Abstract
Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are essential for combating the coronavirus disease 2019 (COVID-19) pandemic. Neutralizing antibody responses to the original Wuhan-Hu-1 strain that were generated during infection and vaccination showed lower effectiveness against variants of concern. Here, we demonstrated that mouse plasma induced by protein nanoparticles that present rationally designed S2GΔHR2 spikes can neutralize the B.1.1.7, B.1.351, and P.1 variants with comparable titers. The mechanism of nanoparticle vaccine-induced immunity was examined in mice for an I3–01v9 60-mer that presents 20 stabilized spikes. Compared with the soluble spike, this nanoparticle showed 6-fold longer retention, 4-fold greater presentation on follicular dendritic cell dendrites, and 5-fold higher germinal center reactions in lymph node follicles. Intact nanoparticles in lymph node tissues were visualized by transmission electron microscopy. In conclusion, spike-presenting protein nanoparticles that induce robust long-lived germinal centers may provide a vaccine solution for emerging SARS-CoV-2 variants.
Keywords: Coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), variant of concern (VOC), vaccine, lymph node, germinal center, broadly neutralizing antibody (bNAb)
ONE-SENTENCE SUMMARY
With prolonged lymph node retention and robust germinal centers, nanoparticles elicit neutralizing antibodies to diverse SARS-CoV-2 variants.
The COVID-19 pandemic has led to more than 124 million infection cases and 2.7 million deaths globally. It has been reported that the human antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients can be sustained for several months after infection (1–5). However, recently identified variants of concern (VOCs) exhibit higher transmissibility and resistance to prior immunity as SARS-CoV-2 continues to adapt to the human host (6, 7). One such variant, B.1.1.7, emerged from southeast England in October 2020 and accounted for two-thirds of new infections in London in December 2020, with a higher transmission rate (43–90%) and risk of mortality (32–104%) than previously circulating strains (8–11). Other variants, such as B.1.351 and P.1, also became prevalent in the Eastern Cape, Western Cape, and KwaZulu-Natal provinces in South Africa and in Manaus, Brazil, respectively (7, 12–14). The rise of SARS-CoV-2 VOCs and their rapid spread worldwide will likely result in more infection cases, hospitalizations, and potentially more deaths, further straining healthcare resources (14).
To date, seven COVID-19 vaccines have been approved for emergency use in humans, with more than 100 candidates being assessed in various phases of clinical trials (15). With the exception of inactivated whole-virion vaccines, diverse platforms have been used to deliver the recombinant SARS-CoV-2 spike, such as mRNA-encapsulating liposomes (e.g., BNT162b2 and mRNA-1273), adenovirus vectors (e.g., ChAdOx1 nCoV-19 [AZD1222], CTII-nCoV, Sputnik V, and Ad26.COV2.S), and nanoparticles (e.g., NVX-CoV2373). These vaccines demonstrated 65–96% efficacies in Phase 3 trials, with reduced morbidity and mortality associated with COVID-19 disease (16–20). However, a notable loss of vaccine efficacy against new SARS-CoV-2 variants was reported because of spike mutations in the receptor-binding domain (RBD; e.g., K417N, E484K, and N501Y), N-terminal domain (NTD; e.g., L18F, D80A, D215G, and Δ242–244), and other regions that are critical to spike stability (e.g., D614G) (7, 21–28). Among circulating VOCs, the B.1.351 lineage appeared to be most resistant to neutralization by convalescent plasma (9.4-fold) and vaccine sera (10.3- to 12.4-fold) (29), while a lesser degree of reduction was observed for an early variant, B.1.1.7 (30, 31). Based on these findings, it was suggested that vaccines would need to be updated periodically to maintain protection against rapidly evolving SARS-CoV-2 (24, 32, 33). This raises the concern that herd immunity may not be achievable with current vaccines and highlights the necessity of developing vaccines that can elicit a broadly neutralizing antibody (bNAb) response to SARS-CoV-2 variants (24, 28). As previously reported (34–40), the production of a bNAb response relies on long-lived germinal center (GC) reactions to activate precursor B cells, stimulate affinity maturation, and form long-term immune memory. Antigen retention and presentation within lymph node follicles are key to the induction of long-lived GC reactions (34, 36) and thus should be a main criterion in the development of bNAb-producing vaccines.
We previously investigated the cause of SARS-CoV-2 spike metastability and rationally designed the S2GΔHR2 spike, which was displayed on three self-assembling protein nanoparticle (SApNP) platforms, including ferritin (FR) 24-mer and multilayered E2p and I3–01v9 60-mers, as COVID-19 vaccine candidates (41) (Fig. 1A). Notably, the I3–01v9 SApNP that presents 20 stabilized spikes induced a potent NAb response against both SARS-CoV-1 and SARS-CoV-2, in addition to critically needed T cell responses (41). Here, we first examined the neutralizing activity of mouse plasma induced by these rationally designed vaccine constructs against representative SARS-CoV-2 variants. We then investigated how SApNPs “behave” in lymph nodes and induce GCs in the mouse model by characterizing vaccine delivery and immunological responses at the intraorgan, intracellular, and intercellular levels. Our findings suggest that a spike-presenting protein nanoparticle vaccine may confer broad protection against SARS-CoV-2 VOCs.
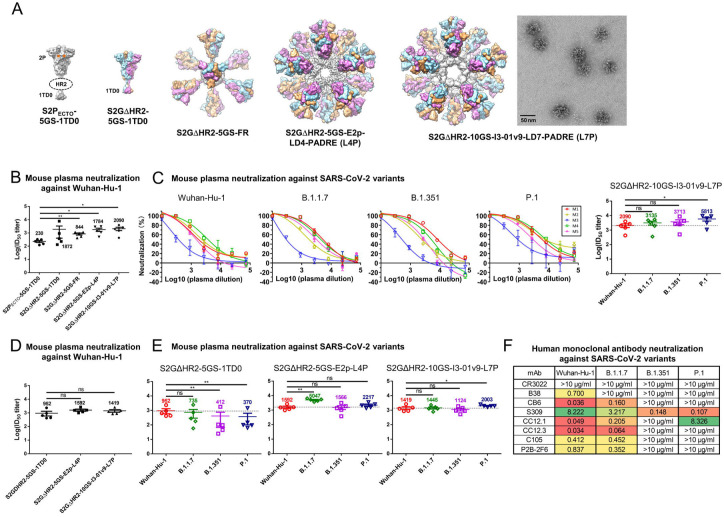
Fig. 1. SARS-CoV-2 SApNP vaccines induce broadly neutralizing antibody responses to three variants of concern.
