Abstract
The swede midge, Contarinia nasturtii, is a cecidomyiid fly that feeds specifically on plants within the Brassicaceae. Plants in this family employ a glucosinolate-myrosinase defense system, which can be highly toxic to nonspecialist feeders. Feeding by C. nasturtii larvae induces gall formation, which can cause substantial yield losses thus making it a significant agricultural pest. A lack of genomic resources, in particular a reference genome, has limited deciphering the mechanisms underlying glucosinolate tolerance in C. nasturtii, which is of particular importance for managing this species. Here, we present an annotated, scaffolded reference genome of C. nasturtii using linked-read sequencing from a single individual and explore systems involved in glucosinolate detoxification. The C. nasturtii genome is similar in size and annotation completeness to that of the Hessian fly, Mayetiola destructor, but has greater contiguity. Several genes encoding enzymes involved in glucosinolate detoxification in other insect pests, including myrosinases, sulfatases, and glutathione S-transferases, were found, suggesting that C. nasturtii has developed similar strategies for feeding on Brassicaceae. The C. nasturtii genome will, therefore, be integral to continued research on plant-insect interactions in this system and contribute to effective pest management strategies.
Keywords: Cecidomyiidae, Diptera, insect pest, genomic resources, transcriptome, detoxification genes
Significance
The swede midge is a serious pest of Brassicaceae, however, few genomic resources exist for the species. Here, we generated an annotated, scaffolded, genome assembly and used this genome to characterize adaptations needed for herbivore specialization on Brassicaceae. This assembly provides the foundation for applying genomics to the study of host plant manipulation and response to defenses and will undoubtedly have a significant impact on swede midge management.
Introduction
Among insect herbivores, phytophagous flies within family Cecidomyiidae are considered to be among the most tightly linked to host plant biology because of their ability to manipulate growth of their hosts (Shorthouse et al. 2005). Cecidomyiid larvae secrete saliva onto plant tissues and feed via extra-oral digestion. Elements within the secretions cause swelling and deformation of plant tissue, and in some species, galls—structures composed of plant tissues created in response to stimuli produced by the gall-inducer (Mamaev 1975; Stone and Schönrogge 2003; Giron et al. 2016). Given that many cecidomyiids are agricultural and forestry pests (Hall et al. 2012), genome sequencing of these species can provide insight into host specialization, host manipulation, and traits that can inform pest management strategies.
The swede midge, Contarinia nasturtii (Kieffer) (Diptera: Cecidomyiidae), is native to Eurasia, and a significant pest of crops in the family Brassicaceae (Chen et al. 2011). Since arriving in North America (Hallett and Heal 2001), it has expanded its geographic range to include Eastern Canada, and Northeastern and Midwestern USA (Chen et al. 2011; Philips et al. 2017). In addition to feeding on cultivated crucifers, such as cabbage, cauliflower, broccoli (Brassica oleraceae L.), and canola (B. napus L., and B. rapa L.), it is found on a wide range of other Brassicaceae (Barnes 1946; Stokes 1953a, 1953b; Hallett 2007; Chen et al. 2009). Crop losses >80% have occurred due to C. nasturtii damage in broccoli (Hallett and Heal 2001) and canola (Hallett 2017), and larval feeding is particularly problematic on fresh vegetables as a single larva is capable of rendering them unmarketable (Stratton et al. 2018).
