Abstract
The proliferation and migration of pulmonary artery endothelial cells are the pathological basis of pulmonary vascular remodeling with pulmonary hypertension. Recent studies have shown that circular RNA (circRNA) regulates biological processes in various vascular diseases, including pulmonary arterial hypertension. It has been reported that circRNA regulates the vascular endothelial cells’ function. Therefore, circRNA may have crucial roles in human pulmonary artery endothelial cells (hPAECs) proliferation, migration, and tube formation in pulmonary arterial hypertension. In this study, we aimed to discover the role and mechanism of circular RNA HIPK3 (circHIPK3) in the proliferation and migration of pulmonary hypertension hPAECs. First, we used platelet-derived growth factor—stimulated hPAECs as a cellular model of pulmonary arterial hypertension. The results showed that platelet-derived growth factor promoted hPAECs proliferation, migration, and tube formation. Notably, platelet-derived growth factor upregulated the expression of circHIPK3 in hPAECs and regulated their proliferation, migration, and angiogenesis. Mechanistically, we confirmed miR-328-3p was copiously pulled down by circHIPK3 in hPAECs. Luciferase reporter and RNA immunoprecipitation assays further indicated the cytoplasmic interactions between circHIPK3 and miR-328-3p. Subsequently, we found that circHIPK3 might increase the expression of STAT3 by sponging miR-328-3p. Collectively, our results demonstrated that the circHIPK3-miR-328-3p-STAT3 axis contributed to the pathogenesis of pulmonary arterial hypertension by stimulating hPAECs proliferation, migration, and angiogenesis. The circHIPK3 has an accelerated role in pulmonary arterial hypertension development, implicating the potential values of circHIPK3 in pulmonary arterial hypertension therapy.
Keywords: pulmonary arterial hypertension, circHIPK3, miR-328-3p, STAT3, hPAECs
Introduction
Pulmonary arterial hypertension (PAH), also known as World Health Organization Group I pulmonary hypertension, is an insidious disease that is associated with a poor long-term prognosis.1 This pulmonary vascular disease is characterized by a sustained increase in pulmonary arterial pressure, increased pulmonary vascular resistance, and remodeling of small pulmonary vessels.2 The proliferation and migration of endothelial cells are essential for angiogenesis, and endothelial dysfunction is the key triggering factor of PAH pathophysiology.3 Recent research work has provided insights into the pathogenesis of PAH, and several potential mechanisms have been identified, including changes in growth factor pathways, cytokines, inflammation, and mitochondrial metabolism.4
Pulmonary arterial remodeling plays an essential role in PAH progression, especially in the late phase of the disease. Previous studies have suggested that platelet-derived growth factor (PDGF) and fibroblast growth factor (FGF) signaling contribute to vascular cell proliferation, migration, differentiation, and apoptosis.5 PDGF has been regarded as a novel potential therapeutic target in PAH.6 Here, PDGF-treated human pulmonary artery endothelial cells (hPAECs) were used to construct cellular models.
Circular RNAs (circRNAs) are a class of noncoding RNAs widespread in mammals and are mainly involved in gene regulation in organisms.7,8 Most circRNAs are derived from exonic regions of genes, and a small proportion also has intron splicing formation.9 Unlike long noncoding RNAs and microRNAs, circRNAs do not have 5′-end and 3′-end structures but are formed by covalently closed-loop structures.10 circRNAs are widely involved in physiological and pathological regulation in humans. Circular RNA HIPK3 (circHIPK3) is highly expressed in the liver, brain, and lungs, mainly originating from the second exon of the gene HIPK3. There are three spliceosomes of HIPK3-derived circRNAs, which are circHIPK3, circHIPK3.1, and circHIPK3.2. In contrast, only circHIPK3 is highly expressed and has a significant function in cells. CircHIPK3 has been found to play essential roles in various vascular diseases.11–13 PAH is a disease associated with vascular remodeling.14 However, the expression and regulatory mechanism of circHIPK3 in PAH have not been clarified. Therefore, in this study, we explored the involvement of circHIPK3 in the progress of PAH. Studies have confirmed that circHIPK3 is located in the cytoplasm.15 circRNA competitively binds to miRNAs and regulates the expression of downstream target genes when it is located in the cytoplasm.16 MicroRNAs (miRNAs) are a class of small noncoding RNAs, including 20∼25 nucleotides, which regulate the expression levels of target genes by binding to the 3′-untranslated region (UTR) of mRNAs, degrading mRNAs, or inhibiting their translation process.17 Multiple studies have suggested that miRNAs play an essential regulatory role in the development of the pathophysiological process. Besides, miRNAs have gained considerable attention as regulators of gene expression and are known to play crucial roles in the prevention and treatment of PAH.18,19
This study used PDGF-treated hPAECs to investigate the effects of PDGF on proliferation, migration, and angiogenesis. We mainly reported the function of circHIPK3 in PAH and preliminarily proposed that circHIPK3 could promote the proliferation, migration, and angiogenesis of hPAECs through miR-328-3p/STAT3. This study may provide a theoretical basis for PAH treatment.
Materials and methods
Cell culture and PDGF treatment
hPAECs and 293T cells were obtained from CCTCC (China Center for Type Culture Collection, Wuhan, China). Cells were cultured with sterile endothelial growth medium (EGM-2, Clonetics, Walkersville, MD, USA) in a 37°C humidified atmosphere containing 5% CO2. PDGF can act on vascular endothelial cells as the incentive of PAH.20 We used 10 ng/mL, 20 ng/mL, and 100 ng/mL PDGF (Sigma-Aldrich, Shanghai, China) to treat hPAECs for 4 h, then the hPAECs were collected for further experiments.
Quantitative real-time polymerase chain reaction
RNAiso (Takara, Otsu, Japan) was utilized to extract total RNA from hPAECs. The complementary DNA (cDNA) was acquired by reverse transcription using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and all-in-one miRNA Reverse Transcription Kit (GeneCopoeia, USA). Then, quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the FastStart Universal SYBR Green Master (Roche, Hangzhou, China) or SYBR Green Human miRNA Assay Kit (GeneCopoeia, USA) on an ABI 7500 PCR instrument to quantitate circRNAs, miRNAs, and mRNA expression. Glycerldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control for circRNAs and mRNA. U6 was used as the internal reference for miRNAs. The primer sequences used for qRT-PCR are shown in Table 1.
