The YARS2KO cell lines were assessed for the effects of YARS2 mutations on the biogenesis of OXPHOS, by western blot analysis using the subunits of OXPHOS and activity of respiratory chain complexes. To examine whether YARS2 interacts with the OXPHOS, we performed the immunoprecipitation assay using FLAG and HA antibodies in mitochondria of HeLa cell lines. To investigate whether defects in yars2 caused the visual impairment in vivo, we investigated the yars2 knockout zebrafish produced by genome editing using the CRISPR/Cas9 system.
Results
Generation of YARS2 knockout HeLa cell lines
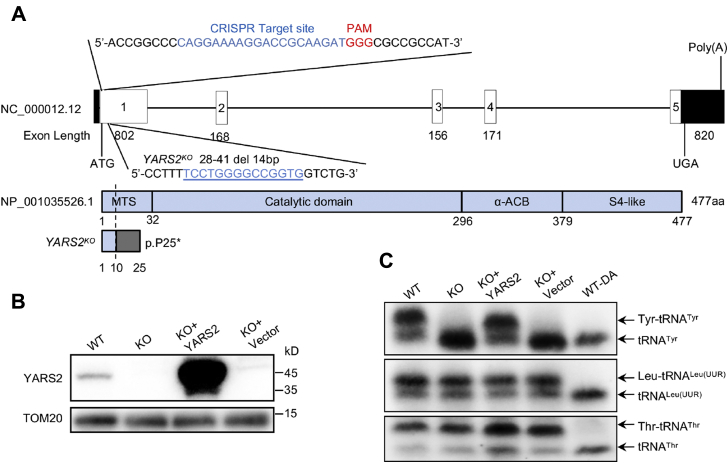
To gain overall information on how the YARS2 deficiency affected the mitochondrial functions, we used CRISPR/Cas9 genomic editing approach to produce the targeted deletion in YARS2 gene in the HeLa cells. As a result, an allele, YARS2del14bp, was generated by introducing a 14 bp deletion in the exon 1 of YARS2 (homozygous cells were described as YARS2KO and wild type as WT). The 14 bp deletion resulted in a frameshift from codon 10, the introduction of a premature stop at codon 25 (p.Pro25∗), and truncated protein with 24 amino acids (Fig. 1A, Fig. S1). This allele was confirmed by Sanger sequencing, restriction fragment length polymorphism (RFLP) with digested with BstNI (Fig. S1) and western blot analysis (Fig. 1B).
Figure 1.
Generation of YARS2 knockout HeLa cell lines using CRISPR/Cas9 system.A, schematic representation of CRISPR/Cas9 target site at exon 1 as used in this study. An allele, YARS2del14bp was produced by a 14 bp delete in the exon 1 and a truncated nonfunctional protein with 24 amino acids. B, western blot analysis of YARS2 in various cells. Twenty micrograms of total cellular proteins of each cell line was electrophoresed through and hybridized with antibodies specific for YARS2 (1:1000 dilution) and with TOM20 (1:2000 dilution) as a loading control. WT, wild-type cells; KO, YARS2KO; KO+YARS2, exogenous YARS2 expression in YARS2KO; KO+vector, vector transfected in YARS2KO. Three independent experiments were performed. C, in vivo aminoacylation of mitochondrial tRNA assays. Ten micrograms of total cellular RNAs purified from various cell lines under acid conditions was electrophoresed through an acid (pH 5.2) 10% polyacrylamide-7 M urea gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probe specific for the tRNATyr, tRNALeu(UUR), and tRNAThr, respectively. Samples for WT cells were deacylated (DA) by heating for 10 min at 60 °C at pH 8.3, electrophoresed, and hybridized with DIG-labeled oligonucleotide probes as described above. Three independent experiments were performed.
Defects in tRNA aminoacylation of tRNATyr were assessed by using electrophoresis in an acid polyacrylamide/urea gel system to separate uncharged tRNA species from the corresponding charged tRNA, electroblotting and hybridizing with DIG-labeled probes specific for tRNATyr, tRNALeu(UUR), and tRNAThr. As shown in Figure 1C, the slower migrating band (upper band) in WT cells represents the charged tRNA, and the faster migrating band (lower band) represents uncharged tRNA. However, only one lower band in the tRNATyrs but two bands of tRNALeu(UUR) and tRNAThr were detected in the YARS2KO cells, indicating that the ablation of YARS2 led to the complete loss of aminoacylation of tRNATyr but did not affect the aminoacylation of tRNALeu(UUR) and tRNAThr. To further distinguish nonaminoacylated tRNA from aminoacylated tRNA, samples of tRNAs were deacylated after heating for 10 min at 60 °C (pH 8.3) and then run in parallel. As shown in Figure 1C, the deacylated samples gave only one band (uncharged tRNA) in both mutant and control cell lines. To further confirm the ablation of YARS2 in HeLa cells, we transferred a plasmid carrying the full-length YARS2 cDNA into the YARS2KO cells. Indeed, the overexpression of YARS2 reversed the deficient aminoacylation of tRNATyr in the YARS2KO cells.
YARS2KO cells exhibited the reductions in the levels in the subunits of OXPHOS, reversible by overexpression of YARS2
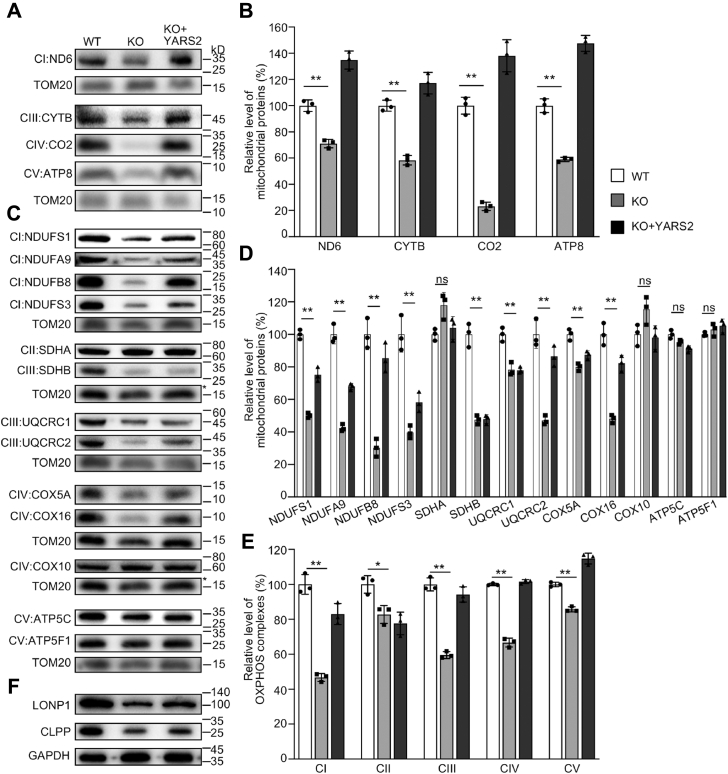
To assess the effect of YARS2 mutation on the OXPHOS biogenesis, we carried out the western blotting analysis to examine the levels in 17 subunits of OXPHOS complexes in YARS2KO and WT cells using TOM20 as a loading control. These subunits included four mtDNA-encoding polypeptides (ND6, CYTB, CO2, and ATP8), 13 nucleus-encoding proteins: NDUFS1, NDUFS3, NDUFA9, and NDUFB8 (subunits of complex I), SDHA and SDHB [subunits of succinate dehydrogenase (complex II)], UQCRC1 and UQCRC2 [subunits of ubiquinol-cytochrome c reductase (complex III)], COX5A, COX10, and COX16 (subunits of complex IV), and ATP5C and ATP5F1 [subunits of H+-ATPase (complex V)] (1). As shown in Figure 2, A and C, the various decreases in the levels of 13 mitochondrial proteins (but not of SDHA, COX10, ATP5C, and ATP5F1) were observed in the YARS2KO cells, as compared with the WT cells. As shown in Figure 2B, the levels of ND6, CYTB, CO2, and ATP8 in the YARS2KO cells were 71%, 58%, 23%, and 59%, with an average of 53%, relative to the mean values measured in the WT cells. As shown in the Figure 2D, the levels of NDUFS1, NDUFS3, NDUFA9, NDUFB8, SDHA, SDHB, UQCRC1, UQCRC2, COX5A, COX10, COX16, ATP5C, and ATP5F1 in YARS2KO cells were 50%, 40%, 42%, 30%, 118%, 48%, 78%, 47%, 80%, 115%, 48%, 95%, and 103%, relative to the mean values measured in the WT cells, respectively. Notably, the average levels in the subunits of complexes I, II, III, IV, and V in the YARS2KO cells were 47%, 82%, 60%, 66%, and 85% of average values measured in the WT HeLa cells, respectively (Fig. 2E). However, the overexpression of YARS2 elevated the levels of these subunits.
Figure 2.
Western blotting analysis of mitochondrial proteins. A and C, twenty micrograms of total cellular proteins from various cell lines was electrophoresed through a denaturing polyacrylamide gel, electroblotted, and hybridized with antibodies for 17 subunits of OXPHOS (4 encoded by mtDNA and 13 encoded by nuclear genes), and TOM20 as a loading control, respectively. B and D, quantification of mitochondrial proteins: four mtDNA-encoding subunits (B) and 13 nucleus-encoding subunits (D). Average relative each polypeptide content per cell was normalized to the average content per cell of Tom20 in each cell line. The values for the latter are expressed as percentages of the average values for the WT HeLa cell line. The calculations were based on three independent determinations. The error bars indicate two standard deviations (SD) of the means. p indicates the significance, according to the t-test, of the differences between mutant and control cell lines. ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001; ns, not significant. E, average levels of subunits from each complex of OXPHOS (5 of complexes I, 2 of II, 3 of III, 4 of IV, and 3 of V). The calculations were based on three independent determinations. Graph details and symbols are explained as above. F, western blot analysis of Clpp involved in mitochondrial ribosome assembly and LONP1 that is a nuclear-encoded mitochondrial protease.
To test whether the YARS2 deficiency affected the mitochondrial proteostasis, we measured the levels of Clpp involved in mitochondrial ribosome assembly (33) and LONP1 that is a nuclear-encoded mitochondrial protease crucial for organelle homeostasis (34) in the YARS2KO and WT cells. As shown in Figure 2F, the levels of Clpp and LONP1 in the YARS2KO cells were 63% and 64% of those in the WT cells, respectively. These indicated that YARS2 deletion impaired mitochondrial proteostasis.
