Abstract
In this communication, we report on the genomic surveillance of SARS-CoV-2 using wastewater samples in Jefferson County, KY. In February 2021, we analyzed seven wastewater samples for SARS-CoV-2 genomic surveillance. Variants observed in smaller catchment areas, such as neighborhood manhole locations, were not necessarily consistent when compared to associated variant results in downstream treatment plants, suggesting catchment size or population could impact the ability to detect diversity.
The successful viral detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater at various pooled scales (1–4) and discovery in the USA of B.1.1.7, B.1.351 and P.1 variants (5), has led to an interest in developing reliable population-level wastewater viral genomic surveillance.
The diversity of SARS-CoV-2 sequences reported to be circulating in the USA, have been determined by sequencing clinical samples; however, these variants can also be surveilled by sequencing wastewater samples (6–9). As of March 2021, the variants of concern - B.1.1.7, B.1.351, and P.1 have been widely detected in clinical samples from 47 states in the USA. In Kentucky, only five clinical cases have been linked to the presence of these variants (5), which could indicate incomplete surveillance. Broadening the application of genomic surveillance using wastewater in the community could enhance SARS-CoV-2 variant population monitoring.
In this communication, we report on the genomic surveillance of SARS-CoV-2 using wastewater samples in Jefferson County, KY. Samples were collected from manholes and treatment facilities, covering populations of 8,000 to 350,000 people (Table 1). RNA isolated from wastewater samples was used to quantify SARS-CoV-2 and analyze the genetic variation through high-throughput sequencing (See Supplementary Methods). Bioinformatics approaches were used to rapidly identify single nucleotide genetic alterations, which were compared with known variants of interest and concern.
Table 1.
Summary of wastewater SARS-CoV-2 samples sequenced in this study, Louisville, KY
| Sample ID | Sewershed population | Location | N1 (Ct) | Sequencing BWA Alignment Rate (%) |
|---|---|---|---|---|
| 833 | 35,956 | Street line manhole leading to Treatment Plant #3a | 28 | 28.02 |
| Treatment Plant #1 | 55,928 | Treatment Plant | 30 | 21.09 |
| 847 | 10,739 | Street line manhole leading to Treatment Plant #2 | 29 | 15.08 |
| 849 | 35,956 | Street line manhole leading to Treatment Plant #3a | 28 | 12.61 |
| 884 | 46,659 | Street line manhole leading to Treatment Plant #3a | 29 | 23.98 |
| 891 | 8,071 | Street line manhole leading to Treatment Plant #2 | 29 | 26.03 |
| Treatment Plant #2 | 349,850 | Treatment Plant | 31 | 19.96 |
Treatment Plant #3 samples had SARS-CoV-2 was detected but were below the threshold for individual mutations for review.
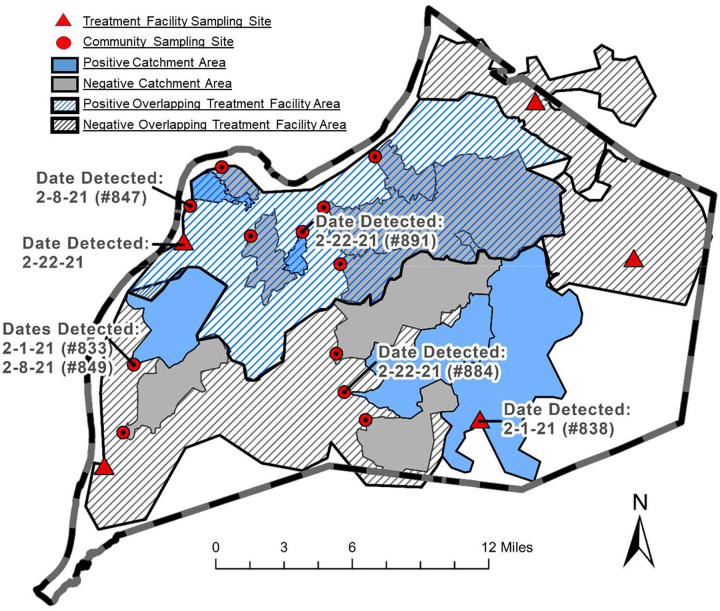
In February 2021, we analyzed seven wastewater samples for SARS-CoV-2 genomic surveillance (Figure 1). We did not detect genetic variations indicative of any current variant of concern, beyond the widespread D614G spike protein mutation (Supplementary Methods Tables 2–5). In all samples, we identified at least four of ten mutations consistent with the presence of the variant of interest B.1.429, and one sample contained seven of ten mutations (Table 2). The B.1.429 variant was confirmed in patient samples in Kentucky in January 2021 (10), and a single patient in the study area was reported to be positive for B.1.1.7 on February 9, 2021 (11). With our current metrics we flagged sites 833, 891, and Treatment plant #2 for potential presence of variant B.1.429 (3/7 sites). Differences in the scale of sample pooling in the community revealed unanticipated inconsistencies in variant representation. Specifically, variants observed in smaller catchment areas, such as neighborhood manhole locations, were not observed in downstream treatment plants, suggesting catchment size or population could impact the ability to detect diversity.