Table 1.
Sequences of the primers for real-time PCR.
| Primers | Sequences (5′-3′) |
|---|---|
| hsa-circHIPK3 forward | TTCAACATGTCTACAATCTCGGT |
| hsa-circHIPK3 reverse | ACCATTCACATAGGTCCGT |
| hsa-circASXL1 forward | TAAACTGCCTGGCCGAATC |
| hsa-circASXL1 reverse | TCCTTCTGCCTCTATGACCTG |
| hsa-circSMARCA5 forward | ACAATGGATACAGAGTCAAGTGTT |
| hsa-circSMARCA5 reverse | CCACAAGCCTCCCTTTTGTTTT |
| hsa-circSETD3 forward | TGAAGAAGATGAAGTTCGGTAT |
| hsa-circSETD3 reverse | GTGCCAGATTTCTGAGTTTT |
| hsa-circFAT3 forward | AATCTGAGATTACAGCCTGCTTCCC |
| hsa-circFAT3 reverse | TTCTCCACACAAGGATCGTTGGACC |
| hsa-circNCX1 forward | GACAGCTTTCATTGGAGACCTGGCT |
| hsa-circNCX1 reverse | GCTCTCCCAGGATGGGGCGCCCCAT |
| hsa-circRHOBTB3 forward | GAAGTTGAAAGATTCTGGGGA |
| hsa-circRHOBTB3 reverse | ACTGGCAGCAGAACAGCAAG |
| hsa-circ-0124644 forward | TGCCTTGGAGTTATGGAGACAGA |
| hsa-circ-0124644 reverse | TCTGGCCGGTGAGATACAAGT |
| hsa-circZNF292 forward | AAGAGACTGGGGTGTGGAAA |
| hsa-circZNF292 reverse | TCTGAAGTTTTCCATTTCTCTGC |
| hsa-circ-001569 forward | TCCCCTGAACATTCTCCCCAT |
| hsa-circ-001569 reverse | GAAAGCACTTGGTGAAGTCGG |
| has-miR-328-3p forward | CCTGGCCCTCTCTGCCCT |
| has-miR-328-3p reverse | GTGCAGGGTCCGAGGTATTC |
| Has-miR-199a-5p forward | CGCGCCCAGTGTTCAGACTAC |
| has-miR-199a-5p reverse | AGTGCAGGGTCCGAGGTATT |
| has-miR-143 forward | CGCGTGAGATGAAGCACTG |
| has-miR-143 reverse | AGTGCAGGGTCCGAGGTATT |
| has-let-7b forward | GCGCGTGAGGTAGTAGGTTGT |
| has-let-7b reverse | AGTGCAGGGTCCGAGGTATT |
| has-miR-124 forward | CGTAAGGCACGCGGTGAA |
| has-miR-124 reverse | AGTGCAGGGTCCGAGGTATT |
| has-miR-138 forward | GCGAGCTGGTGTTGTGAATC |
| has-miR-138 reverse | AGTGCAGGGTCCGAGGTATT |
| has-miR-25 forward | GCGCATTGCACTTGTCTCG |
| has-miR-25 reverse | AGTGCAGGGTCCGAGGTATT |
| has-miR-34a forward | CGCGTGGCAGTGTCTTAGCT |
| has-miR-34a reverse | AGTGCAGGGTCCGAGGTATT |
| bFGF forward | GCGACCCTCACATCAAGCTA |
| bFGF reverse | AGCCAGGTAACGGTTAGCAC |
| VEGF forward | ACAACAAATGTGAATGCAGACCA |
| VEGF reverse | GAGGCTCCAGGGCATTAGAC |
| STAT3 forward | CAGCAGCTTGACACACGGTA |
| STAT3 reverse | AAACACCAAAGTGGCATGTGA |
| GAPDH forward | CGCTCTCTGCTCCTCCTGTTC |
| GAPDH reverse | ATCCGTTGACTCCGACCTTCAC |
| U6 forward | GCTTCGGCAGCACATATACT |
| U6 reverse | GTGCAGGGTCCGAGGTATTC |
Cell transfection
For transient transfection, circHIPK3 siRNAs, scrambled siRNAs, miR-328-3p mimics, negative control (NC) mimics, miR-328-3p antisense oligonucleotides (miR-328-3p inhibitor), NC inhibitor, STAT3 overexpression vector pcDNA-STAT3 (STAT3), and empty vector pcDNA (vector) were purchased from GenePharma (Shanghai, China). Lipofectamine® 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) was used for cell transfection. Cells of 104 were seeded in six-well plates and cultured for 24 h. When the confluence reached up to 50% to 60%, transfection was performed using Lipofectamine 3000 following the manufacturer’s protocol. The sequences for these siRNAs are shown in Table 2.
Table 2.
The siRNAs, miRNA mimics, and inhibitor sequences.
| Name | Sequences (5′-3′) |
|---|---|
| circHIPK3 siRNA1 | GGUACUACAGGUAUGGCCUTT |
| AGGCCAUACCUGUAGUACCTT | |
| circHIPK3 siRNA2 | CUACAGGUAUGGCCUCACATT |
| UGUGAGGCCAUACCUGUAGTT | |
| circHIPK3 siRNA3 | CCCGGUAUUAUAGGUAUGGTT |
| CCAUACCUAGAAUACCGGGTT | |
| miR-328-3p mimics | CUGGCCCUCUCUGCCCUUCCGU |
| GGAAGGGCAGAGAGGGCCAGUU | |
| NC mimics | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT | |
| miR-328-3p inhibitor | CUGGCCCUCUCUGCCCUUCCGU |
| NC inhibitor | UUGUACUACACAAAAGUACUG |
To establish a stable overexpression cell line, lentiviral circHIPK3 plasmid and negative lentiviral vector were designed and produced by GenePharma (Shanghai, China). Cells were transfected with lentiviral at a multiplicity of infection of 20. The stable overexpressed cells were selected by using Puromycin methods.
CCK-8 assay
The Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to measure cell viability. Briefly, hPAECs were cultured in 96-well plates at a density of 1 × 104 cells per well. Subsequently, the cells were treated with 10 μL CCK-8 solution at 37°C for 90 min. At indicated time points, the absorbance was assessed at 450 nm.