To further examine the effect of YARS2 mutation on the OXPHOS, we undertook the western blotting analysis to determine the levels in 15 subunits of OXPHOS complexes using lymphoblastoid cell line (I-1) bearing homozygous YARS2 p.191Gly>Val mutation and control cell line (A61) lacking the mutation (32). These subunits included four mtDNA-encoding polypeptides (ND6, CYTB, CO2, and ATP8), 13 nucleus encoding proteins: NDUFS1, NDUFS3, NDUFB8, NDUFA9, SDHA, SDHB, UQCRC1, UQCRC2, COX5A, COX16, and ATP5F1. As shown in Fig. S2, A and C, the various decreases in the levels of 11 mitochondrial proteins (but not of NDUFB8, SDHA, COX16, and ATP5F1) were observed in the mutant cell line carrying the p.191Gly>Val mutation, as compared with those in control line. As shown in Fig. S2B, the levels of ND6, CYTB, CO2, and ATP8 in the mutant cell line were 75%, 84%, 72%, and 89%, with an average of 80%, relative to the mean values measured in the control line. As shown in the Fig. S2D, the levels of NDUFS1, NDUFS3, NDUFB8, NDUFA9, SDHA, SDHB, UQCRC1, UQCRC2, COX5A, COX16, and ATP5F1 in the mutant cell line were 72%, 66%, 106%, 84%, 106%, 54%, 98%, 87%, 126%, 78%, and 131%, relative to the mean values measured in the control line, respectively. Notably, the average levels in the subunits of complexes I, II, III, IV, and V in the mutant cell line were 82%, 80%, 90%, 92%, and 110% of average values measured in the control line, respectively (Fig. S2E).
Deletion of YARS2 altered the stability and activity of OXPHOS complexes
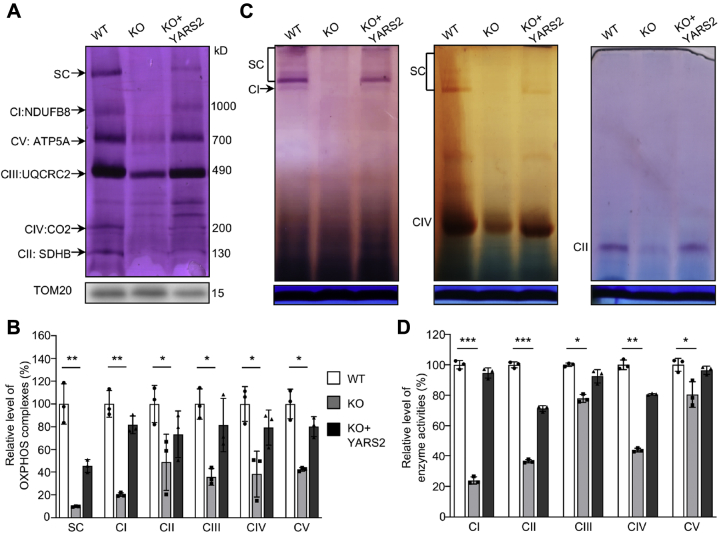
We analyzed the consequence of YARS2 deletion on the oxidative phosphorylation machinery. Mitochondrial membrane proteins isolated from YARS2KO and WT cell lines were separated by BN-PAGE, electroblotting, and hybridizing with human OXPHOS antibody cocktail consisting of NDUFB8, ATP5A, UQCRC2, CO2, and SDHB. As illustrated in Figure 3A, the YARS2KO cells exhibited the instability of intact supercomplexes, complexes I, II, III, IV, and V. As shown in Figure 3B, the levels of supercomplexes, complexes I, II, III, IV, and V in the YARS2KO cells were 9.8%, 20.2%, 48.7%, 35.7%, 38.4%, and 42.3% of those average values in WT cells, respectively. Furthermore, we carried out the in-gel activity assays to verify the YARS2 deletion–induced alterations in complexes I, II, and IV (35, 36). As shown in Figure 3C, the in-gel activities of complexes I, II, and IV were drastically decreased in the YARS2KO cells, as compared with the WT cells. We further measured the activities of OXPHOS complexes by the use of isolating mitochondria from mutant and control cell lines (36, 37, 38). The activity of complex I (NADH ubiquinone oxidoreductase) was determined through the oxidation of NADH with ubiquinone as the electron acceptor. The activity of complex II (succinate ubiquinone oxidoreductase) was examined the activity of complex II through the artificial electron acceptor DCPIP. The activity of complex III (ubiquinone cytochrome c oxidoreductase) was assessed through the reduction of cytochrome c (III) by using D-ubiquinol-2 as the electron donor. The activity of complex IV (cytochrome c oxidase) was monitored through the oxidation of cytochrome c (II). The activity of complex V (F1-ATP synthase) was explored through the NADH oxidation via conversion of phosphoenolpyruvate to lactate by two step reaction. As shown in Figure 3D, the average activities of complexes I, II, III, IV, and V in YARS2KO cells were 24%, 37%, 78%, 44%, and 80% of the mean value measured in WT cells, respectively. However, the overexpression of YARS2 cDNAs in the YARS2KO cells restored the defects in the activities of these complexes.
Figure 3.
Analysis of stability and activity of OXPHOS complexes.A, assembly and stability of OXPHOS complexes. Mitochondria extracted from various cell lines were solubilized with n-dodecyl-β-D-maltoside, electroblotted, and hybridized with antibody cocktail specific for subunits of five OXPHOS complexes and with TOM20 as a loading control. B, quantification of the levels of complexes I, II, III, IV, V and supercomplexes (SC) in mutant and wild-type cell lines. The calculations were based on three independent determinations. C, in-gel activity of respiratory chain complexes I, II, and IV. A total of 20 μg of mitochondrial proteins (8 g/g digitonin/protein ratio) from various cell lines was used for BN-PAGE, and the activities of complexes were measured in the presence of specific substrates (NADH and NTB for complex I, sodium succinate, phenazine methosulfate, and NTB for complex II, DAB and cytochrome c for complex IV). Coomassie staining was used as a loading control. D, enzymatic activities of respiratory chain complexes. The activities of respiratory complexes were investigated by enzymatic assay on complexes I, II, III, IV, and V in mitochondria isolated from various cell lines. The calculations were based on three independent experiments. Graph details and symbols are explained in the legend to Figure 2.
Deficient oxidative phosphorylation
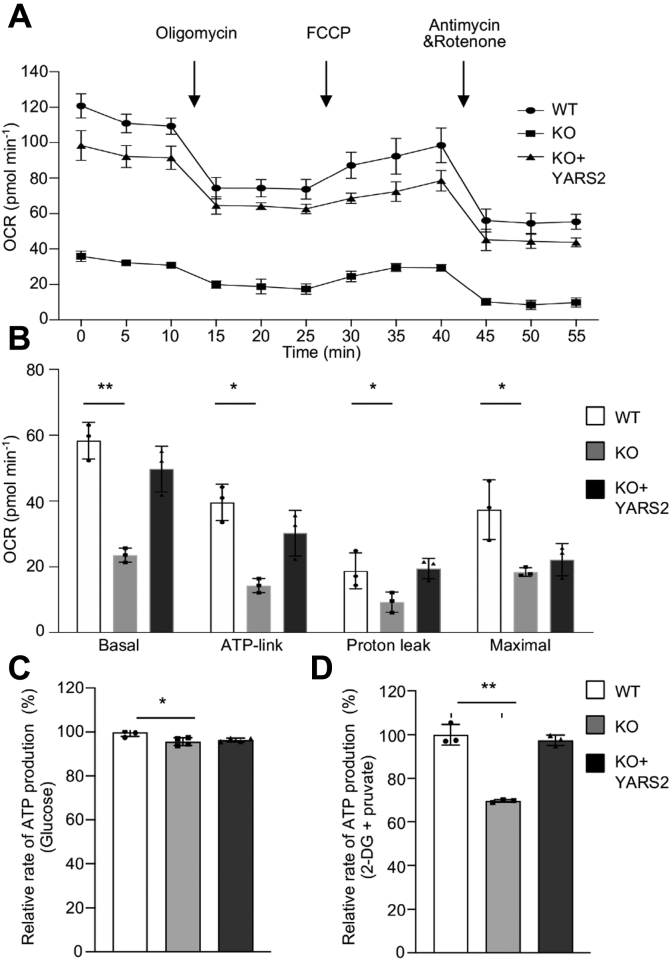
Oxygen consumption rate (OCR) is an indicator of mitochondrial respiration (39). Using the Seahorse Bioscience XF-96 Extracellular Flux Analyzer, we measured the OCRs of various cell lines, including basal respiration, O2 consumption attributing to ATP production, proton leak, maximum respiratory rate, reserve capacity, and nonmitochondrial respiration (39, 40). As shown in Figure 4A, the average basal OCRs in the YARS2KO cells were 40% (p < 0.001) of the mean values measured in WT cells. To further investigate which of the enzyme complexes of the respiratory chain was affected in the YARS2KO cells, OCR was monitored after the sequential addition of oligomycin (to inhibit the ATP synthase), FCCP (to uncouple the mitochondrial inner membrane and allow for maximum electron flux through the ETC), antimycin A (to inhibit complex III), and rotenone (to inhibit complex I). As shown in Figure 4B, the ATP linked OCR, proton leak OCR, and maximal OCR in YARS2KOcells were 36% (p < 0.01), 50% (p < 0.05), and 49% (p < 0.05) relative to the mean values measured in the WT cells, respectively. As expected, the overexpression of YARS2 cDNAs in the YARS2KO cells rescued the respiratory deficiency.
Figure 4.
Deficient oxidative phosphorylation.A, an analysis of O2 consumption in the various cell lines using different inhibitors. The rates of O2 (OCR) were first measured on 2 × 104 cells of each cell line under basal condition and then sequentially added to oligomycin (1.5 mM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) (0.5 mM), rotenone (1.0 mM), and antimycin A (1.0 mM) at indicated times to determine different parameters of mitochondrial functions. B, graphs presented the basal OCR, ATP-linked OCR, proton leak OCR, and maximal OCR in cell lines. Basal OCR was determined as OCR before oligomycin minus OCR after rotenone/antimycin A. ATP-lined OCR was determined as OCR before oligomycin minus OCR after oligomycin. Proton leak was determined as Basal OCR minus ATP-linked OCR. Maximal was determined as the OCR after FCCP minus nonmitochondrial OCR. C and D, measurement of whole-cell and mitochondrial ATP levels using bioluminescence assay. Cells were incubated with 10 mM glucose or 5 mM 2-deoxy-d-glucose plus 5 mM pyruvate to determine ATP generation under mitochondrial ATP synthesis. Average rates of ATP level per cell line and are shown: (C) ATP levels in total cells. D, ATP levels in mitochondria. Three independent experiments were made for each cell line. Graph details and symbols are explained in the legend to Figure 2.