(A) Molecular surface representations of vaccine constructs, including two spikes (S2PECTO-5GS-1TD0 and S2GΔHR2-5GS-1TD0) and three spike-presenting SApNPs (S2GΔHR2-5GS-ferritin (FR), S2GΔHR2-5GS-E2p-LD4-PADRE (E2p-L4P), and S2GΔHR2-10GS-I3-01v9-LD7-PADRE (I3-01v9-L7P)). Representative negative-stain EM (nsEM) image of S2GΔHR2-10GS- I3-01v9-L7P SApNPs is shown on the right. (B) Neutralization of the original Wuhan-Hu-1 strain by mouse plasma induced by 5 different vaccines at week 5 after two intraperitoneal injections. ID50 titers derived from SARS-CoV-2-pp neutralization assays are plotted, with average ID50 values labeled on the plots. (C) Mouse plasma neutralization against the original Wuhan-Hu-1 strain and three variants, B.1.1.7, B1.351, and P.1, at week 5 after two intraperitoneal injections of the adjuvanted S2GΔHR2-10GS-I3-01v9-L7P vaccine. Left panels 1-4: percent neutralization plots of individual mice against 4 SARS-CoV-2 strains; Right panel: ID50 plot. In (B) and (C), the plasma samples were generated in the previous study (41), where mice were immunized with 50 μg of adjuvanted vaccine antigen. (D) Neutralization of the original Wuhan-Hu-1 strain by mouse plasma induced the S2GΔHR2 spike and two large S2GΔHR2-presenting SApNPs. Vaccines were administered via footpad injections (0.8 μg/injection, for a total of 3.3 μg/mouse). (E) Mouse plasma neutralization against the original Wuhan-Hu-1 strain and three variants, B.1.1.7, B1.351, and P.1, at week 5 after two footpad injections of the S2GΔHR2 spike and two large S2GΔHR2-presenting SApNPs. In (B)-(E), the ID50 values are plotted as mean ± SEM. The data were analyzed using two-tailed unpaired Student’s t-test for comparison between different vaccine groups or two-tailed paired Student’s t-test for comparison of ID50 titers against SARS-Cov-2 variants using the same plasma samples from a mouse. *p < 0.05, **p < 0.01. (F) Neutralization of four SARS-CoV-2 strains by human monoclonal antibodies including CR3022, B38, CB6, S309, CC12.1, CC12.3, C105, and P2B-2F6. IC50 values are listed and color-coded (white: no neutralization; green to red: low to high). The IC50 values were calculated with the %neutralization range constrained within 0.0-100.0%.
We first assessed the neutralizing activity of polyclonal plasma elicited by various spike and SApNP vaccine formulations in pseudoparticle (pp) neutralization assays (42). Mouse plasma at week 5 after intraperitoneal (i.p.) injections of adjuvanted vaccine antigens (50 μg) from our previous study were analyzed against the original SARS-CoV-2 strain, Wuhan-Hu-1, as a baseline for comparison (Fig. 1B). The soluble S2PECTO spike elicited the lowest 50% inhibitory dilution (ID50) titers, whereas the soluble S2GΔHR2 spike induced a stronger NAb response, showing a 7.1-fold higher (or 8.1 times) average ID50 titer, which did not reach statistical significance because of within-group variation. All three spike SApNP vaccines elicited superior neutralizing antibody responses compared with the soluble S2PECTO spike (41, 43). Notably, the I3–01v9 SApNP vaccine showed the most potent NAb response, with an average ID50 titer of 2090, which was 8.1-fold higher than the soluble S2PECTO spike. Despite the differences in ID50 titers, the overall pattern remained the same as observed in our previous study (41). These differences can be attributed to the inherent variation of pseudovirus neutralization assays (42) (see Methods).Next, we assessed neutralizing activity against three major SARS-CoV-2 variants (Fig. 1C, fig. S1A, B). The I3–01v9 SApNP induced a stronger NAb response against three VOCs, with 0.5-fold (B.1.1.7), 0.8-fold (B.1.351), and 1.8-fold (P.1) higher ID50 titers compared with the Wuhan-Hu-1 strain (Fig. 1C). These results were confirmed in a second experiment, in which mice were intraperitoneally immunized with low doses (5 and 15 μg) of I3–01v9 SApNP. Remarkably, all three SARS-CoV-2 VOC-pps were neutralized by mouse plasma, with ID50 titers comparable to the high-dose group (45 μg; fig. S1C, D). To examine whether routes of injection affect the elicitation of NAb responses against VOCs, we performed a third experiment, in which a low dose (3.3 μg) of adjuvanted antigen was intradermally administered into four footpads (i.e., 0.8 μg/footpad). The large (~55–60 nm) E2p and I3–01v9 SApNPs that present 20 S2GΔHR2 spikes yielded higher ID50 titers than the soluble S2GΔHR2 spike (Fig. 1D, fig. S1E, F), whereas a significant reduction of neutralizing titers against the variants was observed for mouse plasma from the S2GΔHR2 group (Fig. 1E, fig. S1E, F), suggesting that nanoparticle display is critical for eliciting a bNAb response. The E2p and I3–01v9 SApNP groups exhibited comparable or stronger NAb responses against three variants relative to the original Wuhan-Hu-1 strain (Fig. 1E). A panel of human NAbs was previously used to validate the SARS-CoV-2-pp neutralization assays, in addition to evaluating the antigenicity of rationally designed spikes and SApNPs (41). This panel of human NAbs was tested against SARS-CoV-2-pps that carry various VOC spikes (Fig. 1F, fig. S1G). Lower potency, measured by the 50% inhibitory concentration (IC50), was observed for all human NAbs against B.1.351 and P.1 variants, with the exception of NAb S309, which was identified from a SARS-CoV-1 patient (44). This finding is consistent with previous reports on convalescent patient plasma (29–31). As a negative control, mouse plasma induced by the S2GΔHR2-presenting I3–01v9 SApNP was tested against pseudoviruses carrying the murine leukemia virus (MLV) Env, or MLV-pps. Nonspecific neutralization was not detected (fig. S1H, I). Altogether, these results demonstrated a clear advantage of spike-presenting SApNPs in eliciting a strong bNAb response against SARS-CoV-2 VOCs compared with soluble spikes.
Next, we studied in vivo behaviors of the S2GΔHR2 spike and two large SApNPs to understand vaccine-induced immunity and why nanoparticles outperform soluble spikes in terms of bNAb elicitation. We first examined the distribution of S2GΔHR2-presenting I3–01v9 SApNPs in mouse lymph nodes via footpad injection (10 μg). The mice were sacrificed 12 h after single-dose (Fig. 2A) and prime-boost (Fig. 2B) regimens. The axillary, brachial, and popliteal sentinel lymph nodes were isolated for histological analysis. The lymph node tissues were stained with the human anti-spike antibody P2B-2F6 (45) to characterize SARS-CoV-2 spikes presented on the I3–01v9 SApNPs. Consistent with our previous study (46), SApNPs accumulated in lymph node follicles, regardless of the number of doses. SApNPs were sequestrated in the center of lymph node follicles after a single-dose (Fig. 2A, images on the left, schematics on the right), but were located along the outer layer of expanded lymph node follicles after the second injection due to preexisting humoral immunity (i.e., GC reactions) that was induced by the first dose (Fig. 2B, images on the left, schematics on the right). Overall, the majority of SApNPs accumulated in lymph node follicles, while their distribution differed slightly depending on the doses.