Wound healing
A wound-healing assay was performed to measure the migration of hPAECs. A sterile Eppendorf pipette tip was used to scratch a wound along the plate diameter when hPAECs reached 80% to 90% confluence. Being washed with phosphate-buffered saline, the phase-contrast image was taken with an Olympus inverted microscope (IX51, Olympus, Japan). After 24 h, the second image was captured. Relative migration was characterized as the percentage of wound area compared to the initial wound area.
Cell migration assay
Transwell assay was performed using 24-well Transwell chambers (3422, Corning, Toledo, USA). At 24 h after transfection, we placed hPAECs at a density of 5 × 104 cells into the upper chamber with a serum-free medium and filled the bottom chambers with a medium with 10% fetal bovine serum (FBS). After incubation at 37°C for 48 h, hPAECs on the underside were first fixed with 1% formaldehyde solution and then stained with crystal violet. Cells of ten fields were randomly selected and counted using an inverted microscope. Experiments were conducted in triplicate.
Tube formation assay
The hPAECs were used in the tube formation assay. A total of 50 μL of matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added to each well of a prechilled 96-well plate and then incubated for 30 min at 37°C to form a gel. Next, the hPAECs were digested and transferred to the wells. For each well, 3 × 104 cells were plated in 100 μL medium. The cells were incubated at 37°C for 4 h, and the endothelial tubes were observed using a light microscope. Photographs were captured with a Nikon inverted microscope (Nikon, Tokyo, Japan), and the tubes were counted and analyzed.
Western blot analysis
Protein extracts were separated by sodium dodecyl sulfate (SDS)-acrylamide and transferred to the nitrocellulose membrane. The proteins on the membranes were combined with primary antibodies anti-bFGF (ab92337, Abcam, 1:1000), anti-vascular endothelial growth factor (anti-VEGF; ab32152, Abcam, 1:1000), anti-STAT3 (ab68153, Abcam, 1:1000), and anti-GAPDH (ab8245, Abcam, 1:3000). The membranes were incubated with horseradish peroxidase-conjugated secondary antibody. GAPDH was used as a control. The enhanced chemiluminescence (ECL) Substrate Kit (Abcam, Shanghai, China) was applied to examine the conjugated signals, and the data analysis was implemented by the ImageJ program (Verse1.52v, National Institutes of Health).
Dual-luciferase reporter assay
To further confirm that miR-328-3p could directly target circHIPK3 and STAT3, a dual-luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The luciferase reporter plasmids pGL3-circHIPK3 wild-type (circHIPK3-WT) and pGL3-STAT3 3′-UTR wild-type (STAT3-WT) containing the binding sequence of miR-328-3p were designed and synthesized in Invitrogen. pGL3-circHIPK3 mutant type (circHIPK3-MUT) and pGL3-STAT3 3′-UTR mutant type (STAT3-MUT) containing the mutated miR-328-3p binding sequence were designed and synthesized. Experiments were performed on 24-well plates. The cells were transfected with a mixture of 0.5 μg circHIPK3 luciferase reporter plasmid or STAT3 luciferase reporter plasmid, 0.5 μg Renilla luciferase reporter vectors (pRL-TK), and 100 pmol miR-328-3p mimics or inhibitor. At 30 h posttransfection, cells were lysed by 100 μL of 1× passive lysis buffer with vigorous shaking. According to the manufacturer’s instructions, the cells’ luciferase activities were measured after transfecting for 48 h by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The Renilla luciferase activity was used as an internal control.
RNA immunoprecipitation assay
Following the manufacturer’s instructions, the Magna RNA immunoprecipitation (RIP) RNA-Binding Protein Immunoprecipitation Kit (Millipore, Merck, Darmstadt, Germany) was utilized to perform RIP assay. Briefly, hPAECs were lysed in RIP lysis buffer, and the magnetic beads with AGO2 or IgG antibody were prepared. The immunoprecipitation reactions were carried out by incubating the RIP lysate and the beads-antibody complex together with rotation overnight at 4°C. Immunoprecipitated RNAs were purified and analyzed by qRT-PCR using the respective primers. The results normalized to the input control.
Statistical analysis
Data were expressed as means ± standard deviation (SD) of results from three or more determinations. Data were analyzed using GraphPad Prism software (Verse 8.0, San Diego, CA, USA). When only two groups were compared, a two-tailed t-test was used to determine statistical significance. When more than two groups were compared, a one-way or two-way analysis of variance was utilized. P values <0.05 were considered statistically significant.
Results
Effect of PDGF on the proliferation, migration, and angiogenesis of hPAECs
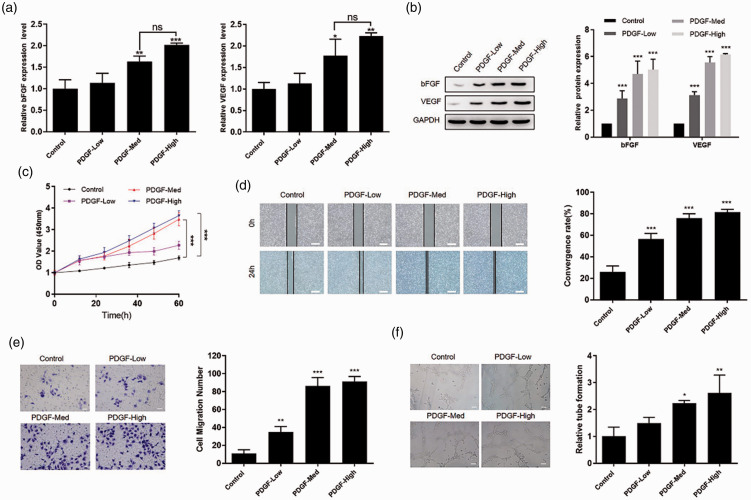
hPAECs were treated with PDGF to induce the PAH model. Basic FGF (bFGF) levels and VEGF as the marker of angiogenesis were increased in PAH patients.21,22 Therefore, we detected the bFGF levels and VEGF levels by qPCR and western blot to confirm the successful model construction. The results showed that PDGF increased bFGF and VEGF expressionin a dose-dependent manner (Fig. 1a and b). To explore the roles of PDGF in hPAECs proliferation, migration, and angiogenesis, CCK-8 assay was used to detect cell proliferation, wound-healing assay and transwell assay were used to detect cell migration, and tube formation assay was employed to detect angiogenesis. The CCK-8 assay revealed that cell proliferation was remarkably increased in a dose-dependent manner after the PDGF treatment with different concentrations (Fig. 1c). Similarly, the wound-healing and transwell assay results showed that cell migration was more significantly increased in PDGF-treated hPAECs than control, and cell migration increased with the increase in PDGF concentration (Fig. 1d and e). Additionally, PDGF increased tube formation of hPAECs in a dose-dependent manner (Fig. 1f). All these data suggested that PDGF promoted cell proliferation, migration, and tube formation in hPAECs. The middle concentration of PDGF (20 ng/mL) was identified and had an evident effect on cell proliferation, cell migration, and tube formation. There were no significant changes in the effects of middle and high PDGF concentrations; therefore, the middle concentration of PDGF was used for further experiments.