Investigation of the C. nasturtii genome can provide insight into host manipulation and adaptations needed for specialization on Brassicaceae that could have significant impacts on C. nasturtii management. Brassicaceae are well-known for their glucosinolate-myrosinase system, often called the “mustard-oil bomb” (Matile 1980; Barco and Clay 2019). Upon herbivore damage, glucosinolates are hydrolyzed by plant-derived myrosinases to form isothiocyanates, nitriles, and other compounds (Kissen et al. 2009), which are toxic to and/or deter many insect herbivores (Halkier and Gershenzon 2006). Nevertheless, some insect herbivores have developed strategies to avoid, sequester, or detoxify glucosinolates and their byproducts (reviewed in Winde and Wittstock 2011; Jeschke et al. 2016). Nitrile-specifier proteins in Pieris butterflies (Wittstock et al. 2004) and glucosinolate-specific sulfatases in the diamondback moth, Plutella xylostella (Ratzka et al. 2002) and cabbage stem flea beetle, Psylloides chrysocephala (Beran et al. 2018; Ahn et al. 2019) help prevent the formation of highly toxic glucosinolate-hydrolysis products. Other insects sequester glucosinolates and use them for defense in combination with insect-derived myrosinases (Jones et al. 2002; Kazana et al. 2007; Beran et al. 2014). Further, some insects employ general detoxification mechanisms, such as glutathione S-transferase (GST), to detoxify isothiocyanates and excrete them (Gloss et al. 2014; Beran et al. 2018), and additional mechanisms of detoxification are still being discovered (Friedrichs et al. 2020).
Overall, very little is known about the genomes of cecidomyiids. Until now, the Hessian fly, Mayetiola destructor (Say), a major pest of wheat (Triticum aestivum L.), was the only cecidomyiid genome sequenced (Zhao et al. 2015). Here, we describe an annotated genome assembly of C. nasturtii using linked-read sequencing, and identify several potential glucosinolate detoxification systems of this Brassicaceae specialist.
Results and Discussion
Genome Assembly, Annotation, and Quality Assessment
In total, 88 million paired-end reads were generated from a linked-read library of a single C. nasturtii pupa. The highest quality de novo assembly (77 million reads, 57.2× raw coverage) had an effective coverage of 45× based on Supernova’s estimated genome size (supplementary table 1, Supplementary Material online). Based on k-mer distribution and analysis the sequencing error rate was 1.03% and heterozygosity was low (0.34%) (supplementary fig. 1, Supplementary Material online). The assembly consisted of 5,545 scaffolds with an N50 of 4.7 Mb (fig. 1), and was 185.9 Mb in total length, which was comparable to the average female genome size estimated by flow cytometry (female: 183.5 ± 0.9 Mb, male = 145.0 ± 0.5 Mb). The 174.9 Mb genome size estimated with k-mer spectra using Jellyfish/GenomeScope was complementary to these estimates (supplementary fig. 1, Supplementary Material online). Only 8.6% of the assembly was comprised of gaps. Completeness of the assembly using BUSCO revealed 79.9% of the core Insecta orthologs were complete and single copy, 0.6% complete and duplicated, 3.2% fragmented, and 16.3% missing (fig. 1, supplementary table 2, Supplementary Material online). Compared with M. destructor with a similar size genome, the C. nasturtii assembly was more contiguous (fig. 1, supplementary table 2, Supplementary Material online). These results suggest that linked-read sequencing is a viable option for minute insects and provides a cost-effective alternative to traditional approaches (Zhao et al. 2015). Additionally, linked-read technology allowed sequencing of a single individual with low heterozygosity without the need for inbreeding. This is of great practical importance for cecidomyiids as most are monogenous, that is, females generally lay eggs of only one sex (Barnes 1950; Dorchin and Freidberg 2004), which makes mating of offspring from a single individual impractical.
Fig. 1.
Contarinia nasturtii life stages (A) and comparison of continuity and completeness of the C. nasturtii (left) and M. destructor (right, Zhao et al. 2015) genome assemblies (B). (A): Adult male C. nasturtii (scale bar indicates 500 µm) (left); damage to canola, Brassica napus L. (va. AC Excel) caused by C. nastrurtii larval feeding (middle); and C. nasturtii larva (right). (B): Plots consist of scaffolds indicated by red and grey sections, sorted by descending length along the radius of each plot. The radius of each plot represents scaffold length with the scale marked at the vertical position (0%). The circumference of each plot (and percentage scale along the outside) indicates the percentage of the genome assembled into cumulative scaffolds, with N90, N50, and the longest scaffold indicated by light orange, dark orange, and red, respectively. Blue and light blue along the circumference represent relative GC/AT content. The cumulative number of scaffolds within a fraction of the genome is indicated by a purple spiral following the radial scale in thousands. Scaffolds of <1,000 bp were removed from the M. destructor assembly to match that of the C. nasturtii assembly. Complete (Comp.), duplicated (Dup.), and fragmented (Frag.) BUSCO annotations and assembly statistics are provided below.