We then used the luciferin/luciferase assay to examine the capacity of oxidative phosphorylation in mutant and wild-type cell lines. Populations of cells were incubated in the media in the presence of glucose and 2-deoxy-D-glucose with pyruvate (40). As shown in Figure 4C, the level of total ATP production in YARS2KO cells, with the presence of glucose, was comparable with those measured in WT cells. In contrast, the level of mitochondrial ATP production in YARS2KO cells, with the presence of 2-deoxy-D-glucose and pyruvate to inhibit the glycolysis, was 70% relative to the mean value measured in WT cells (Fig. 4D).
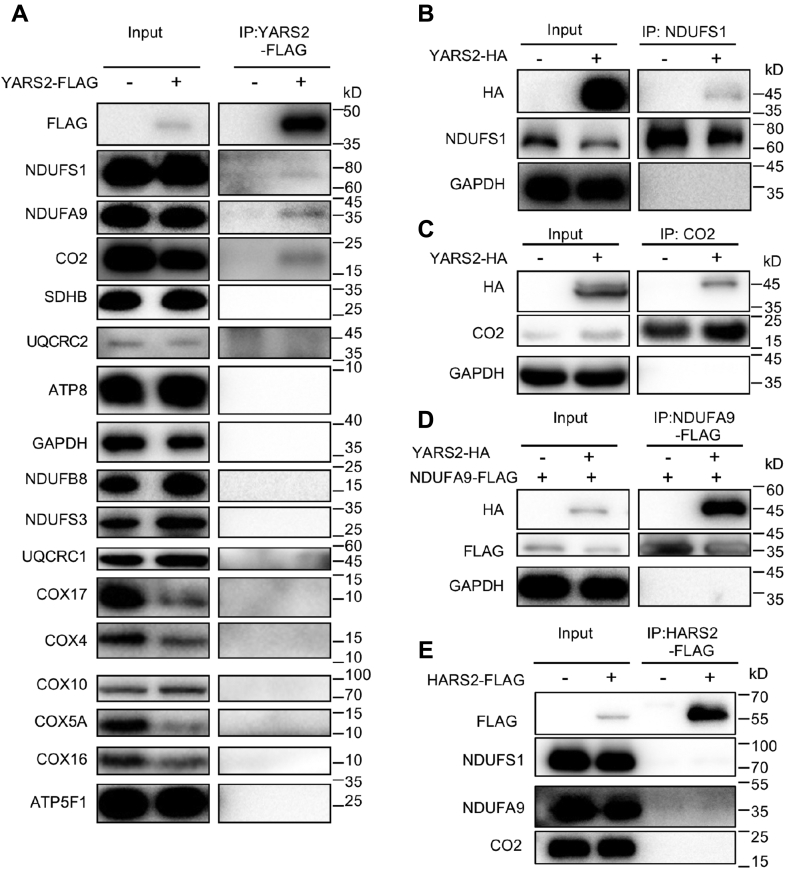
YARS2 may interact with NDUFS1, NDUFA9 of complex I, and CO2 of complex IV
To test whether YARS2 interacted with the OXPHOS complex, we performed the immunoprecipitation assay using FLAG antibodies in mitochondria of HeLa cell lines overexpressed with human FLAG-tagged YARS2. As shown in Figure 5A, the YARS2-FLAG antibody reciprocally immunoprecipitated to NDUFS1, NDUFA9 of CI subunits, and CO2 of CIV subunit, but did not bind to NDUFB8 and NDUFS3 of CI subunits, SDHB of CII subunit, UQCRC1 and UQCRC2 of CIII subunits, COX4, COX5A, COX10, COX16, and COX17 of CIV subunits, ATP8 and ATP5F of CV subunits, respectively. These data indicated the potential interactions of YARS2 with complex I and complex IV by binding to NDUFS1, NDUFA9, and CO2, respectively. These interactions of YARS2 with these three subunits were verified by the fact that antibodies against NDUFS1, FLAG-NDUFA9, and CO2 co-immunoprecipitated with the HA-tagged YARS2. However, the CO-IP signal of NDUFS1 with YARS2 was much weaker than those of CO2, suggesting that the interaction of YARS2 with NDUFS1 may be very weak and unstable. Furthermore, Flag-tagged mitochondrial histidyl-tRNA synthetase (HARS2) was overexpressed in HeLa cells for immunoprecipitation as a negative control. As shown Figure 5E, FLAG-HARS2 did not co-immunoprecipitate NDUFS1, NDUFA9, or CO2, indicating no interaction of HARS2 to these subunits. Moreover, immunofluorescence staining experiments showed that a carboxy terminus FLAG or HA-tagged YARS2 displayed the overlap with CO2, NDUFS1, NDUFA9, and NDUFS3, respectively, with a significant correlation (Pearson’s coefficient of 0.59, 0.71, 0.73, and 0.45, respectively) (Fig. S3). These data suggested the potential direct interaction of YARS2 with NDUFS1, NDUFA9, and CO2, respectively.
Figure 5.
Immunoprecipitation analysis of YARS2 with subunits of OXPHOS complexes. A, co-immunoprecipitation of YARS2-FLAG and subunits of OXPHOS. HeLa cells transiently expressing with or without YARS2-FLAG were solubilized with a lysis buffer and lysate proteins were immunoprecipitated with immunocapture buffer (left) (input) and FLAG-antibody (right) (IP), respectively. Immunoprecipitates were analyzed by SDS-PAGE and western blotting using anti-FLAG, anti-subunits of OXPHOS (NDUFS1, NDUFS3, NDUFA9 and NDUFB8 for complex I, SDHB for complex II, UQCRC1 and UQCRC2 for complex III, CO2, COX4, COX5A, COX10, COX16, and COX17 for complex IV, ATP8 and ATP5F1 for complex V) and GAPDH antibodies, respectively. B and C, co-immunoprecipitation of YARS2-HA and NDUFS1, CO2 using anti-HA, anti-NDUFS1, CO2, and GAPDH antibodies, respectively. D, co-immunoprecipitation of YARS2-HA and NDUFS9A-FLAG using anti-HA, anti-FLAG, and GAPDH antibodies, respectively. E, co-immunoprecipitation of HARS2-FLAG and NDUFS1, NDUFA9, CO2 in lysates of HeLa cells transfected with plasmids containing HARS2 using anti-FLAG, anti-NDUFS1, NDUFA9, and CO2 antibodies respectively.
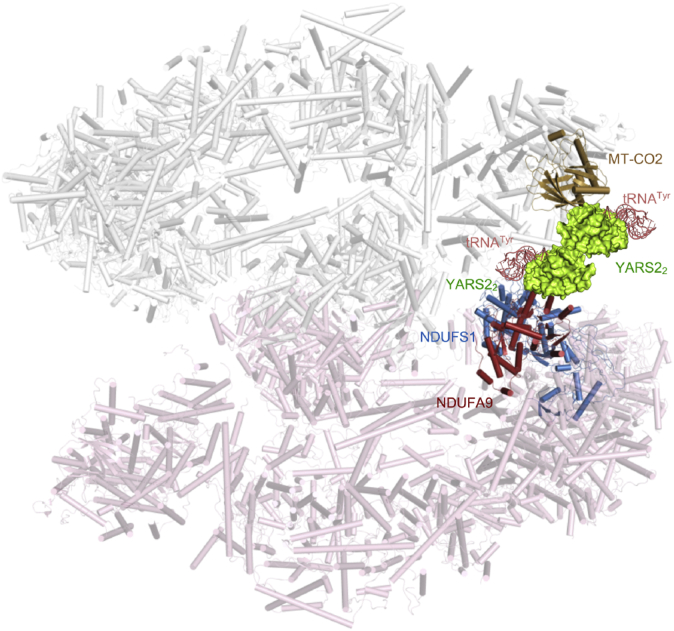
The overall structure of supercomplexes (MCI2III2IV2, PDB: 5XTI) showed that the NDUFS1, interacting with NDUFA9, were the components of matrix arm of CI (41). As shown in Figure 6, there was a pocket formed by CO2 of CIV together with NDUFS1 and NDUFA9 of membrane arm of CI in the megacomplexes I2III2IV2. This pocket may provide an interspace for the interactions of YARS2 with complex I and complex IV. As YARS2 functioned usually as a homodimer (PDB: 2PID) (42), we proposed that the YARS2 homodimer with tRNATyr complex resided at the pocket as a component of supercomplexes by interacting with CI and CIV. Therefore, YARS2 may play a critical role in the formation of the megacomplex in mitochondrial respiratory chain.
Figure 6.
The proposed model of YARS2 interacting with the OXPHOS supercomplex. The partial OXPHOS supercomplex was derived from (MCI2III2IV2, PDB: 5XTI) (41). The structure of Thermus thermophilus tryosyl-tRNA synthetase and tRNATyr complex (PDB ID: 1H3E) was used as an original templet for construction of the model of humanYARS2-tRNATyr complex. The structure of YARS2 was extracted from the crystal structures of human mitochondrial tyrosyl-tRNA synthetase (PDB ID: 2PID) (42). The model of human YARS2-tRNATyr complex was generated using PyMOL molecular visualization system (PyMOL Molecular Graphics System, Version 2.3.3, Schrödinger, LLC). A model of YARS2 interacting with the OXPHOS supercomplex was proposed as the YARS2 homodimer–tRNATyr complex interacts with CO2 of CIV, NDUFS1 and NDUFA9 of membrane arm of CI in the megacomplexes I2III2IV2, respectively.