Fig. 1.
Effect of PDGF on the function of hPAECs. (a) qRT-PCR for bFGF and VEGF mRNA expression in hPAECs 24 h after PDGF treatment. (b) Western blot for bFGF and VEGF expression in hPAECs 24 h after PDGF treatment. (c) hPAECs were treated with the indicated concentration of PDGF; a CCK-8 assay was used to detect cell proliferation. (d) Representative images and quantitative data of scratch wound-healing migration in hPAECs, scale bar = 25 μm. (e) Transwell assays examined the migration of hPAECs, scale bar = 25 μm. (f) Tube formation assay on hPAECs, scale bar = 25 μm. Data were expressed as mean ± SD in three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. PGDF: platelet-derived growth factor; VEGF: vascular endothelial growth factor; bFGF: basic fibroblast growth factor; ns: not significant.
CircHIPK3 was upregulated in hPAECs under PDGF treatment
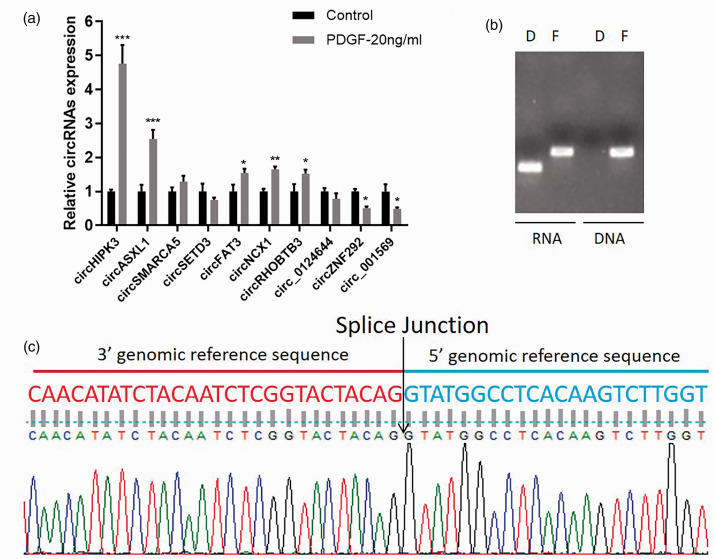
To detect the expression of circRNAs in control and PDGF-treated hPAECs, qRT-PCR was performed, and the results showed that the expression of circHIPK3 was significantly upregulated in PDGF-treated hPAECs compared with the control group (Fig. 2a). Subsequently, we confirmed the head-to-tail splicing of circHIPK3 through RT-PCR and the Sanger sequencing. RT-PCR results revealed that positive amplification products were obtained from cDNA rather than gDNA by circRNA-specific primers, and DNA amplified from cDNA and gDNA using linear primers (Fig. 2b). The sequence of circHIPK3 head-to-tail site is shown in Fig. 2c.
Fig. 2.
CircHIPK3 was upregulated in hPAECs under PDGF treatment. (a) The qPCR verification of 10 randomly selected differentially expressed circRNAs. (b) RT-PCR validation of circHIPK3 in hPAECs cells using circHIPK3 specific primers and HIPK3 linear primers. The circHIPK3 specific primers amplified circHIPK3 from cDNA but not from gDNA. (c) Sanger sequencing peak map of the circHIPK3. Data were expressed as mean ± SD in three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. PGDF: platelet-derived growth factor.
Knockdown of circHIPK3 inhibited PDGF-induced proliferation, migration and angiogenesis of hPAECs
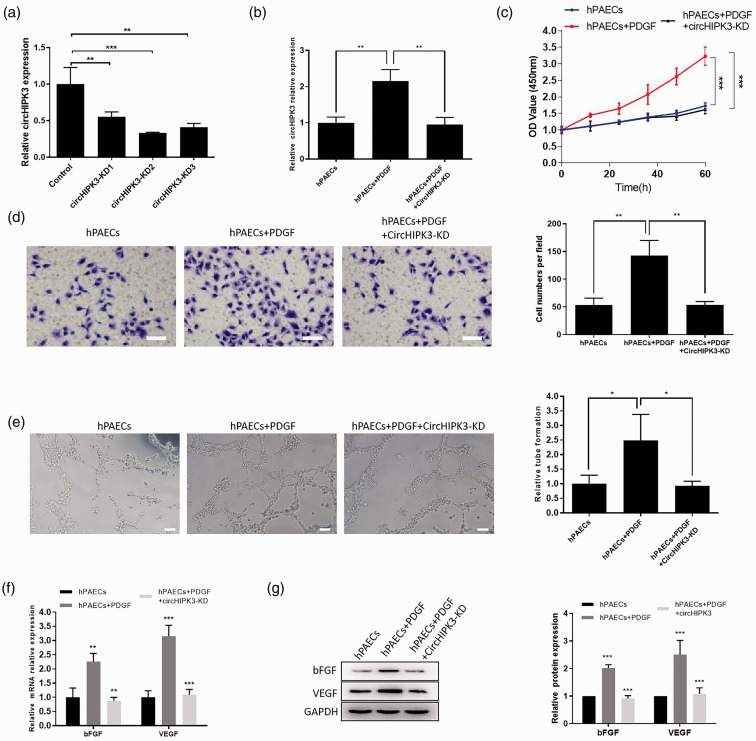
To identify the function of circHIPK3 in PDGF-induced hPAECs, we designed three siRNAs to decrease the expression of circHIPK3. qRT-PCR results showed that the knockdown effect of siRNA-2 is better than other siRNA (Fig. 3a). Therefore, siRNA-2 was used for subsequent experiments. We transfected circHIPK3 siRNA with PDGF treatment. Consistently, circHIPK3 siRNA significantly downregulated the expression of circHIPK3 in PDGF-treated hPAECs (Fig. 3b). Subsequently, a CCK-8 assay was performed to reveal the fact that circHIPK3 knockdown could suppress PDGF-induced hPAECs proliferation (Fig. 3c). Similarly, the transwell assay confirmed that knockdown of circHIPK3 inhibited the increase of cell migration induced by PDGF in hPAECs (Fig. 3d). Moreover, the tube formation assay showed that knockdown of circHIPK3 significantly inhibited the ability of PDGF-treated hPAECs to form capillary-like structures (Fig. 3e). Following, qRT-PCR and western blot were used to detect bFGF and VEGF expression, and the results showed that knockdown of circHIPK3 inhibited the effect of PDGF on bFGF and VEGF expression at mRNA and protein level (Fig. 3f and g). These assays showed that circHIPK3 knockdown inhibited the PDGF-induced proliferation, migration, and angiogenesis abilities of hPAECs.