Annotation of the genome through NCBI’s eGAP identified 16,017 genes, with 14,889 containing protein-coding regions. This gene number is similar to other flies, including Drosophila melanogaster (Adams 2000), Anopheles gambiae (Holt et al. 2002), and Musca domestica (Scott et al. 2014). In total, 26,752 transcripts were annotated with a mean of 1.68 transcripts/gene; 23,265 (93.2%) transcripts were fully supported by experimental evidence. In addition to protein-coding genes, 1,789 noncoding RNAs were identified (1,537 fully supported), including tRNA, lncRNAs, and others. 84% of all aggregated reads from RNA-Seq libraries aligned to the genome indicating high reliability of the assembly. The full annotation report is available online (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Contarinia_nasturtii/100/, last accessed March 4, 2021). BUSCO analysis against the Insecta and Diptera gene sets suggested highly complete annotation with 98.8% and 92.0% complete (single and duplicated) matches, respectively (supplementary table 2, Supplementary Material online). Compared with the original gene set of M. destructor, the C. nasturtii RefSeq gene set is on average 7.3% more complete (across Insecta and Diptera odb10) (supplementary table 2, Supplementary Material online). Comparison of k-mer spectra using KAT indicated that haploid representation of the assembly was successful and free of diploid content (supplementary fig. 2, Supplementary Material online). Prior to annotation, WindowMasker masked 30.01% of the assembly (supplementary table 3, Supplementary Material online); this level of repetitive sequences is less than that observed (52%) in M. domestica and several other insect species (Scott et al. 2014). k-mer analysis estimated 13.9% of the genome is made up of repetitive content (supplementary fig. 1, Supplementary Material online), which is similar to the repeat content in the M. destructor genome (12%) (Zhao et al. 2015). Despite differences in estimates between the two programs likely due to assembly vs. raw read inputs, the respective comparison to other species (e.g. Scott et al. 2014, Zhao et al. 2015) supports the high quality of this assembly.
Identification of Glucosinolate Detoxification Systems
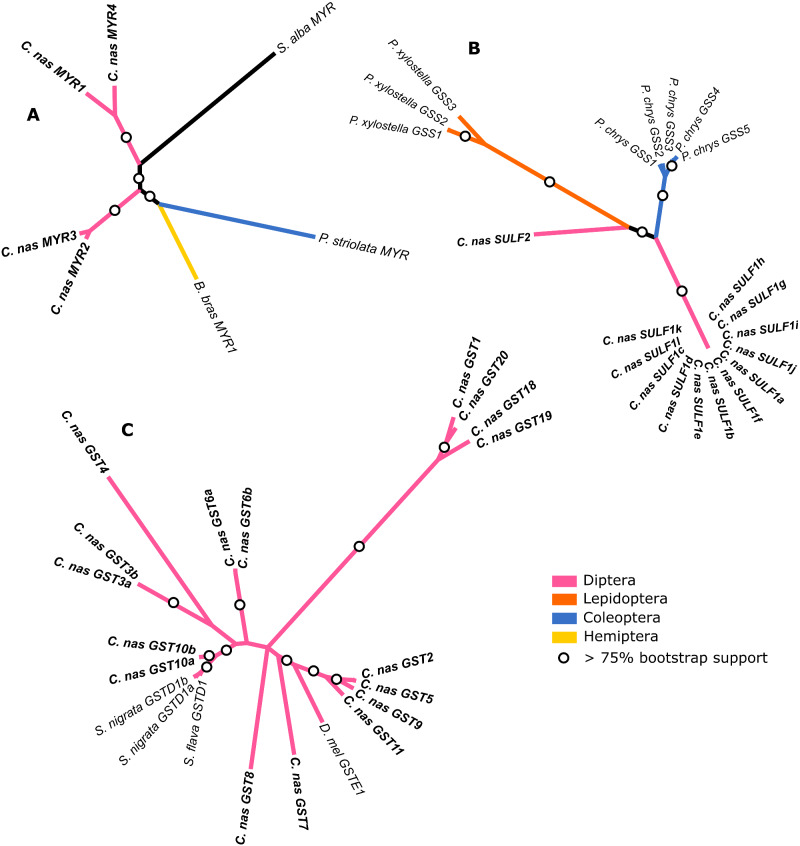
Several genes encoding components of glucosinolate detoxification systems functionally characterized in other insects were identified in the C. nasturtii genome. Both Phyllotreta striolata (Beran et al. 2014) and Brevicoryne brassicae (Kazana et al. 2007) are capable of sequestering glucosinolates and possess myrosinases that are distinct from plant myrosinases (Jones et al. 2002, Beran et al. 2014). The four full-length C. nasturtii myrosinase genes identified were also unique from plant myrosinases (fig. 2, supplementary fig. 3 and supplementary table 4, Supplementary Material online). C. nasturtii myrosinase-2 and 3 clustered with those from P. striolata and B. brassicae, but not with C. nasturtii myrosinase-1 and 4 (fig. 2A). The C. nasturtii myrosinases contain conserved glucose-binding and catalytic sites, as in P. striolata and B. brassicae myrosinases, and are likely functional (supplementary fig. 3, Supplementary Material online).
