Abstract
Semaphorins and plexins are cell surface ligand/receptor proteins that affect cytoskeletal dynamics in metazoan cells. Interestingly, they are also present in Choanoflagellata, a class of unicellular heterotrophic flagellates that forms the phylogenetic sister group to Metazoa. Several members of choanoflagellates are capable of forming transient colonies, whereas others reside solitary inside exoskeletons; their molecular diversity is only beginning to emerge. Here, we surveyed genomics data from 22 choanoflagellate species and detected semaphorin/plexin pairs in 16 species. Choanoflagellate semaphorins (Sema-FN1) contain several domain features distinct from metazoan semaphorins, including an N-terminal Reeler domain that may facilitate dimer stabilization, an array of fibronectin type III domains, a variable serine/threonine-rich domain that is a potential site for O-linked glycosylation, and a SEA domain that can undergo autoproteolysis. In contrast, choanoflagellate plexins (Plexin-1) harbor a domain arrangement that is largely identical to metazoan plexins. Both Sema-FN1 and Plexin-1 also contain a short homologous motif near the C-terminus, likely associated with a shared function. Three-dimensional molecular models revealed a highly conserved structural architecture of choanoflagellate Plexin-1 as compared to metazoan plexins, including similar predicted conformational changes in a segment that is involved in the activation of the intracellular Ras-GAP domain. The absence of semaphorins and plexins in several choanoflagellate species did not appear to correlate with unicellular versus colonial lifestyle or ecological factors such as fresh versus salt water environment. Together, our findings support a conserved mechanism of semaphorin/plexin proteins in regulating cytoskeletal dynamics in unicellular and multicellular organisms.
Keywords: choanoflagellates, plexin, semaphorin, axon guidance
Significance
Semaphorins and plexins are cell surface ligand receptor pairs that affect cytoskeletal dynamics in animal cells. Interestingly, they are also present in choanoflagellates, a class of unicellular flagellates that forms the sister group to animals. We describe here the diversity and unique features of choanoflagellate semaphorins and plexins, which may have contributed to the evolutionary transition from unicellular organisms to multicellular animals.
Introduction
Semaphorins and plexins, originally identified as axon guidance molecules, play an essential role in a wide range of cellular processes in development, adult physiology, and cancer through regulation of cytoskeletal and cell adhesion components (Jongbloets and Pasterkamp 2014; Koropouli and Kolodkin 2014; Gurrapu and Tamagnone 2016). In our previous study on the evolution of semaphorins and plexins, we described their diversity and conservation in Metazoa (Junqueira Alves et al. 2019). Interestingly, semaphorins and plexins are also present in choanoflagellates, a class of unicellular flagellates that forms the sister group to Metazoa (Brunet and King 2017). As semaphorins/plexins were not found in two other related clades of unicellular organisms—Ichtyosporea and Filasterea—, we posited that semaphorins and plexins may have evolved in a common ancestor of Metazoa and Choanoflagellata, more than 600 Ma (Junqueira Alves et al. 2019).
The domain structure of plexins is well conserved between metazoan and choanoflagellate species. Plexins are type I transmembrane proteins with an extracellular Sema domain that binds to semaphorin ligands. The Sema domain is followed by a long extracellular stalk comprised of an array of domains—three PSI (domain found in plexins/semaphorins/integrins) and six IPT (found in integrins/plexins/transcription factors) domains, in the order PSI-IPT-PSI-IPT-PSI-IPT-IPT-IPT-IPT (Jones 2015). Intriguingly, structural analyses revealed that these extracellular domains form a ring-shaped structure oriented in parallel to the cell surface (Kong et al. 2016; Suzuki et al. 2016). The intracellular part of plexins contains a Rho-binding domain (RBD) that can regulate plexin activity, and a Ras-GTPase activating protein (Ras-GAP) domain that can inactivate small G proteins of the Ras superfamily (Hota and Buck 2012). The Ras-GAP domain is activated by dimerization of plexins upon semaphorin binding (Pascoe et al. 2015).
Semaphorins can be either type I transmembrane proteins, GPI-anchored proteins, or secreted proteins. Like plexins, semaphorins contain an extracellular Sema domain that binds to the Sema domain of plexins. The extracellular stalk region of semaphorins is quite distinct between paralog groups, containing, for example, fibronectin type III (FNIII), thrombospondin, or immunoglobulin-like (Ig) domains (Junqueira Alves et al. 2019). The intracellular part of semaphorins contains, in general, no conserved domains.
Despite intensive study, critical aspects of semaphorin/plexin function remain unresolved. We reasoned that analysis of the evolutionary history and diversity of semaphorins/plexins in choanoflagellates might provide additional insights into the principles of semaphorin/plexin biology. Our initial evolutionary study had included two choanoflagellate species, Monosiga brevicollis and Salpingoeca rosetta (Junqueira Alves et al. 2019). Here, we extended our analysis to 20 additional choanoflagellate species, including 19 species that have transcriptome data available from a comprehensive evolutionary study (Richter et al. 2018) and the species Choanoeca flexa (Brunet et al. 2019).
Choanoflagellates comprise two orders that are distinguished by distinct lifestyles and extracellular structures (Carr et al. 2017). Whereas species of the Acanthoecida order primarily live a solitary lifestyle and are encased in an inorganic silica-based “armor” (lorica), species of the Craspedida order have either solitary or colony-forming lifestyles and are often associated with adherent matrices and “cups” (theca) synthesized from organic material (Hoffmeyer and Burkhardt 2016). It should be noted, however, that some Acanthoecida species, such as Diaphanoeca sphaerica, can form colonies, although cells remain solitary inside their lorica (Nitsche et al. 2017). Here, we report the expression of semaphorins and plexins in both orders of choanoflagellates. We describe the unique and common features of semaphorins/plexins in Choanoflagellata and Metazoa, thus providing insights into their shared roles in unicellular and multicellular organisms.
Materials and Methods
Sequence Search
To retrieve semaphorin and plexin protein sequences of choanoflagellates, we searched the FASTA files of translated transcriptomes that were published as supplemental datasets in recent studies (Richter et al. 2018; Brunet et al. 2019) with the BLASTP algorithm (version 2.2.26+; default parameters) (Altschul et al. 1997), using the polypeptide sequences of Plexin-1 and Sema-FN1 of S. rosetta and M. brevicollis (Junqueira Alves et al. 2019) as input queries. Relevant semaphorin and plexin hits in the BLASTP search results were identified by the long length of the alignments, which extended typically over more than 1,000 amino acids. The identity of BLASTP hits as semaphorins or plexins was confirmed by annotating the Sema domain and other parts of their characteristic domain architectures.
The accession codes and assigned names for all plexins and semaphorins analyzed in this study are listed in supplementary table S1, Supplementary Material online, and annotated Genpept sequence files (*.gp) are included in supplementary data, Supplementary Material online. Of note, the study of Richter et al. (2018) covered two strains of Stephanoeca diplocostata, termed FRANCE and AUSTRALIA. The amino acid sequence identity between the Plexin-1 proteins of these two strains is 99.1%, and it is 98.8% between their Sema-FN1 proteins. We used for our studies only the sequences of the strain S. diplocostata FRANCE.
