Abstract
Noncoding RNAs are exquisitely titrated by the cellular RNA surveillance machinery for regulating diverse biological processes. The RNA exosome, the predominant 3′ RNA exoribonuclease in mammalian cells, is composed of nine core and two catalytic subunits. Here, we developed a mouse model with a conditional allele to study the RNA exosome catalytic subunit DIS3. In DIS3-deficient B cells, integrity of the immunoglobulin heavy chain (Igh) locus in its topologically associating domain is affected, with accumulation of DNA-associated RNAs flanking CTCF-binding elements, decreased CTCF binding to CTCF-binding elements and disorganized cohesin localization. DIS3-deficient B cells also accumulate activation-induced cytidine deaminase–mediated asymmetric nicks, altering somatic hypermutation patterns and increasing microhomology-mediated end-joining DNA repair. Altered mutation patterns and Igh architectural defects in DIS3-deficient B cells lead to decreased class-switch recombination but increased chromosomal translocations. Our observations of DIS3-mediated architectural regulation at the Igh locus are reflected genome wide, thus providing evidence that noncoding RNA processing is an important mechanism for controlling genome organization.
The RNA surveillance machinery ensures the appropriate titration of coding and noncoding RNAs (ncRNAs) in cells to allow RNA-dependent biological functions while avoiding RNA-induced pathophysiologies1,2. Among the various factors that participate in RNA surveillance, the RNA exosome complex is responsible for degradation and/or 3′-end processing of a variety of ncRNAs3, although the biological relevance of the transcription and subsequent degradation of exosome-sensitive ncRNAs is ambiguous. The 11-subunit exosome complex contains two RNase subunits, EXOSC10 and DIS3, although the function of EXOSC10 in RNA degradation has been shown to be limited4. We and others have previously identified subsets of RNA-exosome-sensitive ncRNAs4–7, including RNAs associated with transcription start sites (aTSS-RNAs), enhancer RNAs (eRNAs, specifically expressed from enhancers), antisense RNAs (asRNAs) and long ncRNAs (lncRNAs, including all ncRNAs longer than 200 bp), but these substrates were identified ex vivo, raising questions about their relevance during development and homeostasis in vivo. In addition, although some of these RNA-exosome-sensitive ncRNAs have been associated with biological functions8–10, the requirement for their rapid decay has remained an important question in the field of RNA biology.
A remarkable set of developmentally programmed DNA rearrangements occurs during B-cell maturation, allowing them to generate high-affinity antibodies to the innumerable antigens encountered11. First, B cells generate the antigen-binding sites of the B-cell receptor by V(D)J recombination in the bone marrow. Thereafter, B cells traverse to secondary lymphoid organs to undergo two additional rounds of genetic diversification by class-switch recombination (CSR) and somatic hypermutation (SHM), altering the B-cell-receptor constant region and increasing antigen affinity. These last processes implicate the roles of activation-induced cytidine deaminase (AID), transcription, DNA repair factors and epigenetics in the Igh and immunoglobulin light chain (Igl) loci12, while recent studies highlight a crucial role for cohesin and loop extrusion mechanisms during CSR13, implicating a set of CTCF-binding elements (CBEs) at the Igh 3′ super-anchor14. During affinity maturation, how AID induces mutations on both sense and antisense DNA strands of V genes and whether this process requires RNA processing remain unanswered questions12 in the domain of antibody biology.
Genome architecture relies on several layers of organization. Chromosomes are found in chromosomal territories, inside which active and inactive domains form A and B compartments, respectively, while substructures create topologically associating domains (TADs) containing different loop interactions15. These layers of genome organization are dependent on many factors, including architectural proteins, such as CTCF, YY1, the cohesin complex, the mediator complex and LDB1 (ref. 15). CTCF binds to specific genomic sequences (CBEs), while the cohesin complex scans long DNA distances before creating a stable complex with two convergent CBEs by the proposed mechanism of loop extrusion16–18. Anomalies in CTCF/cohesin-mediated TAD regulation are associated with pathologies15 (for example, Cornelia de Lange syndrome, developmental defects and activation of oncogenic sequences), defects in V(D)J recombination19,20 and altered CSR13. Thus, an evolving topic of investigation is the factors regulating loop extrusion leading to TAD formation and dissolution.
In this study, we demonstrate that massive accumulation of RNAs and chromatin-associated RNAs perturbs CTCF/cohesin localization and associated mechanisms, and exposes B cells to DNA translocations. DNA:RNA hybrid accumulation alters mutational distribution by overexposing the sense single-stranded DNA to AID-mediated deamination. This affects the patterns of mutational distribution during SHM and increases microhomology-mediated DNA repair during CSR. Taken together, our results reveal the critical function of DIS3-mediated ncRNA processing in two distinct processes: (1) architectural organization of the B-cell genome and (2) distribution of somatic mutations at the Igh locus.
Results
An experimental mouse model to study DIS3 RNase activity.
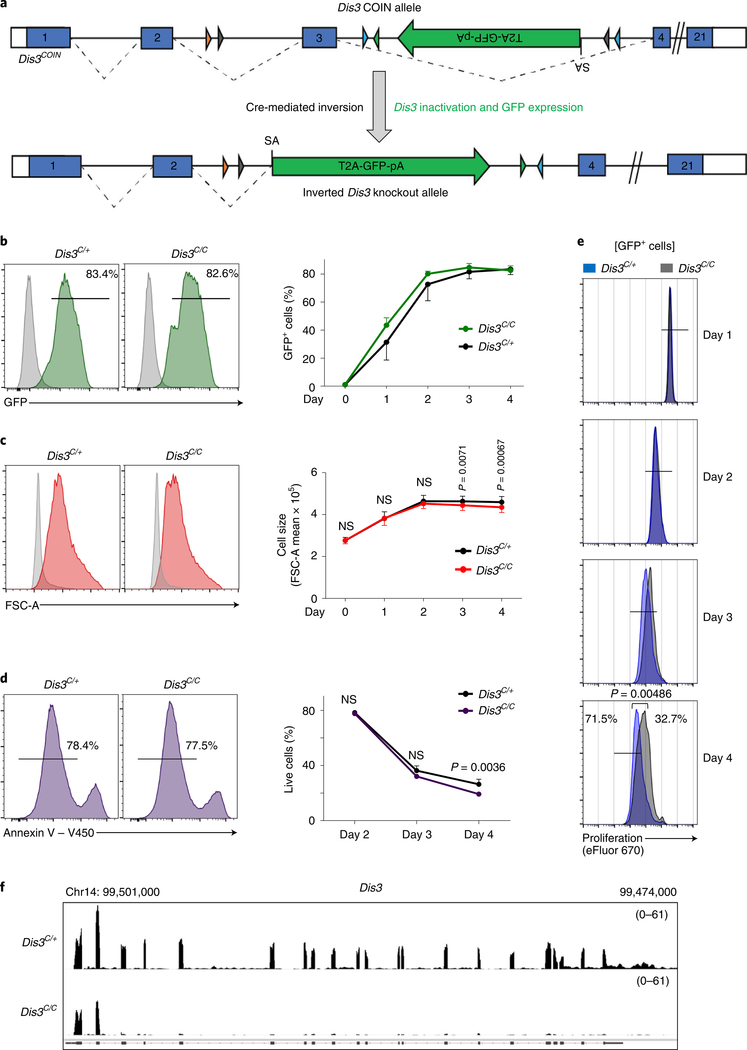
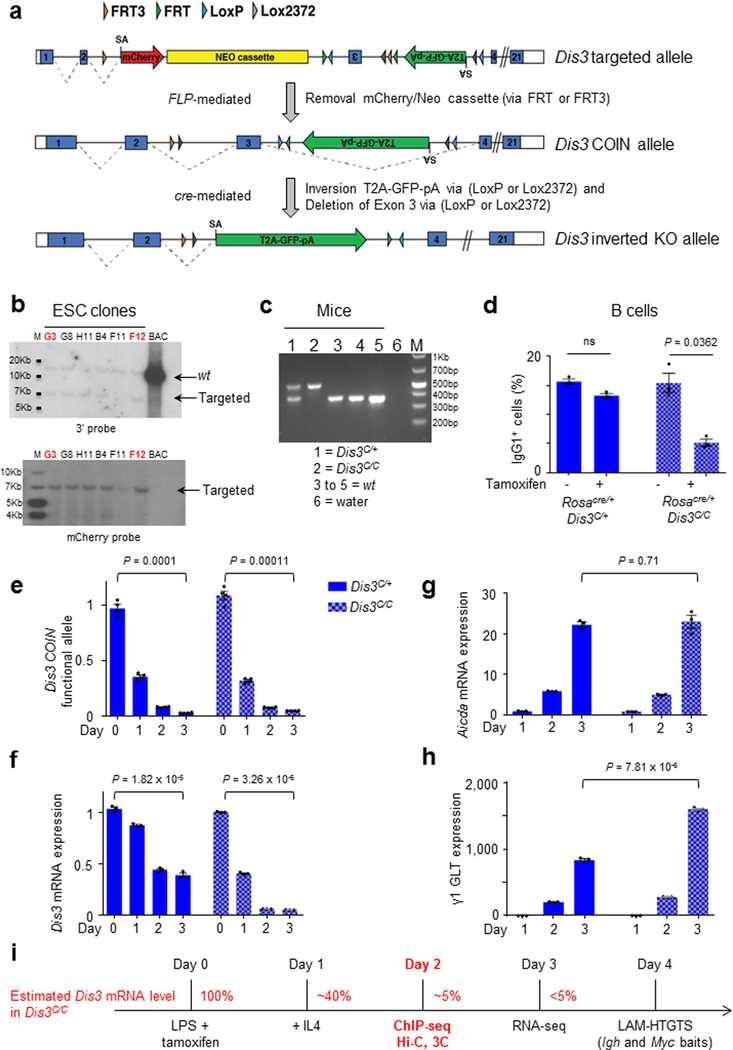
We created a conditional-inversion (COIN) allele of the Dis3 gene, named Dis3COIN (Dis3C; Fig. 1a and Extended Data Fig. 1a), that expresses green fluorescent protein (GFP) following Cre-mediated inversional deletion of the Dis3COIN allele (Fig. 1b). We validated the Dis3 targeting in embryonic stem cells (ESCs) by Southern blot (Extended Data Fig. 1b) and in mice by PCR (Extended Data Fig. 1c), and generated appropriate models to study B cells in vitro and in vivo. Rosacre/+Dis3COIN mice expressing a tamoxifen-dependent Cre were used for B-cell stimulations in ex vivo culture conditions, while AIDcre/+Dis3COIN were used for studies of B-cell germinal center (GC) activation in vivo. VHB1–8KI/KAIDcre/+Dis3COIN mice were used to evaluate in vivo SHM specifically. In the absence of a functional RNA exosome complex, a defect in CSR is observed in activated B cells4,7, so we tested the potential of DIS3-deficient cells to undergo class switching. We stimulated Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C B cells in vitro in the presence or absence of tamoxifen. We observed a CSR defect only in homozygous cells in the presence of tamoxifen, but not in heterozygous cells or Dis3C/C cells in the absence of tamoxifen treatment (Extended Data Fig. 1d). We therefore chose Dis3C/+ cells treated with tamoxifen as the control for Dis3C/C tamoxifen-treated cells. Initial experiments demonstrated that inversion-coupled deletion of the Dis3COIN allele in B-cell culture leads to expression of GFP without significant changes in cell activation, viability or proliferation at relevant experimental time points (Fig. 1c–e). At both the DNA and RNA levels, Dis3 allelic inversion and inactivation were approximately 95% after 1 d of interleukin-4 (IL-4) treatment in Dis3C/C culture (Extended Data Fig. 1e,f). As controls, Aicda mRNA expression (corresponding to AID protein) was not affected, but γ1 germline transcripts were increased due to lack of processing by the RNA exosome (Extended Data Fig. 1g,h). These cells were collected 2 d after IL-4 application, and RNA sequencing (RNA-seq) illustrates efficient loss of Dis3 mRNA in DIS3-deleted (Dis3C/C) B cells (Fig. 1f). Accordingly, we successfully generated a new mouse model to specifically inactivate the RNA exosome catalytic subunit DIS3 and defined the experimental kinetics for B-cell studies, when cell physiology is not affected but Dis3 loss is efficient (Extended Data Fig. 1i).
Fig. 1 |. Specific deletion of the RNA exosome catalytic subunit DIS3.
a, Schematic of the Dis3 COIN allele. Lox sites located in introns 2 and 3 allow an inversion of the GFP gene coupled to the deletion of exon 3 and Dis3 inactivation after Cre activity. Dashed lines show the splicing between the endogenous Dis3 exons or the GFP gene after inversion (Extended Data Fig. 1.). SA, splice acceptor. b, Representative histogram (left) of GFP expression before (gray) and after (green) 4 d of B-cell stimulation, using Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C cells. Quantification (right) of GFP expression following Dis3 allelic inversion during the kinetics of B-cell stimulation (four independent experiments, n = 5; data are the mean ± s.e.m.). c, Flow cytometric analysis of Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C activated B cells. Representative histograms demonstrating the size increase during in vitro B-cell stimulation (day 4; red) compared with resting cells (day 0; gray). Size quantification (right) during the kinetics (four independent experiments, n = 5; data are the mean ± s.e.m.; two-tailed paired t-test). d, Representative histograms showing live cells (annexin V−) at day 2. Viability was quantified at day 2, day 3 and day 4 after B-cell stimulation (two independent experiments performed in triplicate; data are the mean ± s.e.m.; two-tailed unpaired t-test). e, Representative histograms of eFluor 670 dye dilution to monitor cell proliferation at day 1, day 2, day 3 and day 4 after B-cell stimulation. Histograms are gated on GFP+ cells. Rosacre/+ Dis3C/+ cells are in blue, while Rosacre/+ Dis3C/C cells are in gray (two independent experiments performed in duplicate; data are the mean ± s.e.m.; two-tailed unpaired t-test). f, RNA-seq tracks showing Dis3 inactivation. B cells were stimulated in vitro and collected 48 h after application of IL-4 (n = 2). Reads per genomic content (RPGC) values are shown.
DIS3 controls RNA level at CBEs in vitro and in vivo.
