Abstract
Chemoreceptors help insects to interact with their environment, to detect and assess food sources and oviposition sites, and to aid in intra- and interspecific communication. In Hymenoptera, species of eusocial lineages possess large chemoreceptor gene repertoires compared with solitary species, possibly because of their additional need to recognize nest-mates and caste. However, a critical piece of information missing so far has been the size of chemoreceptor gene repertoires of solitary apoid wasps. Apoid wasps are a paraphyletic group of almost exclusively solitary Hymenoptera phylogenetically positioned between ant and bee, both of which include eusocial species. We report the chemosensory-related gene repertoire sizes of three apoid wasps: Ampulex compressa, Cerceris arenaria, and Psenulus fuscipennis. We annotated genes encoding odorant (ORs), gustatory, and ionotropic receptors and chemosensory soluble proteins and odorant-binding proteins in transcriptomes of chemosensory tissues of the above three species and in early draft genomes of two species, A. compressa and C. arenaria. Our analyses revealed that apoid wasps possess larger OR repertoires than any bee lineage, that the last common ancestor of Apoidea possessed a considerably larger OR repertoire (∼160) than previously estimated (73), and that the expansion of OR genes in eusocial bees was less extensive than previously assumed. Intriguingly, the evolution of pollen-collecting behavior in the stem lineage of bees was associated with a notable loss of OR gene diversity. Thus, our results support the view that herbivorous Hymenoptera tend to possess smaller OR repertoires than carnivorous, parasitoid, or kleptoparasitic species.
Keywords: Apoidea, Ampulicidae, Crabronidae, Philanthidae, chemoreceptor gene repertoires, eusociality evolution
Significance
We analyzed chemosensory genes in three solitary apoid wasp species closely related to bees. The genomes of the analyzed wasps contain significantly more chemoreceptor genes than any previously sequenced genome of eusocial bees. Our results suggest that the last common ancestor of Apoidea possessed more chemoreceptor genes than were previously assumed and that the evolution of the pollen-collecting behavior of bees was associated with a significant chemoreceptor gene repertoire size reduction.
Introduction
Most insects rely on the detection of volatile compounds to find and evaluate resources over spatial distance, and they additionally exploit less-volatile compounds in short-range intra- and interspecific interactions (Hansson and Stensmyr 2011; Oi et al. 2015; Couto et al. 2017). Perception of these compounds (semiochemicals) is mediated by proteins of different gene families, collectively referred to as chemosensory-related genes (CRGs). In insects, CRGs encode: 1) membrane-bound receptor proteins (chemoreceptors), such as gustatory receptors (GRs), odorant receptors (ORs), and ionotropic receptors (IRs), and 2) nonreceptor proteins, such as chemosensory soluble proteins (CSPs) and odorant-binding proteins (OBPs). Insect GRs and ORs likely share a common phylogenetic origin (Robertson et al. 2003; Thoma et al. 2019), whereas IRs are not phylogenetically related to GRs and ORs (Robertson and Wanner 2006; Croset et al. 2010), but are a variant of ionotropic glutamate receptor receptors, iGluRs (Benton et al. 2009). The repertoire sizes of these three gene families differ significantly between insect species (Sanchez-Gracia et al. 2009; Sánchez-Gracia et al. 2011; Eyun et al. 2017). Eusocial ants and eusocial bees possess comparatively large OR gene sets. For instance, genomes of eusocial ants contain between 300 and 500 OR-coding genes (Wurm et al. 2011; Zhou et al. 2012, 2015; Oxley et al. 2014; McKenzie and Kronauer 2018), the genome of the honey bee, Apis mellifera, contains 177 OR-coding genes (Robertson and Wanner 2006), and the genome of the bumble bee, Bombus terrestris, contains 166 OR-coding genes (Brand et al. 2015). Many of the OR-coding genes within a given species, and especially in Hymenoptera, represent closely related paralogs that arose by tandem gene duplication, resulting in extensive tandem blocks (Sadd et al. 2015; McKenzie and Kronauer 2018).
One hypothesis posits that the large diversity of ORs found in eusocial Hymenoptera (wasps, ants, and bees) is linked to the eusocial lifestyle of these insects, because chemical communication plays a critical role for establishing eusociality (e.g., enabling nest-mate and caste recognition). Intriguingly, however, the solitary wasp Nasonia vitripennis was found to also possess a comparatively large number of OR-coding genes (301; Robertson et al. 2010), indicating that large chemoreceptor gene repertoire sizes per se are not unique to eusocial species (Karpe et al. 2017).
Some ORs are used to sense cuticular hydrocarbons (CHCs) (McKenzie et al. 2016; Pask et al. 2017), which are an integral part of the cuticle of possibly all insects. It is generally assumed that CHCs initially served insects as desiccation barrier (reviewed by Kather and Martin 2015). However, CHCs additionally gained semiochemical function (Blomquist and Begneres 2010; Leonhardt et al. 2016). CHCs are generally long-chained saturated or unsaturated hydrocarbons; but the CHCs profile of an individual insect consists of a complex mixture of compounds that are often species-, gender-, and age-specific. In addition, in eusocial species, CHCs typically also differ between the individuals of different castes and between individuals living in different nests (Kather and Martin 2015; Pask et al. 2017). Research on ants has led to the identification of CHC-responsive ORs (McKenzie et al. 2016; Pask et al. 2017; Slone et al. 2017). Intriguingly, the CHC-responsive OR clade was found to be more diverse than any other clade of CRGs in the respective ant species, thus significantly contributing to the extraordinarily large OR gene repertoire size of ants (Smith et al. 2011; Pask et al. 2017).
One problem in assessing hypotheses explaining the large size of OR repertoires in eusocial Hymenoptera has been the lack of knowledge on the size of chemosensory gene repertoires in solitary Hymenoptera closely related to eusocial species with large repertoire sizes. This information would allow better judging whether the evolution of eusociality indeed led to chemoreceptor gene repertoire expansions or whether a large gene repertoire size was already present in solitary ancestors of the eusocial lineages. In order to fill this knowledge gap, we studied the repertoire sizes of CRGs in three solitary apoid wasps: 1) Ampulex compressa, a representative of the apoid wasp family Ampulicidae, which has been identified as the extant sister lineage of all remaining Apoidea; 2) Cerceris arenaria, a representative of the apoid wasp family Philanthidae, which is phylogenetically positioned between Ampulicidae and Psenidae (and bees); 3) Psenulus fuscipennis, a representative of the apoid wasp family Psenidae, which is the species most closely related to bees in our taxon sampling (Branstetter et al. 2017; Peters et al. 2017; Sann et al. 2018).
Ampulex compressa is a parasitoid of the American cockroach, Periplaneta americana (Blattodea: Blattidae) (Keasar et al. 2006). Ampulex compressa females use olfactory cues to locate their host and use their taste to assess the suitability of the host (i.e., determining the level of juvenile hormone; Veltman and Wilhelm 1990). Cerceris arenaria is a predator of weevils (Coleoptera: Curculionidae; Blösch 2000; Polidori et al. 2006), and their females show an opportunistic behavior of raiding and usurping nests (incl. any provision stored in the nests) of con-specific females (Field and Foster 1995; Polidori et al. 2006). Intriguingly, the usurping females sometimes share the same nest and even jointly provision the same brood cells (Field and Foster 1995). It has been shown that C. arenaria females use olfactory cues to locate the nests of con-specific females (Polidori et al. 2006). We deemed inclusion of a species of Cerceris in our taxonomic sampling important, as species in the genus Cerceris exhibit primarily solitary lifestyles; however, some species in the genus are known to cooperate when provisioning brood cells (Polidori et al. 2006). Psenulus fuscipennis is a predator of aphids (Hemiptera: Sternorrhyncha; Blösch 2000). Information on P. fuscipennis chemosensation is missing. Although all bee species (larvae and adults) feed on extra- and intrafloral plant parts (i.e., pollen, nectar, and floral oils), the larvae of the three apoid wasp species feed on preyed animals provided by their mothers, and their adults feed on nectar and honeydew excreted by aphids (Gould and Bolton 1988). Adult A. compressa females are known to additionally feed on the hemolymph of their preyed animals (Piek et al. 1984; Arvidson et al. 2018; Gnatzy et al. 2018).
Here we report results from annotating CRGs in chemosensory transcriptomes and in draft genomes of apoid wasps and from comparing the annotated CRGs to each other and to CRGs of closely related eusocial Hymenoptera. We show that apoid wasps possess larger OR-coding gene repertoires than closely related eusocial bees. At least one of the three apoid wasps possesses a more extensive tandem array of OR-coding genes than of any other species of Apoidea, ranking among all Hymenoptera studied in this respect, only second to the clonal raider ant (McKenzie and Kronauer 2018). Finally, we show that the genome of A. compressa harbors a larger number of putative CHC-sensing OR genes than all previously studied genomes of eusocial bees.
Materials and Methods
Insect Sample Collection and Preparation
We collected chemo-sensitive tissue (i.e., antennae, palps, and tarsi) from adult wasps. Samples of A. compressa originated from a laboratory stock cultured in the Aquazoo Löbbecke in Düsseldorf, Germany. Specimens of C. arenaria were field-collected at various locations in Germany, whereas those of P. fuscipennis were reared from trap nests set up in Epe, The Netherlands. Sample collection information is summarized in supplementary data A1, Supplementary Material online (doi: 10.17632/z2mzyr74br.1). We killed all specimens by freezing them at −80 °C. To minimize degradation of RNA, we kept all samples on dry ice when sampling of chemo-sensitive tissue with a pair of sterile forceps. We stored all tissues in RNAlater (QIAGEN, Hilden, Germany) before extracting their RNA.