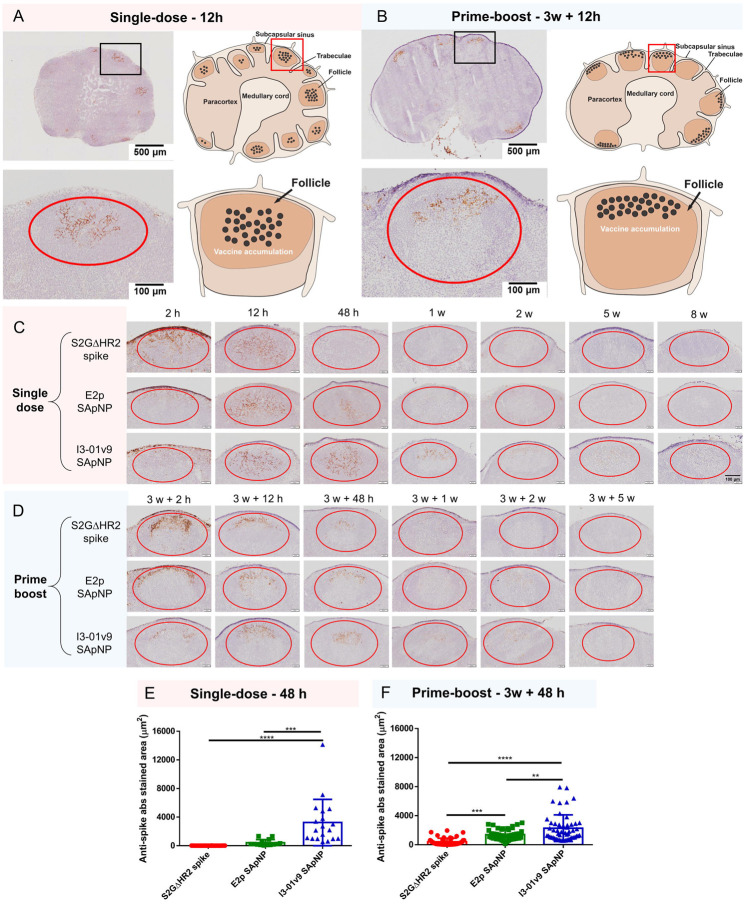
Fig. 2. SARS-CoV-2 SApNP vaccines induce long-term lymph node follicle retention.
(A, B) S2GΔHR2-presenting I3–01v9 SApNP vaccine distribution in a lymph node 12 h after (A) a single-dose or (B) prime-boost footpad injections (10 μg/injection, 40 μg/mouse). Schematic illustration of SApNPs in lymph node follicles is shown. (C, D) Histological images of the S2GΔHR2 spike and S2GΔHR2-presenting E2p and I3–01 SApNP vaccine trafficking and retention in lymph node follicles 2 h to 8 weeks after (C) a single-dose or (D) prime-boost injections. (E, F) Quantification of vaccine accumulation in lymph node follicles 48 h after (E) a single-dose or (F) prime-boost injections. Data were collected from more than 10 lymph node follicles (n = 3–4 mice/group). The data points are expressed as mean ± SD. The data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison post hoc test. **p < 0.01, ***p < 0.001, ****p < 0.0001.
In this context, we examined patterns of trafficking and lymph node follicle retention for soluble S2GΔHR2 spike vs. the S2GΔHR2-presenting E2p and I3–01v9 SApNPs. To facilitate this analysis, the mice were sacrificed 2 h to 8 weeks after a single dose (Fig. 2C) and 2 h to 5 weeks after the boost (Fig. 2D). The injection dose was normalized to the total amount of protein (10 μg) per injection into each footpad (40 μg/mouse). As shown in Fig. 2C, the S2GΔHR2 spikes that trafficked into lymph node follicles at 2 h cleared within 48 h. In contrast, the two large SApNPs accumulated in the subcapsular sinus at 2 h and then trafficked into follicles 12 h after the single-dose injection. Remarkably, I3–01v9 SApNPs remained detectable in lymph node follicles after 2 weeks, suggesting 6-fold longer retention than the S2GΔHR2 spike (Fig. 2C). The results for these protein nanoparticles are thus consistent with the pattern of size dependency that was observed for ovalbumin-conjugated gold nanoparticles in our previous study (46), in which small (5–15 nm) nanoparticles cleared shortly after the injection, whereas large (50–100 nm) nanoparticles were retained in lymph node follicles for weeks. Similar patterns of antigen retention were observed after the second injection, although the boost appeared to exert a more positive effect on the soluble spike, which could be detected in lymph node follicles at 48 h (Fig. 2D). Nonetheless, prolonged retention was observed for both E2p and I3–01v9 SApNPs 2 weeks after the boost injection. Overall, the multivalent display of S2GΔHR2 spikes on the I3–01v9 SApNP resulted in 325- and 4-fold greater accumulation in lymph node follicles compared with the soluble spike 48 h after the single-dose (Fig. 2E) and prime-boost (Fig. 2F) injections, respectively. These findings reveal the advantage of a leading vaccine candidate identified in our previous study, S2GΔHR2–10GS-I3–01v9-L7P (41), in terms of spike antigen retention in lymph node follicles.
Next, we determined the involvement of resident cells in antigen retention in lymph node follicles. Recent studies reported that follicular dendritic cells (FDCs) are resident stromal cells in follicles and can retain immune complexes, virus-like particles, virus, and bacteria (46–51). Follicular dendritic cells are key to naive antigen retention, GC initiation and maintenance, and B cell affinity maturation (39, 52–55). Here, we hypothesized that FDCs comprise the major cell population in lymph node follicles that retain SARS-CoV-2 spikes and spike-presenting SApNPs. To test this hypothesis, we administered vaccines via footpad injections and collected mouse lymph nodes at the peak of accumulation (12 h) after single-dose (Fig. 3A) and prime-boost (Fig. 3B) injections. Lymph node tissue samples were stained with the anti-spike antibody P2B-2F6 (45) for the spike antigen and anti-CD21 and CD169 antibodies for FDCs and subcapsular sinus macrophages, respectively. The S2GΔHR2 spike and SApNP (E2p or I3–01v9) signals colocalized with FDC (CD21+) networks in lymph node follicles (Fig. 3A, B). This result confirmed the critical role of FDC networks in mediating vaccine retention in lymph node follicles.
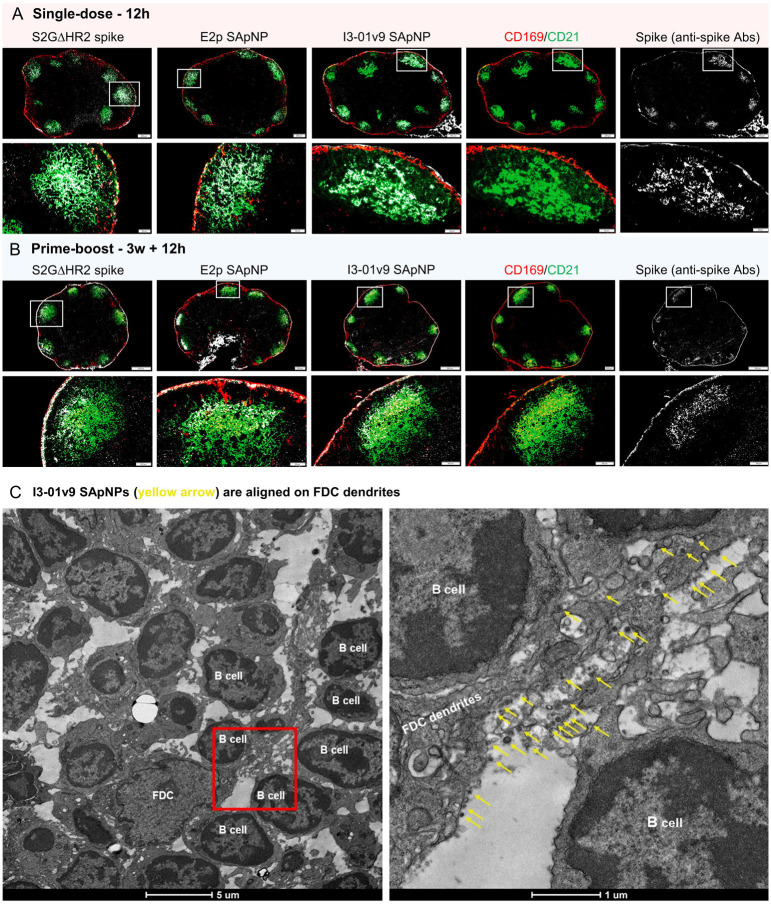
Fig. 3. SARS-CoV-2 SApNP vaccines interact with follicular dendritic cells (FDCs) and are presented on FDC dendrites to B cells.