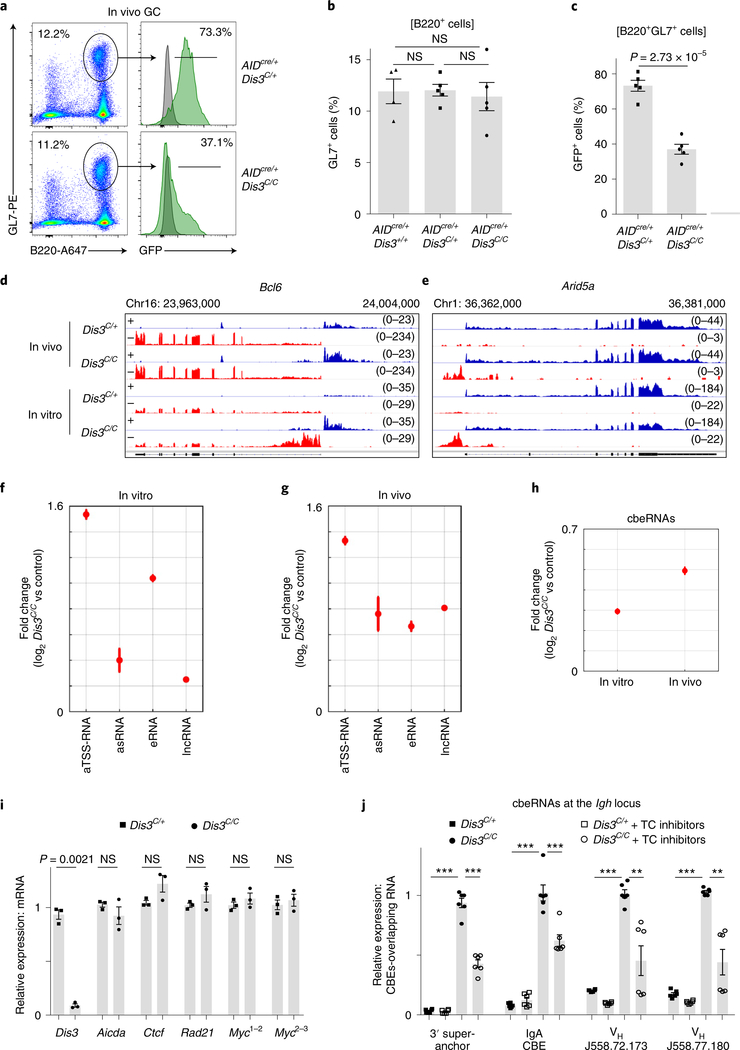
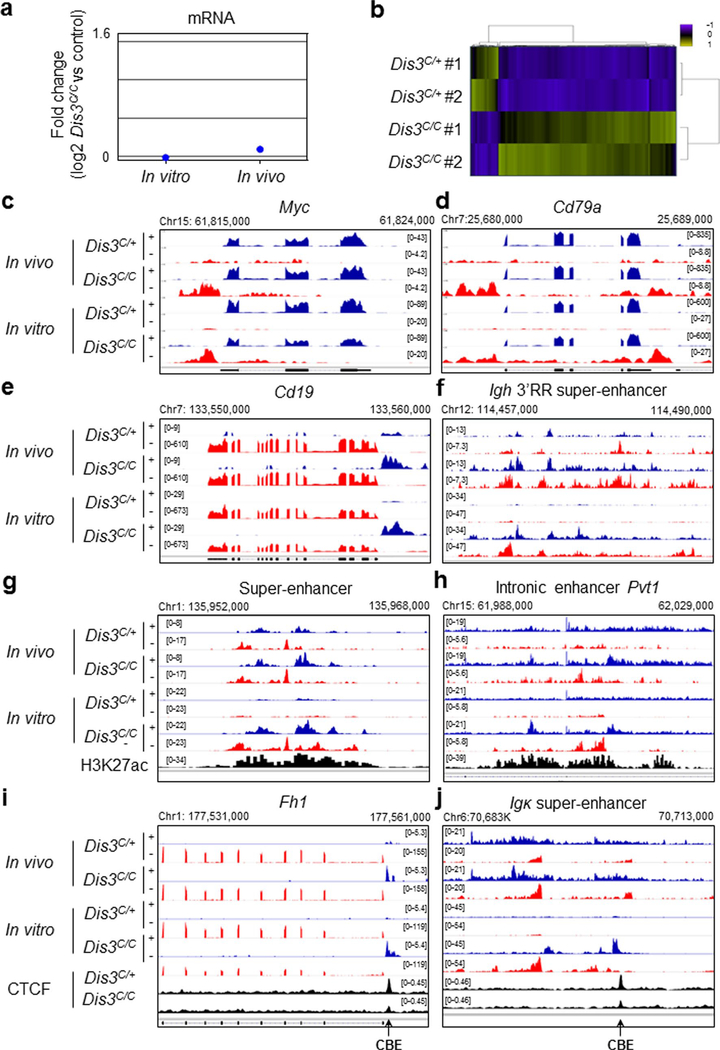
Although RNA exosome substrates have been described previously7, an experimental system to identify exosome-sensitive ncRNAs in vivo was lacking. Here, we directly isolated activated B cells from GCs in Peyer’s patches to perform RNA-seq. The percentage of GC B cells was similar in control, AIDcre/+Dis3C/+ and AIDcre/+Dis3C/C mice (Fig. 2a,b), while the counterselection of DIS3-deficient GFP+ cells occurred in vivo during the ongoing GC reaction (Fig. 2c). To evaluate the RNA processing activity of DIS3 in vitro and in vivo, we sequenced the RNAs from both Rosacre/+Dis3COIN stimulated B cells and GC B cells. RNA-seq analyses revealed DIS3-sensitive substrates, principally aTSS-RNAs, eRNAs, asRNAs and lncRNAs (Fig. 2d–g), while steady-state messenger RNA (mRNA) levels were mainly unaffected at these immediate time points (Extended Data Fig. 2a,b). This strategy also revealed insights into DIS3 substrates at genes that are poorly expressed in vitro, with a pertinent example at the GC-specific Bcl6 gene21,22 (Fig. 2d). Many DIS3-sensitive ncRNAs were found to be present in vivo at important regions of B-cell activation and translocation23–25, including Arid5a, Myc, Cd19, Cd79a and the Igh 3′ regulatory region (RR) super-enhancer26 (Fig. 2e and Extended Data Fig. 2c–f). DIS3-sensitive eRNAs from GC B cells could also be visualized, as shown for super-enhancer and intronic enhancer eRNAs (Extended Data Fig. 2g,h). Relevant to this study, we found that CBEs that are occupied with CTCF protein in B cells are frequently overlapped by DIS3-sensitive RNAs (named cbeRNAs; Fig. 2h and Extended Data Fig. 2i,j).
Fig. 2 |. RNA-seq from GC and in vitro activated B cells reveals new DIS3-sensitive RNAs overlapping CBEs.
a, Flow cytometry analysis of Peyer’s patches from AIDcre/+Dis3C/+ and AIDcre/+Dis3C/C mice. GFP expression was evaluated from GC B cells (B220+ and GL7+), with wild-type cells (gray) used as the control, and compared to Dis3COIN cells (green) (representative from three experiments). Dot plots show the gating strategy to study and sort GC B cells. b, Quantification of GC (B220+ and GL7+) cells by FACS from AIDcre/+Dis3+/+ (n = 4), AIDcre/+Dis3C/+ (n = 5) and AIDcre/+Dis3C/C (n = 5) Peyer’s patches cells (three independent experiments; data are the mean ± s.e.m.; two-tailed unpaired t-test). c, Quantification of GFP expression from GC B cells (n = 5, mean ± s.e.m., two-tailed unpaired t-test). d–g, DIS3-sensitive RNA substrates from in vivo GC and in vitro activated B cells. B cells were collected directly from in vivo GC cells (AIDcre/+Dis3C/+ and AIDcre/+Dis3C/C; n = 2) or from in vitro stimulations (Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C; n = 2) for RNA-seq. RPGC values are shown. Owing to the use of strand-specific RNA-seq, + (blue) or − (red) indicates the two strands of DNA corresponding to the RNA being identified. d, Example of DIS3-sensitive antisense TSS RNAs and antisense intragenic RNAs at the Bcl6 locus, a hallmark of the GC reaction. e, Example of DIS3-sensitive antisense TSS RNAs and antisense intragenic RNAs at Arid5a, frequently translocated in B cells. f, Genome-wide analysis of DIS3-sensitive RNA substrates from in vitro activated B cells (n = 2; data are the mean ± standard error (s.e.)). As control, Rosacre/+Dis3C/+ in vitro activated B cells were used, which do not express ncRNAs. g, RNA-seq of AIDcre/+ Dis3C/C GC B cells (B220+, GL7+ and GFP+). The increased expression of the different ncRNA classes is shown (n = 2; data are the mean ± s.e.). As control, we used Rosacre/+Dis3C/+ in vitro activated B cells. h, Genome-wide analysis of CBE-overlapping RNAs (cbeRNAs) in vitro and in vivo. i, RT–qPCR evaluation of Ctcf, Rad21 and Myc mRNA expression at the time of Hi-C and ChIP–seq experiments (24 h after IL-4 treatment). Aicda (AID) and Dis3 were used as controls (three experiments with qPCR performed in triplicate; each point is the mean of one experiment, ± s.e.m., paired two-tailed t-test). Myc expression was evaluated using two different pairs of primers located on exons 1 and 2 (Myc1,2) or exons 2 and 3 (Myc2,3), with similar results. j, RT–qPCR evaluation of CBE-overlapping ncRNA expression 24 h after IL-4 treatment and 4 h after transcription (TC) inhibitors treatments (two experiments with qPCR performed in triplicate; each point is one qPCR value, ± s.e.m., unpaired two-tailed t-test. P = 2.03 × 10−6; P = 2.31 × 10−6; P = 2.8 × 10−5; P = 0.00091; P = 2.05 × 10−6; P = 0.0053; P = 2.54 × 10−13; P = 0.0026, from left to right).
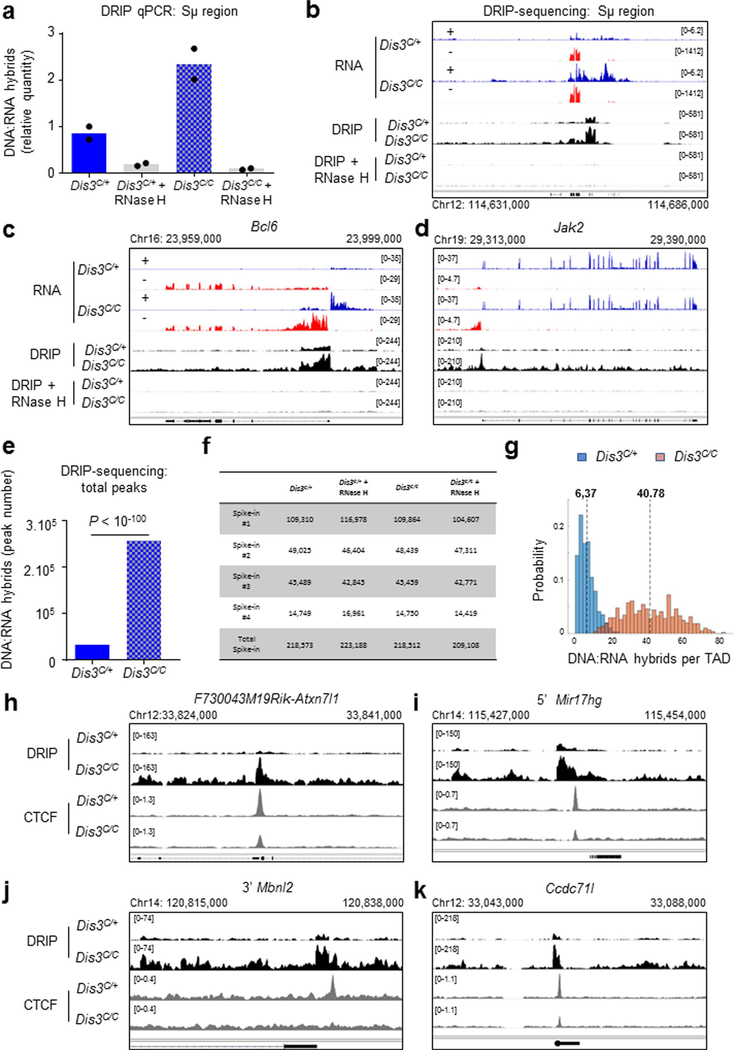
Accumulation of RNA exosome substrates also has been correlated with increased chromatin-associated RNAs in the genome7,27–29. Using a DNA:RNA hybrid immunoprecipitation (DRIP) approach, we confirmed the predisposition of DIS3-deficient cells to accumulate DNA:RNA hybrids, while RNase H treatment efficiently degraded these chromatin-associated RNAs (Extended Data Fig. 3a–d). Deep DRIP sequencing (DRIP-seq) captures DNA-associated RNAs at many places, as shown for Sμ, Bcl6 and Jak2 (Extended Data Fig. 3b–d), and reveals a massive accumulation of DNA:RNA hybrids over the genome in the absence of DIS3, with more than 250,000 peaks detected (Extended Data Fig. 3e,f), a fold increase of >7 compared to the control, corresponding to >40 DNA:RNA hybrids per TAD in the absence of DIS3 (Extended Data Fig. 3g). We further noted that DIS3-sensitive RNAs often form robust DNA:RNA hybrids in proximity to CBEs (Extended Data Fig. 3h–k), although not all DNA:RNA hybrids are in TAD neighbor CBEs.
We confirmed the overall expression of Aicda, Ctcf, Rad21 and Myc mRNAs are comparable between Dis3C/+ and Dis3C/C B cells (Fig. 2i) and the existence of these new CBE-overlapping RNAs by quantitative PCR with reverse transcription (RT–qPCR) at multiple sites in the Igh locus (Fig. 2j). We noted that these cbeRNAs are sensitive to both transcription inhibitors and DIS3 activity (Fig. 2j).
DIS3 activity controls optimal CTCF and cohesin binding in the genome.
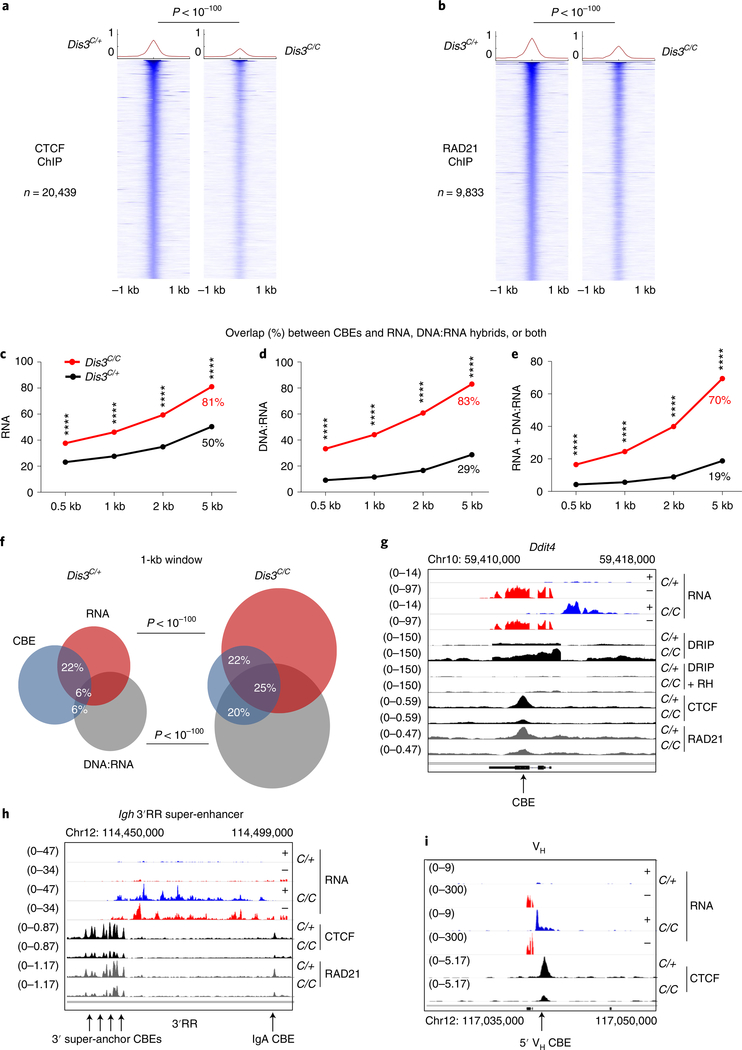
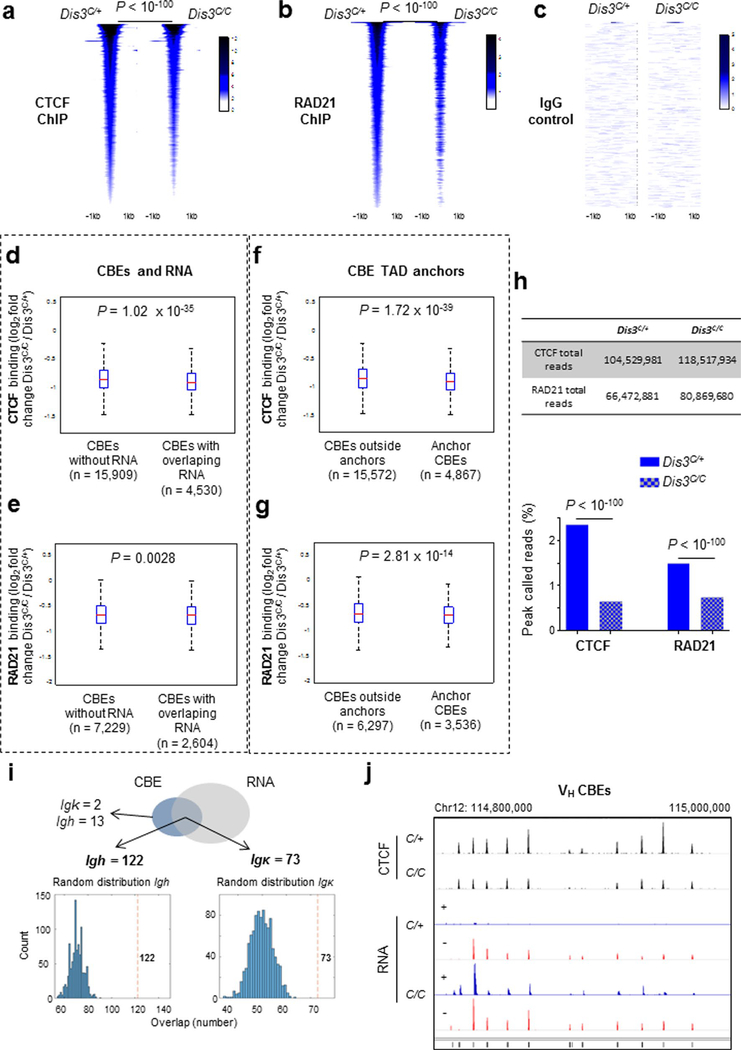
We hypothesized that these DIS3-sensitive cbeRNAs and DNA-associated RNAs could influence CTCF binding and/or cohesin-mediated loop extrusion, and ultimately genomic architecture30,31. Accordingly, we activated B cells to perform chromatin immunoprecipitation followed by sequencing (ChIP–seq) for CTCF and RAD21 at early experimental time points (Extended Data Fig. 1i). As shown, CTCF and RAD21 binding were visibly and statistically decreased genome wide in DIS3-deficient cells (Fig. 3a,b and Extended Data Fig. 4a–c). We analyzed the overlap between CBEs and RNAs, CBEs and DNA:RNA hybrids or CBEs and both RNAs and DNA:RNA hybrids in Dis3C/+ and Dis3C/C cells, using different window sizes. These overlaps were strongly increased in Dis3C/C cells (Fig. 3c–e), with more than 83% of CBEs containing a DNA:RNA hybrid in a 5-kb window in Dis3C/C cells, compared to 29% in control cells. These data suggest an inverse correlation between CTCF/RAD21 binding and DIS3-sensitive RNAs and/or DNA-associated RNA accumulation at CBEs.