RNA Preparation, Sequencing, and Assembly
We collected 28 samples of A. compressa (11 females, 17 males), ten samples of C. arenaria (five females, five males), and 69 samples of P. fuscipennis (38 females, 31 males) (supplementary data A1, doi: 10.17632/z2mzyr74br.1). We pooled all sampled chemo-sensitive tissues of a given species and sex for RNA extraction. We extracted and purified RNA using TRIzol reagent (Life Science Technologies-Invitrogen, ThermoFisher Scientific) with the Direct-zol RNA Miniprep kit (Zymo Research, Freiburg, Germany) following the manufacturer’s protocol. We assessed the quantity and quality of the obtained RNA extracts by agarose gel electrophoresis and with a NanoDrop spectrometer (ThermoFisher Scientific). After library preparation following standard protocols, all samples were 150-bp paired-end sequenced with an Illumina HiSeq3000 sequencer (Illumina Inc., CA) at the Max Planck-Genome-Center Cologne, Germany. We quality-trimmed the resulting raw reads with Trimmomatic version 0.36 (Bolger et al. 2014) implemented in the trinityrnaseq toolbox, applying the software’s default parameters (Grabherr et al. 2011; Haas et al. 2013).
We de novo assembled the quality-filtered raw reads of each species with the software Trinity version 2.4.0 (Grabherr et al. 2011; Haas et al. 2013). We subsequently assessed the quality of the assemblies by mapping the quality-filtered raw reads onto the assembled contigs and scaffolds with the software Bowtie version 2.3.4.1 (Langmead and Salzberg 2012). We additionally studied the presence of single-copy gene transcripts predicted to be present in the analyzed species with the Benchmarking Universal Single-Copy Orthologs software, BUSCO (http://busco.ezlab.org, last accessed Semptember 2017; gene set Hymenoptera, n = 4,415; Simão et al. 2015; Waterhouse et al. 2018; supplementary fig. S1, Supplementary Material online). In addition to the Trinity-inferred de novo transcriptome assembly, we generated genome-guided transcriptome assemblies from the raw reads of A. compressa and C. arenaria, whose genome scaffolds were available to us. We indexed the genome scaffolds using Bowtie2 version 2.3.4.1, assembled the raw reads using TopHat2 version 2.1.1 (Kim et al. 2013), and predicted transcripts using the splice-junction mapper tool Cufflinks version 2.2.1 (Trapnell et al. 2012). The draft genome assemblies of these two species and the automatically inferred protein-coding gene models were kindly provided to us by the Leibniz Graduate School of Genomic Biodiversity Research, Germany. The genome assembly statistics are given in table 1 (note that the two apoid wasp draft genomes have not been published yet).
Table 1.
The Statistics for the Genome Assemblies of Ampulex compressa and Cerceris arenaria
| Statistics | A. compressa | C. arenaria |
|---|---|---|
| Total number of scaffolds | 18,453 | 182,826 |
| Assembly size (bp) | 279,244,254 | 358,163,941 |
| Amount of Ns per 100 kb (bp) | 877 | 7,422 |
| Size of smallest scaffold (bp) | 93 | 83 |
| Size of largest scaffold (bp) | 16,314,322 | 9,578,785 |
| Total number of nucleotide base counts | ||
| Count of A | 79,307,969 | 102,095,833 |
| Count of C | 59,083,877 | 63,903,708 |
| Count of G | 59,096,860 | 63,923,668 |
| Count of N | 2,447,908 | 26,581,550 |
| Count of T | 79,307,640 | 101,659,182 |
| GC content (%) | 42.70 | 38.55 |
| N and L statistics | ||
| N50 (bp) | 9,128,341 | 2,093,498 |
| N75 (bp) | 2,712,415 | 743,455 |
| L50 (bp) | 12 | 40 |
| L75 (bp) | 28 | 109 |
| Number of sequences (bp) | ||
| ≥0 | 18,453 | 182,826 |
| ≥1,000 | 668 | 5,547 |
| ≥5,000 | 215 | 623 |
| ≥10,000 | 170 | 399 |
| ≥25,000 | 146 | 323 |
| ≥50,000 | 133 | 278 |
| Assembly size based only on sequences (bp) | ||
| ≥0 | 279,244,254 | 358,163,941 |
| ≥1,000 | 275,197,192 | 328,427,061 |
| ≥5,000 | 274,378,842 | 320,283,610 |
| ≥10,000 | 274,062,538 | 318,839,227 |
| ≥25,000 | 273,713,023 | 317,700,425 |
| ≥50,000 | 273,276,827 | 316,179,035 |
Note.—Both genomes were sequenced using Illumina HiSeq sequencer. Four libraries were generated (i.e., 250-bp paired-end, 800-bp paired-end, 3-kb mate pair, 8-kb mate pair) and sequenced to an estimated coverage depth of 62×, 21×, 23×, and 18× (Cerceris arenaria), and 94×, 19×, 25×, and 13× (Ampulex compressa). The genome assemblies were inferred with Platanus (Kajitani et al. 2019).
A Posteriori Sex Confirmation
We inferred the sex of one sample (C. arenaria male, for which we were unsure of the identity of the sex due to loss of information on vials prior to RNA extraction), a posteriori by identification and analysis of the transcripts (and predicted peptides) of the sex-determining genes transformer and doublesex in the assembled transcriptome of this sample. Transformer peptides in males are truncated, whereas full-length peptides in females contain the highly distinctive CAM-domain (Hediger et al. 2010; Verhulst et al. 2010). Doublesex can be categorized in female- and male-specific peptide sequences that contain sex-specific C-terminal regions (An et al. 1996; Shukla and Nagaraju 2010). We identified mRNA coding for transformer and doublesex in the assembled transcriptome by searching the Ap. mellifera amino acid sequences of these genes (ABW99105, NP_001128300) against the nucleotide sequences of the transcriptome using TBlastN of the BLAST+ version 2.7.1 (Altschul et al. 1990; Camacho et al. 2009) software suite. We considered the putative transformer transcripts that translated into an uninterrupted CAM-domain as indicative of the female sex. We considered the sample to be a male as only truncated transformer transcripts were detected. This categorization was confirmed by the presence of doublesex transcripts encoding for a homologous C-terminal region to the Ap. mellifera male-specific Doublesex isoform (Cho et al. 2007).
Identification of CRGs
We compiled amino acid sequences of ORs, GRs, IRs, CSPs, and OBPs of Hymenoptera, targeting those species with sequenced and well-annotated genomes (references in table 3). The sequences were used to set up reference search databases for each of the above protein families utilizing the software suite BLAST+ version 2.7.1 (Altschul et al. 1990; Camacho et al. 2009).
Table 3.
Size of Chemosensory-Related Gene Repertoires in Hymenoptera
| Species | Family | Lifestyle | ORs | GRs | IRs | CSPs | OBPs | Reference |
|---|---|---|---|---|---|---|---|---|
| Ampulex compressa | Ampulicidae | sol. para. | 311 | 17 | 29 | 7 | 17 | This study (genomic data) |
| Cerceris arenaria | Crabronidae | sol. pred. | 241 | 10 | 31 | 6 | 12 | This study (genomic data) |
| Psenulus fuscipennis | Crabronidae | sol. pred. | 122 | 13 | 29 | 14 | 25 | This study (transcriptomic data) |
| Apis mellifera | Apidae | eus. herb. | 177 | 14 | 21 | 6 | 21 | Brand and Ramirez (2017); Robertson and Wanner (2006); GenBank |
| Bombus terrestris | Apidae | eus. herb. | 166 | 25 | 22 | 5 | 16 | Brand and Ramirez (2017); Sadd et al. (2015) |
| Habropoda laboriosa | Apidae | sol. herb. | 151 | NA | NA | NA | NA | Karpe et al. (2017) |
| Dufourea noveangliae | Halictidae | sol. herb. | 112 | NA | NA | NA | NA | Karpe et al. (2017) |
| Lasioglossum albipes | Halictidae | pol. eus. | 159 | 23 | NA | NA | NA | Zhou et al. (2012) |
| Nasonia vitripennis | Pteromalidae | sol. para. | 301 | 58 | 111 | 7 | 93 | Robertson et al. (2010, 2018); Zhou et al. (2012); GenBank |
| Harpegnathus saltator | Formicidae | eus. pred. | 377 | 21 | 23 | 11 | 11 | Zhou et al. (2012, 2015); Kulmuni et al. (2013) |
| Solenopsis invicta | Formicidae | eus. pred. | 392 | 219 | NA | NA | NA | Zhou et al. (2012) |
Note.—ORs, odorant receptors; GRs, gustatory receptors; IRs, ionotropic receptors; CSPs, chemosensory proteins; OBPs, odorant-binding proteins; NA, not annotated; sol., solitary; para., parasitoid; pred., predatory; herb., herbivorous; eus., eusocial; pol., polymorphic. Colors correspond to those in tables 4 and 5, and figures 1 and 2.
We searched the de novo and the genome-guided transcriptome assemblies for candidate CRG-coding DNA sequences using the following procedures: 1) BlastX search of all contigs and scaffolds against the reference databases with an e-value cutoff of 1 × 10−2 when searching the OR database and an e-value cutoff of 1 × 10−6 when searching the remaining databases; 2) BlastX search of all contigs and scaffolds with at least one hit in a reference database (step 1) against the NCBI nonredundant protein database (nr) (as of June 6, 2017) using the same e-value cutoff values as in step 1, masking low complexity regions, specifying an HSP length cutoff of 33 amino acids, hit coverage of 60%, and considering only the top ten positive hits; 3) confirmatory search of all candidate CRG-coding DNA sequences retained from step 2 against the Interproscan version 5 (Jones et al. 2014) and gene ontology classification using Blast2GO version 4 (Conesa et al. 2005).