(A, B) S2GΔHR2 spike and S2GΔHR2-presenting E2p and I3–01 SApNP vaccine interaction with FDC networks in lymph node follicles 12 h after (A) a single-dose or (B) prime-boost injections (10 μg/injection, 40 μg/mouse). Vaccine antigens (the S2GΔHR2 spike and S2GΔHR2-presenting E2p and I3–01 SApNPs) colocalized with FDC networks. Immunostaining is color-coded (Green: CD21; Red: CD169; White: anti-spike). (C) Representative TEM images of an FDC surrounded by multiple B cells. S2GΔHR2-presenting I3–01 SApNPs (yellow arrows) presented on FDC dendrites.
The induction of potent bNAb responses by spike-presenting SApNPs (Fig. 1) suggests the effective activation of naïve B cells and subsequent recalls by crosslinking B cell receptors (56–58). However, still unclear are the manners in which FDC networks present SApNPs to B cells to trigger such a bNAb response. Here, we analyzed the interface between FDC networks and B cells by transmission electron microscopy (TEM). Briefly, fresh lymph nodes were isolated and directly immersed in fixative. The processed tissue samples were sectioned and stained on copper grids for TEM analysis. We first determined whether SApNPs, such as the S2GΔHR2-presenting I3–01v9 SApNP, remain intact in vivo (fig. S2). Mouse lymph nodes were isolated 2 h after the injection of a high dose (50 μg) of the non-adjuvanted I3–01v9 SApNP. The TEM images revealed that round-shape granules corresponding to intact SApNP aligned on the macrophage surface or inside endolysosomes of the macrophage in a lymph node (fig. S2). We next studied the relative location between FDCs and I3–01v9 SApNPs and how FDCs present SApNPs to B cells. Mouse lymph nodes were collected 2, 12, and 48 h after a single-dose (50 μg) and 12 h after the boost of the I3–01v9 SApNP vaccine. The FDCs exhibited the characteristic morphology of long dendrites that surrounded and interacted with B cells in lymph node follicles (Fig. 3C, fig. S3). While few I3–01v9 SApNPs were observed on FDC dendrites at 2 h (fig. S3D), notably more nanoparticles migrated to and aligned on FDC dendrites at 12 and 48 h (Fig. 3C, figs. S3A–C, yellow arrows). The TEM images indicated that FDCs can present many SApNPs to neighboring B cells in this “hugging mode”, in which their long dendrites brace B cells to maximize interactions between multivalently displayed spikes and B cell receptors. These results demonstrated the intrinsic nature of FDCs as a reservoir for the sequestration, retention, and presentation of virus-like particles, or SApNPs with similar molecular traits, to initiate GC reactions.
Lastly, we investigated whether the prolonged retention of S2GΔHR2-presenting E2p and I3–01v9 SApNPs induce more robust GCs in lymph node follicles than the soluble S2GΔHR2 spike. Immunohistological analysis was performed to characterize GC B cells (GL7+) and T follicular helper (Tfh) cells (CD4+Bcl6+). For the I3–01v9 SApNP, 2 weeks after immunization, we observed robust GCs in lymph node B cell follicles (B220+) with well-formed dark zone (DZ) and light zone (LZ) compartments, which contain GC B cells, FDCs, and Tfh cells (35, 37, 59–62) (Fig. 4A). We then extended the analysis to the S2GΔHR2 spike and spike-presenting SApNPs 2, 5, and 8 weeks after the single-dose injection (Fig. 4B, fig. S4A–C) and 2 and 5 weeks after the boost (Fig. 4C, fig. S4D, E). Two semi-quantitative metrics, the GC/FDC ratio (i.e., whether GC formation is associated with an FDC network, %) and GC size (i.e., occupied area), were used in this analysis. Overall, the soluble S2GΔHR2 spike and both large SApNPs induced robust GCs 2 weeks after immunization (Fig. 4B, fig. S4A). The E2p and I3–01v9 SApNPs that present 20 S2GΔHR2 spikes induced robust, long-lived GCs, whereas the spike alone failed to sustain robust GCs at week 8 with either the single-dose (Fig. 4B, D) or prime-boost (Fig. 4C, E) injections. The I3–01v9 SApNP generated larger GCs than the soluble spike, 2.0-fold larger after the single-dose (Fig. 4B, D) and 2.4-fold larger after the boost (Fig. 4C, E), measured at week 8.
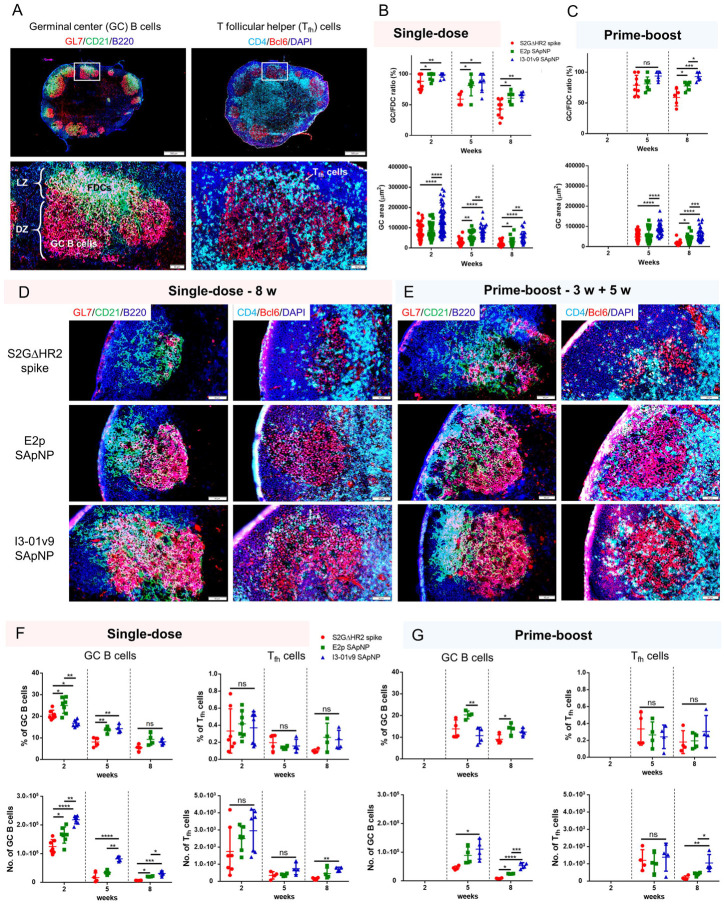
Fig. 4. SARS-CoV-2 SApNP vaccines induce robust long-lived germinal centers.