Fig. 3 |. DIS3 regulates optimal CtCF and RAD21 binding to the genome during B-cell activation.
a–i, Evaluation of CTCF and RAD21 binding in the absence of DIS3 activity. Cells were collected at an early time point (24 h of IL-4 treatment) and ChIP peaks were analyzed in parallel to DIS3 RNA substrates in Rosacre/+Dis3C/+ (C/+) and Rosacre/+Dis3C/C (C/C) activated B cells. Examples of noncoding transcription overlapping CTCF-binding sites and decreasing CTCF/RAD21 binding in the absence of DIS3 activity are shown in g–i. a, Genome-wide analysis of CTCF ChIP–seq in Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C activated B cells. Two-tailed Wilcoxon rank-sum test was used to compare the 20,439 CTCF peaks in Dis3C/+ and Dis3C/C B cells. b, Genome-wide analysis of RAD21 ChIP–seq in Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C activated B cells. Two-tailed Wilcoxon rank-sum test was used to compare the 9,833 RAD21 peaks in Dis3C/+ and Dis3C/C B cells. c–e, Overlaps between CBEs and RNAs (c), CBEs and DNA:RNA hybrids (d) or CBEs and both (e), using different windows sizes for analyses. χ2 two-tailed proportions test was used (P < 10−100 for each test). f, Venn diagrams showing the overlap between CBEs (using a 1-kb window from the CTCF peaks determined from ChIP–seq experiment; blue circles), RNAs from RNA-seq (red circles) and DNA:RNA hybrids from DRIP-seq (gray circles) in control and DIS3-deficient cells. χ2 two-tailed proportions tests were used to determine the overlaps between CBEs and DNA:RNA hybrids or between CBEs and RNAs. g, Decreased CTCF and RAD21 binding at the Ddit4 locus, where antisense noncoding transcripts are expressed and DNA:RNA hybrids accumulate in the absence of DIS3. RH, RNase H. h, Decreased CTCF and RAD21 binding at the Igh 3′ super-anchor CBEs and IgA CTCF-binding sites, while eRNAs were strongly expressed from the Igh 3′RR super-enhancer. i, Decreased CTCF binding at the VHJ558.77.180 gene, while ncRNAs were strongly expressed from the VH promoter. a–i, CTCF and RAD21 ChIP–seq was performed twice, and DRIP products were sequenced three times with similar results (Extended Data Figs. 3 and 4).
In the absence of DIS3, more than two-thirds of CBEs had RNAs and/or DNA-associated RNAs within a 1-kb window (Fig. 3f), with accompanying decreased CTCF/RAD21 binding (Fig. 3g). Furthermore, CTCF and RAD21 binding were more affected at CBEs that expressed DIS3-sensitive RNAs (Extended Data Fig. 4d,e). Notably, CTCF and RAD21 binding were more decreased at TAD anchor CBEs than at other CBEs (Extended Data Fig. 4f,g). While similar library sizes were sequenced, the percentage of CTCF and RAD21 reads corresponding to ChIP peaks was significantly lower in the absence of DIS3, with fold decreases of >3 and >2, respectively (Extended Data Fig. 4h), suggesting these proteins could be located outside their cognate interacting sites, particularly for RAD21 if DNA:RNA hybrids impede its scanning.
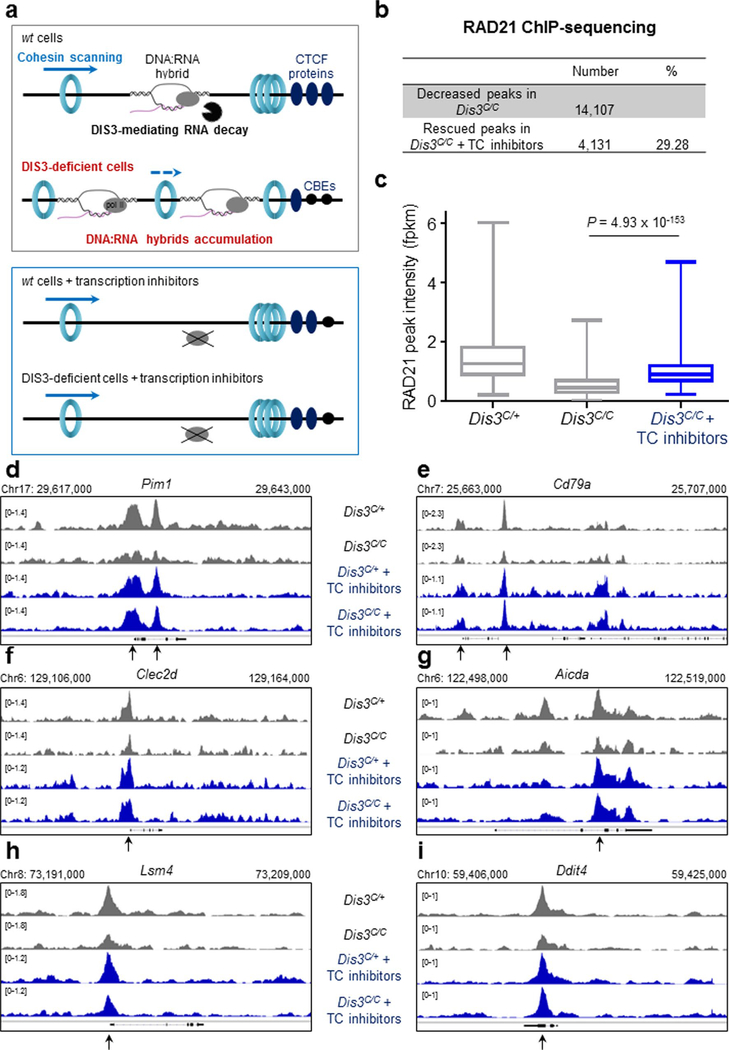
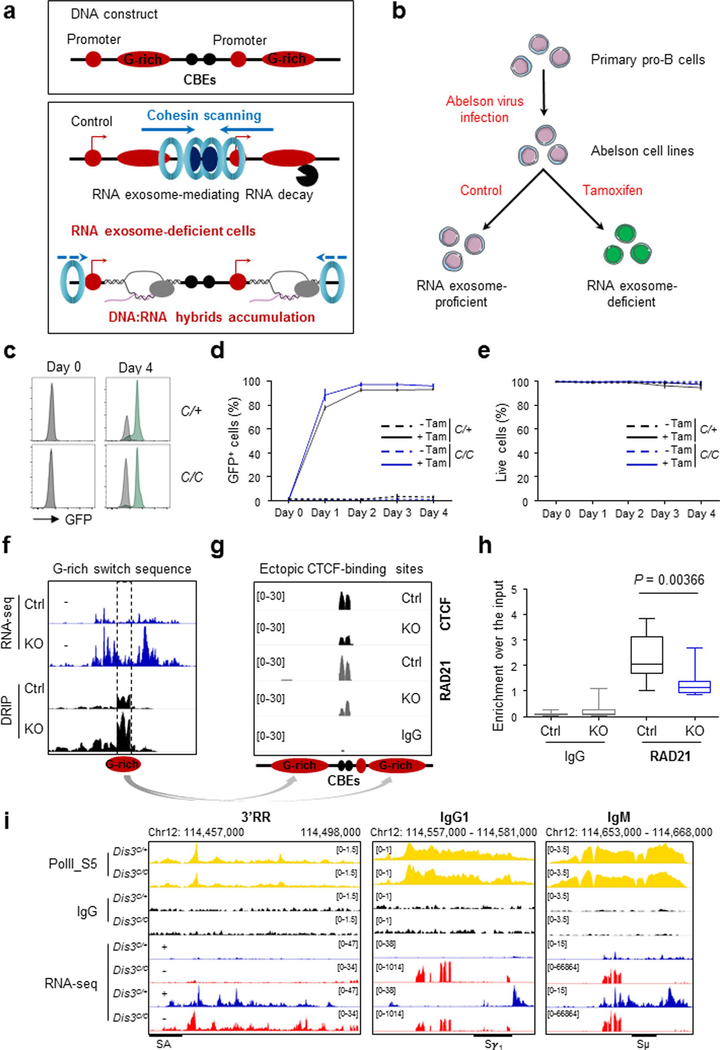
Strikingly, co-transcriptional DNA:RNA hybrid inhibition through transcription inhibitor treatments rescued RAD21 binding at more than 4,000 sites in Dis3C/C B cells (Extended Data Fig. 5a–i), demonstrating that the presence of ncRNAs causally influences RAD21 recruitment in the B-cell genome. Thus it is conceivable that DIS3 regulates the localization of RAD21 by directly preventing DNA:RNA hybrid accumulation that otherwise would impede RAD21 scanning. Finally, G-rich sequences from the Sμ region can directly inhibit the binding of CTCF and RAD21 scanning to ectopic CBEs flanked by these structures prone to forming DNA:RNA hybrids (Extended Data Fig. 6a). Clearly, in the absence of RNA exosome activity, there is a decrease in the recruitment of CTCF and RAD21 at introduced CBEs neighboring G-rich sequences (Extended Data Fig. 6b–h). We also noted that ncRNA and DNA:RNA hybrid accumulation is a posttranscriptional effect that does not perturb accumulation of RNA polymerase II (RNA pol II) itself, as measured by the levels of transcription-initiating S5-phosphorylated RNA pol II occupancy distribution at an early time point after Dis3 deletion (Extended Data Fig. 6i). Gene expression might be expected to change at delayed time points due to accumulation of DNA:RNA hybrids, alterations in genome architecture, effects on transcription termination and other reasons.
At immunoglobulin genes, we observed many defects in CTCF binding, as exemplified by the Igk super-enhancer (Extended Data Fig. 2j). Importantly, at the 3′ end of the Igh locus, the 3′ super-anchor14 CBEs and the IgA CBE showed decreased CTCF and RAD21 occupancy while they accumulated eRNAs in DIS3-deficient B cells (Fig. 3h). Similarly, at the 5′ extremity of the Igh locus, a subset of VH genes displayed increased sense and antisense transcripts with a concomitant decrease of CTCF binding to surrounding sites (Fig. 3i and Extended Data Fig. 3i,j). So, the Igh TAD, which is bordered by a cluster of CBEs forming the 3′ super-anchor and by individual CBEs encompassing the VH genes at the 5′ end, is affected regarding CTCF binding and cohesin localization by loss of DIS3 activity.
DIS3 supervises Igh topologically associating domain integrity.
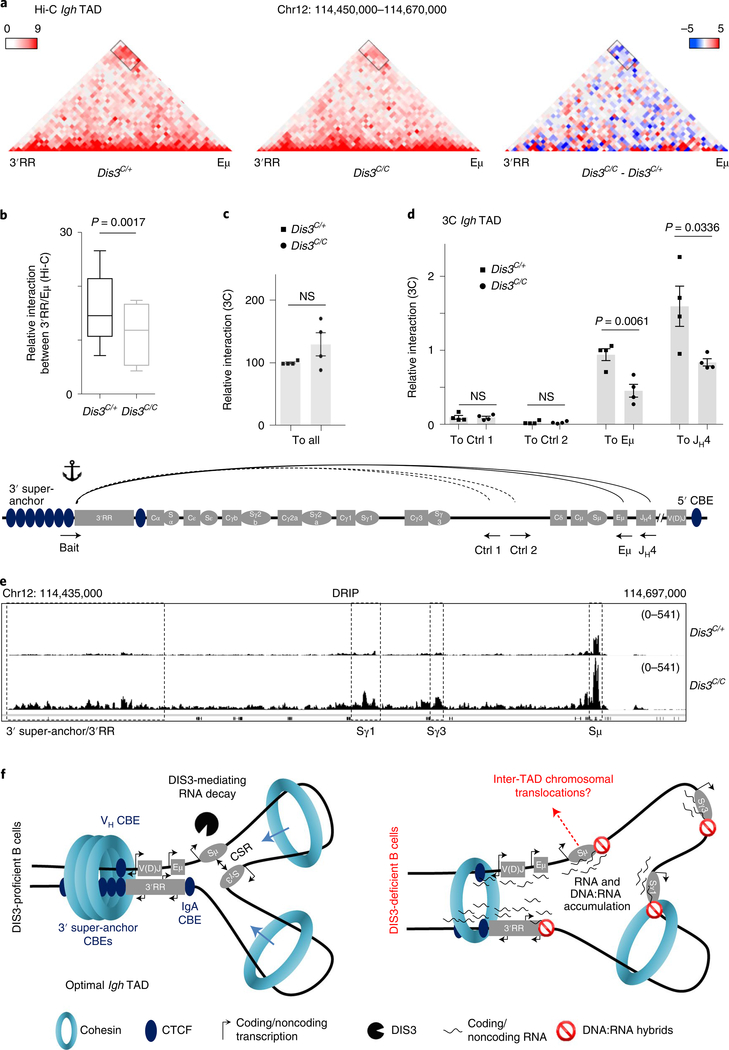
Following B-cell activation, the 3′RR induces germline transcription32,33 and forms a loop with Eμ34, while switch regions align via loop extrusion13,14 to promote DNA breaks and recombination inside the Igh TAD. Since CTCF/RAD21 binding is reduced in Dis3C/C B cells at important CBEs in the Igh locus, we evaluated Igh TAD integrity using high-throughput chromosome conformation capture (Hi-C) and chromosome conformation capture (3C) experiments. We stimulated primary B cells to generate Hi-C libraries 24 h after IL-4 treatment (Extended Data Fig. 1i), minimizing secondary effects that might be caused by prolonged DIS3 depletion. We sequenced three independent experiments and merged them to obtain sufficient resolution at the Igh TAD. We found alteration of the Igh TAD interactions, with the 3′RR/V(D)J-Eμ interaction frequency reduced in Dis3C/C B cells (Fig. 4a). Using the differential analysis feature in ‘Juicebox’, we also observed these decreased interactions between the 3′RR and Eμ regions, as visualized with the cluster of blue squares corresponding to attenuated interactions (Fig. 4a). We extracted the interaction values between the two bins encompassing the Eμ/Sμ fragment and the four bins encompassing the 3′super-anchor/3′RR (eight interactions in total) and found a statistically significant decreased interaction frequency (Fig. 4b). To confirm and quantify these interactions, we performed 3C experiments at the Igh TAD from the 3′RR/3′ super-anchor point of view, followed by qPCR (Fig. 4c). The overall 3C interactions (including all possible interactions) were similar in Dis3C/+ and Dis3C/C cells (Fig. 4c), validating our samples, and the interactions between the bait and intergenic control regions were also comparable (Fig. 4d). In contrast, we confirmed the decreased interactions between 3′RR and Eμ, and between 3′RR and JH4, in multiple biological replicates (Fig. 4d).