We translated all retained candidate CRG-coding DNA sequences, choosing the reading frame suggested by the BlastX hit, using the standard genetic code. Only those resulting sequences comprising more than 50 amino acids were analyzed further. To minimize redundancy, we searched all amino acid sequences against each other and retained only the longest sequence of those exhibiting below 98% identity with each other. We added to the final compilation the CRG protein orthologs from various bee species with sequenced genomes: Ap. mellifera (all CRGs, Robertson and Wanner 2006), B. terrestris (all CRGs, Brand and Ramirez 2017), Habropoda laboriosa, Dufourea noveangliae (only ORs, Karpe et al. 2017), and Lasioglossus albipes (ORs and GRs, Zhou et al. 2012, 2015). Subsequently, we built a local BLAST+ search database of Apoidea CRGs to interrogate and identify protein-coding DNA sequences (CDS) in the draft genomes of A. compressa and C. arenaria. The genomes were interrogated repeatedly until no more new target gene loci discovered.
Manual Annotation of Candidate Genes in Available Draft Genomes
We indexed by position coordinates the genome-based predicted transcripts against genome scaffold nucleotide indices to ensure that coordinates correspond to each other using Samtools faidx version 1.5 (Li et al. 2009). This step enabled us to match and merge the genome gene model predictions generated by AUGUSTUS version 3.0 (Stanke et al. 2006) with the transcript predictions generated by Cufflinks version 2.2.1 (Trapnell et al. 2012) into a single gene transfer file for each target contig and scaffold using CLC Genomics Workbench version 9.11.0 (http://www.qiagenbioinformatics.com). The transfer file in GenBank format stores both scaffold nucleotide sequences and the gene feature models predicted via AUGUSTUS and Cufflinks.
We manually annotated all target gene models in the two available draft genome scaffolds, using the gene features in the GenBank files as guides. We mapped and used as supporting evidence the transcriptome raw reads assembled by TopHat2. To the respective target GenBank record bearing a single contig or scaffold, the RNAseq raw reads were read into and manually annotated using Artemis genome viewer tool, development version 16.0.17 (Berriman and Rutherford 2003). We used the coordinates of BLAST hits (from Apoidea-only search database interrogation) to locate the target CRG features in the opened/read target scaffolds and used them as seed gene models to start manual annotation. In many instances, we edited the predicted splice junctions and split or merged some automated gene models based on our knowledge of the CRG structures. In other cases, we modeled new gene structures ab initio following the coverage depth of the mapped RNA-seq raw reads as guide by searching the flanking DNA regions using the online NCBI ORF finder version v 0.4.3 and the online version of the AUGUSTUS version 3.0 gene prediction tool (Stanke and Morgenstern 2005). We additionally also compared the amino acid sequences with the NCBI nonredundant conserved domain database (CDD) via the Domain Enhanced Lookup Time Accelerated BLAST (DeltaBlast) algorithm, BLAST+ version 2.2.26 (Boratyn et al. 2012). The workflow of our annotation procedure is visualized in supplementary figure S2, Supplementary Material online.
Candidate CRGs Validation and Nomination
During the annotation process, we assessed all predicted gene models for their completeness (i.e., presence of start codon, of stop codon, and of canonical splice sites) and for the presence of conserved structural features (i.e., gene-family specific domains), and we edited the predicted gene models when necessary (see supplementary fig. S3B, Supplementary Material online, for the splice site signals). Specifically, we conducted a confirmatory search for the presence of the domains 7tm_6 (PF02949), 7tm_7 (PF08393), Lig_chan (PF00060), OS-D (PF03392), and PBP_GOBP (PF01395) in the encoded OR, GR, IR, CSP, and OBP proteins respectively. The presence of secondary and of tertiary conserved domains was assessed via the SUPERFAMILY and the Pfam database, respectively, bundled with InterProScan version 5 (Jones et al. 2014) and via the NCBI CDD version 3.16 server (Marchler-Bauer et al. 2015). For CSPs and OBPs, we additionally assessed the presence of cellular export signal peptides with the SignalP version 4.1 server (Nielsen 2017). Finally, we assessed the presence of transmembrane helices in the predicted OR, GR, and IR proteins with the software TMHMM version 2 (Krogh et al. 2001).
Due to the high number and the high nucleotide divergence of OR genes, we assigned index names based primarily on the order of the annotated genes’ location on the scaffolds. The only exception was the naming of the putative Orco-like gene, which we consistently named Or1 (supplementary data A2, Supplementary Material online; doi: 10.17632/6zpp8tgz5c.1). Specifically, we applied the following nomenclature: the first two letters indicate the genus (uppercase letter) and species (lowercase letter) name, in which the gene was annotated, followed by the Or gene family acronym (e.g., AcOr2 designates the putative odorant receptor 2 in the species A. compressa). We indicated incomplete sequences from partial gene models by adding a hyphen (-) to the index name, followed by either the acronym cte (for incomplete sequence on c-terminal), the acronym nte (for incomplete sequence on n-terminal), or inc (if incomplete on both ends; e.g., AcOr92-inc). For a likely pseudogenized gene, we added the -pse acronym to the gene index name (e.g., AcOR224-pse). We distinguished splice variants of a gene by adding an uppercase letter, preappended by a hyphen, to the gene index name (e.g., AcOr15-A, AcOr15-B).
We assigned the GR- and IR-coding genes names and index numbers following the names and symbols of their orthologs in the honey bee and the bumble bee (Robertson and Wanner 2006; Brand and Ramirez 2017) and, in a few instances, by following the nomenclature that had been used to name these genes in ants (Zhou et al. 2012, 2015; Kulmuni et al. 2013). A few names were assigned a period (.) followed by a number to distinguish between paralogs (e.g., AcIR25a.1 and AcIR25a.2). The naming of CSPs and OBPs followed the nomenclature used for naming these genes in the honey bee (Forêt and Maleszka 2006) and in fruit flies (Pelosi et al. 2005, 2006). All candidate gene names of P. fuscipennis were prefixed with Pfu to indicate that the candidate transcripts or predicted proteins are derived exclusively from de novo-assembled transcripts. All CRG annotated contigs and scaffolds are provided as supplementary data A3, Supplementary Material online (doi: 10.17632/npzkyn6sb6.1).
Phylogenetic Analysis
We translated the nucleotide sequences of all the annotated CRGs to amino acid sequences using the standard genetic code. If splice variants of a gene were predicted, we considered only the amino acid sequence resulting from the longest splice variant in the phylogenetic analysis. We inferred the phylogenetic relationships of ORs using the sequence data from the three apoid wasps as well as of the following bees, ants, and wasps: Ap. mellifera (Robertson and Wanner 2006), B. terrestris (Brand and Ramirez 2017), H. laboriosa, D. novaeangliae (Karpe et al. 2017), L. albipes (Kocher et al. 2018), H. saltator, S. invicta (Oxley et al. 2014; Zhou et al. 2015), and N. vitripennis (Robertson et al. 2010). Nasonia vitripennis served as an outgroup. However, we performed an additional extended analysis by sampling sequences from additional species to enable us to assign the OR gene clades previously established in Hymenoptera (Zhou et al. 2015; McKenzie and Kronauer 2018).
We applied the same taxon sampling as outlined above for phylogenetically analyzing the remaining gene families, except that we additionally included all amino acid sequences for the respective gene family found in Drosophila melanogaster (Hekmat-Scafe et al. 2002; Robertson et al. 2003; Pelosi et al. 2005, 2006) and Bombyx mori (Guo et al. 2017) for outgroup comparison.
We analyzed a total of 2,420 ORs, 530 GRs, 246 IRs, 70 CSPs, and 271 OBPs. The amino acid sequences of a given gene family were aligned with the software MAFFT version 7.310, using the L-INS-i algorithm (Katoh and Standley 2013). For phylogenetically analyzing ORs, we studied the OR amino acid sequence alignment with IQTree version 1.6.6 (Nguyen et al. 2015). We used Modelfinder (Kalyaanamoorthy et al. 2017), implemented in IQTree, to find the most suitable substitution model, considering all amino acid substitution models for nuclear-encoded genes integrated in the IQTree (i.e., BLOSUM62, Dayhoff, DCMut and JTTDCMut, JTT, LG, PMB, VT, WAG) and additionally considered the LG4M and LG4X mixture models (Le et al. 2012). We tested the models with different parameters for heterogeneity rate (+I, +G, +I + G, +R, +I + R) and considered empirical state frequencies in the data (+F). The corrected Akaike information criterion (AICc; Hurvich and Tsai 1989) was used to select the most suitable model and model parameters, which turned out to be JTT+G + I. We computed standard nonparametric bootstraps replicates using the IQTree tool by testing every 50 bootstraps for convergence with RAxML (Stamatakis 2014) using the integrated a posteriori bootstop function (Pattengale et al. 2010) with the extended majority rule convergence criterion.
We phylogenetically analyzed the remaining CRG families with the software FastTree version 2.15 (Price et al. 2009), applying again the maximum likelihood optimality criterion and conducting 1,000 nonparametric bootstrap replicates to assess tree robustness. We selected the applied most suitable substitution model, which turned out to be WAG+G + I, with the aid of the software ProtTest version 3 (Darriba et al. 2011).
We rooted the OR protein tree by specifying the canonical odorant receptor coreceptor (ORCo) clade subtree as outgroup (Thoma et al. 2019). We rooted the GR protein tree by specifying the CO2-like (Gr21a and Gr63a) subtree clade of fruit fly as outgroup (the GRs that are expressed on olfactory receptor neurons in other insects and yet to be reported as present in Hymenoptera). We rooted the IRs protein tree by specifying the IR8a and IR25a subtree clades as outgroups (the two clades occur in all arthropods and function as coreceptors to other IRs; Croset et al. 2010; Eyun et al. 2017) and we rooted the protein trees of CSP and OBP by specifying the OBP clade B and CSP4 clade, respectively, as arbitrary outgroups. We viewed and rendered the trees with the software FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and further edited the draft tree figures using the software Inkscape (Bah 2011).
Evolutionary Turnover of ORs
We used the software NOTUNG version 2.9 (Chen et al. 2000) to estimate ancestral OR repertoire sizes at nodes of the phylogeny of the investigated species and to quantify gene family expansions and constrictions along branches of the species phylogeny. We based the species tree on the phylogenetic trees published by Peters et al. (2017) and Sann et al. (2018).