(A) Top: Representative immunohistological images of germinal centers at week 2 after a single-dose injection of the S2GΔHR2-presenting I3–01 SApNP vaccine (10 μg/injection, 40 μg/mouse). Bottom: germinal center B cells (GL7+, red) adjacent to FDCs (CD21+, green) in lymph node follicles (left), and Tfh cells in the light zone (LZ) of germinal centers (right). (B, C) Quantification of germinal center reactions using immunofluorescent images: GC/FDC ratio and sizes of germinal centers 2, 5, and 8 weeks after (B) a single-dose or (C) prime-boost injections (n = 4–7 mice/group). The GC/FDC ratio is defined as whether germinal center formation is associated with an FDC network (%). Representative immunohistological images of germinal centers in mice immunized using S2GΔHR2 spike or S2GΔHR2-presenting E2p and I3–01 SApNP vaccines at week 8 after (D) a single-dose or (E) prime-boost injections. (F, G) Quantification of germinal center reactions using flow cytometry: percentage and number of germinal center B cells and T follicular helper cells 2, 5, and 8 weeks after (F) a single-dose or (G) prime-boost injections. The data points are shown as mean ± SD. The data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison post hoc test for each timepoint. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We further characterized GC reactions by flow cytometry. Fresh mouse lymph nodes were disaggregated into a single cell suspension and stained with an antibody cocktail to quantify GC B cells and Tfh cells (fig. S5A). The results were consistent with the immunohistological analysis, in which all spike-based vaccine antigens, including the S2GΔHR2 spike and SApNPs, showed robust GCs at week 2 after the injection that declined over time, as measured at weeks 5 and 8 (Fig. 4F). The E2p and I3–01v9 SApNPs generated a larger population of GC B cells than both the S2PECTO and S2GΔHR2 spikes at week 2 (fig. S5B, C). Although the boost dose had little impact on the frequency of GC B cells and Tfh cells, it appeared to extend GC formation within lymph nodes (Fig. 4F, G), which may promote B cell development toward bNAbs. Notably, the GC B cell and Tfh cell populations elicited by the soluble S2GΔHR2 spike were barely detectable 5 weeks after immunization (Figs. 4F, G). This result was reminiscent of a recent study of an mRNA vaccine, in which GC reactions diminished to baseline levels at week 4 after a single-dose injection (63). The S2GΔHR2-presenting I3–01v9 SApNP generated 3.7/5.2-fold more GC B cells and 3.7/4.4-fold more Tfh cells than the soluble S2GΔHR2 spike after one/two-dose immunization at week 8 (Fig. 4G). Therefore, SApNPs that were retained on FDC dendrites could present SARS-CoV-2 neutralizing epitopes to enable more effective B cell recognition than the soluble spike, and consequently induce more robust and long-lived GCs in lymph nodes.
To end the COVID-19 pandemic, vaccines need to effectively block current and emerging SARS-Cov-2 variants that evade NAb responses by mutating key epitopes on the viral spike (24). Such vaccines must induce long-lasting GCs to facilitate the development of bNAbs. Effective vaccine retention and presentation are critical for inducing and sustaining GC reactions, which in turn promote the proliferation and affinity maturation of antigen-specific B cells. Here, we found that the S2GΔHR2-presenting I3–01v9 SApNP, a vaccine candidate from our recent study (41), elicited 6-fold longer retention and 4-fold greater accumulation in lymph node follicles than the stabilized S2GΔHR2 spike alone with a prime-boost regimen (Fig. 2D, F). This can be attributed to the intrinsic physiological properties of lymph nodes that mediate vaccine trafficking and retention in follicles in a size-dependent manner, which would favor retaining large (> 50 nm) virus-like nanoparticles (46–48, 64, 65). Supporting this notion are the images of retained I3–01v9 SApNPs aligned on long FDC dendrites, suggesting that such protein nanoparticles can present spike antigens to B cells for rapid initiation and then sustain GC reactions in lymph node follicles for an extended period of time (Fig. 3, 4). Specifically, the S2GΔHR2-presenting I3–01v9 SApNP generated 2.4-fold larger GCs and greater numbers of GC B cells (5.2-fold) and Tfh cells (4.4-fold) than the soluble S2GΔHR2 spike with the prime-boost regimen (Fig. 4). The findings in this study provide quantitative evidence that spike-presenting SApNP vaccines are uniquely suited for inducing long-lived robust GCs in lymph node follicles. Our analyses also provide potential explanations for the mechanism by which the S2GΔHR2-presenting I3–01v9 SApNP can elicit a more effective bNAb response than the soluble spike (Fig. 1B) (32). Further investigation at the monoclonal level will reveal how bNAbs recognize conserved epitopes and facilitate the rational design of SARS-CoV-2 spike antigens (24). Superior NAb responses have been reported for other COVID-19 vaccine candidates that take advantage of particulate display (66–78). Importantly, our S2GΔHR2-presenting I3–01v9 SApNP vaccine elicited bNAb responses to three SARS-CoV-2 VOCs (Fig. 1), thus meeting a major criterion for next-generation COVID-19 vaccines.
Protein vaccines have well-established records of both safety and effectiveness (79–82), but they have yet to be deployed in the global campaign against the COVID-19 pandemic. Through formulation with adjuvants to further enhance immune responses (83–88), protein vaccines can be used either alone or as a booster for currently used nucleic acid vaccines. One such protein vaccine, NVX-CoV2373 (full-length S2P spikes formulated with Matrix-M™ adjuvant), showed 96.4% efficacy in human trials (89). In our studies, we formulated the spike-presenting E2p and I3–01v9 SApNPs with the AddaVax and aluminum phosphate adjuvants (41), which induced stronger GC reactions than non-adjuvanted SApNPs at week 2 (fig. S6). Further development of these SApNP vaccines may benefit from a systematic analysis of adjuvants that target diverse immune pathways. An in-depth understanding of how different vaccine platforms (e.g., inactivated virions, mRNAs, viral vectors, and nanoparticles) behave in vivo, such as trafficking and retention in lymph nodes, interactions with immune cells, antigen processing and presentation, and GC reactions and NAb responses, will provide insights into vaccine-induced immunity (82, 90–92). This knowledge will in turn accelerate vaccine development against SARS-CoV-2 and other emerging pathogens.
MATERIALS AND METHODS
SARS-CoV-2 spike and SApNP vaccine antigens
The design, expression, and purification of a stabilized SARS-CoV-2 spike, S2GΔHR2, and three SApNPs that present either 8 or 20 S2GΔHR2 spikes were described in our recent study (41). Briefly, the spike gene of the SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank accession no. MN908947) was modified to include the mutations 682GSAGSV687 and K986G/V987G, in addition to truncation of the HR2 stalk (ΔE1150-Q1208). The viral capsid protein SHP (Protein Data Bank: 1TD0) was added as a C-terminal trimerization motif to stabilize the S2GΔHR2 trimer, resulting in a soluble S2GΔHR2–5GS-1TD0 spike (41). The S2GΔHR2 spike was genetically fused to FR, multilayered E2p, and multilayered I3–01v9 with 5GS, 5GS, and 10GS linkers, respectively, resulting in three S2GΔHR2-presenting SApNPs (41). An S2PECTO-5GS-1TD0 spike construct that contained the mutations 682GSAGSV687 and K986G/V987G but without HR2 deletion (41) was included for comparison. All of the vaccine antigens were transiently expressed in ExpiCHO cells and purified by a CR3022 antibody column and size-exclusion chromatography (SEC) on a Superose 6 10/300 GL column as described previously (41).