Fig. 2.
Unrooted maximum likelihood consensus trees of amino acid sequences for enzymes involved in glucosinolate detoxification that were identified in the C. nasturtii RefSeq annotations, with additional sequences from Genbank. Myrosinases are shown in panel (A), sulfatases in (B), and GSTs in (C). The color of each branch indicates the insect Order of the sequences in each phylogeny; the only noninsect taxon, S. alba (Brassicaceae) is pictured in black in (A). Branch labels indicate the taxon name followed by the gene ID, and the following taxon names are abbreviated on the trees: C. nas, C. nasturtii; B. bras, B. brassicae; D. mel, D. melanogaster; and P. chrys, P. chrysocephala. Contarinia nasturtii sequences are indicated in bold text.
Sulfatases have evolved in insects that feed on Brassicaceae plants to modify glucosinolates so they are no longer recognized by myrosinases and converted into toxic derivatives (Jeschke et al. 2016). Insect sulfatase was first discovered in P. xylostella (Ratzka et al. 2002) and, more recently, in P. chrysocephala (Beran et al. 2018; Ahn et al. 2019). No C. nasturtii genes were annotated as encoding sulfatases. However, Ahn et al. (2019) found five arylsulfatase-like enzymes with sulfatase activity in P. chrysocephala and two genes encoding arylsulfatase-like enzymes were found in C. nasturtii, one of which had five isoforms (fig. 2B, supplementary table 5, Supplementary Material online). The two putative C. nasturtii arylsulfatase-like enzymes clustered with sulfatases from P. chrysocephala. Furthermore, there was high conservation in amino acid residues of C. nasturtii arylsulfatase-like enzymes and both possessed sulfatase signature features and catalytic residues (supplementary fig. 4, Supplementary Material online). Interestingly, sulfatases in P. xylostella and P. chrysocephala contain a signal peptide and are secreted into the midgut (Ratzka et al. 2002; Ahn et al. 2019), while those in C. nasturtii lack a signal peptide indicating they are not secreted (supplementary table 5, Supplementary Material online). As the feeding behavior of P. xylostella and P. chrysocephala are markedly different from larval C. nasturtii, it is possible that glucosinolate detoxification occurs after uptake by the cell or that C. nasturtii sulfatases have a different biological function.
GSTs are known for their role in xenobiotic detoxification and are represented by several classes within the larger family (Ranson et al. 2001), some of which play a role in glucosinolate detoxification (Gloss et al. 2014, 2019). Gloss et al. (2014) found the Brassicaceae-specialist-drosophilids Scaptomyza flava and S. nigrita use delta-class glutathione S-transferase 1 (GSTD1) for glucosinolate detoxification. Twenty genes encoding GST-like genes were found in the C. nasturtii genome, 17 of which were complete and were within the expected size range (fig. 2C, supplementary table 6, Supplementary Material online). C. nasturtii GST10a and GST10b clustered with GSTD1s from S. flava and S. nigrita (fig. 2) and residues in the aromatic zipper motif (H-site and α8-helix) were well-conserved (supplementary fig. 5, Supplementary Material online); these were identified by Gloss et al. (2014) as important for isothiocyanate detoxification. Recently, Gloss et al. (2019) found epsilon class GSTs (GSTE) were also involved in glucosinolate detoxification and four C. nasturtii GSTs clustered with D. melanogaster GSTE1 (fig. 2). Based on our combined results, C. nasturtii myrosinases, arylsulfatases, and GSTs should be explored further to examine their roles in glucosinolate detoxification.