Generation of yars2 knockout zebrafish using CRISPR/Cas9 system
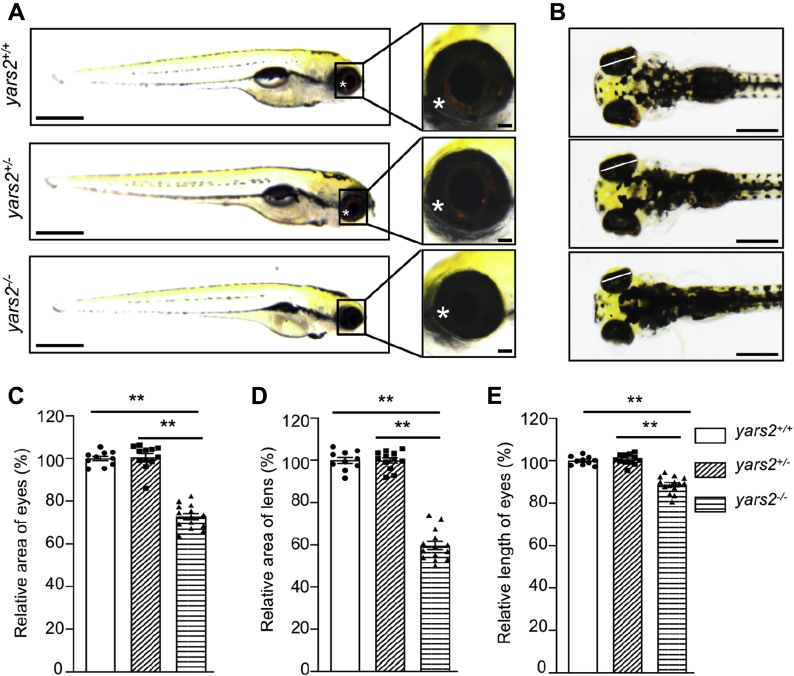
To investigate the pathological impact of yars2 deficiency in vivo, we used the CRISPR/Cas9 technology to produce zebrafish mutant lines as described elsewhere (43, 44). A single-guide RNA (sgRNA) targeting exon 1 of yars2 was coinjected with Cas9 mRNA into wild-type one-cell stage embryos (Fig. S4). As a consequence, a 5 bp deletion at positions (c.117 T to c.121 G) in the exon 1 was produced (heterozygous and homozygous zebrafish were described as yars2+/−, yars2−/−, and wild type as yars2+/+). In fact, this deletion introduced a premature stop at codon 42 (p.L42∗) and subsequently propagated after the confirmation of the mutation by Sanger sequencing and western blot analysis (Fig. S4). The yars2−/− zebrafish were larval lethal before 10 dpf, while yars2+/− zebrafish were adult-viable. As shown in Figure 7, A and B, marked reductions of black-stained organ in the abdomen were observed in the yars2−/− larva at 5 dpf, as compared with those in the WT and yars2+/− larva. However, we did not see significant changes of the body lengths in yars2−/− and yars2+/− larval at 5 dpf (Fig. 7A, Fig. S5). As an assay of function, we also analyzed heart beat rates in 32 s at yars2−/−, yars2+/−, and wild-type larval at 5 dpf. As shown in Fig. S5B, there were no significant differences in the heart beat rates between yars2+/+and yars2−/− or yars2+/− larval.
Figure 7.
Deletion of yars2 caused the eye defects in zebrafish.A, the lateral views of yars2−/−, yars2+/−, and yars2+/+ zebrafish at 5 dpf, and the eye morphologies of zebrafish were illustrated under a Leica microscope with an objective magnification of 20×. Asterisk indicated the eye pigments. B, the dorsal views of the yars2+/+, yars2+/−, and yars2−/− zebrafish at 5 dpf. C–E, quantification of the area and length of eyes as well as area of lens of yars2+/+ (n = 10), yars2+/− (n = 12), and yars2−/− (n = 14) zebrafish, as detailed elsewhere (47). The values for the mutants were expressed as percentages of the average values for the wild type. Graph details and symbols are explained in the legend to Figure 2.
Loss of yars2 caused the retinal defects in zebrafish at 5 dpf
Zebrafish is visually responsive by 72 h post fertilization and has functional retina at 5 dpf (45). The retina of zebrafish is composed of five layers [outer nuclear layer (ONL), outer plexiform layer (OPL), inner nuclear layer (INL), inner plexiform layer (IPL), and ganglion cell layer (GCL)] (46). As shown in Figure 7A, yaars2−/− larvae at 5 dpf exhibited faint pigments, as compared with those in WT larvae. We then evaluated the sizes of eyes and lens in the zebrafish at 5 dpf (46). As shown in Figure 7, C, D and E, yaars2−/− larvae revealed significant decreases in the average areas (72.8%) of eyes, average areas (59.7%) of lens, and average lengths (88.7%) of eyes, as compared with those in the yars2+/+ larvae, respectively. However, there were no differences of those parameters between yars2 +/− and yars2+/+ larvae.
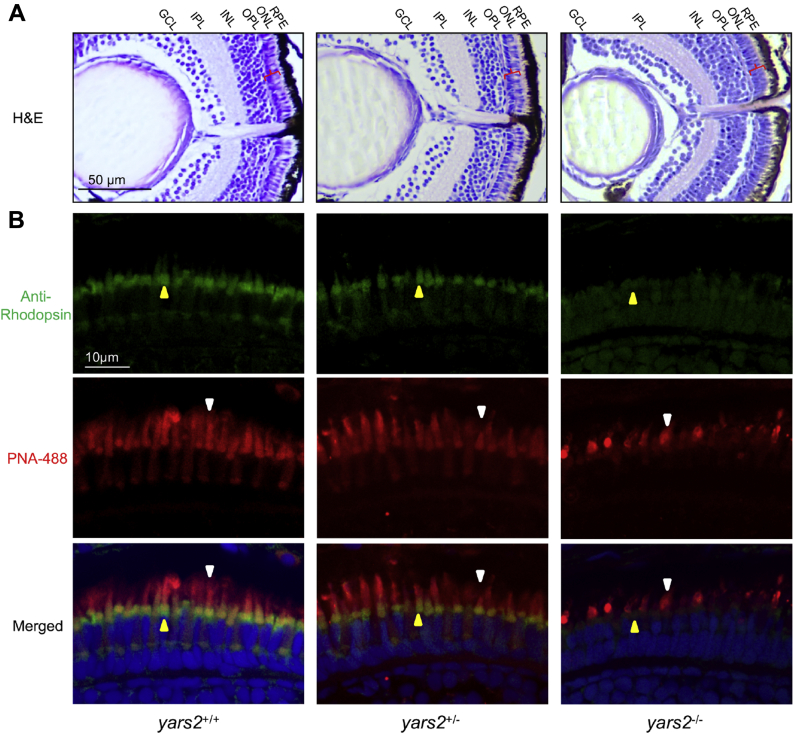
We then assessed whether the loss of yars2 altered the thickness of layers in zebrafish retina using hematoxylin and eosin (H&E)-stained retinal sections from larvae at 5 dpf. As shown in Figure 8A, the ablation of yars2 did not change the thickness in layers of pigmented epithelium (RPE), INL, IPL, and GCL, whereas the ONLs (containing the rod and cone granules: photoreceptor cells) in yars2−/− larvae were reduced 20%, as compared with those in the yars2+/+ larvae.
Figure 8.
Retina defects in zebrafish. A, hematoxylin and eosin (HE) staining of retinas in the yars2+/+, yars2+/−, and yars2−/− zebrafish at 5 dpf. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium. The angle brackets colored in red denoted the ONL. Scale bar, 50 μm. Shorter outer segments in the cones and rods were indicated. B, cones and rods in the retinal cross sections of yars2+/+, yars2+/−, and yars2−/− zebrafish at 5 dpf, stained with lectin PNA-488 (green, yellow arrows) and 1D4/Rhodopsin (red, white arrows) and DAPI (blue) staining for nuclei. Scale bar, 10 μm.
We further examined the morphology of cones and rods by immunolabeling with their specific markers, peanut agglutinin lectin (PNA-488) for cone and rhodopsin (prototypical G-protein-coupled receptor) for rods, respectively. As shown in Figure 8B, the outer segments of cones (staining with PNA-488) in the yars2−/− larvae were much shorter than those in the yars2+/+ larvae (47). Moreover, the outer segments of rods (staining with rhodopsin) in the yars2−/− larvae were much less than those in yars2+/+ larvae, indicating that the loss of yars2 may affect the cone and rod developments (47). Furthermore, drastic changes of the layer below the rods were observed in the yars2−/− larval, as compared with those in the yars2+/+ larval. However, there were no significant differences of morphology of cones and rods between yars2+/− and yars2+/+ larvae. These data implied that the yars2 deficiency led to the defects of retina, especially for photoreceptor degeneration.
Mitochondrial defects in retina
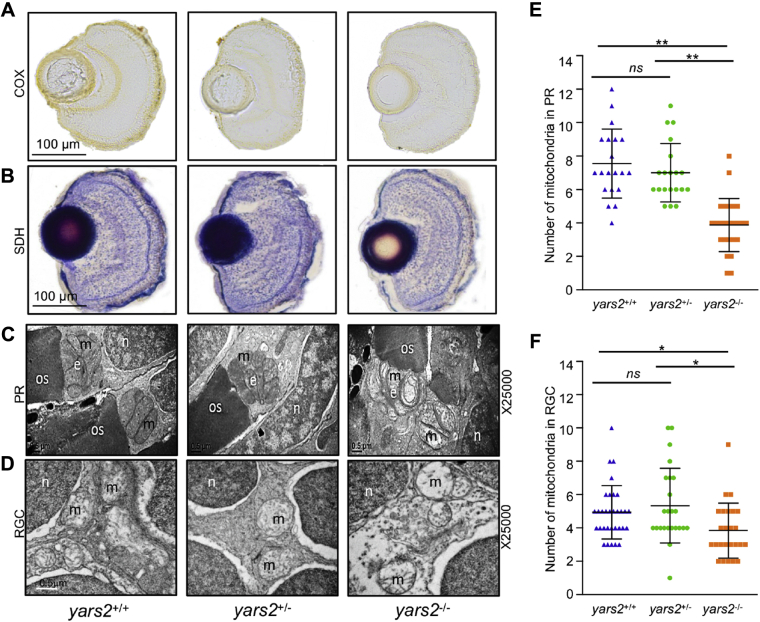
Mitochondrial dysfunctions in retina were assessed by the enzyme histochemistry (EHC) staining for SDH and COX in the frozen sections of retina in the yars2−/−, yars2+/−, and yars2+/+ larvae at 5 dpf. As shown in Figure 9, A and B, markedly reduced staining of COX but mildly decreased staining of SDH were observed in yars2−/−, as compared with those in the yars2+/+ larvae. However, the activities of COX and SDH in yars2−/− were much lower than those in yars2+/−. These results indicated that the loss of yars2 caused the pronounced deficiency of complex IV.
Figure 9.