Protein Domain Annotations
Signal peptide sequences were predicted with SignalP 5.0 (www.cbs.dtu.dk/services/SignalP) (URLs in this study were last accessed September 1, 2020). Where applicable, alternative start codons were tested to identify the most likely start codon for the signal peptide. Transmembrane domains were predicted using SOSUI (harrier.nagahama-i-bio.ac.jp/sosui). Initial protein domain annotations were performed by homology searches with HHPred (toolkit.tuebingen.mpg.de/tools/hhpred), SMART (smart.embl.de), NCBI CCD (www.ncbi.nlm.nih.gov/cdd), Expasy Prosite (prosite.expasy.org), and EBI HHMER (www.ebi.ac.uk/Tools/hmmer). Domain annotations were refined by manually comparing sequences with the hidden Markov model (HMM) sequence-conservation logos of protein domains (obtained from PFAM database), by performing secondary structure prediction with JPred (www.compbio.dundee.ac.uk/jpred/), and by analyzing multiprotein alignments (Clustal Omega). For domain assignments in plexins, we also used the crystal structures of the extracellular part of mouse Plexin-A1 (PDB 5L56) (Kong et al. 2016) and the intracellular part of mouse Plexin-A3 (PDB 3IG3) (Wang et al. 2013) as guides for the setting of domain boundaries. To annotate serine/threonine (S/T)-rich domains in semaphorins, peptide sequences were scanned with NetOGlyc (www.cbs.dtu.dk/services/NetOGlyc/) to calculate probability scores for O-linked glycosylation (Steentoft et al. 2013). Results were plotted to graphically identify regions with a high density of S/T-sites with probability scores > 0.9, and these regions were assigned as S/T-rich domains.
Sequence Conservation Plots
To generate sequence conservation plots, multiple alignment files (*.msf) for plexin and semaphorin sequences were created with Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/). The conservation scores were calculated and plotted from the msf alignments with the web-based Plotcon algorithm (emboss.bioinformatics.nl/cgi-bin/emboss/plotcon), using a window size of 75 aa for similarity scoring.
Structural Modeling
The monomer and dimer conformations of H. balthica (Hbalt) Plexin-1 were modeled using a stepwise comparative modeling technique (supplementary fig. S10, Supplementary Material online).
Monomer Templates
First, we removed the signal peptide fragment (from M1 to P27). Next, attempting to obtain a high coverage rate to model the extracellular, transmembrane, and intracellular regions of the protein, we submitted the Hbalt Plexin-1 sequence to the MHOLline web service, version 2.0 (Capriles et al. 2010; Rossi et al. 2020) to select templates considering the BATS (Blast Automatic Targeting for Structures) score, which ranks template sequences from the BLAST results file by sequence alignment score, expectation value, identity, sequence similarity, number of gaps, and the alignment coverage. Applying a BATS score threshold of 0.70, we chose the following set of structures from Protein Data Bank (PDB): 5L5C: A, 5L56: A, 5L59: A, 5L5N: A, 5L5M: A, 5L5L: A, 5L5K: A, 5L5G: A, 3H6N: A, 5V6T: A, 5V6R: A, 3IG3: A, 3RYT: A, 5E6P: A, 2REX: A, 3HM6: X, 3SU8: X, 3SUA: C. Supplementary Table S2, Supplementary Material online lists the identity, similarity, gaps, coverage, and reference of each structure. Finally, we used the Clustal Omega server (Sievers et al. 2011) for multiple sequence alignment between target sequence and templates.
Dimer Templates
To construct a dimeric conformation of active Hbalt Plexin-1, we searched for multimeric plexin structures in PDB. We found the dimeric structure of Danio rerio (zebrafish) Plexin-C1 (PDB 4M8N: A: D), with an intracellular coverage level of 78% (supplementary tables S2 and S3, Supplementary Material online). We used the PyMOL program v. 2.3.4 (Schrödinger) to construct a dimeric structure for the extracellular domain based on the monomeric model. We applied rotations and translations considering both the orientation of mouse Plexin-A2 (PDB 3AL9: A: B), Ramachandran plots obtained with Molprobity web service, and alternative conformations obtained with SymmDock web service (Schneidman-Duhovny et al. 2005). We used Clustal Omega to obtain the multiple sequence alignment between the target sequence and templates.
Model Construction
First, we determined secondary structure predictions (supplementary table S3, Supplementary Material online) with the following programs: NetSurfP (Klausen et al. 2019), Porter (Mirabello and Pollastri 2013), Jufo (Leman et al. 2013), Frag1D (Zhou et al. 2010), PSIPRED (Buchan and Jones 2019), Scratch (Pollastri et al. 2002), and Stride (Frishman and Argos 1995). Supplementary Table S3, Supplementary Material online also presents the alignment between the Hbalt Plexin-1 structural model and the structures of the templates. Additionally, we generated residue-residue contact prediction via CMAPpro (Di Lena et al. 2012), SVMcon (Cheng and Baldi 2007), and RaptorX (Wang et al. 2017), as well as the prediction of transmembrane helices with TMHMM (Krogh et al. 2001) and HMMTOP (Tusnády and Simon 1998).
Next, we used the program Modeller v9.24 (Webb and Sali 2016) to construct the 3D models using multiple templates and restraints of secondary structure and residue-residue contact, as defined in the previous step. We then evaluated the energy quality of the structures of all models using a series of different algorithms: molpdf, DOPE (Discrete Optimized Protein Energy), DOPE-HR, and normalized DOPE. Moreover, we assessed the stereochemical quality via Ramachandran plots generated by Procheck (Laskowski et al. 1996) and Molprobity (Davis et al. 2007).
After selecting the best model, we performed structural optimization using Molprobity and Swiss-PdbViewer program version 4.1 (Guex and Peitsch 1997). The Molprobity software was used to add hydrogens and to flip the violated side chains. With the Swiss-PdbViewer software, we reoriented the transmembrane helix, according to the Ramachandran plot, and minimized the energy with the GROMOS96 force field. Lastly, to assess the energy distribution for each residue in the final 3D model, we generated the DOPE profiles of the monomer and dimer structures and of the relevant template sequences (see supplementary fig. S7, Supplementary Material online).
All structure images were generated using PyMOL version 2.3.4 (Schrödinger). The illustrative phosphatidylcholine (POPC) model membrane was constructed with membrane v1.1 plugin of the VMD program (Humphrey et al. 1996).
Alignments and Phylogenetic Analysis
We performed separate alignments of semaphorin and plexin protein sequences using the algorithm M-Coffee of the T-Coffee software platform (tcoffee.crg.cat), which combines different alignment algorithms to produce optimized alignments. Phylogenetic trees were inferred with MrBayes 3.2 software (Ronquist et al. 2012). For each tree, the program was set to find the best model of substitution during analysis and found that WAG (Whelan and Goldman 2001) is the model that best describes the evolution of both proteins.
For the phylogenetic inference of the 22 Choanoflagellata species evaluated in this study, we generated a partial phylogeny by using a set of six conserved gene sequences (SSU, LSU, tubA, hsp90, EFL, EF-1A) (Carr et al. 2017). We performed separate alignments using the Muscle algorithm (Edgar 2004a,b) implemented in Mega X (Kumar et al. 2016) and combined the aligned sequences with the FASTA alignment joiner (users-birc.au.dk/palle/php/fabox/alignment_joiner.php). We divided the alignment into 14 partitions (three for each of the four coding sequences and one for each of the two no-coding sequences), and used the software MrModelTest (github.com/nylander/MrModeltest2) to search for the best nucleotide substitution model for each partition.
All Bayesian trees were inferred with MrBayes 3.2 software, with the aid of the CIPRES Portal (www.phylo.org). We obtained consensus trees after 50 million steps of Markov chains in two runs of four chains each (with 25% of burning). We examined parameter convergence using Tracer software (Rambaut et al. 2018) and visualized the trees using FigTree software (tree.bio.ed.ac.uk/software/figtree/). The input files containing the alignments and scripts used here are included as supplementary data files, Supplementary Material online.
We also inferred semaphorin/plexin phylogenies using Parsimony and Maximum-Likelihood methods. We used the PAUP* software (Swofford 2003) to infer the Parsimony tree. We started trees via stepwise sequence addition in a random order, with 30 replicates, and swapped the obtained tree using the tree-bisection reconnection (TBR) algorithm with 500 bootstrap replications. We used MEGA X (Kumar et al. 2018) to infer the best substitution model and the Maximum-Likelihood tree with 10,000 bootstrap replications (the best model was WAG, with corrections for the amino acid frequencies, gamma distribution, and the proportion of invariable sites).