Results
Annotation of Putative Chemosensory Genes from Genomic and Transcriptomic Data
We obtained 74,249 (A. compressa), 136,527 (C. arenaria), and 112,054 (P. fuscipennis) transcripts in the de novo-assembled RNA-seq data. A significantly lower count of transcripts was obtained when applying a genome-assisted assembly strategy: 29,018 (A. compressa) and 49,242 (C. arenaria) (note that it was not possible to apply this approach to the P. fuscipennis RNA-seq data due to the lack of a draft genome assembly for this species). We identified CRG models in 53 of the 18,453 genome scaffolds in A. compressa and in 85 of the 182,826 genome scaffolds in C. arenaria. The details are summarized in table 2.
Table 2.
Summary Statistics of CRGs Annotated in Genome Scaffolds and Chemosensory Transcriptomes of Three Species of Apoid Wasps
| Species | Ampulex compressa | Cerceris arenaria | Psenulus fuscipennis |
|---|---|---|---|
| Number of scaffolds with annotated CRGsa | |||
| OR-coding genes | 21 | 50 | NA |
| GR-coding genes | 9 | 7 | NA |
| IR-coding genes | 14 | 17 | NA |
| CSP-coding genes | 4 | 5 | NA |
| OBP-coding genes | 5 | 6 | NA |
| Transcriptome of chemosensory tissues (genome-guided assembly)b | |||
| Assembled paired reads | 54,297,198 | 81,619,921 | |
| Contig count | 94,602,641 | 125,197,722 | |
| Transcript count | 29,018 | 49,242 | |
| Mapped reads (%) | 87.1 | 87.4 | |
| Avg. fragment length (bp) | 349.2 | 337.7 | |
| Std. dev | 88.9 | 91.8 | |
| N50 (contigs) | 1,510 | 4,839 | |
| G + C (%) | 44.37 | 42.52 | |
| Transcriptome of chemosensory tissues (de novo assembly)c | |||
| Assembly size (bp) | 299,500,853 | 261,520,523 | 249,077,981 |
| Assembled paired reads | 10,368,996 | 41,558,667 | 12,431,360 |
| Mapped reads (%) | 95.33 | 89.57 | 92.83 |
| Predicted genes | 74,249 | 136,527 | 112,054 |
| Predicted transcripts | 151,692 | 239,673 | 210,850 |
| N50 | 2,490 | 1,238 | 2,569 |
| G + C (%) | 43.63 | 40.91 | 39.83 |
Note.—NA, genome draft not available.
The number of scaffolds does not per se indicate the spread of CRGs across the genome, as the draft genomes of the two studied species have not been assembled to chromosome level. The genome of A. compressa is more contiguous than that of C. arenaria, indicated by the high number of CRG regions located at scaffold ends in C. arenaria (annotated as incomplete, see supplementary data A2, Supplementary Material online).
Tophat2 was used to map reads onto the draft genome scaffolds, and Cufflinks was used to assemble the mapped reads onto the transcriptome scaffolds.
Statistics inferred with the trinityrnaseq plugin scripts bundled with Trinity.
Manual annotation of the transcripts in the genome scaffolds and considering de novo gene models of A. compressa and C. arenaria led to the identification of 674 OR-coding genes (311 in A. compressa, 241 in C. arenaria, and 122 in P. fuscipennis). Likewise, we identified 40 (17, 10, 13) GR-coding genes, 79 (29, 31, 29) IR-coding genes, 27 (7, 6, 14) CSP genes, and 54 (17, 12, 25) OBP-coding genes in the three apoid wasps (table 3). All translated amino acid model sequences from the genomes of A. compressa and C. arenaria, and from the chemosensory transcriptome of P. fuscipennis are given in supplementary data A4, Supplementary Material online (doi: 10.17632/nv7chmrz7k.1).
Genomic Organization of Chemosensory Genes
We found most OR-coding genes (and a few GR-, IR-, and OBP-coding genes) congregated in clusters on a few contigs and scaffolds, with genes in a given cluster sharing the same gene structure (supplementary fig. S3, Supplementary Material online). We found in this study, in the species A. compressa, the largest tandem array cluster of ORs, comprising 78 genes (AcOr69–147), located on scaffold 24 (see supplementary data A2, sheet 1, Supplementary Material online, doi: 10.17632/6zpp8tgz5c.1). We identified a few tandem clusters of OR-coding genes in both A. compressa and C. arenaria sharing flanking genes with the same orientation on either side of the clusters, indicating structural microsynteny between the two genomes. The largest microsyntenic cluster, with an array size of 43 OR genes (A. compressa scaffold 115_cov79) and 34 OR genes (C. arenaria scaffold 117_cov65), respectively, are both flanked upstream by an “Aquaporin-like” gene on the complementary strand and downstream by a “Spatascin-like” gene on the OR-coding strand. However, in contrast to A. compressa, C. arenaria has another OR-coding gene (CaOr207) located at the 5′-end of the Aquaporin-like gene (supplementary fig. S3, Supplementary Material online). A second cluster of ten OR-coding genes in either species (A. compressa scaffold 16901_cov81, C. arenaria scaffold 55_cov65) also seems conserved regarding its location based on flanking genes (supplementary fig. S3, Supplementary Material online).
We found GR-coding genes located on nine (A. compressa) and eight (C. arenaria) scaffolds, respectively, of the draft genomes. In both species, the putative sugar receptor-coding genes GR1 (encoded by nine exons) and GR2 (encoded by 12 exons) are clustered and arranged facing each other at one site in the respective genome scaffolds. Most of the IR-coding genes are spatially spread in the draft genomes of A. compressa and C. arenaria, except eight genes that are congregated in four clusters, each comprising two genes on their respective coding strands. Finally, although some OBP-coding genes are spatially clustered on A. compressa and C. arenaria genome contigs and scaffolds, we found CSP-coding genes consistently spatially isolated from each other on separate scaffolds (see supplementary data A2, Supplementary Material online).
The Phylogeny of ORs
Detailed information on the inferred phylogenetic relationships of the analyzed OR amino acid sequences (with branch support values inferred from 250 bootstrap replicates) is given in supplementary data A5, Supplementary Material online (http://doi.org/10.17632/h4tdmjkg5r.1). We identified 32 OR clades (ortholog groups) relative to the ORCo clade subtree, declared as root (ortholog groups defined here as lineages that already existed in the last common ancestor of Hymenoptera) (supplementary fig. S4, Supplementary Material online and table 3). All of the clades had previously been delineated and recognized in wasp-waisted Hymenoptera, Apocrita, and are labeled A–Z, ZA, ZB, XA, ORCo, 9-exon, with some entries “Unclassified” (Zhou et al. 2012, 2015; McKenzie and Kronauer 2018). Using the clonal raider ant ortholog CbirOr5-XA1 as reference, we identified clade-XA with amino acid sequences exclusively from apoid wasps. We did not find representative amino acid sequences for clade-O, clade-Y, and clade-ZB in any apoid wasps, they were exclusively present in the outgroup species N. vitripennis.
The OR repertoire sizes of the clades across the species analyzed differ substantially from each other. The 9-exon subtree clade was the largest in each investigated species and comprises a total of 860 amino acid sequences (∼35% of all analyzed ORs) (row highlight in magenta in table 4). The second largest clade, L, contains 377 ORs (∼15% of all ORs) and represents exclusively genes with five exons each. Except for the clades with conserved single sequence copies (clades ORCo, B, C, I, Q, W, Z, and ZA), the clades with multiple sequences from a given species formed monophyletic subclades in our phylogenetic analysis.
Table 4.
Repertoire Sizes of OR Phylogenetic Clades (Subfamilies) in Hymenoptera
| Clade | Bootstrap Support (%) | Total | Ac | Ca | Pfu | Nv | Am | Bt | La | Dn | Hl | Hs | Si | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORCo | 100 | 11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Root clade |
| A | 96 | 47 | 2 | 2 | 5 | 3 | 3 | 1 | 1 | 2 | 1 | 17 | 10 | Expansions in ants |
| B | 100 | 10 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Conserved and single copy |
| C | 99 | 9 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Conserved single copy |
| D | 100 | 25 | 3 | 1 | 2 | 12 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | Only in ants and wasps |
| E | 98 | 171 | 10 | 9 | 7 | 35 | 6 | 9 | 24 | 8 | 11 | 27 | 25 | Expansions across species |
| F | 100 | 58 | 1 | 15 | 3 | 28 | 1 | 1 | 2 | 1 | 0 | 1 | 5 | Expansions in wasps |
| G | 99 | 24 | 5 | 4 | 7 | 2 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | Expansions in solitary wasps |
| H | 94 | 79 | 7 | 12 | 2 | 1 | 14 | 11 | 8 | 3 | 4 | 9 | 8 | Expansions across species |
| I | 100 | 11 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Conserved and single copy |
| J | 91 | 105 | 5 | 4 | 3 | 3 | 23 | 18 | 14 | 12 | 19 | 3 | 1 | Expansions in bees |
| K | 100 | 20 | 3 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | Conserved duplicates |
| 9-exon | 97 | 859 | 176 | 109 | 41 | 91 | 42 | 38 | 41 | 21 | 54 | 128 | 118 | Expansions across species |
| L | 93 | 377 | 45 | 38 | 20 | 8 | 59 | 46 | 23 | 14 | 28 | 55 | 41 | Expansions across species |
| M | 97 | 16 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | Expansion in ants |
| N | 98 | 26 | 18 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | Expansion in Ampulex, absent in bees |
| O | 69 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Nasonia only |
| P | 100 | 52 | 0 | 0 | 5 | 0 | 5 | 10 | 8 | 1 | 0 | 11 | 12 | Expansions in eusocial species |
| Q | ? | 13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | Single copy, but expansion in ants |
| R | 100 | 22 | 7 | 4 | 0 | 0 | 0 | 0 | 6 | 1 | 1 | 2 | 1 | Absent in eusocial bees |
| S | 100 | 13 | 1 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Specific to wasps and ants |
| T | 100 | 71 | 3 | 4 | 6 | 23 | 2 | 10 | 5 | 3 | 5 | 8 | 2 | Expansions across species |
| U | 99 | 132 | 7 | 10 | 4 | 7 | 1 | 1 | 2 | 5 | 1 | 38 | 56 | Expansions in wasps and ants |
| V | 100 | 156 | 7 | 8 | 5 | 11 | 7 | 6 | 13 | 8 | 7 | 53 | 31 | Expansions across species |
| W | 87 | 11 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Absent in Ampulex |
| X | 100 | 26 | 2 | 2 | 1 | 16 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | Expansion in Nasonia |
| Y | ? | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Ant only |
| Z | 100 | 25 | 0 | 1 | 1 | 19 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | Expansion in Nasonia |
| ZA | 96 | 6 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Specific to wasps and ants |
| ZB | 100 | 24 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Nasonia only |
| XA | 100 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Apoid wasp only |
| Unclassified | 100 | 14 | 1 | 1 | 1 | 2 | 1 | 4 | 1 | 1 | 2 | 0 | 0 | Absent in ants |
| Totals | 2,420 | 311 | 241 | 122 | 301 | 177 | 166 | 159 | 90 | 144 | 377 | 332 |
Note.—Ac, Ampulex compressa; Am, Apis mellifera; Bt, Bombus terrestris; Ca, Cerceris arenaria; Dn, Dufourea noveangliae; Hl, Habropoda laboriosa; Hs, Harpegnathos saltator; La, Lasioglossum albipes; Nv, Nasonia vitripennis; Pfu, Psenulus fuscipennis; Si, Solenopsis invicta. Clade bootstrap support values are indicated in the second column (the underlyingmaximum likelihood phylogenetic tree is shown in supplementary fig. S4, Supplementary Material online). Colors correspond to those in tables 3 and 5 and figures 1 and 2.