Mitochondrial defects in the zebrafish retina.A and B, assessment of mitochondrial function in the retina by enzyme histochemistry (EHC) staining for COX (A) and SDH (B) in the frozen sections of retina in the yars2+/+, yars2+/−, and yars2−/− larvae at 5 dpf. Scale bar, 100 μm. C and D, mitochondrial morphology from photoreceptors (PR) (C) and RGC (D) of transmission electron microscopy. Ultrathin sections were visualized with 25,000× magnification. Scale bar, 1 μm. e, ellipsoid; m, mitochondrion; n, nucleus; os, outer segment of photoreceptors. E and F, quantification of mitochondrial numbers of photoreceptors (E) from yars2+/+ (n = 20), yars2+/− (n = 20), and yars2−/− (n = 25) and RGC (F) from yars2+/+ (n = 32), yars2+/− (n = 24), and yars2−/− (n = 25) zebrafish at 5dpf. Graph details and symbols are explained in the legend to Figure 2.
Mitochondrial defects in the retina of larvae at 5 dpf were further evaluated by using transmission electron microscope. Mitochondria were usually confined solely to the ellipsoid region of photoreceptors in the zebrafish (48). As shown in Figure 9, C and D, the photoreceptor cells and retinal ganglion cells (RGC) in the yars2−/− zebrafish displayed less numbers and abnormal morphology of mitochondria, including enlarged mitochondria and the partial loss of cristae, as compared with those in the yars2+/+ larvae. As shown in Figure 9, C and E, mitochondrial numbers of photoreceptor cells in the yars2−/− (n = 25) and yars2+/− (n = 20) mutant larvae were 52% and 92%, related to the mean values of yars2+/+ larvae (n = 20), respectively. As shown in Figure 9, D and F, mitochondrial numbers of RGC cells in the yars2−/− (n = 25) and yars2+/− (n = 24) zebrafish larvae were 77% and 108%, related to the average values of those in yars2+/+ larvae (n = 32), respectively.
Discussion
The impacts of YARS2 on the mitochondrial translation and the stability of OXPHOS
YARS2, like other human mitochondrial tRNA synthetases, has a central role in the aminoacylation of tRNATyr and mitochondrial translation. In fact, human cytoplasmic aminoacyl-tRNA synthetases have a number of noncanonical functions, including transcriptional regulation, inflammation, angiogenesis, apoptosis, and mitochondrial bioenergetics (11, 12, 13, 49, 50). These aminoacyl-tRNA synthetases were present in a free form or part of a high-molecular-weight multisynthetase complex and three scaffold proteins (11, 51). However, the nontranslational functions of human mitochondrial aminoacyl-tRNA synthetases remain poorly understood. In the present investigation, we demonstrated that the YARS2 deficiency caused the pleiotropic effects in the HeLa mutant cells: impairment of mitochondrial translation and instability of OXPHOS complexes. As expected, the primary defects in the YARS2 deficiencies were the aberrant aminoacylation of tRNATyr and impaired synthesis of mtDNA-encoding 13 OXPHOS subunits (24, 29, 31, 32). The reduced levels among 13 mtDNA-encoding OXPHOS subunits in the cells bearing the YARS2 mutations or deletions were correlated with the tyrosine codon usages of polypeptides, especially marked reductions in the ND1, ND4, ND5, ND6, CO1, and CO2 with high contents of tyrosine codon (24, 29, 31, 32). Strikingly, YARS2 mutant cells displayed various reductions in nucleus-encoding OXPHOS subunits, NDUFS1, NDUFS3, NDUFA9, NDUFB8, SDHB, UQCRC1, UQCRC2, COX5A, and COX16. The mitochondrial translational defects may result in the imbalances between the increased levels of de novo protein synthesis and decreased folding capacity for the mtDNA- and nucleus-encoded mitochondrial proteins (52, 53). It was worthwhile to note that YARS2 deficiency impaired mitochondrial proteostasis, evidenced by the decreased levels of Clpp and LONP1 in the YARS2KO cells. Furthermore, the ablation of YARS2 could affect the association of YARS2 and mRNA and regulate the translation of nucleus-encoding OXPHOS subunits (54). Alternatively, YARS2 may interact with NDUFS1, NDUFA9 of complex I and CO2 of complex IV to provide further stabilization of OXPHOS complexes (41, 55, 56). Notably, WARS2 deficiency abolished its interaction with COX11 and led to noncanonical effects on mitochondrial function (57). These deficiencies gave rise to the instability of complexes I, II, III, IV, and V as well as intact supercomplexes observed in YARS2−/− cell lines. In fact, each OXPHOS complex is a multisubunit machine integrated into the mitochondrial inner membrane, comprising mtDNA-encoded subunit(s) and nuclear-encoded subunits, except complex II (56). These mtDNA-encoded subunits appear to act as seeds for building new complexes, which requires nuclear-encoded subunit import and assembly with the assistance of assembly factors (55, 56). As a consequence, these defects yielded the reduced activities of these respiratory chain enzyme complexes. Strikingly, the variations in the average lower concentrations of OXPHOS (complexes I, II, III, IV, and V) proteins in the YARS2KO line showed a significant correlation with the variations in the reduced activities in the respiratory chain complexes (r = 0.98, p = 0.0034) (Fig. S6). Therefore, our data demonstrated that YARS2 has profound impacts on the stability of OXPHOS complexes.
The critical role of YARS2 in the vision function
The retina is a part of the central nervous system and shares the characteristically high metabolism of the brain (58, 59). In particular, the photoreceptor cells and RGCs are the biggest ATP demand cells in the retina (58, 60). In fact, aberrant mitochondrial tRNA metabolism, caused by mutations in mitochondrial tRNA or mitochondrial tRNA synthetases, is associated with visual impairment as sole phenotype or associated with other clinical presentations (31, 61, 62, 63, 64, 65). Notably, defects in mt-aaRSs resulted in the respiratory deficiency, deficient ATP synthesis, and subsequent failure of cellular energetic process (14, 15). In fact, inhibition of wars2 resulted in trunk vessel deficiencies, disordered endocardial–myocardial contact, and impaired heart function in the zebrafish (57). In this study, it was hypothesized that the mitochondrial dysfunctions caused by YARS2 deficiency particularly ablated the function of retina in zebrafish. These mitochondrial dysfunctions were evidenced by pronounced reduced activities of COX in the retina, abnormal morphology, and reduced numbers of mitochondria in the photoreceptor cells and RGCs of yars2−/− mutant zebrafish. Indeed, zebrafish at 5 dpf have functional retina, composed of five layers (ONL, OPL, INL, IPL, and GCL) (45, 46). These mitochondrial dysfunctions caused by the deletion of yars2 led to the pathological consequence of vision function. At 5 dpf, yars2−/− mutant larvae exhibited significant decreases in the average area and length of eyes, indicating that the loss of Yars2 altered the embryonic eye development. The vision deficiencies were further evidenced by the fact that the yars2−/− mutant zebrafish exhibited the reduced thickness of ONL containing the rod and cone granules: photoreceptor cells. Furthermore, yars2−/− larvae displayed shortened outer segments of cones and rods, indicating that the loss of yars2 affected the development and functions of cones and rods. Therefore, we demonstrated that the yars2 deficiency led to the retinal defects, as in the case of tissue-specific effects in the DARS2 and WARS2 mutations (52, 53).
In summary, our findings demonstrated that YARS2 has the critical roles in the OXPHOS stability and vision functions. Aside from its central function in the mitochondrial protein synthesis, YARS2 may regulate the mitochondrial proteostasis and activity of OXPHOS complexes via its potential interacting with NDUFS1, NDUFA9, and CO2. The mitochondrial dysfunctions induced by the loss of Yars2 led to the alterations in the eye development and retina defects in the zebrafish. Our findings highlight the critical roles of mitochondrial aminoacyl-tRNA synthetase in the stability and activity of OXPHOS complex and their pathological consequences in visual impairment.
Experimental procedures
Generation of YARS2KO HeLa cell line and culture condition
The HeLa cell lines were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies) (containing 4.5 mg of glucose, 0.11 mg pyruvate/ml, and 50 μg of uridine/ml), supplemented with 5% FBS. Lymphoblastoid cell lines (I-1) bearing the homozygous YARS2 p.191Gly>Val mutation and control (A61) lacking these mutations were grown in the RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (32).
YARS2 KO HeLa cell lines were generated by utilizing the Lenti CRISPR 458 plasmid (Addgene) containing the sgRNAs (66). sgRNAs were designed using the CRISPR design tool (http://crispr.mit.edu) to minimize potential off-target effects. The sequences of sgRNAs that ultimately produced successful deletion clones were 5’CAGGAAAAGGACCGCAAGAT3’. HeLa cells were transfected with Lenti CRISPR 458 plasmid (Addgene) containing the sgRNAs using jetPRIME (Polyplus-transfection SA, Illkirch, France), according to the manufacturer’s instructions. After 24 h, the cell cultures in the same media were treated with 1 μg/ml of puromycin for 3 days. The cells were then collected and plated in DMEM (containing 4.5 mg of glucose, 0.11 mg pyruvate/ml, and 50 μg of uridine/ml), supplemented with 10% FBS. Subsequently, cells were cloned by limiting dilution, and individual clones were isolated.
Fragments spanning five exons and flanking sequences in each clone were PCR amplified, purified, and subsequently analyzed by Sanger sequencing, as detailed previously (31). These sequence results were compared with the YARS genomic sequence (RefSeq NC_000012.12). The genotyping for the YARS214bpdel mutation in each clones was PCR amplified for 354 bp fragment spanning partial promoter region and exon 1 and followed by Sanger sequence analysis and subsequently RFLP analysis by the digestion of BstNI. In fact, the YARS214bpdel mutation abolished the site of BstNI in this fragment. The forward and reverse primers for this genotyping analysis were 5’-ACCTTCCCTAGGAGCTGTAAGTAG-3’ and 5’AGATGACCCACATGAAGCGAGTC-3’, respectively.
For the rescuing of YARS2KO cells, the full-length coding region of YARS2 cDNA was obtained by RT-PCR amplification using the high-fidelity Pfu DNA polymerase (Promega) and total RNA isolated from HeLa cells as template, with primers with EcoRI site: 5’-GGAATTCCATGGCGGCGCCCAT-3’(nt.24–38) and BamHI site: CGGGATCCCGTCACAACTGAAGCCATTTTATAA-3’(nt.1435–1457) (GenBank accession no. NM_001040436.3) (67). The PCR products were cloned by using the TA Cloning Kit (TAKARA) and analyzed by Sanger sequencing and then subcloned into a pCDH-puro-cMyc Vector (Addgene plasmid 46970). The YARS2KO HeLa cell lines were transfected with pCDH-puro-cMyc Vector with YARS2 cDNA, pMD2.G (Addgene), and psPAX2 (Addgene) plasmids in DMEM (10569-010) using Lipofectamine 3000 (Life Technologies) (68, 69).