Fig. 3.
CircHIPK3 regulated the function of PDGF-treated hPAECs. (a) The expression level of CircHIPK3 transfected with siRNA-1, siRNA-2, and siRNA-3 was verified by qRT-PCR. (b) Effect of PDGF treatment on circHIPK3 expression. CircHIPK3 siRNA reduced the expression of circHIPK3 in PDGF-treated hPAECs. (c) CCK-8 assay was performed to detect the proliferation of hPAECs under and circHIPK3 knockdown. (d) The hPAECs migration was measured using a Transwell assay, scale bar = 25 μm. (e) The effect of circHIPK3 on tube formation assay in hPAECs treated with PDGF, scale bar = 25 μm. (f and g) The qRT-PCR and western blot were used to detect bFGF and VEGF expression at mRNA and protein level. Data were expressed as mean ± SD in three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. PGDF: platelet-derived growth factor; VEGF: vascular endothelial growth factor; bFGF: basic fibroblast growth factor; hPAEC: human pulmonary artery endothelial cell; GADPH: glycerldehyde-3-phosphate dehydrogenase.
CircHIPK3 binding with miR-328-3p
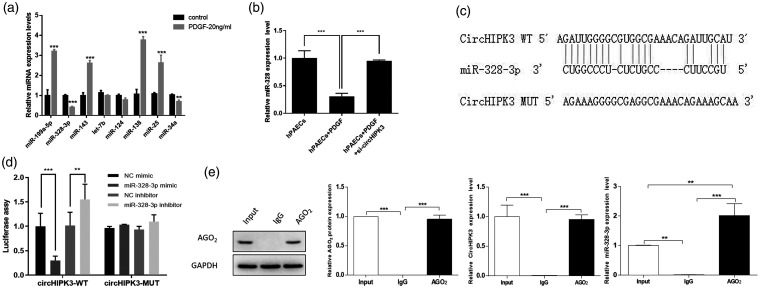
Based on previous research, we learned that circHIPK3 was localized in the cytoplasm.23,24 Given that circRNA has been shown to act as a miRNA sponge in the cytoplasm,25 we next investigated the ability of circHIPK3 to bind to miRNAs. We selected some miRNAs related to PAH according to previous reports and detected the expression of these miRNAs in PDGF-treated hPAECs. The results displayed that miR-328-3p expression was more significant in PDGF-treated cells than control cells (Fig. 4a). In order to detect the effects of circHIPK3 on miR-328-3p expression, qRT-PCR was performed. It was observed that PDGF decreased the expression of miR-328-3p in hPAECs, and circHIPK3 knockdown could increase miR-328-3p expression in PDGF-treated hPAECs (Fig. 4b). The binding sequence of circHIPK3 and miR-328-3p is shown in Fig. 4c. For the sake of confirming the binding sites between circHIPK3 and miR-328-3p, a dual-luciferase reporter assay was performed. The results showed that the luciferase activity was decreased in circHIPK3-WT and miR-328-3p mimics cotransfected 293T cells. The luciferase activity was increased in circHIPK3-WT and miR-328-3p inhibitor cotransfected cells, compared with corresponding control and circHIPK3-WT cotransfected cells. However, the luciferase activity had no significant change in circHIPK3-MUT transfected cells (Fig. 4d). The results implied the combination of circHIPK3 and miR-328-3p. RNA-induced silencing complex (RISC) contained the catalytic subunit AGO2, which was a critical component for the miRNA-sponge role of circRNAs.26 Furthermore, we used the Ago2 antibody to pull down the RISC and then detected the expression of circHIPK3 and miR-328-3p in precipitation through qRT-PCR. IgG antibody was used as the control antibody. Input sample was collected as the positivity control. As shown in Fig. 4e, both circHIPK3 and miR-328-3p were enriched by the Anti-Ago2 compared to Anti-IgG. Hence, circHIPK3 negatively regulated its target miR-328-3p.
Fig. 4.
CircHIPK3 binding with miR-328-3p. (a) The expression level of miRNAs in PDGF-treated hPAECs was detected by qRT-PCR. (b) The expression level of miR-328-3p in hPAECs was downregulated after PDGF treatment. CircHIPK3 knockdown reversed this effect. (c) The putative binding sites between circHIPK3 and miR-328-3p and the mutant sites in circHIPK3-MUT reporter were displayed. (d) Luciferase activity was detected in 293T cells cotransfected with circHIPK3-WT or circHIPK3-MUT reporter and miR-328-3p mimics or miR-328-3p inhibitor. (e) RNA immunoprecipitation assays demonstrated that circHIPK3 was directly binding to miR-328-3p. Data were expressed as mean ± SD in three independent experiments. **P < 0.01, ***P < 0.001. PGDF: platelet-derived growth factor; hPAEC: human pulmonary artery endothelial cell; GADPH: glycerldehyde-3-phosphate dehydrogenase; NC: negative control; IgG: immunoglobulin.