The 9-exon OR clade genes have been reported to code for receptors sensitive to CHC (Pask et al. 2017; Slone et al. 2017). We refer to these genes as CHC-ORs in the following. Among Apoidea, the A. compressa genome encodes 177 putative CHC-ORs, whereas the C. arenaria genome encodes 108 putative CHC-ORs (table 4). The genomes (or transcriptomes) of all other species considered in the present study encode less than 100 putative CHC-ORs, particularly the pollen-collecting bees encode smaller CHC-ORs repertoires. Analysis of the 9-exon (CHC-ORs) subclade phylogeny revealed species-specific expansions, in correspondence to the diversity of the CHCs among the species (fig. 1; see also Zhou et al. 2015).
Fig. 1.
Phylogeny of the 9-exon OR clade of odorant receptor (OR) amino acid sequences inferred under the optimality criterion maximum likelihood. The 9-exon OR clade is the largest OR sublineage in Hymenoptera. The subclades are labeled 9-exon_a to _u. The dendrogram was rooted using the ORCo clade. The subclades reveal diversified expansions across the species represented. Previously, Zhou et al. (2015) distinguished three 9-exon subclades: alpha, beta, and gamma lineages, shown as node labels in green font.
The Evolutionary Turnover of ORs
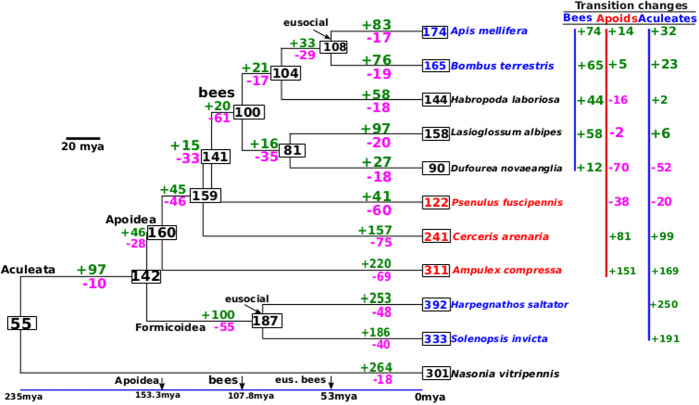
We found two expansions of ORs along the branches leading to Formicoidea (ants) and to Apoidea (apoid wasps and bees), as well as an OR repertoire contraction in the lineage leading to the ancestor of extant bees (fig. 2). The apoid wasps historically experienced rapid expansion episodes in their OR-coding gene repertoires at a higher rate than eusocial bees. Overall, apoid wasp OR-coding genes experienced more episodes of rapid gene births than gene deaths.
Fig. 2.
Evolutionary turnover of OR genes in representative Hymenoptera species. Inference of ancestral OR repertoire in Apoidea via reconciliation of gene tree and species tree using NOTUNG 2.9 (Chen et al. 2000). The gene tree was inferred based on an alignment of all annotated ORs with MAFFT 7.3.07 (Katoh and Standley 2013). IQ-Tree 1.6.6 (Nguyen et al. 2015) was used in order to find a suitable substitution model for the data (JTT+G) and to infer the phylogeny. The topology and divergence time of the species tree are based on results by Peters et al. (2017) and Sann et al. (2018). Both Formicoidea and apoid wasps exhibit a more rapid increase in OR genes than eusocial bees. The last common ancestor of Apoidea had a larger OR repertoire size than modern species, suggesting that the bee lineages—eusocial and solitary—experienced a significant OR gene loss.
The Phylogeny of GRs
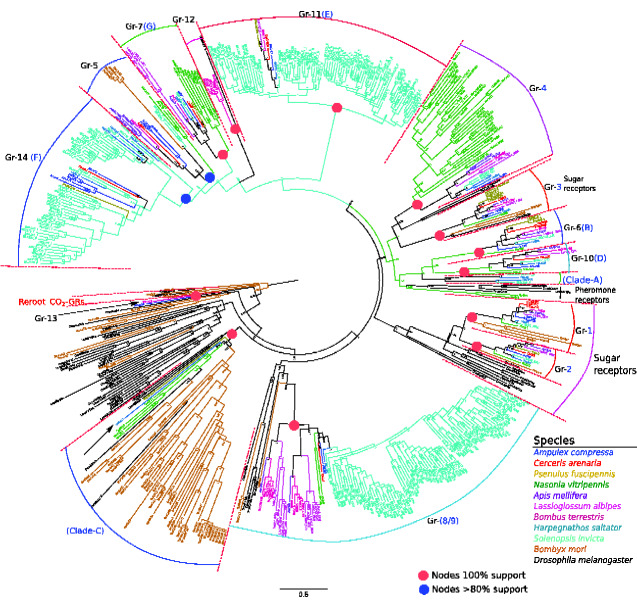
We recovered all of the 16 GR clades previously delineated by Zhou et al. (2012) in Hymenoptera also in our study (fig. 3 and table 5). However, we also recovered additional clades. We found the following two major groups of GR lineages:
Fig. 3.
Phylogeny of gustatory receptor (GR) amino acid sequences inferred under the optimality criterion maximum likelihood. The tree was reconstructed from 289 amino acid sequences and was subsequently rooted with CO2-detecting GRs from Drosophila melanogaster and Bombyx mori. All GR clades identified by us containing at least one sequence of Apoidea and supported by at least 80% bootstrap support are labeled Gr-1 to -15 and clade A; labels in blue fonts follow the nomenclature established by Zhou et al. (2012).
Table 5.
GR Repertoire Sizes in Hymenoptera and Selected Outgroup Species
| Clades | Boostrap Support (%) | Total | Ac | Ca | Pfu | Am | Bt | La | Nv | Hs | Si | Dm | Bm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gr-1 | 100 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | 4 |
| Gr-2 | 100 | 10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Gr-3 | 100 | 16 | 1 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 |
| Gr-4 | 100 | 45 | 1 | 0 | 0 | 2 | 1 | 2 | 35 | 2 | 2 | 0 | 0 |
| Gr-5 | 99 | 17 | 2 | 1 | 0 | 3 | 1 | 1 | 3 | 0 | 2 | 0 | 4 |
| Gr-6(B) | 100 | 11 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Gr-7(G) | 100 | 18 | 1 | 1 | 1 | 1 | 1 | 3 | 8 | 1 | 1 | 0 | 0 |
| Gr-8/9 | 100 | 117 | 1 | 1 | 0 | 2 | 10 | 11 | 2 | 1 | 82 | 4 | 3 |
| Gr-10(D) | 100 | 9 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 0 |
| Gr-11(E) | 100 | 84 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 79 | 0 | 0 |
| Gr-12 | 100 | 6 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| Gr-13 | 100 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gr-14(F) | 98 | 58 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 4 | 47 | 0 | 0 |
| Gr-15(C) | 100 | 52 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 1 | 0 | 5 | 41 |
| (Clade A) | 99 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 |
| Others | — | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 21 |
| Totals | 530 | 17 | 10 | 13 | 13 | 25 | 23 | 58 | 17 | 219 | 59 | 76 |
Note.—Ac, Ampulex compressa; Am, Apis mellifera; Bm, Bombyx mori; Bt, Bombus terrestris; Ca, Cerceris arenaria; Dm, Drosophila melanogaster; Hs, Harpegnathus saltator; La, Lasioglossum albipes; Nv, Nasonia vitripennis; Pfu, Psenulus fuscipennis; Si, Solenopsis invicta. Clade bootstrap support values are those given in the phylogenetic tree shown in figure 3. Colors correspond to those in tables 3 and 4 and figures 1 and 2. Clade names printed in bold correspond to those reported by Zhou et al. (2012).
Sugar Receptor GR Orthologs
This group contains three GR lineages of sugar receptors (SRs): Gr-1, Gr-2, and Gr-3. In terms of presence, and not considering P. fuscipennis, all Hymenoptera studied possess all three sugar receptor genes, and the coding genes are consistently present in single copy. The SR GRs are also present in the studied species outside Hymenoptera.