Results and Discussion
Domain Architectures of Choanoflagellate Semaphorins and Plexins Reveal Conversed and Unique Features Compared with Metazoan Counterparts
Our previous analysis of the genome sequences M. brevicollis and S. rosetta had identified a pair of Sema-FN1 and Plexin-1 in each of these species (Junqueira Alves et al. 2019). To examine the evolutionary history and diversity of semaphorins and plexins in choanoflagellates on a broader scale, we extended our study to 20 additional choanoflagellate species for which transcriptome data have recently been generated (Richter et al. 2018; Brunet et al. 2019). We searched the predicted protein sequences based on translation of mRNA sequences with the BLASTP algorithm, using the semaphorin and plexin sequences of M. brevicollis and S. rosetta as queries. We discovered 14 additional Sema-FN1 and Plexin-1 proteins in both orders of choanoflagellates–the Craspedida and the Acanthoecida (supplementary table S1; fig. S1–S3, Supplementary Material online; all protein sequence files are included in supplementary data, Supplementary Material online).
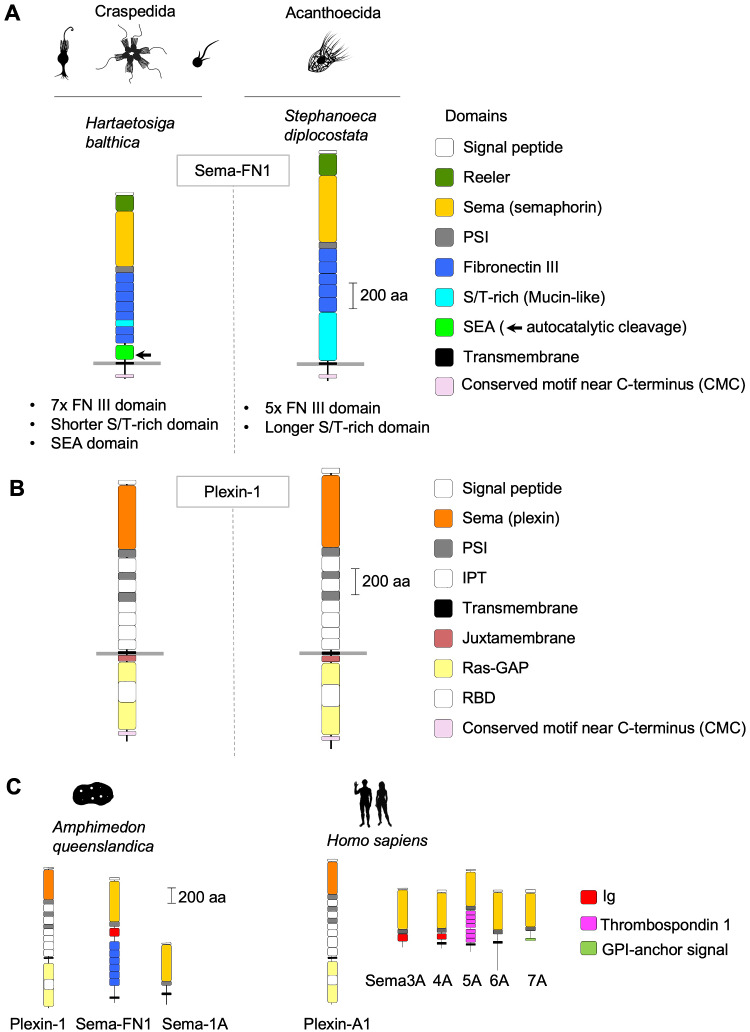
The choanoflagellate Sema-FN1 proteins are structurally more diverse than the corresponding plexins (fig. 1A). Following the signal peptide (which is removed in the secretory pathway, thus no longer part of the mature protein), choanoflagellate Sema-FN1 begins with an N-terminal Reeler domain, a unique structural arrangement not found in metazoan semaphorins (Junqueira Alves et al. 2019). Interestingly, several mammalian extracellular matrix proteins, such as F-spondin and reelin also contain an N-terminal Reeler domain, and they play important roles in axon guidance and neural precursor migration (Klar et al. 1992; D'Arcangelo et al. 1995). As the Reeler domain can form weak homophilic dimers (Tan et al. 2008), it is plausible that it may help stabilize Sema-FN1 dimers, thus facilitating dimerization and activation of Plexin-1 receptors.
Fig. 1.
Domain architecture of choanoflagellate semaphorins and plexins. (A) Domain architecture of Sema-FN1, shown for one Craspedida and one Acanthoecida species. The N-terminal Reeler domain is unique for choanoflagellate semaphorins and not found in metazoan semaphorins. It is followed by a Sema and a PSI domain. The extracellular stalk contains 7 FNIII domains in Craspedida and 5 in Acanthoecida. A SEA domain with autocatalytic cleavage site (arrow) is only present in Craspedida Sema-FN1. The serine/threonine (S/T)-rich domain of variable size is predicted to be the site of O-linked glycosylation. A CMC motif containing ∼10 conserved amino acid positions is found close to the C-terminus. (B) Domain architecture of Plexin-1 is conserved in choanoflagellates and nearly identical to Metazoa plexins, except for the presence of a CMC motif with similarity to the CMC of Sema-FN1. Note that the Signal peptides (N-terminal), and IPT (extracellular) and RBD domains (intracellular) are colored in white. (C) Representative examples of plexin and semaphorin orthologs of Metazoa, shown for a sponge species (Amphimedon queenslandica) and for man (Homo sapiens). Silhouette pictures of organisms are from phylopic.org.
The Reeler domain of Sema-FN1 is followed by the stereotypical Sema plus PSI domain arrangement found in all semaphorins and plexins. The next part of the extracellular stalk of Sema-FN1 consists of an array of fibronectin type III (FNIII) domains: 7 in Craspedida Sema-FN1, and 5 in Acanthoecida Sema-FN1.
Following the FNIII domains, Craspedida and Acanthoecida semaphorins showed further divergence. First, in Sema-FN1 of Craspedida, the FNIII array is followed by a SEA domain, which is not present in Sema-FN1 of Acanthoecida or in metazoan semaphorins. During protein synthesis, the SEA domain undergoes autoproteolysis at the motif G↓S[V/I]VV (Pelaseyed et al. 2013; Pei and Grishin 2017), and this motif is indeed also conserved in the SEA domain of Craspedida Sema-FN1. The SEA domain has been suggested to act as a “breaking point” in various transmembrane proteins, enabling them to dissociate under mechanical pull in order to prevent breaching of the plasma membrane (Pelaseyed et al. 2013). In this context, it could be speculated that the SEA domain of Craspedida Sema-FN1 may act similarly as a protective mechanism against excessive pulling forces.
A second divergence between the semaphorin orthologs of the two orders of choanoflagellates is the length of the serine/threonine (S/T)-rich domains: ∼20–100 amino acids (aa)-long for most Craspedida Sema-FN1 and ∼50–600 aa-long for Acanthoecida Sema-FN1. The S/T-rich domains are predicted sites for O-linked glycosylation. Interestingly, in mammalian Mucin proteins, the S/T-rich domains are similarly found near a SEA domain, thus the S/T-rich domain of Craspedida Sema-FN1 is likely of the Mucin-like type (Pei and Grishin 2017). O-linked glycosylation of Mucin-like S/T-rich domains can provide stability to protein structures and lubrication and protection of cell surfaces (Van den Steen et al. 1998).
No previously known domains were found in the intracellular part of choanoflagellate Sema-FN1. However, based on sequence alignments, we detected a conserved motif near the C-terminus (termed CMC), a motif stretching over ∼20–70 aa, with approximately 10 conserved positions (supplementary fig. S4, Supplementary Material online). The CMC was detected in all choanoflagellate Sema-FN1 proteins with the exception of Didymoeca costata (a species belonging to the Acanthoecida order). So far, the CMC has no identifiable homology to other known protein sequences.
The choanoflagellate plexins (Plexin-1) share a highly conserved domain structure with metazoan plexins (fig. 1B and C). The extracellular part of Plexin-1 consists of a Sema domain followed by an array of alternating PSI and IPT domains. The intracellular part begins with a juxtamembrane helix, followed by the Ras-GAP domain, which contains an insert of a RBD domain (Wang et al. 2013). The high conservation between choanoflagellate Plexin-1 and metazoan plexins on the domain architecture level—in both extracellular and intracellular parts—suggests a conserved fundamental signaling mechanism in regulating cellular processes via Ras small GTPases.