Fig. 4 |. DIS3 regulates DNA:RNA accumulation and Igh tAD formation during B-cell activation.
a–d, Evaluation of Igh TAD architecture in DIS3-deficient cells. Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C B cells were stimulated and analyzed at an early time point (24 h of IL-4 treatment) for Hi-C (four independent experiments with three biological replicates) and 3C (two independent experiments with four biological replicates) experiments. Combined data were used for Hi-C analyses and Hi-C visualization was performed using Juicebox. a, Decreased DNA interactions at the Igh TAD in the absence of DIS3. Interactions between the 3′RR super-enhancer and the Eμ intronic enhancer (black rectangle) in Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C activated B cells are shown (left and middle; relative interaction values are in red). The decreased interaction between the 3′RR and Eμ is shown using the subtraction method from Juicebox, calculated by subtracting the control value from the observed value (right; relative gained interactions are in red and the relative lost interactions are in blue). b, The interaction values between the 3′ super-anchor/3′RR region and Eμ/Sμ regions were extracted from Hi-C data and quantified using a two-tailed paired t-test. The box and whiskers plots represent these values, the bottom and top whiskers represent the minimum and maximum scores, respectively, outside the interquartile range. The box plots show the lower quartile, median and upper quartile. c, Schematic of the 3C experiment (bottom, not on scale). Bait located at the end of the 3′RR and beginning of the 3′ super-anchor was used, in combination with control intergenic regions, Eμ, or JH4 interacting regions. Due to the use of LAM-HTGTS to specifically enrich Igh TAD interactions, the interacting regions contain a universal reverse adaptor at their 3′ end. The interaction between the bait and all these interactions using the universal reverse primer is shown as control (ctrl) in Dis3C/+ (squares) and Dis3C/C (dots), two-tailed unpaired t-test. d, The 3′RR/intergenic control 1, 3′RR/intergenic control 2, 3′RR/Eμ and 3′RR/JH4 relative interactions were quantified by 3C experiments followed by qPCR. Bar graphs from c and d represent the mean ± s.e.m. from four biological replicates; each point is the mean for one biological replicate, two-tailed unpaired t-test. Note the JH4 restriction fragment can be either germline or create a V(D)J exon, because the restriction site is located upstream of the recombination signal sequence. e, Integrative Genomics Viewer (IGV) tracks showing the accumulation of DNA:RNA hybrids at the Igh locus. DNA:RNA hybrids strongly accumulate at switch (S) regions, particularly at Sμ donor region, Sγ1, Sγ3 and at the 3′ super-anchor/3′RR super-enhancer. f, Model postulating DIS3-mediated processing of ncRNAs is important for efficient Igh TAD formation and CSR. Left: the Igh locus contains a 3′ super-anchor composed of multiple CBEs, which is critical for loop extrusion during CSR. At the 5′ extremity of VH genes different CBEs are found and stabilize the Igh TAD in concert with the 3′ super-anchor. Inside the Igh TAD, critical interactions allow an optimal CSR, including Eμ−3′RR super-enhancer interaction. DIS3 is necessary to process the different ncRNAs that are strongly expressed as eRNAs, germline transcripts and CTCF-overlapping RNAs during B-cell activation, ultimately contributing to the formation of an optimal Igh TAD necessary for CSR. Right: in the absence of DIS3, an accumulation of ncRNAs and DNA:RNA hybrids is observed, which decreases CTCF and RAD21 localization to their cognate DNA targets. We propose that these ncRNAs and DNA:RNA hybrids impede cohesin-mediated scanning/loop extrusion and perturb Igh TAD stabilization, in particular the 3′RR/Eμ interaction. This phenomenon contributes to decreased CSR and potentially exposes Sμ DNA breaks to inter-TAD recombination, that is, translocations.
CSR to some isotypes partially relies on the loading of the cohesin complex near the 3′ super-anchor/3′RR and subsequent scanning to reach the Eμ/V(D)J region followed by switch regions alignment through loop extrusion before DNA recombination13, although the 3′RR can stimulate transcription independently of the Igh TAD35, and even SHM, involving trans-chromosomal interactions via transvection36. ncRNAs are strongly expressed at switch regions, and stalled RNA pol II facilitates the formation of DNA:RNA hybrids37, while the RNA exosome degrades these ncRNAs4,7, resulting in a dynamic equilibrium of DNA:RNA hybrid formation and resolution in wild-type B cells. The presence of switch regions prone to forming DNA:RNA hybrids could impede the loop extrusion process that allows switch regions alignment in the Igh TAD. Consistent with this possibility, we observed a strong accumulation of DNA:RNA hybrids in the Igh TAD in the absence of DIS3 at critical sites, including the 3′ super-anchor/3′RR and switch regions Sμ, Sγ1 and Sγ3 (Fig. 4e and Extended Data Fig. 3a,b). We propose that DNA:RNA hybrid accumulation impairs CTCF binding at CBEs and interferes with cohesin scanning and stabilization of the CTCF–cohesin complex, as exemplified at the Igh locus in DIS3-deficient activated B cells (Fig. 4f). We note that, in our experimental conditions, 3′RR-mediated transcriptional activity at switch regions was not perturbed to drastically reduce germline transcription, allowing us to evaluate AID activity in the Igh locus.
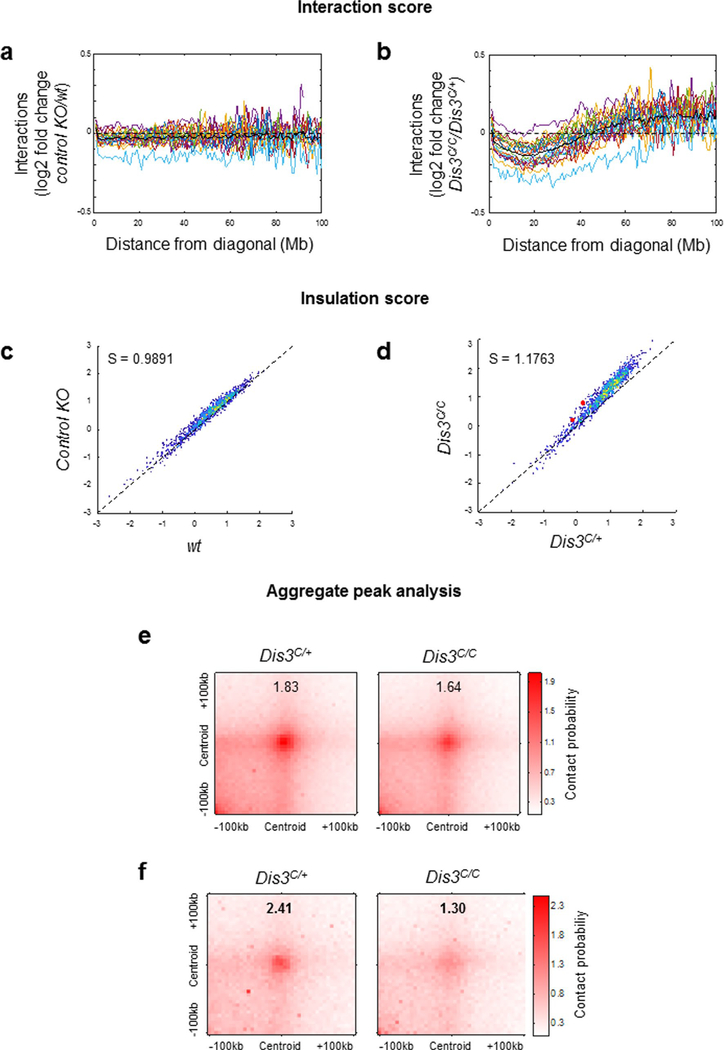
We also note that DIS3-deficient B cells demonstrate altered chromosome architecture genome wide, with visible distinct interaction patterns (Extended Data Fig. 7a,b). These alterations are reproducible (Supplementary Fig. 1a,b) and show specific defects corresponding to decreased long-range chromatin interactions (Extended Data Fig. 8a,b). Furthermore, short-range chromosome interactions were also affected in the absence of DIS3, as measured by insulation score analyses (Extended Data Fig. 8c,d) and aggregate peak analyses (APA; Extended Data Fig. 8e,f), while chromatin A/B compartmentalization was not affected (Supplementary Fig. 2). These observations are consistent with our model (Fig. 4f) where DIS3-deficient B cells accumulate RNAs and DNA:RNA hybrids while undergoing decreased CTCF–cohesin binding at many CBEs across the genome, likely disturbing chromosome interactions. Our data suggest that, in addition to transcription38, posttranscriptional DIS3-mediated RNA processing plays an important role in homeostasis of genome architecture. DIS3-deficient B-cell genome architecture alterations are unlikely due to perturbation of RNA pol II accumulation, as shown in the Igh TAD (Extended Data Fig. 6i).
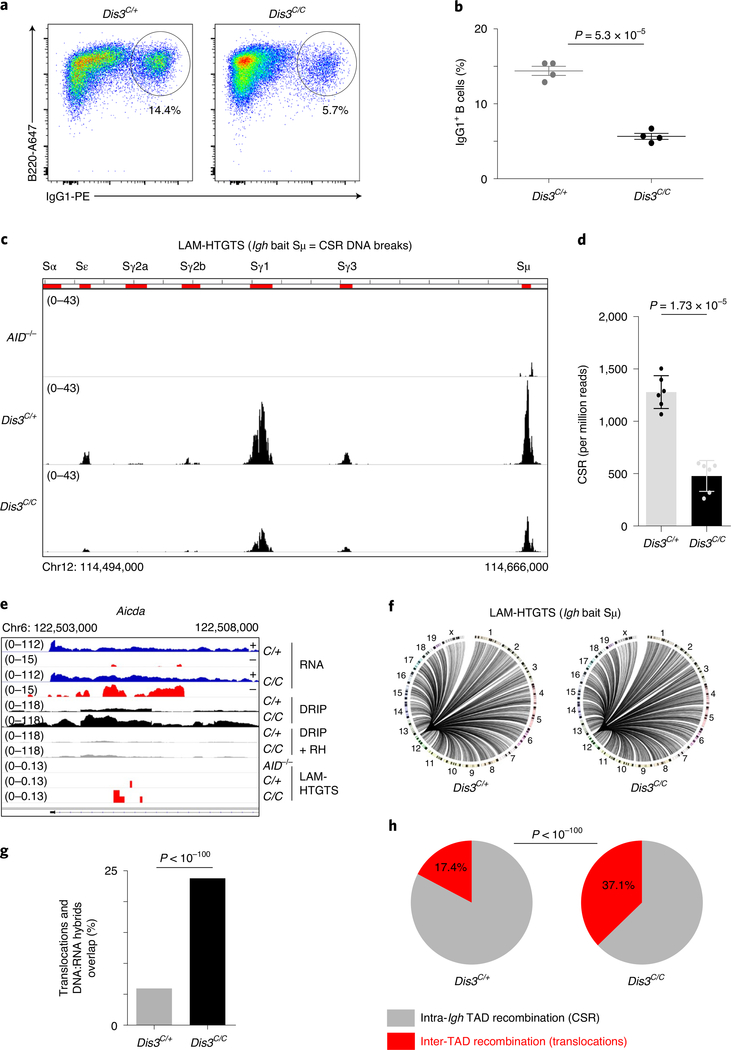
Decreased CSR and increased aberrant DNA recombination in the absence of DIS3.
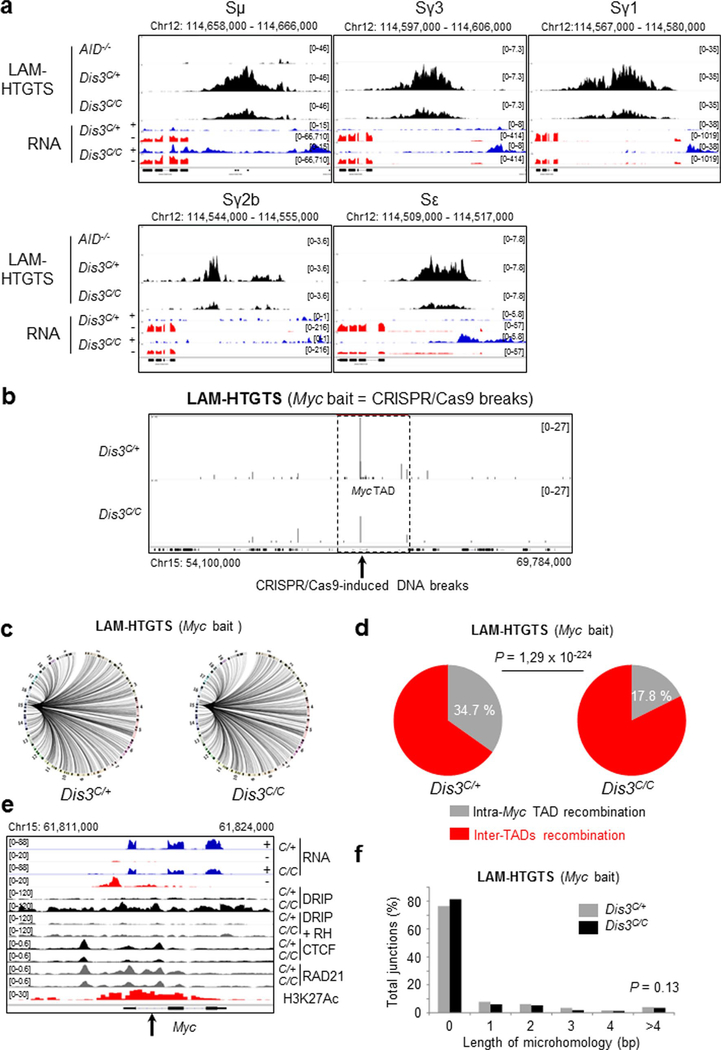
In DIS3-deficient B cells, we confirmed a strong reduction in CSR to IgG1 by flow cytometry analyses (Fig. 5a,b). We used linear amplification-mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS)39 for deep sequencing of the DNA junctions in these cells. This assay showed a strong decrease in DNA breaks at all the S regions of the Igh locus in DIS3-deficient cells, a definitive demonstration that S sequence breaks are DIS3 dependent (Fig. 5c,d and Extended Data Fig. 9a). DNA junctions overlapping well-defined AID hotspots were captured (Fig. 5e) and a large number of translocations identified genome wide (Fig. 5f). Moreover, these translocations tended to accumulate at regions of DNA:RNA hybrid accumulation, as shown in the Aicda locus (Fig. 5e) and other regions of the genome (Fig. 5g). Importantly, the percentage of intra-Igh TAD recombination (that is, CSR) to inter-TAD recombination was altered in DIS3-deficient cells, with a twofold increase in aberrant translocations (Fig. 5h).
Fig. 5 |. DIS3 deficiency decreases intra-tAD class-switch recombination but increases inter-tAD aberrant translocations.
a, Flow cytometry analysis of IgG1 class switching after B-cell stimulation from Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C mice (representative of four independent experiments). b, Quantification of IgG1 class switching after B-cell stimulation (four independent experiments, n = 4; data are the mean ± s.e.m., two-tailed unpaired t-test). c, LAM-HTGTS evaluation of DNA breaks at the Igh locus. Rosacre/+Dis3C/+ (n = 3), Rosacre/+Dis3C/C (n = 3) and AID−/− (n = 1) B cells were activated in vitro and treated with lipopolysaccharide (LPS) and IL-4 for 3 d before DNA extraction. Endogenous DNA recombination was evaluated using Igh bait Sμ. The different switch regions are indicated by red bars. d, Quantification of CSR DNA breaks obtained by LAM-HTGTS (three independent experiments were performed with two sequencings per experiment, each dot represents the result from one sequencing; data are the mean ± standard deviation, two-tailed paired t-test). e, Example of DNA translocations at the AID hotspot Aicda super-enhancer (intron 1 is shown). The concomitant increase of ncRNA, DNA:RNA hybrids and translocations in the absence of DIS3 is shown. Note that the RNA-seq tracks are in log scale to show all the sense and antisense accumulated RNAs. f, Circos plots showing the global DNA junctions, including translocations detected by LAM-HTGTS at the Igh locus. DNA breaks from three independent experiments are shown. g, The overlap between translocation breakpoints and DNA:RNA hybrids in Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C is shown. Bar graphs represent the percentage of overlap; χ2 two-tailed proportions test. h, The proportions of intra-Igh TAD recombination (that is, CSR) and inter-TAD recombination (that is, translocation) are shown. Data were obtained from three independent experiments (with total DNA junctions of 26,750 in Dis3C/+ and 13,761 in Dis3C/C); χ2 two-tailed proportions test.
We confirmed these results using a complementary strategy. We stimulated primary B cells in vitro, infected them with a retrovirus for expression of a CRISPR–Cas9 guide RNA targeting the Myc intron and collected these cells to performed LAM-HTGTS at the Myc locus. A similar imbalance of intra-TAD versus inter-TAD recombination was seen in the Myc TAD, containing the Myc locus, using these double-strand breaks (DSBs) induced by CRISPR–Cas9 (Extended Data Fig. 9b–d). The Myc locus accumulated ncRNAs, DNA:RNA hybrids, and had decreased CTCF/RAD21 binding (Extended Data Fig. 9e), probably overexposing the cells to translocations in the absence of DIS3. Taken together, our observations suggest that architectural integrity of TADs functions as a caretaker for genome stability, and RNA-induced aberrations of TAD architecture can expose cells to DNA translocations.
DIS3-mediated RNA processing is necessary for physiological DNA recombination and mutations.