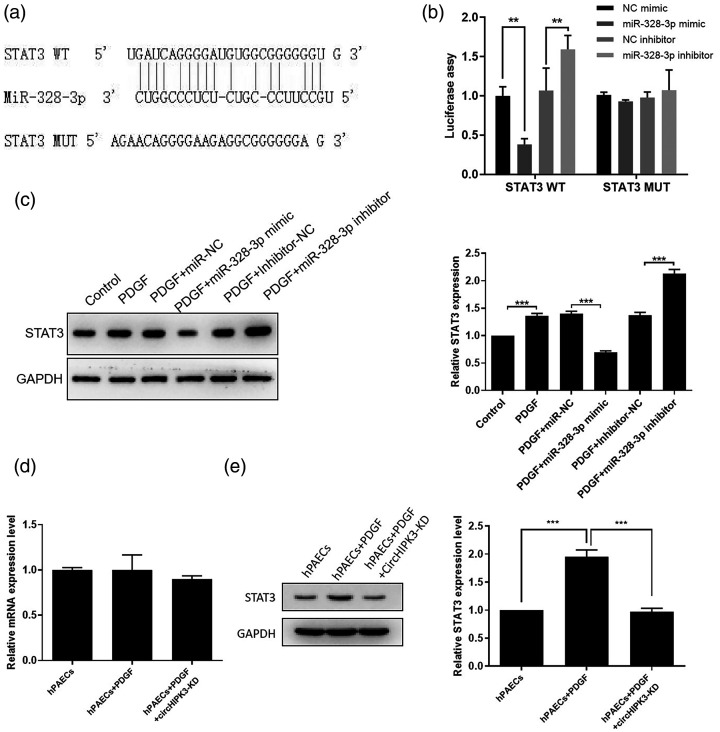
STAT3 is a novel target of miR-328-3p
To explore the target gene of miR-328-3p, we utilized a bioinformatics program. The results showed that STAT3 is a predicted target gene of miR-328-3p. Previous studies have shown that STAT3 plays a critical role in pulmonary hypertension.27–29 The predicted interaction between miR-328-3p and the STAT3 3′-UTR is illustrated in Fig. 5a. Subsequently, we confirmed that miR-328-3p directly targeted the predicted binding sites in the STAT3 3′-UTR by a luciferase reporter assay (Fig. 5b). miR-328-3p overexpression resulted in reduced STAT3 protein levels and inhibited miR-328-3p-induced STAT3 protein levels increase in PDGF-treated hPAECs (Fig. 5c). Furthermore, we observed that the STAT3 protein levels were significantly decreased in the circHIPK3 knockdown and PDGF-treated hPAECs than PDGF-treated cells, whereas STAT3 mRNA remained unchanged (Fig. 5d and e). The above results indicated that miR-328-3p downregulated STAT3 expression through binding to its 3′-UTR directly, and STAT3 expression was indirectly regulated by circHIPK3.
Fig. 5.
Identification of STAT3 as a novel target for miR-328-3p. (a) Schematic representation of the putative miR-328-3p binding site in the STAT3 3′-UTR. (b) Luciferase assays were conducted to assess miR-328-3p targeting of the STAT3 3′-UTR. (c) The STAT3 expression level was downregulated by miR-328-3p overexpression but upregulated by miR-328-3p inhibitor in PDGF-induced hPAECs. (d and e) Detection of effects of circHIPK3 knockdown on mRNA and protein expression of STAT3 in PDGF-induced hPAECs via qPCR and western blot. Data were expressed as mean ± SD in three independent experiments. **P < 0.01, ***P < 0.001. PGDF: platelet-derived growth factor; hPAEC: human pulmonary artery endothelial cell; GADPH: glycerldehyde-3-phosphate dehydrogenase; STAT3: STAT3 overexpression vector pcDNA-STAT3; NC: negative control.
CircHIPK3-miR-328-3p-STAT3 axis promoted the proliferation, migration, and angiogenesis of hPAECs
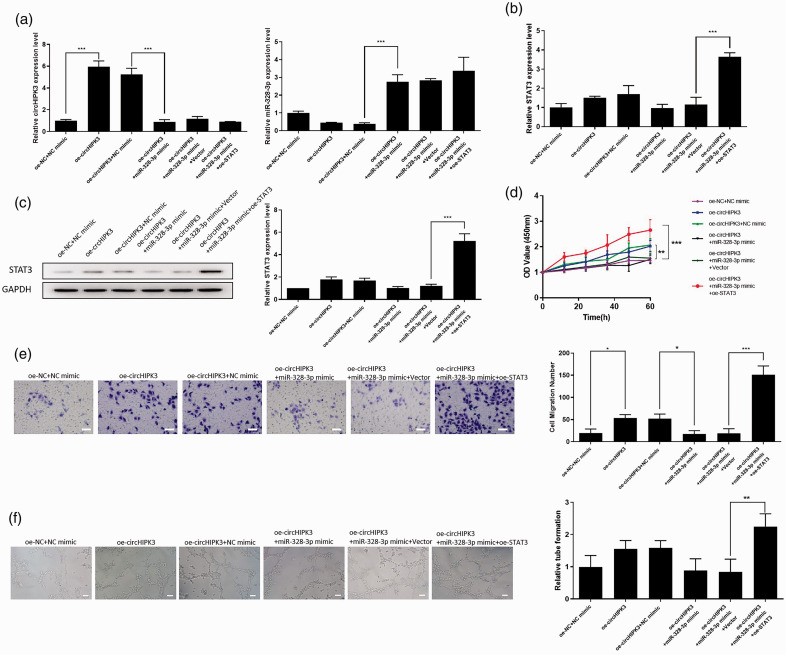
Finally, we explored whether circHIPK3 exerts its biological function by regulating miR-328-3p and STAT3 expression. A rescue assay between miR-328-3p and STAT3 was performed in circHIPK3 overexpression cells. MiR-328-3p mimics and STAT3 vector were transfected in cells, and the expression of circHIPK3, miR-328-3p, and STAT3 was detected by qRT-PCR. It was found that overexpression of circHIPK3 significantly increased the expression of circHIPK3 and decreased the expression of miR-328-3p. The inhibitory effect of circHIPK3 on miR-328-3p levels could be restored by overexpressing miR-328-3p (Fig. 6a). CircHIPK3 or miR-328-3p did not induce significant changes in the expression of STAT3 at mRNA levels (Fig. 6b). However, overexpression of circHIPK3 remarkably induced the protein expression of STAT3. The effect of circHIPK3 on STAT3 expression was attenuated by overexpression of miR-328-3p (Fig. 6c). Moreover, the CCK-8 assay and Transwell assay revealed that circHIPK3 promoted cell proliferation and migration of hPAECs. miR-328-3p overexpression attenuated the effect of circHIPK3 on cell proliferation and migration. STAT3 overexpression significantly reversed the miR-328-3p overexpression-induced proliferation and migration (Fig. 6d and e). Besides, tube formation assay showed that cirHIPK3 promoted capillary-like network formation, and the roles were inhibited by miR-328-3p. STAT3 overexpression significantly increased the ability of hPAECs to form capillary-like structures (Fig. 6f). Taken together, circHIPK3 promoted the proliferation, migration, and angiogenesis of hPAECs by competitive binding miR-328-3p and negatively modulated the STAT3 expression.