Mitochondrial tRNA analysis
Total RNAs were obtained from HeLa cell lines (∼1.0 × 108 cells), as described previously (70). For tRNA aminoacylation analysis, 10 μg of total RNAs was electrophoresed at 4 °C through an acid (pH 5.2) 10% polyacrylamide/8 M urea gel to separate the charged and uncharged tRNA as detailed elsewhere (71, 72, 73). The gels were electroblotted onto a positively charged nylon membrane (Roche) for the hybridization analysis with oligodeoxynucleotide probes specific tRNATyr, tRNALeu(UUR), and tRNAThr, as detailed elsewhere (71, 72, 73).
Experimental fish and maintenance
AB wild-type strain and myocardium-specific transgenic Tg (cmlc2: egfp) zebrafish (Danio rerio) were used for this investigation. The animal protocols used in this investigation were approved by the Zhejiang University Institutional Animal Care and Use Committee. All fishes were kept in recirculating water at 28 °C and fed with commercial pellets at a daily ration of 0.7% of their body weight. Embryos were reared at 28.5 °C according to standard protocols (74). Embryos were staged by hours post fertilization (hpf) and days post fertilization (dpf) (75).
Yars2-knockout zebrafish line generated by CRISPR/Cas9 system
The zCas9 expression plasmid pSP6-2sNLS-spCas9 was linearized by Xba I and used as a template for Cas9 mRNA in vitro synthesis with mMESSAGE mMACHINE mRNA transcription synthesis kits (Ambion). The sequence of sgRNAs was designed according to criteria as described previously (66, 76). The gRNA transcription plasmid was pT7-gRNA. We used the CRISPR/Cas9 design tool (http://zifit.partners.org) to select specific targets to minimize off-target effects. The sequences of sgRNAs that ultimately produced successful deletion clones were: 5’GGACTCTTTCCCGGAGGTCGCGG3’ (GenBank accession no. NM_001143920.1). Cas9-encoding mRNA (300 ng/μl) and gRNA (200 ng/μl) were coinjected into one-cell-stage wild-type embryos. Injected embryos were incubated at 28.5 °C and collected for making genomic DNA for genotyping at 50 hpf. Genomic DNA of the 50 hpf injected embryo was used as template to amply DNA segments carrying the partial exo1 sequence in the yars2 carrying the 5 bp deletion at position 117 by using the following two primers: 5’-:AAACATCCGCCAAACCTCCC-3’ (forward) and 5’-AGGAGACCCCGGTTATGGAG-3’ (reverse) (GenBank accession no. NM_001143920.1). The genotyping for the yars25bpdel mutation in each fish was PCR amplified for 170 bp fragment in the partial exon 1 and followed by Sanger sequence analysis and subsequently PAGE analysis. The forward and reverse primers for this genotyping analysis were described as above.
Western blotting analysis
Western blot analysis was performed on samples from the human cell lines and zebrafish larvae at 5 dpf, as detailed elsewhere (31, 32, 43). Fishes were sacrificed after anesthesia and homogenized in RIPA reagent (Invitrogen) using a homogenizer. In total, 20 μg of proteins was electrophoresed through 10% bis-Tris SDS-polyacrylamide gels and then transferred to a polyvinyl difluoride membrane. The antibodies used for this investigation were from Abcam [YARS2 (ab127542), ND6 (ab81212), NDUFB8 (ab110242), TOM20 (ab56783), SDHB(ab14714), UQCRC2 (ab14745), total human OXPHOS antibody cocktail (ab110411)], Proteintech [NDUFS1 (12444-1-AP), NDUFA9 (20312-1-AP), NDUFS3 (15066-1-AP), SDHA (14865-1-AP), CYTB (55090-1AP), UQCRC1 (21705-1-AP), CO2 (55070-1-AP), COX17 (11464-1-AP), COX16 (19425-1-AP), COX10 (10611-2-AP), COX5A (11448-1-AP), ATP8 (26723-1-AP), ATP5C (10910-1-AP), ATP5F1 (15999-1-AP) and Clpp (15698-1-AP)], LONP1 (15440-1-AP) and COX4 (diagbio, db15), GAPDH (Hangzhou Goodhere Biotechnology Co, Ltd, AB-M-M 001), Anti-HA antibody (Rakland, 600-401-334), and Flag-tagged monoclonal antibody (GNI, GNI4110)]. Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Jackson) were used as a secondary antibody. Quantification of density in each band was performed as detailed previously (31).
Immunoprecipitation analysis
The pcDNA3.1-FLAG and pcDNA3.1-HA constructs were gifts from Dr E. Wang (Chinese Academy of Sciences). The human YARS2 cDNAs were cloned into the pcDNA3.1-FLAG and pcDNA3.1-HA vector. The human NDUFA9 cDNA was amplified with primers: 5’- GG AATTCCATGGCGGCTGCCGCA-3’ (forward) and 5’- GGCGATATCGCCAAATG TTGACGGTCTTGGCCG-3’ (reverse) and cloned into the pcDNA3.1-FLAG vector. HeLa cells were transfected with constructs using jetPRIME (Polyplus -transfection SA). After transfection for 24 h, the cells were collected, washed by phosphate-buffered saline (PBS), and lysed with 1 ml of ice-cold lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% (w/v) sodium deoxycholate, 1%(v/v) NP-40] supplemented with a protease inhibitor cocktail(B14001, Selleck) for 15 min on ice. The whole-cell lysates were collected by centrifugation at 12,000g for 30 min at 4 °C and incubated with anti-FLAG tag monoclonal antibody (GNI4110, GNI) or CO2, or NDUFS1 overnight at 4 °C. The Protein A-Agarose beads (Roche) were washed four times with 1 ml of cold PBS buffer (pH 7.4). The beads-bound proteins were eluted and denatured with an SDS loading buffer and subjected to SDS-PAGE and western blot analyses (77, 78).
Blue native electrophoresis analysis
Blue native gel electrophoresis was performed by isolating mitochondrial proteins from various cell lines, as detailed previously (35, 36). Samples containing 30 μg of mitochondrial proteins were separated on 3 to 11% Bis-Tris Native PAGE gel. The primary antibodies applied for this experiment were total human OXPHOS antibody cocktail and TOM20 as a loading control. Alkaline phosphatase labeled goat anti-mouse IgG and goat anti-rabbit IgG (Beyotime) were used as secondary antibodies and protein signals were detected using BCIP/NBT alkaline phosphatase color development kit (Beyotime).
Assays of activities of OXPHOS complexes
For evaluating the activity of complexes I, II, and IV in gel, 30 μg of freshly prepared mitochondrial proteins from various cell lines was loaded on 3 to 11% Bis-Tris Native PAGE gel, run at 150 V in dark blue cathode buffer for 1 h, and then at 250 V in light cathode buffer for 2 h. The native gels were prewashed in cold water and then incubated with the substrate of with the fresh substrates of complex I [0.1 mg/ml NADH, 2.5 mg/ml Nitrotetrazolium Blue chloride (NTB), 2 mM Tris-HCl, pH 7.4], complex II (20 mM sodium succinate, 2.5 mg/ml NTB, 0.2 mM phenazine methosulfate, 5 mM Tris-HCl, pH 7.4), and complex IV [0.5 mg/ml diaminobenzidine (DAB), 1 mg/ml cytochrome c, 45 mM phosphate buffer, pH 7.4] for 2 h at room temperature, respectively. The gel was washed with 50% methanolthree times (36, 37, 38).
EHC staining for SDH and COX in the frozen sections from yars2+/−, yars2−/−, and WT zebrafish larvae at 5 dpf was performed as detailed elsewhere (44, 78). Briefly, freshly dissected retina tissues were embedded in OCT compound (Tissue-Tek), frozen on dry ice, and sectioned to 8 μm. For SDH staining, samples were incubated in 0.1 M phosphate buffer, pH 7.6, containing 5 mM EDTA, 1 mM potassium cyanide, 0.2 mM phenazine methosulfate, 50 mM succinic acid, 1.5 mM nitro blue tetrazolium at 37 °C for 25 min. For COX staining, samples were incubated in 5 mM phosphate buffer, pH 7.6, containing 5 mM EDTA, 1 mM potassium cyanide, 0.2 mM phenazine methosulfate, 50 mM succinic acid, 1.5 mM nitro blue tetrazolium at 37 °C for 60 min.
Hematoxylin and eosin (H&E) staining
For H&E staining, zebrafish larvae at 5 dpf were anesthetized and fixed in 4% paraformaldehyde with 0.1 M phosphate buffer (PFA) for 24 h at room temperature. Retinal sections were then dehydrated, infiltrated, embedded in paraffin, sliced 3 μm thick by pathologic microtome (RM2016, Leica), and finally stained with H&E (Thermo) to observe tissue architectures (78, 79, 80).
Immunohistochemistry
Zebrafish larvae at 5 dpf were fixed with 4% PFA and permeabilized with 0.01% Triton X-100. Retinal sections were cryoprotected in 8 μM and dried onto frost-free slides at 37 °C overnight, and next blocked (1% BSA, 5% normal goat serum, 0.2% Triton-X-100, 0.1% Tween-20 in 1X PBS) in a humidified chamber for 2 h. Cones and rods OS were labeled respectively with anti-1D4/Rhodopsin (1:250, Abcam ab5417) and anti-lectin PNA-488 (1:2000, Molecular Probes L21409) antibodies overnight at 4 °C.
Transmission electron microscopy
The ultrastructure of zebrafish retina at 5 dpf was observed using transmission electron microscopy (47). Retina tissues were fixed in 2.5% glutaraldehyde, embedded in Epon 812, and cut into 100-nm-thick slices using UC7 ultramicrotome (Leica), then stained with 2% uranyl acetate and alkaline lead citrate. Finally, the images of retina ultrastructure were captured by a Hitachi-7650 transmission electron microscope (Hitachi).
Statistical analysis
All statistical analysis was performed using the unpaired, two-tailed Student’s t-test contained in the Graphpad prism 8 program (Graphpad software) and Microsoft-Excel program (version 2016). A p value <0.05 was considered statistically significant.
Data availability
Representative experiments are shown in the figures and supplemental materials. For any additional information, please contact the corresponding author.