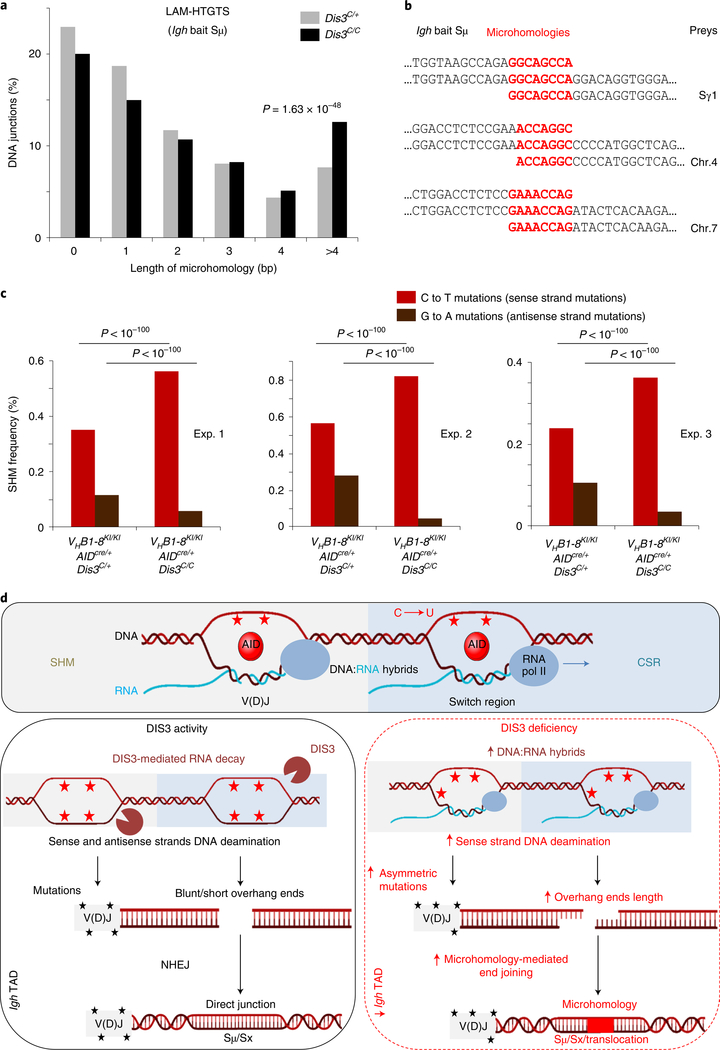
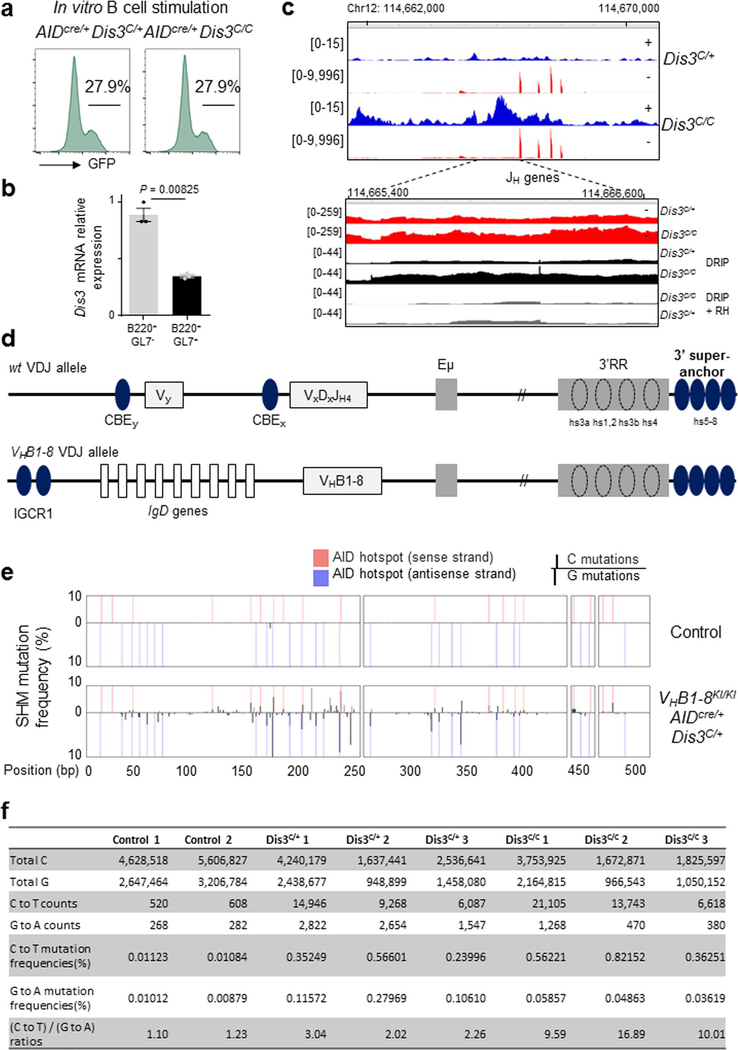
One crucial aspect of the antibody diversity mechanism is the distribution of AID mutagenic activity to S regions and V(D)J genes, causing programmed DNA DSBs11,12,40 and inducing physiological distribution of mutations of variable gene sequences41. We considered the decrease in IgG1 CSR (Fig. 5a) as possibly independent of changes in Igh architecture since the deletion of the 3′RR or 3′CBEs does not totally affect IgG1 CSR42,43. Evaluation of IgG1 CSR provides us an opportunity to disconnect the function of DIS3 in protecting genome architecture from its role in regulating mutational activity of AID inside the Igh locus (Supplementary Discussion). We wanted to evaluate the role of DIS3-mediated 3′-end processing of ncRNAs overlapping S regions and V(D)J in promoting physiological distribution of mutations that cause proper CSR and SHM. Accordingly, we analyzed the LAM-HTGTS DNA junctions to evaluate the contribution of DIS3 to AID-mediated, coordinated single-strand breaks on both DNA strands of S sequences to promote DNA DSBs. The sequence analyses show that DIS3 deficiency caused an increase in microhomology-mediated44 DNA junctions in B cells (Fig. 6a). Many examples of microhomology junctions were found at the Igh locus and translocation partners on different chromosomes (Fig. 6b). In contrast, CRISPR–Cas9-induced DNA junctions revealed indistinguishable levels of microhomology lengths between Dis3C/+ and Dis3C/C cells (Extended Data Fig. 9f), as expected from exogenously generated blunt DSBs. These observations show a dysregulation of coordinated and closely spaced AID-mediated single-strand breaks in Dis3C/C B cells during CSR, leading to an increase in staggered breaks that caused microhomology-mediated end joining (Fig. 6d).
Fig. 6 |. DIS3-sensitive RNA processing influences DNA repair mechanisms and distribution of physiological mutations in activated B cells.
a,b, Microhomology-mediated DNA repair. a, Global analysis of the DNA junctions from Igh LAM-HTGTS showing the length and frequencies of microhomologies overlapping the Igh bait (Sμ) and the prey sequences. All DNA junctions from three independent experiments were combined and the distributions of microhomology-mediated repair evaluated by a χ2 two-tailed proportions test (comparing DNA junctions with microhomology length of >4 bp to the total junctions between Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C cells). b, Examples of microhomology-containing DNA junctions from Rosacre/+Dis3C/C B cells at physiological CSR and translocation junctions. SHM analyses in DIS3-deficient cells. c, Quantification of C and G mutation frequencies in GC B cells. Three pairs of VHB1–8KI/KIAIDcre/+Dis3C/+ and VHB1–8KI/KIAIDcre/+Dis3C/C mice were used. Analyses were performed at the JH gene from the VHB1–8 allele. χ2 two-tailed proportions tests were used to compare the number of C to T mutations relative to the total number of C’s sequenced and the number of G to A mutations compared to the total number of G’s sequenced. d, Proposed model. Top: during SHM and CSR, germline/sense transcripts are expressed at the Igh locus, leading to exposure of the sense single-stranded DNA to AID mutagenic activity (C to U deamination; red stars) as a result of DNA:RNA-associated single-stranded DNA exposure. Bottom left: DIS3 processed sense RNAs, providing AID accessibility to both DNA strands during CSR and SHM. At V(D)J regions, the ratio of sense to antisense mutations (black stars) is physiologically distributed. At switch regions, AID-initiated DNA breaks are equally distributed and frequently generate blunt ends or short overhang DNA extremities. NHEJ is the main DNA repair mechanism, resulting in direct DNA junctions (or short microhomologies). Bottom right: DIS3-deficient cells showed defects in the ability to process DNA-associated RNAs, which increases DNA:RNA hybrids and sense strand DNA deamination. At V(D)J regions, the SHM frequencies were increased on the sense DNA strand and decreased on the antisense strand. At switch regions, this DNA:RNA hybrid accumulation facilitated the creation of DNA with overhang ends and subsequent ligation between complementary sequences, resulting in increased microhomology-mediated DNA repair between switch regions (Sμ and Sx acceptor) or Sμ and translocation partners. Note that the Igh TAD organization was optimal in the presence of DIS3 activity, while intra-Igh TAD interactions between the 3′RR super-enhancer and Eμ were decreased in DIS3-deficient cells.
A direct examination of strand asymmetric DNA mutagenesis can be undertaken at the B-cell V(D)J exons during the GC reaction, marked by robust cellular competition and selection for the best high-affinity antibody-producing B cells45. While DIS3 deficiency did not affect cell viability, proliferation or activation ex vivo at early time point points of culture (Fig. 1c–e), the counterselection of AIDcre/+Dis3C/C GFP+ GC cells eventually occurred in vivo in Peyer’s patches, either by lack of selection or because of DNA damage46, while the overall pool of GC B cells was not affected (Fig. 2a–c and Extended Data Fig. 10a,b). Interestingly, an increase in sense transcripts and DNA:RNA hybrids originating from the V(D)J allele was observed in these cells, as well as a strong accumulation of antisense RNAs (Extended Data Fig. 10c). To interrogate SHM specifically in a single V(D)J allele, we generated VHB1–8 gene knock-in (VHB1–8KI/KI) AIDcre/+Dis3COIN mice (Extended Data Fig. 10d), immunized them to induce SHM47 and collected GC B cells from Peyer’s patches to perform V(D)J sequencing. The overall mutation frequencies in GC-derived B cells were strikingly higher compared to the levels observed in the control DNA samples (Extended Data Fig. 10e). We analyzed the hallmark of AID activity—C to T mutations on the sense DNA strand and the G to A mutations on the antisense strand—in a defined region of the VHB1–8 sequence that accumulated mutations at AID hotspots (Extended Data Fig. 10e). In the absence of DIS3 activity, the C to T mutation frequency was significantly increased, and inversely, the G to A frequency decreased (Fig. 6c and Extended Data Fig. 10f). While the frequencies of sense to antisense DNA strand mutations were consistent between the controls, the ratios were greatly increased in DIS3-deficient cells (approximately fivefold; Fig. 6c and Extended Data Fig. 10f). These observations demonstrate that noncoding transcription processing contributes to strand-specific physiological DNA mutation distribution in B cells (Fig. 6d and Supplementary Fig. 3). Complete deletion of the 3′RR is required to reduce SHM frequency48, while its partial deletion does not affect SHM since V gene transcription occurs sufficiently49. We note that DIS3 deletion does not abolish 3′RR-mediated V gene transcription, allowing us to unravel effects of template strand-specific SHM through the posttranscriptional activity of DIS3.
Discussion
We propose that accumulation of DNA:RNA hybrids in the genome prevents CTCF and cohesin association with CBEs, thus suggesting a direct cis inhibition of CTCF binding to its CBEs, although other mechanistic possibilities exist. For example, a set of RNAs that are important for CTCF clustering at CBEs50,51 could be occluded by accumulation of DIS3-sensitive RNAs in trans, or CTCF could be titrated out from the chromosomes or from the nucleus due to accumulation of cbeRNAs or ncRNAs52,53. Usually convergent CBEs are stabilized by the cohesin complex54 and the decrease of CTCF binding at one site, coupled with a defect of cohesin scanning, could affect the CTCF-binding stability of its CBE pair. The altered pattern of cohesin localization is likely due to the accumulation of DNA:RNA hybrids, as inhibiting these structures with transcription inhibitors rescues their recruitment to large numbers of sites. In addition, ectopically introduced CBEs in the mammalian genome flanked by DNA:RNA hybrid-forming G-rich sequences demonstrate reduced CTCF and cohesin occupancy in the absence of RNA exosome activity. Cohesin complex scanning is normally rapid55, inducing dynamic DNA loop interactions and TAD stabilization. The regulation of cohesin scanning is unknown, but our data suggest that accumulation of DNA-associated RNAs is a factor that could delay the kinetics of loop extrusion. While DNA:RNA hybrids are found widespread in the genome56, the equilibrium between formation and resolution of DNA:RNA hybrids could be a regulatory mechanism for cohesin progression and subsequent loop organization, at least at some specific regions of the genome.
In particular, at the Igh locus, we postulate that in the absence of DIS3 and ncRNA turnover, accumulation of DNA:RNA hybrid structures formed by ncRNA aggregation on transcribed DNA impedes cohesin-mediated loop extrusion and CTCF–cohesin complex stabilization at the 3′ super-anchor and VH CBEs. An important question is whether DIS3 activity regulates the genome architecture outside the Igh TAD. Extending our observations genome wide, we could evaluate effects of DIS3 deletion, RNAs and chromatin-associated RNA accumulation on chromosome architecture. We found both long-range and short-range chromosome interactions were perturbed in Dis3C/C cells. Consistent with these observations of altered chromosome interactions, we observed increased inter-TAD translocations both from the Igh TAD and Myc TAD in DIS3-deficient B cells. Taken together, we provide evidence that, both at the Igh TAD and genome wide, DIS3 is important for stabilization of chromosome architecture. We note that these experiments were performed at early time points after DIS3 depletion, and perhaps underestimate the full effect of RNA and DNA:RNA hybrid accumulation on CTCF/cohesin recruitment, genome organization and overall transcription. Our results also suggest that although the loop extrusion mechanism may not control gene expression all across the genome20,57,58, but might be important for the regulation of individual loci that developmentally define cellular identity like the Igh TAD during B-cell activation. Finally, our study implies that loop extrusion may have unappreciated biological functions including a continuous monitoring mechanism of mammalian genomic integrity59.
The corollary of DNA-associated RNA accumulation would be some variations in AID-mediated mutagenic activity by overexposure of single-strand DNA, independently of Igh TAD structure. While this effect has been demonstrated at AID off-targets60, its contribution to physiological mutations during affinity maturation is unknown. We show that DIS3 efficiently regulates AID activity at the V(D)J exon during SHM in the GC reaction in vivo, thus including RNA processing as an important factor in generating high-affinity antibodies both during immune responses and following vaccination59. Different mechanisms have been proposed for repair of AID-induced DNA DSBs; the major one is classical nonhomologous end joining (NHEJ)61,62, while microhomology-directed repair is usually a minor factor in normal cells63. Here we provide the first example of how ncRNA processing redirects classical NHEJ repair toward microhomology-driven DNA repair. The overexposure of sense DNA strands at switch regions to deamination by AID facilitates the creation of longer overhang DNA ends during CSR, and their eventual joining. We observed this increase in microhomology-mediated DNA repair between two S regions, but also between Sμ and translocation partners. This unbalanced distribution of mutagenesis and subsequent consequences should be investigated further, and could be an important diagnostic and evolutionary parameter of human tumors harboring DIS3 mutations, particularly considering that DIS3 is frequently mutated in cancers64, including multiple myeloma65.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41588-020-00772-0.
Methods
Generation of Dis3COIN mice.
To make the Dis3 COIN targeting vector, a bacterial artificial chromosome clone (bMQ137G10) containing the mouse Dis3 gene locus was modified using bacterial homologous recombination. After a series of cloning steps, a construct consisting of the mCherry gene cassette with a 5′ splice acceptor and a 3′ poly A signal sequence, and a neo gene cassette with the human ubiquitin promoter and a 3′ poly A signal sequence was inserted (for selection of positive ESC clones) near the inverted exon 3. Two FRT3 and two FRT sites were inserted for ultimate removal of the mCherry and the neo cassette, and for reverting exon 3 by Flp/FRT recombination. An inverted eGFP gene with a 3′ poly A signal, a 5′ splice acceptor and T2A was inserted as a reporter gene of the knockout (KO) allele, and two Lox2372 and two LoxP sequences were inserted to invert the eGFP cassette by the Cre/lox recombination machinery. A 13-kb DNA fragment containing the Dis3 mCherry/neo/eGFP modification with 5′ and 3′ homology arms was subcloned into a diphtheria toxin A vector. The linearized targeting vector was electroporated into ESCs (129S6/SvEv × C57BL/6 hybrid), and positive ESC clones were screened using PCR with primer sets for both 5′ and 3′ homology arm loci. Only positive clones produced the expected 3.5-kb DNA fragments. The following oligonucleotides were used for the 5′ homology arm locus: Dis3_HA_5′_for and Dis3_HA_5′_rev; and for the 3′ arm: Dis3_HA_3′_for and Dis3_HA_3′_rev (Supplementary Table 1). After PCR selection, genomic DNA fragments of positive ESC clones were validated using external and internal Southern blot probes. Genomic DNA was digested by XbaI. Two positive ESC clones (G3 and F12) were injected into blastocysts, and chimeric mice were crossed with ACTB:FLPe B6J transgenic mice (B6.Cg-Tg(ACTFLPe)9205Dym/J) to delete the mCherry/neo cassette and revert the direction of exon 3. Dis3 COIN mice were backcrossed and genotyped using the primers Dis3_geno_for and Dis3_geno_rev (Supplementary Table 1), and then crossed with different Cre-expressing mouse strains to obtain the various Dis3 conditional KO mice for in vitro and in vivo studies. We used heterozygous mice as controls (that is, Dis3COIN/+; named here as Dis3C/+) and homozygous mice as conditional KO (that is, Dis3COIN/COIN; named here as Dis3C/C), from littermate pairs.