Fig. 6.
CircHIPK3 promoted the function of hPAECs through regulating the miR-328-3p/STAT3 axis. (a) qRT-PCR analysis for circHIPK3 and miR-328-3p in hPAECs from each group. (b) qRT-PCR detected the expression of STAT3 in hPAECs from each group at the mRNA level. (c) Western blot analysis for STAT3 protein in hPAECs from each group. (d) CCK-8 assay was performed to detect cell proliferation in different groups. (e) Transwell assay was used to measure cell migration in different groups, scale bar = 25 μm. (f) Representative images and graph of the numbers of tubules formed in each group, scale bar = 25 μm. Data were expressed as mean ± SD in three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. STAT3: STAT3 overexpression vector pcDNA-STAT3; NC: negative control; OD: optical density.
Discussion
In this study, we revealed that circHIPK3 was remarkably upregulated in hPAECs of the PAH cell model. Functional assays showed that knockdown of circHIPK3 inhibited PDGF-induced hPAECs proliferation, migration, and angiogenesis. Also, we demonstrated that high expression of circHIPK3 suppressed the expression of miR-328-3p. Mechanistically, we found that the signal transducer and activator of transcription 3 (STAT3) could be a target of miR-328-3p, and circHIPK3 overexpression indirectly caused STAT3 upregulation. hPAECs promotes the proliferation, migration, and angiogenesis of hPAECs through the miR-328-3p/STAT3 pathway.
The role of circRNAs in PAH has been extensively studied in recent years. For example, circ_0016070 is associated with PAH by promoting pulmonary artery smooth muscle cells (PASMC) proliferation.30 Elevated serum circ_0068481 levels are a potential diagnostic and prognostic indicator of idiopathic PAH.31 On the other hand, the role of circRNA in endothelial cells is also gradually becoming clear. For instance, circRNA-001175 promoted cell proliferation and angiogenesis and inhibited apoptosis of the human umbilical vein endothelial cells induced by high glucose.32 HIPK3 is reported to produce abundant back spliced products, which are deemed as circRNAs. CircHIPK3 regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/insulin-like growth factor 1 (IGF-1) pathway.33 It has been reported that CircHIPK3 mediates retinal vascular dysfunction in diabetes mellitus.11 In this study, we explored the expression of circHIPK3 in hPAECs of PAH cell model and the effects of circHIPK3 on hPAECs function. The expression of circHIPK3 was upregulated in the PAH cell model and was positively associated with cell functions.
Notably, circRNAs can function as endogenous competitive RNAs and play sponge roles, regulating gene expression.34 After the online prediction and experimental analysis, we identified that circHIPK3 could directly regulate the miR-328-3p level as a miRNA sponge. MiR-328-3p has been reported to be involved in the proliferation of PASMC in relevant hypoxic pulmonary hypertension.35 The target genes of miR-328-3p were explored, and STAT3 was confirmed to be a target gene of miR-328-3p. STAT3 has been reportedly associated with PAH. For example, lincRNA-Cox2 promotes PAH development by regulating the let-7a/STAT3 pathway.36 MiR-125-5p ameliorates monocrotaline-induced PAH by targeting the transforming growth factor (TGF)-beta1 and interleukin-6/STAT3 signaling pathways.37 Moreover, our rescue assays proved that miR-328-3p/STAT3 axis was responsible for regulating circHIPK3 in PAH pathogenesis.
However, there are some deficiencies in this study. First, the roles and mechanism of circHICK3 in PAH were confirmed through in vitro experiments. The roles of circHIPK3 in PAH pathogenesis need to be further explored by in vivo studies. Second, the circRNA-miRNA-mRNA network is elusive. Individual circRNA sponges to multiple miRNAs and then regulates the expression of multiple target genes.38 Thus, whether circHIPK3 sponges to other miRNAs and regulates other genes expression in PAH need further exploration.
In summary, this study showed that circHIPK3 could promote the proliferation, migration, and angiogenesis of hPAECs through the miR-328-3p/STAT3 pathway, and circHIPK3 might be a potentially valid target for the diagnosis and treatment of PAH. This study provides a new perspective in research on circRNAs in PAH, as well as a scientific basis for novel developments in the diagnosis and treatment of PAH.
Acknowledgments
The authors thank Unimed Scientific Inc. for assistance in data analysis.
Footnotes
Conflict of interest: The author(s) declare that there is no conflict of interest.
Contributorship statement: Liuqing Hong and Jincai Lin performed the research implementation and Methodology. Jiyuan Liu performed the data analysis. Xiaoying Ma reviewed this article. Yinzhu Luo validated the study. Ying Shen wrote the original draft. Liyan Zhang designed and organized the experiments.
Funding: This work was supported by Fuzhou Key Clinical Specialty Construction Project (201912007) and Fuzhou Science and Technology Plan Project (2020-WS-62).