Supporting information
This article contains supporting information.
Conflict of interest
All the authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
M. -X. G. and P. J. designed the experiments, monitored the project progression, data analysis, and interpretation. X. J., Z. N., C. W., F. M., and Q. Y. contributed to the experiments and contributed to data analysis Figure 1, Figure 2, Figure 3, Figure 4, Figure 5. Z. Z., M. C., J. S., and J. Z. performed the experiments and contributed to data analysis in Figure 6, Figure 7, Figure 8. P. J. and M.-X. G. acquired funds. P. J. prepared the initial draft of the article. M.-X. G. made the final version of the article. All the authors reviewed the article.
Funding and additional information
This work was supported by a grant from the National Key Research and Development Program of China, China (2018YFC1004802 to M.-X. G.), grants from National Natural Science Foundation of China, China (31970557 to M.-X. G., 31671303 to P. J.) and Zhejiang Provincial Medical and Health Research Program, China (2019RC149 to X. J.).
Edited by Karin Musier-Forsyth
Contributor Information
Pingping Jiang, Email: ppjiang@zju.edu.cn.
Min-Xin Guan, Email: gminxin88@zju.edu.cn.
Supporting information
References
- 1.Wallace D.C. Mitochondrial genetic medicine. Nat. Genet. 2018;50:1642–1649. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- 2.Craven L., Alston C.L., Taylor R.W., Turnbull D.M. Recent advances in mitochondrial disease. Annu. Rev. Genomics Hum. Genet. 2017;18:257–275. doi: 10.1146/annurev-genom-091416-035426. [DOI] [PubMed] [Google Scholar]
- 3.DiMauro S., Schon E.A. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T., Nagao A., Suzuki T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 5.Guan M.X. Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Mitochondrion. 2011;11:237–245. doi: 10.1016/j.mito.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Boczonadi V., Ricci G., Horvath R. Mitochondrial DNA transcription and translation: Clinical syndromes. Essays Biochem. 2018;62:321–340. doi: 10.1042/EBC20170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng F., Zhou M., Xiao Y., Mao X., Zheng J., Lin J., Lin T., Ye Z., Cang X., Fu Y., Wang M., Guan M.X. A deafness-associated tRNA mutation caused pleiotropic effects on the m1G37 modification, processing, stability and aminoacylation of tRNAIle and mitochondrial translation. Nucleic Acids Res. 2021;49:1075–1093. doi: 10.1093/nar/gkaa1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sissler M., González-Serrano L.E., Westhof E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 2017;23:693–708. doi: 10.1016/j.molmed.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Fine A.S., Nemeth C.L., Kaufman M.L., Fatemi A. Mitochondrial aminoacyl-tRNA synthetase disorders: An emerging group of developmental disorders of myelination. J. Neurodev. Disord. 2019;11:29. doi: 10.1186/s11689-019-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ognjenović J., Simonović M. Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 2018;15:623–634. doi: 10.1080/15476286.2017.1330245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo M., Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang P., Guo M. Structural characterization of human aminoacyl- tRNA synthetases for translational and nontranslational functions. Methods. 2017;113:83–90. doi: 10.1016/j.ymeth.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Son K., You J.S., Yoon M.S., Dai C., Kim J.H., Khanna N., Banerjee A., Martinis S.A., Han G., Han J.M., Kim S., Chen J. Nontranslational function of leucyl-tRNA synthetase regulates myogenic differentiation and skeletal muscle regeneration. J. Clin. Invest. 2019;129:2088–2093. doi: 10.1172/JCI122560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Serrano L.E., Chihade J.W., Sissler M. When a common biological role does not imply common disease outcomes: Disparate pathology linked to human mitochondrial aminoacyl-tRNA synthetases. J. Biol. Chem. 2019;294:5309–5320. doi: 10.1074/jbc.REV118.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyynismaa H., Schon E.A. Mixing and matching mitochondrial aminoacyl synthetases and their tRNAs: A new way to treat respiratory chain disorders? EMBO Mol. Med. 2014;6:155–157. doi: 10.1002/emmm.201303586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oprescu S.N., Griffin L.B., Beg A.A., Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finsterer J., Zarrouk-Mahjoub S. Phenotypic spectrum of DARS2 mutations. J. Neurol. Sci. 2017;376:117–118. doi: 10.1016/j.jns.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Steenweg M.E., Ghezzi D., Haack T., Abbink T.E., Martinelli D., van Berkel C.G., Bley A., Diogo L., Grillo E., Te Water Naudé J., Strom T.M., Bertini E., Prokisch H., van der Knaap M.S., Zeviani M. Leukoencephalopathy with thalamus and brainstem involvement and high lactate 'LTBL' caused by EARS2 mutations. Brain. 2012;135:1387–1394. doi: 10.1093/brain/aws070. [DOI] [PubMed] [Google Scholar]
- 19.Simon M., Richard E.M., Wang X., Shahzad M., Huang V.H., Qaiser T.A., Potluri P., Mahl S.E., Davila A., Nazli S., Hancock S., Yu M., Gargus J., Chang R., Al-Sheqaih N. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce S.B., Gersak K., Michaelson-Cohen R., Walsh T., Lee M.K., Malach D., Klevit R.E., King M.C., Levy-Lahad E. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet. 2013;92:614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diodato D., Melchionda L., Haack T.B., Dallabona C., Baruffini E., Donnini C., Granata T., Ragona F., Balestri P., Margollicci M., Lamantea E., Nasca A., Powell C.A., Minczuk M., Strom T.M. VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum. Mutat. 2014;35:983–989. doi: 10.1002/humu.22590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMillan H.J., Humphreys P., Smith A., Schwartzentruber J., Chakraborty P., Bulman D.E., Beaulieu C.L., Majewski J., Boycott K.M., Geraghty M.T. Congenital visual impairment and progressive microcephaly due to lysyl-transfer ribonucleic acid (RNA) synthetase (KARS) mutations: The expanding phenotype of aminoacyl-transfer RNA synthetase mutations in human disease. J. Child. Neurol. 2015;30:1037–1043. doi: 10.1177/0883073814553272. [DOI] [PubMed] [Google Scholar]
- 23.Coughlin C.R., Scharer G.H., Friederich M.W., Yu H.C., Geiger E.A., Creadon-Swindell G., Collins A.E., Vanlander A.V., Coster R.V., Powell C.A., Swanson M.A., Minczuk M., Van Hove J.L., Shaikh T.H. Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J. Med. Genet. 2015;52:532–540. doi: 10.1136/jmedgenet-2015-103049. [DOI] [PubMed] [Google Scholar]
- 24.Riley L.G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S.C., Thorburn D., Ryan M.T., Giegé R., Bahlo M., Christodoulou J. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia—MLASA syndrome. Am. J. Hum. Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommerville E.W., Ng Y.S., Alston C.L., Dallabona C., Gilberti M., He L., Knowles C., Chin S.L., Schaefer A.M., Falkous G., Murdoch D., Longman C., de Visser M., Bindoff L.A., Rawles J.M. Clinical features, molecular heterogeneity, and prognostic implications in YARS2-related mitochondrial myopathy. JAMA Neurol. 2017;74:686–694. doi: 10.1001/jamaneurol.2016.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima J., Eminoglu T.F., Vatansever G., Nakashima M., Tsurusaki Y., Saitsu H., Kawashima H., Matsumoto N., Miyake N. A novel homozygous YARS2 mutation causes severe myopathy, lactic acidosis, and sideroblastic anemia 2. J. Hum. Genet. 2014;59:229–232. doi: 10.1038/jhg.2013.143. [DOI] [PubMed] [Google Scholar]
- 27.Shahni R., Wedatilake Y., Cleary M.A., Lindley K.J., Sibson K.R., Rahman S. A distinct mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA) phenotype associates with YARS2 mutations. Am. J. Med. Genet. A. 2013;161A:2334–2338. doi: 10.1002/ajmg.a.36065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley L.G., Heeney M.M., Rudinger-Thirion J., Frugier M., Campagna D.R., Zhou R., Hale G.A., Hilliard L.M., Kaplan J.A., Kwiatkowski J.L., Sieff C.A., Steensma D.P., Rennings A.J., Simons A., Schaap N. The phenotypic spectrum of germline YARS2 variants: From isolated sideroblastic anemia to mitochondrial myopathy, lactic acidosis and sideroblastic anemia 2. Haematologica. 2018;103:2008–2015. doi: 10.3324/haematol.2017.182659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasarman F., Nishimura T., Thiffault I., Shoubridge E.A. A novel mutation in YARS2 causes myopathy with lactic acidosis and sideroblastic anemia. Hum. Mutat. 2012;33:1201–1206. doi: 10.1002/humu.22098. [DOI] [PubMed] [Google Scholar]
- 30.Riley L.G., Menezes M.J., Rudinger-Thirion J., Duff R., de Lonlay P., Rotig A., Tchan M.C., Davis M., Cooper S.T., Christodoulou J. Phenotypic variability and identification of novel YARS2 mutations in YARS2 mitochondrial myopathy, lactic acidosis and sideroblastic anaemia. Orphanet. J. Rare Dis. 2013;8:193. doi: 10.1186/1750-1172-8-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M., Zhao F., Sun Y.H., Zhang M., Zhou X., Chen Y. The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet. 2016;25:584–596. doi: 10.1093/hmg/ddv498. [DOI] [PubMed] [Google Scholar]
- 32.Fan W., Zheng J., Kong W., Cui L., Aishanjiang M., Yi Q., Wang M., Cang X., Tang X., Chen Y., Mo J.Q., Sondheimer N., Ge W., Guan M.X. Contribution of a mitochondrial tyrosyl-tRNA synthetase mutation to the phenotypic expression of the deafness-associated tRNASer(UCN) 7511A>G mutation. J. Biol. Chem. 2019;294:19292–19305. doi: 10.1074/jbc.RA119.010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczepanowska K., Maiti P., Kukat A., Hofsetz E., Nolte H., Senft K., Becker C., Ruzzenente B., Hornig-Do H.T., Wibom R., Wiesner R.J., Krüger M., Trifunovic A. CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 2016;35:2566–2583. doi: 10.15252/embj.201694253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truscott K.N., Lowth B.R., Strack P.R., Dougan D.A. Diverse functions of mitochondrial AAA+ proteins: Protein activation, disaggregation, and degradation. Biochem. Cell Biol. 2010;88:97–108. doi: 10.1139/o09-167. [DOI] [PubMed] [Google Scholar]