Choanoflagellate Plexin-1 also contains a CMC motif with homology to that of Sema-FN1 (supplementary fig. S4, Supplementary Material online). The C-terminal location of the CMC leads us to speculate that it may anchor both Plexin-1 and Sema-FN1 to scaffold proteins of similar types to control subcellular localization.
Diversification of Semaphorins/Plexins in Choanoflagellate Species
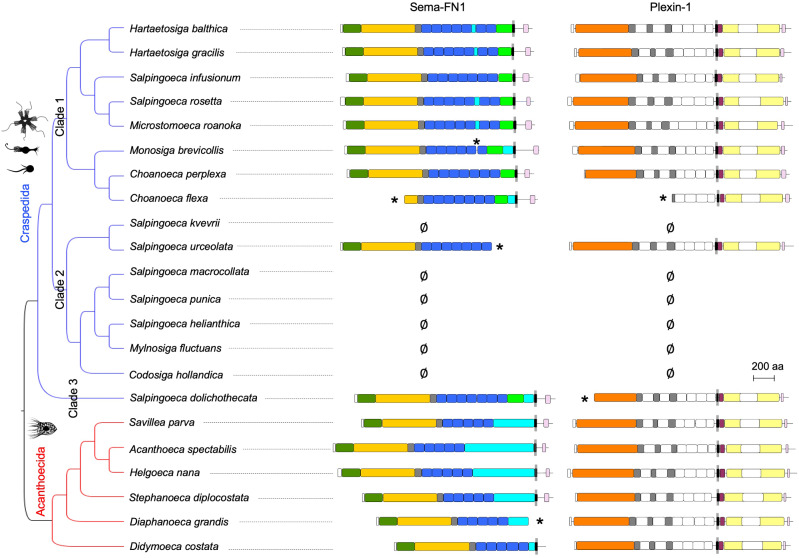
As mentioned above, out of the 22 choanoflagellate species that we surveyed, we detected semaphorin/plexin pairs in 16 species, including members in both Craspedida and Acanthoecida orders (fig. 2). Six species have no detectable semaphorin or plexin sequences in their translated transcriptomes, and they all belong to clade 2 of Craspedida (Carr et al. 2017), forming two groups: one comprised only Salpingoeca kvevrii and the other included Codosiga hollandica and four other related species (fig. 2). We cannot currently distinguish if this absence is due to gene loss or transcriptional silencing. The high sequencing coverage of the transcriptomes makes it unlikely that technical artifacts play a role (supplementary fig. S5, Supplementary Material online).
Fig. 2.
Semaphorins and plexins expressed in choanoflagellates. Protein domain structures of semaphorins and plexins, plotted next to the topology of the Bayesian phylogenetic hypothesis of the choanoflagellates analyzed in this study based on 10,816 aligned nucleotides from partial sequences of the genes SSU, LSU, tubA, EFL, and EF-1A (after Carr et al. 2017). Asterisks (*) indicate incomplete sequence and ∅ indicates that the respective semaphorin or plexin mRNA was not detectable. See fig. 1 for colors of domains. See fig. 6 for branch lengths and posterior probabilities of nodes.
Interestingly, another choanoflagellate plexin has recently been detected in the uncultured species UC1 (in addition to the 22 species detailed here). This uncultered species was identified by single-cell genome sequencing of microorganisms in oceanic waters (López-Escardó et al. 2019). We found that the Plexin-1 of UC1 also has the typical domain architecture of plexins, and falls into the order Craspedida (supplementary fig. S6, Supplementary Material online). No semaphorin has yet been identified for UC1, possibly due to limited genome coverage of the single-cell sequencing technology.
Sequence Conservation of Choanoflagellate Semaphorins and Plexins
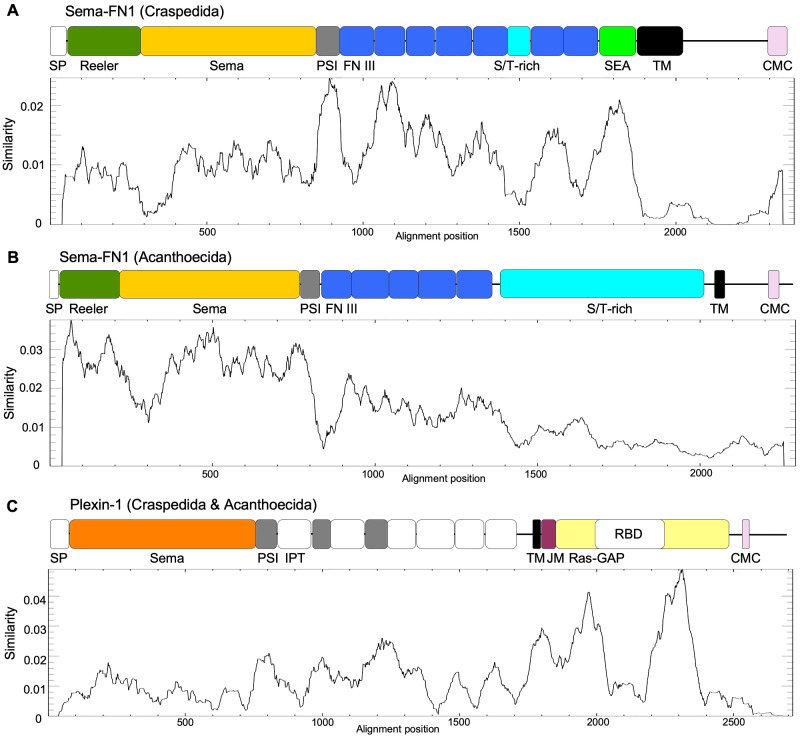
We next performed protein sequence alignments of all choanoflagellate semaphorins and plexins described here and plotted similarity scores below their domain structures. This revealed that for Craspedida Sema-FN1, the stalk area displayed the highest conservation, whereas for Acanthoecida Sema-FN1, the N-terminal domains were the best conserved regions (fig. 3A and B). For choanoflagellates plexins, the highest degree of conservation is in the intracellular Ras-GAP domain (fig. 3C), underscoring the importance of this catalytic domain for plexin signaling.
Fig. 3.
Protein sequence conservation of choanoflagellate semaphorins and plexins. (A) Sequence conservation plot for Craspedida Sema-FN1. The highest conserved parts are the PSI domain and the SEA domain, whereas the intracellular part exhibits lowest sequence conservation, except for the CMC motif. The y axis represents similarity score calculated by plotcon algorithm, based on protein alignments (window size of 75 aa). Note that some protein domains stretched out according to alignments and are not to scale. SP, signal peptide; FN III, fibronectin type III domain; TM, transmembrane domain. CMC, conserved motif near C-terminus. (B) Conservation plot of Sema-FN1 sequences of Acanthoecida shows high sequence conservation of the Reeler and the Sema domains, but low conservation of the S/T-rich domain. (C) Sequence conservation plot for choanoflagellate Plexin-1. The highest conservation is in the intracellular Ras-GAP domain, which is interrupted by the insert of a less conserved RBD domain. JM, juxtamembrane helix. See also fig. 1 for domain names.
3D Modelling Reveals Structural Conservation of Choanoflagellate Plexin-1 and Metazoan Plexins
The high conservation of the domain order of choanoflagellate and metazoan plexins prompted us to carry out 3D structural modeling, using the structural data of vertebrate plexins as templates. We modeled Plexin-1 of Hartaetosiga balthica (Hbalt) as an example for a choanoflagellate plexins.