Mice.
AIDcre/+ (previously named Aicdacre) have been described previously7; VHB1–8high knock-in47 mice (named in this study as VHB1–8KI/KI) were kindly provided by M. Nussenzweig (Rockefeller University). Rosa26creERT2 (that is, tamoxifen-dependent cre) mice, which we refer to as Rosacre/+, were supplied by P. Chambon (University of Strasbourg); AID−/− mice were obtained from T. Honjo (Kyoto University). Mice aged 6 to 12 weeks were used for experiments. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Columbia University. The housing conditions were 22 °C, 50% of relative air humidity and 12-h light cycles (from 7:00 to 19:00). Dis3 COIN allele mice are available upon request following signing of a Uniform Biological Material Transfer Agreement.
Immunization.
VHB1–8KI/KIAIDcre/+Dis3C/+ and VHB1–8KI/KIAIDcre/+Dis3C/C littermate mice were immunized with 100 μg of NP-chicken γ-globulin, ratio 30–39 (Biosearch Technologies, N-5055D-5), resuspended in 100 μl of PBS and mixed with the same volume of alum adjuvant (Thermo Scientific, 77161) by intraperitoneal injection. Peyer’s patches were collected 7 d later for GC B-cell sorting.
B-cell isolation and in vitro stimulation.
CD43− cells were isolated from spleens after red blood cell lysis using CD43 magnetic beads and LS columns (Miltenyi Biotec). In vitro B-cell stimulations were performed as previously described7 using Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C CD43− splenocytes. Cre-mediated activation was achieved by tamoxifen treatment (4-OHT: (Z)-4-hydroxytamoxifen; Sigma-Aldrich); FBS (Atlanta Biologicals), LPS from Escherichia coli O111:B4 (Sigma) and IL-4 from Peprotech. Cells were routinely analyzed by flow cytometry to monitor GFP expression, cell activation and viability.
Flow cytometry and cell sorting.
Cells were washed in PBS, suspended in FACS buffer (PBS with 2% FBS) containing Fc block (clone 2.4G2; BD Biosciences; 1:100 dilution) for 5 min at room temperature, and then incubated for 30 min at 4 °C with antibodies (anti-B220, clone RA3–6B2; anti-GL7, clone GL7; anti-IgG1, clone A85–1; BD Biosciences; 1:100 dilution), washed in cold PBS and resuspended in FACS buffer for analysis or complete medium for cell sorting. Lymphocytes were analyzed as singlet, live cells (generally using either the dead cell exclusion dye 7AAD or DAPI; BD Biosciences). Cells were analyzed on a BD LSR Fortessa or BD Accuri C6 Plus. Cell sorting was performed using a BD FACS Aria II or Bio-Rad S3e cell sorter. Flow cytometry data were analyzed using FlowJo.
Proliferation.
Cells were initially incubated for 24 h with LPS and tamoxifen to eliminate Dis3 expression. Cells were subsequently stained with the cell proliferation dye eFluor 670 (eBioscience) and treated with LPS and IL-4 for a further 72 h for analysis.
Apoptosis.
Cells were stained with Annexin V BV421 (BD Biosciences) following the manufacturer’s instruction. For viability analyses, the ‘lymphocyte gate’ included all cells (live and dead).
Transcription inhibitors.
Cells were treated at day 2 with DRB (5,6-dichlorobenzimidazole 1-β-D-ribofuranoside; Sigma) at 100 μM for 4 h and triptolide (InvivoGen) at 1 μM for 1 h, or with dimethylsulfoxide as control. Cells were collected for RNA extraction or fixed for ChIP experiments.
RNA sequencing.
For direct in vivo RNA-seq, GC B cells from Peyer’s patches were sorted as B220+GL7+GFP+ and resuspended in TRIzol (Invitrogen TRIzol Reagent). In vitro, B cells were stimulated for 48 h with LPS + IL-4 before RNA extraction. RNA quality was evaluated by the Bioanalyzer system (Agilent), and RNA samples were sequenced using the RNA Ribozero 90M 100PE sequencing kit (Illumina) at the Columbia University Genome Center.
Quantitative PCR with reverse transcription and quantitative PCR.
RNA was extracted with TRIzol (Invitrogen TRIzol Reagent) solution following the manufacturer’s instructions at the time points indicated. Complementary DNA was synthesized using Superscript IV (Invitrogen) and random hexamers. qPCR was performed on 20 ng of complementary DNA using Maxima SYBR green (Thermo Scientific) and run in triplicate on a LightCycler 480 II apparatus (Roche). Oligonucleotides were purchased from IDT (Supplementary Table 1).
Linear amplification-mediated high-throughput genome-wide translocation sequencing.
Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C B cells were stimulated for 4 d before DNA extraction. The LAM-HTGTS method was adapted from the previously published protocol39. Briefly, genomic DNA was sonicated with three pulses of 5 s and 25% amplitude using the Sonic Dismembrator model 500 (Fisher Scientific). Linear amplification-mediated PCR was performed using Phusion High-Fidelity DNA Polymerase (NEB) and Sμ 5′ biotinylated bait. Eight independent PCRs were run on ~5 μg of sonicated DNA each, with 90 cycles of primer elongation. These biotinylated DNA fragments were purified using Dynabeads C1 streptavidin beads (Invitrogen) on a rotisserie overnight at room temperature. On-bead ligation was performed with T4 DNA ligase (NEB) and annealed bridge adaptors for 1 h at 25 °C, 2 h at 22 °C and overnight at 16 °C. Around 15 cycles of nested PCR were performed on the DNA–bead complexes using an Sμ nested primer and a reverse universal primer I7. These pairs of primers also contain barcodes and adaptors for the next steps in the procedure. PCR products were cleaned with the QIAquick Gel Extraction Kit (QIAGEN) and incubated with NspI (NEB) for 1 h at 37 °C to digest and remove germline DNA. The final tagged PCR was performed using P5-I5 and P7-I7 primers for 15 cycles and purified using Select-a-Size DNA Clean & Concentrator Kit (Zymo Research) with a cutoff of 200 bp. Library quality was evaluated by Bioanalyzer (Agilent) and sequencing performed using the Illumina Miseq Reagent Kit v2 (500 cycles) following the manufacturer’s instructions.
Linear amplification-mediated high-throughput genome-wide translocation sequencing at Myc locus.
We targeted Myc intron 1 with the Myc_CRISPR_bait, cloned into pMSCV-Cas9–2A-GFP-sgRNA (Addgene plasmid no. 124889) using BbsI and the golden gate strategy. Plat-E packaging cells were transfected using polyethylenimine and washed after 24 h. Retrovirus-containing supernatant was collected after 48 h. Primary splenocytes were pre-activated for 1 d with LPS and tamoxifen to delete the Dis3 gene, and then infected by spinofection for 90 min at 30 °C with the retroviral supernatant supplemented with polybrene (8 μg ml−1; Sigma). Cells were resuspended in LPS + IL-4 and grown for three additional days. The DNA was extracted to perform LAM-HTGTS, using the Myc bait and the Myc nested primer (Supplementary Table 1). SphI was used as blocking enzyme to remove germline DNA.
DRIP sequencing.
We followed a published DRIP-seq protocol66. Briefly, we stimulated Rosacre/+Dis3C/+ and Rosacre/+Dis3C/C cells with IL-4 for 2 d, performed DNA extraction and used a cocktail of restriction enzymes (BamHI, EcoRI, HindIII, NcoI, SpeI and XbaI) overnight at 37 °C in buffer 2.1 (NEB). RNase H (NEB) was added in negative controls. Then we performed S9.6 immunoprecipitation (IP; using 10 μg of S9.6 antibody per IP) and elution. The immunoprecipitated DNA fragments were validated by qPCR at the Sμ region. DNA fragments were sonicated using Bioruptor Plus sonication system (Diagenode; three pulses at low intensity for 30 s) before library preparation (see below). The same volume of each library was used for sequencing. For the spike-in controls, we used PCR products from bacterial DNA amplified with the following primers: Segmented Filamentous Bacteria_for, Segmented Filamentous Bacteria_rev, Helicobacter Pylori_for, H. Pylori_rev, Universal Bacterial Primers_for, Universal Bacterial Primers_rev, Lactobacillus_for and Lactobacillus_rev (Supplementary Table 1) as previously described8. We mixed the same ratios of PCR products in the different samples and quantified the spike-in read numbers after sequencing (Extended Data Fig. 3e). We validated our DRIP-seq on the four different samples (Dis3C/+ versus Dis3C/C, and Dis3C/+ versus Dis3C/C treated with RNase H) and then performed deeper sequencing of Dis3C/+ and Dis3C/C samples. Finally, we combined the data from three independent sequencing reads to obtain high-resolution maps for Dis3C/+ and Dis3C/C cells.
Chromosome conformation capture experiments.
We adapted our 3C experiments from the 3C-HTGTS method published by the Alt Laboratory67. Briefly, cells were crosslinked using 1% formaldehyde, neutralized with glycine (0.2 M final) and washed twice with cold PBS. Cells were lysed on ice with lysis buffer (10 mM Tris-HCl (pH 8), 10 mM NaCl and 0.2% NP40) in the presence of protease inhibitor (Sigma). Nuclei were resuspended in 0.5% SDS for 10 min at 62 °C and neutralized with Triton X-100 for 20 min at 37 °C. DNA restriction was performed using CviQ1 (Thermo Fisher) in buffer B overnight at 37 °C, before heat inactivation for 20 min at 62 °C. Ligation was performed for 4 h at room temperature. Reverse crosslinking was performed in resuspension buffer (10 mM Tris-HCl (pH 8), 0.5 M NaCl and 1% SDS) overnight at 68 °C. Next, DNA was treated with RNase A and proteinase K and cleaned by phenol/chloroform. After this 3C step, we followed the LAM-HTGTS protocol (see above) to specifically enrich the 3C interactions at the Igh TAD. Briefly, ligated DNA was sonicated using the Bioruptor Plus sonication system (Diagenode; two pulses at low intensity for 30 s), and 16 μg was used for the LAM-HTGTS step. A 3′ super-anchor/3′RR biotin bait was used for primer elongation. These single-stranded DNA fragments were incubated with streptavidin beads (Dynabeads C1 streptavidin beads; Invitrogen) overnight at room temperature and washed in binding buffer. A universal I7 adaptor was ligated before performing nested PCR using 3′ super-anchor/3′RR nested primer and universal I7 reverse, and PCR products were cleaned using QIAquick Gel Extraction Kit (QIAGEN). qPCR was performed in triplicate using 3′ super-anchor/3′RR forward primer in combination with 3′ super-anchor/3′RR total reverse to normalize the data. The different control and interaction frequencies were quantified using the super-anchor/3′RR forward primer in combination with the following reverse primers: universal I7 for total interactions, intergenic control 1, intergenic control 2, Eμ-rev and JH4_rev (Supplementary Table 1).
Somatic hypermutation sequencing.
VHB1–8KI/KIAIDcre/+Dis3C/+ and VHB1–8KI/KI AIDcre/+Dis3C/C mice were immunized with NP-chicken γ-globulin (see above), and total GC B cells B220+ GL7+ were sorted for genomic DNA extraction. The VHB1–8 V(D)J gene was amplified by PCR (VHB1–8_for and VHB1–8_rev) using the Q5 high-fidelity DNA polymerase (NEB) and low cycle numbers (20 cycles). PCR products were cleaned using the QIAquick Gel Extraction Kit (QIAGEN) and used for library preparation. Tail DNA from the same animals was used as controls.
Cell fixation.
For ChIP–seq and Hi-C experiments, B cells were stimulated in vitro for 24 h with LPS and tamoxifen treatment, and then treated with LPS + IL-4 for 24 h. These cells were fixed using 1% formaldehyde for 10 min at room temperature. Formaldehyde was quenched with 2.5 M glycine (final concentration of 0.2 M), and cells were washed with cold PBS. Cell pellets were used immediately for the next step (lysis) or stored at −80 °C before lysis.
Chromatin immunoprecipitation.
Approximately 5 × 106 fixed cells were incubated in lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris-Cl (pH 8.0)) on ice for 20 min, and genomic DNA was sheared using a Bioruptor Plus sonication system (Diagenode). Sonication was performed at 4 °C for 30 cycles in total, using the high-intensity setting for 30 s followed by 30 s of rest. After centrifugation, the supernatant was diluted tenfold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl (pH 8.0) and 167 mM NaCl). CTCF (Abcam, ab70303) and RAD21 (Abcam, ab992) antibodies were added to chromatin, and samples were rotated at 4 °C for 16 h. Prewashed magnetic beads (Thermo Fisher, PI26162) were added to the lysate and incubated with rotation at room temperature for 1 h. Magnetic beads and chromatin fragment complexes were washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl (pH 8.0) and 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl (pH 8.0) and 500 mM NaCl), LiCl buffer (250 mM LiCl, 1% IGPAL-CA630, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris-Cl (pH 8.0)) and TE buffer. DNA fragments were eluted with 300 μl of elution buffer (1% SDS and 0.1 M NaHCO3) and reverse crosslinked at 65 °C for 16 h. After proteinase K treatment and phenol/chloroform extraction, DNA was purified by ethanol precipitation and dissolved in 50 μl of water. The same protocol was used for RNA pol II S5 ChIP (ab5131). In total, 5 μg of antibody was used per IP.
Creation of Abelson cell lines.
Abelson cell lines were generated as previously described68. Briefly, Plat-E cells were grown in DMEM (Life Technologies) with 10% FBS in the presence of blasticidin (10 μg μl−1) and puromycin (1 μg μl−1), and 80% confluent cells were transfected by polyethylenimine treatment with the Abelson virus plasmid. Supernatant was collected 2 d later. Bone marrow cells were collected from Rosacre/+Exosc3C/+Bcl2Tg and Rosacre/+Exosc3C/CBcl2Tg mice, and suspended at 1 million cells per ml in complete medium (DMEM with 15% FBS, NEAA, sodium pyruvate, penicillin–streptomycin and 50 μM β-mercaptoethanol) in six-well plates, and 1 ml of viral supernatant was added. Five days later, fresh medium was added, and cells were then split every 2 d. After immortalization, cells were validated by flow cytometry for GFP inversion and viability.
Ectopic CTCF-binding sites DNA construct.
We synthesized (Gene Universal) a ~1-kb region containing two CTCF-binding sites in convergent orientation, followed by a CMV promoter (to drive the transcription of the second switch sequence), and included restriction sites for subsequent cloning steps (AgeI/PacI and NotI/XhoI). We cloned this cassette into pcDNA5/TO vector (Thermo Fisher Scientific). The G-rich sequences were amplified by PCR with restriction sites containing oligonucleotides (Sμ_for_AgeI, Sμ_rev_PacI, Sμ_for_NotI and Sμ_rev_XhoI) from the PJ14 plasmid (Addgene no. 25912) and cloned into the first vector using the adapted restriction sites. This construct was used to transfect Abelson cell lines and to generate stable clones. Cells were selected using hygromycin (800 μg ml−1) for 10 d and then split in two as control (ethanol) or tamoxifen-treated cells over 2 d. Five million cells were used for the ChIP experiments. qPCRs were performed in triplicates using these oligonucleotides (eCBE_for and eCBE_rev), with 1:10 diluted inputs used to calculate the enrichment (2−ΔCt) for RAD21 binding.
High-throughput chromosome conformation capture.