GRs Only Found in Hymenoptera
This group contains GR lineages that are only found in Hymenoptera (fig. 3). Unlike the situation in other solitary wasps (Zhou et al. 2015), the GR orthologs in apoid wasps are consistently present in single copy. The only notable exceptions are two instances of GR paralogs of clade Gr-5 and Gr-14 in A. compressa. In apoid wasps, the paralogs in the Gr-5 clade are seemingly complete; the orthologous genes in the honey bee are pseudogenized (AmGrX-, AmGrY-, and AmGrZpse). Both A. compressa and C. arenaria possess the GR lineages GR-11 and 13, which we labeled based on their closest homologs in the bumble bee lineages BtGr11–13 (Park et al. 2015; Sadd et al. 2015). GRs of the clades A, Gr-14, and Gr-15 occur exclusively in ants and in a few wasps.
We did not recover orthologs of the fruit fly’s carbon dioxide-like gustatory receptors DmelGr21a and Gr63a in any apoid wasp or in any other apocritan wasp.
The Phylogeny of IRs
We identified 24 IR clades in the inferred phylogenetic tree rooted with the IR8a and IR25a subtree clades (fig. 4). The evolution of IR-coding genes across Protostomia has been described and the IR homologs designated as either “antennal IRs” or “divergent IRs” (Croset et al. 2010). In our analysis, we further subdivided the “antennal IRs” into two groups: the “classical antennal IRs” and the “Hymenoptera-only antennal IRs.” Another group of IRs was named here “divergent-like IRs,” because its IRs share an internal node with the “divergent” IRs of D. melanogaster (table 6). The three groups are described in the following:
Fig. 4.
Phylogeny of IR amino acid sequences inferred under the optimality criterion maximum likelihood. The tree was rooted with the IR8a and IR25a clades. All identified clades clustered with 100% bootstrap support. The IR25a clade has duplicates (blue highlight). Hymenoptera-specific IR clades are highlighted in gray. The IR lineages labeled in red are antennal IRs, which are highly conserved across Arthropoda. IR-X clade comprises exclusively of IR sequences from apoid wasps. Orthologs of IR21a, IR31a, IR40a, IR64a, IR75a/b/c/d, and IR84a are absent in the analyzed apoid wasps draft genomes.
Table 6.
IR Repertoire Sizes in Hymenoptera and Outgroup Species
| Clades | Boostrap Support % | Total | Ac | Ca | Pfu | Nv | Am | Bt | Hs | Dm | Bm | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IR8a | 100 | 9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Classic antennal IRs |
| IR21a | 100 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| IR25a | 100 | 17 | 2 | 2 | 4 | 2 | 1 | 2 | 2 | 1 | 1 | |
| IR31a | — | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| IR40a | 100 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| IR64a | 42 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| IR68a | 100 | 9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| IR75a/b/c | 100 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | |
| IR75d | 100 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| IR76b | 100 | 9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| IR84a | — | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| IR93a | 100 | 10 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |
| IR56e | 98 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | Fruit fly “divergent” IR |
| IR75f | 100 | 16 | 3 | 3 | 1 | 0 | 3 | 3 | 3 | 0 | 0 | Hymenoptera-only antennal IRs |
| IR75u | 100 | 10 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 0 | 0 | |
| IR218 | 100 | 7 | 1 | 1 | 2 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR309 | 100 | 3 | 1 | 1 | 0 | ? | 0 | 0 | 1 | 0 | 0 | |
| IR310 | 100 | 3 | 1 | 1 | 0 | ? | 0 | 0 | 1 | 0 | 0 | |
| IR328 | 100 | 7 | 1 | 1 | 2 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR329 | 100 | 6 | 1 | 1 | 1 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR330 | 100 | 9 | 2 | 1 | 3 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR331 | 100 | 9 | 1 | 2 | 3 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR332 | 100 | 4 | 0 | 1 | 1 | ? | 1 | 1 | 0 | 0 | 0 | |
| IR333 | 100 | 3 | 1 | 1 | 0 | ? | 0 | 1 | 0 | 0 | 0 | |
| IR334 | 100 | 4 | 1 | 1 | 0 | ? | 1 | 1 | 0 | 0 | 0 | Hymenoptera “divergent” IRs |
| IR335 | 100 | 5 | 1 | 1 | 0 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR336 | 100 | 7 | 2 | 1 | 0 | ? | 1 | 1 | 2 | 0 | 0 | |
| IR337 | 100 | 5 | 1 | 1 | 0 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR338 | 100 | 4 | 1 | 2 | 0 | ? | 1 | 0 | 0 | 0 | 0 | |
| IR339 | 100 | 6 | 1 | 2 | 0 | ? | 1 | 1 | 1 | 0 | 0 | |
| IR-X | 100 | 3 | 1 | 1 | 1 | ? | 0 | 0 | 0 | 0 | 0 | Novel clade: unique to apoid wasps only |
| Others | 73 | 0 | 0 | 0 | ? | 0 | 0 | 0 | 56 | 17 | Sequences found only in outgroup species D. melanogaster and B. mori in our analysis. | |
| Total | 256 | 27 | 29 | 26 | 10 | 21 | 22 | 23 | 71 | 27 |
Note.—Ac, Ampulex compressa; Am, Apis mellifera; Bm, Bombyx mori; Bt, Bombus terrestris; Ca, Cerceris arenaria; Dm, Drosophila melanogaster; Hs, Harpegnathos saltator; Nv, Nasonia vitripennis; Pfu, Psenulus fuscipennis. Clade bootstrap values are those given in the phylogenetic tree shown in figure 4. Clades highlighted in gray are not found in Hymenoptera, except Nasonia which has IR21a and 64a. The “?” represent unnamed IRs described by Robertson et al. (2018). Clade names typed in red are classic antennal IRs, those typed in blue are only found in Hymenoptera, those typed in green are here referred to as “divergent IRs.” IR-X is restricted to apoid wasps. “Others” represent IRs found only in outgroup species.
Classical Antennal IRs
This group comprises five clades (IR8a, IR25a, IR68a, IR76b, and IR93a) out of the seven currently known antennal IR clades whose encoded proteins function as coreceptors (Croset et al. 2010). Except for clade IR25a, all other clades have one IR ortholog from each of the species analyzed (fig. 4). Except for the honey bee, Ap. mellifera, all species possess two paralogous genes of clade IR25a (i.e., IR25a.1 and IR25a.2). The honey bee lacks IR25a.2. The result of an additional phylogenetic analysis of the IR25a clade is presented in supplementary figure S5, Supplementary Material online.
Hymenoptera-Only Antennal IRs
This group comprises 11 IR clades: IR75f (IR75f.1, -2, -3), IR75u, IR218, IR310, and IR328–333, similar to reported repertoires in ants (Smith et al. 2011). Ampulex compressa and C. arenaria each possess three IRs belonging to clade IR75f (named IR75f.1–3), equivalent to other Hymenoptera. The two apoid wasps, A. compressa and C. arenaria, each possess one IR ortholog of clade IR75u, also found in bees. In contrast, H. saltator, N. vitripennis, and P. fuscipennis each have two IR orthologs of clade IR75u (i.e., IR75u.1 and IR75u.2).
Hymenoptera “Divergent”-Like IRs
This group comprises eight IR clades (i.e., IR309, IR334–339, IR-X), each containing between three and seven IRs in the species analyzed by us. Most of the Hymenoptera “divergent”-like IR clades are more closely related to each other than to “divergent” IRs of Drosophila (Benton et al. 2009; Croset et al. 2010). The only notable exception was clade IR335, which was sister to the “divergent” IRs IR10a/100a of D. melanogaster. The IR-X clade, which is sister to IR clade IR339, comprises exclusively IRs of apoid wasps.
The Phylogenies of CSPs and OBPs
We identified seven CSP subfamilies (clades CSP1–7, considering only the subclades with sequences from apoid wasps) (supplementary fig. S6A, Supplementary Material online). Proteins of clades CSP2 and CSP5 each contain 5-α helices domains, whereas the proteins of the remaining clades each contain 6-α helices domains. Clade CSP7 contains only genes of apoid wasps and H. saltator; CSP8 contain only genes of Nasonia and Drosophila. In the OBP phylogenetic tree, we identified 13 subfamily clades, labeled clades A–M (supplementary fig. S6B, Supplementary Material online).
Discussion
It has been hypothesized that the evolution of a eusocial lifestyle in Hymenoptera has led to an expansion of the OR-coding gene repertoire in the corresponding eusocial lineages (Robertson et al. 2010; Zhou et al. 2015). However, the validity of this hypothesis has been questioned by some authors (Fischman et al. 2011; Karpe et al. 2017). In the present study, we revisited this hypothesis to assess whether or not the transition from solitary to a eusocial lifestyle in the superfamily Apoidea was associated with a noteworthy increase in the number of CRGs. To this end, we combined genomic and transcriptomic sequence data to first-time characterize protein-coding DNA sequences (CDS) of various CRG families (i.e., CSPs, GRs, IRs, OBPs, and ORs) in apoid wasps.
We found that the genomes of the studied solitary apoid wasps (A. compressa and C. arenaria) harbor a larger number of OR-coding genes than any previously investigated species of Apoidea, including eusocial species. Only some ants (which represent the sister lineage of Apoidea; Peters et al. 2017) possess larger OR gene repertoires (Smith et al. 2011; Zhou et al. 2012, 2015; McKenzie and Kronauer 2018). Apoid wasps also possess larger IR repertoires than any bee investigated so far. Coincidentally, larger ORs and IRs sizes have recently been reported in the genomes of two braconid wasps: Aphidius ervi (228 ORs and 42 IRs) and Lysiphlebus fabarum (156 ORs and 40 IRs) (Dennis et al. 2020). In contrast, the repertoire sizes of GRs, CSPs, and OBPs vary only little among Apoidea (incl. bees) (see table 3). Although our findings are congruent with those reported for N. vitripennis, another solitary parasitoid wasp (Robertson et al. 2010, 2018), they also revealed that a large chemoreceptor gene repertory is not restricted to eusocial species.