Conclusions
The sequencing of the C. nasturtii genome provides the foundation necessary to explore plant-gall insect interactions at the molecular level. Due to the intimate interaction between cecidomyiid larvae and their host plants during feeding, they are thought to follow a gene-for-gene model of coevolving “effectors” (Hatchett and Gallun 1970), which was further supported in M. destructor based on analysis of its genome (Zhao et al. 2015). These gene-for-gene interactions have yet to be explored in C. nasturtii; however, we have begun to investigate insect adaptation in this system by identifying potential glucosinolate detoxification systems characterized in other insects. Given the nature of C. nasturtii infesting plants in the Brassicaceae, which have formidable defenses in the form of glucosinolate-myrosinase systems, this genome should foster exploration of both broader questions of insect-plant coevolutionary dynamics and potential use of these genes in pest management.
Materials and Methods
Genome Sequencing, Assembly, and Size Estimation
DNA was extracted from a single C. nasturtii pupa according to the 10× Genomics’ (Pleasanton, CA) “DNA extraction from single insects” protocol with homogenization by razor blades (details in Supplementary Material online). DNA concentration was adjusted to 0.65 ng/µl and loaded onto a Chromium Genome Chip. Libraries were prepared using the Chromium Genome Library & Gel Bead Kit v.2 (10× Genomics) and Chromium controller according to manufacturer’s recommendations, but with additional shearing (Major et al. 2020). The library was quantified by qPCR using a Kapa Library Quantification Kit (Kapa Biosystems-Roche) and sequenced on a partial lane of NovaSeq6000 (Illumina, San Diego, CA) with paired-end 150 bp reads.
Raw reads were demultiplexed with mkfastq in Supernova v2.1.1 (10× Genomics) by the UC Davis Genome Centre (details in Supplementary Material online). Genome characteristics from the k-mer distribution of debarcoded raw reads were estimated with Jellyfish v2.2.3 (Marçais and Kingsford 2011) and GenomeScope v1.0 (Vurture et al. 2017) (k = 21, k-mer coverage cutoff = 10,000).
A de novo genome assembly was constructed with mkoutput (style = pseudohap2) in Supernova v2.1.1. Reads were subsampled to create and compare several assemblies (48–60× coverage) (supplementary table 1, Supplementary Material online). The highest quality assembly (based on a balance between scaffold N50, phase block N50, contig N50, and predicted genome size) was obtained with 77 million randomly selected reads (∼57×) (56× recommended) and was submitted to NCBI (GCA_009176525). During processing and quality checks, NCBI identified 1,115 sequences that may have originated from bacterial contaminants, ecto-, or endo-symbionts; these were masked in the updated assembly AAFC_1.1 (GCA_009176525.2) (supplementary table 7, Supplementary Material online).
Genome size was estimated by flow cytometry from DNA isolated from individual adult male (n = 13) and female (n = 10) C. nasturtii heads. The head of a female Drosophila virilis was used as an internal standard (1 C = 328 Mb genome) (Johnston et al. 2019).
Genome Annotation and Quality Assessment
Structural and functional annotation of genes was conducted with NCBI’s Eukaryotic Genome Annotation Pipeline (eGAP) v.8.3 (Thibaud-Nissen et al. 2013). To aid in annotation, RNA-Seq was conducted on pooled samples of each C. nasturtii life stage (eggs, first-third instar larvae, pupae, and adult males and females) (NCBI SRA: SRX6853817-SRX6853823) (details in Supplementary Material online). Additional transcripts available from salivary glands (NCBI SRA: SRS5439046) were also used. Prior to annotation, eGAP uses RepeatMasker (http://www.repeatmasker.org, last accessed March 4, 2021) and/or WindowMasker (Morgulis et al. 2006) to mask repeats in the genome assembly; after which, eGAP uses the RNA-Seq data and several NCBI RefSeq protein sets to inform gene model prediction. The full eGAP process can be accessed online at: https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/#process (last accessed March 4, 2021).