ORCID iD: Liyan Zhang https://orcid.org/0000-0003-4582-2414
References
- 1.Hoeper MM, Bogaard HJ, Condliffe R, et al. Definitions and diagnosis of pulmonary hypertension. J Am Coll Cardiol 2013; 62: D42–D50. [DOI] [PubMed] [Google Scholar]
- 2.Leopold JA, Maron BA. Molecular mechanisms of pulmonary vascular remodeling in pulmonary arterial hypertension. Int J Mol Sci 2016; 17: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya R, Kwon J, Li X, et al. Distinct role of PLCbeta3 in VEGF-mediated directional migration and vascular sprouting. J Cell Sci 2009; 122: 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schermuly RT, Ghofrani HA, Wilkins MR, et al. Mechanisms of disease: pulmonary arterial hypertension. Nat Rev Cardiol 2011; 8: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noskovicova N, Petrek M, Eickelberg O, et al. Platelet-derived growth factor signaling in the lung. From lung development and disease to clinical studies. Am J Respir Cell Mol Biol 2015; 52: 263–284. [DOI] [PubMed] [Google Scholar]
- 6.Perros F, Montani D, Dorfmuller P, et al. Platelet-derived growth factor expression and function in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med 2008; 178: 81–88. [DOI] [PubMed] [Google Scholar]
- 7.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 8.Alhasan AA, Izuogu OG, Al-Balool HH, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016; 127: e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabo L, Morey R, Palpant NJ, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015; 16: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X, Li X, Zhang P, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform 2017; 18: 547–557. [DOI] [PubMed] [Google Scholar]
- 11.Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017; 136: 1629–1642. [DOI] [PubMed] [Google Scholar]
- 12.Bai M, Pan CL, Jiang GX, et al. CircHIPK3 aggravates myocardial ischemia-reperfusion injury by binding to miRNA-124-3p. Eur Rev Med Pharmacol Sci 2019; 23: 10107–10114. [DOI] [PubMed] [Google Scholar]
- 13.Deng Y, Wang J, Xie G, et al. Circ-HIPK3 strengthens the effects of adrenaline in heart failure by MiR-17-3p-ADCY6 axis. Int J Biol Sci 2019; 15: 2484–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson AAR, Lawrie A. Targeting vascular remodeling to treat pulmonary arterial hypertension. Trends Mol Med 2017; 23: 31–45. [DOI] [PubMed] [Google Scholar]
- 15.Han B, Shaolong E, Luan L, et al. CircHIPK3 promotes clear cell renal cell carcinoma (ccRCC) cells proliferation and metastasis via altering of miR-508-3p/CXCL13 signal. Onco Targets Ther 2020; 13: 6051–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun 2019; 10: 3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rama AR, Perazzoli G, Cabeza L, et al. Novel microRNA sponges to specifically modulate gene expression in colon cancer cells. Nucleic Acid Ther. Epub ahead of print 21 May 2020. DOI: 10.1089/nat.2020.0861. [DOI] [PubMed]
- 18.Luo L, Xiao L, Lian G, et al. miR-125a-5p inhibits glycolysis by targeting hexokinase-II to improve pulmonary arterial hypertension. Aging (Albany NY) 2020; 12: 9014–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L, Lin P, Chen Y, et al. miR-182-3p/Myadm contribute to pulmonary artery hypertension vascular remodeling via a KLF4/p21-dependent mechanism. Theranostics 2020; 10: 5581–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morii C, Tanaka HY, Izushi Y, et al. 3D in vitro model of vascular medial thickening in pulmonary arterial hypertension. Front Bioeng Biotechnol 2020; 8: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benisty JI, McLaughlin VV, Landzberg MJ, et al. Elevated basic fibroblast growth factor levels in patients with pulmonary arterial hypertension. Chest 2004; 126: 1255–1261. [DOI] [PubMed] [Google Scholar]
- 22.Seyfarth HJ, Sack U, Gessner C, et al. [Angiogenin, bFGF and VEGF: angiogenic markers in breath condensate of patients with pulmonary hypertension]. Pneumologie 2015; 69: 207–211. [DOI] [PubMed] [Google Scholar]
- 23.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018; 9: 417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chen D, Lu X, Yang F, et al. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res 2019; 11: 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumtaz PT, Taban Q, Dar MA, et al. Deep insights in circular RNAs: from biogenesis to therapeutics. Biol Proced Online 2020; 22: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 2017; 8: 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian Z, Li Y, Yang H, et al. PDGFBB promotes proliferation and migration via regulating miR-1181/STAT3 axis in human pulmonary arterial smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2018; 315: L965–L976. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Shao R, Gao W, et al. Inhibition of miR-361-5p suppressed pulmonary artery smooth muscle cell survival and migration by targeting ABCA1 and inhibiting the JAK2/STAT3 pathway. Exp Cell Res 2018; 363: 255–261. [DOI] [PubMed] [Google Scholar]
- 29.Song S, Carr SG, McDermott KM, et al. STIM2 (stromal interaction molecule 2)-mediated increase in resting cytosolic free Ca(2+) concentration stimulates PASMC proliferation in pulmonary arterial hypertension. Hypertension 2018; 71: 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S, Jiang H, Li M, et al. Circular RNA hsa_circ_0016070 is associated with pulmonary arterial hypertension by promoting PASMC proliferation. Mol Ther Nucleic Acids 2019; 18: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Chen Y, Yao H, et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ 2019; 9: 2045894019888416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei X, Ye S, Jin G, et al. Overexpression of circRNA-001175 promotes proliferation and angiogenesis and inhibits apoptosis of the human umbilical vein endothelial cells (HUVECs) induced by high glucose. Int J Clin Exp Pathol 2018; 11: 359–366. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhao R, Liu W, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev 2019; 2019: 7954657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016; 238: 42–51. [DOI] [PubMed] [Google Scholar]
- 35.Xing Y, Zheng X, Fu Y, et al. Long noncoding RNA-maternally expressed gene 3 contributes to hypoxic pulmonary hypertension. Mol Ther 2019; 27: 2166–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng G, He L, Zhang Y. LincRNA-Cox2 promotes pulmonary arterial hypertension by regulating the let-7a-mediated STAT3 signaling pathway. Mol Cell Biochem 2020; 475: 239–247. [DOI] [PubMed] [Google Scholar]
- 37.Cai Z, Li J, Zhuang Q, et al. MiR-125a-5p ameliorates monocrotaline-induced pulmonary arterial hypertension by targeting the TGF-beta1 and IL-6/STAT3 signaling pathways. Exp Mol Med 2018; 50: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F, Zhang R, Zhang X, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY) 2018; 10: 2266–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]