Animal immunization and sample collection
Similar immunization protocols were reported in our previous vaccine studies (41, 93–95). Briefly, Institutional Animal Care and Use Committee (IACUC) guidelines were followed for all of the animal studies. BALB/c mice (6 weeks old) were purchased from The Jackson Laboratory and kept in ventilated cages in environmentally controlled rooms at The Scripps Research Institute. The mouse studies were conducted according to Association for the Assessment and Accreditation of Laboratory Animal Care guidelines, and the protocols were approved by the IACUC. For the immunogenicity study, the mice were intraperitoneally immunized at either weeks 0 or 3 with 200 μl of antigen/adjuvant mix containing 5–50 μg of vaccine antigen and 100 μl of adjuvant (41) or immunized at weeks 0 and 3 with 20 μl of antigen/adjuvant mix containing 0.8 μg of vaccine antigen per injection (3.3 μg/mouse) and 10 μl of adjuvant per injection (four footpads were injected with a total of 80 μl [20 μl/footpad]) via intradermal footpad injections. For the mechanism study of vaccine trafficking, retention, and induced GCs, the mice were immunized at weeks 0 and 3 with 20 μl of antigen/adjuvant mix containing 10 μg of vaccine antigen and 10 μl of adjuvant per injection. To visualize S2GΔHR2-presenting I3–01v9 SApNPs in lymph node tissues using TEM, the mice were immunized at weeks 0 and 3 with 70 μl of antigen/adjuvant mix containing 50 μg of vaccine antigen and 20 μl of adjuvant per injection. Vaccines were administered into intradermal footpads of mice using a 29-gauge insulin needle under 3% isoflurane anesthesia with oxygen. Blood was drawn from the maxillary/facial vein into an ethylenediaminetetraacetic acid (EDTA)-coated tube 2 weeks after each immunization. Blood plasma was isolated after centrifugation at 14000 rotations per minute (rpm) for 10 min. Plasma was heated and inactivated at 56°C for 30 min. The supernatant was then collected after centrifugation at 8000 rpm for 10 min. Plasma was used for the neutralization assay to determine neutralizing antibody responses. The axillary, brachial, and popliteal sentinel lymph nodes were collected at the end timepoint for further analysis.
SARS-CoV-2 pseudovirus neutralization assay
The SARS-CoV-2-pp neutralization assays were described in our previous study (41). Briefly, SARS-CoV-2-pps were generated by the co-transfection of HEK293T cells with the HIV-1 pNL4-3.lucR-E- plasmid (obtained from the National Institutes of Health AIDS reagent program; https://www.aidsreagent.org/) and the expression plasmid encoding the S gene of various SARS-CoV-2 strains, including three variants: B.1.1.7, B.1.351, and P.1 (GISAID accession no. EPI_ISL_601443, EPI_ISL_678597, and EPI_ISL_792680, respectively). The HEK293T-hACE2 cell line (catalog no. NR-52511) and pcDNA3.1(–) vector containing the S gene of the SARS-CoV-2 isolate Wuhan-Hu-1 (catalog no. NR52420) were requested from the BEI Resources (https://www.beiresources.org/) on September 23, 2020 and used in the pseudovirus neutralization assays (42). Based on sequence alignment, spike mutations were incorporated into the S gene of the original Wuhan-Hu-1 isolate (catalog no. NR52420) to create respective expression plasmids for B.1.1.7, B.1.351, and P.1. SARS-CoV-2-pp neutralization by immunized mouse plasma and human monoclonal antibodies (mAbs) was performed according to a previously described protocol (41). Using the same co-transfection expression system as described above for the SARS-CoV-2-pps, we produced pseudoviruses carrying the murine leukemia virus (MLV) Env, MLV-pps, for use as a negative control (41). Percent neutralization data were analyzed using GraphPad Prism 9.0.2 software. ID50/IC50 values were calculated using constraints for the percent neutralization (0–100%), whereas unconstrained neutralization plots were shown in Fig 1 and fig S1.
Histology, immunostaining, and imaging
The mice were sacrificed 2 h to 8 weeks after a single-dose and 2 h to 5 weeks after the boost immunization. The axillary, brachial, and popliteal sentinel lymph nodes were isolated for histological analysis. Fresh lymph nodes were rapidly merged into frozen section compound (VWR International, catalog no. 95057–838) in a plastic cryomold (Tissue-Tek at VWR, catalog no. 4565) using liquid nitrogen to preserve the antigens on the cell membrane and spike. Lymph node samples were stored at −80°C and sent to the Centre for Phenogenomics on dry ice for further sample processing. Tissue sections (8 μm) were cut on a cryostat (Cryostar NX70) and collected on charged slides. Sections were post-fixed in 10% neutral buffered formalin and permeabilized in phosphate-buffered saline containing 0.5% Triton X-100 before immunostaining. Protein Block (Agilent) was used to block nonspecific antibody binding before incubating the sections with primary antibody overnight at 4°C. After washing in TBST, the sections were incubated in fluorophore-conjugated secondary antibodies for 1 h at room temperature. Lymph node tissue sections were stained with human anti-spike antibody P2B-2F6 (45) (1:50) and biotinylated goat anti-human secondary antibody (Abcam, catalog no. ab7152, 1:300), followed by streptavidin-horseradish peroxidase reagent (Vectastain Elite ABC-HRP Kit, Vector, catalog no. PK-6100) then DAB (ImmPACT DAB, Vector, catalog no. SK-4105) to study the distribution and retention of the S2GΔHR2 spike alone and S2GΔHR2-presenting E2p and I3–01v9 SApNPs. For immunofluorescent staining, tissue sections were stained for FDCs using anti-CD21 antibody (Abcam, catalog no. ab75985, 1:1800) followed by anti-rabbit secondary antibody conjugated with Alexa Fluor 555 (Thermo Fisher, catalog no. A21428; 1:200), for B cells using anti-B220 antibody (eBioscience, catalog no. 14-0452-82, 1:100) followed by anti-rat secondary antibody conjugated with Alexa Fluor 674 (Thermo Fisher, catalog no. A21247; 1:200), and for subcapsular sinus macrophages using anti-sialoadhesin (CD169) antibody (Abcam, catalog no. ab53443, 1:600) followed by anti-rat secondary antibody conjugated with Alexa Fluor 488 (Abcam, catalog no. ab150165; 1:200). Germinal center B cells were labeled using rat anti-GL7 antibody (FITC; BioLegend, catalog no. 144604, 1:250). T Follicular helper cells were labeled using CD4 antibody (Biolegend, catalog no. 100402, 1:100) followed by anti-rat secondary antibody conjugated with Alexa Fluor 488 (Abcam, catalog no. ab150165; 1:1000) and Bcl6 antibody (Abcam, catalog no. ab220092, 1:300) followed by anti-rabbit secondary antibody conjugated with Alexa Fluor 555 (Thermo Fisher, catalog no. A21428; 1:1000). Nuclei were then counterstained with DAPI (Sigma-Aldrich, catalog no. D9542, 100 ng/ml). The stained tissue sections were scanned using an Olympus VS-120 slide scanner and imaged using a Hamamatsu ORCA-R2 C10600 digital camera for all bright-field and fluorescent images. The bright-field images of stained S2GΔHR2 spike and S2GΔHR2-presenting SApNPs in lymph node follicles and fluorescent images of GCs were quantified using ImageJ software (National Institutes of Health) (96).
Electron microscopy analysis of protein nanoparticles and lymph node tissues
Electron microscopy (EM) analysis was performed by the Core Microscopy Facility at The Scripps Research Institute. For negative-staining EM analysis of protein nanoparticles, the S2GΔHR2-10GS-I3-01v9-L7P SApNP samples were prepared at the concentration of 0.01 mg/ml. Carbon-coated copper grids (400 mesh) were glow-discharged and 10 μL of each sample was adsorbed for 2 min. Excess sample was wicked away and grids were negatively stained with 2% uranyl formate for 2 min. Excess stain was wicked away and the grids were allowed to dry. For EM analysis of mouse tissues, the lymph nodes were dissected from each animal and immersed in oxygenated 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1M Na cacodylate buffer (pH 7.4) fixative overnight at 4°C (97). After washing in 0.1 M sodium cacodylate buffer, the tissue samples were post-fixed in buffered 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1-1.5 h at 4°C, rinsed in the same buffer, and then stained en bloc with 0.5% uranyl acetate overnight at 4°C. The tissue samples were washed in double-distilled H2O and dehydrated through a graded series of ethanol followed by acetone, infiltrated with LX-112 (Ladd) epoxy resin, and polymerized at 60°C. Ultrathin lymph node sections (at 70-nm thickness) were prepared for imaging. Samples were analyzed at 80 kV with a Talos L120C transmission electron microscope (Thermo Fisher) and images were acquired with a CETA 16M CMOS camera.