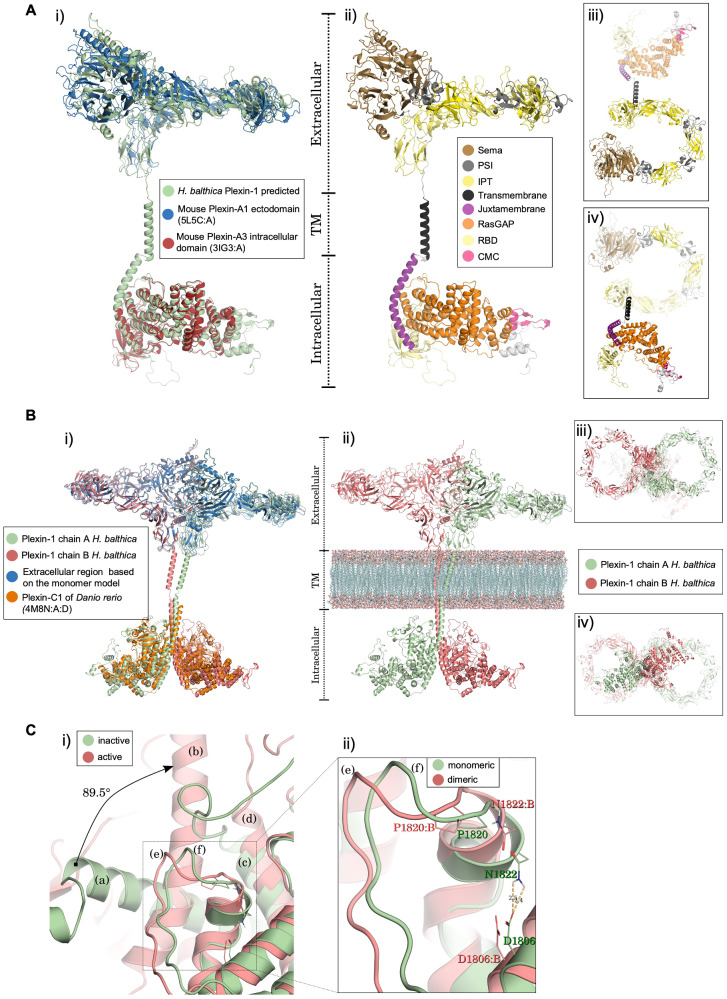
We first predicted the monomeric 3D structure of Hbalt Plexin-1 using comparative modeling with multiple templates of vertebrate plexins to ensure as much coverage as possible (supplementary tables S2 and S3, Supplementary Material online). We found that the best structural model for Hbalt Plexin-1 presented a high level of architectural conservation compared with the vertebrate templates (fig. 4A-i). Hbalt Plexin-1 has the typical domain architecture found in all metazoan plexins: a large extracellular multidomain in a ring-shaped arrangement, a single transmembrane helix, and an intracellular signaling domain (fig. 4A-ii; supplementary table S4, Supplementary Material online). The energy profiles further demonstrated structural consistency and conservation between Hbalt Plexin-1 and the main vertebrate plexin templates (i.e., mouse Plexin-A1 ectodomain and -A3 intracellular domain) based on similar energy presented by the amino acid residues (supplementary fig. S7A, Supplementary Material online). The stereochemical quality examined by Ramachandran plots showed that the vast majority of the residues were in favorable or allowed regions (97.4% and 97.0%, according to Molprobity and Procheck, respectively).
Fig. 4.
Structural 3D models of choanoflagellate Plexin-1. (A) Monomer model of H. balthica (Hbalt) Plexin-1. (i) Structural alignment of monomeric Hbalt Plexin-1 with the ectodomain of mouse Plexin-A1 (PDB 5L5C: A) and the intracellular domain of mouse Plexin-A3 (3IG3: A). (ii–iv) Domain organization of Hbalt Plexin-1, shown as front view (ii), top view (iii), and bottom view (iv). Extracellular domains: SEMA, PSI, and IPT. Transmembrane region: TM. Intracellular domains: JM (juxtamembrane helix), Ras-GAP, RBD, and CMC (conserved motif near C-terminus). (B) 3D model of dimeric Hbalt Plexin-1. (i) Structure of Hbalt Plexin-1 dimer model (chains A and B) aligned with extracellular region of monomer model and intracellular regions of dimeric zebrafish Plexin-C1 (4M8N: A: D). (ii) Overview of extracellular, transmembrane, and intracellular domains (front view): chains A and chain B traversing model membrane (phosphatidylcholine) constructed with VMD software. (iii) Top view of the dimeric model. (iv) Bottom view of the dimeric model. (C) Structural changes predicted upon dimerization of Hbalt Plexin-1. (i) Comparison between the inactive (monomeric) and active (after dimerization) structures. The juxtamembrane helix rotates 89.5° from the inactive (a) to the active structure (b) (chain A). The helix 11 (according to Wang et al., 2013) remains stable in both conformations (c and d). The activation segments (e) and (f) also undergo conformational changes. (ii) Detailed view of conformational changes of activation segment between monomeric and dimeric structures. The displacement in the activation segment of dimeric conformation (chain B) promotes breakdown of the hydrogen bond between D1806: B and N1822: B.
Specifically, the ring-shaped extracellular region of Hbalt Plexin-1 consists of a Sema domain, followed by three consecutive PSI-IPT domains, three additional IPT domains, and a transmembrane region (TM) (fig. 4A-iii). The intracellular region of Hbalt Plexin-1 contains a juxtamembrane helix followed by a Ras-GAP domain with an inserted RBD domain (fig. 4A-iv), also in agreement with vertebrate plexin structures (He et al. 2009; Wang et al. 2013; Kong et al. 2016). Following the Ras-GAP domain is a CMC motif, composed of one helix followed by two antiparallel beta-strands based on structural prediction (see fig. 4A-ii).
We next modeled dimeric Hbalt Plexin-1 protein structures to gain insights into signaling activation mechanisms of choanoflagellate plexins. Earlier studies showed that the Ras-GAP domain of plexins in the monomeric state adopts an inactive conformation (He et al. 2009; Tong et al. 2009; Wang et al. 2012; Wang and Xu 2013); but upon dimerization, conformation changes in the intracellular region of plexins lead to Ras-GAP activation (Wang et al. 2013; Kuo and Zhang 2016). We therefore constructed a 3D structure model for dimeric Hbalt Plexin-1, which showed convergence of energy profiles for all amino acid residues as compared with the main vertebrate templates, suggesting structural consistency and conservation (fig. 4B and supplementary fig. S7B, Supplementary Material online). The stereochemical quality analysis by Molprobity and Procheck indicated that 97.0% of the residues were in favorable or allowed regions. The predicted dimerization of Hbalt Plexin-1 involves multiple segments at the dimer interface: juxtamembrane helix (L1351-E1368), helix 11 (I1702-T1716), and a loop-helix segment (C1808-K1828), termed “activation segment,” that extrudes from the Ras-GAP domain (fig. 4C-i). In the juxtamembrane helix, upon dimerization, the last two turns (G1369-S1377) unwind, converting to an extended loop that allows the remaining N-terminal helical portions (R1335-E1368) to straighten and rotate by 89.5° (fig. 4C-i; supplementary movie S1, Supplementary Material online). Meanwhile, in the Ras-GAP domain, there is a slight adjustment of helix 11 to accommodate the movement of the juxtamembrane helix (fig. 4C-i).
Notably, in the structural model of Plexin-1, a hydrogen bond between an asparagine residue in the activation segment (N1822) and an aspartate residue in the following helix (D1806) stabilize the inactive structure. Additionally, a neighboring proline residue (P1820) buries the asparagine residue (N1822) and prevents its access to the Rap substrate (fig. 4C-ii). Concordantly, Wang et al. have also shown steric clashes between this proline residue and the Rap substrate in an inactive Plexin-C1/Rap complex using molecular docking (Wang et al. 2013). Upon dimerization, the new conformation of the juxtamembrane helix and helix 11 leads to conformational changes in the activation segment, pulling it away from the Ras-GAP active site, thus preventing the hydrogen bond formation and permitting the catalytic mechanism for signal transduction (fig. 4C-ii).