Phylogenetic analysis of the OR gene family in Apoidea revealed that the large size of OR-coding gene repertoires in the investigated apoid wasps is primarily due to extensive expansion of two clades of ORs: the 9-exon clade and the L clade. ORs of the 9-exon clade have been implicated with the detection of CHCs, considered particularly important for the communication of individuals sharing a nest and hence the establishment of eusocial societies (Pask et al. 2017; Slone et al. 2017). However, most apoid wasps are solitary, suggesting that CHC-detecting ORs may serve other important functions, such as host or prey recognition (Kather and Martin 2015; Wurdack et al. 2015). Studies on bees suggest that OR members of the L clade are used to identify conspecific sexual partners (Robertson et al. 2010; Sadd et al. 2015). Overall, our results imply that, at least in the superfamily Apoidea, eusociality alone is not a very good predictor for whether or not the genome of a species encodes a large OR gene repertoire.
The ultimate reasons for large OR and IR gene repertoire sizes in apoid wasps remain enigmatic. If the number of chemoreceptor genes is indicative of chemosensory abilities, our results could suggest that solitary apoid wasps are better adapted to using olfactory sense in a chemically diverse environment than eusocial bees. However, there is currently no evidence substantiating this assumption. Either way, it is worth considering the necessity for parasitoids and kleptoparasites to track and properly identify their host species. The ability to do so very likely also requires a finely tuned chemosensory system, in which a large number of chemoreceptors would be highly beneficial (Oeyen et al. 2020).
Flowering plants are known to emit floral scents and to produce nectar with sugars and phytochemicals in varying proportions to attract and reward flower-visiting insects (Chalcoff et al. 2006; Wright and Schiestl 2009). At least six ORs in honey bee foragers (Karpe et al. 2017) are implicated in the detection of floral scent signatures from a variety of herbaceous plant species (Gong et al. 2015; Milet-Pinheiro et al. 2015). In addition, the GR lineages that contain GRs capable of detecting sugars in nectar are known to occur in all major groups of Arthropoda (Kent and Robertson 2009; Freeman et al. 2014). As most (if not all) adult apoid wasps visit flowers for nectar feeding, it appears plausible that apoid wasps possess orthologs of the OR clade H (see supplementary data A5, Supplementary Material online [doi: 10.17632/h4tdmjkg5r.1] and table 4) and orthologs of GR sugar receptors (SR) (see fig. 3). Our data are thus consistent with apoid wasps having acquired nutrition from plant sources during their early diversification.
The Evolutionary Mechanisms of OR Diversification in Apoid Wasps
Compared with GRs and IRs (Eyun et al. 2017), ORs are a family of rapidly evolving genes that exhibit a high turnover (high birth and death rates) over short evolutionary time (McKenzie and Kronauer 2018). Our results on the OR gene turnover among the studied Hymenoptera (see fig. 2) revealed that the last common ancestor of Apoidea had a significantly larger OR repertoire (∼160 OR-coding genes) than previously assumed (73; Robertson et al. 2010; Sadd et al. 2015), with the inferred repertoire size of the ancestor of bees being significantly smaller. Consistent with earlier reports (Engsontia et al. 2015; Zhou et al. 2015; McKenzie and Kronauer 2018), the expansions and contractions occurred in specific OR lineages, leading to gene copy number differences among the extant Apoidea (see supplementary fig. S3, Supplementary Material online and table 4). At least in the extant Apoidea species studied by us, the high turnover of OR-coding genes is due to higher rate of gene births rather than gene deaths.
One of the most intriguing results obtained by us is the ancestral reduction of the OR gene repertoire size in the stem lineage of bees (Anthophila). Since neither the last common ancestor of bees and Psenidae, nor the more recent common ancestor of bees was eusocial, the reduction is possibly linked to the transition from a predatory to a pollen-collecting behavior. Consistent with this interpretation of the data is the insight that also nonaculeate Apocrita that secondarily switched to a herbivorous lifestyle are characterized by a small OR gene repertoire (Xiao et al. 2013; Zhou et al. 2015). The specific mechanisms responsible for the reduction in repertoire sizes of some OR lineages in both eusocial and solitary bees compared with apoid wasps remain unclear.
Local tandem duplication is a characteristic phenomenon of rapidly evolving OR genes in Hymenoptera (Zhou et al. 2015; McKenzie et al. 2016; Brand and Ramirez 2017). In the analyzed Hymenoptera, A. compressa possesses the largest block of ORs (78) in one genome location among Apoidea, only second to the clonal raider ant with 89 ORs (McKenzie and Kronauer 2018). We interpret the high number of OR-coding genes in apoid wasps as a result of extensive local tandem duplication. We suggest that this may have happened by unequal crossing-over (McKenzie and Kronauer 2018), probably facilitated by increased preponderance of transposable elements (TEs) around locally duplicated OR-coding genes (Schrader et al. 2014; Sadd et al. 2015). However, we did not document the TEs prevalence in the apoid wasps to support this suggestion. Unlike the situation in eusocial bees (Robertson et al. 2010; Sadd et al. 2015), the tandem-arrayed OR-coding genes in apoid wasps translate to full-length proteins (in A. compressa) with very few partial genes or pseudogenes. However, the few partial genes and the few pseudogenes are located nonrandomly at the flanks of specific blocks of arrayed ORs, suggesting a mixture of older and younger OR-coding genes within the arrayed blocks. In the draft genomes of A. compressa and C. arenaria, a few of the arrayed OR blocks that we analyzed appear organized in microsynteny when considering coding strand orientation, gene copies, and gene structure (see supplementary fig. S3 and data A2, Supplementary Material online, doi: 10.17632/6zpp8tgz5c.1).
Conclusion
Comparative analysis of the chemoreceptor gene sizes between eusocial bees and their closest relatives, solitary apoid wasps, has so far not been possible due to a lack of information about the chemosensory gene repertoires of solitary apoid wasps. Here we provide such information by presenting manually annotated CRG sets from genomes and transcriptomes (of chemosensory tissues) of A. compressa, C. arenaria, and P. fuscipennis. Compared with the genomes of eusocial bees, we found the genomes of solitary apoid wasps to be characterized by comparatively small GR, CSP, and OBP repertoire sizes, and these repertoire sizes to vary comparatively little among Apoidea. In contrast, we found the genomes of apoid wasps to possess remarkably large OR and IR repertoire sizes. The large size of OR repertoires in apoid wasps (compared with bees) indicate that eusocial Apoidea do not per se possess more ORs than solitary Apoidea. The large OR repertoire sizes in solitary apoid wasps suggest that the wasps’ chemosensory abilities at the molecular level could have been underestimated and that advanced chemosensory abilities are paramount for predatory or parasitoid wasps to find their prey or hosts in a chemically complex environment (see also Oeyen et al. 2020). Our results further suggest that the last common ancestor of Apoidea possessed a larger OR gene repertoire than previously thought. Finally, our data indicate that the transition from a predatory to a pollen-collecting behavior during the evolution of bees was associated with a significant loss of OR diversity.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dieter Schulten for providing A. compressa cultured in the Aquazoo Löbbecke in Düsseldorf, Germany. We are indebted to the Leibniz Graduate School on Genomic Biodiversity Research, Germany, for generously providing access to the unpublished draft genomes of A. compressa and of C. arenaria and for allowing publication of those scaffolds on which we manually annotated chemosensory-related genes. G.F.O. thanks the Alexander von Humboldt foundation for a postdoctoral stipend (KEN_1184840_GF-P) and the Max Planck Society for additional funds. Parts of the study were supported by the German Research Foundation (DFG; NI 1387/3, NI 1387/5). E.G.W. was additionally supported by EXTEMIT - K (CZ.02.1.01/0.0/0.0/15_003/0000433).
Author Contributions
Conceived the study: G.F.O., E.G.W., O.N., and T.P. Collected insects in the field: O.N., R.V., and T.P. Identified insect species: O.N. and T.P. Generated the data and/or performed gene annotations: G.F.O., T.P., and E.G.W. Analyzed the data: G.F.O. and T.P. Contributed materials and reagents: E.G.W. and O.N. All authors contributed to the writing of the manuscript with G.F.O., E.G.W., O.N., and T.P. taking the lead.