- 35.Jha P., Wang X., Auwerx J. Analysis of mitochondrial respiratory chain super-complexes using blue native polyacrylamide gel electrophoresis (BN-PAGE) Curr. Protoc. Mouse Biol. 2016;6:1–14. doi: 10.1002/9780470942390.mo150182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y., Zhang J., Yu J., Wang Y., Lu Y., Liang M., Li Q., Jin X., Wei Y., Meng F., Gao Y., Cang X., Tong Y., Liu X., Zhang M. Contribution of mitochondrial ND1 3394T>C mutation to the phenotypic manifestation of Leber’s hereditary optic neuropathy. Hum. Mol. Genet. 2019;28:1515–1529. doi: 10.1093/hmg/ddy450. [DOI] [PubMed] [Google Scholar]
- 37.Birch-Machin M.A., Turnbull D.M. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 38.Meng F., He Z., Tang X., Zheng J., Jin X., Zhu Y., Ren X., Zhou M., Wang M., Gong S., Mo J.Q., Shu Q., Guan M.X. Contribution of the tRNAIle 4317A>G mutation to the phenotypic manifestation of the deafness-associated mitochondrial 12S rRNA 1555A>G mutation. J. Biol. Chem. 2018;293:3321–3334. doi: 10.1074/jbc.RA117.000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J., Landar A., Darley-Usmar V.M. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong S., Peng Y., Jiang P., Wang M., Fan M., Wang X., Zhou H., Li H., Yan Q., Huang T., Guan M.X. A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res. 2014;42:8039–8048. doi: 10.1093/nar/gku466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo R., Zong S., Wu M., Gu J., Yang M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257.e1212. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 42.Bonnefond L., Frugier M., Touze E., Lorber B., Florentz C., Giege R., Sauter C., Rudinger-Thirion J. Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features. Structure. 2007;15:1505–1516. doi: 10.1016/j.str.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 43.Chen D., Zhang Z., Chen C., Yao S., Yang Q., Li F., He X., Ai C., Wang M., Guan M.X. Deletion of Gtpbp3 in zebrafish revealed the hypertrophic cardiomyopathy manifested by aberrant mitochondrial tRNA metabolism. Nucleic Acids Res. 2019;47:5341–5355. doi: 10.1093/nar/gkz218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q., Zhang L., Chen D., He X., Yao S., Zhang Z., Chen Y., Guan M.X. Deletion of Mtu1 (Trmu) in Zebrafish revealed the essential role of tRNA modification in mitochondrial biogenesis and hearing function. Nucleic Acids Res. 2018;46:10930–10945. doi: 10.1093/nar/gky758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fadool J.M., Dowling J.E. Zebrafish: A model system for the study of eye genetics. Prog. Retin. Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collery R.F., Veth K.N., Dubis A.M., Carroll J., Link B.A. Rapid, accurate, and non-invasive measurement of Zebrafish axial length and other eye dimensions using SD-OCT allows longitudinal analysis of myopia and emmetropization. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Cao S., Yu M., Hu H. TMEM216 deletion causes mislocalization of cone opsin and rhodopsin and photoreceptor degeneration in Zebrafish. Invest. Ophthalmol. Vis. Sci. 2020;61:24. doi: 10.1167/iovs.61.8.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarboush R., Chapman G.B., Connaughton V.P. Ultrastructure of the distal retina of the adult Zebrafish, Danio Rerio. Tissue Cell. 2012;44:264–279. doi: 10.1016/j.tice.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Park S.G., Ewalt K.L., Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: New perspectives on housekeepers. Trends Biochem. Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Akaike T., Ida T., Wei F.Y., Nishida M., Kumagai Y., Alam M.M., Ihara H., Sawa T., Matsunaga T., Kasamatsu S., Nishimura A., Morita M., Tomizawa K., Nishimura A., Watanabe S. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017;8:1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyeon D.Y., Kim J.H., Ahn T.J., Cho Y., Hwang D., Kim S. Evolution of the multi-tRNA synthetase complex and its role in cancer. J. Biol. Chem. 2019;294:5340–5351. doi: 10.1074/jbc.REV118.002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agnew T., Goldsworthy M., Aguilar C., Morgan A., Simon M., Hilton H., Esapa C., Wu Y., Cater H., Bentley L., Scudamore C., Poulton J., Morten K.J., Thompson K., He L. A Wars2 mutant mouse model displays OXPHOS deficiencies and activation of tissue-specific stress response pathways. Cell Rep. 2018;25:3315–3328. doi: 10.1016/j.celrep.2018.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E.I., Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 2014;19:458–469. doi: 10.1016/j.cmet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Levi O., Arava Y. mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee S., Ghosh A. Molecular mechanism of mitochondrial respiratory chain assembly and its relation to mitochondrial diseases. Mitochondrion. 2020;53:1–20. doi: 10.1016/j.mito.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Signes A., Fernandez-Vizarra E. Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem. 2018;62:255–270. doi: 10.1042/EBC20170098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M., Sips P., Khin E., Rotival M., Sun X., Ahmed R., Widjaja A.A., Schafer S., Yusoff P., Choksi P.K., Ko N.S., Singh M.K., Epstein D., Guan Y., Houštěk J. Wars2 is a determinant of angiogenesis. Nat. Commun. 2016;7:12061. doi: 10.1038/ncomms12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Country M.W. Retinal metabolism: A comparative look at energetics in the retina. Brain Res. 2017;1672:50–57. doi: 10.1016/j.brainres.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Wong-Riley M.T. Energy metabolism of the visual system. Eye Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ames A. Energy requirements of CNS cells as related to their function and to their vulnerability to ischemia: A commentary based on studies on retina. Can. J. Physiol. Pharmacol. 1992;70:S158–S164. doi: 10.1139/y92-257. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J., Ji Y., Liu X., Chen J., Wang B., Zhang M., Guan M.X. Leber's hereditary optic neuropathy caused by a mutation in mitochondrial tRNAThr in eight Chinese pedigrees. Mitochondrion. 2018;42:84–91. doi: 10.1016/j.mito.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 62.Qu J., Li R., Tong Y., Lu F., Qian Y., Hu Y., Mo J.Q., West C.E., Guan M.X. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Invest. Ophthalmol. Vis. Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- 63.Smith P.R., Bain S.C., Good P.A., Hattersley A.T., Barnett A.H., Gibson J.M., Dodson P.M. Pigmentary retinal dystrophy and the syndrome of maternally inherited diabetes and deafness caused by the mitochondrial DNA 3243 tRNALeu(UUR) A to G mutation. Ophthalmology. 1999;106:1101–1108. doi: 10.1016/S0161-6420(99)90244-0. [DOI] [PubMed] [Google Scholar]
- 64.Peragallo J.H., Keller S., van der Knaap M.S., Soares B.P., Shankar S.P. Retinopathy and optic atrophy: Expanding the phenotypic spectrum of pathogenic variants in the AARS2 gene. Ophthalmic Genet. 2018;39:99–102. doi: 10.1080/13816810.2017.1350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMillan H.J., Humphreys P., Smith A., Schwartzentruber J., Chakraborty P., Bulman D.E., Beaulieu C.L., au fnm, Majewski J., Boycott K.M., Geraghty M.T. Congenital visual impairment and progressive microcephaly due to lysyl–transfer ribonucleic acid (RNA) synthetase (KARS) mutations: The expanding phenotype of aminoacyl–transfer RNA synthetase mutations in human disease. J. Child. Neurol. 2015;30:1037–1043. doi: 10.1177/0883073814553272. [DOI] [PubMed] [Google Scholar]
- 66.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonnefond L., Fender A., Rudinger-Thirion J., Giege R., Florentz C., Sissler M. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- 68.Cheng Z., Gong Y., Ma Y., Lu K., Lu X., Pierce L.A., Thompson R.C., Muller S., Knapp S., Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodina A., Wang T., Yan P., Gomes E.D., Dunphy M.P., Pillarsetty N., Koren J., Gerecitano J.F., Taldone T., Zong H., Caldas-Lopes E., Alpaugh M., Corben A., Riolo M., Beattie B. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.King M.P., Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 1993;126:10228–10237. [PubMed] [Google Scholar]
- 71.Enriquez J.A., Attardi G. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996;264:183–196. doi: 10.1016/s0076-6879(96)64019-1. [DOI] [PubMed] [Google Scholar]
- 72.Jia Z., Zhang Y., Li Q., Ye Z., Liu Y., Fu C., Cang X., Wang M., Guan M.X. A coronary artery disease-associated tRNAThr mutation altered mitochondrial function, apoptosis and angiogenesis. Nucleic Acids Res. 2019;47:2056–2074. doi: 10.1093/nar/gky1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou M., Xue L., Chen Y., Li H., He Q., Wang B., Meng F., Wang M., Guan M.X. A hypertension-associated mitochondrial DNA mutation introduces an m1G37 modification into tRNAMet, altering its structure and function. J. Biol. Chem. 2018;293:1425–1438. doi: 10.1074/jbc.RA117.000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Westerfield M. University of Oregon Press; Eugene: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 75.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the Zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 76.Gonzales A.P., Yeh J.R. Cas9-based genome editing in Zebrafish. Methods Enzymol. 2014;546:377–413. doi: 10.1016/B978-0-12-801185-0.00018-0. [DOI] [PubMed] [Google Scholar]
- 77.Escobar-Alvarez S., Gardner J., Sheth A., Manfredi G., Yang G., Ouerfelli O., Heaney M.L., Scheinberg D.A. Inhibition of human peptide deformylase disrupts mitochondrial function. Mol. Cell. Biol. 2010;30:5099–5109. doi: 10.1128/MCB.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu J., Liang X., Ji Y., Ai C., Liu J., Zhu L., Nie Z., Jin X., Wang C., Zhang J., Zhao F., Mei S., Zhao X., Zhou X., Zhang M. PRICKLE3 linked to ATPase biogenesis manifested Leber's hereditary optic neuropathy. J. Clin. Invest. 2020;130:4935–4946. doi: 10.1172/JCI134965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips M.J., Webb-Wood S., Faulkner A.E., Jabbar S.B., Biousse V., Newman N.J., Do V.T., Boatright J.H., Wallace D.C., Pardue M.T. Retinal function and structure in ant-deficient mice. Invest. Ophthalmol. Vis. Sci. 2010;51:6744–6752. doi: 10.1167/iovs.10-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berdougo E., Coleman H., Lee D.H., Stainier D.Y.R., Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in Zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Representative experiments are shown in the figures and supplemental materials. For any additional information, please contact the corresponding author.