Figure 1. Study sites within Louisville, KY.
Distribution of the sewershed area, treatment plants and community locations, in Jefferson County with corresponding dates, sampled. SARS-CoV-2 was detected at all sites. Samples that contained at least 50% of the single amino acid mutations for a variant with a nucleotide frequency above a 5 % threshold for individual mutations are flagged for review. This relatively low threshold serves the purpose of identifying geographic (sewershed) areas for heightened public health surveillance. With our current metrics we flagged sites 833, 891, and Treatment plant #2 for potential presence of variant B.1.429.
Table 2.
Summary of B.1.429 specific mutation prevalence by sample
| Ref Pos | Gene/ORF | Ref Allele | Alt Allele | Variant Desc | 833 | Treatment Plant #1 | 847 | 849 | 884 | 891 | Treatment Plant #2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1059 | ORF1ab1 | C | T | T265I | 0.9309 | 0.8798 | 0.9823 | 0.8906 | 0.9844 | 0.9773 | 0.7382 |
| 12878 | ORF1ab1 | A | G | 14205V | 0.2084 | 0 | 0 | 0.0015 | 0 | 0.0504 | 0.9968 |
| 14408 | ORF1ab2 | C | T | P314L | 1 | 1 | 0.9975 | 1 | 0.909 | 0.8757 | 0.8537 |
| 17014 | ORF1ab2 | G | T | D1183Y | 0.049 | 0.0051 | 0 | 0.025 | 0.0024 | 0.0026 | 0.0027 |
| 21600 | S | G | T | S13I | 0 | 0 | 0 | 0 | 0 | 0.0025 | 0 |
| 22018 | S | G | T | W152C | 0.1287 | 0 | 0 | 0 | 0.002 | 0.0022 | 0.0016 |
| 22917 | S | T | G | L452R | 0.1297 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23403 | S | A | G | D614G | 0.9972 | 1 | 0.9969 | 1 | 0.9969 | 0.9977 | 0.9981 |
| 25563 | ORF3a | G | T | Q57H | 0.9893 | 0.6967 | 0.9621 | 0.9987 | 0.8682 | 0.7933 | 0.4046 |
| 28887 | N | C | T | T205I | 0.0422 | 0.0426 | 0 | 0.0017 | 0 | 0 | 0 |
Given the highly variable viral genome sequence coverage recovered from wastewater samples, there is an urgent need to develop a set of consistent thresholds constituting positive/negative presence of a variant. Monitoring SARS-CoV-2 variants in wastewater may warn of an emerging variant of concern and identify variant dominance occurring when a new variant is introduced in a community. Wastewater genetic monitoring may be particularly useful in the context of limited clinical sample sequencing capacity because a broad perspective on the genetic diversity can be obtained from a few samples. To develop comprehensive epidemiological frameworks required to guide policy, population-level wastewater surveillance of viral genetic diversity should be complemented by clinical sample testing.
Supplementary Material
Acknowledgments:
This project was supported by funding from Louisville Metro Public Health and Wellness. Part of this work was performed with assistance of the UofL Genomics Facility, which was supported by NIH/NIGMS KY-INBRE P20GM103436, the J.G. Brown Cancer Center, University of Louisville, and user fees. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: Quantitative PCR (qPCR) Control RNA from Heat-Inactivated SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR 52347.
Footnotes
Ethics: The University of Louisville Institutional Review Board classified this project as Non-Human Subjects Research (NHSR) (reference #: 717950).
Competing Interests: The authors have no conflicts to report related to the submitted work.
References
- 1.Betancourt WW, et al. (2020) Wastewater-based Epidemiology for Averting COVID-19 Outbreaks on The University of Arizona Campus. medRxiv:2020.2011.2013.20231340. [Google Scholar]
- 2.Wu F, et al. (2020) SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weidhaas J, et al. (2021) Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci Total Environ 775:145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeager RA, et al. (2020) Wastewater sample site selection to estimate geographically-resolved community prevalence of COVID-19: A research protocol. medRxiv:2020.2008.2023.20180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC (Updated March 8, 2021) Variant Cases. (https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html). [Google Scholar]
- 6.Fontenele RS, et al. (2021) High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahn K, et al. (2021) Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv:2021.2001.2008.21249379. [Google Scholar]
- 8.Crits-Christoph A, et al. (2021) Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemudryi A, et al. (2020) Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep Med 1(6):100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadfield J, et al. (2018) Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34(23):4121–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louisville Metro Public Health & Wellness (2021) Louisville COVID-19 Briefing. (https://louisvilleky.gov/government/louisville-covid-19-resource-center).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.