Phylogenetic Analysis of Choanoflagellate Semaphorins and Plexins
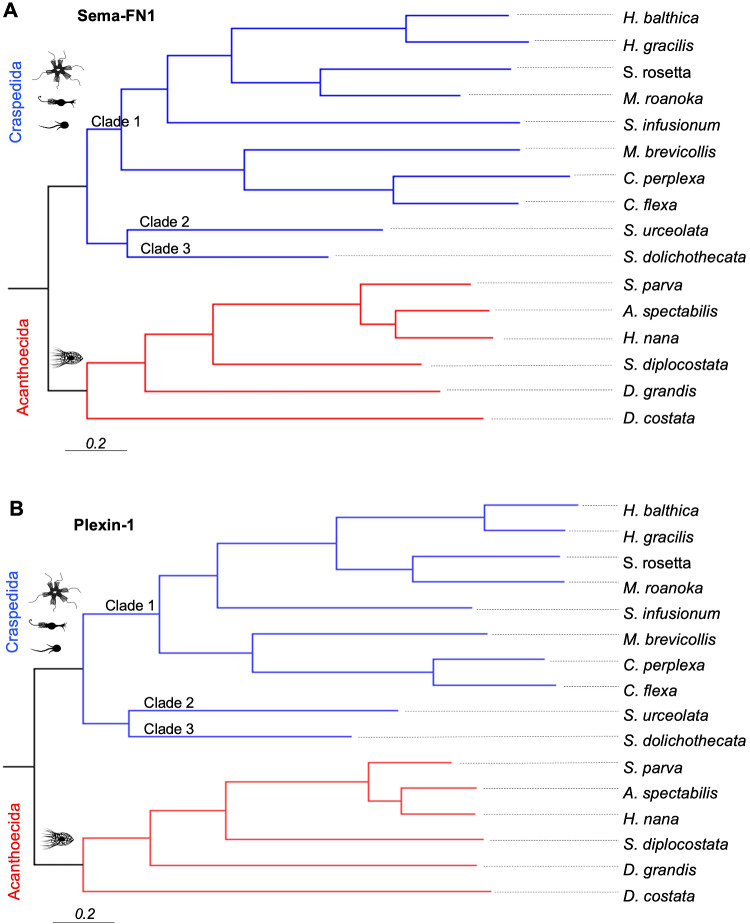
To gain further insights into the evolutionary relationship of semaphorins and plexins in choanoflagellates, we inferred phylogenetic trees based on Sema-FN1 and Plexin-1 protein sequences. The topologies of the Bayesian phylogenetic trees were identical for semaphorins and plexins, and both were well supported, as shown by the posterior probabilities of 1.0 for all internal nodes and the absence of polytomies (fig. 5A and B). Parsimony and Maximum-Likelihood analyses revealed similar topologies (supplementary fig. S8, Supplementary Material online). The phylogenetic trees of Plexin-1 and Sema-FN1 were also highly similar to the Choanoflagellata phylogenetic hypothesis based on a six-gene approach (Carr et al. 2017), with one exception regarding the position of Salpingoeca urceolata (a member of the Craspedida clade 2): in the semaphorin/plexin phylogenetic trees, it is in the sister group of clade 3, whereas in the six-gene phylogeny tree, it is in the sister group of clade 1 (figs. 5A, B, and 6).
Fig. 5.
Phylogenetic trees of semaphorins and plexins. Phylogenetic trees of choanoflagellate Sema-FN1 (A) and Plexin-1 (B) proteins. Trees based on 50% majority-rule consensus Bayesian inference from 2,902 aligned amino acids from available Plexin-1 sequences and 3,022 aligned amino acids of Sema-FN1 sequences. Branch lengths are proportional to the number of amino acids substitutions per site, indicated by the scale bar. Posterior probability values reached 1 for all nodes and are omitted for simplicity.
Semaphorin/Plexin Expression Shows No Correlation with Ecological or Morphological Characteristics of Choanoflagellates
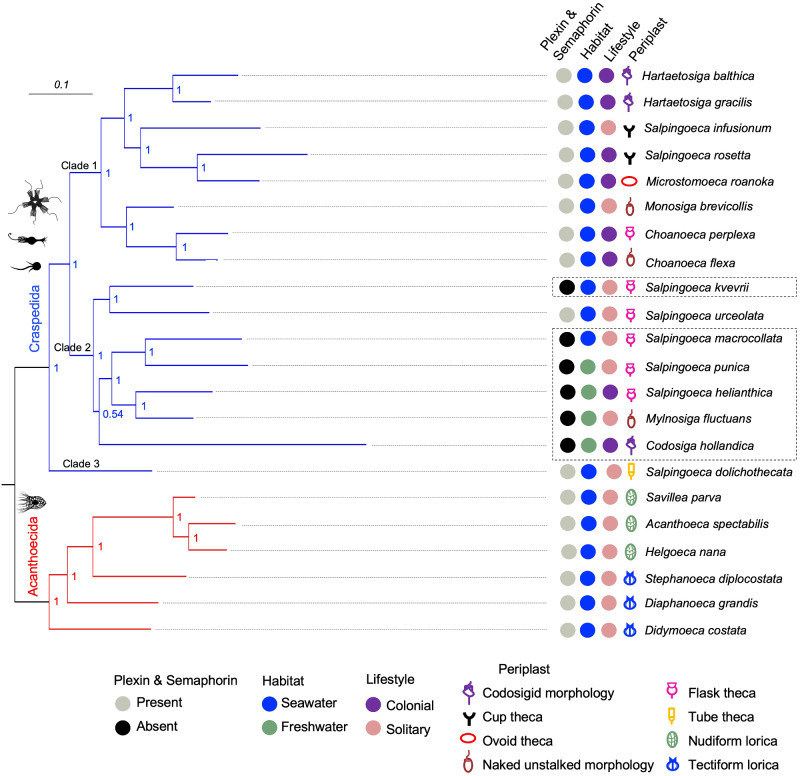
To investigate putative correlations between the expression of semaphorin/plexin and the ecological or morphological traits of choanoflagellates, we plotted semaphorin/plexin expression status, preferred habitat (seawater vs. saltwater), potential lifestyle (colonial vs. solitary), and periplast characteristics next to the phylogenetic tree of Choanoflagellata (fig. 6 and supplementary table S5, Supplementary Material online).
Fig. 6.
Absence of correlation of semaphorin/plexin expression with ecological or morphological characteristics of choanoflagellates. Left, Bayesian phylogenetic hypothesis of choanoflagellates (based on 10,816 aligned nucleotides from partial sequences of the genes SSU, LSU, tubA, EFL, and EF-1A) (after Carr et al., 2017). Branch lengths are proportional to the number of nucleotide substitutions per site, indicated by the scale bar (top left). Values at internal nodes denote posterior probabilities. Right, colored dots and symbols indicate the presence or absence of semaphorin/plexin expression, preferred habitats (seawater vs. freshwater), potential lifestyles (colonial vs. solitary), and the type of periplast. The species lacking plexin and semaphorin expression are boxed with dashed lines.
Considering a more comprehensive sample of Choanoflagellata, it is apparent that Craspedida species can be found in both seawater and freshwater environments, whereas Acanthoecida live mainly in seawater, with only two species found in freshwater lakes (Paul 2012; Nitsche 2014). We found that none of the four freshwater species of Craspedida in our study expressed semaphorin/plexin. However, two saltwater species, S. kvevrii and S. macrollata, also lacked semaphorin/plexin expression, suggesting that the expression of semaphorin/plexin is not essential for life in either freshwater or saltwater.
We next investigated whether semaphorin/plexin expression might be potentially associated with colonial or solitary lifestyle, however, no correlations were apparent (fig. 6). Interestingly, analysis of Sema-FN1 and Plexin-1 expression in S. rosetta (Fairclough et al. 2013) revealed that even though both genes were expressed in all stages of colonial and solitary life forms, the expression level was about 2–3-fold higher in colonial forms and in solitary swimmers (which can initiate colony formation) than in other life forms (supplementary fig. S9, Supplementary Material online). This raises the possibility that semaphorin/plexin may contribute to the process of colony formation for S. rosetta. We wish to emphasize the uncertainty of the coloniality status for many species due to incomplete data, thus future analyses of lifestyle in expanded datasets may provide additional information on this question.