The in situ Hi-C protocol from the 4DN Nucleosome Consortium was followed. Briefly, five million cells were crosslinked for each experiment and DpnII was used for DNA digestion followed ligation. To shear the DNA after ligation, ultra-sonication (Sonic Dismembrator model 500, Fisher Scientific) was used with ten timed on-pulses of 5 s each. Subsequently, to generate the library, each end of the isolated DNA fragments (isolation performed using Dynabeads MyOne Streptavidin T1 beads) was trimmed and ligated with the following adaptor oligonucleotide duplexes: Hi-C1_for and Hi-C1_rev, Hi-C2_for and Hi-C2_rev, Hi-C3_for and Hi-C3_rev, Hi-C4_for and Hi-C4_rev, and then amplified using the oligonucleotides HiC_for and HiC_rev (Supplementary Table 1). Alternatively, the NEBNext Ultra II Kit (NEB E7645) and Multiplex Oligos (NEB E7600S) were used for making the library. Four independent experiments from three biological replicates were combined for analyses. As controls, we used in vitro stimulated wild-type B cells and control KO B cells as previously published8.
Library preparation.
DNA concentration was evaluated using the Qubit 1X dsDNA HS Assay Kit (Life Technologies). DNA fragments from ChIP, Hi-C, DRIP and SHM-seq were prepared for final library preparation following the protocol for the NEBNext Ultra II DNA Library Prep Kit for Illumina, manufactured by NEB. Adaptor-ligated and final DNA products were cleaned up using AMPure XP beads (Beckman Coulter). Barcoding was performed using NEBNext Multiplex Oligos from Illumina (NEB).
Statistical analysis.
Statistical analyses were performed using Prism 6 software (GraphPad) and confirmed using Python. Statistical significance was expressed as a P value: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 and NS (P > 0.05).
Data visualization.
All analyzed data were visualized using the IGV51 and mapped on the reference genome mm9.
RNA sequencing analysis.
RTA (Illumina) was used for base calling and bcl2fastq2 (version 2.17) for converting BCL to fastq format, coupled with adaptor trimming. The sequenced reads were mapped to the reference genome (mm9) by using hisat2 (version 2.1.0)69 with default parameters. We used StringTie70 (version 1.3.3b) to assemble the alignments into potential transcripts. Finally, RNA-seq replicates were combined for visualization in IGV. The normalization method for IGV71 tracks is RPGC; that is, the read count was normalized as if 1× depth sequencing was performed for all samples. The peak annotation module in Homer72 was used to annotate RNA-seq transcripts. Exosome substrate ncRNAs were defined as noncoding transcripts with twofold higher fragments per kilobase of transcript per million mapped read (FPKM) values in Dis3C/C than in Dis3C/+. Since transcripts with extremely low expression levels may demonstrate large fold changes, transcripts with FPKM values < 0.1 in both samples were filtered out.
The overall expression changes of different RNA classes were characterized: enhancer RNAs (RNA expressed in enhancer regions), antisense RNAs (expressed in antisense of exons), lncRNAs (ncRNAs that are greater than 200 nucleotides in length), anti-TSS RNAs (expressed in the antisense orientation from transcription start sites) and mRNAs. Potential enhancer regions of activated B cells were obtained by analyzing public H3K4me1 and H3K27ac ChIP–seq data14 in the Gene Expression Omnibus dataset GSE62063. Those peaks possessing both histone markers were defined as potential enhancers.
For CTCF-overlapping RNAs, we defined a 1-kb window surrounding CTCF peaks from our ChIP–seq experiments and determined whether the RNAs are overlapping with these regions. The same strategy was used for CTCF-overlapping DNA:RNA hybrids.
ChIP sequencing analysis.
Sequenced read pairs were mapped to the reference genome (mm9) using Bowtie 2 (version 2.2.9)73 with the parameter ‘-X 2000--local’. Duplicate alignments were removed using the MarkDuplicates module in Picard (http://broadinstitute.github.io/picard/index.html). Alignments with low mapping quality (q < 10) or that were not properly paired were also removed. MACS2 (version 2.1.3)74 with the parameter ‘--f BAMPE --B’ was used for peak calling. The reads per million mapped values were derived based on the bedgraph files generated by MACS2. To analyze the peaks that differed between Dis3C/+ and Dis3C/C, featureCount75 (version 1.5.3) was used to calculate the fragment counts of each peak in Dis3C/+ and Dis3C/C. These fragment counts were then converted into FPKM values. The peak summit surrounding signal tracks was generated by extracting and plotting the normalized pile-up signal (number of reads mapped to the specific genome site) within 1-kb upstream and downstream of each peak summit from the bigwig files that were generated in the data processing step.
DRIP sequencing analysis.
Similar to ChIP–seq data processing, read pairs were mapped to the reference genome (mm9) using Bowtie 2 (version 2.2.9)73 with the parameter ‘--X 2000 --local’. Duplicate alignments were marked and discarded using the MarkDuplicates module in Picard. Properly paired alignments with high mapping quality (q > 10) were filtered for peak calling (MACS2 (ref. 74) with the parameter ‘--f BAMPE --B’). FeatureCount75 was used to calculate the counts of fragments that mapped to each peak in Dis3C/+ and Dis3C/C.
High-throughput chromosome conformation capture analysis.
Raw fastq files were demultiplexed by Barcode Splitter (version 0.18.0; Leach Robert and Parsons Lance; https://bitbucket.org/princeton_genomics/barcode_splitter/; 2017) with the parameter ‘--mismatches 2’. Read pairs with barcodes detected in at least one end were retained and subjected to adaptor trimming by Cutadapt76 with the parameters ‘--trim-n --q 10,10 --u 6 --U 6 --m 5 --e 0.1’. Juicer pipeline77 was used to align the chimeric reads to the reference genome (mm9), build the Hi-C map, which can be visualized by Juicebox78, and detect the notable loops and contact domains genome wide. Vanilla coverage79 normalized observation over expectation values were used to compare the contact intensity between genomic regions in different conditions and to quantify the intensity of chromatin contacts.
High-throughput chromosome conformation capture insulation scores.
The insulation scores were evaluated using a strategy similar to that previously published80. The bin size was set to 10 kb. We calculated the mean signal of a 250 kb × 250 kb (25 bins × 25 bins) square in the upper right of each diagonal bin and assigned the value to the bin. These values were normalized for each chromosome by calculating the log2 ratio of the value of each bin to the mean of all values of that chromosome.
High-throughput chromosome conformation capture data identify A/B compartments.
The compartments were calculated using the Eigenvector module in Juicer at 50-kb resolution. The directions of the resultant eigenvectors were assigned to compartment A and B according to the gene density.
Interaction score.
To evaluate the impact of DIS3 deletion on intrachromosomal interactions from our Hi-C data, we evaluated the ‘interaction score’, where the mean difference (log2 fold change of Dis3C/C versus Dis3C/+) was calculated for each diagonal of the Hi-C contact matrix of each chromosome. We used the matrix observed values normalized by the KR matrix balancing method54 and 1-Mb resolution. The mean differences were plotted in response to the distance from the main diagonal. As control, we calculated this score in wild-type and control KO cells (log2 fold change of control KO versus wild-type).
Aggregate peak analysis.
APA were performed on all loops detected in Dis3C/+ as previously described54, and we also followed a previously published50 strategy to perform APA on the top 25% differential loops. Other parameters used were: 5-kb resolution, KR normalization, 100-kb window and 200-kb minimum centroid distance. The APA score is defined as the ratio of the central pixel to the mean of the pixels in the lower-left corner, that is, P2LL value.
Comparison of 3′RR–Eμ interaction frequency from high-throughput chromosome conformation capture data.
The Hi-C data were binned into 50-kb bins to contact frequency matrix and normalized using the Knight–Ruiz normalization method. Four bins of 10 kb from the 3′ super-anchor/3′RR (chr12: 114,450,000–114,490,000) and two bins of 10 kb from Eμ/Sμ (chr12: 114,650,000–114,670,000) were extracted and normalized by the total detected contacts, evaluating eight different interactions. The interaction frequency between 3′RR and Eμ in Dis3C/+ and Dis3C/C were compared using a paired t-test.
Somatic hypermutation analysis.
Alignment of the reads to reference sequences was performed using the Smith–Waterman algorithm built into the MATLAB (R2017b) Bioinformatics toolbox with parameters of match bonus = 1, mismatch penalty = 3, gap opening penalty = 6 and gap extension penalty = 3. Alignments with a score of >160 were kept for further analysis. We only analyzed data from the read 1 of paired-end sequencing, which are known to have the best quality81. To reduce sequencing noise, the nucleotide from the reference sequence was masked if the corresponding per-base mutation frequency was >0.1. Finally, an analysis was undertaken of the C to T and G to A mutations in AID hotspot regions (WRCY/RGYW motif, where W = adenine or thymine, R = purine, C = cytosine, Y = pyrimidine and G = guanine), calculating the mutation frequency for each sequence. For statistical analyses, the proportion of C to T mutations compared to the total sequenced number of Cs in the control mouse (VHB1–8KI/KIAIDcre/+Dis3C/+) was compared to the Dis3C/C mouse (VHB1–8KI/KI AIDcre/+ Dis3C/C) and a χ2 two-tailed proportions test was applied between these two proportions. The same method was used for the G to A mutation frequencies.
Linear amplification-mediated high-throughput genome-wide translocation sequencing analysis.
We used FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to examine the sequencing quality and Cutadapt76 to remove adaptor sequences at the 3′ end of the raw reads. The Transloc pipeline39 was used with default parameters to obtain all possible translocation events. For Sμ LAM-HTGTS, these events were categorized as either CSR (that is, intra-Igh TAD recombination, inside chr12: 114,466,607–114,668,078) or non-CSR (that is, inter-TADs, outside chr12: 114,466,607–114,668,078). For Myc LAM-HTGTS, we separated translocation events into intra-Myc TAD recombination (inside chr15: 60,920,000–63,620,000) and inter-TAD recombination (outside chr15: 60,920,000–63,620,000). The microhomology events were determined by whether the 3′ end of bait and 5′ end of prey sequences shared the identical nucleotides. Circos82 (version 0.69–6) plots were used to visualize the translocation events.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data are available via the NCBI under accession number PRJNA544488. Source data are provided with this paper.
Code availability
Code is available at https://github.com/basulab-cu/Dis3-project.
Extended Data
Extended Data Fig. 1 |. Generation of Dis3COIN mouse model and efficient Dis3 deletion in activated B cells.
a, Dis3 targeted allele includes mCherry and Neo resistance cassettes, inverted Dis3 exon 3, GFP gene, and loxP/FRT sites (top). After FLP-mediated removal of mCherry/Neo cassettes and exon 3 reversion, Dis3COIN allele is functional (middle). Dis3 KO allele after cre-mediated inversion (bottom). b, ESC screening by Southern blot with 3′ or mCherry probes. Clones G3 and F12 were selected for microinjection. Screening was performed one time, each clone tested with different probes. c, Mouse genotyping. Dis3 wt allele generates a band of 369 bp, Dis3COIN allele 489 bp. This gel is representative of our genotyping, performed at least 10 times. d, Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C B cells were stimulated in the presence (+) or in the absence (−) of tamoxifen, and CSR to IgG1 was quantified (3 independent experiments, mean is shown +/− s.e.m., two-tailed paired t-test). e, qPCR quantification of Dis3 allelic inversion on genomic DNA, normalized to the GFP gene. This qPCR is specific of the Dis3 COIN allele before inversion, and shows loss of the Dis3 functional allele after inversion. f, g, and h, RT-qPCR quantification of Dis3, Aicda (AID) mRNAs, and γ1 germline transcript expression on total RNA, normalized to Gapdh. Kinetic in panels e to h was performed one time in triplicate, mean is shown +/− s.e.m., two-tailed unpaired t-test). i, Resting B splenocytes were collected and stimulated with LPS and tamoxifen to inactivate Dis3COIN alleles, IL4 was added at day 1. ChIP, Hi-C, and 3C experiments were performed at early time points. RNA-seq were performed 2 days after IL4 treatment, while cells were harvested at late time-point to study the accumulated DNA junctions from CSR or translocations by LAM-HTGTS.
Extended Data Fig. 2 |. Accumulation of DIS3-sensitive ncRNAs in B cells isolated from the germinal center in vivo and stimulated in vitro.
a, mRNA expression in vitro and in vivo. RNA-sequencing from Rosacre/+ Dis3C/+ versus Rosacre/+ Dis3C/C in vitro activated B cells (n = 2), and in vivo AIDcre/+ Dis3C/+ versus AIDcre/+ Dis3C/C (n = 2) were analyzed. b, Differential gene expression in DIS3-deficient B cells. RNA-sequencing from Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells (n = 2) were analyzed using a hierarchical unsupervised clustering method and results are shown as heat maps. Up-regulated genes are shown in green and down-regulated genes are shown in blue. A total of 160 genes showed differential RNA expression at this early time-point. c to e, Examples of DIS3-sensitive aTSS and antisense RNAs at various genes, including Myc, Cd19, and Cd79a, genes frequently translocated in B cells. f, eRNAs expressed from the Igh 3′RR super-enhancer. g, Example of DIS3-sensitive enhancer RNAs (eRNAs) at super-enhancer, with the active enhancer mark H3K27ac. h, Example of DIS3-sensitive eRNAs at intronic enhancer, from Pvt1 locus, with the active enhancer mark H3K27ac. i, Example of DIS3-sensitive CBE-overlapping RNAs at Fh1 locus. j, Example of DIS3-sensitive CBE-overlapping RNAs downstream the Igk super-enchancer.
Extended Data Fig. 3 |. Accumulation of DNA-associated RNAs in the absence of DIS3.
a DNA:RNA immunoprecipitation (DRIP). DRIP was performed on Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells. DNA was extracted, digested by restriction enzymes, and treated with RNase H for the negative controls before immunoprecipitation using S9.6 antibody. qPCR quantification of DNA:RNA hybrids at Sμ region. DRIP products were quantified directly by qPCR, relative to the input. One experiment performed in duplicate, each dot represents one qPCR value. b to j, DRIP-sequencing: these DRIP products were prepared for deep sequencing. 3 deep-sequencings were performed with similar results. Coding and non-coding transcription determined by RNA-sequencing is shown (‘RNA’ tracks). b, DNA:RNA hybrids accumulation at Sμ region, relative to a. c. Example of DNA:RNA hybrids accumulation at the Bcl6 gene, correlating with the accumulation of intronic ncRNA in DIS3-deficient cells. d, Example of DNA:RNA hybrids accumulation at the Jak2 gene. e, Peak calling was applied to quantify the different peaks corresponding to DNA:RNA hybrids in DRIP sequencing experiments. Quantification is shown in Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells (bar graphs show the number of peaks, χ2 two-tailed proportions test). f, Read numbers from the different spike-in controls used in DRIP experiments are shown. g, Distribution of DNA:RNA hybrids per TAD in Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells, means are indicated by dashed lines. h to k, Multiple examples of accumulation of DNA:RNA hybrids overlapping CBEs and decreasing CTCF binding in the absence of DIS3.
Extended Data Fig. 4 |. Genome-wide decrease of CtCF and RAD21 binding in the absence of DIS3.
a and b. CTCF and RAD21 ChIP–seq from Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells (biological replicates number two). Common peaks (n = 19,042 and n = 6,873 respectively) between two replicates were used in the heat map, two-tailed Wilcoxon rank-sum test. c. IgG negative control. d and e. CTCF binding and RAD21 localization to CBEs overlapped by RNA are more affected than other CBEs of the genome in the absence of DIS3. f and g. CTCF binding and RAD21 localization to TAD anchor CBEs are more affected than other part of the genome in the absence of DIS3. Panels d to g: box and whiskers plots represent ChIP values (fold changes), bottom and top whiskers represent LQ – 1.5*(UQ−LQ) and UQ + 1.5*(UQ−LQ) where LQ and UQ are lower and upper quartiles, outside interquartile range. Box plots show lower quartile, median and upper quartile, two-tailed Wilcoxon rank-sum test. h. Percentages of CTCF and RAD21 ChIP reads found inside peaks. Total number of reads was similar between Dis3C/+ and Dis3C/C (top), while reads found in peaks were decreased for CTCF and RAD21. Bar graphs show the percentage of reads inside the peaks, χ2 two-tailed proportions test. i. Analysis of ncRNAs and CBE peaks overlap. CTCF occupancy in the B cell genome was determined from control B cells (Dis3C/+) and overlapped with all ncRNAs found in Dis3C/C B cells (1 kb window). The Venn diagram shows 73 CBEs located at the Igκ and 122 CBEs at the Igh loci have overlap ncRNAs. A simulated random distribution demonstrates that the numbers are superior to what would be observed from random genomic overlaps. j. Decreased CTCF binding at Igh V CBEs. IGV tracks show some example of decreased CTCF-binding to VH CBEs where RNAs are accumulated in the absence of DIS3.