Lymph node disaggregation, cell staining, and flow cytometry
Germinal center reactions, including the percentage of GC B cells (GL7+B220+) and T follicular helper cells (CD3+CD4+CXCR5+PD1+), and the number of GC B cells and T follicular helper cells were studied by flow cytometry (fig. S5A). The mice were sacrificed 2, 5, and 8 weeks after a single-dose and 2 and 5 weeks after the boost immunization. Fresh axillary, brachial, and popliteal sentinel lymph nodes were collected and mechanically disaggregated. These lymph node samples were merged in enzyme digestion solution containing 958 μl of Hanks’ balanced salt solution (HBSS) buffer (Thermo Fisher Scientific, catalog no. 14185052), 40 μl of 10 mg/ml collagenase IV (Sigma-Aldrich, catalog no. C5138), and 2 μl of 10 mg/ml of DNase (Roche, catalog no. 10104159001) in an Eppendorf tube. After incubation at 37°C for 30 min, lymph node samples were filtered through a 70 μm cell strainer and spun down at 400 × g for 10 min. The supernatant was discarded, and the cell pellet was resuspended in HBSS blocking solution containing 0.5% (w/v) bovine serum albumin and 2 mM EDTA. The nonspecific binding of Fc receptors was blocked using anti-CD16/32 antibody (BioLegend, catalog no. 101302) on ice for 30 min. Cocktail antibodies, Zombie NIR live/dead stain (BioLegend, catalog no. 423106), Brilliant Violet 510 anti-mouse/human CD45R/B220 antibody (BioLegend, catalog no. 103247), FITC anti-mouse CD3 antibody (BioLegend, catalog no. 100204), Alexa Fluor 700 anti-mouse CD4 antibody (BioLegend, catalog no. 100536), PE anti-mouse/human GL7 antibody (BioLegend, catalog no. 144608), Brilliant Violet 605 anti-mouse CD95 (Fas) antibody (BioLegend, catalog no. 152612), Brilliant Violet 421 anti-mouse CD185 (CXCR5) antibody (BioLegend, catalog no. 145511), and PE/Cyanine7 anti-mouse CD279 (PD-1) antibody (BioLegend, catalog no. 135216) were then mixed with the cells and placed on ice for 30 min. After washing the cell with HBSS blocking solution after antibody staining, the samples were fixed using 1.6% paraformaldehyde (Thermo Fisher Scientific, catalog no. 28906) in HBSS on ice for 30 min. The cell samples were stored in HBSS blocking solution for the flow cytometry study. Sample events were acquired by a 5-laser BD Biosciences LSR II analytical flow cytometer with BD FACS Diva 6 software at the Core Facility of The Scripps Research Institute. The data were further processed using FlowJo 10 software.
Statistical analysis
Data were collected from 4–7 mice per group. All of the statistical analyses were performed and graphs were generated using GraphPad Prism 6.01 software. For the antibody analysis, comparisons between different vaccine groups were performed using two-tailed unpaired Student’s t-test. Comparisons of neutralizing antibody titers against SARS-CoV-2 variants were performed using the same plasma samples and analyzed using two-tailed paired Student’s t-test. For the vaccine accumulation and GC study, comparisons between different vaccine groups were performed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison post hoc test. Statistical significance was determined as ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://xxx/xxx/xxx.
fig. S1. Spike and spike-presenting SApNP vaccine-induced neutralizing antibody responses against SARS-CoV-2 variants of concern (VOCs).
fig. S2. SARS-CoV-2 spike-presenting SApNP interaction with macrophages in a lymph node.
fig. S3. TEM images of SARS-CoV-2 spike-presenting I3–01v9 SApNP interaction with FDCs in a lymph node.
fig. S4. Immunohistological analysis of SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.
fig. S5. Flow cytometry analysis of SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.
fig. S6. Adjuvant effect on SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.
Acknowledgements
Funding:
This work was funded by National Institutes of Health grants AI137472, AI139092 (to J.Z.), Ufovax/SFP-2018–0416, Ufovax/SFP-2018–1013, and Ufovax/SFP-2020–0111 (to J.Z.). Y.-N.Z. thanks the Natural Sciences and Engineering Research Council of Canada (NSERC) for a postdoctoral fellowship. We thank V. Bradaschia, Kyle Duffin, and M. Ganguly at the Centre for Phenogenomics for their expertise in histology and immunostaining. We acknowledge the expert assistance of S. Henderson, K. Vanderpool, and T. Fassel at the Core Microscopy Facility at The Scripps Research Institute. We thank A. Saluk, B. Seegers, and B. Monteverde at the Flow Cytometry Core Facility of The Scripps Research Institute for their expertise in flow cytometry. The authors thank M. Arends for proofreading the manuscript.
Footnotes
Competing interests: The authors declare no competing interests.
Data and material availability: All data are available in the main text or in the supplementary materials. Additional data related to this paper may be requested from the corresponding author.
References
- 1.Dan J. M. et al. , Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isho B. et al. , Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 5, eabe5511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer A. S. et al. , Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 5, eabe0367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y. et al. , Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell 183, 1496–1507.e1416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marot S. et al. , Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat. Commun. 12, 844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burioni R., Topol E. J., Assessing the human immune response to SARS-CoV-2 variants. Nat. Med., Published Ahead-of-Print (2021). [DOI] [PubMed] [Google Scholar]
- 7.Tegally H. et al. , Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature, Published Ahead-of-Print (2021). [Google Scholar]
- 8.Challen R. et al. , Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 372, n579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirby T., New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 9, e20–e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies N. G. et al. , Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science, eabg3055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washington N. L. et al. , Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv, 2021.2002.2006.21251159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabino E. C. et al. , Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 397, 452–455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faria N. R. et al. , Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv, 2021.2002.2026.21252554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascola J. R., Graham B. S., Fauci A. S., SARS-CoV-2 viral variants—tackling a moving target. JAMA, Published Ahead-of-Print (2021). [DOI] [PubMed] [Google Scholar]
- 15.Zimmer C., Corum J., S.-L. W., in Coronavirus vaccine tracker. (https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html).