Finally, no correlation was detected between semaphorin/plexin expression and the periplast type (i.e., the extracellular structures of choanoflagellates) (fig. 6). It will be interesting to examine in future studies with more comprehensive data sets if a particular trait or ecological feature of choanoflagellates can be associated with semaphorin/plexin expression.
Functional Convergence of Semaphorin and Plexin Evolution
We observed two noteworthy instances of structural similarities between choanoflagellate and metazoan semaphorin/plexin proteins that may reflect convergent evolution due to shared functions. They may also provide information regarding a link of semaphorin/plexin function with mechanobiology.
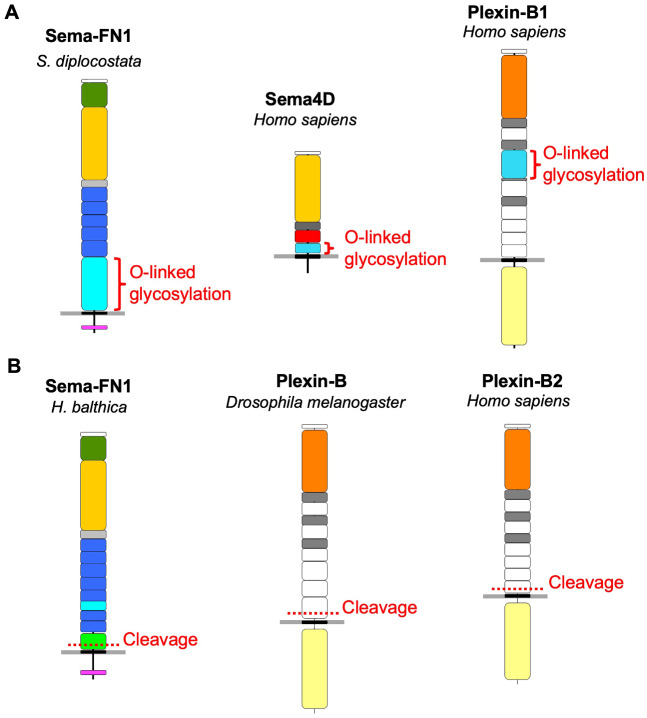
First, the S/T-rich domain, a potential site for O-linked glycosylation, is present in both choanoflagellate Sema-FN1 and mammalian Sema4D and Plexin-B1 (fig. 7A). As we did not find such domains in semaphorins and plexins of other metazoan clades, it seems likely that these S/T-rich domains may have evolved twice independently. The O-glycosylation of the S/T-rich region in human Sema4D and Plexin-B1 has been biochemically confirmed (Steentoft et al. 2013; King et al. 2017) (see glycodomain.glycomics.ku.dk). Such O-glycosylation may support the embedding of semaphorins and plexins in the glycocalyx, a carbohydrate-enriched protective coating that surrounds mammalian cells and also choanoflagellates (Leadbeater 2008). Interestingly, the glycocalyx has been linked to the regulation of cell adhesion and mechano-transduction (DuFort et al. 2011).
Fig. 7.
Potential convergence in plexin and semaphorin evolution. (A) S/T-rich domains (red brackets), which are sites for O-linked glycosylation likely contributing to a glycocalyx surrounding cells, are present in choanoflagellate Sema-FN1 and also in mammalian Sema4D and Plexin-B1. These domains are likely acquired independently during evolution of semaphorin/plexin proteins. (B) Peptide cleavage (dotted red lines) of extracellular domain in the juxtamembrane area of the extracellular domain occurs in choanoflagellate Sema-FN1 by autoproteolysis of the SEA domain, and in Drosophila Plexin-B and human Plexin-B2 (and Plexin-B1, not shown) by proteolytic cleavage by pro-protein convertases. This cleavage may provide a “breaking point’ as a protective mechanism against excessive mechanical pulling forces. See fig. 1 for colors of domains.
A second interesting similarity between choanoflagellate and metazoan semaphorins/plexins is the peptide cleavage in the extracellular domain near the plasma membrane. In the case of mammalian Plexin-B and Drosophila Plexin-B plexins, it occurs through proteolytic processing, for example, by pro-protein convertases (Artigiani et al. 2003); but in the case of choanoflagellate Sema-FN1, it likely occurs in the SEA domain by autoproteolytic cleavage (fig. 7B). It appears that these independently evolved cleavage points in the peptide chain could provide mechanical protection as a “breaking point” if an excessive force would stretch plexin protein. In this context, it is interesting to note that a recent study has described a mechanosensing role for the mammalian Plexin-D1 (Mehta et al. 2020), further supporting a link of plexins with biomechanical processes.
Conclusion
The analysis of cell surface proteins of choanoflagellates can provide insights into the origin of multicellular organisms. For example, choanoflagellates express several cell adhesion proteins, for example, cadherins, that are thought to have been repurposed for the development of multicellularity in Metazoa (King et al. 2003). The current study has identified semaphorins and plexins as additional examples of cell surface proteins shared by Choanoflagellata and Metazoa.
Interestingly, choanoflagellate plexins are well conserved, both in domain order and in 3D structure, supporting their conserved function in affecting cytoskeletal dynamics. In contrast, choanoflagellate semaphorins are structurally diverse, consistent with the model that the primary function of semaphorins is to present the Sema domain to plexin receptors, leading to their dimerization. The three unique domain features of choanoflagellate Sema-FN1 not found in metazoan semaphorins may represent adaptations to specific requirements in choanoflagellates: the N-terminal Reeler domain may support dimerization, the S/T-rich domain may enable association with glycocalyx, and the juxtamembrane SEA domain may allow peptide cleavage as a protective mechanism against mechanical rupture of plasma membrane. Remarkably, the feature of O-linked glycosylation is also found in mammalian Sema4D and Plexin-B1, whereas the feature of juxtamembrane peptide cleavage is also found in fly and mammalian Plexin-Bs, suggesting that these protein properties may have independently evolved as a result of functional convergence related to shared biomechanical functions.
As Metazoa of all clades express plexins and semaphorins (Junqueira Alves et al. 2019), we hypothesize that plexin function is indispensable for multicellular development. In metazoan evolution, several genome duplication events may have allowed copies of plexins and semaphorins to evolve into newly specialized forms, likely promoting diversification of cellular interactions in a wide range of developmental and physiological contexts.
Interestingly, the expression of semaphorin/plexin genes was not detected in two groups of species of clade 2 of Craspedida. This may have been caused by loss of chromosomal segments or silencing of gene expression due to mutations or epigenetic changes; further whole-genome reconstructions will be needed to answer this question. Based on phylogenies, two independent chromosome segment losses may have occurred in the ancestors of each of the two groups; alternatively, a single gene silencing event may have occurred in an ancestor of clade 2 species, which was reverted in the lineage of S. urceolata. At this point, we can conclude that semaphorin/plexin expression is not essential for choanoflagellate survival, but may provide additional fitness.
The prototypic function of plexins may not lie in the direct regulation of cell–cell interactions, but more likely in the control of cytoskeletal dynamics. We may add here as a speculation that a potential function of choanoflagellate semaphorin/plexin activity could be associated with the collar feeding apparatus, an evolutionary hallmark of choanoflagellates, which displays remarkable cytoskeletal structural plasticity during prey capture (Dayel and King 2014). In this context, it is worth noting that sponges also express Sema-FN1 that is structurally related to choanoflagellate Sema-FN1 (see fig. 1). The choanocyte cells of sponges are suspected to be homologous to choanoflagellates, representing a potential evolutionary link between Metazoa and Choanoflagellata (Brunet and King 2017). It will be interesting to examine if the Sema-FN1 of sponges is indeed expressed by the choanocyte cells and whether it executes a similar function as in choanoflagellates.