BUSCO (Benchmarking Universal Single-Copy Orthologs) v.3.0.2 was used to assess the completeness of the genome/annotated gene set against the Insecta and Diptera odb10 data sets (Simão et al. 2015; Waterhouse et al. 2018). BUSCO results were compared with the M. destructor genome (GCA_000149195.1) and the original gene set (OGS1.0) from the i5K initiative [https://i5k.nal.usda.gov/data/Arthropoda/maydes-(Mayetiola_destructor)/, last accessed March 4, 2021]. BlobToolKit was used to visualize quality metrics of the C. nasturtii and M. destructor genomes (Challis et al. 2020). To validate the assembly and ensure it was largely free of diploid content, KAT v2.4.1 (Mapleson et al. 2016) in Comp mode (with default settings, k = 27) was used to compare k-mer spectra of raw reads (debarcoded) to those of the assembly.
Identification of Glucosinolate Detoxification Systems
Genes encoding elements of glucosinolate detoxification systems that had been functionally characterized in other insects (i.e., myrosinases, sulfatases including arylsulfatase, and GSTs) were identified from the C. nasturtii RefSeq gene set annotations and the genome assembly and manually curated (details in Supplementary Material online, supplementary tables 4–6, Supplementary Material online). SignalP 5.0 (Armenteros et al. 2019) and TMHMM 2.0 (Krogh et al. 2001) were used to predict signal peptides and transmembrane domains, respectively. ProtoParam (Gasteiger et al. 2005) was used to predict molecular weight and isoelectric points for each protein. Amino acid sequences were aligned in MAFFT (https://mafft.cbrc.jp/alignment/server/, last accessed March 4, 2021; L-INS-i algorithm, Mafft homologs—on). Maximum likelihood phylogenies for genes of interest were constructed using IQ-TREE’s web server (Trifinopoulos et al. 2016) and resulting extended consensus trees were visualized with FigTree 1.4.4 (Rambaut and Drummond 2010) (details in Supplementary Material online).
Supplementary Material
Acknowledgments
We would like to thank Shane Hladun, Jennifer Holowachuk, Doug Baldwin, and Stephanie Harris for their technical support, and Dr Scott Geib for advice and assistance. We would also like to thank the Reviewer for constructive comments. The gDNA extraction and sequencing were carried out at the DNA Technologies and Expression Analysis Core Facility at the UC Davis Genome Center (CA, USA) supported by a NIH Shared Instrumentation Grant 1S10OD010786-01. Dr Spencer Johnston (Texas A & M University, TX, USA) kindly provided genome size estimation via flow cytometry. This work was supported by the Agriculture and Agri-Food Canada Genomic Resources and Development Initiative (Grant J-001581 to B.A.M., M.A.E., and D.D.H.) and the Natural Sciences and Engineering Research Council of Canada-Industrial Research Chair Program (Grant 545088 to B.A.M.).
Data Availability
The data underlying this article are available in GenBank at https://www.ncbi.nlm.nih.gov/ and can be accessed with PRJNA565389 and PRJNA579966. Data are also available in the USDA National Agricultural Library I5K workspace at https://i5k.nal.usda.gov/contarinia-nasturtii.