Data Availability
The data set accompanying this publication are deposited in Mendeley Data portal under My Datasets at https://data.mendeley.com/drafts/ and can be accessed via the following doi links: supplementary data A1, Supplementary Material online (doi: 10.17632/z2mzyr74br.1)—field raw sample collection data of the insects under study, supplementary data A2, Supplementary Material online (doi: 10.17632/6zpp8tgz5c.1)—full annotation details of all the CRGs in the three apoid wasps, supplementary data A3, Supplementary Material online (doi: 10.17632/npzkyn6sb6.1)—the CRG annotated scaffolds and contigs, supplementary data A4, Supplementary Material online (doi: 10.17632/nv7chmrz7k.1)—the annotated amino acid sequences of all CRGs, and supplementary data A5, Supplementary Material online (doi: 10.17632/h4tdmjkg5r.1)—the inferred OR amino acid sequence phylogenetic tree and a newick tree file of the analyzed OR sequences.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- An W, Cho S, Ishii H, Wensink PC. 1996. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol. 16(6):3106–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidson R, Landa V, Frankenberg S, Adams ME. 2018. Life history of the emerald jewel wasp Ampulex compressa. J Hym Res. 63:1–13. [Google Scholar]
- Bah T. 2011. Inkscape: guide to a vector drawing program. 4th ed.NY: Prentice Hall Press. [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136(1):149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Rutherford K. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform. 4(2):124–132. [DOI] [PubMed] [Google Scholar]
- Blomquist G, Bagneres A. 2010. Insect hydrocarbons: biology, biochemistry, and chemical ecology. New York: Cambridge University Press. [Google Scholar]
- Blösch M. 2000. Die Grabwespen Deutschlands (Sphecidae s. str., Crabronidae). Lebensweise, Verhalten, Verbreitung. (Tierwelt Deutschlands 71). Keltern: Goecke & Evers. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, et al. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct. 7(1):12–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, et al. 2015. Rapid evolution of chemosensory receptor genes in a pair of sibling species of orchid bees (Apidae: Euglossini). BMC Evol Biol. 15:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Ramirez SR. 2017. The evolutionary dynamics of the odorant receptor gene family in corbiculate bees. Genome Biol Evol. 9(8):2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branstetter MG, et al. 2017. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr Biol. 27(7):1019–1025. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalcoff VR, Aizen MA, Galetto L. 2006. Nectar concentration and composition of 26 species from the temperate forest of South America. Ann Bot. 97(3):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-colton M. 2000. NOTUNG: a program for dating gene duplications. J Comput Biol. 7(3–4):429–447. [DOI] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Zhang J. 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177(3):1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Couto A, Mitra A, Thiéry D, Marion-Poll F, Sandoz J-C. 2017. Hornets have it: a conserved olfactory subsystem for social recognition in Hymenoptera? Front Neuroanat. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, et al. 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6(8):e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis AB, et al. 2020. Functional insights from the GC-poor genomes of two aphid parasitoids, Aphidius ervi and Lysiphlebus fabarum. BMC Genomics 21(1):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Robertson HM, Satasook C. 2015. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res Notes. 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyun SI, et al. 2017. Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol Biol Evol. 34(8):1838–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Foster WA. 1995. Nest co-occupation in the digger wasp Cerceris arenaria: cooperation or usurpation? Anim Behav. 50(1):99–112. [Google Scholar]
- Fischman BJ, Woodard SH, Robinson GE. 2011. Molecular evolutionary analyses of insect societies. Pub Nat Ann Sci. 108(Suppl 2):10847–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, Maleszka R. 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16(11):1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EG, Wisotsky Z, Dahanukar A. 2014. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc Natl Acad Sci U S A. 111(4):1598–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatzy W, Volknandt W, Dzwoneck A. 2018. Egg-laying behavior and morphological and chemical characterization of egg surface and egg attachment glue of the digger wasp Ampulex compressa (Hymenoptera, Ampulicidae). Arthropod 47(1):74–81. [DOI] [PubMed] [Google Scholar]
- Gong WC, et al. 2015. Floral scent composition predicts bee pollination system in five butterfly bush (Buddleja, Scrophulariaceae) species. Plant Biol J. 17(1):245–255. [DOI] [PubMed] [Google Scholar]
- Gould J, Bolton B. 1988. The Hymenoptera. British Museum (Natural History). Oxford: Oxford University Press. [Google Scholar]
- Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, et al. 2017. Expression map of a complete set of gustatory receptor genes in chemosensory organs of Bombyx mori. Insect Biochem Mol Biol. 82:74–82. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC. 2011. Evolution of insect olfaction. Neuron 72(5):698–711. [DOI] [PubMed] [Google Scholar]
- Hediger M, et al. 2010. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184(1):155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, Mckinney AJ, Mark A. 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12(9):1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvich C, Tsai C. 1989. Regression and time series model selection in small samples. Biometrika 76(2):297–307. [Google Scholar]
- Jones P, et al. 2014. Sequence analysis InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, et al. 2019. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin L. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe SD, Dhingra S, Brockmann A, Sowdhamini R. 2017. Computational genome-wide survey of odorant receptors from two solitary bees Dufourea novaeangliae (Hymenoptera: Halictidae) and Habropoda laboriosa (Hymenoptera: Apidae). Sci Rep. 7(1):10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather R, Martin S. 2015. Evolution of cuticular hydrocarbons in the Hymenoptera: a meta-analysis. J Chem Ecol. 41(10):871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasar T, Sheffer N, Glusman G, Libersat F. 2006. Host-handling behavior: an innate component of foraging behavior in the parasitoid wasp Ampulex compressa. Etholog 112(7):699–706. [Google Scholar]
- Kent LB, Robertson HM. 2009. Evolution of the sugar receptors in insects. BMC Evol Biol. 9:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, et al. 2018. The genetic basis of a social polymorphism in halictid bees. Nat Commun. 9(1):4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson È, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kulmuni J, Wurm Y, Pamilo P. 2013. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 110(6):538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-red alignment with Bowtie2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Dang CC, Gascuel O. 2012. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol Biol Evol. 29(10):2921–2936. [DOI] [PubMed] [Google Scholar]
- Leonhardt SD, Menzel F, Nehring V, Schmitt T. 2016. Ecology and evolution of communication in social insects. Cell 164(6):1277–1287. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Fetter-Pruneda I, Ruta V, Kronauer DJC. 2016. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc Natl Acad Sci U S A. 113(49):14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Kronauer DJC. 2018. The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 28(11):1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milet-Pinheiro P, Ayasse M, Dötterl S. 2015. Visual and olfactory floral cues of Campanula (Campanulaceae) and their significance for host recognition by an oligolectic bee pollinator. PLoS One 10(6):e0128577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol Biol. 1611:59–73. [DOI] [PubMed] [Google Scholar]
- Oeyen JP, et al. 2020. Sawfly Genomes Reveal Evolutionary Acquisitions That Fostered the Mega-Radiation of Parasitoid and Eusocial Hymenoptera. Genome Biol Evol. 12(7):1099–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oi CA, et al. 2015. Dual effect of wasp queen pheromone in regulating insect sociality. Curr Biol. 25(12):1638–1640. [DOI] [PubMed] [Google Scholar]
- Oxley PR, et al. 2014. The genome of the clonal raider ant Cerapachys biroi. Curr Biol. 24(4):451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, et al. 2015. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics 16(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask GM, et al. 2017. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat Commun. 18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. 2010. How many bootstrap replicates are necessary? RECOMB 2009:184–200. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Calvello M, Ban L. 2005. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses. 30:291–292. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Zhou J, Ban LP, Calvello M. 2006. Review soluble proteins in insect chemical communication. Cell Mol Life Sci. 63(14):1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RS, et al. 2017. Evolutionary history of the Hymenoptera. Curr Biol. 27(7):1013–1018. [DOI] [PubMed] [Google Scholar]
- Piek T, Visser JH, Veenendaal RL. 1984. Change in behaviour of the cockroach, Periplaneta americana, after being stung by the sphecid wasp Ampulex compressa. Entomol Exp Appl. 35(2):195–203. [Google Scholar]
- Polidori C, Federici M, Papadia C, Andrietti F. 2006. Nest sharing and provisioning activity of females of the digger wasp, Cerceris rubida (Hymenoptera, Crabronidae). Ital J Zool. 73(1):55–65. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, et al. 2018. Genome sequence of the wheat stem sawfly, Cephus cinctus, a primitive hymenopteran and wheat pest, illuminates evolution of hymenopteran chemoreceptors. Genome Biol Evol. 10:2997–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Gadau J, Wanner KW. 2010. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 19:121–136. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16(11):1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100(Suppl 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd BM, et al. 2015. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J. 2011. Comparative genomics of the major chemosensory gene families in arthropods. Chichester: Encycl Life Sci John Wiley & Sons Ltd. [Google Scholar]
- Sanchez-Gracia A, Vieira FG, Rozas J. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103(3):208–216. [DOI] [PubMed] [Google Scholar]
- Sann M, et al. 2018. Phylogenomic analysis of Apoidea sheds new light on the sister group of bees. BMC Evol Biol. 18:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, et al. 2014. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla JN, Nagaraju J. 2010. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J Genet. 89(3):341–356. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slone JD, et al. 2017. Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proc Natl Acad Sci U S A. 114(32):8586–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, et al. 2011. Draft genome of the globally widespread and invasive argentine ant (Linepithema humile). Proc Natl Acad Sci U S A. 108(14):5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RaxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Web Server):W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma M, et al. 2019. Transcriptome surveys in silverfish suggest a multistep origin of the insect odorant receptor gene family. Front Ecol Evol. 7:1–13. [Google Scholar]
- Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman J, Wilhelm W. 1990. Husbandry and display of the Jewel wasp. Int Zoo Yearbook. 30(1):118–126. [Google Scholar]
- Verhulst EC, van de Zande L, Beukeboom LW. 2010. Insect sex determination: it all evolves around transformer. Curr Opin Genet Dev. 20(4):376–383. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GA, Schiestl FP. 2009. The evolution of floral scent: the influence of olfactory learning by insect pollinators on the honest signaling of floral rewards. Funct Ecol. 23(5):841–851. [Google Scholar]
- Wurdack M, et al. 2015. Striking cuticular hydrocarbon dimorphism in the mason wasp Odynerus spinipes and its possible evolutionary cause (Hymenoptera: Chrysididae, Vespidae). Proc R Soc Biol Sci. 282:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm Y, et al. 2011. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci U S A. 108(14):5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JH, et al. 2013. Obligate mutualism within a host drives the extreme specialization of a fig wasp genome. Genome Biol. 14(12):R141–R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8(8):e1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. Chemoreceptor evolution in Hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol. 7(8):2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data set accompanying this publication are deposited in Mendeley Data portal under My Datasets at https://data.mendeley.com/drafts/ and can be accessed via the following doi links: supplementary data A1, Supplementary Material online (doi: 10.17632/z2mzyr74br.1)—field raw sample collection data of the insects under study, supplementary data A2, Supplementary Material online (doi: 10.17632/6zpp8tgz5c.1)—full annotation details of all the CRGs in the three apoid wasps, supplementary data A3, Supplementary Material online (doi: 10.17632/npzkyn6sb6.1)—the CRG annotated scaffolds and contigs, supplementary data A4, Supplementary Material online (doi: 10.17632/nv7chmrz7k.1)—the annotated amino acid sequences of all CRGs, and supplementary data A5, Supplementary Material online (doi: 10.17632/h4tdmjkg5r.1)—the inferred OR amino acid sequence phylogenetic tree and a newick tree file of the analyzed OR sequences.