In summary, our study on choanoflagellate semaphorins and plexins revealed shared but also unique features compared with their metazoan counterparts. Our findings support a conserved function of semaphorin/plexin signaling in the mechanoregulation of cellular processes in both unicellular and multicellular organisms.
Supplementary Material
Supplementary material are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
Acknowledgments
We thank Martin Carr, University of Huddersfield, Thibaut Brunet, UC Berkeley, and Jimin Pei, UT Southwestern, for scientific advice. This work was supported by National Institutes of Health (R01 NS092735 to R.H.F.).
References
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigiani S, et al. 2003. Functional regulation of semaphorin receptors by proprotein convertases. J. Biol. Chem. 278(12):10094–10101. [DOI] [PubMed] [Google Scholar]
- Brunet T, King N. 2017. The origin of animal multicellularity and cell differentiation. Dev. Cell 43(2):124–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet T, et al. 2019. Light-regulated collective contractility in a multicellular choanoflagellate. Science 366(6463):326–334. [DOI] [PubMed] [Google Scholar]
- Buchan DWA, Jones DT. 2019. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47(W1):W402–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriles PV, et al. 2010. Structural modelling and comparative analysis of homologous, analogous and specific proteins from Trypanosoma cruzi versus Homo sapiens: putative drug targets for chagas’ disease treatment. BMC Genomics 11:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M, et al. 2017. A six-gene phylogeny provides new insights into choanoflagellate evolution. Mol. Phylogenet. Evol. 107:166–178. [DOI] [PubMed] [Google Scholar]
- Cheng J, Baldi P. 2007. Improved residue contact prediction using support vector machines and a large feature set. BMC Bioinformatics 8(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G, et al. 1995. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374(6524):719–723. [DOI] [PubMed] [Google Scholar]
- Davis IW, et al. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35(web server issue):W375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, King N. 2014. Prey capture and phagocytosis in the choanoflagellate Salpingoeca rosetta. PLoS One 9(5):e95577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lena P, Nagata K, Baldi P. 2012. Deep architectures for protein contact map prediction. Bioinformatics 28(19):2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM. 2011. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell. Biol. 12(5):308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004a. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004b. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, et al. 2013. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14(2):R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Argos P. 1995. Knowledge-based protein secondary structure assignment. Proteins 23(4):566–579. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723. [DOI] [PubMed] [Google Scholar]
- Gurrapu S, Tamagnone L. 2016. Transmembrane semaphorins: multimodal signaling cues in development and cancer. Cell Adh. Migr. 10(6):675–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Yang T, Terman JR, Zhang X. 2009. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc. Natl. Acad. Sci. U.S.A. 106(37):15610–15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer TT, Burkhardt P. 2016. Choanoflagellate models - Monosiga brevicollis and Salpingoeca rosetta. Curr. Opin. Genet. Dev. 39:42–47. [DOI] [PubMed] [Google Scholar]
- Hota PK, Buck M. 2012. Plexin structures are coming: opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions. Cell. Mol. Life Sci. 69(22):3765–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14(1):33–38. [DOI] [PubMed] [Google Scholar]
- Jones EY. 2015. Understanding cell signalling systems: paving the way for new therapies. Philos. Trans. A Math. Phys. Eng. Sci. 373:20130155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloets BC, Pasterkamp RJ. 2014. Semaphorin signalling during development. Development 141(17):3292–3297. [DOI] [PubMed] [Google Scholar]
- Junqueira Alves C, Yotoko K, Zou H, Friedel RH. 2019. Origin and evolution of plexins, semaphorins, and Met receptor tyrosine kinases. Sci. Rep. 9(1):1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. 2003. Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301(5631):361–363. [DOI] [PubMed] [Google Scholar]
- King SL, et al. 2017. Characterizing the O-glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Adv. 1(7):429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A, Baldassare M, Jessell TM. 1992. F-spondin: a gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell 69(1):95–110. [DOI] [PubMed] [Google Scholar]
- Klausen MS, et al. 2019. NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins 87(6):520–527. [DOI] [PubMed] [Google Scholar]
- Kong Y, et al. 2016. Structural basis for plexin activation and regulation. Neuron 91(3):548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropouli E, Kolodkin AL. 2014. Semaphorins and the dynamic regulation of synapse assembly, refinement, and function. Curr. Opin. Neurobiol. 27:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YC, Zhang X. 2016. Regulation of plexin: a ring of structural twists and turns. Neuron 91(3):497–499. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8(4):477–486. [DOI] [PubMed] [Google Scholar]
- Leadbeater BSC. 2008. Choanoflagellate evolution: the morphological perspective. Protistology. 5:256–267. [Google Scholar]
- Leman JK, Mueller R, Karakas M, Woetzel N, Meiler J. 2013. Simultaneous prediction of protein secondary structure and transmembrane spans. Proteins 81(7):1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Escardó D, et al. 2019. Reconstruction of protein domain evolution using single-cell amplified genomes of uncultured choanoflagellates sheds light on the origin of animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374(1786):20190088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V, et al. 2020. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature 578(7794):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello C, Pollastri G. 2013. Porter, PaleAle 4.0: high-accuracy prediction of protein secondary structure and relative solvent accessibility. Bioinformatics 29(16):2056–2058. [DOI] [PubMed] [Google Scholar]
- Nitsche F. 2014. Stephanoeca arndti spec. nov.–first cultivation success including molecular and autecological data from a freshwater acanthoecid choanoflagellate from Samoa. Eur. J. Protistol. 50(4):412–421. [DOI] [PubMed] [Google Scholar]
- Nitsche F, Thomsen HA, Richter DJ. 2017. Bridging the gap between morphological species and molecular barcodes - exemplified by loricate choanoflagellates. Eur. J. Protistol. 57:26–37. [DOI] [PubMed] [Google Scholar]
- Pascoe HG, Wang Y, Zhang X. 2015. Structural mechanisms of plexin signaling. Prog. Biophys. Mol. Biol. 118(3):161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. 2012. Acanthocorbis mongolica nov. spec.: description of the first freshwater loricate choanoflagellate (Acanthoecida) from a Mongolian lake. Eur. J. Protistol. 48(1):1–8. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. 2017. Expansion of divergent SEA domains in cell surface proteins and nucleoporin 54. Protein Sci. 26(3):617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, et al. 2013. Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell-protective device. FEBS J. 280(6):1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P. 2002. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 47(2):228–235. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DJ, Fozouni P, Eisen MB, King N. 2018. Gene family innovation, conservation and loss on the animal stem lineage. Elife 7:e34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi AD, et al. 2020. MHOLline 2.0: workflow for automatic large-scale modeling and analysis of proteins. Rev. Mund. Engen Tecnol. Gestão 5:1–14. [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33(web server issue):W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, et al. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. Embo J. 32(10):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, et al. 2016. Structure of the plexin ectodomain bound by semaphorin-mimicking antibodies. PLoS One. 11(6):e0156719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *Swofford DL. 2003. PAUP. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland (MA): Sinauer Associates.
- Tan K, Duquette M, Liu JH, Lawler J, Wang JH. 2008. The crystal structure of the heparin-binding reelin-N domain of f-spondin. J. Mol. Biol. 381(5):1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, et al. 2009. Structure and function of the intracellular region of the plexin-b1 transmembrane receptor. J. Biol. Chem. 284(51):35962–35972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády GE, Simon I. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283(2):489–506. [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. 1998. Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33(3):151–208. [DOI] [PubMed] [Google Scholar]
- Wang S, Sun S, Li Z, Zhang R, Xu J. 2017. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13(1):e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 5(207):ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pascoe HG, Brautigam CA, He H, Zhang X. 2013. Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin. Elife 2:e01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu J. 2013. Predicting protein contact map using evolutionary and physical constraints by integer programming. Bioinformatics 29(13):i266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18(5):691–699. [DOI] [PubMed] [Google Scholar]
- Zhou T, Shu N, Hovmöller S. 2010. A novel method for accurate one-dimensional protein structure prediction based on fragment matching. Bioinformatics 26(4):470–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.