Literature Cited
- Adams MD. 2000. The genome sequence of Drosophila melanogaster. Science 287(5461):2185–2195. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, et al. 2019. Identification and evolution of glucosinolate sulfatases in a specialist flea beetle. Sci Rep. 9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenteros JJA, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotech. 37:420–423. [10.1038/s41587-019-0036-z] [DOI] [PubMed] [Google Scholar]
- Barco B, Clay NK. 2019. Evolution of glucosinolate diversity via whole-genome duplications, gene rearrangements, and substrate promiscuity. Annu Rev Plant Biol. 70:585–604. [DOI] [PubMed] [Google Scholar]
- Barnes HF, editor. 1946. Gall midges of economic importance. In: Gall midges of root and vegetable crops. Vol. I. London: Crosby, Lockwood & Son. [Google Scholar]
- Barnes HF. 1950. The identity of the swede midge, with notes on its biology. Ann Appl Biol. 37(2):241–248. [Google Scholar]
- Beran F, et al. 2018. One pathway is not enough: the cabbage stem flea beetle Psylliodes chrysocephala uses multiple strategies to overcome the glucosinolate-myrosinase defense in its host plants. Front Plant Sci. 9:1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran F, et al. 2014. Phyllotreta striolata flea beetles use host plant defense compounds to create their own glucosinolate-myrosinase system. Proc Natl Acad Sci U S A. 111(20):7349–7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. 2020. BlobToolKit–Interactive quality assessment of genome assemblies. G3 (Bethesda). 10(4):1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, et al. 2009. Occurrence of the new invasive insect Contarinia nasturtii (Diptera: Cecidomyiidae) on cruciferous weeds. J Econ Entomol. 102(1):115–120. [DOI] [PubMed] [Google Scholar]
- Chen M, et al. 2011. Swede midge (Diptera: Cecidomyiidae), ten years of invasion of crucifer crops in North America. J Econ Entomol. 104(3):709–716. [DOI] [PubMed] [Google Scholar]
- Dorchin N, Freidberg A. 2004. Sex ratio in relation to season and host plant quality in a monogenous stem‐galling midge (Diptera: Cecidomyiidae). Ecol Entomol. 29(6):677–684. [Google Scholar]
- Friedrichs J, et al. 2020. Novel glucosinolate metabolism in larvae of the leaf beetle Phaedon cochleariae. Insect Bioch Mol Biol. 124:103431. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, et al. 2005. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Springer protocols handbooks. Totowa: Humana Press. p. 571–607. [Google Scholar]
- Giron D, Huguet E, Stone GN, Body M. 2016. Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J Insect Physiol. 84:70–89. [DOI] [PubMed] [Google Scholar]
- Gloss AD, et al. 2014. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol Biol Evol. 31(9):2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss AD, et al. 2019. Evolution of herbivory remodels a Drosophila genome. bioRxiv: 767160.
- Halkier BA, Gershenzon J. 2006. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 57:303–333. [DOI] [PubMed] [Google Scholar]
- Hall DR, et al. 2012. The chemical ecology of cecidomyiid midges (Diptera: Cecidomyiidae). J Chem Ecol. 38(1):2–22. [DOI] [PubMed] [Google Scholar]
- Hallett RH. 2007. Host plant susceptibility to the swede midge (Diptera: Cecidomyiidae). J Econ Entomol. 100(4):1335–1343. [DOI] [PubMed] [Google Scholar]
- Hallett RH. 2017. The challenge of swede midge management in canola. In: Reddy GVP, editor. Integrated management of insect pests on canola and other Brassica oilseed crops. Wallingford: Centre for Agriculture and Biosciences International. p. 44–67. [Google Scholar]
- Hallett RH, Heal JD. 2001. First Nearctic record of the swede midge (Diptera: Cecidomyiidae), a pest of cruciferous crops from Europe. Can Entomol. 133(5):713–715. [Google Scholar]
- Hatchett JH, Gallun RL. 1970. Genetics of the ability of the Hessian fly, Mayetiola destructor, to survive on wheats having different genes for resistance. Ann Entomol Soc Am. 63(5):1400–1407. [Google Scholar]
- Holt RA, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298(5591):129–149. [DOI] [PubMed] [Google Scholar]
- Jeschke V, Gershenzon J, Vassão DG. 2016. Insect detoxification of glucosinolates and their hydrolysis products. In: Kopriva S, editor. Advances in botanical research. London: Elsevier. p. 199–245. [Google Scholar]