Extended Data Fig. 5 |. Co-transcriptional DNA:RNA hybrids accumulation in DIS3-deficient cells impairs optimal RAD21 localization.
a. Model suggesting impaired cohesin scanning in the presence of DNA:RNA hybrids in Dis3C/C cells. While DIS3-proficient cells resolve DNA:RNA hybrids using DIS3 activity, DIS3-deficient cells accumulate these structures (and associated proteins) which impair cohesin scanning (and/or decreases the processing speed of the cohesin complex) and ultimately perturb loop extrusion. Transcription inhibitors decrease the amount of co-transcriptional DNA:RNA hybrids and potentially restore cohesin positioning, even in the absence of DIS3. RAD21 ChIP–sequencing in activated B cells using transcription inhibitors. Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C cells were treated with IL4 for 24 h and with transcription (TC) inhibitors or DMSO as control for 4 h before fixation (one experiment). b. Number and percentage of RAD21 peaks decreased in the absence of DIS3 and rescued after transcription inhibitors treatments. All peaks with a fold increase >1 after TC inhibitors treatment were considered. c. Genome-wide analysis of RAD21 binding with or without transcription inhibitors in Dis3C/C. Here we focused on RAD21 peaks which were decreased by 2-fold compare to the control Dis3C/+ without treatment. In these conditions 1,012 peaks show efficient rescue. The box and whiskers plots represent these values, the bottom and top whiskers represent the minimum and maximum scores, respectively, outside the interquartile range. The box plots show the lower quartile, median and upper quartile. This experiment was performed one time, two-tailed Wilcoxon rank-sum test. d to i. Multiple examples of RAD21 restoration after transcription inhibitor treatment. IGV tracks show the defect of RAD21 positioning in the absence of DIS3 (Dis3C/+ versus Dis3C/C, grey tracks) while transcription inhibitors restore RAD21 localization even in the absence of DIS3 (blue tracks). Black arrows indicate the RAD21 peaks which are strongly affected by DIS3-deficiency and rescued after transcription inhibitors treatments.
Extended Data Fig. 6 |. G-rich sequences directly inhibit CtCF/RAD21 binding in the absence of RNA exosome activity.
a. CBEs were cloned into an expression vector containing G-rich regions corresponding to Sμ. Expected phenotypes in the presence (control) or in the absence of RNA exosome (KO) activity are shown. b. We generated Abelson cell lines. After tamoxifen treatment C/C cells inverse Exosc3COIN alleles, inactivating RNA exosome activity and expressing GFP. c. Abelson cell lines were treated with tamoxifen during 4 days, GFP expression was quantified by flow cytometry. Grey histograms show untreated cells and green histograms show tamoxifen-treated cells. d and e. Kinetic of GFP expression and viability in Abelson cell lines, performed 2 times in triplicates with similar results (mean is shown +/− s.e.m.). f. RNA-seq and DRIP-seq tracks showing ncRNAs and DNA:RNA hybrids accumulation at Sμ region in activated B cells (see Fig. 4e and Extended Data Fig. 3b). The G-rich sequence (dotted line region) was cloned into our construct, flanking CBEs on both sides. g. Abelson cell lines (C/C) were transfected and selected for the presence of the construct containing ectopic CBEs. Cells were split in two, as control and tamoxifen-treated, and CTCF and RAD21 ChIP were performed. h. Reproducible defect in RAD21 recruitment at ectopic CBEs. We isolated independent Abelson cell lines (C/C) clones to perform ChIP in the presence (ctrl) or in the absence (KO) of RNA exosome and evaluated RAD21 DNA binding directly by qPCR in these biological replicates (2 independent experiments performed on 3 clones, qPCR were done in triplicates, two-tailed unpaired t-test). The box and whiskers plots represent these values, the bottom and top whiskers represent the minimum and maximum scores, respectively, outside the interquartile range. The box plots show the lower quartile, median and upper quartile. i. Primary B cells were stimulated to perform RNA polymerase II S5 phosphorylated ChIP.
Extended Data Fig. 7 |. Genome-wide alteration in chromosome organization in DIS3-deficient B cells.
Combined data from 4 independent were used for these analyses. Hi-C data visualization using ‘Juicebox’ and DNA interaction changes on all chromosomes (1 to 19 and chromosome X). Interaction frequencies are calculated by ‘control value divided by observed value (observed/control)’, the relative gained interactions are shown in red and the relative lost interactions are shown in blue. Each square represents a full chromosome, beginning on the top left corner and finishing on the bottom right corner. a. Control KO/wt interactions are shown. An irrelevant KO (located on chromosome 12) was used to evaluate the global impact on genomic organization. Random patterns were obtained implying random gain and loss of interaction equivalent to no change. b. Dis3C/C/Dis3C/+ interactions are shown, with an obvious specific pattern of altered genomic interactions.
Extended Data Fig. 8 |. Quantification of chromosome alterations in DIS3-deficient B cells.
a and b. ‘Interaction score’: the average log2 fold-change of interactions along each diagonal of Hi-C maps is shown for each chromosome to evaluate the distance-dependent changes in interactions. Each colored curve represents one chromosome, and the black curve shows the average curve. These data are related to Extended Data Fig. 7. a. No differences were detected when dividing control KO by wt interactions. b. DIS3-deficiency induces a clear shift in this score, reflecting the altered patterns shown in Extended Data Fig. 7. c. Insulation scores at TAD boundaries are shown for control KO and wt activated B cells. S: slope of the fitted linear model. 1,224 insulation scores are shown. d. Insulation scores at TAD boundaries are shown for Dis3C/C and Dis3C/+ activated B cells. S: slope of the fitted linear model. 1,224 insulation scores are shown. The Igh 5′ and 3′ ends are shown as red dots. e. APA analysis showing the genome-wide loop accumulation, in control Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C cells. A total of 800 loops were analyzed. f. APA analysis showing the loop accumulation in the top 25% differential loops in control Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C cells. Loop interactions are defective genome-wide in the absence of DIS3, and much pronounced for the top 200 differential loops, as previously described.
Extended Data Fig. 9 |. Decrease physiological DNA double strand breaks at Igh switch regions, and increase chromosomal translocations at the Myc tAD in the absence of DIS3.
a. LAM-HTGTS from AID−/−, Rosacre/+ Dis3C/+, and Rosacre/+ Dis3C/C activated B cells at Sμ, Sγ3, Sγ1, Sγ2b, and Sε regions with corresponding RNA-seq (combined data from 3 independent experiments). b to f. CRISPR/Cas9-induced DNA breaks and processing at the Myc locus Primary B cells (Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C) were infected during in vitro stimulation with CRISPR/Cas9 retrovirus to induce DNA breaks at the Myc locus. DNA junctions were analyzed by LAM-HTGTS using Myc bait (2 independent experiments with 2 or 3 biological replicates and 3 independent libraries used for sequencing). b. DNA breaks induced by CRISPR/Cas9 at Myc intron 1 and captured using Myc bait. Black arrow indicates the position of CRISPR/Cas9-induced DNA breaks. Myc TAD is outlined by dashed lines. c. Circos plots showing global DNA translocations. d. 2 fold decrease in intra-TAD recombination in the absence of DIS3. The proportion of intra-Myc TAD recombination and inter-TADs translocations is shown (total DNA junctions of 8,122 in Dis3C/+ and 5,529 in Dis3C/C), χ2 two-tailed proportions test. e. NcRNAs and DNA:RNA hybrids accumulation at the Myc locus decrease CTCF/RAD21 binding. RNA-seq data show aTSS and intronic ncRNAs accumulation in Dis3C/C cells, DRIP displays increased DNA:RNA hybrids, while CTCF and RAD21 ChIP-seq reveal decreased CTCF/RAD21 binding. Black arrow indicates CRISPR/Cas9-induced DNA breaks. f. Global analysis of the DNA junctions from Myc LAM-HTGTS, showing length and frequencies of microhomology overlapping bait and prey sequences. All DNA junctions from 3 experiments were combined and the distributions of insertions were evaluated by χ2 two-tailed proportions test (comparing DNA junctions with microhomology length >4 to the total junctions between Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C cells).
Extended Data Fig. 10 |. In vivo somatic hypermutation exploration in Dis3C/C B cells.
a. B splenocytes from one pair of AIDcre/+ Dis3C/+ and AIDcre/+ Dis3C/C mice were stimulated. GFP expression was examined after 4 days. In ex vivo culture conditions AIDcre/+ Dis3C/C cells do not encounter selection and maintain numbers of GFP+ cells. b. RT-qPCR quantification of Dis3 mRNA expression in AIDcre/+ Dis3C/C Peyer’s patches B cells. B220+ GL7+ and B220+ GL7− cells were sorted to show the specific deletion of the Dis3COIN allele in activated B cells (one experiment performed in triplicate, mean is shown +/− s.e.m., two-tailed unpaired t-test). c. RNA-seq showing increased ncRNAs overlapping JH genes in the absence of DIS3 activity during B cell stimulation. Top. RNA-seq tracks encompassing the Eμ intronic enhancer and JH genes. Bottom. RNA-seq tracks showing increased sense transcription (red) and DNA:RNA hybrid accumulation (black) downstream of the JH4 gene in the absence of DIS3 activity (2 RNA-sequencing and 3 DRIP-sequencing) d. Top: Schematic of wt recombined VDJ genes in the context of CBEs. Bottom: VHB1–8KI alleles have different configuration, conserving the major CTCF-binding region IGCR1 (IGCR1 is deleted from the functional allele in physiological conditions). e. DNA mutation analyses of GC (B220+ GL7+) B cells from VHB1–8KI/KI AIDcre/+ Dis3C/+ (n = 3) mice. Tail DNA from the same animals was used as control (n = 2). AID hot-spots are indicated, and C or G mutations refer to the sense DNA strand. f. Quantification of C and G mutation frequencies in GC B cells. 3 pairs of VHB1–8KI/KI AIDcre/+ Dis3C/+ and VHB1–8KI/KI AIDcre/+ Dis3C/C mice were used, analyses were performed at the JH gene from the VHB1–8 allele. χ2 two-tailed proportions tests were used to compare the number of C to T mutations relatively to the total C sequenced, and the number of G to A mutations compared to the total G sequenced.
Supplementary Material
Acknowledgements
We thank M. Nussenzweig (Rockefeller University) for providing VHB1–8 mice, T. Honjo (Kyoto University) for AID−/− mice, F. Alt (Harvard University) for the LAM-HTGTS protocol, A. Schooley and J. Dekker for help in analyses of insulation scores, B. Sleckman (University of Alabama) for sharing the Abelson cell line system and R. Pavri (IMP, Vienna) for discussions regarding this study. Research in the Basu laboratory is supported by grants to B.L. (EMBO fellowship, ALTF 906–2015) and U.B. (NIAID 1R01AI099195, RO1AI134988 and RO1AI143897), Leukemia & Lymphoma Society, and the Pershing Square Sohn Cancer Research Alliance. This study utilized facilities at Columbia University Irving Medical Center flow cytometry core facility and genome center (P30CA013696).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41588-020-00772-0.
Supplementary information is available for this paper at https://doi.org/10.1038/s41588-020-00772-0.
Peer review information Nature Genetics thanks Yves Denizot, Ralph Stadhouders and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Laffleur B & Basu U Biology of RNA surveillance in development and disease. Trends Cell Biol. 29, 428–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair L, Chung H & Basu U Regulation of long noncoding RNAs and genome dynamics by the RNA surveillance machinery. Nat. Rev. Mol. Cell Biol 21, 123–136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilchert C, Wittmann S & Vasiljeva L The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Pefanis E et al. RNA exosome-regulated long noncoding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preker P et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 39, 7179–7193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almada AE, Wu X, Kriz AJ, Burge CB & Sharp PA Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pefanis E et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothschild G et al. Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class-switch recombination. Sci. Immunol 5, eaay5864 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh I et al. MiCEE is a ncRNA–protein complex that mediates epigenetic silencing and nucleolar organization. Nat. Genet 50, 990–1001 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Alt FW, Zhang Y, Meng F-L, Guo C & Schwer B Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152, 417–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Methot SP & Di Noia JM Molecular mechanisms of somatic hypermutation and class-switch recombination. Adv. Immunol 133, 37–87 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Zhang X et al. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature 575, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vian L et al. The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfelder S & Fraser P Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet 20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Nora EP et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao SSP et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ba Z et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586, 305–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen HM, Peters A, Baron B, Zhu X & Storb U Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280, 1750–1752 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Mlynarczyk C, Fontán L & Melnick A Germinal center-derived lymphomas: the darkest side of humoral immunity. Immunol. Rev 288, 214–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiarle R et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein IA et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng F-L et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159, 1538–1548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birshtein BK Epigenetic regulation of individual modules of the immunoglobulin heavy chain locus 3′ regulatory region. Front. Immunol 5, 163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojima T et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress and cellular senescence. Mol. Cell 72, 970–984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahba L, Amon JD, Koshland D & Vuica-Ross M RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingo-Prim J et al. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun 10, 2135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merkenschlager M & Nora EP CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet 17, 17–43 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Dekker J & Mirny L The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saintamand A et al. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nat. Commun 6, 7084 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Fitz J et al. Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction. Nat. Genet 52, 505–515 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Wuerffel R et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27, 711–722 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denizot Y et al. Polymorphism of the human α1 immunoglobulin gene 3′ enhancer hs1,2 and its relation to gene expression. Immunology 103, 35–40 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Noir S et al. The IgH locus 3′ cis-regulatory super-enhancer co-opts AID for allelic transvection. Oncotarget 8, 12929–12940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavri R et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143, 122–133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steensel Bvan & Furlong, E. E. M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol 20, 327–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc 11, 853–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews AJ, Zheng S, DiMenna LJ & Chaudhuri J Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination and long-range DNA repair. Adv. Immunol 122, 1–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gearhart PJ, Johnson ND, Douglas R & Hood L IgG antibodies to phosphorylcholine exhibit more diversity than their IgM counterparts. Nature 291, 29–34 (1981). [DOI] [PubMed] [Google Scholar]
- 42.Vincent-Fabert C et al. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b and hs4) dramatically affects class-switch recombination and Ig secretion to all isotypes. Blood 116, 1895–1898 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Volpi SA et al. Germline deletion of IgH 3′ regulatory region elements hs 5, 6, 7 (hs5–7) affects B cell-specific regulation, rearrangement and insulation of the Igh locus. J. Immunol 188, 2556–2566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panchakshari RA et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc. Natl Acad. Sci. USA 115, 762–767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesin L, Ersching J & Victora GD Germinal center B-cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer CT et al. The microanatomic segregation of selection by apoptosis in the germinal center. Science 358, eaao2602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih T-AY, Meffre E, Roederer M & Nussenzweig MC Role of BCR affinity in T-cell dependent antibody responses in vivo. Nat. Immunol 3, 570–575 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Rouaud P et al. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J. Exp. Med 210, 1501–1507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morvan CL, Pinaud E, Decourt C, Cuvillier A & Cogné M The immunoglobulin heavy-chain locus hs3b and hs4 3′ enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood 102, 1421–1427 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Hansen AS et al. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell 76, 395–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saldaña-Meyer R et al. RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell 76, 412–422.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garland W et al. A functional link between nuclear RNA decay and transcriptional control mediated by the polycomb repressive complex 2. Cell Rep. 29, 1800–1811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen AS, Amitai A, Cattoglio C, Tjian R & Darzacq X Guided nuclear exploration increases CTCF target search efficiency. Nat. Chem. Biol 16, 257–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao SSP et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davidson IF et al. Rapid movement and transcriptional re‐localization of human cohesin on DNA. EMBO J. 35, 2671–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niehrs C & Luke B Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol 21, 167–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley MJ & Corces VG Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stik G et al. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat. Genet 52, 655–661 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Senigl F et al. Topologically associated domains delineate susceptibility to somatic hypermutation. Cell Rep. 29, 3902–3915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim J et al. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell 169, 523–537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan CT et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Boboila C, Alt FW & Schwer B Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol 116, 1–49 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Daniel JA & Nussenzweig A The AID-induced DNA damage response in chromatin. Mol. Cell 50, 309–321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lionetti M et al. A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget 6, 26129–26141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker BA et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132, 587–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 66.Sanz LA & Chédin F High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc 14, 1734–1755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain S, Ba Z, Zhang Y, Dai H-Q & Alt FW CTCF-binding elements mediate accessibility of RAG substrates during chromatin scanning. Cell 174, 102–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bredemeyer AL et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature 456, 819–823 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pertea M et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson JT et al. Integrative Genomics Viewer. Nat. Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B-cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 77.Durand NC et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durand NC et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crane E et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schirmer M et al. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krzywinski M et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available via the NCBI under accession number PRJNA544488. Source data are provided with this paper.