- 16.Dagan N. et al. , BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med., Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baden L. R. et al. , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams T. C., Burgers W. A., SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir. Med., Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logunov D. Y. et al. , Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 397, 671–681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polack F. P. et al. , Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q. et al. , The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182, 1284–1294.e1289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rees-Spear C. et al. , The impact of spike mutations on SARS-CoV-2 neutralization. bioRxiv, 2021.2001.2015.426849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wibmer C. K. et al. , SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv, 2021.2001.2018.427166 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Burton D. R., Topol E. J., Variant-proof vaccines - invest now for the next pandemic. Nature 590, 386–388 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Voysey M. et al. , Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreano E. et al. , SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv, 2020.2012.2028.424451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M. et al. , SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell, Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Beltran W. F. et al. , Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell, Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P. et al. , Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. bioRxiv, 2021.2001.2025.428137 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Supasa P. et al. , Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell, Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collier D. A. et al. , SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv, 2021.2001.2019.21249840 (2021). [Google Scholar]
- 32.Wang Z. et al. , mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature, Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q. et al. , No higher infectivity but immune escape of SARS-CoV-2 501Y.V2 variants. Cell, Published Ahead-of-Print (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A., Eliciting B cell immunity against infectious diseases using nanovaccines. Nat. Nanotechnol. 16, 16–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Rappuoli R., Glycoconjugate vaccines: principles and mechanisms. Sci. Transl. Med. 10, eaat4615 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Cyster J. G., Allen C. D. C., B cell responses: cell Interaction dynamics and decisions. Cell 177, 524–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHeyzer-Williams M., Okitsu S., Wang N., McHeyzer-Williams L., Molecular programming of B cell memory. Nat. Rev. Immunol. 12, 24–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akkaya M., Kwak K., Pierce S. K., B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. 20, 229–238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Silva N. S., Klein U., Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 15, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L. et al. , Single-component, self-assembling, protien nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 7, eabf1591 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford K. H. D. et al. , Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wrapp D. et al. , Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto D. et al. , Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Ju B. et al. , Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y.-N. et al. , Nanoparticle size influences antigen retention and presentation in lymph node follicles for humoral immunity. Nano Lett. 19, 7226–7235 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Heesters B. A., Myers R. C., Carroll M. C., Follicular dendritic cells: dynamic antigen libraries. Nat. Rev. Immunol. 14, 495–504 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Cyster J. G., B cell follicles and antigen encounters of the third kind. Nat. Immunol. 11, 989–996 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Batista F. D., Harwood N. E., The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 9, 15–27 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Kuka M., Iannacone M., Viral subversion of B cell responses within secondary lymphoid organs. Nat. Rev. Immunol. 18, 255–265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rappuoli R., Serruto D., Self-assembling nanoparticles usher in a new era of vaccine design. Cell 176, 1245–1247 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Allen C. D. C., Okada T., Cyster J. G., Germinal-center organization and cellular dynamics. Immunity 27, 190–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen C. D. C., Cyster J. G., Follicular dendritic cell networks of primary follicles and germinal centers: Phenotype and function. Semin. Immunol. 20, 14–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mesin L., Ersching J., Gabriel D. Victora, Germinal center B cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumgarth N., The shaping of a B cell pool maximally responsive to infections. Annu. Rev. Immunol. 39, (2021). [DOI] [PubMed] [Google Scholar]
- 56.López-Sagaseta J., Malito E., Rappuoli R., Bottomley M. J., Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 58–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irvine D. J., Hanson M. C., Rakhra K., Tokatlian T., Synthetic nanoparticles for vaccines and immunotherapy. Chem. Rev. 115, 11109–11146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irvine D. J., Swartz M. A., Szeto G. L., Engineering synthetic vaccines using cues from natural immunity. Nat. Mater. 12, 978–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crotty S., Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Szakal A K, Kosco a. M H, Tew J. G., Microanatomy of lymphoid tissue during humoral immune responses: structure function relationships. Annu. Rev. Immunol. 7, 91–109 (1989). [DOI] [PubMed] [Google Scholar]
- 61.Merkenschlager J. et al. , Dynamic regulation of TFH selection during the germinal centre reaction. Nature 591, 458–463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shulman Z. et al. , T follicular helper cell dynamics in germinal centers. Science 341, 673–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lederer K. et al. , SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53, 1281–1295.e1285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokatlian T. et al. , Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller S. N., Tian S., DeSimone J. M., Rapid and persistent delivery of antigen by lymph node targeting PRINT nanoparticle vaccine carrier to promote humoral immunity. Mol. Pharm. 12, 1356–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B. et al. , A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 10, 18149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walls A. C. et al. , Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e1317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Powell A. E. et al. , A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 7, 183–199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brouwer P. J. M. et al. , Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell 184, 1188–1200.e1119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park K. S. et al. , Lipid-based vaccine nanoparticles for induction of humoral immune responses against HIV-1 and SARS-CoV-2. J. Control. Release 330, 529–539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang Y.-F. et al. , Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano 15, 2738–2752 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Lam J. H. et al. , Next generation vaccine platform: polymersomes as stable nanocarriers for a highly immunogenic and durable SARS-CoV-2 spike protein subunit vaccine. bioRxiv, 2021.2001.2024.427729 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Cohen A. A. et al. , Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang W. C. et al. , SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv. Mater. 32, e2005637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan T. K. et al. , A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 12, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lainšček D. et al. , Immune response to vaccine candidates based on different types of nanoscaffolded RBD domain of the SARS-CoV-2 spike protein. bioRxiv, 2020.2008.2028.244269 (2020). [Google Scholar]
- 77.Ma X. et al. , Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 53, 1315–1330.e1319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalvie N. C. et al. , Engineered SARS-CoV-2 receptor binding domain improves immunogenicity in mice and elicits protective immunity in hamsters. bioRxiv, 2021.2003.2003.433558 (2021). [Google Scholar]
- 79.Rappuoli R. et al. , Vaccinology in the post−COVID-19 era. Proc. Natl. Acad. Sci. U.S.A. 118, e2020368118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeyanathan M. et al. , Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 20, 615–632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corey L., Mascola J. R., Fauci A. S., Collins F. S., A strategic approach to COVID-19 vaccine R&D. Science 368, 948–950 (2020). [DOI] [PubMed] [Google Scholar]
- 82.DeFrancesco L., Whither COVID-19 vaccines? Nat. Biotechnol. 38, 1132–1145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arunachalam P. S. et al. , Adjuvanting a subunit SARS-CoV-2 nanoparticle vaccine to induce protective immunity in non-human primates. bioRxiv, 2021.2002.2010.430696 (2021). [Google Scholar]
- 84.Hotez P. J., Corry D. B., Strych U., Bottazzi M. E., COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat. Rev. Immunol. 20, 399–400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kasturi S. P. et al. , Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470, 543–547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reed S. G., Orr M. T., Fox C. B., Key roles of adjuvants in modern vaccines. Nat. Med. 19, 1597–1608 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Kuo T.-Y. et al. , Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci. Rep. 10, 20085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian J.-H. et al. , SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 12, 372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keech C. et al. , Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quast I., Tarlinton D., B cell memory: understanding COVID-19. Immunity 54, 205–210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Irvine D. J., Read B. J., Shaping humoral immunity to vaccines through antigen-displaying nanoparticles. Curr. Opin. Immunol. 65, 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shin M. D. et al. , COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 15, 646–655 (2020). [DOI] [PubMed] [Google Scholar]
- 93.He L. et al. , Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines. Sci. Adv. 6, eaaz6225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He L. et al. , Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines. bioRxiv, 2020.2008.2022.262634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He L. et al. , HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 4, aau6769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson T. J., Glutaraldehyde fixation chemistry: oxygen-consuming reactions. Eur. J. Cell Biol. 45, 160–169 (1987). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://xxx/xxx/xxx.
fig. S1. Spike and spike-presenting SApNP vaccine-induced neutralizing antibody responses against SARS-CoV-2 variants of concern (VOCs).
fig. S2. SARS-CoV-2 spike-presenting SApNP interaction with macrophages in a lymph node.
fig. S3. TEM images of SARS-CoV-2 spike-presenting I3–01v9 SApNP interaction with FDCs in a lymph node.
fig. S4. Immunohistological analysis of SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.
fig. S5. Flow cytometry analysis of SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.
fig. S6. Adjuvant effect on SARS-CoV-2 spike/spike-presenting SApNP vaccine-induced GCs.