- Johnston JS, Bernardini A, Hjelmen CE. 2019. Genome size estimation and quantitative cytogenetics in insects. In: Brown SJ, Pfrender ME, editors. Insect genomics. New York, NY: Humana Press. p. 15–26. [DOI] [PubMed] [Google Scholar]
- Jones AME, Winge P, Bones AM, Cole R, Rossiter JT. 2002. Characterization and evolution of a myrosinase from the cabbage aphid Brevicoryne brassicae. Insect Bioch Mol Biol. 32(3):275–284. [DOI] [PubMed] [Google Scholar]
- Kazana E, et al. 2007. The cabbage aphid: a walking mustard oil bomb. Proc Biol Sci. 274(1623):2271–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R, Rossiter JT, Bones AM. 2009. The ‘mustard oil bomb’: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem Rev. 8(1):69–86. [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Major KM, et al. 2020. Early life exposure to environmentally relevant levels of endocrine disruptors drive multigenerational and transgenerational epigenetic changes in a fish model. Front Mar Sci. 7:471. [Google Scholar]
- Mamaev BM. 1975. Evolution of gall forming insects-gall midges. Leeds: W. S. Maney Ltd. [Google Scholar]
- Mapleson D, Accinelli GG, Kettleborough G, Wright J, Clavijo BJ. 2016. KAT: a K-mer Analysis Toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33:574–576. [10.1093/bioinformatics/btw663] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile PH. 1980. The mustard oil bomb-compartmentation of the myrosinase system. Biochem Physiol Pflanz. 175(8-9):722–731. [Google Scholar]
- Morgulis A, Gertz EM, Schäffer AA, Agarwala R. 2006. WindowMasker: window-based masker for sequenced genomes. Bioinformatics 22(2):134–141. [DOI] [PubMed] [Google Scholar]
- Philips C, Ambourn A, Christianson L. 2017. First detections of swede midge (Diptera: Cecidomyiidae) in Minnesota. J Entomol Sci. 52(3):297–300. [Google Scholar]
- Rambaut A, Drummond AJ. 2010. FigTree v1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (last accessed Oct 3, 2020).
- Ranson H, et al. 2001. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem J. 359(Pt 2):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. 2002. Disarming the mustard oil bomb. Proc Natl Acad Sci U S A. 99(17):11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JG, et al. 2014. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 15(10):466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stokes BM. 1953a. Biological investigations into the validity of Contarinia species living on the Cruciferae, with special reference to the swede midge, Contarinia nasturtii (Kieffer). Ann Appl Biol. 40(4):726–741. [Google Scholar]
- Stokes BM. 1953b. The host plant range of the swede midge (Contarinia nasturtii Kieffer) with special reference to types of plant damage. Tijdschr Plantenziekten 59(3):82–90. [Google Scholar]
- Stone GN, Schönrogge K. 2003. The adaptive significance of insect gall morphology. Trends Ecol Evol. 18(10):512–522. [Google Scholar]
- Stratton CA, Hodgdon EA, Zuckerman SG, Shelton AM, Chen YH. 2018. A single swede midge (Diptera: Cecidomyiidae) larva can render cauliflower unmarketable. J Insect Sci. 18(3):24. [10.1093/jisesa/iey062] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorthouse JD, Wool D, Raman A. 2005. Gall-inducing insects–Nature's most sophisticated herbivores. Basic Appl Ecol. 6(5):407–411. [Google Scholar]
- Thibaud-Nissen F, et al. 2013. Eukaryotic Genome Annotation Pipeline. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda: National Center for Biotechnology Information.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurture GW, et al. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33(14):2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winde I, Wittstock U. 2011. Insect herbivore counteradaptations to the plant glucosinolate–myrosinase system. Phytochemistry 72(13):1566–1575. [DOI] [PubMed] [Google Scholar]
- Wittstock U, et al. 2004. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc Natl Acad Sci U S A. 101(14):4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, et al. 2015. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr Biol. 25(5):613–620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in GenBank at https://www.ncbi.nlm.nih.gov/ and can be accessed with PRJNA565389 and PRJNA579966. Data are also available in the USDA National Agricultural Library I5K workspace at https://i5k.nal.usda.gov/contarinia-nasturtii.